Abstract
Cadaverine is an important C5 platform chemical with a wide range of industrial applications. However, the cadaverine inhibition on the fermenting strain limited its industrial efficiency of the strain. In this study, we report an engineered Escherichia coli strain with high cadaverine productivity that was generated by developing a robust host coupled with metabolic engineering to mitigate cadaverine inhibition. First, a lysine producing E. coli was treated with a combination of radiation (ultraviolet and visible spectrum) and ARTP (atmospheric and room temperature plasma) mutagenesis to obtain a robust host with high cadaverine tolerance. Three mutant targets including HokD, PhnI and PuuR are identified for improved cadaverine tolerance. Further transcriptome analysis suggested that cadaverine suppressed the synthesis of ATP and lysine precursor. Accordingly, the related genes involved in glycolysis and lysine precursor, as well as cadaverine exporter was engineered to release the cadaverine inhibition. The final engineered strain was fed-batch cultured and a titer of 58.7 g/L cadaverine was achieved with a yield of 0.396 g/g, both of which were the highest level reported to date in E. coli. The bio-based cadaverine was purified to >99.6% purity, and successfully used for the synthesis of polyurethane precursor 1,5-pentamethylene diisocyanate (PDI) through the approach of carbamate decomposition.
Keywords: Cadaverine, End-product inhibition, Escherichia coli, Transcriptome analysis, Metabolic engineering, Bio-based diisocyanate
1. Introduction
Polymers are an indispensable material, and have been widely used in plastics, rubbers, fibers, coatings, adhesives, foams and specialty polymers [1,2]. With increasing concerns about the shortage of fossil resources and global warming [3], the bio-based production of polymers from renewable resources has become a promising alternative to the traditional petrochemical approach [4]. This trend triggers on the huge market for the biobased chemicals that can serve as a monomer for synthesizing bio-based polymers, including polyurethane, polyamide and other macromolecule materials. Among them, polyurethanes are one of the most important polymers and widely used in foam plastics, adhesives, coatings and fibers [5,6], which are mainly produced from diamines upon diamine-derived diisocyanate. 1,6-Hexanediamine is the most widely used monomer for the synthesis of polyurethane or polyamide. While no biobased 1,6-hexanediamine exists at present, bio-based cadaverine is an ideal altenative to synthesize 1,6-hexanediamine-derived polymers [7]. As an essential bio-based platform chemical, the efficient bio-production of cadaverine has become a research focus in recent years [8].
Cadaverine has been produced via fermentation or whole cell bioconversion processes by engineered Corynebacterium glutamicum or Escherichia coli [9,10]. For example, by expressing lysine decarboxylases (LDCs) originated from different organism sources in E. coli, whole-cell biocatalysts for the efficient production of cadaverine from lysine have been obtained, and the highest level of cadaverine (205 g/L) was achieved from 400 g/L lysine with a molar yield of 92.1% [11]. For the fermentative production of cadaverine from renewable carbon sources, such as glucose, multiple metabolic engineering strategies have been studied and applied to engineer C. glutamicum or E. coli to improve production, such as overexpressing or engineering LDC [12,13]; enhancing metabolic flux towards the precursor lysine by overexpressing key genes or deleting an undesired competing pathway; eliminating the pathway of byproduct accumulation; transporter engineering to improve cadaverine export or increase the intracellular lysine level; and improving cofactor supply [14,15]. These systematic metabolic engineering strategies have resulted in improved production strains. However, the yield of cadaverine from glucose is still insufficient for the industrial scale.
Recently, some researchers reported that the inhibitory effect of cadaverine significantly limits cadaverine production [16]. In E. coli, a high level of intracellular cadaverine was confirmed to induce closing of porins, causing inadequate cell absorption of nutrients, and reducing cell growth and metabolism [17]. Additionally, when the amount of extracellular cadaverine reached 20.4 g/L, the growth rate of E. coli decreased by 35%. Although the strain of C. glutamicum exhibited a higher tolerance to cadaverine, its growth rate was decreased by 67% in the presence of 102 g/L cadaverine. To mitigate the toxic effect of cadaverine in fermentative production, Wang et al. [18] developed a synthetic microbial consortium consisting of two engineered E. coli strains, a l-lysine-producing strain and a 1,5-DAP-producing strain. These observations suggested that mitigating end-product inhibition would be useful for increasing cadaverine production. However, there have been few reports on metabolic engineering to improve cadaverine production through the release of end-product inhibition.
In this study, E. coli was engineered for improved cadaverine production. First, to alleviate cadaverine toxicity, a lysine-producing E. coli strain was treated with a combination of radiation and ARTP mutagenesis, and the mutants with enhanced cadaverine tolerance were isolated. Using a specific isolated mutant strain as a chassis cell, cadaverine production was improved by 31%. Genome sequencing of the tolerant strain revealed three interesting mutants crucial for cadaverine tolerance and synthesis of precursor lysine. Furthermore, the inhibitory effect of cadaverine accumulation on glucose consumption and cadaverine production was investigated by transcriptome analysis. Several gene targets were identified and engineered, and cadaverine production was improved by 53.8%. The final engineered strain produced more than 58.7 g/L cadaverine in a 7.5 L fermenter, the highest reported fermentative cadaverine production by E. coli. The fermentation-derived cadaverine was purified to polymer-grade and bio-based 1,5-pentamethylene diisocyanate was finally produced.
2. Material and methods
2.1. Strain and media
The strains used in this study are listed in Supplementary Table S1. E. coli strain that used for seed and plasmid construction were cultivated in Luria-Bertani (LB) medium (containing 10 g/L peptone, 5 g/L yeast extract, 5 g/L NaCl) with appropriate concentration of antibiotics (ampicillin 100 μg/mL, 50 μg/mL streptomycin), and incubated in 37 °C with shaking at 200 rpm, pH value that was kept constant. After incubation, 1 mL of seed culture was added into 30 mL of fermentation medium which contained (per liter): 20 g Glucose, 10 g (NH4)2SO4, 5 g yeast extract, 5 g peptone, 0.5 g KCL, 1 g MgSO4•7H2O, 32 mg FeSO4•7H2O, 32 mg MnSO4•H2O, 86 mg ZnSO4•7H2O, 77 mg CuSO4, 60 mg/L Vitamin B1, 10 mg Nicotinamide, 30 μg Biotin, 0.3 g l-Threonine, 0.1 g l-Methionine. The fermentation medium for strains additionally contained antibiotics (ampicillin 100 μg/mL, 50 μg/mL streptomycin) and incubated in 37 °C with shaking at 200 rpm. The initial pH was adjusted to 7.0 by NH4OH (30%, v/v), 20 g/L glucose and 10 g/L (NH4)2SO4 was supplemented at every 8 h to fermentation medium until glucose is no longer consumed. All measurements were performed on three technical replicates.
2.2. Construction of plasmids and the metabolically engineered strain
The plasmids used in this study are listed in Supplementary Table S1. The primers used for gene amplifications are listed in Table S1. DNA manipulations were performed according to standard protocols. Plasmids and oligonucleotide primers used in the study are listed in Supplementary Table S1 and Supplementary Table S2. The initial cadaverine-producing strain NT1003/CadA was constructed from E. coli NT1003 by transformed plasmid pCDF-cadA. The recombinant plasmids and strains construction process were as follows: In the case of gene dapA, fragment encoding by gene dapA from E. coli MG1655 prepared by PCR with the primer pairs was introduced into the BamHI/XbaI site of pTrc99A. The resultant plasmid was designated pTrc99A-dapA and used for overexpression. By the same token, plasmids used for overexpression of other genes were prepared . Subsequently, plasmid pTrc99A-X (X: fragments) transformed into E. coli NT1003 to construct recombinant strains for further research.
