Highlights
-
•
Droplet digital PCR was applied to detect circulating tumor-derived DNA (ctDNA) in ovarian cancer (OC) follow-up.
-
•
Five out of six recurrent OC cases were positive in ddPCR analyses before increased plasma CA125.
-
•
Increased allele frequency of the ctDNA is associated with increased tumor volume after OC recurrence.
-
•
No ctDNA was detected in the plasma of recurrence-free OC cases (maximum, 439 PFS days)
Keywords: ctDNA, Ovarian cancer, ddPCR, CA125
Abstract
Objective
Ovarian cancer (OC) is an intractable gynecological tumor, and frequent recurrence is experienced within a few years even after the complete eradication of tumor tissues by radical resection and neo-adjuvant chemotherapies. The conventional recurrence marker, CA125, is widely used for follow-up after resection of OC, but CA125 has a long half-life in blood and lacks dynamic responses to tumor recurrence. Recent developments in liquid biopsy procedures are expected to overcome the difficulties in early diagnosis of OC recurrence after surgery.
Methods
We applied droplet digital PCR (ddPCR) technology to detect circulating tumor-derived DNA in OC patients’ plasma during follow-up. Exome sequencing of 11 tumor–normal pairs of genomic DNA from consecutive OC patients identified tumor-specific mutations, and ddPCR probes were selected for each sample.
Results
Six of 11 cases showed apparent recurrence during follow-up (mean progression-free survival was 348.3 days) and all six cases were positive in ddPCR analyses. In addition, ddPCR became positive before increased plasma CA125 in five out of six cases. Increased allele frequency of circulating tumor DNA (ctDNA) is associated with increased tumor volume after recurrence. ddPCR detected ctDNA signals significantly earlier than increased CA125 in the detection of OC recurrence by imaging (49 days and 7 days before, respectively: p < 0.05). No ctDNA was detected in the plasma of recurrence-free cases.
Conclusions
Our results demonstrate the potential of identifying ctDNA by ddPCR as an early detection tool for OC recurrence.
1. Introduction
In Japan, approximately 9,000 people are diagnosed with ovarian cancer (OC) and 4,500 people die of OC each year (Shoji et al., 2018). Furthermore, OC is increasing in Eastern/Southern European countries and Asia (Coburn et al., 2017). Most OC cases (90%) are epithelial OC, and the main histological type is high-grade serous ovarian carcinoma (HGSOC), which has a poor prognosis (Torre et al., 2018). Despite advances in surgical procedures and the development of effective chemotherapeutic drugs for the complete eradication of OC, the survival outcomes in patients with advanced OC remain unfavorable because of recurrence within a few years (Bowtell et al., 2015). Early OC lacks subjective symptoms, and thus many cases (40%) are diagnosed at stages III and IV. The reported 5-year survival rates of stages III and IV OCs are 49.5% and 30.8%, respectively in Japan (Nagase et al., 2020).
To detect recurrence of OC, CA125 is often measured as a marker in the follow-up after OC treatment. The National Institutes of Health statement recommends conducting a medical interview, physical initial examination, consultation, and CA125 measurement for the follow-up of OC after treatment completion (Colombo et al., 2019). Furthermore, in the US, many gynecologists measure CA125 to check for OC recurrence (Harmandayan et al., 2011). CA125 has a high positive predictive value but low sensitivity (Rustin et al., 1996), and is useful for continuous clinical measurement to show trends in treatment response because of its long biological half-life in serum. However, false negatives cannot be ruled out with a single measurement of CA125 (Gu et al., 2009). Its value can be heterogeneous within and between OC patients because CA125 does not directly reflect absolute tumor volume (Van Gorp et al., 2011). Therefore, early diagnoses of OC recurrence on the basis of periodical changes rather than absolute values of CA125 are being investigated. The Japanese guidelines (Tokunaga et al., 2021) for the detection of OC recurrence recommend the following criteria as indicators of OC recurrence: three consecutive elevations even within the normal range (Wilder et al., 2003); an elevation of 10 U/ml or more (Santillan et al., 2005); or an elevation of 25 U/ml or more in 1 month (Meier et al., 1997). However, the specifications for evaluation are sometimes not applicable because of the health insurance regulations in Japan that state that the CA125 test can only be performed once in a month. Thus, there is an urgent need to identify a biomarker that is more reliable and rapid than CA125 for improving the prognosis of OC.
A new technology called liquid biopsy is being developed for cancer diagnosis. Using liquid samples (including plasma and urine) that can be taken from the body using minimally invasive methods, liquid biopsy can be a useful biomarker for the detection of tumor formation and progression (Crowley et al., 2013). Circulating plasma contains fragmented DNA derived from various tissues including tumors (Lo et al., 2021). Conventional biopsy techniques, taking tissues with endoscopes or needles, are painful and risky, and because only a small portion of tumor tissue can be collected, only fragmentary profile information can be detected. It has become clear that the circulating blood of patients with early stage cancer contain mutated circulating tumor DNA (ctDNA) from cancer cells. In particular, ctDNA is being actively studied as a potential detection tool of recurrence after surgery (Wan et al., 2017).
In colorectal cancer, Diehl et al. showed that ctDNA from patients with metastatic colorectal cancer after liver metastasectomy outperformed the serum marker CEA in detecting small lesions (Diehl et al., 2008). Furthermore, Allegretti et al. reported that residual and non-residual ctDNA at the first (3-month) postoperative follow-up was associated with earlier recurrence and disease-free status in three and seven patients, respectively. In addition, their study reported an increased sensitivity of ctDNA results when analyzed in combination with serum CEA levels (58.8% to 63.6%). Therefore, these studies strongly suggest that ctDNA is a powerful biomarker for micro-residual disease of colorectal cancer (Allegretti et al., 2020). In non-small cell lung cancer, ctDNA has enabled the identification and monitoring of several actionable genomic mutations, including EGFR, ERBB2, MET, ALK, ROS1, and RET (Thress et al., 2015).
