Abstract
Intrinsically disordered proteins and protein regions (IDRs) make up around 30% of the human proteome where they play essential roles in dictating and regulating many core biological processes. While IDRs are often studied as isolated domains, in naturally occurring proteins most IDRs are found adjacent to folded domains, where they exist as either N- or C-terminal tails or as linkers connecting two folded domains. Prior work has shown that charge properties of IDRs can influence their conformational behavior, both in isolation and in the context of folded domains. In contrast, the converse scenario is less well-explored: how do the charge properties of folded domains influence IDR conformational behavior? To answer this question, we combined a large-scale structural bioinformatics analysis with all-atom implicit solvent simulations of both rationally designed and naturally occurring proteins. Our results reveal three key takeaways. Firstly, the relative position and accessibility of charged residues across the surface of a folded domain can dictate IDR conformational behavior, overriding expectations based on net surface charge properties. Secondly, naturally occurring proteins possess multiple charge patches that are physically accessible to local IDRs. Finally, even modest changes in the local electrostatic environment of a folded domain can substantially modulate IDR-folded domain interactions. Taken together, our results suggest that folded domain surfaces can act as local determinants of IDR conformational behavior.
Keywords: Intrinsically disordered proteins, Conformational ensemble, All-atom simulations, Sequence design
Graphical abstract
Highlights
-
•
Intrinsically disordered regions (IDRs) are mostly found adjacent to folded domains.
-
•
Here we propose that the folded domain surface properties influence IDR behavior.
-
•
We combine all-atom simulations and sequence design of IDRs and folded domains.
-
•
IDR conformational behavior is determined by a complex combination of factors.
-
•
Folded domains can substantially alter IDR conformational biases.
1. Introduction
Intrinsically disordered proteins and protein regions (IDRs) are highly prevalent in eukaryotic proteomes and are associated with diverse biological functions ranging from transcriptional regulation and signal transduction to facilitating formation of biomolecular condensates (Wright and Dyson, 1999, 2015; van der Lee et al., 2014). Unlike folded domains, IDRs lack a precise three-dimensional structure and are instead characterized by broad conformational fluctuations. As a result, IDRs are frequently described in terms of a conformational ensemble (Mittag and Forman-Kay, 2007; Receveur-Bréchot et al., 2006). The vast majority of disordered sequences – roughly 90% in the human proteome – are adjacent to folded domains (FDs) (van der Lee et al., 2014). As a result, most IDRs can be considered either disordered tails (N- or C-terminal IDRs connected to a folded domain) or disordered linkers (IDRs that connect two folded domains) (Mittal et al., 2018).
IDR conformational behavior has emerged as a key determinant of IDR function (Mittag and Forman-Kay, 2007; Das et al., 2015; Sherry et al., 2017; Dyla and Kjaergaard, 2020; Borcherds et al., 2014). Given that IDR conformational behavior is encoded at least in part by amino acid sequence, understanding the physicochemical principles that relate sequence to ensemble is an ongoing topic of interest (Mittag and Forman-Kay, 2007; Das et al., 2015; Zerze et al., 2015; Sawle and Ghosh, 2015; Mao et al., 2013; Martin and Holehouse, 2020; Lin et al., 2018). Systematic studies have yielded a quantitative understanding of the sequence determinants of IDR conformational behavior (van der Lee et al., 2014; Borcherds et al., 2014; Marsh and Forman-Kay, 2010; Sørensen and Kjaergaard, 2019; Müller-Späth et al., 2010; Mao et al., 2010; Martin et al., 2016; Das and Pappu, 2013; Portz et al., 2017; Zheng et al., 2020; Bowman et al., 2020); however, most of these studies have focused IDRs in isolation as opposed to systems consisting of IDRs tethered to FDs.
Disordered tails can stabilize proteins, prevent aggregation, determine binding affinities, and facilitate a range of inhibitory and activating interactions (Uversky, 2013; Staby et al., 2020; Graña-Montes et al., 2014; Keul et al., 2018; Sankaranarayanan et al., 2021). Disordered linkers determine the orientations that tethered domains can interact with one another, thereby influencing allosteric communication in multidomain proteins (Sørensen and Kjaergaard, 2019; Yanez Orozco et al., 2018; McCann et al., 2014; Huang et al., 2020; Ma et al., 2011). Traditional sequence determinants of IDR conformational behavior (including charge content, charge patterning, hydrophobicity, and proline content) can regulate the behavior of IDRs tethered to FDs either as linkers or tails (Mittal et al., 2018; Sherry et al., 2017; Sørensen and Kjaergaard, 2019; Keul et al., 2018; Sankaranarayanan et al., 2021; Sun et al., 2018; Ganguly et al., 2012; Lecoq et al., 2017; Vuzman and Levy, 2010; Krois et al., 2018; Martin et al., 2021). Most prior work has emphasized the role of electrostatics in governing the interaction between IDRs and FDs, and FDs have been shown to influence IDR behavior in a manner that depends on the charge properties of IDR sequences (Mittal et al., 2018). However, the converse question is less well-explored: how do the surface charge properties of folded domains alter FD-IDR interaction and the resulting conformational biases of the IDR (Martin et al., 2021; Patil and Nakamura, 2006)?
In this work, we sought to gain an understanding of the combined role that IDR and FD properties play in governing the behavior of disordered tails tethered to FDs. To do so, we first performed all-atom implicit solvent simulations of rationally designed proteins consisting of negatively or positively charged low-complexity IDRs attached to charge variants of the superfolder Green Fluorescent Protein (sfGFP). We titrated the length and net charge per residue (NCPR) of the IDR and the net charge and surface charge distribution of the FD. This combination of proteins allowed us to explicitly model how distinct sequence features influence IDR conformational behavior.
These simulations revealed that the emergent interactions between disordered tails and FDs can be unintuitive due to the conformational flexibility of tails, the heterogenous distribution of charged residues on the surface, and the competing and long-range nature of electrostatic interactions. Furthermore, even in our relatively simple systems, conformational behavior cannot necessarily be determined by simple metrics, such as the net charge of either domains.
Given these results, we next performed a structural bioinformatics analysis to more precisely characterize the surface charge distribution characteristics of human FDs with N/C-terminal tails. We observed that FDs tend to contain multiple correlated regions of charge (charge patches) that are relatively large and physically accessible to the IDR. Finally, to tie our observations to a real protein system, we performed simulations of homologous proteins with variable surface charge distribution but conserved structural features. Our simulations revealed that even modest changes in surface charge distribution can significantly influence IDR conformational behavior and IDR-FD interactions. Taken together, our results suggest that the local electrostatic environment of a FD can impact the conformational behavior of an IDR in concrete and unintuitive ways.
2. Results
2.1. Sequence properties of disordered tails are similar to those of all disordered regions
Prior work has established that charge-associated sequence features play a key role in determining IDR conformational behavior (Sørensen and Kjaergaard, 2019; Müller-Späth et al., 2010; Mao et al., 2010; Das and Pappu, 2013; Mittag et al., 2010a; Das et al., 2020). With this in mind, we first sought to establish if the distribution of charge properties of IDRs adjacent to FDs differs from that of all IDRs. While other types of interactions will undoubtedly contribute to IDR-FD interactions, we chose to focus initially on charge-mediated interactions owing to their established importance in IDR conformational behavior.
We undertook a structural bioinformatics analysis to identify FDs with known structures which had N- or C-terminal disordered tails (see Methods). To contextualize our analysis, we generated three subsets of IDRs for further analysis: (1) all IDRs in the human proteome (2) all N- or C-terminal IDRs regardless of if structural data on adjacent folded domains is known, and (3) N- or C-terminal IDRs for which structural information about the adjacent folded domains is known (see Methods, Supplementary Fig. S1, and Supplementary Fig. S2). These three sets allow us to ask if IDRs that are adjacent to folded domains represent a special subclass of IDRs, or if they are statistically equivalent to all IDRs in the human proteome.
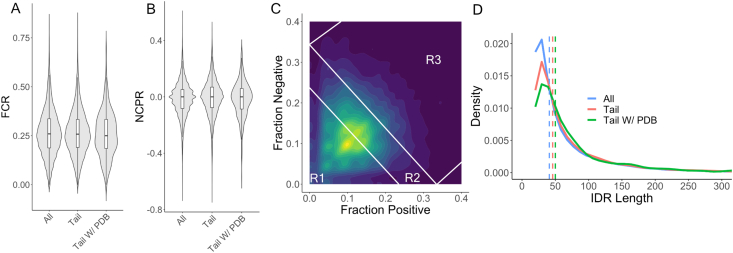
Given the importance of charge interactions in dictating IDR conformational behavior, we first characterized the charge properties of our three sets of IDRs. A comparison of the fraction of charged residues (FCR) and mean net charge per residue (NCPR) across these three sets revealed no statistical differences, with a mean FCR of 0.26 and a near-neutral NCPR of 0.01 (Fig. 1A and B). Similarly, analysis of other sequence properties including charge patterning (κ) (Das and Pappu, 2013), presence of phosphosites, hydrophobicity, and aromaticity showed no statistical difference between the three sets of IDRs (Supplementary Fig. S3). Finally, we compared the distribution of IDRs across the diagram of states, a classification tool developed by Das and Pappu to categorize IDRs in terms of the relative density of positively and negatively charged residues (Fig. 1C) (Das and Pappu, 2013). Again, no marked differences were observed among the three sets of IDRs (Supplementary Fig. S4). In conclusion, in terms of charge properties, tail IDRs with structural data are not statistically different from non-tail IDRs (Supplementary Table 1).
Fig. 1.