Genes of E. coli MG1655 were knocked out by CRISPR-Cas9 system as the following procedures. E. coli was cultured in 5 mL of LB medium at 30 °C overnight. One milliliter of seed culture was inoculated into 50 mL of LB medium. After incubation at 30 °C until the OD600 of 0.2, l-arabinose (30 mM) was added and continue to incubation until the OD600 of 0.6. The cell suspension was centrifuged at 4000 g for 10 min, washed twice with 40 mL sterile water, and washed once with 20 mL of 10% glycerol. The cells were resuspended in 600 μL of 10% glycerol. Transformation of E. coli was carried out by electroporation with a 2.5 kV, 200 Ω, 25 μF electric pulse in a 0.2 cm cuvette using a Gene Pulser (Bio-Rad Laboratories, Hercules, CA, USA). Gene deletion were constructed as follows. In the case of speG deletion, the upstream and downstream regions were amplified by PCR using the primer pair speG_1Fw/1Rv and speG_2Fw/2Rv, respectively. The two fragments were conjugated by overlap PCR using the primer pair speG_1Fw/2Rv. Other genes were constructed similarly. Primers used for gene knockout were designed with the help of Guide RNA Target Design Tool. Transformation of E. coli by electroporation was carried out using the Gene Pulser XCell (Bio-Rad, Hercules, USA).
2.3. Determination the cadaverine tolerance of strain
Seed culture was prepared in LB medium (containing ampicillin 100 μg/mL, 50 μg/mL streptomycin), and it was grown overnight at 37 °C with shaking. The final concentrations of cadaverine in the fermentation broth were adjusted to 0 g/L, 20 g/L, 30 g/L, 40 g/L, 50 g/L, respectively, when the cell mass reach 0.4 (OD600). Samples were taken at regular intervals to detect the growth of cell and the production of cadaverine in the fermentation broth. The culture was taken up 40 h.
2.4. UV and ARTP mutation of strains
For UV mutation, the power of the UV lamp is 15 W, and the irradiation distance is 30 cm. Strains were collected using sterile physiological saline (washed 2 to 3 times), when cultured to logarithmic growth phase, and the collected cell were diluted with sterile physiological saline to prepare a cell suspension (OD600 of 0.4). Take 10 mL of the bacteria suspension in a glass plate, place the glass plate on a magnetic stirrer for ultraviolet mutagenesis, and set the irradiation time to 20–120 s. Cell suspension with different irradiation times were coated in solid medium and placed in a dark environment for simultaneous cultivation, respectively. The culture conditions were 30 °C, 24–36 h, dark. After recording the number of colonies grown on the plate, calculate the fatality of different UV mutagenesis time was calculated. For ARTP mutation, strain cultured to logarithmic growth phase were collected using sterile physiological saline (washed 2 to 3 times), and the collected cell were diluted with sterile physiological saline to prepare a cell suspension (OD600 of 0.6–0.8). 10 μL of the cell suspension adheres to a sterile slide, evenly. Mutation was performed by ARTP mutagenesis breeder: the power was 100 W, the gas flow was 10 SLM, the irradiation distance was 2 mm, the carrier gas was helium, and the strain was subjected to plasma mutagenesis with 20–60 s as the irradiation time. The calculation of fatality at different ARTP mutagenesis times is consistent with UV.
The fatality calculation formula is as follows:
| Fatality rate = (1-m) / n × 100% |
In the formula: m is the average number of colonies on the plate after the mutagenesis treatment; n is the number of colonies on the plate without the mutagenesis treatment.
2.5. Transcriptome analysis
RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA) and RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). After the sample is qualified, mRNA was purified from total RNA using Ribo-zero kit for Illumina® (NEB, USA). Sequencing libraries were generated using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. At last, we used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways.
2.6. Fed-batch culture
The seed culture and fed-batch culture medium in the bioreactor were the same as for shake flask culture. Feeding medium (per liter): 600 g Glucose, 300 g Ammonium sulfate. Seed culture was continued until OD600 of 5.0, an appropriate number of cells (10%, v/v) was tranferred into a 7.5 L fermenter (BioFio 115, New Brunswick Scientific, Edison, NJ, United States) with a liquid loading of 3 L. The pH was kept constant at 7.0 by automated addition of NH4OH (30%, v/v). Dissolved oxygen was maintained above 25% by variation of the stirrer and aeration rate. The pH was kept at 37 °C. When glucose was exhausted, glucose was added at an appropriate rate to maintain glucose concentration above 5 g/L by manual feeding. All the experiments were carried out in triplicate.
2.7. Purification of cadaverine and the synthesis of bio-based 1,5-pentamethylene diisocyanate
The fermentation broth contains substances such as cadaverine, bacteria, inorganic salts, soluble proteins and pigments. To purify the cadaverine, bacteria and protein in the fermentation broth are removed by membrane filtration. Saturated sodium hydroxide is added to convert the cadaverine in the fermentation broth into molecular states, and then the cadaverine and water are vaporized (separated from inorganic salts, soluble proteins and pigments). The vaporized gas is condensed to obtain the aqueous pentamethylenediamine solution and refined Distillate to remove the water, and finally obtain the product.
PDI was synthesized by the approach of carbamate decomposition as described by Sun et al. with some modifications [19]. Nitrogen was introduced into the reaction to maintain the pressure at 1.4 Mpa, and the reaction temperature was 210 °C. First, urea and cadaverine generate pentanediamine diurea, and then 1-butanol is added for alcoholysis to produce 1,5-pentanedicarbamate (PDU) [20]. Zirconium tetrachloride can be used as a catalyst. Finally, 1,5-pentamethylene diisocyanate (PDI) is obtained from PDU pyrolysis. In the reaction, the heat carrier is dioctyl sebacate and zinc picolinate is used as a catalyst. The reaction temperature is 280 °C and the vacuum degree is 0.08 Mpa.
2.8. Analytical methods
The DCW was computed from a curve of OD600 with respect to dry weight. An OD600 of 1.0 represented 400 mg dry weight/L. The concentrations of glucose and l-lysine were analyzed by using an SBA-40C biosensor analyzer (Shandong Province Academy of Sciences, China). Acetic acid was evaluated by using high performance liquid chromatography (HPLC) (1290, Agilent Technologies, Santa Clara, CA, United States) equipped with an on-exchange column (prevail organic acid 5 m, 250 × 4.6 mm, Grace, Columbia, MD, United States) and 25 mM KH2PO4 (adjusted to a pH of 2.5 by H3PO4) was sed as a mobile phase with a flow rate of 1 mL/min. Cadaverine was detected by HPLC after derivatization with dansyl chloride, using an Agilent 1290 Infinity System (Santa Clara, CA, United States) equipped with a fluorescence detector (FLDG1321B).
Fourier-transform infrared (FT-IR) spectroscopy was conducted on a Varian 3100 spectrometer. All spectra were collected from 4000 to 400 cm−1 with a resolution of 4 cm−1 and 32 scans.
