Abstract
Autologous chimeric antigen receptor (CAR) T-cell therapies targeting CD19 have high efficacy in large B-cell lymphomas (LBCL), but long-term remissions are observed in less than half the patients and treatment-associated adverse events such as immune effector cell-associated neurotoxicity syndrome (ICANS) are a clinical challenge. We performed single-cell RNA-sequencing with capture-based cell identification on autologous axicabtagene ciloleucel (axi-cel) anti-CD19 CAR T-cell infusion products to identify transcriptomic features associated with efficacy and toxicity in 24 patients with LBCL. Patients that achieved a complete response by PET/CT at their 3-month follow-up had 3-fold higher frequencies of CD8 T-cells expressing memory signatures compared to patients with partial response or progressive disease. Molecular response measured by cell-free DNA (cfDNA) sequencing at day 7 post-infusion was significantly associated with clinical response (p=0.008), and a signature of CD8 T-cell exhaustion was associated (q=2.8×10−149) with a poor molecular response. Furthermore, a rare cell population with monocyte-like transcriptional features was associated (p=0.0002) with high-grade ICANS. Our results suggest that heterogeneity in the cellular and molecular features of CAR T-cell infusion products contribute to variation in efficacy and toxicity after axi-cel therapy in LBCL, and that day 7 molecular response may serve as an early predictor of CAR T-cell efficacy.
INTRODUCTION
LBCL patients who remain in complete (CR) or partial response (PR) by imaging at 3-months following autologous anti-CD19 CAR T-cell therapy have a significantly better outcome than those with stable (SD) or progressive disease (PD) prior to this point1,2. This is likely due to a combination of factors, such as the in vivo expansion and activity of CAR T-cells following infusion and the development of tumor-intrinsic mechanisms of escape or resistance3–6. Cellular features associated with response have been explored in the context of B-cell leukemia and identified phenotypes within the apheresis product7,8 and CAR T-cell infusion product9,10 as being important determinants. However, this has not been thoroughly investigated in the context of LBCL11 and is yet to be systematically examined using single cell transcriptomics.
Cytokine release syndrome (CRS) and immune effector cell-associated neurotoxicity syndrome (ICANS) are common adverse events associated with CAR T-cell therapy12,13. In adults with lymphoma treated with axi-cel, CRS is mostly low-grade, but ICANS is more commonly high-grade, with grade 3 or 4 (ICANSgr3–4) occurring in 30–45% of patients12,13. Although reversible in most cases, ICANS prolongs hospitalization, requires intensive care in a subset of patients, delays recovery, and increases the cost of care. Less commonly, seizures and cerebral edema have been noted with ICANS and have been fatal in some patients. While the pathophysiology of CRS has been studied extensively and appropriate management strategies with anti-IL-6 therapy have been developed12,14–16, the pathophysiology of ICANS and its optimal management are less clear. The development of ICANS is associated with a high level of inflammatory cytokines within circulation and in the cerebrospinal fluid17–19, with monocyte-derived IL-1 being recently highlighted as a key driver of neurotoxicity20. Increased permeability of the blood-brain-barrier may allow these cytokines and immune cells to enter the central nervous system and contribute to neurological inflammation18,19. However, the features of CAR T-cells that drive this hyper-inflammatory state in some patients and not others are currently unknown.
We hypothesized that the heterogeneity of autologous CAR T-cell infusion products may contribute to the inter-patient variability in efficacy and toxicity, and investigated this by performing single-cell RNA-sequencing (scRNA-seq) of standard-of-care axi-cel infusion products administered to LBCL patients. We thereby identified features associated with clinical response, molecular response, and the development of ICANS grade 3–4. These data provide new insights into the cellular and molecular basis for inter-patient variability in CAR T-cell efficacy and toxicity.
RESULTS
Gene expression signatures of CAR T-cell infusion products determined by scRNA-seq and CapID+.