The charge sequence properties of disordered tails are similar to those of all IDRs. Distributions of relevant IDR properties. Three sets of IDRs were included in this analysis: all IDRs with or without structural data (N = 34,095), all tails with or without structural data (N = 11,254), and all tails with structural data (N = 619). A) Violin/box plots of fraction of charged residues (FCR) across the three sets of IDRs. The horizontal black line in the box plot refers to the median, while the bottom/top of the box refers to the first/third quartile. B) Violin/box plots of net charge per residue (NCPR) across the three sets of IDRs. C) 2D density plot of fraction of positive charges and fraction of negative charges for all tails with or without structural data. Diagonal white lines separate the diagram of states into three regions: R1 (weak polyelectrolytes and polyampholytes), R2 (intermediate polyampholytes), and R3 (strong polyampholytes). D) Density plots of IDR length across the three sets of IDRs. Dashed line refers to the median IDR length for a given group.
Given that chain length plays a major role in determining IDR global dimensions, we also characterized the length distribution of disordered tails and compared it to the length distribution of all IDRs for context (Fig. 1D). The median length of disordered tails (46 residues) is similar to that of all lDRs (41 residues), and there is not a substantial difference in the length distribution between these two groups.
Considering charge interactions are not limited to intra-IDR interactions but can also include IDR-FD interactions, we next wondered how FD surface charge properties might tune IDR-FD interactions. To explore this idea, we first turned to all-atom implicit solvent simulations of synthetic proteins consisting of N-terminal IDRs attached to FDs. We specifically sought to probe the effects of varying FD surface charge distributions when attached to IDRs of varying lengths and charge. Studying synthetic proteins enables us to explicitly model the effects of FD surface charge distribution under varying contexts and probe if complex conformational behavior can emerge from relatively simple systems.
2.2. Rational design of synthetic IDR-folded domain proteins
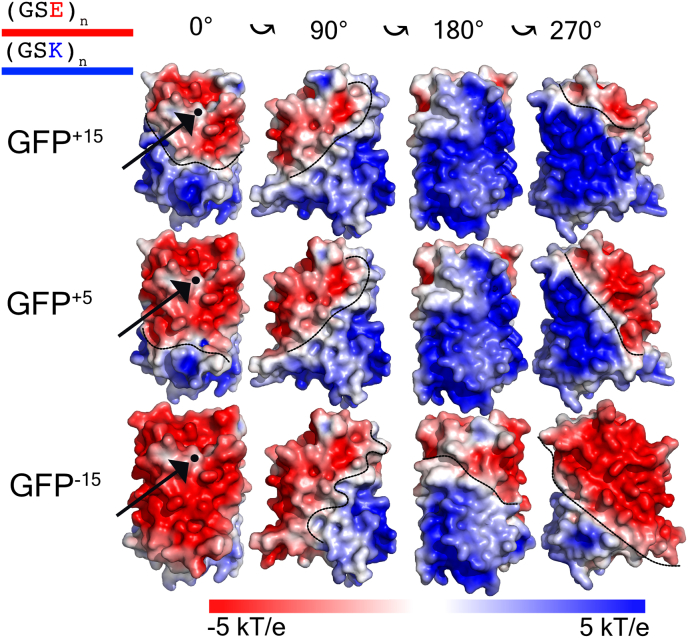
Taking inspiration from the experimentally-tested supercharged GFP, we designed in silico GFP charge variants that systematically varied the number and position of charged residues on the protein surface (Lawrence et al., 2007; Pak et al., 2016; Cummings and Obermeyer, 2018; Laber et al., 2017). We generated computational models for three GFP variants: a highly net positive variant (GFP+15) a moderately net positive variant (GFP+5) and a highly net negative variant (GFP−15) (Fig. 2 and Supplementary Fig. S5). The distribution of the charged residues divides the surface of each GFP variant into two patches of opposite charge polarity, where the area of each patch varies with overall net charge (Fig. 2).
Fig. 2.
Overview of the IDR-FD systems constructed. Three variants of the sfGFP were designed (GFP+15, GFP+5, and GFP−15). For each variant, a subset of residues were mutated according to a ruleset such that each variant had a negative ‘patch’ of varying sizes emanating from the N-terminus. Each negative patch was then surrounded by a positive ‘patch’ encompassing the remainder of the protein. The black dot present on each of the GFP variants in their initial orientation corresponds to where the N-terminal tail is attached to. Black dotted lines superimposed on each GFP variant demarcate the boundary between their respective negative and positive patch. For each variant, we also report its mean electrostatic potential and patchiness value.
In addition, we designed ten low-complexity polyelectrolytic disordered sequences using the repetitive base unit of (GSE)n or (GSK)n where n ranges between 4 and 12 in increments of 2. These repetitive polyelectrolytic GS-rich sequences were designed to minimize secondary structure, avoid the confounding contribution of other sidechains, and to control for charge patterning. Each IDR was connected to the N-terminus of the GFP charge variants, allowing us to co-vary IDR and folded domain charge properties independently. Further details regarding protein design are provided in the Methods.
To explore how varying FD and IDR charge properties impacts the conformational behavior of the tails we performed all-atom implicit solvent simulations using the CAMPARI simulation engine with the ABSINTH implicit solvent model (Vitalis and Pappu, 2009). The FD backbone dihedral angles were held fixed, while in the IDRs backbone dihedral angles were fully sampled. All sidechain degrees of freedom were sampled freely. In addition to simulating IDRs in the context of FDs, we also performed simulations of the IDRs in isolation.
2.3. Overall net charge does not necessarily determine the interaction between disordered tails and folded domains
To gain an initial sense of the conformational behavior of each IDR-FD system, we quantified the average inter-residue distance between each pair of residues on the FD and IDR normalized by the distances expected if the IDR behaved as a self-avoiding random coil (the “excluded volume” limit, see Methods) (Fig. 3). The resulting heatmap of this normalized distance (scaling map) offers a way to visualize IDR-FD interaction. In addition, we quantified additional sequence context information and the IDR radius of gyration in isolation and in the context of the FDs (Supplementary Fig. S6 and S7).
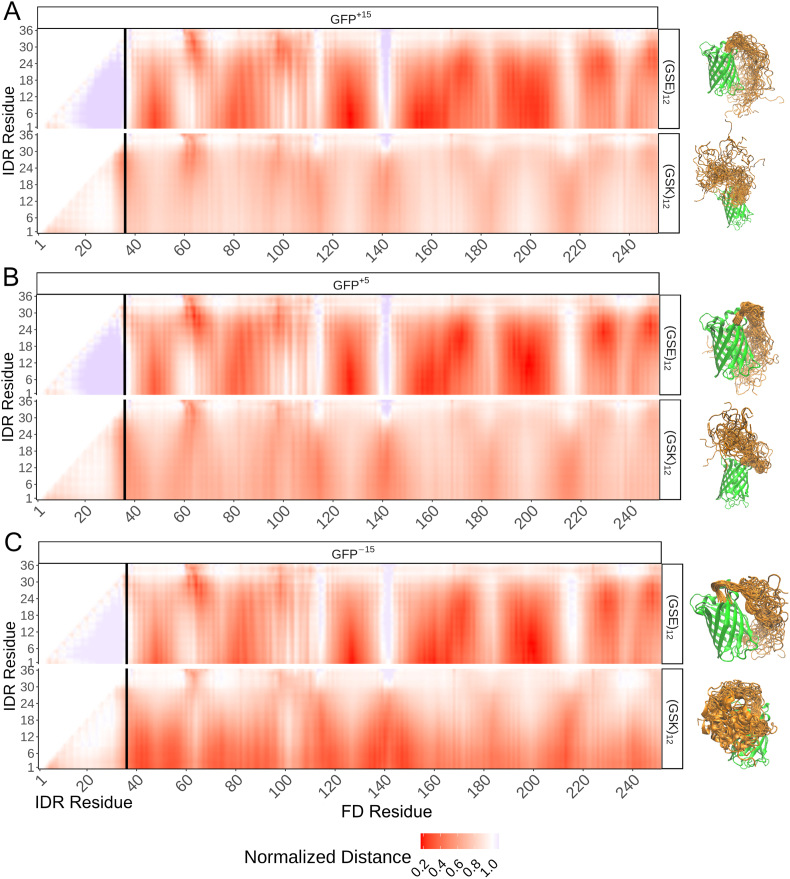
Fig. 3.
The conformational behavior of (GSE)12-GFPXand (GSK)12-GFPXis dependent on the tail's net charge. In addition, the conformational behavior of (GSK)12-GFPX is dependent on surface charge properties. Each panel refers to a scaling map for a specific system: A) (GSE/K)12-GFP+15. B) (GSE/K)12-GFP+5. C) (GSE/K)12-GFP−15. Each element on the scaling map refers to the average distance between residues i and j from all-atom implicit solvent simulations divided by the average distance between residues i and j in the corresponding excluded volume simulation. Inter-residue distances were calculated between all residues of the tail (residues 1–36) and the entire protein. To the right of each panel, we show representative snapshots of the resulting conformational ensemble.
Our expectation was that we would see charge complementarity between the tail and the folded domain. In all simulations the IDR remains fully solvent exposed and any interacts that occur are intrinsically fuzzy (Tompa and Fuxreiter, 2008). Specifically, we anticipated that negatively charged IDRs would be attracted to positively charged FDs and repelled from negatively charged FDs (and vice versa). To our surprise, this was not the case. While the (GSK)12 tail matches this expectation, the (GSE)12 tail interacts even with the most negatively charged GFP−15 variant. These trends were robust as a function of tail length (Supplementary Fig. S8). As such, our simulations of a set of deliberately simple rationally designed proteins reveal an unanticipated layer of conformational complexity.
