Abstract
The pathophysiological mechanism(s) driving non-alcoholic fatty liver disease, the most prevalent chronic liver disease globally, have yet to be fully elucidated. Here, we identify regulator of G protein signaling 6 (RGS6), up-regulated in the livers of NAFLD patients, as a critical mediator of hepatic steatosis, fibrosis, inflammation, and cell death. Human patients with high hepatic RGS6 expression exhibited a corresponding high inflammatory burden, pronounced insulin resistance, and poor liver function. In mice, liver-specific RGS6 knockdown largely ameliorated high fat diet (HFD)-driven oxidative stress, fibrotic remodeling, inflammation, lipid deposition and cell death. RGS6 depletion allowed for maintenance of mitochondrial integrity restoring redox balance, improving fatty acid oxidation, and preventing loss of insulin receptor sensitivity in hepatocytes. RGS6 is both induced by ROS and increases ROS generation acting as a key amplification node to exacerbate oxidative stress. In liver, RGS6 forms a direct complex with ATM kinase supported by key aspartate residues in the RGS domain and is both necessary and sufficient to drive hyperlipidemia-dependent ATM phosphorylation. pATM and markers of DNA damage (γH2AX) were also elevated in livers from NAFLD patients particularly in samples with high RGS6 protein content. Unsurprisingly, RGS6 knockdown prevented ATM phosphorylation in livers from HFD-fed mice. Further, RGS6 mutants lacking the capacity for ATM binding fail to facilitate palmitic acid-dependent hepatocyte apoptosis underscoring the importance of the RGS6-ATM complex in hyperlipidemia-dependent cell death. Inhibition of RGS6, then, may provide a viable means to prevent or reverse liver damage by mitigating oxidative liver damage.
Keywords: RGS proteins, ATM, NAFLD, Steatosis, Oxidative stress
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) describes a spectrum of hepatic disorders that range from the comparatively benign nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH), which may progress to cirrhosis or hepatocellular carcinoma (HCC). Globally, the prevalence of NAFLD is on the rise and currently hovers around 25% worldwide [1]. The obesity epidemic is believed to be a primary driver of NAFLD with associated oxidative stress, insulin resistance, chronic inflammation and hyperlipidemia contributing to hepatocyte dysregulation. Similarly, the rate of NAFLD is higher in patients diagnosed with type II diabetes mellitus (T2DM) [2]. Although progress has been made in elucidating the mechanisms underlying NAFLD and the identification and development of novel therapies, the highly diverse rate of disease progression and lack of predictive biomarkers have stymied efforts to combat the contributions of NAFLD to global morbidity and mortality [2].
Accumulation of toxic lipid species is a critical nexus in the pathogenesis of NAFLD and results breakdown of hepatic capacity for fatty acid (FA) metabolism. Adipocyte insulin resistance can contribute by increasing the supply of FAs to the liver [3]. Excess glucose or fructose consumption can further drive FA formation via de novo lipogenesis [4]. Liver FAs are metabolized via β-oxidation in the mitochondria or peroxisomes or via esterification to triglycerides. Regardless of the source, excess FAs serve as substrates for generation of lipotoxic species that trigger recruitment of hepatocellular stress responses, carcinogenic genomic instability, release of inflammatory cytokines, stimulation of apoptotic and necrotic cell death cascades, and pro-fibrotic activation of resident hepatic stellate cells (HSCs) [2].
An ideal drug target for the treatment of NAFLD would sit at a critical nexus of multiple pathogenic signaling cascades. One potential candidate is the multi-functional serine/threonine kinase ataxia–telangiectasia mutated (ATM). Canonically, ATM controls the DNA damage checkpoint initiating cell cycle arrest and DNA repair or apoptosis depending on the extent of the genotoxic insult. ATM is directly activated via oxidation [5] and promotes ROS-dependent cell death across multiple cell types and organ systems. ATM as well as markers of DNA damage are up-regulated in the livers of NAFLD patients [6], and ATM promotes hepatocyte loss and fibrotic remodeling following high fat diet (HFD) feeding in mice [7]. In isolated hepatocytes, ATM drives mitochondrial dysregulation, DNA damage, and cell growth inhibition [6] and may represent a key link between NAFLD and hepatocellular carcinogenesis. Human ataxia–telangiectasia patients, who harbor loss-of-function ATM variants, also display progressive liver disease [8], a phenotype recapitulated in ATM knockout mice, which display impaired liver regeneration [9].Though global ATM de-regulation is characterized by a multi-system degenerative disease, identification of hepatocyte-specific ATM regulators could facilitate the development of novel therapies for NAFLD.
Global deletion of regulator of G protein signaling 6 (RGS6) mitigates alcohol-dependent liver damage [10]. RGS6 is expressed at low levels in liver but can be induced by hepatotoxic stimuli such as alcohol [10].Although originally discovered as a G-protein regulator with the demonstrated capacity to modulate multiple GPCR-dependent physiological processes via its GTPase-activating protein (GAP) activity [[10], [11], [12], [13]], RGS6 also functions in G protein-independent cellular signaling including promotion of oxidative stress and ATM activation in response to the chemotherapeutic agent doxorubicin [14,15].Further, requisite RGS6 binding partner Gβ5, which forms a co-stabilizing complex with all members of the R7 family of RGS proteins [16], is up-regulated in the livers of patients presenting with drug-induced liver injury (DILI), and hepatic Gβ5 knockdown protects the liver against acetaminophen-dependent liver damage providing an additional link between RGS-Gβ5 complexes and hepatotoxicity in vivo [17].Together, these observations led us to hypothesize that RGS6 might be involved in multiple liver pathologies via its ability to increase reactive oxygen species (ROS) and trigger apoptosis. Here, we demonstrate that RGS6, up-regulated in the livers of NAFLD patients and high fat diet-fed (HFD) mice, forms a complex with ATM in liver, facilitates ATM phosphorylation, and drives HFD-induced hepatic steatosis, fibrosis, and apoptosis.
2. Materials and methods
Antibodies and Reagents – The source/catalog information for all reagents (Table S1), antibodies (Table S2), assay kits (Table S3), and cell lines (Table S4) has been provided in Supplementary Tables 1–4. Table S2 provides information on antibody dilutions for all applications (immunoblotting, immunohistochemistry, and immunoprecipitation).Table S3 includes information regarding kits used for measuring collagen, hydroxyproline, NAD+/NADH, Ca2+ flux, lipid peroxidation (MDA), fatty acid oxidation (FAO), cell death (cytoplasmic histone-associated DNA fragments), Terminal deoxynucleotidyl transferase dUTP Nick-End Labelling (TUNEL), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and triglycerides. All samples were processed and analyses performed according to the manufacturer's protocol.
Animals – Mouse experiments were performed at the Department of Pharmaceutical Sciences, Babasaheb Bhimrao Ambedkar University, Lucknow in association with S.D. College of Pharmacy & Vocational Studies (Ref No: SDCOP&VS/AH/CPCSEA/34). Male Swiss albino mice were procured from Biological E. Limited (Registration #38/99/CPCSEA) and were handled following the Guide for the Care and Use of Laboratory Animals (NIH, USA). Mice (24–30 g, 8–12 weeks of age) were maintained on a balanced laboratory diet as per NIN, Hyderabad, India and provided with tap water ad libitum. Animal housing facilities were kept at 20 ± 2 °C, 65–70% humidity, and on a 12h/12h day/night cycle. Animals were group housed with 3–5 mice/cage.
RGS6 gene silencing via small hairpin RNA (shRNA) delivery in vivo – shRNA against RGS6 was purchased from Santacruz Biotechnology (Paso Robles, CA, USA). A scrambled shRNA served as the control for all the experiments. Invivofectamine 3.0 (Thermo Fisher Scientific, Waltham, MA, USA) was used to deliver scramble (WT) or RGS6-targeted (RGS6 KD) shRNA by tail vein injection. Briefly, 6–8 μg of selected shRNA was diluted in 60 μL of 5% glucose and mixed with the transfection reagent. The mixture was incubated to form the complexes at room temperature for 15 min. Mice received 3 injections with a 3-day interval between injections. The efficiency of in vivo delivery of shRNA was assessed by immunoblotting. Following shRNA administration, body weight (1X/week) and food intake (1–2X/week) were monitored. No notable alterations in animal weight, food intake or general wellbeing were noted (data not shown).
HFD treatment regimens – All experimental groups were fed with normal chow (NC; 4.4% fat) for the first 7–8 weeks of life. Subsequently, mice were either left on the NC diet (control) or switched to high-fat diet (HFD; Harlan Teklad TD93075; Indianapolis, IN, USA) for an additional 12 weeks. Animals were allowed ad libitum access to control or high-fat food. The HFD contained a total of 4.8 kcal/g with 54.8% of caloric content derived from fat (28% saturated, 30% trans, 28% monounsaturated and 14% polyunsaturated). The control chow contained 3.1 kcal/g and 17% fat. Unless otherwise noted, animals were sacrificed, and samples were isolated for downstream histological and biochemical analyses at the 12 week time point. shRNA delivery was performed prior to initiation of HFD feeding. Animals were sacrificed via cervical dislocation, blood was collected; and tissues were dissected and subdivided for histological and biochemical analyses.
Histology & Immunohistochemistry – Paraffin-embedded formalin-fixed mouse and human liver tissue sections were stained with Hematoxylin and Eosin (H&E) to assess macroscopic changes in tissue architecture. The lysochromediazo dye, oil red o (Sigma, St. Louis, MO, USA), was used to detect neutral triglycerides and lipids in tissue sections. The Masson Trichrome staining kit (Sigma) and Sirius red stain were used to detect collagen deposition for the determination of liver fibrosis as per manufacturers’ protocol. Detection of cytotoxicity in liver tissue was achieved using a TUNEL kit from Biovision (Milpitas, CA, USA). Immunohistochemical staining of both mouse and human liver tissue sections was performed as per a standard protocol. Briefly, sections were dewaxed in xylene (2 times, 15 min each), treated with graded series of alcohol solutions for 10 min each, immersed in 3% hydrogen peroxide in methanol to block endogenous peroxide activity and washed with distilled water (2 times, 5 min each). For antigen retrieval, slides were dipped in citrate buffer for 15 min at 100 °C. Sections were then washed with 1X PBS buffer for 5 min and blocking was done with 5% BSA in PBS for 1 h at room temperature. Next, slides were incubated overnight at 4 °C with primary antibodies in 3% BSA in PBS within a moist chamber. Following washing (3 × 10 min) in PBS at room temperature, sections were incubated for 1 h at room temperature with peroxidase-conjugated secondary antibodies. A positive reaction was detected by exposure to stable diaminobenzidene (Abcam, Cambridge, UK) for 3 min. The sections were counterstained in Harris hematoxylin, treated with graded series of alcohol for 10 min each grade (30%, 50%, 70%, 90% and 100%), dipped in xylene and left to incubate overnight. Slides were mounted with DPX (Sigma) and observed under the microscope. For proliferating cell nuclear antigen (PCNA), F4/80, CD68 and TUNEL staining, 7–10 sections were stained from each animal with 5 pictures randomly selected from each slide. Each image was scored for positive stained (brown color) nuclei and averaged. For RGS6 staining in mouse and human liver sections, the histoscore was determined from the average stain intensity using Image J (NIH) for 7–10 slides/animal, 5 images per slide.