The products mixture collected was analyzed on a Thermo scientific trace 1310/ISQ 7000 gas chromatography–mass spectrometry (GC–MS) instrument and a Bruker AV400 nuclear magnetic resonance (NMR) spectrometer. A capillary column of HP5 (30 m × 0.25 mm × 0.25 μm) and mass spectrometry detector operated in an electron impact mode was employed, with ionization voltage being 70 eV, electron source temperature being 250 °C and acquisition being performed from 50 to 800 m/z. Helium was used as the carrier gas of the instrument.
Standardization was conducted under the same conditions. The sample was dissolved in deuterated DMSO (Dimethyl sulfoxide) and performed at room temperature on NMR spectrometer. The proton resonance frequency is 400.13MHZ, the number of data points is 4096, the number of accumulations is 12, and the relaxation time is 2 s.
For the quantitative analysis, products were evaluated by using high performance liquid chromatography (HPLC) (1260, Agilent Technologies, Santa Clara, CA, United States) equipped with a column (HC–C18, 250 × 4.6 mm, Grace, Columbia, MD, United States). Acetonitrile and H2O (The ratio is 6:4) was sed as a mobile phase with a flow rate of 1 mL/min. The column temperature is 30 °C, detection wavelength is 205 nm (ultraviolet detector). Products were also evaluated by using gas chromatography system (7890B, Agilent Technologies, Santa Clara, CA, United States) equipped with a column HP5 (30 m × 0.25 mm × 0.25 μm). The detector temperature is set to 220 °C, the air flow is 300 mL/min, the hydrogen flow is 45 mL/min, and the nitrogen purge flow is 25 mL/min.
3. Results
3.1. Cadaverine inhibition in the lysine-producing E. coli
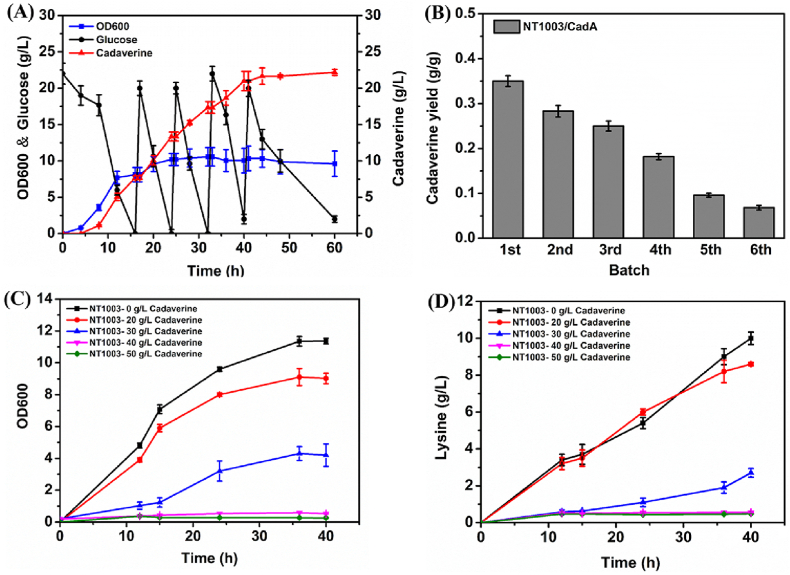
Lysine is a direct precursor for cadaverine biosynthesis. Using a lysine producer stain as the host, a cadaverine-producing strain can be easily developed by overexpressing heterogenous or endogenous LDC [21]. Here, the lysine producer E. coli NT1003 was employed [22]. Overexpressing the endogenous LDC encoded by the cadA gene in E. coli NT1003 resulted in successful production of cadaverine. As shown in Fig. 1A, 22.2 g/L of cadaverine was produced in fed-batch culture of the E. coli NT1003/CadA strain after 60 h. However, the cadaverine yield from glucose was only 0.22 g/g. Moreover, we found that the cadaverine yield of each batch decreased as fermentation progressed, suggesting end-product inhibition by cadaverine (Fig. 1B).
Fig. 1.
Inhibitory effects of end-product cadaverine on fermentation profiles of cadaverine producing strain E. coli N1003/CadA. (A) Fermentation profiles during fed-batch cultivation by E. coli N1003/CadA. (B) Cadaverine yields of E. coli NT1003/CadA in each batch. (C) Growth profiles of the host strain E. coli NT1003 at different initial cadaverine concentrations. (D) Lysine titers of the host strain E. coli NT1003 at different initial cadaverine concentrations.
The production of precursor lysine is one of the most important factors affecting cadaverine production. To determine the inhibitory effect of cadaverine, the capacity of E. coli NT1003 for cell growth and precursor lysine production at different initial cadaverine concentrations was evaluated. In the absence of extracellular cadaverine, the cells grew to the stationary phase by about 12 h, and the lysine production of the NT1003 strain was approximately 10 g/L after 40 h of fermentation (Fig. 1C and D). However, in the presence of 30 g/L cadaverine, the cell growth and lysine production were significantly decreased, and only 2 g/L of lysine was detected at 40 h. With further increasing cadaverine concentration (up to 40 and 50 g/L), cell growth and lysine production were absolutely inhibited (Fig. 1C and D). These results clearly revealed that extracellular cadaverine inhibits cell growth and lysine production in the host cell of E. coli NT1003. It is obvious that such inhibition would further affect the synthesis of end-product cadaverine.
3.2. Improving cadaverine tolerance of the lysine producer E. coli
Due to the toxic effect of cadaverine, developing a superior host strain with improved cadaverine tolerance might benefit to cadaverine production. Both UV and ARTP mutagenesis have been widely used for microbial engineering due to their outstanding features of rapid mutation rates and high operational flexibility [23,24]. To determine the appropriate exposure time of UV and ARTP, we first evaluated the survival rate of E. coli NT1003 using a gradient of irradiation times. In UV mutagenesis, the optimal exposure time was 60 s with a fatality rate of 70%. The selected irradiation time for ARTP was 40 s and the fatality rate was 90% (Fig. 2A). After several rounds of combined UV and ARTP mutagenesis, a mutant strain A-03, which was named E. coli NT1005, was isolated (Supplementary Fig. 1). In the presence of different concentrations of cadaverine ranging from 20 g/L to 40 g/L, E. coli NT1005 grew faster and had higher lysine production than the wild-type E. coli NT1003 (Fig. 2B and C, Supplementary Fig. 2). Under 40 g/L cadaverine, the growth and lysine production of E. coli NT1005 was still slightly affected by the presence of cadaverine, while the growth of E. coli NT1003 was completely suppressed (Fig. 1C). The genetic stability of E. coli NT1005 was also evaluated using six successive subculture tests. Lysine production levels ranged from 7.8 to 8.2 g/L and the growth rate ranged from 6.8 to 6.9, suggesting that E. coli NT1005 was genetically stable (Fig. 2D).
Fig. 2.
Screening of cadaverine tolerant strain by combining UV and ARTP mutagenesis. (A) Determination of the optimal exposure time of UV and ARTP based on the fatality rate. (B) Growth profiles of the mutant stain E. coli NT1005 at different cadaverine concentrations. (C) Lysine titers of the mutant stain E. coli NT1005 at different cadaverine concentrations. (D) The genetic stability of E. coli NT1005.
3.3. Identification of mutations affecting cadaverine tolerance using whole-genome sequencing
To identify genetic targets that can be exploited to alleviate cadaverine toxicity in E. coli, genomic DNA was isolated from the mutant E. coli NT1005 and sequenced to obtain the whole genome. Mutations were identified by comparing each sequenced genome to that of the parental strain, which was also sequenced. All gene mutations for E. coli NT1005 are listed in Table 1. The results revealed 16 single-nucleotide variants, which included 13 synonymous mutations and three non-synonymous mutations. Among the three non-synonymous mutations, one mutant site, I176T, was found in the protein PuuR. Two mutant sites, P49S and G51L, were found in the toxic protein HokD, which has been reported to be a homologue of the hok (host killing) gene. A frame shift mutation was detected in the phnI gene.