Several studies have reported comparisons between CA125 and ctDNA as a recurrence biomarker in OC (Asante et al., 2020). Pereira and colleagues reported that ctDNA was elevated prior to computed tomography scan (CT) findings using droplet digital PCR (ddPCR) technology (Pereira et al., 2015), while Parkinson et al. reported that ctDNA in the TP53 region of HGSOC was measured as a specific biomarker (Parkinson et al., 2016). However, these studies investigated the ctDNA of HGSOC only and did not include ovarian clear cell carcinoma.
In Japan, two cancer gene profiling tests (Foundation One CDx (Frampton et al., 2013) and NCC Oncopanel (Sunami et al., 2019) were approved by the government in December 2018, and since June 2019, these tests have been covered by the national health insurance. More than one hundred genes are covered by the two tests, and thus it is relatively easy to identify cancer-specific mutations for ddPCR probes that can be used for liquid biopsy. Considering this current trend, the discovery of biomarkers using genomic information and the establishment of detection methods are urgently required in the treatment of OC after surgery. The usefulness of ctDNA detection with ddPCR should be demonstrated by comparisons with CA125 and imaging tests in the follow-up of OC after treatment.
The purpose of this study was to establish a method to identify individual mutations, regardless of histology, that could be used in all OC patients. In this study, we included the two major histological types of OC, HGSOC and clear cell carcinoma, and conducted experiments using commercially available probes for ddPCR to detect ctDNA in the plasma of OC patients after treatment. Furthermore, we attempted to analyze the relationship of the onset of recurrence, recurrent tumor size, and the duration of progression-free survival with CA125 and ctDNA.
2. Materials & methods
2.1. Patient background
Eleven patients with advanced OC were enrolled in this study, all of whom had completed treatment and were confirmed to have no residual tumor by imaging and CA125 detection. We selected consecutive OC cases using two criteria. First, advanced OC cases who underwent complete resection from 2016 to 2019 at the Miyagi Cancer Center (MCC) Hospital were selected. Second, preoperative CA125-positive (≥35 Units/ml) cases were included. Eleven OC patients undergoing follow-up after treatment at MCC hospital were enrolled in this study; their OC histology and patient backgrounds are described in Table 1. This study was approved by the ethics committee of MCC (registration ID: 2019–038). Written informed consent was obtained from all patients. The timing of recurrence was defined by the detection of new lesions by imaging, either via contrast enhanced CT scan or positron emission tomography by expert radiologists. Clinical data were obtained from the patients’ medical records, including treatment history, CA125 level, and histological type.
Table 1.
Summary of the cases in this study.
| Parameters | Case number |
|---|---|
| Total | 11 |
| Median age in years (range) | 54.7(42–68) |
| Stage | |
| IIB | 2 |
| iiiA | 3 |
| iiiB | 2 |
| iiiC | 3 |
| ⅣB | 1 |
| Primary site histology | |
| Serous | 6 |
| Clear cell | 4 |
| Mixed (Serous + Clear cell) | 1 |
2.2. DNA extraction from FFPE specimens
Tumor samples were collected at the time of initial surgery and stored as formalin-fixed paraffin-embedded (FFPE) specimens. Surgically removed cancer tissue with a non-cancerous component was treated with 10% neutral buffered formalin solution (060–01667; Fujifilm Wako Pure Chemical Co., Osaka, Japan). Paraffin embedding and preparation of tissue slides were undertaken using standard protocols for genome analyses. Tissues were cut from paraffin blocks (5 µM thick, 10 pieces) and we separately scratched cancer and non-cancerous tissue specimens on the same glass slides. By doing so, we could remove potential false positives caused by formalin fixation (Shibuya et al. (Shibuya et al., 2018), Ito et al (Ito et al., 2021). Eleven pairs of tumor and normal tissue specimens were subjected to dissection using the AVENIO Millisect System (Roche, Basel, Switzerland). The choice of region for DNA extraction was macroscopically selected on the basis of the corresponding hematoxylin–eosin staining. DNA was extracted with the Maxwell RSC DNA FFPE Kit- PKK, Custom (AX2500; Promega, Madison, WI) and Maxwell RSC Instrument (Promega) according to the manufacturer's instructions. In total, 590 ng to 4,480 ng whole genomic DNA was obtained.
2.3. Plasma collection and DNA extraction from plasma
Ten milliliters of blood were collected when the enrolled patients had blood drawn for their routine CA125 testing (every 1 to 2 months) during the follow-up period. Sample blood was collected in a PAX gene Blood ccfDNA Tube (Becton Dickinson, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. The blood was then centrifuged at 1900 × g for 15 min at room temperature, and the supernatant plasma was transferred to a separate DNA LoBind Tube (0030108051; Eppendorf Germany). The plasma was further centrifuged at 1900 × g for 10 min at room temperature to remove remaining leukocytes. Obtained plasma was frozen and stored at − 80 °C until purification of circulating cell-free DNA (ccfDNA). The ccfDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (55114; Qiagen Gmbh, Hilden, Germany) according to the manufacturer's instructions.
2.4. Whole exome sequencing and bioinformatics analyses
Genomic DNA extracted from both the tumorous and non-tumorous portions of each patient FFPE specimen was subjected to whole exome sequencing (WES). WES was outsourced to Macrogen Japan Corp and the Novaseq 6000 was used for the WES. The bioinformatics procedures (Figure S1A) were performed as described previously (Ito et al., 2021). Briefly, after the raw reads were trimmed, the paired reads were mapped onto hg38 references with BWA-MEM (Liu et al., 2013). The variant calling was performed with Mutect2 in the Genome Analysis Toolkit 4.1 (McKenna et al., 2010) and the normal panel consisted of 11 non-cancerous tissue data. The filtering conditions were as follows: false discovery rate = 0.01; unique alt read count = 20; min allele fraction = 0.1; and minimum depth = 100. The annotations for the called variants were performed with Annovar (Wang et al., 2010) with a custom-made 4.7 KJPN variant dataset and COSMIC 90 (Tate et al., 2019). To do so, we converted hg38 to hg19 with liftOver software (Hinrichs et al., 2006).