An overview of our approach is shown in Figure 1. We performed whole-transcriptome scRNA-seq of 137,326 residual cells obtained from washing the standard-of-care axi-cel product bags following infusion of the CAR T-cells into 24 LBCL patients (16 DLBCL, 6 tFL, 2 PMBCL; Supplementary Table 1). Each sample was run fresh for library preparation, thus each sample is its own batch and ‘batch effect’ could not be formally assessed. However, clusters were defined by genes associated with cell states (Extended Data 1), cells from all samples were interspersed across multiple clusters and all clusters contained cells from multiple samples (Figure 2a–b, Supplementary Figure 1).
Figure 1: Single cell analysis of standard of care (SOC) CAR T-cell infusion products.
A schematic overview of the experimental design and bioinformatics flow for scRNA-seq analysis of 137,276 residual cells from CAR T-cell infusion products of 24 LBCL patients. Our approach incorporated single cell transcriptome profiling of CAR T-cell infusion products boosted by CapID+, correlation of single cell functional states and gene expression signatures with efficacy assessed by positron emission tomography/computed tomography (PET/CT) and by cfDNA sequencing, and with toxicity assessed by clinical grading.
Figure 2: Single cell analysis of standard of care (SOC) CAR T-cell infusion products.
a) An overview of the 133,405 cells that passed quality control (QC) for subsequent analyses in this study. Cells are color coded by the corresponding patient origin (sample ID) in the tSNE plot and a bar graph showing the number of cells per patient that passed QC. b) Cells are color coded by tSNE cluster number and a bar graph showing the distribution of cells from each patient among clusters. c) Individually scaled density plots show the normalized expression for the CAR, CD4 and CD8B transcripts in 10X scRNA-seq data and CapID hybrid capture sequencing data derived from the same scRNAseq libraries. Histogram overlays with identical scaling showing the relative fraction of cells with zero counts are shown in Extended Data 2b. d) The tSNE overview and bar graph summary of the cells identified as being CAR-positive (left), CD4 T-cells (middle) and CD8 T-cells (right) using 10X scRNA-seq data and those rescued by CapID+.
Cell types and functional states were defined according to previously described marker genes21. To improve this classification, we employed hybrid-capture sequencing of marker genes and the CAR construct sequence from scRNA-seq libraries, and refined CD4 and CD8 classification by shared nearest neighbor (SNN) clustering (CapID+; Supplementary Table 2, Methods). This approach does not rescue transcript dropout that results from failure of an mRNA molecule to be reverse transcribed in the GEMs (gel bead in emulsion). Rather, CapID increases the sequencing saturation of informative genes (Extended Data 2), thereby increasing the sensitivity of detection and signal-to-noise ratio for these transcripts within a given library (Figure 2c). This rescued cell identities (Figure 2d) and led to a more significant correlation with flow cytometry-based measurement of cell frequencies (Extended Data 3). Our CapID+ methodology therefore allows for the accurate assignment of CAR status and cellular gene expression. CAR expression was interspersed across the clusters (Figure 2d) and was associated with few significant differences in gene expression compared to non-transformed T-cells (Extended Data 4). We therefore did not separate CAR-positive and CAR-negative cells for subsequent analyses.
Cell signatures associated with clinical response
At the 3-month follow-up by PET/CT, a clinically-relevant surrogate time point to predict long-term durability of response after axi-cel therapy in LBCL1, 13 patients had progressive disease (PD, 50%), 1 was in partial response (PR, 4%), 9 were in complete response (CR, 38%) and 1 was not evaluable (NE). We compared the representation of cell types and functional states between the infusion products from patients in CR and those in PR/PD. This revealed a significant enrichment of exhausted CD8 and CD4 T-cells within the infusion products of patients with PR/PD, and a significant enrichment of memory CD8 T-cells within the infusion products of patients who achieved CR (Figure 3a).