Immunoblotting – Tissues were promptly dissected and flash frozen using liquid nitrogen. Tissue homogenates and cell pellets were prepared in 1X RIPA buffer containing protease (p8340) and phosphatase (#3) inhibitor cocktails (Sigma), and protein content quantified by BCA assay. 20 μg of protein per sample was subjected to SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were washed 1X in TBST and blocked for 1 h with 5% BSA in TBST. After one wash with 1X TBST for 5 min, the membranes were incubated overnight in primary antibodies dissolved in 3% BSA in TBST at 4 °C. Next day, membranes were washed 3 times in TBST at room temperature and probed with respective horseradish peroxidase-labelled secondary antibodies dissolved in 3% BSA in TBST. Membranes were washed three times using 1X TBST for 5 min duration at room temperature. Immunoblots were developed (UVP ChemStudio Analytik Jena) using the chemiluminescence method and densitometric quantification of immunoblots was performed using Image J software (U.S. NIH).
Measurement of fatty acid oxidation (FAO) – FAO assay was performed with 20 mg mice liver tissue as a whole or in the isolated mitochondria using the assay kit from Biomedical Research & Clinical Application (BMR, NY, USA). For this colorimetric assay, oxidation of the substrate octanoyl-coA generates NADH that is coupled to the reduction of the tetrazolium salt INT to formazan [18].
Determination of liver NADH/NAD+ ratios – The ratio of NADH to NAD+ was determined using an assay kit according to the manufacturer's instructions (Abcam, Cambridge, MA). Briefly, 20 mg of liver tissue was washed with cold PBS and homogenized in 400 μL of NAD+/NADH extraction buffer. The supernatant was collected after centrifugation at 14,000 rpm for 5 min at 4 °C. The collected supernatant was filtered through a 10 kD Spin Column (Abcam) to remove the enzymes that rapidly consume NADH before performing the assay. For determination of total NAD+ and NADH, 50 μL of supernatant was transferred onto a 96-well plate in triplicate. For measuring NADH levels, 200 μL of the supernatant was heated at 60 °C for 30 min to decompose NAD+, and 50 μL to the plate in triplicate. A NADH cycling enzyme/buffer mix (100 μL) was added to each sample and standard, mixed, and incubated at room temperature for 5 min to convert NAD+ to NADH. The NADH developer (10 μL) was added to each reaction and incubated at room temperature for 5 h before reading the plate at OD 450 nm. The standard was prepared according to the manufacturer's protocol. The NAD + level was calculated as the total NADH/NAD+ with the level of NADH subtracted. Data are expressed as millimoles per kilogram of liver.
Determination of liver pyruvate and lactate levels – The concentrations of lactate and pyruvate in the liver were determined by a colorimetric method using lactate and puruvate assay kits, respectively, according to the manufacturer's instructions (Abcam, Cambridge, MA). Briefly, 10 mg of liver tissues was washed with cold PBS and homogenized in 500 μL of pyruvate or lactate assay buffer. The supernatant was collected after centrifugation at 14,000 rpm for 5 min at 4 °C to remove any insoluble material. Since endogenous lactate dehydrogenase (LDH) may degrade lactate, the collected supernatant was deproteinized as described in the manufacturer's protocol. For measurement of pyruvate and lactate, 50 μL of supernatant was transferred onto 96-well plates in triplicate. The assay buffer/probe or substrate/enzyme mix (50 μL) was added to each sample and standard, mixed, and incubated at room temperature for 30 min protected from light. The mixed samples were measured at OD 570 nm (for pyruvate) and OD 450 nm (for lactate).
Lipid peroxidation assays - As a reflection of hepatic levels of lipid peroxidation, malondialdehyde (MDA) levels were determined by colorimetric assay (Abcam, Cambridge, MA). 30 mg of liver tissue was homogenized and processed according to the manufacturer's protocol.
RGS6 cloning and construct generation – The full-length RGS6L coding sequence was amplified by PCR from human blood cDNA using Phusion Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific) with compatible cloning sites (XhoI/HindIII). RNA was isolated from human blood using Trizol (Invitrogen, Carlsbad, CA, USA) and cDNA was prepared by reverse transcription using a cDNA synthesis kit (Thermo Fisher Scientific) following the manufacturers’ protocol. The resultant PCR product was loaded onto a 1% agarose gel, the gel was visualized under UV light gel doc (UVP chemStudio, Analytik Jena, Jena, Germany), and a band was observed, subsequently cut, and eluted using a gel extraction kit (Qiagen, Hilden, Germany). To clone the PCR product into pEGFP-N1, the PCR product and vector were double digested with XhoI/HindIII (NEB) and digestion products were subjected to agarose gel electrophoresis and extracted. Ligation was achieved by mixing the digested amplified product and vector in appropriate buffer with T4 DNA ligase enzyme (NEB) at 4 °C overnight. The resultant construct was transformed into DH5α cells, plated on LB agar containing (50 mg/ml Kanamycin) and grown overnight at 37 °C. Colonies were picked and grown in LB broth overnight at 37 °C. Positive clones of RGS6 in pEGFP-N1 were confirmed by restriction digestion of ligated plasmids. RGS6 deletion & point mutation sequences were generated by overlapping primer-based PCR amplification method and cloned into pEGFP-N1 vector. Information regarding the sequence of primers has been included in Table S5.
The full-length mouse RGS6 sequence was isolated from mouse brain and cloned into the PMD20 vector. The lentiviral vector for mRGS6 was generated via subcloning into the pLenti CMV Puro DEST cloning vector (Addgene, Watertown, MA, USA) and packaged using the pMD2.G VSV-G envelope expressing plasmid (Addgene) and psPAX2 (Addgene). Lentiviral particles were generated in HEK293 cells as per a standard protocol. 70 μL of lentivirus containing 2 X 108 particles of either mRGS6-Lenti or a control empty vector virus with the addition of invivofectamine was injected into the tail vein of mice. Two weeks after lentiviral injection, the mice were subjected to HFD treatment as above. After 12 weeks mice were euthanized by cervical dislocation and blood/multiple tissues were collected for downstream analysis.
Culture of HepaRG and LX2 cell lines – The human hepatocyte cell line HepaRG was cultured in William's E Medium with the GlutaMAX™ Supplement (Thermo Fisher Scientific) and 10% FBS (Gibco, Waltham, MA, USA) in a 37 °C incubator at 5% CO2. Cells were treated with PA (400 μM, 24 h), H2O2 (0.4 mM or 200 mM) for 3, 6 or 24 h, N-acetylcysteine (NAC; 5 mM; 1 h pre-treatment), ATM inhibitor KU55933 (5 μM, 1 h pre-treatment), cyclosporin A (0.2 μM, 1 h pre-treatment), Ru360 (50 μM, 1 h pre-treatment), Carbonyl cyanide m-chlorophenyl hydrazone (CCCP; 50 μM; 6 h) or polyethylene glycol catalase (Peg-cat; 200 U/mL, 1 h pre-treatment) where indicated. The human hepatic stellate cell line LX2 was cultured in DMEM with 3% FBS in a 37 °C incubator at 5% CO2. Cells were treated with PA (400 μM, 24 h) where indicated. HepaRG and LX2 cells were also transfected with RGS6-targeted or scramble shRNA (Santa Cruz Biotechnology). Cells were plated at low density (approximately 1 X 105 cells/60 mm dish) prior to transfection and allowed to grow at least 60%–70% confluence. Cells were transfected after 24–26 h with lipofectamine 3000 (Thermo Fisher Scientific) or via electroporation (Neon Electroporator, Thermo Fisher Scientific) as per the manufacturer's protocols.
Isolation and culture of murine hepatocytes – Primary hepatocytes were isolated from 2-month-old mice according to a standard collagenase perfusion protocol. The liver was perfused first with EGTA solution (5.4 mM KCl, 0.44 mM KH2PO4, 140 mM NaCl, 0.34 mM Na2HPO4, 0.5 mM EGTA, and 25 mM Tricine, pH 7.2) and then with DMEM (Gibco) containing 0.075% type I collagenase (Sigma), followed by an additional digestion step (0.009% collagenase at 37 °C with agitation for 15 min) and centrifugation as described previously [19]. The isolated hepatocytes were then cultured in hepato-ZYME-SFM media (Thermo Fisher Scientific) on collagen-coated plates and maintained at 37 °C in a humidified cell culture incubator (5% CO2). Cells were not disturbed for at least 16–24 h before drug treatment. Post isolation, cells were transfected with RGS6-targeted or scramble shRNA (Santa Cruz Biotechnology) and treated with palmitic acid (PA, 400 μM, 24 h) where indicated.
Isolation and culture of primary human hepatocytes – Human livers were removed at autopsy from deceased individuals within 1–4 h after cessation of cardiac function. Medical history was collected from the relatives of the deceased through a questionnaire. Samples were collected from individuals with no reported medical complications (hepatic or cardiac pathology, diabetes, or kidney disorders). Relatives were briefed about the goal and design of the study and written consent was obtained. The condition of organs was further confirmed by studying gross architectural changes by H&E. Primary human hepatocytes were isolated from human liver tissue essentially as previously described [20,21]. Briefly, liver tissue samples were collected on ice in MEME solution with 0.5% fatty acid free BSA by a clinician of the Forensic Medicine Department, Sagore Dutta Medical College & Hospital, Kolkata, West Bengal. Immediately, under aseptic conditions, tissues were first diced, washed in cold HBSS, and then minced thoroughly in MEME. EGTA (0.5 mM) was added in the cell slurry and it was placed in a shaking water bath for 10–15 min at 37 °C. After centrifugation, the cell slurry was washed twice in MEME. Pre-warmed digestion buffer (HBSS, 0.05% collagenase IV, 0.5% fatty acid free BSA, 10 mM CaCl2) was added and the slurry placed in a shaking water bath again for 30 min at 37 °C. BSA was included in the digestion process to minimize cell damage & prevent hemolysis of red blood cells (RBCs). The solution was gently vortexed by repeated pipetting and passed through a metal strainer to remove lumps. The resultant supernatant was filtered again through a 100 μm cell strainer and then placed on ice. Resulting cell suspensions were centrifuged (3000 rpm for 5 min, 4 °C) and the supernatant was discarded. The hepatocyte pellet was gently re-suspended in a minimal amount of MEME, and RBC lysis buffer added to completely remove RBCs. After 3 min, cells were centrifuged again (3000 rpm for 5 min, 4 °C), washed with MEME twice and finally resuspended in William's E medium. Cells were counted for viability and diluted to 1 × 106 cells/ml in medium containing 1% non-essential amino acids, 1% GlutaMAX™, 2% human serum, 100 nM dexamethasone, 100 nM insulin and 0.375% fatty acid free BSA. Isolated hepatocytes were plated on type 1 collagen coated plates, at a density of 250,000/cm2. After adherence (overnight undisturbed), cells were transfected with RGS6 targeted or scramble shRNA using a Neon electroporator. Cells were then treated with PA (400 μM, 24 h) or exposed to media containing 10% serum collected from patients with reported NAFLD (or control serum).