2.4. Interaction between IDR tails and FD surfaces depend on the locally accessible surface from the perspective of the IDR
To better understand the molecular basis for the unexpected IDR-FD interactions, we generated 3D contour volume plots that project the terminal tail residue for a given IDR:FD system (Fig. 4, see Methods). Roughly speaking, these plots can be thought of as the three-dimensional analogue of a two-dimensional density plot, where contour levels reflect probability density (Supplementary Fig. S9). To complement these plots we also generated scaling maps for each system using a subset of evenly spaced FD surface residues (Supplementary Figs. S10 and S11).
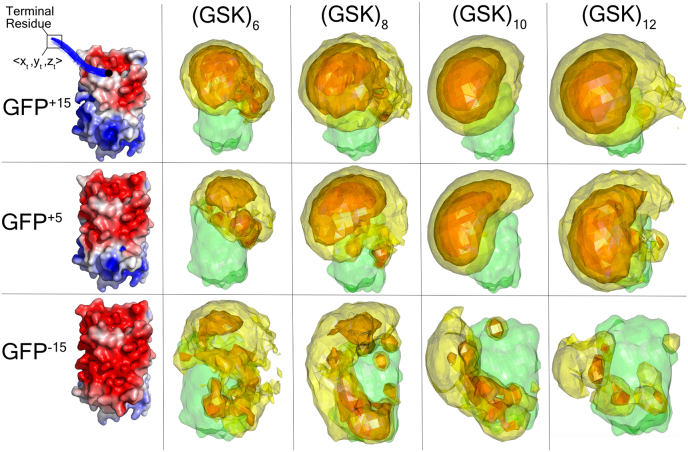
Fig. 4.
3D contour volume plots for (GSK)n-GFP+15, (GSK)n-GFP+5, and (GSK)n-GFP−15at tail lengths 18, 24, 30, and 36 illustrate that conformations tend to cluster closer to the negative patch of the GFP in a manner dependent on tail length and surface charge distribution. The green surface refers to the GFP. Each colored region (excluding the green one) represents a 3D contour volume of the <x,y,z> coordinates of the terminal residue (notated as <xt,yt,zt>) across all frames for each IDR-FD system. Each region was plotted to encapsulate 25% (red), 50% (orange), or 75% (yellow) of the data.
We first examined the positively charged (GSK)n tail. While GFP+15 and GFP+5 repel the (GSK)n from the FD surface (Fig. 4 top and middle rows), with GFP−15 we observed heterogeneous FD-IDR interaction (Fig. 4 bottom row). These results are consistent with our naive expectation, in which the positively charged IDR is repelled by a positively charged surface but attracted once the surface is sufficiently negative. In the case of GFP−15, as the (GSK)n becomes longer, new parts of the FD surface become accessible leading to the enhancement and depletion of residue-specific FD-IDR interactions (Supplementary Fig. 11 [compare along rows] and Supplementary Fig. 12). In this system IDR length can tune the IDR-accessible surface area of the FD, dictating where and how a tail can interact with the surface.
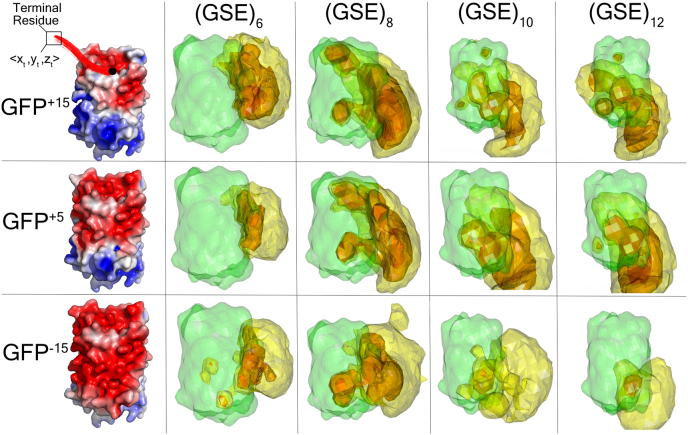
We next performed the same analysis for the negatively charged (GSE)n tail. As noted above and in contrast to the (GSK)n, the (GSE)12 tail interacts with all three GFP variants (Figs. 3 and 5, Supplementary Figs. S13 and S14). What explains this unexpected result? The surface of the GFP+15 includes a positively charged patch distal from the FD:IDR junction (Fig. 4). As the GFP net charge is titrated from +15 to −15 this positive patch shrinks, but does not disappear (Fig. 4). To gain some intuition regarding how the tail is interacting with the small positive patch, it is worth referring back to the snapshot of frames illustrated in Fig. 3 and Supplementary Fig. S15. Essentially, the tail is able to orient away from the adjoining negative patch at the FD:IDR junction and then loop across the GFP to either orient towards or be in close contact with the positive patch. This suggests FD-IDR interaction is determined by the locally accessible positive patch, as opposed to the overall FD charge. As with the (GSK)n tail, as (GSE)n becomes longer, more of this patch becomes accessible to the IDR. Finally, we note that the trends we observe for (GSE)12-GFPX and (GSK)12-GFPX persist at higher NaCl concentrations (Supplementary Figs. S16 and S17).
Fig. 5.
3D contour volume plots for (GSE)n-GFP+15, (GSE)n-GFP+5, and (GSE)n-GFP−15at tail lengths 18, 24, 30, and 36 illustrate that conformations cluster towards or near the positive patch of the GFP as tail length increases. The green surface refers to the GFP. Each colored region (excluding the green one) represents a 3D contour volume of the <x,y,z> coordinates of the terminal residue (notated as <xt,yt,zt>) across all frames for each IDR-FD system. Each region was plotted to encapsulate 25% (red), 50% (orange), or 75% (yellow) of the data.
These results neatly highlight two key features that determine the IDR:FD interactions. Firstly, FD-IDR interactions are not necessarily driven by the overall net charge of the FD and the IDR but by a balance of the intra-IDR interactions with FD:IDR interactions. Secondly, the surface-accessible residues from the perspective of the IDR are the key determinant of FD:IDR interaction. As such, local charge patches can have a profound impact on FD:IDR interactions.
2.5. Folded-domain-induced changes to the conformational behavior of an IDR can seem superficially modest
Having assessed IDR-FD interaction we next considered the impact that a FD can have on the intrinsic conformational behavior of an IDR. We used internal scaling profiles to quantify the intra-IDR interaction in both the (GSK)n and (GSE)n cases. Internal scaling profiles measure the average distance across all sets of inter-residue pairs separated by the same number of residues, providing an ensemble-average measure of the intra-IDR attraction.
For the (GSK)n tails we observed a systematic diminution in IDR dimensions as net charge on the folded domain was titrated from +15 to −15 (Fig. 6, Supplementary Fig. S18). This trend is explained by the reduction in FD-IDR electrostatic repulsion and the ultimately attractive FD-IDR interaction that leads to local polyelectrolyte condensation. Additionally, the charge surface at the FD:IDR junction is relatively acidic, further enabling IDR compaction through favorable electrostatic interaction with (GSK)n.
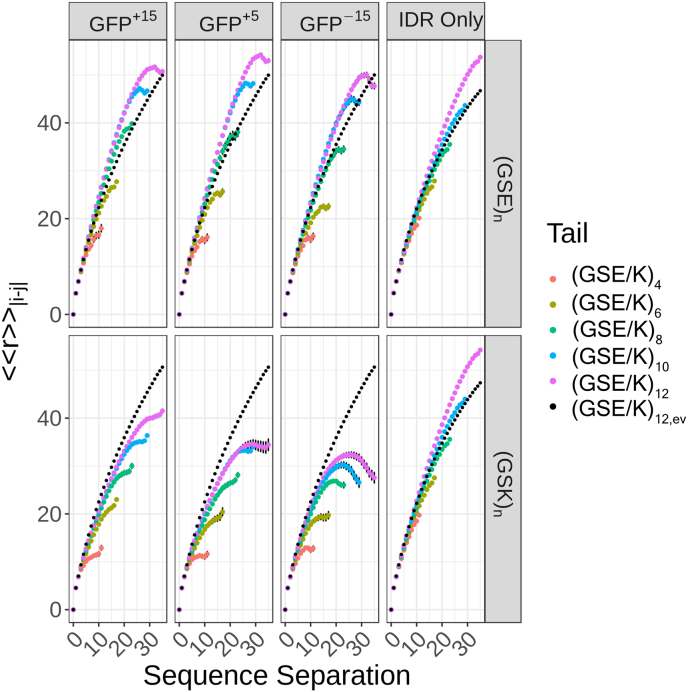
Fig. 6.
The conformational ensemble of tails is influenced by the interplay of the tail's length and net charge and the FD's net charge and surface charge distribution. Internal scaling profiles for each tail ((GSE)n or (GSK)n) attached to each GFP variant (first, second, and third columns) and each tail in isolation (fourth column) for the full Hamiltonian simulations. The black dotted line represents the internal scaling profile for (GSE/K)12 attached to each GFP variant and in isolation for the excluded volume simulations. The spatial separations for all pairs of IDR residues that are |j−i| apart in the linear sequence are calculated for each of the conformations in the relevant ensemble.The patterns of intra-IDR distances can be summarized in terms of so-called internal scaling profiles. The ensemble-averaged spatial distance for each sequence separation, denoted as is plotted against |j−i|.