Figure 3: Molecular phenotypes of CAR T-cell infusion products associated with response determined by PET/CT.
a) Cell types and functional states that were significantly more frequent in CAR T-cell products from patients with continued CR at 3 months (blue) or those from patients with PR/PD (red). Q-values were calculated by a two-sided Fisher exact test with a Benjamini-Hochberg correction. b) Heatmap of four CD8+ T cell clusters (C1-C4) generated from unsupervised clustering of genes that were differentially expressed in CD8 T-cells from the infusion products of patients with CR compared to those from patients with PR/PD. A color-coded track shows the cells that originated from infusion products of CR patients (blue: CR) and the percentage of these cells within each cluster labelled at the top. Additional tracks show the scGSVA scores of CD8 dysfunction and CD8 memory signatures, respectively, and the inferred cell cycle status. The percentage of cells that originated from infusion products of CR patients is significantly different between clusters (One-way ANOVA p < 2.2×10−16). The corresponding sample origins are labeled at the bottom, colored as per Figure 2a. c) Violin plots show the scGSVA scores of cells from each the four clusters in 2b. C1, n=26,917 cells. C2, n=9,047 cells. C3, n=10,113 cells. C4, n=6,440 cells. Box, median +/− interquartile range. Whiskers, 1.5X interquartile range. Pairwise comparisons were performed using a two-sided Wilcoxon rank-sum test with a Benjamini-Hochberg correction. d) Scatter plots of CCR7+CD27+ CD8 T-cells measured by CapID in the infusion products of patients who achieved CR compared to those who had PR/PD (Two-sided Fisher exact test p<2.2× 10−16). e) Gene sets that are significantly positively (+) or negatively (−) associated with CR in CD8 (above) or CD4 (below) T-cells. For each pathway, a heatmap of the single cell GSVA scores are shown, with % of cells originated from infusion products of CR patients annotated on the top. The origin of the gene set is shown in brackets (B, biocarta; R, reactome; K, KEGG; P, PID). f) A heatmap of five CD4+ T cell clusters (C1-C5) determined by unsupervised clustering of genes that were differentially expressed in CD4 T-cells from infusion products of patients with CR compared to those from patients with PR/PD. The percentage of cells that originated from infusion products of CR patients is shown in a track at the top and is significantly different between clusters (One-way ANOVA p < 2.2×10−16). Cells are annotated by inferred cell cycle state and sample origin as in panel b.
We then identified differentially expressed genes (DEGs) between CD8 T-cells from CR patients compared to PR/PD patients (Supplementary Table 3) followed by unsupervised hierarchical clustering using the DEGs (Figure 3b). Four cell clusters were identified with cells that showed differential expression of the genes associated with CR or PR/PD, respectively. Each cluster contains cells from all patients, but the relative proportions of cells from patients with CR significantly differ across the clusters (P<2.2×10−16). Cells within the C1 cluster express the highest levels of genes associated with PR/PD, the lowest level of genes associated with CR, had the lowest proportion of cells from CR patients (19%), and are characterized by high expression of genes encoding lymphocyte activating 3 (LAG3)22, the exhaustion-associated transcription factors basic leucine zipper ATF-like transcription factor (BATF)23,24 and inhibitor of DNA binding 2 (ID2)25,26, interferon gamma (IFNG), effector molecules (GZMA, GZMB, GNLY), and major histocompatibility class II (MHCII) molecules. These genes are associated with activation and exhaustion of CD8 T-cells, in line with our observed enrichment of exhausted CD8 T-cells in PR/PD patients (Figure 3a). We therefore evaluated the expression of a previously defined CD8 T-cell dysfunction signature27 by single cell gene set variation analysis (scGSVA). This signature was highest in the C1 cluster, with progressively less expression in the C2 and C3 clusters and the lowest expression in the C4 cluster (Figure 3b–c). The features associated with activation and exhaustion in cells predominantly originating from PR/PD patients are therefore linked with a profile of CD8 T-cell dysfunction. Cells within the C2 and C3 clusters have variable expression of genes