Table 1.
Mutations of strains identified by genome sequencing.
| Gene ID | Gene name | Gene product | Nucleotide alteration | Amino acid alteration | Strain |
|---|---|---|---|---|---|
| b4099 | phnI | Carbon-phosphorus lyase core complex subunit | C49A | L18E | NT1003ΔphnI |
| b1299 | puuR | DNA-binding transcriptional repressor | T527C | I176T | NT1003ΔpuuR |
| b1562 b1562 |
hokD hokD |
Toxic protein Toxic protein |
C145T G151A |
P49S E51K |
NT1003ΔhokD NT1003ΔhokD |
To determine whether these mutant genes were involved in cadaverine tolerance, we engineered E. coli NT1003 by deleting puuR, hokD and phnI, respectively. The cadaverine tolerance of the resulting strains was tested and compared to the parental strain and the relevant evolved strain. As shown in Fig. 3A and B, with the addition of 30 g/L cadaverine, the mutant strains NT1003 ΔpuuR and NT1003 ΔphnI were more tolerant to cadaverine than the wild-type strain (Fig. 3A and B), though less tolerant than the evolved strain E. coli NT1005. In contrast, deletion of the hokD gene significantly decreased the cadaverine tolerance and resulted in lower biomass growth and lysine production compared to the wild-type strain (Fig. 3A and B). These results confirmed the important roles of puuR, hokD and phnI in regulating cadaverine tolerance of E. coli. PhnI protein has nucleosidase activity, capable of producing d-ribose-5-triphosphate from GTP and ATP [25]. Inactivation of the phnI gene might reduce ATP consumption, making more ATP available for resisting cadaverine. PuuR is a transcription factor that regulates the putrescine degradation Puu pathway [26]. The first enzyme of the Puu pathway, PuuA, can catalyze the γ-glutamylation of cadaverine [27]. The PuuR mutation may affect the expression of PuuA and alter the intracellular cadaverine level, and consequently improve cadaverine tolerance. In addition, we found that knockout of the puuA gene to block the Puu pathway did not benefit to the final cadaverine production, as well as the interruption of the other cadaverine degration pathway (Supplementary Fig. 3).
Fig. 3.
The effect of mutant genes on cadaverine tolerance. (A) The effect of puuR, hokD or phnI knockout on the growth profile of E. coli NT1003 in the presence of 30 g/L cadaverine. (B) The effect of puuR, hokD or phnI gene knockout on the lysine titer of E. coli NT1003 in the presence of 30 g/L cadaverine.
3.4. Transcriptome analyses of cadaverine tolerant E. coli NT1005 in response to cadaverine
The cadaverine production capacity of E. coli NT1005 as a host was subsequently evaluated by the overexpression of cadA. As shown in Fig. 4 A, a total of 27.53 g/L cadaverine was finally produced at 60 h and the cadaverine yield from glucose was 0.273 g/g (the molar yield of cadaverine from glucose was 48.2%); production and yield were respectively 1.24-fold and 1.23-fold higher than that of E. coli NT1003/CadA (Fig. 4B). These results indicated that the end-product inhibition by cadaverine could be partly released by using a cadaverine tolerant strain as the chassis cell. However, we still found that glucose utilization activity and cadaverine yield were gradually decreased during the fed-batch fermentation (Fig. 4A and B). In the second and third batches, 20 g/L of glucose was totally consumed within 8 h, while the glucose consumption was obviously decreased in the last three batches. In the first batch, the cadaverine yield from glucose reached 0.336 g/g, which was decreased to 0.289 g/g, 0.281 g/g, 0.264 g/g, 0.248 g/g and 0.188 g/g respectively in the following five batches (Fig. 4B).
Fig. 4.
Transcriptome analysis of the mutant strain E. coli NT1005/CadA in response to cadaverine overproduction. (A) Fermentation profiles during fed-batch cultivation by E. coli N1005/CadA. (B) Cadaverine yields of E. coli NT1005/cadA in each batch with E. coli NT1003/cadA as a control. C) Genes related to significantly down-regulated pathways (by transcriptome profiling). Blue, green, yellow and dark blue tables represent down-regulated genes of the oxidative phosphorylation pathway, and blue, green, yellow and dark blue shapes represent the components of the respiratory chain. The gray table represents down-regulated genes of the citrate cycle (TCA) pathway. Genes in the blue table encode the corresponding proteins as follows: nuoF, NADH quinone oxidoreductase subunit F; nuoG, NADH quinone oxidoreductase subunit G; nuoC, NADH quinone oxidoreductase subunit CD; nuoD, NADH-ubiquinone oxidoreductase chain D; nuoE, NADH quinone oxidoreductase subunit E; nuoM, NADH quinone oxidoreductase subunit M; nuoL, NADH quinone oxidoreductase subunit L; nuoH, NADH quinone oxidoreductase subunit H; ndh, NADH quinone oxidoreductase II; nuoN, NADH quinone oxidoreductase subunit N. Genes in the green table encode the corresponding proteins as follows: sdhA, succinate: quinone oxidoreductase, FAD binding protein; sdhD, succinate: quinone oxidoreductase, membrane protein; sdhB, succinate: quinone oxidoreductase, iron-sulfur cluster binding protein. Genes in the yellow table encode the corresponding proteins as follows: cyoD, cytochrome bo3 ubiquinol oxidase subunit 4; cyoB, cytochrome bo3 ubiquinol oxidase subunit 1; cyoC, cytochrome bo3 ubiquinol oxidase subunit 3; cyoA, cytochrome bo3 ubiquinol oxidase subunit 2; cyoE, heme O synthase. Genes in the dark blue table encode the corresponding proteins as follows: atpA, ATP synthase F1 complex subunit alpha; atpG, ATP synthase F1 complex subunit gamma; atpD, ATP synthase F1 complex subunit beta; atpE, ATP synthase Fo complex subunit c; atpF, ATP synthase Fo complex subunit b; ppa, inorganic pyrophosphatase; ppk, polyphosphate kinase. Genes in the white table encode the corresponding proteins as follows: gltA, citrate synthase; icd, isocitrate dehydrogenase; acnB, hypothetical protein; sdhA, succinate: quinone oxidoreductase, FAD binding protein; sdhB, succinate quinone oxidoreductase, iron-sulfur cluster binding protein; fumA, fumarase A; sucA, subunit of E1 (0) component of 2-oxoglutarate dehydrogenase; sucB, dihydrolipoyltranssuccinylase; sucC, succinyl-CoA synthetase subunit beta; sucD, succinyl-CoA synthetase subunit alpha; frdA, fumarate reductase flavoprotein subunit; frdB, fumarate reductase iron-sulfur protein. D) Significantly down-regulated genes related to carbon metabolism, contained in the glycolysis and precursor synthesis pathways. Genes encode the corresponding proteins as follows: glk, glucokinase, pgi, glucose-6-phosphate isomerase; pfkA, 6-phosphofructokinase I; pfkB, 6-phosphofructokinase II; fbaA, fructose-bisphosphate aldolase class II; fbaB, fructose-bisphosphate aldolase class I; gapA, glyceraldehyde-3-phosphate dehydrogenase A; pgk, phosphoglycerate kinase; gpmM, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase; eno, enolase, ppc, phosphoenolpyruvate carboxylase; pck, phosphoenolpyruvate carboxykinase (ATP); pykF, pyruvate kinase I; pykA, pyruvate kinase II; aceE, pyruvate dehydrogenase E1 component; aceF, pyruvate dehydrogenase E2 component; lpd, lipoamide dehydrogenase; aspC, aspartate aminotransferase; asd, aspartate-semialdehyde dehydrogenase; dapA, 4-hydroxy-tetrahydrodipicolinate synthase; dapB, 4-hydroxy-tetrahydrodipicolinate reductase; dapD, tetrahydrodipicolinate succinylase; dapE, succinyl-diaminopimelate desuccinylase; dapF, diaminopimelate epimerase; lysA, diaminopimelate decarboxylase; cadA, lysine decarboxylase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To further uncover the inhibitory effect of cadaverine and identify potential gene targets for improving cadaverine production, transcriptome profiling was conducted to identify changes in global gene transcription levels in response to cadaverine. The cells of E. coli NT1005/CadA in the third and fourth batch were collected and compared. Transcript levels of 218 genes were significantly changed upon cadaverine accumulation, of which 152 genes were significantly down-regulated and 66 were significantly up-regulated (Supplementary Fig. 4). Differentially expressed genes were assigned GO terms, and enrichment analysis was subsequently conducted to identify the significantly induced pathways. Notably, 11 pathways were found to be significantly down-regulated, while only one was up-regulated (Table 2). Within the 11 down-regulated pathways, the largest number of down-regulated genes were enriched in oxidative phosphorylation, citrate cycle, carbon metabolism, and amino acid biosynthesis.