In contrast to (GSK)n and despite engaging in extensive FD-IDR interactions, the (GSE)n tails are highly expanded (Fig. 6, Supplementary Fig. S18). This reflects the balance in intra-IDR repulsion, repulsion between the tail and the negative surface at the FD:IDR junction, and the need of the IDR to ‘reach’ across the FD surface to interact with the positively charged patch. As an intriguing caveat, if IDR dimensions were being probed directly in the presence/absence of a folded domain, a naive conclusion might be that the FD does not alter the IDR behavior. As such, monitoring IDR dimensions alone may mask FD-driven changes in the conformational ensemble through compensatory effects.
In Fig. 7, we summarize the results and potential implications of the GFP simulations. To reiterate, the purpose of these simulations was not to serve as a predictive model of IDR-FD behavior, but rather to probe the effects of FD surface charge distribution under different contexts and determine if complex conformational behavior can emerge from relatively simple systems. Though the behavior of the (GSK)n-(GFP)X systems can be essentially reduced to the net charge of the IDR and GFP (Fig. 7A), the behavior of (GSE)n-(GFP)X is more nuanced. Specifically, it appears that the presence of a relatively small positive patch can facilitate interactions between the tail and FD if the tail is long enough (Fig. 7B). This behavior also underscores that the conformational flexibility of a tail can enable it to ‘find’ an electrostatically complementary patch, even if that patch is distant and the tail is anchored to a large patch inducing repulsive interactions. In addition, the location of an electrostatically complementary patch may be as fundamental as its size or charge in influencing conformational behavior (Fig. 7B).
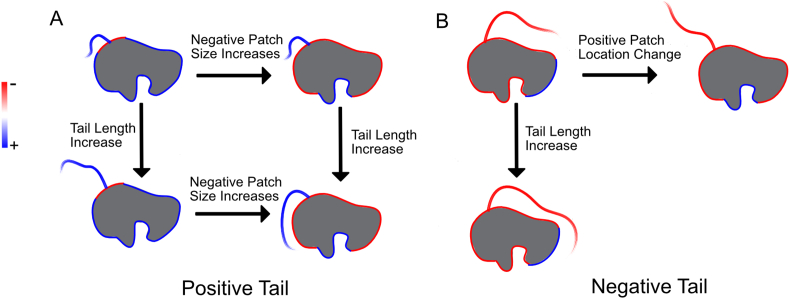
Fig. 7.
Framework for reasoning about how modulating specific properties of tails and folded domains can influence conformational behavior. A) Illustration of how the size of an adjoining, electrostatically complementary patch and the length of a tail can influence a tail's conformational behavior. When the length of the positive tail is small, the size of the negative patch is relatively unimportant in terms of influencing conformational behavior. However, as the length of the tail increases, the size of the negative patch influences the tail's conformation. When the size of the negative patch is small/large, the longer tail does not/does closely interact with it. B) Illustration of how the location of a distant electrostatically complementary patch and the length of a tail can influence a tail's conformational behavior. In the top left configuration, the tail is oriented towards the distant positive patch. As the length of the tail increases, this effect is magnified and the tail makes closer contact with the positive patch. However, when the location of the positive patch changes to being ‘sandwiched’ between two negatively charged regions, the tail no longer orients towards it.
2.6. Surface charge distribution characteristics of natural proteins
Given that the surface charge distribution of a FD can influence IDR-FD interactions and IDR conformational behavior (at least in synthetic proteins), we wondered if this was actually relevant for naturally occurring proteins. To answer this we characterized the surface charge distribution characteristics for 214 proteins with N- or C- terminal tails and 3D structural data that satisfied a particular set of criteria (see Methods).
We first quantified the NCPR of each FD (Supplementary Fig. 19A). On average, FDs had a mean NCPR close to zero (mean NCPR = −0.006). While calculating the net charge of a protein can be thought of as a way to quantify its electrostatics, this bulk metric ignores the spatial orientation of the protein's atoms, the partial charges induced by individual atoms, and the protein's interactions with its neighboring solvent and ions. Given this, we sought to quantify the overall electrostatic potential of each FD. To do so, we calculated the electrostatic potential at each point on the surface of the FD, and then averaged these values to give a scalar parameter that reports on the electrostatic potential generated by the surface in a solvated environment (Supplementary Fig. 19B). We term this quantity the mean electrostatic potential () (see Supplementary Methods). Interestingly, though the NCPR and of the FD are related (r = 0.74), they are not perfectly correlated (Supplementary Fig. 19C). We interpret this to mean that is capturing information that a coarser metric like NCPR is not.
To further quantify the distribution of charge density across the surface of the protein, we determined if regions on the surface of a protein contain correlated regions of charge (such regions are termed ‘patches’, see Methods for details). Given the results of the (GSE)n-GFPX systems, we speculated that the presence, size, electrostatic characteristics, and location of these patches might be an informative description of the FD's surface charge distribution.
To identify patches, we expressed the APBS-calculated electrostatic surface as a graph G and identify connected components of G that consist of nodes whose electrostatic potential is either exclusively greater than or less than a threshold. Each connected component in each subgraph can then be thought of as a ‘patch’ (further details in Methods).
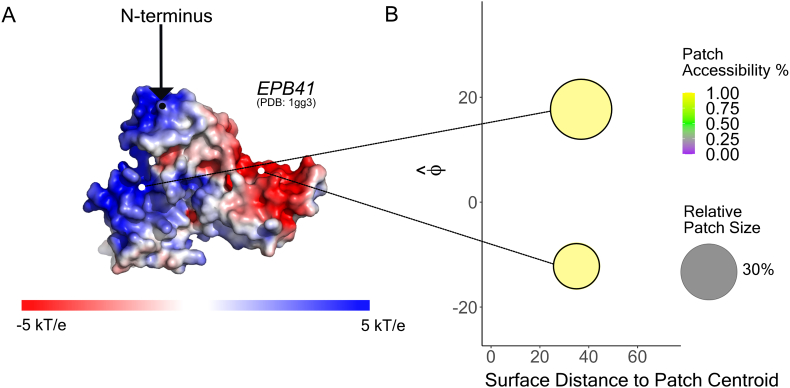
After identifying a patch we quantified three things: 1) its mean electrostatic potential, 2) the relative size of the patch, and 3) the fraction of the patch that is ‘accessible’ to the tail. Accessibility is calculated based on the fraction of nodes comprising a patch that can be reached by the tail assuming that the shortest distance between the FD:IDR junction and that node is less than the mean end to end distance of a length matched self avoiding walk (SAW) (see Methods for details). This distance considers the excluded volume of the folded domain. Below, we show an example of this calculation for the protein EPB41 (Fig. 8) and each of the GFP mutants (Supplementary Fig. 20). In addition, we illustrate the patchiness landscape for all proteins in the analysis and complement it with the sequence charge profile for each corresponding IDR (Supplementary Figs. 21 and 22). This illustration was also supplemented with one including the three GFP mutants to demonstrate their surface charge properties in a relevant context (Supplementary Fig. 23).
Fig. 8.
Patch visualization for membrane protein EPB41. A) APBS surface for the membrane protein EPB41 (PDB1gg3), with the N-terminal junction demarcated. From this surface, it is evident that there is a negative patch and positive patch, roughly equidistant from the N-terminal junction. B) This 3D information is condensed into a 2D representation by quantifying the mean electrostatic potential, relative size, and accessibility fraction of each patch as a function of distance between the N-terminal junction and the patch centroid along the surface of the protein.
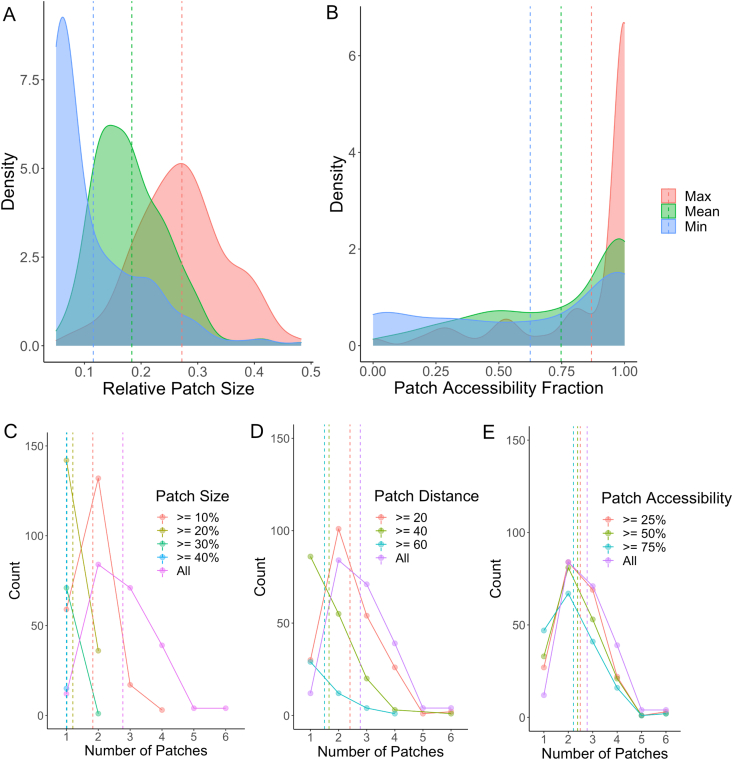
To summarize the patchiness data for all 214 proteins, we quantified the distribution of the mean/maximum/minimum relative patch size and patch accessibility fraction for each protein (Fig. 9A and B). Patchy regions on proteins tend to occupy a relatively significant fraction of the surface and on average are highly accessible. In addition, we quantified the distribution of the number of patches on a protein, and stratified these distributions as a function of relative size, distance between the FD:IDR junction and patch centroid, and accessibility (Fig. 9C–E). While proteins generally have more than one patch (mean of 2.8), the number of patches on a protein tends to decrease as a function of patch size above a certain threshold. This trend also applies when stratifying the number of patches on a protein by distance and accessibility.