2.5. Droplet digital PCR
The BIORAD ddPCR Mutation Assay (BioRad, Hercules, CA) was used to select commercially available probes for the somatic mutations identified in the WES of FFPE samples. The probes for 11 samples were selected (Table 2) and ddPCR was performed with the QX200 AutoDG Droplet Digital PCR IVD System (BioRad). Then, 8 μl eluted ccfDNA (approximate DNA amount was 0.128 μg) was used for each PCR reaction. The total PCR reaction volume was 40 μl (20,000 drops were generated). The PCR conditions were as follows: one cycle at 95 °C for 10 min; 40 cycle steps at 94 °C for 30 sec and 55 °C for 1 min; and finally, 98 °C for 10 min denaturation for enzyme deactivation. Probe fluorophores were FAM/HEX, and wild-type and mutant copy amounts were measured. All the ddPCR experiments were performed at least twice. Detection efficiency of ctDNA by ddPCR was tested with DNA extracted from corresponding tumor specimens for all probes. Allele frequency (AF) was calculated when mutant-type signal dots (FAM) at one time point or more than one mutant-type signal dot at consecutive time points existed in the presence of wild-type signal dots (HEX) using the formula:
Table 2.
Droplet digital PCR probes used in this study.
| Case | Assay Name | Assay ID: | Nucleotide Mutation: | COSMIC ID |
|---|---|---|---|---|
| T1 | TP53 p.R273H | dHsaMDV2010109 | c.818G > A | COSM10660 |
| T2 | PIK3CA p.H1047R | dHsaMDS2512492 | c.3140A > G | COSM775 |
| T3 | TP53 p.T253P | dHsaMDS565152834 | c.757A > C | COSM45980 |
| T4 | TP53 p.R306Efs*39 | dHsaMDS471419499 | c.916delC | COSM44631 |
| T5 | MYH13 p.E1419D | dHsaMDS130976248 | c.4257G > T | COSM1380696 |
| T6 | FGFR2 p.S252W | dHsaMDV2010045 | c.755C > G | COSM36903 |
| T7 | PIK3CA p.T1052K | dHsaMDS2512492 | c.3155C > A | COSM17447 |
| T8 | TP53 p.C242W | dHsaMDS2516194 | c.726C > G | COSM11356 |
| T9 | TP53 pY236C | dHsaMDV2516916 | c.707A > G | COSM10731 |
| T10 | TP53 R175H | dHsaMDV2010105 | c.524G > A | COSM10648 |
| T11 | MYH6 p.R673H | dHsaMDV2516916 | c.707A > G | COSM10731 |
AF (%) = mutant-type dots (FAM) × 100 (%)/mutant-type dots (FAM) + wild-type dots (HEX)
2.6. Image analysis
Imaging studies (contrast-enhanced CT, PET-CT) every 6 months are performed, and additional imaging studies were performed when there was a suspicion of recurrence, such as CA125 level was a continuously increase despite the level within the normal range. Volume analysis of CT images before and after treatment of recurrence was performed on six patients with recurrence. The tumor volume of CT at the time of recurrence was calculated using Aquarius iNtuition Edition version 4 (TeraRecon, Durham, NC). We only analyzed the effects of treatments on recurrent tumors that consisted of single legions but not multiple lesions to avoid complex calculations.
2.7. Statistical analysis
The correlations between tumor volume and AF detected by ddPCR were analyzed using Pearson’s correlation coefficient. The comparison between CA125 and ddPCR with recurrence was performed with Student’s t-test. The Kaplan–Mayer progression-free survival curves were drawn using JMP 15pro software (version 15).