Table 2.
KEGG enrichment of differential pathways (down- and up-regulated).
| Functional category | NO. of genes | NO. of background | P value (<0.05) |
|---|---|---|---|
| Down-regulated | |||
| Oxidative phosphorylation | 26 | 41 | 3.56E-05 |
| Citrate cycle (TCA cycle) | 17 | 26 | 0.000622 |
| Carbon metabolism | 42 | 107 | 0.000969 |
| Biosynthesis of amino acids | 42 | 118 | 0.004248 |
| Valine, leucine and isoleucine biosynthesis | 11 | 16 | 0.004306 |
| 2-Oxocarboxylic acid metabolism | 14 | 26 | 0.006807 |
| C5-Branched dibasic acid metabolism | 8 | 10 | 0.007899 |
| Pyruvate metabolism | 20 | 50 | 0.017012 |
| Glycolysis/Gluconeogenesis | 17 | 40 | 0.017495 |
| Biosynthesis of secondary metabolites | 78 | 287 | 0.038525 |
| RNA degradation | 8 | 15 | 0.038807 |
| Up-regulated: | |||
| Homologous recombination | 4 | 27 | 0.023835 |
Glycolysis and TCA cycle are important pathways for glucose consumption in E. coli. Detailed analyses showed that most genes related to glycolysis and TCA cycle were down regulated (Fig. 4C and D). This suggested that cadaverine accumulation could repress central carbon metabolism and thus further decrease cadaverine production in the cadaverine-producing strain, consistent with the reduced glucose consumption in the fourth batch. The oxidative phosphorylation pathway was another significant category of genes downregulated in response to cadaverine accumulation (Table 2). Of 41 genes in oxidative phosphorylation, 26 genes related to the core oxidative phosphorylation modules of NADH dehydrogenase, succinate dehydrogenase, cytochrome c oxidase, and ATPase were down-regulated by cadaverine between 0.59 and 1.96-fold (Fig. 4C). Combined with the down regulation of glycolysis and the TCA cycle, we inferred that the accumulation of cadaverine might inhibit the synthesis of ATP. A previous study reported that the shortage of intracellular ATP has a significantly negative impact on sugar consumption rate, cell growth and l-lysine production [28]. Accordingly, we found that the expression of genes involved in lysine biosynthesis, such as asd, dapE, dapF, dapD and lysA, was largely repressed by cadaverine accumulation (Fig. 4D).
3.5. Metabolic engineering to improve cadaverine production
Based on the results of transcriptome analysis, a hypothetical model of the cadaverine inhibition mechanism was established, where disrupted oxidative phosphorylation, glycolysis and TCA cycle inhibit glucose consumption and ATP synthesis. Energy breakdown might be induced by cadaverine accumulation and further influence on synthesis of the lysine precursor, thereby reducing final cadaverine production (Fig. 5A). Decreased expression levels of genes related to the disrupted pathways in response to cadaverine were also confirmed by real-time RT-PCR analysis (data not shown). To further release the cadaverine inhibition effect and improve final cadaverine production, genes of the glycolysis and precursor lysine biosynthesis pathways in E. coli NT1005/CadA were targeted. Genes involved in glycolysis were overexpressed under the control of the strong constitutive promoter of trc to examine their effect on cadaverine production. The cadaverine titer of the strains overexpressing the genes gpmM and pgi increased by 1.32-fold and 1.12-fold, respectively, compared with the control strain, and the cadaverine yield of the two strains was also increased by 1.22-fold and 1.17-fold, respectively (Fig. 5B). Overexpressing the pykA gene to increase the metabolic flux into pyruvate significantly reduced cadaverine production (8.36 ± 0.25 g/L) compared to E. coli NT1005 ΔpuuR/CadA (27.53 ± 0.93 g/L), which might be related with the competition of pyruvate metabolism with the synthesis of the key precursor oxaloacetate.
Fig. 5.
Engineering of E. coli NT1005/cadA based on the hypothetical cadaverine inhibition model. (A) The hypothetical cadaverine inhibition model. Accumulation of cadaverine induced energy breakdown and further influenced the function of oxidative phosphorylation, glycolysis and citrate cycle (TCA) pathway. (B) The effect of overexpressing genes involved in glycolysis and lysine synthesis pathway on cadaverine production.
The effect of genes involved in lysine biosynthesis was subsequently tested. First, ppc, which encodes the major anaplerotic enzyme supplying oxaloacetate in E. coli, was overexpressed, and the cadaverine production in the resulting strain reached 34.13 g/L, 1.24-fold higher than that the control strain E. coli NT1005/CadA. Then, genes including aspC, lysC, asd, dapA, dapB, dapD, dapF and lysA were separately overexpressed (Fig. 5B). Among them, overexpression of lysC, dapA, dapB, and lysA showed a positive effect on final cadaverine production, and the highest cadaverine titer was observed in the dapA overexpressing strain. Genes including gpmM, lysA, dapA and dapB, which contributed most to improved cadaverine production based on the above results, were subsequently engineered in combination. The strain that co-overexpressed lysA and dapA in E. coli NT1005/CadA exhibited the highest cadaverine yield of 0.394 g/g from glucose (Supplementary Fig. 5).