Fig. 9.
Patches on a protein are on average relatively large, accessible, numerous. A) Distribution of the mean, maximum, and minimum relative patch size across all 214 proteins. B) Distribution of the mean, maximum, and minimum patch accessibility fraction across all 214 proteins. C) Frequency distribution of the number of patches on a protein stratified by patch size. D) Frequency distribution of the number of patches on a protein stratified by the distance between the FD:IDR junction and patch centroid. E) Frequency distribution of the number of patches on a protein stratified by patch accessibility fraction. Vertical dotted lines correspond to the mean value for a given metric.
Finally, to tie together the IDR and FD bioinformatic analyses, we determined if there were any correlations between electrostatic properties of IDRs and FDs. Though most analyses did not yield significant correlations (Supplementary Table S7), we did observe a statistically significant correlation between the FCR of an IDR and FD and the FCR of an IDR and the total patch size of a FD (Supplementary Fig. S24). In summary, our results reveal that many globular proteins can be reasonably considered as patchy colloids with multiple charged patches.
2.7. Effect of surface charge distribution characteristics in real protein systems
Our GFP simulations suggested that locally accessible charge patches can play a large role in influencing conformational behavior depending on the sequence composition of the tail. Given that proteins on average consist of multiple patches that are relatively large and accessible, we finally wondered how the effect of surface charge distribution manifests in real protein systems.
To test this, we examined coronavirus nucleocapsid (N) proteins from evolutionarily divergent beta-coronaviruses. N protein is a ∼400 residue largely disordered protein essential for packaging of the coronavirus RNA genome (Masters, 2019). In prior combined simulation/single-molecule work, we characterized the solution behavior of the disordered N-terminal domain (NTD) adjacent to the folded RNA binding domain (RBD) and found electrostatic interactions to play a role in influencing its conformational behavior (Cubuk et al., 2021). Intriguingly, while the RBD structure is highly conserved, the surface charge varies across the coronaviridae family. As such, we wondered if this might be a good system to identify functionally orthologous proteins in which folded-domain charge properties are variable.
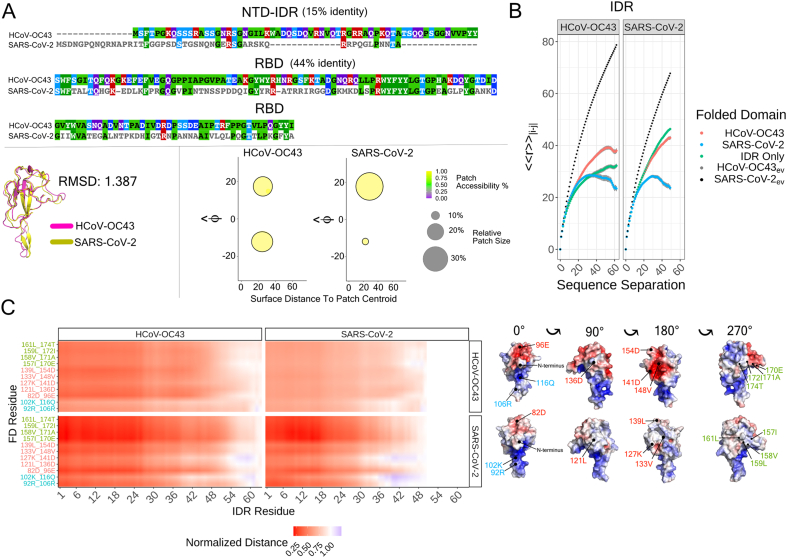
Our work focussed on NTD-RBD constructs comparing variants from SARS-CoV-2 (CoV-2) and Human coronavirus OC43 (HCoV-OC43, which we abbreviate to OC43). Between these two orthologs the RBD structure is almost identical, the NTD sequences relatively similar, but the RBD surface charge distribution is quite different (Fig. 10A, Supplementary Tables 8–9). We performed simulations of the natural NTD-RBD constructs from each virus, and chimeric variants in which the IDRs from the CoV-2 and OC43 N-protein were interchanged (i.e CoV-2NTD attached to OC43RBD and OC43NTD attached to CoV-2RBD).
Fig. 10.
The RBD of OC43 and CoV-2 N protein influences IDR conformational behavior and IDR-FD interactions. A) Aligned sequences of OC43 and CoV-2 N protein (top), aligned structures of OC43RBD and CoV-2RBD (bottom-left), and patch information for OC43RBD and CoV-2RBD (bottom-right). B) Internal scaling profiles for each tail (OC43NTD or CoV-2NTD) attached to each RBD (OC43RBD and CoV-2RBD) and each tail in isolation for the full Hamiltonian simulations. The black dotted line represents the internal scaling profile for the corresponding IDR-FD excluded volume simulations. C) Scaling maps for each of the four proteins; OC43NTD-OC43RBD (top-left), OC43NTD-CoV-2RBD (bottom-left), CoV-2NTD-OC43RBD (top-right), CoV-2NTD-CoV-2RBD (bottom-right). Each entry in the scaling map refers to the average distance between residues i and j in the full Hamiltonian simulation divided by the average distance between residues i and j in the corresponding excluded volume simulation. Inter-residue distances were calculated between all residues of a given tail and 11 specific residues on each RBD. Because there is not an exact one-to-one mapping between the linear sequence and their geometric location for the two RBDs, we determined an isomorphic mapping between 11 specific residues (e.g residue 92 [R] for CoV-2RBD corresponds to residue 106 [R] for OC43RBD, etc.). To the right of the scaling map, we show these residues in context of the electrostatic surface potential maps for OC43RBD and CoV-2RBD.
Internal scaling profiles reveal a pronounced effect of the FD on IDR conformational behavior (Fig. 10B). Both OC43NTD and CoV-2NTD are expanded when they are attached to OC43RBD in comparison to when they are attached to CoV-2RBD (Fig. 10B, Supplementary Fig. S25). Interestingly, OC43NTD is expands when attached to OC43RBD compared to its dimensions in isolation, offering another instance in which FD-derived interactions alter IDR conformational behavior.. To complement these plots, we also generated scaling maps for each system among all residues (Supplementary Fig. S26) and among a subset of ‘isomorphic’ surface residues from OC43RBD and CoV-2RBD (Fig. 10C).
For CoV-2NTD-CoV-2RBD, we observe that the basic beta-strand extension from the RBD repels the arginine-rich C-terminal region of the NTD, while the N-terminal region of the NTD engages with a hydrophobic face on the RBD. However, for CoV-2NTD-OC43RBD, neither of these preferential interactions occur and we instead observe weaker, non-specific interactions with OC43RBD. Similarly for OC43NTD-OC43RBD and OC43NTD-CoV-2RBD the behavior is more like the CoV-2NTD-OC43RBD construct.
What features of OC43RBD explain this behavior? We suspect that the larger negative patch on OC43RBD mitigates the repulsion observed between the positive beta-strand extension from CoV-2RBD and the positive C-terminal region of CoV-2NTD. Likewise, the relative reduction in hydrophobic residues coupled with a more symmetric surface charge distribution (Fig. 10A, bottom right) may prevent the preferential interactions observed between the N-terminal region of CoV-2NTD and the hydrophobic face of CoV-2RBD. In summary, the CoV-2 and OC43 N proteins serve as concrete examples where relatively modest changes in the surface charge distribution of the FD can influence IDR conformational behavior and IDR-FD interaction preferences. Furthermore, the precise degree of influence is dependent on the specific NTD, again highlighting the complexity between the interplay of IDR and FD sequence properties on conformational behavior.
3. Discussion
Disordered tails – disordered regions at the C- or N-termini of proteins with folded domains – make up 33% of human IDRs and are essential regulators of biological function (van der Lee et al., 2014). In this work we combined all-atom implicit solvent simulations and structural bioinformatics to examine how the charge properties of folded domains can influence the conformational behavior of disordered tails.
Simulations of (GSE)n-GFPx and (GSK)n-GFPx allowed us to systematically probe the effects of surface charge, tail charge, and tail length on the conformational behavior of our systems. Overall, the (GSE)n-GFPx simulations differed substantially from that of (GSK)n-GFPx, yielding complementary insights. Importantly, we verified the salt-dependence of these results and observed that while electrostatically-driven interactions are diminished as a function of salt, the same trends and features are observed across all salt concentrations in a system-specific manner.
For the (GSK)n-GFPx systems, we found that tail length and the size/charge of an adjoining patch can provide complementary properties that influence conformational behavior. As a more general model, interactions between polyelectrolytic tails and electrostatically complementary adjoining patches are dictated by a complex balance between energetically favorable IDR-FD interactions vs. unfavorable IDR-IDR interactions, and the conformational entropic cost of interacting with a specific region on the surface (i.e., the entropic cost of “fuzzy” binding) (Tompa and Fuxreiter, 2008; Arbesú et al., 2018; Fuxreiter, 2020). An additional feature not well captured in our work is the energetically (entropically) favorable contribution of ion release that conventionally is considered to drive complex coacervation (Sing and Perry, 2020). Moreover, these tradeoffs are a function of a tail's length and charge, and its adjoining patch size and charge (Fig. 7A). Of practical relevance, we note that in our structural bioinformatics analysis, there was a non-trivial fraction of proteins (∼20%) with patches that were less than roughly 15 Å between the FD:IDR junction and patch centroid (Supplementary Fig. 21).