3. Results
3.1. Exome analyses revealed somatic mutations in OCs to enable selection of personalized ddPCR probes
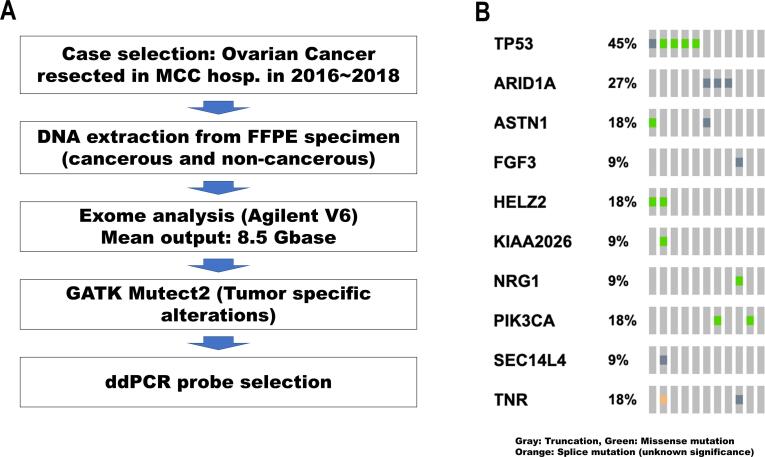
Ovarian tumors in this study consisted of high-grade serous carcinoma, clear cell carcinoma, and a mixture of the two types of histology (six, four, and one case, respectively). The genomic DNA of OC and surrounding non-cancerous tissues was extracted for exome analyses. The exome data are summarized in Supplementary Table 1 and Figure S1B. The average number of tumor-specific genetic alterations was 66, similar to our previous study (Shibuya et al., 2018). Only one out of 11 cases underwent neoadjuvant chemotherapy before surgery and no qualitative differences in genomic DNA among the samples were detected. The aim of the exome sequencing in this study was to identify appropriate tumor-specific mutations for ddPCR analysis, and all cases showed at least one mutation for which ddPCR probes were commercially available (Table 2). The mutation spectra showed the typical OC mutation patterns (Figure S1B); somatic TP53 mutations were identified in high-grade serous carcinoma and ARID1A and PIK3CA mutations were found in the clear cell carcinomas analyzed in this study (Shibuya et al., 2018)
3.2. Tumor-specific ctDNA fragments were clearly detected at OC recurrence before CA125 had increased and were negative in patients without recurrence
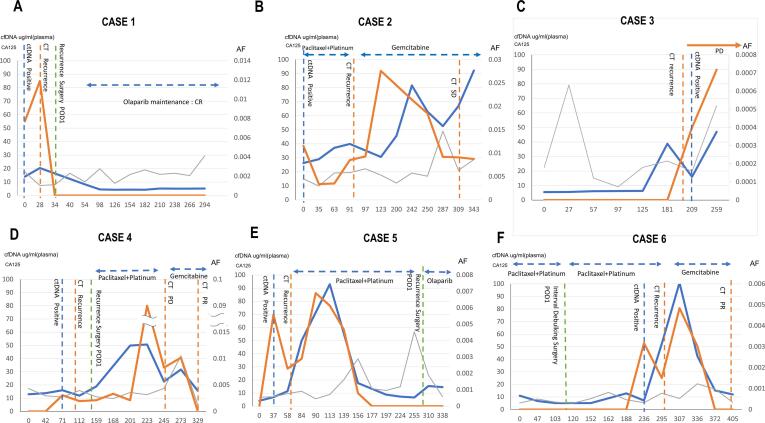
The tumor-specific ctDNA fragments and CA125 levels were compared as follow-up markers of OC. At the time of recurrence detected by CT, both tumor-specific ctDNA fragments and CA125 were positive. During the follow-up periods, six cases showed definite recurrence; Fig. 1 shows the time course of tumor-specific ctDNA fragments with cancer-specific mutations, CA125, and total ccfDNA of these six patients. Among them, cases 1, 4, and 5 underwent surgery, and case 1 underwent successive chemotherapies (Fig. 1A, D, and E). These cases showed an obvious decline in all three tests. Conversely, cases 2 and 3 (Fig. 1 B and C) died of the primary disease and the three markers increased during the observation periods. These results suggested that tumor-specific ctDNA fragments and ctDNA may be good follow-up markers.
Fig. 1.
Time course of CA125, ccfDNA, and ddPCR copy numbers in recurrent OC. Panels A to F indicate the time courses of six recurrent cases during the study. Horizontal axes indicate the date and left vertical axes indicate the score of CA125 (unit/dl: blue lines) and the concentration of cell-free DNA (ng/ml: gray lines). Allele frequencies of tumor-specific variants are indicated with the right vertical axis (%: red lines). The horizontal double-sided arrows with dotted lines indicate the period of chemotherapy. The vertical dotted lines indicate clinical events such as CT scanning or surgical resection.
We next investigated whether tumor-specific ctDNA fragments can detect OC recurrence before increased CA125. Fig. 1 shows that tumor-specific ctDNA fragments became positive earlier than CA125 in five out of six recurrent cases (cases 1, 2, 4, 5, and 6). In the time course of case 1, CA125 was not over 35 U/ml when the recurrence appeared in the CT image during the observation period. In Fig. 1C, case 3 showed a rapid increase in ccfDNA, but by either CT imaging or CA125, tumor recurrence was not detected for 6 months. The cause of increased ccfDNA in this case may be unrelated to the disease state of OC and has not been clearly elucidated. However, case 1 (Fig. 1A) showed the second complete eradication of tumor cells and became negative for all markers (ccfDNA, ctDNA, and CA125) for 10 months. On the contrary, all five recurrence-negative cases did not show an increase in all the markers during the follow-up. In our observation, tumor-specific ctDNA fragment-detection of tumor-specific mutations was more sensitive for five cases than CA125 (Fig. 1A, B, D, E, F).
3.3. Relative changes in AF in tumor-specific ctDNA fragments were correlated with changes in tumor volume
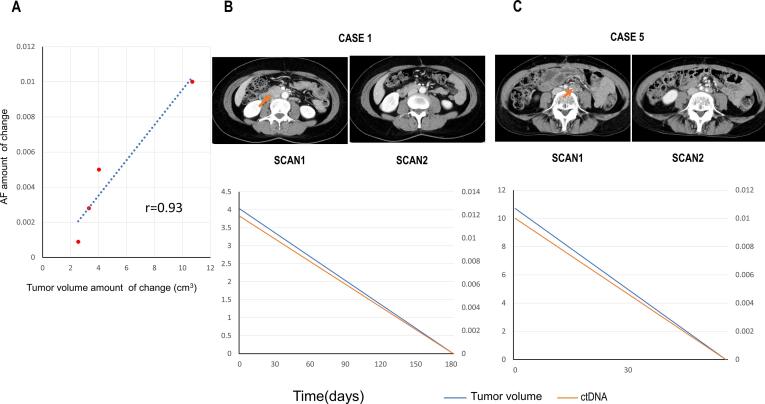
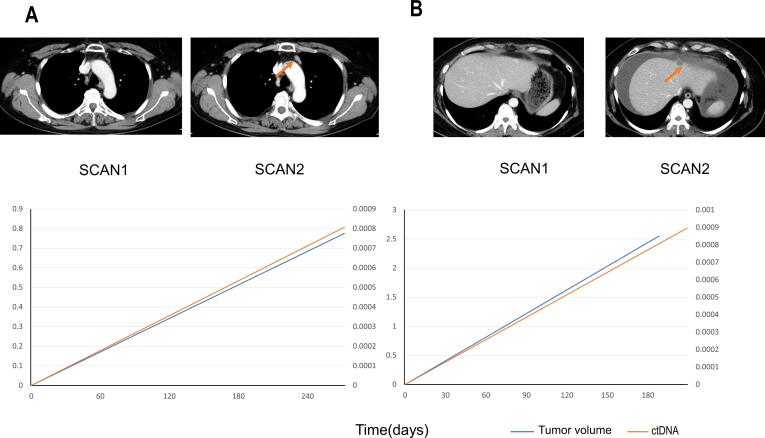
The increases or decreases in tumor volumes in CT images were compared with the AFs of the most recently measured ctDNA of each case. Four patients with a single recurrent lesion and accurate measurements of AF changes were included in the study. The other two cases were eliminated from this analysis because of multiple recurrent lesions. The measured tumor images and the changes in ctDNA and tumor volume are shown in Fig. 2 and Figure S2. The increase or decrease in volume was positively correlated with ctDNA (Pearson r = 0.93). The correlation also indicated that the treatment effect can be measured by ctDNA. In OC, like other types of cancer, tumor volume was shown to be negatively associated with response to treatment (Diehl et al., 2008) (To et al., 2003), suggesting that AF can be a good follow-up marker and could be used to determine the timing of CT imaging for the detection of OC recurrence. Further studies are required to confirm if tumor-specific ddPCR is useful for determining the treatment strategy for recurrence.