Transporter engineering is another area of research emphasis, which has been widely used to improve host tolerance to extreme conditions or increase titers of various products [29]. Intracellular cadaverine accumulation is more toxic to cell growth and metabolism with closed porins [30]. Increasing the secretion of cadaverine is supposed to benefit to the release of cadaverine inhibition. With C. glutamicum as the host, the overexpression of endogenous cadaverine exporter CgmA increased the cadaverine yield by 11.3% [31], and the overexpression of heterologous CadB increased cadaverine yield by 30% [32]. Overexpression of the cgmA gene in a putrescine-producing C. glutamicum was also capable of increasing putrescine production by 24% compared to the control strain [33]. The regulation of polyamine transporter expression has not yet been studied in E. coli. The cadaverine-lysine antiporter CadB is the only known protein showing cadaverine excretion activity in E. coli [34]. By overexpressing cadB in the E. coli NT1005/CadA-lysA-dapA, cadaverine production was increased to 42.35 g/L with a molar yield of 71% in flask (Table 3), indicating that transporter engineering might function as an alternative approach to release cadaverine toxity. Finally, the engineered strain E. coli NT1005/CadA-lysA-dapA-cadB was employed for a fed-batch experiment.
Table 3.
Fermentation result of engineering strain which overexpressed gene cadB.
| Engineered strains | Titer g/L | Yield g/g | OD600 | Folda | Foldb |
|---|---|---|---|---|---|
| E. coli NT1005/CadA (The control) | 27.53 ± 0.93 | 0.273 ± 0.005 | 12.81 ± 0.44 | 1 | 1 |
| E. coli NT1005/CadA-lysA-dapA | 38.79 ± 0.34 | 0.394 ± 0.004 | 11.51 ± 1.1 | 1.41 | 1.44 |
| E. coli NT1005/CadA-lysA-dapA-cadB | 42.35 ± 0.67 | 0.403 ± 0.004 | 12.31 ± 0.83 | 1.49 | 1.48 |
In this table, “a” represents: change fold compared with the control titer; “b” represents: change fold compared with the control yield.
3.6. Cadaverine production by fed-batch cultivation in the 7.5 L fermentor
To comprehensively evaluate the overall production performance of E. coli NT1005/CadA-lysA-dapA-cadB, fed-batch fermentation was carried out in a 7.5 L bioreactor, with the strain E. coli NT1005/CadA as a control. When the glucose concentration was lower than 5 g/L, glucose and ammonium sulfate were fed into the bioreactor, optimization of the fermentation conditions are illustrated in Supplementary Fig. 6. Details of the fermentation results are illustrated in Fig. 6. The cells grew fast from the beginning of fermentation, and OD600 reached the maximum value of 23.68 at 28 h. The accumulation of cadaverine gradually increased at a relatively constant rate after 16 h. At 64 h, the final cadaverine titer was 58.7 g/L, which was 43.9% higher than the control strain (38.3 g/L), with a productivity of 0.92 g/L/h. The overall cadaverine yield from glucose was 0.396 g/g, which is 26.1% higher than the control (0.314 g/g) (Fig. 6).
Fig. 6.
The fermentation profiles by the recombinant strain E. coli NT1005/CadA-lysA-dapA-cadB and the control strain E. coli NT1005/CadA with a fed-batch strategy. Glucose and ammonium sulfate feeding started at 15 h. Black or blue symbols/lines represent growth profiles, orange and gray symbols/lines represent glucose consumption, orange and gray columns/bars represent cadaverine titer. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.7. Purification of cadaverine from fermentation broth
Following the fermentation, cadaverine was purified from the culture broth by membrane separation, vacuum evaporation, crystallization and two-step distillation. After removal of impurities such as cell debris and protein by ultrafiltration and microfiltration, the filtrate was concentrated in a vacuum evaporator. The pH of cadaverine concentrate was subsequently adjusted to 13.5 by sodium hydroxide to make the salt in the solution crystallize, which was separated by filtration. Cadaverine and water in the cadaverine concentrate was then gasified simultaneously in a vacuum evaporator and collected to obtain cadaverine aqueous solution. After two-step distillation, the product of cadaverine with more than 99.6% purity (GC analysis) was obtained (Supplement Figure 7).
Fig. 7.
The identification of PDI. (A) FTIR spectra of cadaverine (PDA) and bio-based PDI. (B) 1H NMR spectra of bio-based PDI; (C) 13C NMR spectra of bio-based PDI.
3.8. The synthesis of bio-based 1,5-pentamethylene diisocyanate (PDI)
In addition to synthesize bio-based polyamide, aliphatic diamines could also have been used to produce polyurethane, where diisocyanate was first synthesized from the diamines, which was then used as the raw material to form polyurethane [5,6]. Among the commercial aliphatic diisocyanates, 1,6-hexamethylene diisocyanate (HDI), which was produced from petroleum-based 1,6-hexanediamine is most widely used and already accounts for 60% of the total amount. PDI is comparable to HDI in its physical and chemical properties PDI, which is an ideal alternative for HDI. To demonstrate the potential of bio-based cadaverine as a polyurethane precursor, we attempted to synthesize PDI from our purified cadaverine. The approach of carbamate decomposition was tested [19] (Scheme 1). Nuclear magnetic resonance (NMR) spectroscopy and fourier-transform infrared (FTIR) spectra showed the successful synthesis of PDI (Fig. 7A, B and 7C). Thus, bio-based cadaverine can be used as raw material for synthesizing diisocyanates. Compared to HDI-derived polyurethane, PDI-derived product reveals similar or even improved properties in terms of flexibility and impact resistance [35,36].
Scheme 1.
Reactions involved in the synthesis of PDU and PDI.
4. Discussion
Due to concerns about limited fossil feedstocks and environmental problems, the development of bio-based processes to produce chemicals from renewable resources has attracted increasing research attention [3]. Cadaverine is an important C5 platform chemical, particularly as a building block to synthesize bio-based polymers [7]. The traditional strategy to develop cadaverine-producing strains has been focused on improving production through reconstructing metabolic pathways. However, the accumulation of cadaverine or other diamines as the end-product is toxic and hampers cell growth, cell viability, and glucose uptake, thus resulting in decreased production of an end-product. Product toxicity has been one of the main bottlenecks in achieving optimal production levels. In this study, the development of a cadaverine-tolerant host combined with metabolic engineering based on transcriptome analyses was performed with the aim to increase final cadaverine production.
Using C. glutamicum or E. coli as the host, several cadaverine producers have been engineered [37]. Although C. glutamicum showed higher tolerance to cadaverine, E. coli possesses a native cadaverine biosynthetic pathway, and the metabolism process and physiological role of cadaverine are relatively well studied in E. coli [38]. Nowadays, the highest titer of cadaverine obtained in engineered E. coli was performed by a consortium of two engineered E. coli, and 28.5 g/L of cadaverine was finally obtained [18]. Here, a lysine producing E. coli was chosen as the host strain in this study. The tolerance of the lysine producing E. coli to cadaverine was first studied by adding varying concentrations of cadaverine. Significant growth inhibition was observed when the cadaverine concentration was above 30 g/L (Fig. 1C). Product toxicity is frequently encountered in metabolic engineering projects [[39], [40], [41]]. Numerous strategies such as in-situ product removal or increasing the robustness of the microbial bio-catalyst have been employed for overcoming sensitivity to the final product [39,42,43]. Strain tolerance can be improved either through an evolutionary approach or through rational engineering that addresses known mechanisms of toxicity [40,44]. Here, UV mutagenesis combined with ARTP mutagenesis was performed to develop a robust host for cadaverine production. Fortunately, a mutant stain NT1005, which could tolerant 40 g/L cadaverine, was obtained. We were pleased to see that this mutant strain had increased cadaverine production ability relative to the parent strain, indicating that increasing strain tolerance was an efficient approach for the improvement of cadaverine production.