Isolation/culture of murine HSCs – HSCs were isolated from Swiss albino mice using a standard perfusion protocol [22]. Mice were perfused with an EGTA solution for 2 min, pronase E (5 min) (Merck, Darmstadt, Germany), and, finally, collagenase D (0.038% for 5–7 min) (Sigma) under anesthesia. Next, we excised the liver and diced the tissue into small pieces to further digest with a HBSS solution containing pronase E and collagenase D supplemented with 1% DNase I (Sisco Research Laboratories, Mumbai, India) 15 min under sterile conditions. The digested solution was then filtered through a 70-μm cell strainer and subjected to low-speed centrifugation (50×g for 3–5 min) to discard pelleted hepatocytes. The single-cell suspension was then mixed with Nycodenz solution (9.6%) and centrifuged again at 1400×g for 25 min. HSCs were collected from the white layer. The number of isolated cells and viability was then determined by Trypan Blue staining and cells plated onto dishes coated with fibronectin. Cell were maintained in DMEM containing 10% FBS, Vitamin A (100 mM), insulin (50 ng/ml) and glutamine (0.5 mM) at 37 °C in a humidified cell culture incubator (5% CO2). Prior to experimental initiation cells were cultured for 2–3 days with daily media changes to ensure the appearance of a suitable morphology. Cells were transfected with RGS6-targeted or scramble shRNA (Santa Cruz Biotechnology) and treated with palmitic acid (PA, 400 μM, 24 h) where indicated.
MitoSox staining – Staining for mitochondrial ROS was performed in PA-treated HepaRG cells ± RGS6 KD as well as primary murine hepatocytes isolated (as above) from control and HFD-fed animals given scramble or RGS6 targeted shRNA. Cells were washed thoroughly, loaded with 5 μM of MitoSox solution (1–2 ml to cover the whole dish), incubated for 15 min at 37 °C, washed 3X with PBS in the dark, mounted using vectashield with DAPI and visualized by fluorescence microscopy. The number of MitoSox positive cells (red stained) were counted on each coverslip.
Measurement of total ROS – ROS generation was estimated in cells using the cell-permeable oxidation-sensitive probe, CM-H2DCFDA (DCFDA, Sigma). Cells were harvested by centrifugation, washed three times with ice-cold PBS, re-suspended in PBS and incubated with 5 μM CM-H2DCFDA for 20 min at 37 °C. After incubation cells were again washed and lysed in PBS with 1% Tween 20. The ROS level in cell lysates was determined at the ratio of dichlorofluorescein excitation at 480 nm to emission at 530 nm. We should note here that the CM-H2DCFDA assay is utilized here as a general oxidative stress indicator and not as a detector of a specific oxidant due to known limitations of the probe [23].
In vitro collagen formation assay – The collagen specific dye Sirius red was utilized to quantitate collagen from cell lysates essentially as previously described [24]. Briefly, a solution of 5 μg/ml Sirius red was prepared by dissolving Sirius red in saturated picric acid. After 1 h, cells were washed with PBS twice and lysed in 0.1 M sodium hydroxide at room temperature. The supernatant was collected and colorimetric measurement done at 530 nm.
Mitochondrial Ca2+and Mitochondrial Membrane Potential (ΔΨM) Measurements – HepaRG cells transfected with scramble or RGS6-targeted shRNA were pretreated with Ru360 (50 mmol/L) or cyclosporin A (0.2 mmol/L) for 1 h to selectively block mitochondrial Ca2+ uptake or opening of the mPTP, respectively. Cells were then challenged with PA (24 h, 400 μM). Mitochondrial membrane potential was measured using a commercially available kit (Abcam). Mitochondria were isolated from cells using a mitochondria isolation kit (Abcam) and then used to measure the levels of Ca2+ using a standard ELISA (Abcam) according to the manufacturer's instructions.
Human serum for cell culture experiments – HepaRG/Lx2/isolated hepatocyte cells/isolated stellate cells were transfected with scramble or RGS6 shRNA and then incubated with the media containing sera of control and NAFLD subjects (10%). The blood was collected from individuals without (control) or with (NAFLD) a confirmed diagnosis of NAFLD. The serum was isolated from the blood with via standard methods. The cell lysates were processed for western blotting as above.
Immunoprecipitation – HepaRG cells (3 × 106) were treated with PA (400 μM) for 6 h, lysed and protein concentration measured via BCA protein assay. 600 μg of protein was equilibrated in IP lysis buffer (50 mM Tris, 5 mM EDTA, 250 mM NaCl and 0.1% Triton X-100) and bait antibodies (ATM or control mouse IgG) for 12 h on a rotor at 4 °C. 30 μl of Protein G sepharose beads (Abcam) were pre-cleared, equilibrated and then added to lysate after 12 h. After a 2 h incubation, protein extracts were immunoprecipitated with respective antibodies followed by washing 3 times with IP buffer. Eluted immuno-complexes were subjected to immunoblotting with prey antibody (RGS6, Abcam). Sample preparation for immunoblotting was done under non-reducing conditions.
Human Samples – Post-mortem human tissue samples (control, NAFLD and NAFLD + co-morbid DM) were acquired after obtaining the ethical clearance from the Centre of Biomedical Research Ethics Committee (Ref: IEC/CBMR/Corr/2018/14/3). We analyzed the baseline cross-sectional data derived from the nonalcoholic fatty liver (NAFLD) patients of the Department of Surgery, Sagore Dutta Medical College & Hospital, Kolkata, West Bengal. Control liver biopsy samples were collected from subjects who underwent liver biopsy in a pre-evaluation for liver transplantation or characterization of solid liver masses suspected to be adenoma or focal nodular hyperplasia based on radiological results without any evidence of hepatic steatosis. The inclusion criteria for subjects with NAFLD are as follows: (i) > 18 years old, (ii) fat infiltration in liver on examination, and (iii) high ALT levels within the 5–6 months. Subjects with the following were excluded: (i) hepatitis B and C virus infection, (ii) hepatitis, (iii) drug-induced liver injury, (iv) Wilson disease or hemochromatosis, (v) excessive alcohol consumption (male >30g/day, female >20g/day), and (vi) diagnosis of malignancy within the past year. In the exploratory analysis, among these biopsy-proven tissues, liver tissues from 50 subjects with or without NAFLD were randomly selected and used for immunohistochemistry (IHC) analysis. The liver histology was reviewed for steatosis grading and NAFLD score by a pathologist and grading has been performed as per a standard clinical method. Steatosis was graded according to the percentage of hepatocytes containing fat droplets as follows: grade 0, none, <5%; grade 1, mild, 5–33%; grade 2, moderate, 34–66%; and grade 3, severe, >66%. In principle, the NASH scoring system comprises the NAFLD Activity Score (NAS), fibrosis stages, and the identification of NASH by pattern recognition. The NAS can range from 0 to 8 and is calculated by the sum of scores of steatosis (0–3), lobular inflammation (0–3), and hepatocytes ballooning (0–2). The liver tissues with or without NAFLD were subjected to Western blot analysis and the tissue slides were stained for noted proteins. Expression scores based on the intensity and distribution of each protein in hepatocytes were obtained. Liver tissues were subsequently divided into low and high RGS6 expression or subdivided according to the HOMA-IR score and subjected to western blot or IHC analysis. For analyses, selected subjects were divided into three groups: no steatosis group (steatosis grade 0) vs. NAFLD group (steatosis grade 1–3) or vs. NAFLD plus T2D group.Summarized clinical data can be found in Table S7.
RGS6-ATM binding simulation – A bioinformatics approach was used to explore the interaction patterns between human ATM and RGS6. The ATM human protein structure with high resolution 7.82 Å (PDBID: 5NP0) was downloaded from RCSB protein databank [25]. RGS6 human regulator of G-protein signaling 6 isoform 1 sequence was collected from the Uniport database [26], and RCSB protein databank was searched for a matching sequence. We found protein structure (PDBID: 2ES0) with query coverage from 325 to 470 amino acids with 100% similarity. Docking studies were performed for 5NP0 A chain and 2ES0 using the Zdock Server [27] and the top 10 interaction models were considered for further molecular dynamics (MD) simulations. Preprocessing for the simulation was done using Pymol and Swiss-PdbViewer [28]. The energy minimization and simulation studies were carried out using GROMACS version 2019.3 for which atom coordinates and topology information was generated using gmx pdb2gmx with GROMOS96 43a1 force field. A simulation box was created by gmxeditconf with periodic boundary conditions in a 2.0-nm cubic box. Solvation was done with the SPC water model and the system was neutralized by genion. Energy minimization was performed using the steepest descent algorithm at 50000 steps. Molecular dynamics simulations were carried out for a period of 10 ns. The simulation results were further analyzed to find best the interaction pattern using Discovery Studio Visualizer (Biovia, San Diego, CA, USA). Table S6 highlights key interacting amino acids in both RGS6 and ATM and the corresponding interaction frequencies in the simulation. Table S8 includes the complete simulation results.
Data acquisition and statistical analyses – Our murine physiology dataset was generated from 2 independent animal cohorts. Cell culture experiments were performed with a minimal experimental N of 3. All immunoblots are representative of at least 3 independent experiments. Data were analyzed by student's t-test, one-, or two-way ANOVA with the post hoc adjustments as appropriate. Statistical analyses were performed using GraphPad Prism Software (La Jolla, CA, USA). Results were considered significantly different at P<0.05. Values are expressed as means ± S.E.M.