Fig. 2.
Comparison between the size of recurrent tumors and allele frequencies of ctDNA. Panel A. The correlation of changes in tumor sizes and allele frequencies (AF). The vertical and horizontal axes indicate the changes in tumor sizes and allele frequencies in each case, respectively. The points of data acquisitions of each case were selected on the basis of the visible changes in metastatic legions in the CT images (see Panel B and C). The denominators and numerators were the data (tumor volume and AF) before and after the changes in metastatic lesion volumes, respectively. Panel B and C. Correlation of tumor volumes and AF after treatment of cases 1 (panel B: surgical resection) and 5 (panel: chemotherapy). In each panel, the top images are enhanced CT before and after the changes. The timings of imaging are indicated just below the images. Bottom graph indicates the changes in AFs (vertical axes) during time course (horizontal axes). Red arrows in the CT images indicate the recurrent tumors.
3.4. Latency of detection of tumor-specific ctDNA fragments for OC recurrence is shorter than that of increased CA125
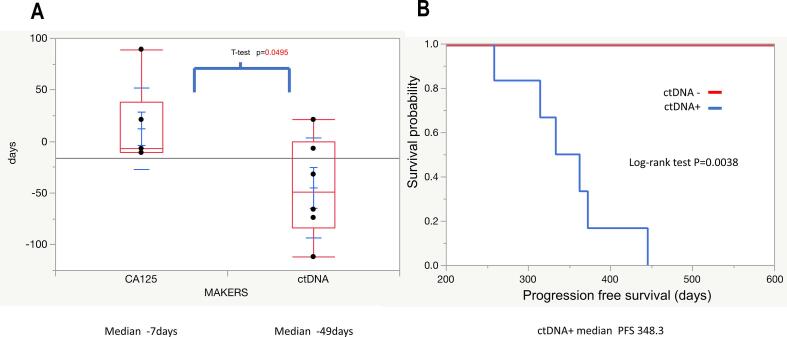
The median values of latency between blood testing and diagnosis of recurrence by imaging were − 49 days and −7 days, although an apparent significant difference was detected (95% CI: −95.8–5.85 and 95% CI: −29.1–53.7, respectively; p = 0.0495, Fig. 3A). The positive value for CA125 is 35 U/ml or higher, but in this study, three consecutive elevations within the normal range, an elevation of 10 U/ml or higher, or an elevation of 25 U/ml or higher in 1 month were additionally considered as positive. The progression-free survival (PFS) of patients whose ctDNA was detected by ddPCR in blood samples collected after treatment was significantly shorter than that of patients whose ctDNA was not detected (median PFS of ctDNA-positive patients 348 days; PFS by Kaplan–Meier analysis; p = 0.0038; Fig. 3B).
Fig. 3.
Clinical significance of detection of ctDNA with ddPCR in the follow-up of OC. Panel A. Comparison of ctDNA and CA125 latency until detection of OC recurrence in CT images. The horizontal axis indicates the markers, and the vertical axis indicates the length of latency of the detection of recurrence in CT images. Each dot indicates the value of the markers and whiskers indicate 95% CI. Panel B. Kaplan–Meier curves for progression-free survival. Red and blue lines indicate patients with and without ctDNA detected by ddPCR, respectively.
4. Discussion
We describe the analysis of ctDNA in the plasma of OC patients during their post-treatment follow-up. Tumor-specific mutations were previously identified by exome analysis of DNA extracted from FFPE specimens of 11 OC patients. ddPCR probes designed against the mutations were selected for each patient and mutated ctDNA fragments in plasma were detected in all six patients with recurrence during the observation period. Quantification of the AF in tumor-specific ctDNA fragments showed high sensitivity in ddPCR in recurrence detection: up to 0.039% ctDNA could be detected. AF changes in ctDNA responded earlier than changes in CA125. Furthermore, tumor-specific ctDNA fragments can indicate OC recurrence significantly earlier than CA125. In addition, the PFS of patients whose ctDNA was detected in blood samples collected after treatment was significantly shorter than that of patients whose ctDNA was not detected. These results indicate that ddPCR for tumor-specific ctDNA is a better marker than CA125 for detection of recurrence and estimation of treatment efficiency.