For the (GSE)n-GFPx systems, we found that all the (GSE)n tails interact with each of the GFP variants to similar extents. While it may be less surprising that the behavior of (GSE)n-GFP+15 and (GSE)n-GFP+5 are similar, the positive patch of GFP−15 is significantly smaller than that of GFP+15 and GFP+5. This counterintuitive behavior underscores that the conformational flexibility of a tail can enable it to ‘find’ an electrostatically complementary patch and that the location of an electrostatically complementary patch can be as fundamental as its size or charge in influencing conformational behavior (Fig. 7B).
Our structural bioinformatics analysis contextualized these results for real proteins. Not only do proteins tend to have multiple patchy regions, these regions tend to be relatively large and physically accessible to their IDRs. Furthermore, if it is more broadly true that IDR-FD interactions cannot necessarily be reduced to coarse charge metrics, it may be more informative to represent the surface charge distribution of a FD as a set of patches (each possessing a mean electrostatic potential, relative size, and accessibility fraction).
Finally, our N protein simulations examined the effect of surface charge distribution on IDR conformational behavior in naturally evolved. The negative patch present on OC43RBD but absent in CoV-2RBD markedly influenced IDR-FD interactions. Whereas OC43NTD and CoV-2NTD interact extensively with CoV-2RBD, they interact more weakly with OC43RBD. The fact that this trend was more pronounced for CoV-2NTD than OC43NTD also highlights that the effect of changes in FD properties can be acutely sensitive to the IDR sequence. Prior work has reported minimal conservation in surface charge across homologous coronavirus proteins, leading to speculation that there may be different mechanisms of RNA recognition and RNP assembly (Saikatendu et al., 2007). To what extent changes in the N-terminal tail influence these differences on RNA binding and viral packaging remain to be seen.
3.1. Experimental observations on the role of electrostatics mediating interactions between disordered regions and folded domains
Prior work by Mittal et al. examined how IDR sequence properties can influence the interaction between two folded domains (in the case of disordered linkers) or IDR-FD interaction (in the case of disordered tails) (Mittal et al., 2018). In that work, IDR sequence properties were shown to dictate if a folded domain would influence IDR conformational behavior. IDR sequences categorized as weak polyampholytes were much more influenced by the presence of folded domains than other types. Here we examine the corollary of that analysis - to what extent can FD surface properties influence IDR conformational behavior for simple, synthetic sequences. In agreement with this prior work, our results here support the conclusion that the balance of intra-IDR and IDR-FD charge interactions will - at a first approximation - determine if and how an IDR will be influenced by the presence of a folded domain. Our results reveal that the surface features of the folded domain can play an equally important role in dictating IDR behavior as IDR sequence.
The intramolecular FD-IDR interactions observed in our study are analogous to the intermolecular interactions described by the polyelectrostatic effect (Mittag et al., 2010a, 2010b; Borg et al., 2007). Under this model, IDR-mediated binding can be driven through a distributed network of transient electrostatic interactions (Borg et al., 2007). Given the equivalence of intra- and inter-molecular interactions for sufficiently long and flexible polymers, it should be unsurprising that physical mechanisms that drive IDR-FD (or indeed IDR:IDR) interactions in trans will manifest as dictating intramolecular interactions (Martin and Holehouse, 2020; Dignon et al., 2018; Martin et al., 2020; Borgia et al., 2018).
Previously, Lotti et al. investigated how disordered and ordered regions interact when covalently linked to one another. To explore this, they performed a series of in vitro experiments in which a set of three different IDRs were N-terminally fused to GFP (Sambi et al., 2010). They observed that different IDRs fused to the same globular protein resulted in polypeptides with secondary structure content distinct from that predicted by the average of the two components in isolation. As in our work, these experiments reveal that the conformational behavior of tails attached to FDs can differ from the behavior observed as autonomous units (Fig. 6).
Martin et al. combined nuclear magnetic resonance (NMR) spectroscopy and small-angle X-ray scattering (SAXS) with coarse-grained simulations to uncover the influence of FD-encoded electrostatic interactions on IDR conformational behavior in the context of the RNA binding protein hnRNPA1 (Martin et al., 2021). Attractive electrostatic interactions between the net-positive C-terminal IDR and negative patches on the surface of the folded RNA recognition motifs (RRMs) engender intramolecular interaction at low (<200 mM NaCl) salt concentrations. However, at higher concentrations (>200 mM NaCl) these interactions are increasingly screened. These results reveal that even for IDR-FD interactions with relatively modest charge properties, the types of charge-dominated interactions we report here are present and pronounced at physiological salt concentrations.
To put the experimental findings of hnRNPA1 in the context of our simulations, a few additional observations are worth nothing: the C-terminal tail is positively charged (NCPR of +0.04), the RRMs are nearly neutral (NCPR of −0.005) and contain two contiguous, oppositely charged faces, and the tail interacts transiently with the negatively charged face of the folded RRMs at low salt concentrations. These observations tie back nicely to the results of our (GSE)n-GFPX simulations. Specifically, hnRNPA1 demonstrates that it is difficult to intuit which regions of a tail will interact with which regions of the surface of a FD based on coarse electrostatic properties alone, and that electrostatically complementary charged regions can drive interactions between a tail and FD. One can imagine that if the net charge of the RRM remained constant but the charge residues were not clustered together along the surface of the RRM, the tail would not interact as strongly with the RRM.
In another relevant example, Krois et al. demonstrated via chemical-shift changes that the disordered N-terminal transactivation domain of p53 (p53-NTAD) interacts with the DNA-binding surface of the DNA-binding Domain (DBD), primarily through electrostatic interactions (Krois et al., 2018). These interactions are thought to inhibit binding of nonspecific DNA to the DBD and therefore enhance selectivity toward target genes. Again, this study highlights that disordered regions can engage in electrostatically-driven FD:IDR interactions at physiological salt conditions.
For p53-NTAD, the N-terminal tail is highly negatively charged (NCPR of −0.21) and the DNA-binding surface is a positively charged region (encompassing roughly 30 residues) in close spatial proximity to the N-terminus. This behavior is analogous to the (GSK)n-GFPX systems, in the sense that a highly charged tail interacts with a proximal, electrostatically complementary charged region. The authors do note however that some contacts made by that tail appear to be weaker and more widely distributed over the DBD surface. This is not entirely surprising given that the tail is 70 residues long, and persistently close contacts with the DNA-binding surface would likely be entropically unfavorable.
4. Conclusions
Our results reveal a complex relationship between the effects of FD surface charge distribution, tail length, and tail net charge on IDR-FD interactions. In addition to charge-mediated interactions, this is growing evidence that additional modes of interaction, such as transient hydrophobically-mediated contacts, can also influence how IDRs interact with folded domains (Sankaranarayanan et al., 2021; Cubuk et al., 2021; Zheng and Castañeda, 2021). In short, the details of FD-IDR interactions are inherently complex and eminently tunable by post-translational modifications or changes in the solution environment (Martin et al., 2021; Moses et al., 2020).
How might FD-IDR interactions contribute to biological function? Possible mechanisms include the ability to alter global dimensions of a multi-domain protein, tune accessibility of binding sites, act as molecular lubricants for protein interaction, or function as locally competitive enzyme inhibitors (Sankaranarayanan et al., 2021; Ortega et al., 2018; Berlow et al., 2017; Davey, 2019). Taken together, our work here and a growing body of extant work by others argues that the interplay between IDRs and folded domains physically connected to one another represents an additional and complex layer of conformational regulation on IDR behavior.
CRediT authorship contribution statement
Ishan Taneja: Conceptualization, Writing – original draft, Visualization, Methodology, Software, Investigation, Data curation. Alex S. Holehouse: Conceptualization, Writing – original draft, Visualization, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dan Griffith and Ryan Emenecker for comments and feedback.
Handling Editor: Bilge San
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.08.002.
Materials and methods
IDR bioinformatic analysis
See Supplementary Figs. 1 and 2 for the specific bioinformatic workflows used to generate the relevant datasets. For each protein of interest, we determined which regions were predicted to be disordered using the MobiDB-lite disorder predictor. Specifically, per-residue consensus prediction was calculated, for each residue in the human proteome, with IDRs defined as contiguous regions of twenty-five residues or more with three or more predictors predicting disorder (ignoring gaps under three residues). A tail was considered N-terminal if it started at residue 1 and ended before the C-terminus of the protein. A tail was considered C-terminal if it started after the first residue and ended on the last residue of the protein. An IDR that was neither N or C-terminal was considered to be a non-tail.
Among IDRs in the reviewed human proteome with an associated PDB structure, a tail was considered N-terminal if it started at residue 1 and was within 10 residues of the residue where the FD began. A tail was considered C-terminal if it ended at the last residue of the protein as was within 10 residues of the residue where the FD ends. The analysis for Fig. 1 was performed using localCIDER (Ginell et al., 2020, Holehouse et al., 2017).
FD bioinformatic analysis
We specifically looked at proteins in the reviewed human proteome with an associated PDB structure with either an N or C terminal tail as defined according to the previously specified criteria. Among these structures, we restricted our analysis to those which had at least 200 residues or had at least 75% of the sequence's non-disordered residues covered. In addition, we only analyzed structures that were determined through X-ray crystallography or Nuclear Magnetic Resonance spectroscopy. Because there may be multiple PDB entries for each protein, we selected the PDB structure that covered the greatest number of residues. If a PDB structure contained multiple chains, we selected the first one sorted alphanumerically. This criterion yielded 214 unique proteins.