3. Results
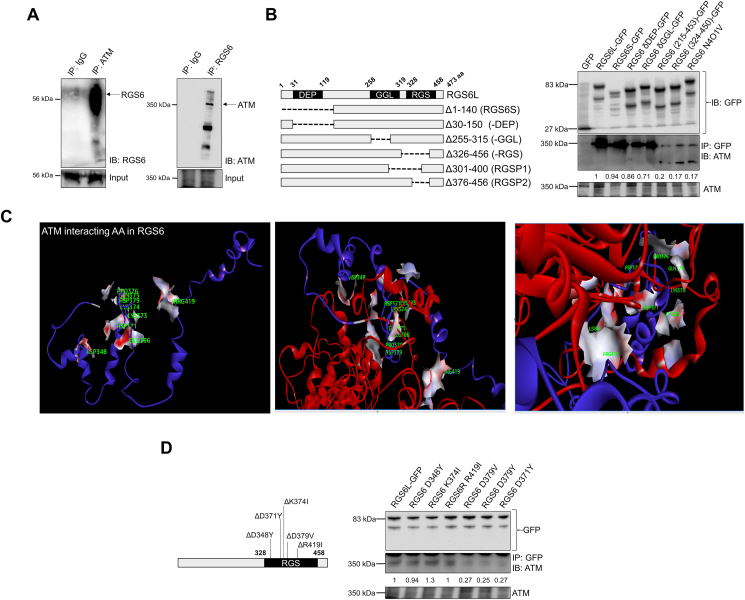
RGS6 and ATM form a complex in liver – Though regulation of G protein signaling necessitates cytoplasmic localization, RGS6 has previously been shown to exist in complex with multiple nuclear proteins including DNA methyltransferase 1 (DNMT1) [29] and histone acetyltransferase Tip60 [30]. Given the functional link between RGS6 and ATM [15], we hypothesized that these proteins might directly bind. Indeed, RGS6 and ATM form a co-immunoprecipitable complex in hepatocytes (Fig. 1A). Mapping of the RGS6-ATM interface identified the RGS domain as required for RGS6-ATM complex formation, specifically amino acids 376–400 (Fig. 1B). In silico modelling predicted a high affinity interaction between RGS6 and ATM supported by key RGS6 residues D348, D371, K374, D379, and R419 (Fig. 1C, Fig. S1, Table S6). Indeed, mutation of aspartate 371 or 379 almost completely abolished RGS6-ATM co-immunoprecipitation (Fig. 1D).
Fig. 1.
RGS6 and ATM form a co-precipitable complex in hepatocytes. (A) Reciprocal ATM and RGS6 co-immunoprecipitation in the human hepatocyte cell line HepaRG. (B) Immunoprecipitation of ATM with RGS6 deletion constructs in HepaRG cells. Densitometric quantification was performed, and the relative quantity of ATM protein pulled down with each RGS6 construct is indicated. (C) In silico modeling of the putative ATM-RGS6 complex revealed key RGS6 residues predicted to support a direct interaction between RGS6 and ATM (D348, D371, K374, D379, and R419 in green). Red = ATM Protein; Blue = RGS6; Surface is shown for ATM-interacting amino acids (AA) of RGS6. (D) Co-immunoprecipitation of ATM with RGS6 point mutants in HepaRG cells. Densitometric quantification was performed, and the relative quantity of ATM protein pulled down with each RGS6 construct is indicated. Data are representative of at least 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
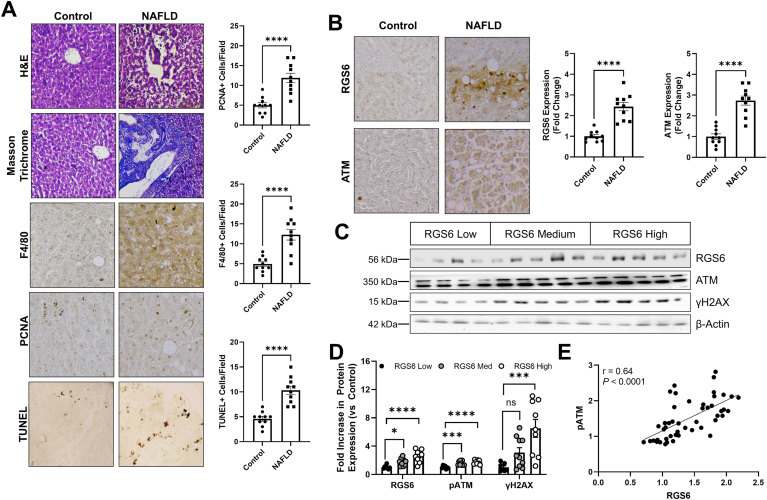
RGS6 and ATM are up-regulated in NAFLD – We acquired liver biopsy samples from human patients (Table S7) with a diagnosis of NAFLD or DILI and age/sex matched controls. Histological evaluation of samples from NAFLD patients revealed widespread fibrosis, immune cell infiltration, cell death and ongoing regeneration (Fig. 2A). This pathology was accompanied by robust ATM and RGS6 upregulation (Fig. 2B). Protein content of RGS6 and ATM as well as ATM phosphorylation and levels of the ATM effector and marker of DNA damage γH2AX were also increased in DILI samples (Fig. S2A) particularly those isolated from patients with high plasma ALT (Fig. S2B). RGS7 and γH2AX immunoreactivity also increased with increasing steatosis grade (Fig. S2C). When we stratified samples based on RGS6 protein content (low, medium, high) we noted a corresponding increase in ATM phosphorylation and γH2AX (Fig. 2C and D) and, indeed, across samples, levels of ATM and RGS6 were positively correlated (Fig. 2E) suggesting a potential functional link between the two proteins.
Fig. 2.
ATM and RGS6 are up-regulated in NAFLD. (A) Histological characterization of NAFLD human liver samples. Detectable disruption of liver architecture (H & E), fibrosis (Masson Trichrome), inflammation (F4/80), regeneration (PCNA) and cell death (TUNEL) [scale bar = 100 μm] was observed in livers from NAFLD patients. F4/80, PCNA, and TUNEL positive cells are quantified (n = 10). (B) Immunohistochemical staining for RGS6 and ATM in liver of control and NAFLD patient liver samples (n = 10). Representative images and quantification across samples are shown. (C) NAFLD samples were stratified based on RGS6 expression (low, n = 7; medium, n-10; high, n = 9) and immunoblotting performed for RGS6, ATM, and γH2AX. (D) Densitometric quantification was performed, and protein content expressed relative to RGS6 low samples. β-Actin is used as a loading control for all immunoblots.(E) Correlation between ATM and RGS6 expression across 48 liver samples detected via immunoblotting. A simple linear regression was used to determine the goodness of fit (r2) and degree to which the slope of the line deviates from zero (P). Data were analyzed by student's t-test or one-way ANOVA with Sidak's post-hoc test. *P < 0.05, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
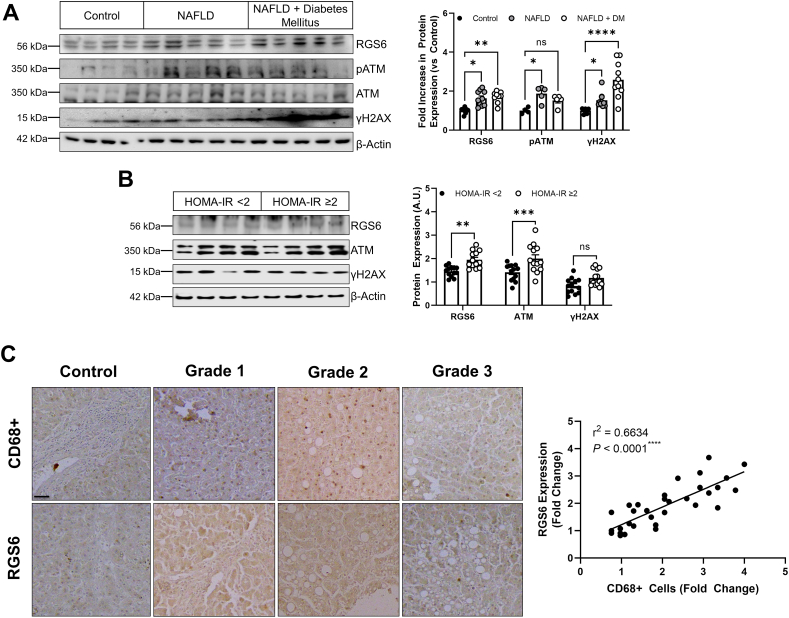
Co-morbid diabetes mellitus, a risk factor for rapid NAFLD progression [31], was associated with increased evidence of DNA damage (Fig. 3A), and, though RGS6 levels were also elevated in these patients RGS6 immunoreactivity was similar to patients with NAFLD alone. However, then we separated our NAFLD samples based on the Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) score for each patient (Table S7), we noted that individuals with the highest levels of insulin resistance also displayed the highest levels of RGS6 and ATM (Fig. 3B). Inflammation is a critical step in the progression of NAFLD to NASH and we noted that RGS6 expression was highly correlated with macrophage marker CD68 in livers from NAFLD patients (Fig. 3C). Taken together, these data suggest that RGS6 up-regulation is accompanied by insulin resistance and a higher inflammatory burden.
Fig. 3.
RGS6 levels are high in the livers of NAFLD patients with co-morbid DM, marked insulin resistance, and a pronounced inflammatory burden. (A) Expression of RGS6, pATM/ATM, and γH2AX in livers from control, NAFLD and NAFLD with co-morbid DM patients (n = 4–10). Representative immunoblots and densitometric quantification are included. (B) Immunoblotting (representative image and quantification) for RGS6, ATM, and γH2AX in NAFLD patient samples stratified based on HOMA-IR score (n = 13). (C) Representative images of RGS6 and CD68 staining in livers [scale bar = 100 μm] from NAFLD samples at each steatosis grade with corresponding correlation (n = 32). A simple linear regression was used to determine the goodness of fit (r2) and degree to which the slope of the line deviates from zero (P). β-Actin is used as a loading control for all immunoblots. Data were analyzed by student's t-test or one-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
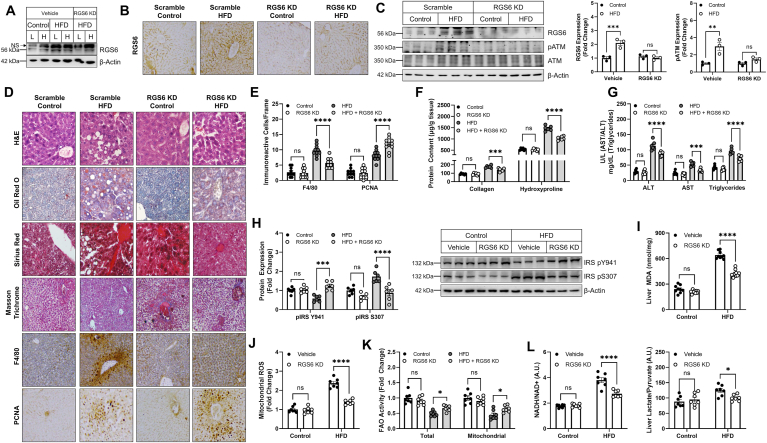
RGS6 depletion protects mice from HFD-induced liver steatosis, inflammation, and fibrosis–Induction of fatty liver via the introduction of a HFD is the most common mouse model of NAFLD. 12 weeks of HFD exposure was sufficient to recapitulate the molecular signature of RGS6, pATM, and γH2AX induction we observed in NAFLD patients (Fig. S3).Thus, we choose this time as the end point for histological and biochemical analysis. We noted that up-regulation of RGS6 following a HFD was confined to the liver and not seen in other tissues such as heart (Fig. 4A). Introduction of RGS6 shRNA led to ã75% reduction in RGS6 expression in liver acutely (Fig. S4) and was sufficient to prevent HFD-dependent RGS6 up-regulation after 12 weeks of HFD feeding (Fig. 4B and C). Notably, hepatic RGS6 knockdown was sufficient to prevent HFD-dependent ATM phosphorylation (Fig. 4C). Histological characterization of livers from HFD-fed mice revealed marked lipid accumulation, fibrotic remodeling, inflammation,and ongoing regeneration (Fig. 4D). Steatosis and inflammation (Fig. 4D and E) were diminished in the livers of RGS6-KD mice and regeneration was enhanced (Fig. 4E). Hepatic RGS6 depletion also decreased fibrosis including collagen and hydroxyproline accumulation (Fig. 4F) following exposure to a HFD. Consistent with maintenance of hepatic functional integrity, levels of ALT, AST, and triglycerides were decreased in livers from animals administered RGS6 shRNA (Fig. 4G).