CA125 and CT imaging are the most frequently used surveillance modalities in ovarian and endometrial cancer. However, CA-125 is considered optional, and CT imaging is recommended only when clinically indicated by the National Comprehensive Cancer Network guidelines (Network, 2017). CT imaging can reliably detect recurrence of OC, but it causes radiation exposure and taking images at a high frequency should be avoided; therefore, good markers that can be detected in blood are necessary for follow-up of OC.
CA125 is widely expressed in other tissues than OC and its expression is altered in endometriosis, uterine fibroids, benign adnexal masses, and other type of malignancies such as cholangiocarcinoma. Moreover, CA125 has a prolonged half-life of 9–44 days (Riedinger et al., 2006). Hence, CA125 responds to tumor changes with some delays and does not reflect the present state of OC recurrence. Our results indicated that tumor-specific ctDNA fragments can reflect the extent of recurrence of OC better than CA125 and supports the idea that tumor-specific ctDNA fragments may be a more ideal marker for OC follow-up.
A study by Pereira (Pereira et al., 2015) and colleagues showed that ctDNA is elevated on average for 7 months (range, 1–11 months) prior to imaging studies, but the superiority of tumor-specific ctDNA fragments to CA125 in plasma was not clearly verified for the detection of OC recurrence in their study. Conversely, our study periodically estimated CA125 and ctDNA in OC follow-up and showed that the latency of the personalized tumor-specific ctDNA fragments appeared to be shorter than that of CA125 in the detection of OC recurrence with statistical significance.
Parkinson et al. showed that the TP53-specific ddPCR probes are useful to detect ctDNA of recurrent high-grade serous OC (Parkinson et al., 2016). However, in Japan, a substantial number of OC cases are clear cell carcinoma in which TP53 mutations are infrequent (Shibuya et al., 2018), and thus their strategy may not be applicable to many OC patients in Japan. Using ddPCR for the detection of tumor-specific ctDNA fragments in this study, it was possible to measure ctDNA according to the individual's specific mutations, rather than being limited to the histological type of OC or a specific range of mutated genes. In Japan, health insurance coverage for two cancer gene profiling tests has been provided by the national health insurance system since June 2019 to achieve precision oncology (Ebi and Bando, 2019;3.10.1200/po.19.00291.), and these profiling tests are commonly used in Japan. In this context, the hurdle of accessing genomic information in individual tumor specimens has been lowered, and the time and cost of designing and validating propensity probes for patient-specific ctDNA detection has been reduced by commercial companies.
In a previous study of untreated lung cancer, there was a significant concordance between ctDNA levels and tumor volume in nine patients (Newman et al., 2014). In our study, a positive correlation between tumor volume and ctDNA AF was confirmed in four patients. It has been highlighted that the correlation with tumor volume may be lower in cases of ascites retention (Parkinson et al., 2016). The two cases in which ctDNA AF could not be compared with recurrent tumor volume did not show ascites in our study. For these cases, it is also important to construe the changes of tumor volumes from liquid biopsy data to respond the disease progression at an appropriate time. Thus, it is important to increase the number of patients who undergo accurate tumor volume measurement in the future.
We found that ctDNA could detect OC recurrence earlier than CA125. However, considering the benefit of the early detection of OC recurrence, a study by Rustin et al. showed that early intervention had no benefit for OC patients (Rustin et al., 2010). At that time, therapeutic agents for recurrent disease were limited. For example, Rustin et al. used only platinum and/or taxane for the treatment of OC recurrence, and early detection of recurrence may not have been effective. Numerous molecular targeted drugs are currently available, and the use of such drugs might improve the prognosis of recurrent OC if combined with the earlier and more accurate detection of OC recurrence by ctDNA. The significance of early therapeutic intervention should be reevaluated in OC follow-up with these new therapeutic agents, with ctDNA detection as a recurrent maker. However, the study design would be difficult because of the varying sensitivity of ddPCR probes in OC patients.
This study demonstrates the usefulness of ctDNA as a predictor of OC recurrence. By incorporating our method into future clinical trials for a new drug, it may be possible to show the effectiveness of ctDNA detection for OC recurrence when the drug is effective. In addition, early detection of recurrence using tumor-specific ctDNA fragments may help OC patients to prepare for the next step, even if additional treatment is not available. The benefits of the early detection of recurrence may be many at this stage, such as identifying a suitable clinical trial for the patient or introducing the possibility of a second surgery that could be a curative treatment for the recurrent lesion. Similarly, ctDNA be a useful biomarker for clinical decision-making such as changing the chemotherapeutic agents. For example, case 1 and 2 in our study (Fig. 1 panel A and B) showed that the effectiveness of the maintaining drug (olaparib) and changing the agents (from Paclitaxel + Platinum to gemcitabine), respectively.
Our study had several limitations. First of all, probes were selected from Bio-Rad’s catalog to match the single nucleotide mutations in each sample. Because we were able to detect ctDNAs in the plasma of recurrence-positive patients using commercial probes, we believe that a certain level of quality was maintained with the commercially available ddPCR probes. However, an off-the-shelf product for all OC patients in the selection of ddPCR probes is not always available. Whether the same performance can be obtained for custom ddPCR probes in the follow up of OC patient remains to be explored. Second weaknesses of this study design: a retrospective study design for small sample size and no scheduled CT scanning. However, ctDNA was detected in the plasma of all patients with relapse and showed a significant difference compared with CA125. In addition, neither ctDNA nor obvious tumor images in contrast enhanced CT or PET-CT were detected in five non-relapsed patients during the observation period. However, it is unclear if the negative results in our study are reproducible. In addition, ctDNA may not be constantly increasing in the positive cases (for example, Fig. 1 panel E). This inconsistency would be caused by the paucity of ctDNA from small, early recurrent OC, indicating that frequent testing of ctDNA with ddPCR may be necessary to precise diagnosis of recurrence of OCs. We did not test multiple probes for the patients without recurrence. Further studies are required to show the sensitivity and specificity of ddPCR. This validation is critical if our approach for the detection of OC recurrence can be applied to a larger number of patients and will be highly useful in trials that link ctDNA as a biomarker to standard treatment.