However, cadaverine inhibition was still observed when the mutant strain was fed-batch cultivated with a gradually decreasing cadaverine yield. To further explore the inhibitory effect of cadaverine, comparative transcriptome analysis was performed. Overproduction of cadaverine resulted in transcriptional downregulation of genes involved in oxidative phosphorylation, glycolysis, the TCA cycle, pyruvate metabolism, and biosynthesis of some amino acids including lysine. The repression of ATP synthesis and lysine synthesis was likely due to cadaverine, thus reducing cell growth and end-product yield. A similar transcriptomic change in response to putrescine was previously reported [45]. By repressing ATP- and NADPH-consuming enzyme coding gene expression to increase the available intracellular ATP and NADPH, putrescine production was successfully enhanced [45]. Here, genes of the glycolysis and lysine synthesis pathways were targeted. We expected to enhance the production of cadaverine by E. coli, thus releasing the cadaverine inhibition on ATP and precursor lysine synthesis. Fortunately, the final cadaverine titer was further improved in the tolerant strain by modifying these genes individually or in combination.
Through the development of a robust host, transcriptome analysis of the strain response to cadaverine, and metabolic engineering of the strain to increase key metabolic flux and cadaverine efflux, the resulting recombinant strain E. coli NT1005/CadA-lysA-dapA-cadB was fed-batch cultivated. During 60-h fermentation, 58.7 g/L of cadaverine was produced with 1.48 g/L/h of maximum productivity and 0.408 g/g of yield, the highest level reported to date in E. coli. So far, relatively few reports have been made to produce polyamine in E. coli, probably due to the toxicity to microorganisms. The inhibition mechanism of cadaverine will likely be elucidated in further studies, providing many opportunities to further improve cadaverine production.
As a bio-based alternative of 1,6-hexanediamine, cadaverine synthesized from the fermentation or the whole cell biocatalyst reaction has been successfully purified to polymer grade (>99.5%) by fractional distillation. Here, a more environmentally friendly separation process was applied and a product purity of more than 99.9% was also achieved. With the bio-based cadaverine, several new bio-based polyamides, such as PA5.2, PA 5.6, and PA 5.10 have been successfully produced by polymerization with dicarboxylic acids [31,46,47]. The production of polyurethane is another important application of cadaverine. High quality of PDI, a precursor of polyurethane, was synthesized from purified cadaverine in our study, providing a promising addition to the portfolio of bio-based polymers for future industrial applications.
5. Conclusions
In this study, a superior host strain E. coli NT1005 with improved cadaverine tolerance was isolated. Transcriptome analysis by tolerant strain revealed the inhibitory effect of cadaverine accumulation on glucose consumption and cadaverine production. Subsequently, several gene targets were identified and engineered, and the final engineered strain E. coli NT1005/CadA-lysA-dapA-cadB produced more than 58.7 g/L cadaverine in a 7.5 L fermenter. Finally, the fermentation-derived cadaverine was purified to polymer-grade and bio-based 1,5-pentamethylene diisocyanate was produced.
Availability of data and materials
All the data generated during the current study are included in the manuscript.
CRediT authorship contribution statement
Xin Wang: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Xing Guo: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Jing Wang: Methodology. Hui Li: Investigation, (part). Feng He: Investigation, (part). Sheng Xu: Investigation, (part). Kequan Chen: Resources, Supervision, Project administration, Funding acquisition. Pingkai Ouyang: Supervision, Project administration, All authors have read and agreed to the published version of manuscript.
Declaration of competing interest
The Authors have no interests to declare.
Acknowledgements
This work is supported by the National Key R&D Program of China (2021YFC2100800), Key Research and Development Program (Social Development) Project of Jiangsu Province (BE2018730), and the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTE1844 and XTB1806).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.09.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Oh I.J., Kim D.H., Oh E.K., Lee S., Lee J. Optimization and scale-up of succinic acid production by Mannheimia succiniciproducens LPK7. J Microbiol Biotechnol. 2009;19:167–171. doi: 10.4014/jmb.0807.447. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y., Kim H., Shin J., Bhatia S.K., Seo H.M., Kim Y. Application of diethyl ethoxymethylenemalonate (DEEMM) derivatization for monitoring of lysine decarboxylase activity. J Mol Catal B Enzym. 2015;115:151–154. [Google Scholar]
- 3.Mekonnen T.H., Mussone P.G., Bressler D.C. Valorization of rendering industry wastes and co-products for industrial chemicals, materials and energy: Review. Crit Rev Biotechnol. 2016;36:120–131. doi: 10.3109/07388551.2014.928812. [DOI] [PubMed] [Google Scholar]
- 4.Steinbuchel A. Non-biodegradable biopolymers from renewable resources: perspectives and impacts. Curr Opin Biotechnol. 2005;16:607–613. doi: 10.1016/j.copbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Udagama R., Degrandi-Contraires E., Creton C., Graillat C., McKenna T.F., Bourgeat-Lami E. Synthesis of acrylic-polyurethane hybrid latexes by miniemulsion polymerization and their pressure-sensitive adhesive applications. Macromolecules. 2011;44:2632–2642. [Google Scholar]
- 6.Feng J., Wang X., Guo P., Wang Y., Luo X. Mechanical properties and wear resistance of sulfonated graphene waterborne polyurethane composites prepared by in situ method. Polymers. 2018;10:75. doi: 10.3390/polym10010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatia S.K., Bhatia R.K., Yang Y. Biosynthesis of polyesters and polyamide building blocks using microbial fermentation and biotransformation. Rev Environ Sci Biotechnol. 2016;15:639–663. [Google Scholar]
- 8.Imao K., Konishi R., Kishida M., Hirata Y., Segawa S., Adachi N. 1,5-Diaminopentane production from xylooligosaccharides using metabolically engineered Corynebacterium glutamicum displaying beta-xylosidase on the cell surface. Bioresour Technol. 2017;245:1684–1691. doi: 10.1016/j.biortech.2017.05.135. [DOI] [PubMed] [Google Scholar]
- 9.Buschke N., Schroder H., Wittmann C. Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J. 2011;6:306–317. doi: 10.1002/biot.201000304. [DOI] [PubMed] [Google Scholar]
- 10.Shin J., Joo J.C., Lee E., Hyun S.M., Kim H., Park S.J. Characterization of a whole-cell biotransformation using a constitutive lysine decarboxylase from Escherichia coli for the high-level production of cadaverine from industrial grade l-lysine. Appl Biochem Biotechnol. 2018;185:909–924. doi: 10.1007/s12010-018-2696-4. [DOI] [PubMed] [Google Scholar]
- 11.Rui J., You S., Zheng Y., Wang C., Gao Y., Zhang W. High-efficiency and low-cost production of cadaverine from a permeabilized-cell bioconversion by a Lysine-induced engineered Escherichia coli. Bioresour Technol. 2020;302 doi: 10.1016/j.biortech.2020.122844. [DOI] [PubMed] [Google Scholar]
- 12.Ma W., Cao W., Zhang H., Chen K., Li Y., Ouyang P. Enhanced cadaverine production from l-lysine using recombinant Escherichia coli co-overexpressing CadA and CadB. Biotechnol Lett. 2015;37:799–806. doi: 10.1007/s10529-014-1753-5. [DOI] [PubMed] [Google Scholar]