Fig. 4.
RGS6 KD in liver protects against hepatosteatosis and fibrosis in HFD-fed mice. Control and HFD (54.8% total fat content)-fed mice were administered scramble or RGS6-targeted shRNA via tail vein injection prior to initiation of the HFD (n = 6). Animals were sacrificed after 12 weeks of HFD feeding. (A) Immunoblotting for RGS6 in liver [L] and heart [H]. (B) Hepatic RGS6 expression measured via immunohistochemistry [scale bar = 100 μm]. (C) Immunblotting for RGS6, pATM and ATM with densitometric quantification. (D) Histological characterization of livers. Liver architecture (H & E), fibrosis (Masson Trichrome, Sirius Red), inflammation (F4/80), regeneration (PCNA) and lipid accumulation (Oil Red O) are depicted [scale bar = 100 μm]. (E) F4/80 and PCNA positive cells per microscope field (n = 10). (F) Hepatic collagen and hydroxyproline content (n = 6/group). (G) Liver enzyme (ALT & AST) and triglyceride levels. (H) Immunoblotting for markers of insulin sensitivity with corresponding densitometric quantification. (I) Liver MDA (n = 7). (J) Mitochondrial ROS (Mitosox) generation (n = 7). (K) Total and mitochondrial fatty acid oxidation (FAO) rate in liver (n = 7). (L) Liver metabolism as indicated by NADH/NAD+ and Liver Lactate/Liver pyruvate (n = 7). β-Actin is used as a loading control for all immunoblots. Data were analyzed by two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01,***P < 0.001, ****P < 0.0001. Data are presented as mean ± SEM. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Chronic HFD feeding led to alterations in the phosphorylation status of Insulin Receptor Substrate-1 (IRS-1), a marker of insulin sensitivity, that was reversed following hepatic RGS6 knockdown (Fig. 4H). Similarly, RGS6 depletion led to reductions in HFD-driven lipid peroxidation (Fig. 4I), mitochondrial ROS accumulation (Fig. 4J), and compromised total and mitochondrial fatty acid oxidation (FAO) (Fig. 4K). Hepatic steatosis was accompanied by hallmarks of dysfunctional cellular metabolism including increased ratios of lactate/pyruvate and NADH/NAD+ (Fig. 4L), indicative of aberrant redox balance. Importantly, RGS6 knockdown in liver diminished the impact of HFD on liver metabolism. Together, these data indicate that RGS6 facilitates aberrant hepatic insulin sensitivity, oxidative stress, and metabolism following HFD feeding.
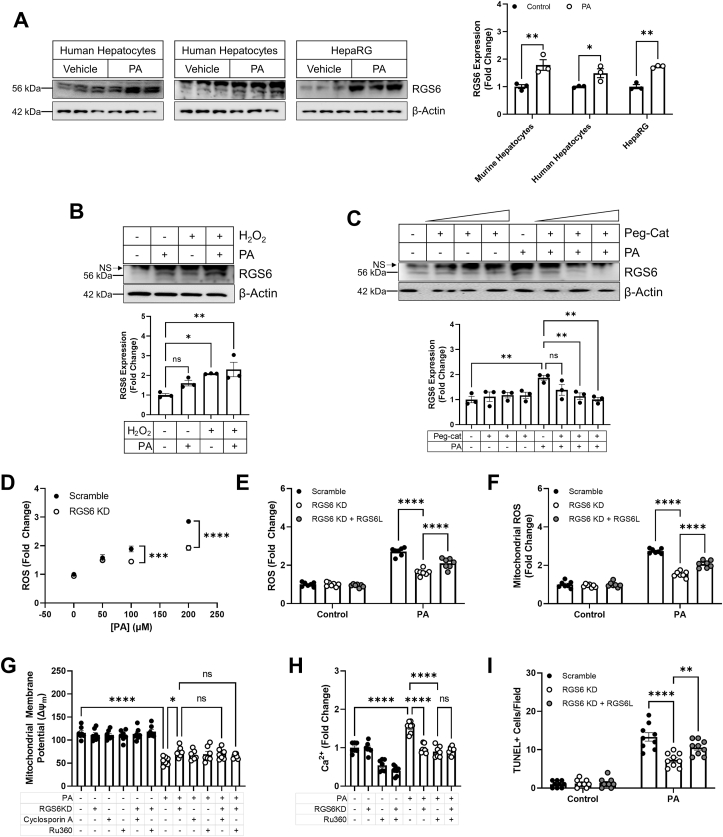
RGS6 drives hyperlipidemia-dependent oxidative stress, mitochondrial dysfunction, and cell death – To further investigate the mechanistic role of RGS6 in hepatic lipotoxicity, we next moved into a cell culture-based system. To mimic a high lipid environment, we treated cells with fatty acid PA, which resulted in up-regulation of RGS6 in human and murine hepatocytes and the human hepatocyte cell line HepaRG (Fig. 5A). Elevation of cellular oxidants is sufficient to drive RGS6 up-regulation in hepatocytes (Fig. 5B) and scavenging of hydrogen peroxide (H2O2) was sufficient to prevent PA-dependent RGS6 protein induction (Fig. 5C) indicating that ROS and RGS6 participate in a feed forward signaling loop that likely drives cyclical amplification of the detrimental impacts of RGS6 in liver. Indeed, RGS6 KD decreased PA-dependent ROS generation in HepaRG cells (Fig. 5D). Further, restoration of RGS6L expression was able to return levels of total ROS (Fig. 5E) and mitochondrial ROS (Fig. 5F) to that observed in control cells effectively counteracting the impact of RGS6 KD. Given the physical and functional interaction between RGS6 and ATM as well as previous data indicating that ATM deletion protects mice against HFD-dependent steatoapoptosis [7], we next sought to investigate the impact of RGS6 on mitochondrial function and cell death. RGS6 KD partially mitigated the impact of PA on mitochondrial membrane potential (Δψm), an effect comparable to inhibition of the mitochondrial Ca2+ uniporter inhibitor with Ru360 or the mitochondrial permeability transition pore with cyclosporin A (Fig. 5G).Similarly, decreasing RGS6 expression was sufficient to prevent mitochondrial Ca2+ accumulation (Fig. 5H). Maintenance of mitochondrial integrity in RGS6 KD cells was also associated with a reduction in cell death following PA exposure (Fig. 5I). Clearly, RGS6 is required for hyperlipidemia-dependent oxidative stress, mitochondrial dysfunction, and cell death in hepatocytes.
Fig. 5.
RGS6, whose up-regulation is ROS driven, promotes mitochondrial dysfunction and cell death in hepatocytes. (A) Immunoblotting for RGS6 in human hepatocytes, murine hepatocytes, and HepaRG cells treated with PA (400 μM, 24 h). Densitometric quantification is provided (n = 3). (B) HepaRG cells were treated with PA (400 μM, 24 h) and/or H2O2 (200 mM, 24 h) and immunoblotting performed for the detection of RGS6. Representative immunoblots and quantification are provided (n = 3). (C) HepaRG cells were treated with PA (400 μM, 24 h) and/or increasing concentrations of the H2O2 scavenger Peg-Cat (up to 200 U/mL, 1 h pre-treatment). Immunoblotting was performed for the detection of RGS6. Representative immunoblots and quantification are provided (n = 3). HepaRG cells were treated with increasing concentrations of PA (24 h) or a fixed drug dose (400 μM) 24 h following introduction of scramble or RGS6-targeted shRNA. The RGS6L isoform was reintroduced into a subset of RGS6 KD cells. (D) and (E) CM-H2DCFDA fluorescence (total ROS; n = 6–7). (F) Mitosox fluorescence (mitochondrial ROS; n = 7). (G) Mitochondrial membrane potential (n = 7) with and without the introduction of mitochondrial permeability transition pore blocker cyclosporin A (0.2 μM, 1 h pre-treatment) or mitochondrial calcium uniporter inhibitor Ru360 (50 μM, 1 h pre-treatment). (H) Mitochondrial calcium (Ca2+) with and without Ru360 (50 μM, 1 h pre-treatment) (n = 7). (I) Cell death (TUNEL positive cells; n = 9). Data were analyzed by one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
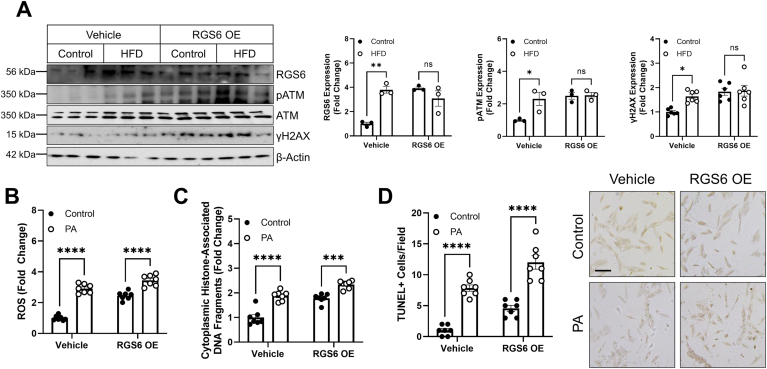
RGS6 overexpression is sufficient to trigger ATM phosphorylation, oxidative stress, and cell death in hepatocytes – While we have, to this point, provided ample evidence that RGS6 is necessary for fulminant HFD-driven hepatotoxicity, we were curious if RGS6 overexpression would be sufficient to trigger hepatic dysfunction in the absence of hyperlipidemia. Intriguingly, overexpression of RGS6 in liver was sufficient to drive ATM phosphorylation and γH2AX induction (Fig. 6A). In cells, the impact of RGS6 overexpression on ROS generation (Fig. 6B) and cell death (Fig. 6C and D) was comparable to PA treatment. These data indicate that RGS6 is both necessary and sufficient to drive hyperlipidemia-dependent activation of ATM and cell death.