Finally, our study suggests that detection of tumor-specific ctDNA fragments by ddPCR can be used in most of the major cancer treatment centers and it is therefore feasible to proceed with a multi-center study of this technology for improvement of prognosis of the ovarian cancers.
CRediT authorship contribution statement
Takamichi Minato: Conceptualization, Investigation, Visualization, Writing – original draft. Shin Ito: Investigation, Methodology. Bin Li: Methodology. Haruna Fujimori: Resources, Data curation, Funding acquisition. Mai Mochizuki: Resources, Data curation, Funding acquisition. Kazunori Yamaguchi: Resources, Data curation. Keiichi Tamai: Resources, Data curation, Funding acquisition. Muneaki Shimada: Writing – review & editing. Hideki Tokunaga: Writing – review & editing. Shogo Shigeta: Writing – review & editing. Ikuro Sato: Resources. Hiroshi Shima: Conceptualization, Resources, Funding acquisition. Hidekazu Yamada: Conceptualization, Resources, Data curation, Supervision. Nobuo Yaegashi: Funding acquisition, Supervision. Jun Yasuda: Conceptualization, Software, Writing – original draft, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Ms. Mika Takeuchi and Ms. Miyuki Ueki for their assistance with the sample preparation. We also thank Ms. Aya Hashimoto for technical assistance and Dr. Hideki Oikawa for his expertise in radiology. We thank H. Nikki March, PhD, from Edanz (https:/jp.edanz.com/ac), for editing a draft of this manuscript.
Funding Information
This work was supported by JSPS KAKENHI with following grant numbers: JP17K07187 (to H. Shima), JP19K08430 (to K. Tamai), JP18K09363 (to M. Mochizuki), JP21K15495 (to H. Fujimori), and JP17K07193 (to J. Yasuda). This work was also supported by JSPS KAKENHI Grant Numbers JP19H03795 (to N. Yaegashi), JP17K11265 (to M. Shimada), and JP19K23904 (to S. Shigeta). This work was also supported by The National Cancer Center Research and Development Fund (29-A-3) and AMED under Grant Number JP19ck0106319 to N. Yaegashi and H. Tokunaga, respectively. This work was also supported by the Foundation for the Promotion of Cancer Research in Japan, Takeda Medical Foundation, and Kobayashi Foundation for Cancer Research (K. Tamai).
Author Contributions
TM, HY, NY, and JY planned the study. TM, BL, and SI performed the experiments. JY performed the bioinformatics analyses. TM collected clinical information and performed the statistical evaluation of the data. MM, KT, TM, and KY checked the data quality, experimental procedures, and interpreted the raw data. MS, HT, and SS proofread the manuscript and critically discussed the scientific findings of this study. HS and IS contributed general management skills and prepared laboratory equipment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gore.2021.100847.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Exome analyses. Panel A. Schematic diagram of exome pipeline. Rectangles indicate the procedures. Panel B. Mutation spectrum of the ovarian tumors analyzed in this study. The diagram was drawn with oncoPrint in CBioportal. The small rectangles indicate the annotation of the mutations, and the color codes are indicated on the bottom of the spectrum.
Supplementary figure 2.
Correlation between the size of recurrent tumors and allele frequencies of ctDNAs. Panels A and B indicate the time courses of cases 2 and 3, respectively. In each panel, the top images are enhanced CT before and after the changes. The timing of images are indicated just below the images. Bottom graph indicates the changes in AF (vertical axes) during time course (horizontal axes). Red arrows in the CT images indicates the recurrent tumors.
References
- Shoji T., Komiyama S., Kigawa J., Tanabe H., Kato K., Itamochi H. An open-label, randomized, phase II trial evaluating the efficacy and safety of standard of care with or without bevacizumab in platinum-resistant epithelial ovarian, fallopian tube, or primary peritoneal cancer patients previously treated with bevacizumab for front-line or platinum-sensitive ovarian cancer: rationale, design, and methods of the Japanese Gynecologic Oncology Group study JGOG3023. BMC Cancer. 2018;18:771. doi: 10.1186/s12885-018-4505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn S.B., Bray F., Sherman M.E., Trabert B. International patterns and trends in ovarian cancer incidence, overall and by histologic subtype. Int. J. Cancer. 2017;140:2451–2460. doi: 10.1002/ijc.30676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell D.D., Böhm S., Ahmed A.A., Aspuria P.J., Bast R.C., Jr., Beral V. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer. 2015;15:668–679. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase S, Ohta T, Takahashi F, Yaegashi N, Board members of the 2020 Committee on Gynecologic Oncology of the Japan Society of Obstetrics and Gynecology. Annual report of the Committee on Gynecologic Oncology, the Japan Society of Obstetrics and Gynecology: Annual patient report for 2017 and annual treatment report for 2012 J. Obstet. Gynaecol. Res. 2021; 47: 1631–1642. 10.1111/jog.14724. [DOI] [PubMed]
- Colombo N., Sessa C., du Bois A., Ledermann J., McCluggage W.G., McNeish I. ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease†. Ann. Oncol. 2019;30:672–705. doi: 10.1093/annonc/mdz062. [DOI] [PubMed] [Google Scholar]
- Harmandayan G.Z., Gao F., Mutch D.G., Virgo K.S., Gibb R.K., Johnson F.E. Ovarian cancer patient surveillance after curative-intent initial treatment. Gynecol. Oncol. 2011;120:205–208. doi: 10.1016/j.ygyno.2010.10.028. [DOI] [PubMed] [Google Scholar]