- 13.Kim H.T., Baritugo K., Oh Y.H., Kang K., Jung Y.J., Jang S. High-level conversion of l-lysine into cadaverine by Escherichia coli whole cell biocatalyst expressing hafnia alvei l-lysine decarboxylase. Polymers. 2019;11:1184. doi: 10.3390/polym11071184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kind S., Wittmann C. Bio-based production of the platform chemical 1,5-diaminopentane. Appl Microbiol Biotechnol. 2011;91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- 15.Li M., Li D., Huang Y., Liu M., Wang H., Tang Q. Improving the secretion of cadaverine in Corynebacterium glutamicum by cadaverine–lysine antiporter. J Ind Microbiol Biotechnol. 2014;41:701–709. doi: 10.1007/s10295-014-1409-4. [DOI] [PubMed] [Google Scholar]
- 16.Wei G., Ma W., Zhang A., Cao X., Shen J., Li Y. Enhancing catalytic stability and cadaverine tolerance by whole-cell immobilization and the addition of cell protectant during cadaverine production. Appl Microbiol Biotechnol. 2018;102:7837–7847. doi: 10.1007/s00253-018-9190-3. [DOI] [PubMed] [Google Scholar]
- 17.Naerdal I., Pfeifenschneider J., Brautaset T., Wendisch V.F. Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microbial Biotechnology. 2015;8:342–350. doi: 10.1111/1751-7915.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Lu X., Ying H., Ma W., Xu S., Wang X. A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli. Front Microbiol. 2018;9:1312. doi: 10.3389/fmicb.2018.01312. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun D.-L., Luo J.-Y., Wen R.-Y., Deng J.-R., Chao Z.-S. Phosgene-free synthesis of hexamethylene-1, 6-diisocyanate by the catalytic decomposition of dimethylhexane-1, 6-dicarbamate over zinc-incorporated berlinite (ZnAlPO4) J Hazard Mater. 2014;266:167–173. doi: 10.1016/j.jhazmat.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Kaminskaia N.V., Kostic N.M. Alcoholysis of urea catalyzed by palladium (II) Complexes. Inorg Chem. 1998;37:4302–4312. doi: 10.1021/ic980065r. [DOI] [PubMed] [Google Scholar]
- 21.Kind S., Jeong W.K., Schroder H., Wittmann C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab Eng. 2010;12:341–351. doi: 10.1016/j.ymben.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Ying H., He X., Li Y., Chen K., Ouyang P. Optimization of culture conditions for enhanced lysine production using engineered Escherichia coli. Appl Biochem Biotechnol. 2014;172:3835–3843. doi: 10.1007/s12010-014-0820-7. [DOI] [PubMed] [Google Scholar]
- 23.Feng W., Liang J., Wang B., Chen J. Improvement of kojic acid production in Aspergillus oryzae AR-47 mutant strain by combined mutagenesis. Bioproc Biosyst Eng. 2019;42:753–761. doi: 10.1007/s00449-019-02079-9. [DOI] [PubMed] [Google Scholar]
- 24.Xueliang Q., Peng X., Xinrui Z., Guocheng D., Juan Z., Juanhua L. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab Eng. 2020;60:66–76. doi: 10.1016/j.ymben.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Kamat S.S., Williams H.J., Raushel F.M. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature. 2011;480:570–573. doi: 10.1038/nature10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto N., Kurihara S., Kitahara Y., Asada K., Kato K., Suzuki H. Mechanism for regulation of the putrescine utilization pathway by the transcription factor PuuR in Escherichia coli K-12. J Bacteriol. 2012;194:3437–3447. doi: 10.1128/JB.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara S., Oda S., Tsuboi Y., Kim H.G., Oshida M., Kumagai H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J Biol Chem. 2008;283:19981–19990. doi: 10.1074/jbc.M800133200. [DOI] [PubMed] [Google Scholar]
- 28.Xu J., Ruan H., Yu H.B., Liu L., Zhang W. Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production from mixed sugar. Microb Cell Factories. 2020;19:39. doi: 10.1186/s12934-020-1294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kell D.B., Swainston N., Pir P., Oliver S.G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015;33:237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Ma W., Chen K., Li Y., Hao N., Wang X., Ouyang P. Advances in cadaverine bacterial production and its applications. Engineering. 2017;3:308–317. [Google Scholar]
- 31.Kind S., Neubauer S., Becker J., Yamamoto M., Volkert M., Von Abendroth G. From zero to hero Production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab Eng. 2014;25:113–123. doi: 10.1016/j.ymben.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Kind S., Kreye S., Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab Eng. 2011;13:617–627. doi: 10.1016/j.ymben.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen A.Q.D., Schneider J., Wendisch V.F. Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J Biotechnol. 2015;201:75–85. doi: 10.1016/j.jbiotec.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Meng S., Bennett G.N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widemann M., Driest P.J., Orecchia P., Naline F., Golling F.E., Hecking A. Structure–property relations in oligomers of linear aliphatic diisocyanates. ACS Sustainable Chem Eng. 2018;6:9753–9759. [Google Scholar]
- 36.Golling F.E., Pires R., Hecking A., Weikard J., Richter F., Danielmeier K. Polyurethanes for coatings and adhesives–chemistry and applications. Polym Int. 2019;68:848–855. [Google Scholar]
- 37.Schneider J., Wendisch V.F. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- 38.Qian Z.G., Xia X.X., Lee S.Y. Metabolic engineering of Escherichia coli for the production of cadaverine: a five carbon diamine. Biotechnol Bioeng. 2011;108:93–103. doi: 10.1002/bit.22918. [DOI] [PubMed] [Google Scholar]
- 39.Royce L.A., Yoon J.M., Chen Y., Rickenbach E., Shanks J.V., Jarboe L.R. Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab Eng. 2015;29:180–188. doi: 10.1016/j.ymben.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay A. Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 2015;23:498–508. doi: 10.1016/j.tim.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Lee K., Bang H.B., Lee Y.H., Jeong K.J. Enhanced production of styrene by engineered Escherichia coli and in situ product recovery (ISPR) with an organic solvent. Microb Cell Factories. 2019;18:79. doi: 10.1186/s12934-019-1129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mckenna R., Moya L., Mcdaniel M., Nielsen D.R. Comparing in situ removal strategies for improving styrene bioproduction. Bioproc Biosyst Eng. 2015;38:165–174. doi: 10.1007/s00449-014-1255-9. [DOI] [PubMed] [Google Scholar]
- 43.Lopez S.D., Griffith D.A., Choi B., Cate J.H.D., Tullmanercek D. Evolutionary engineering improves tolerance for medium-chain alcohols in Saccharomyces cerevisiae. Biotechnol Biofuels. 2018;11:90. doi: 10.1186/s13068-018-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehrs M., Tanjore D., Eng T., Lievense J., Pray T.R., Mukhopadhyay A. Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 2019;27:524–537. doi: 10.1016/j.tim.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Li Z., Liu J. Transcriptomic changes in response to putrescine production in metabolically engineered Corynebacterium glutamicum. Front Microbiol. 2017;8:1987. doi: 10.3389/fmicb.2017.01987. 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H.T., Baritugo K.-A., Oh Y.H., Hyun S.M., Khang T.U., Kang K.H. Metabolic engineering of Corynebacterium glutamicum for the high-level production of cadaverine that can be used for the synthesis of biopolyamide 510. ACS Sustainable Chem Eng. 2018;6:5296–5305. [Google Scholar]
- 47.Kim H.T., Baritugo K.-A., Oh Y.H., Kang K.-H., Jung Y.J., Jang S. High-level conversion of l-lysine into cadaverine by escherichia coli whole cell biocatalyst expressing hafnia alvei L-lysine decarboxylase. Polymers. 2019;11:1184. doi: 10.3390/polym11071184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.