Fig. 6.
RGS6 expression is sufficient to drive hepatic ATM phosphorylation in vivo and oxidative stress and cell death in hepatocytes. (A) Control and HFD-fed mice were administered vehicle or a viral construct encoding RGS6L via tail vein injection prior to initiation of the HFD (n = 3). Animals were sacrificed after 12 weeks of HFD feeding. Immunoblotting was performed to detect RGS6, pATM/ATM, and γH2AX and densitometric quantification provided. β-Actin is used as a loading control for all immunoblots. (B–D) HepaRG cells were treated with PA (400 μM, 24 h) 24 h following transfection with RGS6L. (B) CM-H2DCFDA fluorescence (n = 7) and apoptosis as measured via (C) cytoplasmic histone-associated DNA fragments (n = 7) or (D) TUNEL positive cells (n = 7). Data were analyzed by two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
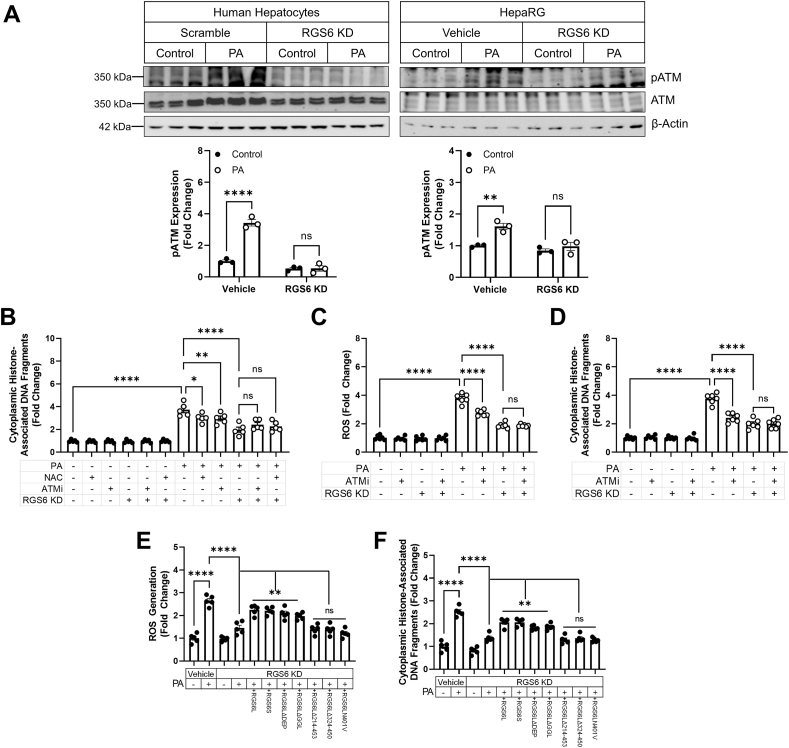
RGS6/ROS/ATM drive PA-dependent apoptosis in hepatocytes– We noted that RGS6 knockdown prevented ATM phosphorylation in both human hepatocytes and HepaRG cells (Fig. 7A).Inhibition of ATM, ROS scavenging with the glutathione donor NAC, or RGS6 KD decreasedcell death in PA-treated human hepatocytes with no additive benefit derived from the combination of interventions indicating that RGS6, ROS, and ATM likely function in the same pathway to promote hepatotoxicity (Fig. 7B). Similarly, in HepaRG cells, RGS6 KD or ATM inhibition decreased ROS generation (Fig. 7C) and cell death (Fig. 7D) following PA treatment. Though the net impact of RGS6 depletion on ROS accumulation was greater in magnitude than observed following ATM inhibition (Fig. 7C), RGS6-dependent cell loss in HepaRG cells appears to proceed primarily via an ATM-dependent mechanism (Fig. 7D). Finally, we noted that RGS6 overexpression in RGS6 KD cells restored PA-dependent ROS accumulation (Fig. 7E) and cell death (Fig. 7F). However, this effect was lost for RGS6 deletion constructs lacking critical portions of the RGS domain essential for RGS6's GAP activity and ATM binding (see Fig. 8).
Fig. 7.
RGS6, ATM and ROS function in the same pathway to promote hyperlipidemia-dependent hepatotoxicity. Scramble or RGS6-targeted shRNA was introduced into human hepatocytes (A, B) or HepaRG cells (A, C–F) 24 h prior to treatment with PA (400 μM, 24 h), NAC (5 mM, 1 h pre-treatment) and/or the ATM inhibitor KU55933 (ATMi; 5 μM, 1 h pre-treatment) where indicated. In a subset of experiments RGS6 deletion constructs were introduced via transfection prior to drug treatments. (A) Immunoblotting for pATM and ATM. Representative westerns and a densitometric quantification are provided (n = 3). β-Actin is used as a loading control for all immunoblots. (B) Cell death (cytoplasmic histone-associated DNA fragments; n = 5) ± RGS6 KD, ATMi, and/or NAC in human hepatocytes. (C) CM-H2DCFDA fluorescence (n = 6) and (D) cell death (cytoplasmic histone-associated DNA fragments; n = 6) ± RGS6 KD and/or ATMi. (E) CM-H2DCFDA fluorescence (n = 5); (F) cell death (cytoplasmic histone-associated DNA fragments; n = 6); and albumin production (n = 5) ± RGS6 KD and/or RGS6 deletion constructs. Data were analyzed by one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
Fig. 8.
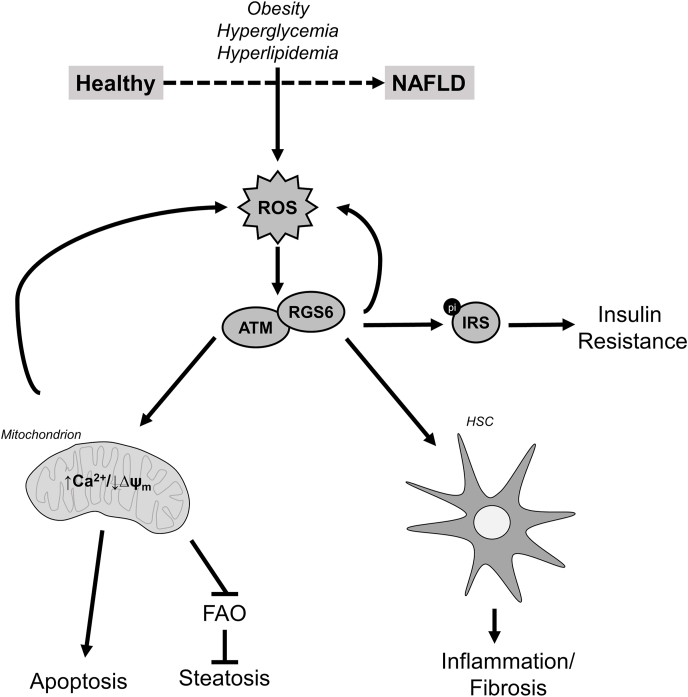
Schematic depicting the role of RGS6 in hyperlipidemia-driven hepatotoxicity. Oxidative stress drives RGS6 up-regulation, which represents an amplification node facilitating further pathogenic ROS generation. By interacting with ATM, RGS6 promotes mitochondrial dysfunction, which compromised fatty acid oxidation leading to hepatic steatosis and, in the face of overwhelming cellular stress, can trigger cell death. RGS6 is also expressed in hepatic stellate cells (HSCs) where RGS6 up-regulation leads to oxidative stress, actions likely also contributing to hepatic inflammation and fibrosis. Finally, RGS6 also suppresses markers of insulin sensitivity, of particular relevance to patients with co-morbid diabetes mellitus.
RGS6 is required for PA-dependent toxicity in HSCs – HSCs play a critical role in promoting extracellular matrix remodeling, progressive fibrosis, inflammation, and cell injury [2]. Though RGS6 clearly participates in hepatocyte-intrinsic mechanisms contributing to hyperlipidemia-driven hepatotoxicity, exposure of both murine (Fig. S5A) and human (Fig. S5B) HSC cell line LX2 also resulted in RGS6 up-regulation and ATM phosphorylation. Lipotoxic factors contributing to substrate-overload in the liver are derived from several peripheral organ systems and reach the liver via the circulation. Regardless of the specific species or source, these extrahepatic factors are likely present in the blood of NAFLD patients. Exposure of either primary hepatocytes (Fig. S6A) or LX2 cells (Fig. S6B) to serum from NAFLD patients was sufficient to increase RGS6, ATM, and γH2AX, a molecular signature remarkably similar to that we observed in the livers of HFD-fed mice or in hepatocytes or HSCs exposed to PA. ATM and γH2AX induction could be reversed via RGS6 knockdown (Figs. S6A and S6B). Similar results were obtained in PA-treated murine HSCs (Fig. 7A) and LX2 cells (Fig. 7B). Finally, RGS6 KD ameliorated oxidative stress (Fig. S7C), cell death (Fig. S7D) and collagen reduction (Fig. S7E) in LX2 cells. Actions of RGS6 in non-hepatocyte liver cell types likely contribute to the reduction in fibrosis and inflammation we observed in RGS6 KD, HFD-fed mice (Fig. 4).
4. Discussion
NAFLD represents a large burden on the global healthcare system. Due to the expected rise in contributing co-morbidities such as obesity and T2D, the demand for therapeutics designed to protect against hepatic lipotoxicity is expected to surge in the coming decades [1]. Here, we identify RGS6 as a critical mediator of hyperlipidemia-dependent liver dysfunction. Though normally expressed at low levels in liver, we noted robust RGS6 up-regulation in livers from NAFLD patients particularly those with co-morbid T2D and severely compromised liver function. High RGS6 expression also correlated with insulin resistance, inflammation, and a molecular signature characterized by elevations in markers for DNA damage, oxidative stress, and induction of pro-apoptotic signaling cascades. In mice, RGS6 knockdown in liver was sufficient to protect against the detrimental impact of a HFD on hepatic integrity and function effectively decreasing steatosis, fibrosis, and cell loss as well as enhancing liver regeneration. The protective impact of RGS6 knockdown likely derives from impacts of RGS6 on several key mechanism(s) both in hepatocytes and neighboring hepatic stellate cells where RGS6 complexes with ATM to modulate ROS generation, mitochondrial function, and cell death. Strikingly, RGS6 overexpression in liver or hepatocytes was sufficient to promote ROS generation and ATM phosphorylation.