- Rustin G.J., Nelstrop A.E., Tuxen M.K., Lambert H.E. Defining progression of ovarian carcinoma during follow-up according to CA 125: a North Thames Ovary Group Study. Ann. Oncol. 1996;7:361–364. doi: 10.1093/oxfordjournals.annonc.a010602. [DOI] [PubMed] [Google Scholar]
- Gu P., Pan L.L., Wu S.Q., Sun L., Huang G. CA 125, PET alone, PET-CT, CT and MRI in diagnosing recurrent ovarian carcinoma: a systematic review and meta-analysis. Eur. J. Radiol. 2009;71:164–174. doi: 10.1016/j.ejrad.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Van Gorp T., Cadron I., Despierre E., Daemen A., Leunen K., Amant F. HE4 and CA125 as a diagnostic test in ovarian cancer: prospective validation of the Risk of Ovarian Malignancy Algorithm. Br. J. Cancer. 2011;104:863–870. doi: 10.1038/sj.bjc.6606092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga H., Mikami M., Nagase S., Kobayashi Y., Tabata T., Kaneuchi M. The 2020 Japan Society of Gynecologic Oncology guidelines for the treatment of ovarian cancer, fallopian tube cancer, and primary peritoneal cancer. J. Gynecol. Oncol. 2021;32:e49. doi: 10.3802/jgo.2021.32.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder J.L., Pavlik E., Straughn J.M., Kirby T., Higgins R.V., DePriest P.D. Clinical implications of a rising serum CA-125 within the normal range in patients with epithelial ovarian cancer: a preliminary investigation. Gynecol. Oncol. 2003;89:233–235. doi: 10.1016/s0090-8258(03)00051-9. [DOI] [PubMed] [Google Scholar]
- Santillan A., Garg R., Zahurak M.L., Gardner G.J., Giuntoli R.L., 2nd, Armstrong D.K. Risk of epithelial ovarian cancer recurrence in patients with rising serum CA-125 levels within the normal range. J. Clin. Oncol. 2005;23:9338–9343. doi: 10.1200/jco.2005.02.2582. [DOI] [PubMed] [Google Scholar]
- Meier W., Baumgartner L., Stieber P., Hasholzner U., Fateh-Moghadam A. CA125 based diagnosis and therapy in recurrent ovarian cancer. Anticancer Res. 1997;17:3019–3020. [PubMed] [Google Scholar]
- Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- Lo Y.M.D., Han D.S.C., Jiang P., Chiu R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372:eaaw3616. doi: 10.1126/science.aaw3616. [DOI] [PubMed] [Google Scholar]
- Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti M., Cottone G., Carboni F., Cotroneo E., Casini B., Giordani E. Cross-sectional analysis of circulating tumor DNA in primary colorectal cancer at surgery and during post-surgery follow-up by liquid biopsy. J. Exp. Clin. Cancer Res. 2020;39:69. doi: 10.1186/s13046-020-01569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K.S., Brant R., Carr T.H., Dearden S., Jenkins S., Brown H. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90:509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Asante D.B., Calapre L., Ziman M., Meniawy T.M., Gray E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020;468:59–71. doi: 10.1016/j.canlet.2019.10.014. [DOI] [PubMed] [Google Scholar]
- Pereira E., Camacho-Vanegas O., Anand S., Sebra R., Catalina Camacho S., Garnar-Wortzel L. Personalized Circulating Tumor DNA Biomarkers Dynamically Predict Treatment Response and Survival In Gynecologic Cancers. PLoS One. 2015;10:e0145754. doi: 10.1371/journal.pone.0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson C.A., Gale D., Piskorz A.M., Biggs H., Hodgkin C., Addley H. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami K., Ichikawa H., Kubo T., Kato M., Fujiwara Y., Shimomura A. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019;110:1480–1490. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya Y., Tokunaga H., Saito S., Shimokawa K., Katsuoka F., Bin L. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer. 2018;57:51–60. doi: 10.1002/gcc.22507. [DOI] [PubMed] [Google Scholar]
- Ito S., Sato I., Mochizuki M., Yamaguchi K., Tamai K., Minato T. Robustness of a Cancer Profiling Test Using Formalin-fixed Paraffin Embedded Tumor Specimens. Anticancer Res. 2021;41:1341–1348. doi: 10.21873/anticanres.14891. [DOI] [PubMed] [Google Scholar]
- Liu H., Bielinski S.J., Sohn S., Murphy S., Wagholikar K.B., Jonnalagadda S.R. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl. Sci. Proc. 2013;2013:149–153. [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To E.W., Chan K.C., Leung S.F., Chan L.Y., To K.F., Chan A.T. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin. Cancer Res. 2003;9:3254–3259. [PubMed] [Google Scholar]
- Network NCC. NCCN Clinical Practice Guidelines in Oncology: OvarianCancer. USA2017.
- Riedinger J.M., Wafflart J., Ricolleau G., Eche N., Larbre H., Basuyau J.P. CA 125 half-life and CA 125 nadir during induction chemotherapy are independent predictors of epithelial ovarian cancer outcome: results of a French multicentric study. Ann. Oncol. 2006;17:1234–1238. doi: 10.1093/annonc/mdl120. [DOI] [PubMed] [Google Scholar]
- Ebi H, Bando H. Precision Oncology and the Universal Health Coverage System in Japan. JCO Precis Oncol. 2019;3.10.1200/po.19.00291. [DOI] [PMC free article] [PubMed]
- Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin G.J., van der Burg M.E., Griffin C.L., Guthrie D., Lamont A., Jayson G.C. Early versus delayed treatment of relapsed ovarian cancer (MRC OV05/EORTC 55955): a randomised trial. Lancet. 2010;376:1155–1163. doi: 10.1016/s0140-6736(10)61268-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.