Among these 214 proteins, we calculated it's surface potential using the adaptive Poisson-Boltzmann solver (APBS) at a pH of 7.0 at an NaCl concentration of 100 mM using the PARSE force field (Jurrus et al., 2018; Dolinsky et al., 2004; Baker et al., 2001). Prior to being analyzed by APBS, each PDB file was converted to a PQR file using the PDB2PQR (Dolinsky et al., 2004). To enable quantitative analysis of the surface potential, a Euclidean distance transform algorithm (EDTSurf) was used first to calculate the molecular surface representation of each protein (Xu and Zhang, 2009). The surface was then overlaid with the APBS potential map and relevant parameters of interest were calculated. This workflow and the code used to calculate relevant parameters is built off the work of Kim et al. (2020).
Patch identification and characterization
The first step is to express the APBS surface (a series of x,y,z coordinates with an electrostatic potential associated with each point) as a graph G where each coordinate is a node and edges are placed between two nodes if the euclidean distance between them is less than 2 Å. Due to this cutoff distance, it is possible that the G is not fully connected and instead contains multiple unique connected components. To ensure G is fully connected, for each connected component ci, an edge is placed between a node in ci and a node in a different connected component cj. The specific pair of nodes chosen is the one that is nearest in euclidean distance among all possible pairs of nodes between ci and each of the other connected components. This procedure is repeated until G is fully connected. Since edges are only placed between nearby nodes during the initial construction of G, the shortest path between two nodes is effectively the distance along the surface of the protein.
To identify a patch, we constrain the graph to nodes whose electrostatic potential is greater than or less than a certain value, yielding the subgraphs Gpos and Gneg. This creates connected components in each subgraph consisting of either ‘positive’ or ‘negative’ nodes. Each connected component in each subgraph can then be thought of as a ‘patch’. Thus, a patch is represented by a set of nodes and edges, with each node having a corresponding electrostatic potential.
For a given patch, its relative size is calculated as the number of nodes in its corresponding connected component ci divided by the total number of nodes in G. Its mean electrostatic potential is calculated by averaging the electrostatic potential across all nodes in ci. The patch accessibility fraction is calculated by determining the fraction of nodes in ci that can be reached by the tail assuming that the shortest distance between the FD:IDR junction and that node is less than the mean end to end distance of a length matched self avoiding walk (SAW). Specifically, this is calculated as where A1 = 5.5 as determined previously (Hofmann et al., 2012; Zheng et al., 2018). Finally, we note that we constrained the structural bioinformatics analysis to patches whose relative size was greater than or equal to 5%.
All code and data used for our complete bioinformatics analysis is available at https://github.com/holehouse-lab/supportingdata/tree/master/2021/taneja_holehouse_tail_fd_2021.
GFP mutant construction
To create a series of GFP variants with varying surface charge distributions, surface residues were mutated in silico to either invert or neutralize the charge (E to K, K to E, D to N, or R to Q). This specific scheme was chosen such that each pair of residues that are being substituted are nearby according to various amino acid distance metrics. Each GFP variant was constructed as follows: 1) Lysine and arginine residues that were within x percent of the nearest surface residues from the FD:IDR junction were mutated (K to E and R to Q) to maximize negative charge. 2) Glutamic acid and aspartic acid residues that were within the remaining 100-x percent of surface residues from the FD:IDR junction were mutated (E to K and D to N) to maximize positive charge. Three GFP variants were specifically constructed where x was either 25 (GFP+15), 50 (GFP+5), or 75 (GFP−15).
The PDB ID of the wtGFP used was 1QYO. In order to isolate the effect of the interactions between the synthetically attached tail and the surface of each GFP variant, we removed 10 residues from the N-terminal tail of the protein (residues 2–11) and 11 residues from the C-terminal tail (residues 227–237) of the protein.Attached N-terminal to each GFP was a sequence of either repeating GSE blocks or GSK blocks. Each low-complexity IDR was predicted to be fully disordered by all predictors tested. Detailed information for each IDR and FD sequence is provided in Supplementary Tables 2, 3, 4, and 5.
SARS-CoV-2 and HCoV-OC43 N-Protein simulations
For SARS-CoV-2RBD, the starting structure used was taken as the first chain extracted from the 6VYO PDB crystal structure. For HCoV-OC43RBD, a homology model was built using SWISS-MODEL (Waterhouse et al., 2018) from the crystal structure of HCoV-OC43RBD (PDB: 4LI4) to account for three missing residues in the RBD extension.
3D contour volume plots
Using the R package ks (Duongks, 2007), we created a 3D contour volume plot of the x, y, z coordinates of the terminal residue of the tail among all frames for each IDR-FD system. Each boundary contains either 25, 50, or 75 percent of the volume of a probability density distribution. On average, the x% volume contour contains x% of the points that were used to generate the kernel density estimate.
All-atom implicit solvent simulations
All-atom Monte Carlo simulations were performed with the ABSINTH implicit solvent model and CAMPARI simulation engine (http://campari.sourceforge.net/) (Vitalis and Pappu, 2009). Simulations were run at 310 K at an ion concentration of 10 mM NaCl (unless specified otherwise). All simulations were performed in sufficiently large box sizes to prevent finite size effects. For simulations with IDRs in isolation, all degrees of freedom available (backbone and sidechain dihedral angles and rigid-body positions) in CAMPARI are sampled. For simulations with folded domains with IDRs, side chains are fully sampled while the backbone dihedral angles in folded domains are not sampled (resulting in the folded domains to remain structurally fixed).
Excluded volume (EV) simulations were performed using the same setup, but with a modified Hamiltonian under which solvation, attractive Lennard-Jones, and polar (charge) interactions are scaled to zero, as described previously (Holehouse et al., 2015). See Supplementary Table 6 for an overview of the simulation input details for each system. All-atom implicit solvent simulations were analyzed using CAMPARITraj (http://camparitraj.com/) and MDTraj (McGibbon et al., 2015).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Arbesú M., Iruela G., Fuentes H., Teixeira J.M.C., Pons M. Intramolecular fuzzy interactions involving intrinsically disordered domains. Front Mol Biosci. 2018;5:39. doi: 10.3389/fmolb.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlow R.B., Dyson H.J., Wright P.E. Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature. 2017;543:447–451. doi: 10.1038/nature21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherds W., Theillet F.-X., Katzer A., Finzel A., Mishall K.M., Powell A.T., Wu H., Manieri W., Dieterich C., Selenko P., Loewer A., Daughdrill G.W. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat. Chem. Biol. 2014;10:1000–1002. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- Borg M., Mittag T., Pawson T., Tyers M., Forman-Kay J.D., Chan H.S. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D., Kragelund B.B., Best R.B., Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman M.A., Riback J.A., Rodriguez A., Guo H., Li J., Sosnick T.R., Clark P.L. Properties of protein unfolded states suggest broad selection for expanded conformational ensembles. Proc. Natl. Acad. Sci. U.S.A. 2020;117(38):23356–23364. doi: 10.1073/pnas.2003773117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk J., Alston J.J., Jeremías Incicco J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1–17. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings C.S., Obermeyer A.C. Phase separation behavior of supercharged proteins and polyelectrolytes. Biochemistry. 2018;57:314–323. doi: 10.1021/acs.biochem.7b00990. [DOI] [PubMed] [Google Scholar]
- Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.K., Ruff K.M., Pappu R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Lin Y.-H., Vernon R.M., Forman-Kay J.D., Chan H.S. Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 2020;117:28795–28805. doi: 10.1073/pnas.2008122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey N.E. The functional importance of structure in unstructured protein regions. Curr. Opin. Struct. Biol. 2019;56:155–163. doi: 10.1016/j.sbi.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Dignon G.L., Zheng W., Best R.B., Kim Y.C., Mittal J. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 2018;115:9929–9934. doi: 10.1073/pnas.1804177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., ks Kernel density estimation and kernel discriminant analysis for multivariate data inR. J. Stat. Software. 2007;21 doi: 10.18637/jss.v021.i07. [DOI] [Google Scholar]
- Dyla M., Kjaergaard M. Intrinsically disordered linkers control tethered kinases via effective concentration. Proc. Natl. Acad. Sci. U.S.A. 2020;117:21413–21419. doi: 10.1073/pnas.2006382117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M. Fuzzy protein theory for disordered proteins. Biochem. Soc. Trans. 2020 doi: 10.1042/BST20200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D., Otieno S., Waddell B., Iconaru L., Kriwacki R.W., Chen J. Electrostatically accelerated coupled binding and folding of intrinsically disordered proteins. J. Mol. Biol. 2012;422:674–684. doi: 10.1016/j.jmb.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginell G.M., Holehouse A.S. In: Intrinsically Disordered Proteins: Methods and Protocols. Kragelund B.B., Skriver K., editors. Springer US; New York, NY: 2020. Analyzing the sequences of intrinsically disordered regions with CIDER and localCIDER; pp. 103–126. [DOI] [PubMed] [Google Scholar]
- Graña-Montes R., Marinelli P., Reverter D., Ventura S. N-terminal protein tails act as aggregation protective entropic bristles: the SUMO case. Biomacromolecules. 2014;15:1194–1203. doi: 10.1021/bm401776z. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Soranno A., Borgia A., Gast K., Nettels D., Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse A.S., Das R.K., Ahad J.N., Richardson M.O.G., Pappu R.V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophysical Journal. 2017;112(1):16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse A.S., Garai K., Lyle N., Vitalis A., Pappu R.V. Quantitative assessments of the distinct contributions of polypeptide backbone amides versus side chain groups to chain expansion via chemical denaturation. J. Am. Chem. Soc. 2015;137:2984–2995. doi: 10.1021/ja512062h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Li M., Lai L., Liu Z. Allostery of multidomain proteins with disordered linkers. Curr. Opin. Struct. Biol. 2020;62:175–182. doi: 10.1016/j.sbi.2020.01.017. [DOI] [PubMed] [Google Scholar]
- Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D.W., Dolinsky T., Konecny R., Koes D.R., Nielsen J.E., Head-Gordon T., Geng W., Krasny R., Wei G.-W., Holst M.J., McCammon J.A., Baker N.A. Improvements to the APBS biomolecular solvation software suite: improvements to the APBS Software Suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keul N.D., Oruganty K., Schaper Bergman E.T., Beattie N.R., McDonald W.E., Kadirvelraj R., Gross M.L., Phillips R.S., Harvey S.C., Wood Z.A. The entropic force generated by intrinsically disordered segments tunes protein function. Nature. 2018;563:584–588. doi: 10.1038/s41586-018-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sureka H.V., Kayitmazer A.B., Wang G., Swan J.W., Olsen B.D. Effect of protein surface charge distribution on protein-polyelectrolyte complexation. Biomacromolecules. 2020;21:3026–3037. doi: 10.1021/acs.biomac.0c00346. [DOI] [PubMed] [Google Scholar]
- Krois A.S., Dyson H.J., Wright P.E. Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E11302–E11310. doi: 10.1073/pnas.1814051115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber J.R., Dear B.J., Martins M.L., Jackson D.E., DiVenere A., Gollihar J.D., Ellington A.D., Truskett T.M., Johnston K.P., Maynard J.A. Charge shielding prevents aggregation of supercharged GFP variants at high protein concentration. Mol. Pharm. 2017;14:3269–3280. doi: 10.1021/acs.molpharmaceut.7b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Phillips K.J., Liu D.R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq L., Raiola L., Chabot P.R., Cyr N., Arseneault G., Legault P., Omichinski J.G. Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-κB and transcription regulatory factors. Nucleic Acids Res. 2017;45:5564–5576. doi: 10.1093/nar/gkx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-H., Forman-Kay J.D., Chan H.S. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry. 2018;57:2499–2508. doi: 10.1021/acs.biochem.8b00058. [DOI] [PubMed] [Google Scholar]
- Ma B., Tsai C.-J., Haliloğlu T., Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A.H., Crick S.L., Vitalis A., Chicoine C.L., Pappu R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A.H., Lyle N., Pappu R.V. Describing sequence-ensemble relationships for intrinsically disordered proteins. Biochem. J. 2013;449:307–318. doi: 10.1042/BJ20121346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.A., Forman-Kay J.D. Sequence determinants of compaction in intrinsically disordered proteins. Biophys. J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.W., Holehouse A.S. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg Top Life Sci. 2020;4:307–329. doi: 10.1042/ETLS20190164. [DOI] [PubMed] [Google Scholar]
- Martin E.W., Holehouse A.S., Grace C.R., Hughes A., Pappu R.V., Mittag T. Sequence determinants of the conformational properties of an intrinsically disordered protein prior to and upon multisite phosphorylation. J. Am. Chem. Soc. 2016;138:15323–15335. doi: 10.1021/jacs.6b10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.W., Holehouse A.S., Peran I., Farag M., Incicco J.J., Bremer A., Grace C.R., Soranno A., Pappu R.V., Mittag T. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367:694–699. doi: 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.W., Thomasen F.E., Milkovic N.M., Cuneo M.J., Grace C.R., Nourse A., Lindorff-Larsen K., Mittag T. Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J.J., Choi U.B., Bowen M.E. Reconstitution of multivalent PDZ domain binding to the scaffold protein PSD-95 reveals ternary-complex specificity of combinatorial inhibition. Structure. 2014;22:1458–1466. doi: 10.1016/j.str.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernández C.X., Schwantes C.R., Wang L.-P., Lane T.J., Pande V.S., Traj M.D. A modern, open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T., Forman-Kay J.D. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Mittag T., Kay L.E., Forman-Kay J.D. Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recogn. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mittag T., Marsh J., Grishaev A., Orlicky S., Lin H., Sicheri F., Tyers M., Forman-Kay J.D. Structure/function implications in a dynamic complex of the intrinsically disordered Sic 1 with the Cdc 4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Holehouse A.S., Cohan M.C., Pappu R.V. Sequence-to-Conformation relationships of disordered regions tethered to folded domains of proteins. J. Mol. Biol. 2018;430:2403–2421. doi: 10.1016/j.jmb.2018.05.012. [DOI] [PubMed] [Google Scholar]
- Moses D., Yu F., Ginell G.M., Shamoon N.M., Koenig P.S., Holehouse A.S., Sukenik S. Revealing the hidden sensitivity of intrinsically disordered proteins to their chemical environment. J. Phys. Chem. Lett. 2020;11:10131–10136. doi: 10.1021/acs.jpclett.0c02822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Späth S., Soranno A., Hirschfeld V., Hofmann H., Rüegger S., Reymond L., Nettels D., Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega E., Rengachari S., Ibrahim Z., Hoghoughi N., Gaucher J., Holehouse A.S., Khochbin S., Panne D. Transcription factor dimerization activates the p300 acetyltransferase. Nature. 2018;562:538–544. doi: 10.1038/s41586-018-0621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C.W., Kosno M., Holehouse A.S., Padrick S.B., Mittal A., Ali R., Yunus A.A., Liu D.R., Pappu R.V., Rosen M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A., Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Portz B., Lu F., Gibbs E.B., Mayfield J.E., Rachel Mehaffey M., Zhang Y.J., Brodbelt J.S., Showalter S.A., Gilmour D.S. Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain. Nat. Commun. 2017;8:15231. doi: 10.1038/ncomms15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Receveur-Bréchot V., Bourhis J.-M., Uversky V.N., Canard B., Longhi S. Assessing protein disorder and induced folding. Proteins: Struct. Funct. Bioinf. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambi I., Gatti-Lafranconi P., Longhi S., Lotti M. How disorder influences order and vice versa - mutual effects in fusion proteins containing an intrinsically disordered and a globular protein: ordered and disordered protein domains. FEBS J. 2010;277:4438–4451. doi: 10.1111/j.1742-4658.2010.07825.x. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan M., Emenecker R.J., Jahnel M., Irmela R.E., Wayland M.T., Alberti S., Holehouse A.S., Weil T.T. The arrested state of processing bodies supports mRNA regulation in early development. Cold Spring Harbor Laboratory. 2021:2021. doi: 10.1101/2021.03.16.435709. 03.16.435709. [DOI] [Google Scholar]
- Sawle L., Ghosh K. A theoretical method to compute sequence dependent configurational properties in charged polymers and proteins. J. Chem. Phys. 2015;143 doi: 10.1063/1.4929391. [DOI] [PubMed] [Google Scholar]
- Sherry K.P., Das R.K., Pappu R.V., Barrick D. Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E9243–E9252. doi: 10.1073/pnas.1706083114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing Charles, Perry Sarah. Recent progress in the science of complex coacervation. Soft Matter. 2020;16(12):2885–2914. doi: 10.1039/d0sm00001a. [DOI] [PubMed] [Google Scholar]
- Sørensen C.S., Kjaergaard M. Effective concentrations enforced by intrinsically disordered linkers are governed by polymer physics. Proc. Natl. Acad. Sci. U.S.A. 2019;116:23124–23131. doi: 10.1073/pnas.1904813116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staby L., Kemplen K.R., Stein A., Ploug M., Clarke J., Skriver K., Heidarsson P.O., Kragelund B.B. Disorder in a two-domain neuronal Ca2+-binding protein regulates domain stability and dynamics using ligand mimicry. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03639-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Cook E.C., Creamer T.P., Kekenes-Huskey P.M. Electrostatic control of calcineurin's intrinsically-disordered regulatory domain binding to calmodulin. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2651–2659. doi: 10.1016/j.bbagen.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. /1. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. The most important thing is the tail: multitudinous functionalities of intrinsically disordered protein termini. FEBS Lett. 2013;587:1891–1901. doi: 10.1016/j.febslet.2013.04.042. [DOI] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., Kim P.M., Kriwacki R.W., Oldfield C.J., Pappu R.V., Tompa P., Uversky V.N., Wright P.E., Babu M.M. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A., Pappu R.V. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuzman D., Levy Y. DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21004–21009. doi: 10.1073/pnas.1011775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang Y. Generating triangulated macromolecular surfaces by euclidean distance transform. PloS One. 2009;4 doi: 10.1371/journal.pone.0008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez Orozco I.S., Mindlin F.A., Ma J., Wang B., Levesque B., Spencer M., Rezaei Adariani S., Hamilton G., Ding F., Bowen M.E., Sanabria H. Identifying weak interdomain interactions that stabilize the supertertiary structure of the N-terminal tandem PDZ domains of PSD-95. Nat. Commun. 2018;9:3724. doi: 10.1038/s41467-018-06133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerze G.H., Best R.B., Mittal J. Sequence- and temperature-dependent properties of unfolded and disordered proteins from atomistic simulations. J. Phys. Chem. B. 2015;119:14622–14630. doi: 10.1021/acs.jpcb.5b08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Castañeda C.A. Previously uncharacterized interactions between the folded and intrinsically disordered domains impart asymmetric effects on UBQLN2 phase separation. bioRxiv. (2021) 2021 doi: 10.1101/2021.02.22.432116. 02.22.432116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zerze G.H., Borgia A., Mittal J., Schuler B., Best R.B. Inferring properties of disordered chains from FRET transfer efficiencies. J. Chem. Phys. 2018;148:123329. doi: 10.1063/1.5006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Dignon G.L., Brown M., Kim Y.C., Mittal J. Hydropathy patterning complements charge patterning to describe conformational preferences of disordered proteins. J. Phys. Chem. Lett. 2020 doi: 10.1021/acs.jpclett.0c00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.