Improving the hepatic antioxidant buffering capacity with interventions such as NAC ameliorates inflammation, steatosis, insulin resistance, and cell death following HFD feeding [32] particularly when administered during an early intervention window initiated concurrently with the HFD [33].Given that oxidative stress precedes the development of key HFD-associated pathological endpoints in liver [34], the impact of RGS6 on hepatic ROS generation, particularly from the mitochondria, likely represents a critical node of RGS6 action in livers exposed to lipotoxic conditions. Notably, H2O2 treatment was sufficient to stimulate RGS6 up-regulation in hepatocytes and scavenging of H2O2 completely prevented hyperlipidemia-dependent RGS6 induction. Thus, RGS6 appears to be recruited to participate in a feed-forward loop that facilitates continual, and likely pathogenic, oxidative stress following hepatic lipid accumulation.
Recent evidence from the livers of NASH patients has revealed robust ATM activation, and ATM depletion from a line of hepatocyte-derived carcinoma cells resulted in mitochondrial dysfunction, enhanced DNA damage, and growth arrest [6]. We now show that RGS6 forms a complex with ATM in liver supported by key aspartate residues (D371 and D379) in the RGS6 domain of RGS6. The corresponding residue in ATM (R2792) lies in the kinase domain. Thus, we predict that RGS6 may influence ATM via modulation of autophosphorylation-dependent ATM monomerization and/or substrate recruitment. RGS6 is, in fact, required for hyperlipidemia-dependent ATM auto-phosphorylation at Ser1981, a modification known to drive ATM activation in response to double stranded DNA breaks [35]. The impact of RGS6 on ROS might represent one potential mechanism linking RGS6 to ATM phosphorylation as ATM can be directly activated via oxidation, though this process appears to require residues in a negative regulatory domain between the kinase domain and FATC domain of ATM [5,36]rather than ATM residues close to the RGS6 binding site. Loss of RGS6-dependent ATM phosphorylation would be expected to impact several cellular processes including mitochondrial function [37], DNA repair, and cell death following ROS accumulation. Notably, RGS6 deletion mutants incapable of ATM binding were unable to counteract the impact of RGS6 KD on PA-dependent hepatotoxicity linking the cytotoxic actions of RGS6 to ATM binding. Metabolism of toxic lipids via β-oxidation occurs both in the mitochondria and peroxisomes depending on the fatty acid structure. ATM localizes to both these compartments where it senses ROS to direct cell fate depending on the severity of stress stimuli [37,38]. In HFD-fed mice, knockdown of hepatic RGS6 is sufficient to rescue compromised total and mitochondrial FAO activity. Similarly, RGS6 KD counterbalanced the impact of HFD on cellular metabolism and mitochondrial ROS generation and prevented PA-dependent loss of mitochondrial membrane potential and calcium accumulation. As oxidative stress and lipid peroxidation drive the transition of steatosis to NASH, RGS6 up-regulation, which we observe in humans with high grade steatosis and severely compromised liver function, represents a critical step in NAFLD pathogenesis. We should note that knockout of ATM has been shown to protect livers against HFD-driven hepatic fibrosis and cell loss but not steatosis or inflammation [7]. ATM deficiency also impairs liver regeneration [9], in contrast to results we obtained with hepatic RGS6 KD where loss of ATM activation was associated with enhanced regeneration. Thus, we hypothesize that RGS6 participates in additional ATM-independent signaling cascades in livers exposed to hyperlipidemia.
In addition to hepatocyte-intrinsic mechanism(s) of hyperlipidemia-dependent hepatic dysfunction, hepatocyte stress and death triggers release of several autocrine and paracrine cytokines that drive recruitment of Kupffer cells (resident hepatic macrophages) and transdifferentiation of HSCs to a myofibroblast-like phenotype characterized by excessive proliferation and generation of extracellular matrix components. These physiological processes are critical for hyperlipidemia-driven inflammation and fibrosis, respectively. Our data demonstrate that circulating factor(s) present in the blood of NAFLD patients are sufficient to drive RGS6 up-regulation and RGS6-dependent ATM phosphorylation in both hepatocytes and HSCs. Knockdown of RGS6 also prevented ATM phosphorylation, ROS generation, and death of cultured HSCs exposed to PA. The impacts of ROS accumulation in HSCs are seemingly paradoxical. On the one hand, NADPH oxidase deficiency protects mice against oxidative stress and decreases HSC activation following chronic liver injury [39], but exposure to free radicals also triggers apoptosis in HSCs [40]. The HSC fate decision following liver injury then depends on a delicate balance between pro- and anti-oxidant mechanisms and only a sub-population of HSCs may display a high degree of susceptibility to oxidative damage [40]. RGS6 appears to drive HSC death in culture, though, given RGS6 knockdown provides substantive protection against HFD-driven hepatic fibrosis, it may be that, in vivo, these actions are counterbalanced by pro-proliferative signals received by HSCs from neighboring cell types.
The exact site of ATM/RGS6 action might also explain differential impacts of global ATM depletion vs liver-specific RGS6 knockdown. For example, the Human Protein Atlas (http://www.proteinatlas.org) predicts a lack of RGS6 expression in monocytes [41]. However, ROS-driven ATM activation in macrophages facilitates the development of a pro-inflammatory phenotype [42]. Impairing host defense mechanisms have often proven deleterious to liver function [43]. Thus, RGS6 depletion may provide some benefit over direct inhibition of ATM action by mitigating the negative consequences of inflammatory mediators on hepatocytes or HSCs without preventing removal of dead cells by macrophages or compromising liver regeneration. In support of this idea is our observation that RGS6 knockdown enhances liver regeneration, actions that may derive from relief of RGS6-dependent growth suppression in either hepatocytes or HSCs as has been previously observed following RGS6 knockout in cancer cells and mouse embryonic fibroblasts exposed to estradiol/heregulin or oncogenic Ras, respectively [30,44,45].
Prior work indicated that the GTPase activating protein (GAP) activity of RGS6, which is directed toward Gαi/o subunits, is dispensable for its pro-apoptotic actions [15,44]. Though numerous GPCRs have been implicated in NAFLD, our results indicate that RGS6 likely participates in GPCR-independent signaling cascades to promote hyperlipidemia-dependent hepatotoxicity. Like RGS6, RGS5 is also up regulated in HSCs in response to pro-fibrogenic stimuli [46]. RGS5 null mice, in contrast, display increased hepatic fibrosis attributed to enhanced signaling via endothelin 1 (ET-1) receptors [46] as well as ectopic hepatic lipid deposition [47]. Hepatocyte-specific RGS5 deletion exacerbates HFD-dependent hepatic steatosis via permissive hyperactivation of transforming growth factor beta-activated kinase 1 (TAK1) [48]. RGS16 also appears to protect against NAFLD via regulation of GPCR-dependent fatty acid oxidation [49]. RGS6 is unique, then, amongst members of the RGS protein family present in liver with regards to both the net impact of RGS protein depletion as well as the underlying mechanism(s).
RGS6 emerges, then, as a potential new therapeutic target in the treatment of NAFLD, which remains a large burden on the global healthcare system. The potential utility of RGS6 inhibition in liver diseases may extend to other conditions as we noted robust up-regulation of RGS6 in liver tissue from drug-induced liver injury and previous data identified RGS6 as a mediator of alcohol-induced hepatic steatosis [10]. Similarly, RGS6 might prove useful as a biomarker for severe hepatic damage predictive of a catastrophic cell fate decision in the presence of insurmountable cellular stress. Given the lack of efficacioustherapies aimed at restoring hepatic function or mitigating ongoing liver damage, elucidation of novel mechanisms of NAFLD pathogenesis provides additional opportunities for intervention.
Financial Support
This work was supported by Department of Biotechnology, India (BT/PR21156/MED/30/1753/2016) and DRDO (DG-(TM)/81/48222/LSRB- 307/SH&DD&BD/2017) to BM and Centre of Biomedical Research (CBMR), Uttar Pradesh Government.
Author contributions
Conception and design: T. Mahata, A. S. Sengar, A. Stewart, B. Maity. Acquisition of data: T. Mahata, A.S. Sengar, M. Basak, K. Das, S.K. Verma, P.K. Singh, S. Saha, S. Biswas, B. Maity. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis, human sample analysis): T. Mahata, A.S. Sengar, M. Basak, K. Das, A. Pramanick, S.K. Verma, P.K. Singh, S. Saha, S. Biswas, S. Sarkar, S. Chatterjee, A. Stewart, B. Maity. Writing, review, and/or revision of the maanuscript: T. Mahata, A.S. Sengar, A. Stewart, B. Maity. Study supervision: B. Maity
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We acknowledge Department of Biotechnology (DBT), DRDO and CBMR for all the support. We also thank Dr. Santosh Chauhan, Institute of Life Science, Bhubaneswar, India for the kind gift of LC3-GFP plasmid and Dr. Sougata Roy Chowdhury, INSPIRE Faculty, University of Calcutta for the intellectual help.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102105.
Contributor Information
Madhusudan Das, Email: madhuzoo@yahoo.com.
Biswanath Maity, Email: bmaity@cbmr.res.in, bmaity28@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2020 doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomonaco R., Ortiz-Lopez C., Orsak B., Webb A., Hardies J., Darland C., Finch J., Gastaldelli A., Harrison S., Tio F., Cusi K. Effect of adipose tissue insulin resistance on metabolic parameters and liver histology in obese patients with nonalcoholic fatty liver disease. Hepatology. 2012;55(5):1389–1397. doi: 10.1002/hep.25539. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz J.M., Noworolski S.M., Erkin-Cakmak A., Korn N.J., Wen M.J., Tai V.W., Jones G.M., Palii S.P., Velasco-Alin M., Pan K., Patterson B.W., Gugliucci A., Lustig R.H., Mulligan K. Effects of dietary fructose restriction on liver fat, de novo lipogenesis, and insulin kinetics in children with obesity. Gastroenterology. 2017;153(3):743–752. doi: 10.1053/j.gastro.2017.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan P., Sharma Y., Maisuradze L., Tchaikovskaya T., Gupta S. Ataxia telangiectasia mutated pathway disruption affects hepatic DNA and tissue damage in nonalcoholic fatty liver disease. Exp. Mol. Pathol. 2020;113:104369. doi: 10.1016/j.yexmp.2020.104369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugherity E.K., Balmus G., Al Saei A., Moore E.S., Abi Abdallah D., Rogers A.B., Weiss R.S., Maurer K.J. The DNA damage checkpoint protein ATM promotes hepatocellular apoptosis and fibrosis in a mouse model of non-alcoholic fatty liver disease. Cell Cycle. 2012;11(10):1918–1928. doi: 10.4161/cc.20259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donath H., Woelke S., Theis M., Hess U., Knop V., Herrmann E., Krauskopf D., Kieslich M., Schubert R., Zielen S. Progressive liver disease in patients with ataxia telangiectasia. Front Pediatr. 2019;7:458. doi: 10.3389/fped.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu S., Shen K.C., Wang Y., Brooks S.C., Wang Y.A. Impaired hepatocyte survival and liver regeneration in Atm-deficient mice. Hum. Mol. Genet. 2005;14(20):3019–3025. doi: 10.1093/hmg/ddi333. [DOI] [PubMed] [Google Scholar]
- 10.Stewart A., Maity B., Anderegg S.P., Allamargot C., Yang J., Fisher R.A. Regulator of G protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc. Natl. Acad. Sci. U. S. A. 2015;112(7):E786–E795. doi: 10.1073/pnas.1418795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart A., Maity B., Wunsch A.M., Meng F., Wu Q., Wemmie J.A., Fisher R.A. Regulator of G-protein signaling 6 (RGS6) promotes anxiety and depression by attenuating serotonin-mediated activation of the 5-HT(1A) receptor-adenylyl cyclase axis. Faseb. J. 2014;28(4):1735–1744. doi: 10.1096/fj.13-235648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maity B., Stewart A., Yang J., Loo L., Sheff D., Shepherd A.J., Mohapatra D.P., Fisher R.A. Regulator of G protein signaling 6 (RGS6) protein ensures coordination of motor movement by modulating GABAB receptor signaling. J. Biol. Chem. 2012;287(7):4972–4981. doi: 10.1074/jbc.M111.297218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Huang J., Maity B., Gao Z., Lorca R.A., Gudmundsson H., Li J., Stewart A., Swaminathan P.D., Ibeawuchi S.R., Shepherd A., Chen C.K., Kutschke W., Mohler P.J., Mohapatra D.P., Anderson M.E., Fisher R.A. RGS6, a modulator of parasympathetic activation in heart. Circ. Res. 2010;107(11):1345–1349. doi: 10.1161/CIRCRESAHA.110.224220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J., Maity B., Huang J., Gao Z., Stewart A., Weiss R.M., Anderson M.E., Fisher R.A. G-protein inactivator RGS6 mediates myocardial cell apoptosis and cardiomyopathy caused by doxorubicin. Canc. Res. 2013;73(6):1662–1667. doi: 10.1158/0008-5472.CAN-12-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J., Yang J., Maity B., Mayuzumi D., Fisher R.A. Regulator of G protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Canc. Res. 2011;71(20):6310–6319. doi: 10.1158/0008-5472.CAN-10-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C.K., Eversole-Cire P., Zhang H., Mancino V., Chen Y.J., He W., Wensel T.G., Simon M.I. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. U. S. A. 2003;100(11):6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pramanick A., Chakraborti S., Mahata T., Basak M., Das K., Verma S.K., Sengar A.S., Singh P.K., Kumar P., Bhattacharya B., Biswas S., Pal P.B., Sarkar S., Agrawal V., Saha S., Nath D., Chatterjee S., Stewart A., Maity B. G protein beta5-ATM complexes drive acetaminophen-induced hepatotoxicity. Redox Biol. 2021;43:101965. doi: 10.1016/j.redox.2021.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong S.C., Jamil A.H.A., Rhodes A., Taib N.A., Chung I. Metabolic role of fatty acid binding protein 7 in mediating triple-negative breast cancer cell death via PPAR-alpha signaling. J. Lipid Res. 2019;60(11):1807–1817. doi: 10.1194/jlr.M092379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni H.M., McGill M.R., Chao X., Du K., Williams J.A., Xie Y., Jaeschke H., Ding W.X. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J. Hepatol. 2016;65(2):354–362. doi: 10.1016/j.jhep.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green C.J., Charlton C.A., Wang L.M., Silva M., Morten K.J., Hodson L. The isolation of primary hepatocytes from human tissue: optimising the use of small non-encapsulated liver resection surplus. Cell Tissue Bank. 2017;18(4):597–604. doi: 10.1007/s10561-017-9641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu I.C., Lipsky M.M., Cole K.E., Su C.H., Trump B.F. vol. 21. 1985. Isolation and culture of hepatocytes from human liver of immediate autopsy; pp. 154–160. (Vitro Cell Dev Biol). 3 Pt 1. [DOI] [PubMed] [Google Scholar]
- 22.Dang T.M., Le T.V., Do H.Q., Nguyen V.T., Holterman A.X.L., Dang L.T.T., Phan N.C.L., Pham P.V., Hoang S.N., Le L.T., Grassi G., Truong N.H. Optimization of the isolation procedure and culturing conditions for hepatic stellate cells obtained from mouse. Biosci. Rep. 2021;41(1) doi: 10.1042/BSR20202514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B., Mann G.E., Moore K., Roberts L.J., 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majumder S., Piguet A.C., Dufour J.F., Chatterjee S. Study of the cellular mechanism of Sunitinib mediated inactivation of activated hepatic stellate cells and its implications in angiogenesis. Eur. J. Pharmacol. 2013;705(1–3):86–95. doi: 10.1016/j.ejphar.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 25.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z., Fisher R.A. RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J. Biol. Chem. 2004;279(14):14120–14128. doi: 10.1074/jbc.M309547200. [DOI] [PubMed] [Google Scholar]
- 30.Huang J., Stewart A., Maity B., Hagen J., Fagan R.L., Yang J., Quelle D.E., Brenner C., Fisher R.A. RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated Dnmt1 degradation and promoting apoptosis. Oncogene. 2014;33(27):3604–3611. doi: 10.1038/onc.2013.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams C.D., Stengel J., Asike M.I., Torres D.M., Shaw J., Contreras M., Landt C.L., Harrison S.A. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y., Gao M., Liu D. N-acetylcysteine protects mice from high fat diet-induced metabolic disorders. Pharm. Res. (N. Y.) 2016;33(8):2033–2042. doi: 10.1007/s11095-016-1941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen F.C., Weng S.W., Tsao C.F., Lin H.Y., Chang C.S., Lin C.Y., Lian W.S., Chuang J.H., Lin T.K., Liou C.W., Wang P.W. Early intervention of N-acetylcysteine better improves insulin resistance in diet-induced obesity mice. Free Radic. Res. 2018;52(11–12):1296–1310. doi: 10.1080/10715762.2018.1447670. [DOI] [PubMed] [Google Scholar]
- 34.Matsuzawa-Nagata N., Takamura T., Ando H., Nakamura S., Kurita S., Misu H., Ota T., Yokoyama M., Honda M., Miyamoto K., Kaneko S. Increased oxidative stress precedes the onset of high-fat diet-induced insulin resistance and obesity. Metabolism. 2008;57(8):1071–1077. doi: 10.1016/j.metabol.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 36.Paull T.T. Mechanisms of ATM activation. Annu. Rev. Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- 37.Valentin-Vega Y.A., Maclean K.H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F.C., Cleveland J.L., Green D.R., Kastan M.B. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119(6):1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., Dere R., Tait-Mulder J., Lee J.H., Paull T.T., Pandita R.K., Charaka V.K., Pandita T.K., Kastan M.B., Walker C.L. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17(10):1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi S.S., Sicklick J.K., Ma Q., Yang L., Huang J., Qi Y., Chen W., Li Y.X., Goldschmidt-Clermont P.J., Diehl A.M. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatology. 2006;44(5):1267–1277. doi: 10.1002/hep.21375. [DOI] [PubMed] [Google Scholar]
- 40.Thirunavukkarasu C., Watkins S., Harvey S.A., Gandhi C.R. Superoxide-induced apoptosis of activated rat hepatic stellate cells. J. Hepatol. 2004;41(4):567–575. doi: 10.1016/j.jhep.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q., Allouch A., Paoletti A., Leteur C., Mirjolet C., Martins I., Voisin L., Law F., Dakhli H., Mintet E., Thoreau M., Muradova Z., Gauthier M., Caron O., Milliat F., Ojcius D.M., Rosselli F., Solary E., Modjtahedi N., Deutsch E., Perfettini J.L. NOX2-dependent ATM kinase activation dictates pro-inflammatory macrophage phenotype and improves effectiveness to radiation therapy. Cell Death Differ. 2017;24(9):1632–1644. doi: 10.1038/cdd.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao B., Tsukamoto H. Inflammation in alcoholic and nonalcoholic fatty liver disease: friend or foe? Gastroenterology. 2016;150(8):1704–1709. doi: 10.1053/j.gastro.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maity B., Yang J., Huang J., Askeland R.W., Bera S., Fisher R.A. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J. Biol. Chem. 2011;286(2):1409–1419. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maity B., Stewart A., O'Malley Y., Askeland R.W., Sugg S.L., Fisher R.A. Regulator of G protein signaling 6 is a novel suppressor of breast tumor initiation and progression. Carcinogenesis. 2013;34(8):1747–1755. doi: 10.1093/carcin/bgt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bahrami A.J., Gunaje J.J., Hayes B.J., Riehle K.J., Kenerson H.L., Yeung R.S., Stempien-Otero A.S., Campbell J.S., Mahoney W.M., Jr. Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury. PloS One. 2014;9(10) doi: 10.1371/journal.pone.0108505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng W., Wang X., Xiao J., Chen K., Zhou H., Shen D., Li H., Tang Q. Loss of regulator of G protein signaling 5 exacerbates obesity, hepatic steatosis, inflammation and insulin resistance. PloS One. 2012;7(1) doi: 10.1371/journal.pone.0030256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Ma J., Nie H., Zhang X.J., Zhang P., She Z.G., Li H., Ji Y.X., Cai J. Hepatic regulator of G protein signaling 5 ameliorates nonalcoholic fatty liver disease by suppressing transforming growth factor beta-activated kinase 1-c-Jun-N-terminal kinase/p38 signaling. Hepatology. 2021;73(1):104–125. doi: 10.1002/hep.31242. [DOI] [PubMed] [Google Scholar]
- 49.Pashkov V., Huang J., Parameswara V.K., Kedzierski W., Kurrasch D.M., Tall G.G., Esser V., Gerard R.D., Uyeda K., Towle H.C., Wilkie T.M. Regulator of G protein signaling (RGS16) inhibits hepatic fatty acid oxidation in a carbohydrate response element-binding protein (ChREBP)-dependent manner. J. Biol. Chem. 2011;286(17):15116–15125. doi: 10.1074/jbc.M110.216234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.