that were associated with CR or PR/PD, respectively, and a mixed representation of cells from CR and PR/PD patients. Cells within the C4 cluster had the lowest expression of genes associated with PR/PD, the highest expression of genes associated with CR, and had the highest proportion of cells originating from patients who achieved a CR (65%). These cells were characterized by high expression of genes associated with a central memory phenotype, such as CCR7, CD27 and SELL (Figure 3b and Supplementary Figure 2a), but not markers associated with a CD8 stem cell memory phenotype (Supplementary Figure 2b). This too was in line with the enrichment of CD8 memory T-cells in products from CR patients when comparing the frequencies of functional states (Figure 3a). Specifically, CD8 T-cells from the infusion products of patients who achieved CR had a ~3-fold higher fraction of CCR7+CD27+ double-positive CD8 T-cells compared to cells from patients with PR/PD (Figure 3d). The frequency of these cells were also low in patients with high stage disease (III/IV) and high international prognostic index (IPI 3–4), suggesting that clinical factors may influence the transcriptional state of CAR T-cells (Supplementary Figure 3a). As CCR7 and CD27 can be expressed on naïve T cells, we used scGSVA to measure the expression of a previously defined CD8 memory signature21. Cells within the C4 cluster had the highest expression of the CD8 memory signature, with progressively lower levels of expression in the C3 and C2 clusters and the lowest expression in the C1 cluster (Figure 3c). Our observations from comparing functional states and DEGs were further corroborated by scGSVA analysis of the association between unselected gene sets and patient outcome, which confirmed the high expression of activation and exhaustion signatures such as PD-1 and IFN-γ signaling in CD8 T-cells from patients with PR/PD, and the high expression of immune memory signatures28 in CD8 T-cells from patients who achieved a CR (Figure 3e, Supplementary Figure 4, Supplementary Table 4).
DEG analysis and unsupervised clustering of CD4 T-cells revealed some similarities with the profiles of CD8 T-cells, such as higher expression of CCL5, GNLY and MHCII genes in clusters with high frequencies of cells from PR/PD patients (C1 and C2) and low frequencies of cells from CR patients (C3–5) (Figure 3f). Cluster C2 consisted only of cells that originated from the infusion products of patients with PR/PD and was characterized by a high expression of TIGIT29 (Supplementary Figure 5). However, in CD4 T-cells there was was higher expression of proliferation-associated genes and cycling cells within clusters with high frequencies of cells from patients who achieved CR (Figure 3f; Supplementary Table 5). This was confirmed by scGSVA analysis, showing an enrichment of proliferation-associated gene sets in CD4 T-cells from patients who achieved CR (Figure 3e, Supplementary Figure 4, Supplementary Table 6).
These data highlight important transcriptional signatures of cells within CAR T-cell infusion products that are associated with clinical efficacy. The mixture of cells from CR and PR/PD patients across multiple clusters is consistent with each infusion product containing cells of both desirable (signatures associated with CR) and undesirable (signatures associated with PR/PD) states, and the relative proportion of these states within each CAR T-cell infusion product being linked with efficacy.
A CD8 T-cell exhaustion signature associated with failure to achieve an early molecular response.
Antigen-driven CAR T-cell expansion reaches its peak approximately 1 week after axi-cel infusion4. We therefore hypothesized that assessment of response dynamics within the first week of treatment may provide insights into CAR T-cell clinical activity. We investigated this by interrogating patterns of early molecular response determined by plasma-derived cell-free DNA (cfDNA) sequencing with a previously-described hybrid-capture panel30 (Methods). Somatic mutations identified at infusion (day 0–1) were used to measure the relative changes in variant allele fractions (VAF) at 1 week (n=22), 2 weeks (n=18) and 1 month (n=16) post infusion. Of 22 patients with available samples, 17 had ≥3 somatic mutations detected at day 0–1 for disease monitoring (Supplementary Table 7). Neither the number of calibrated mutations, nor the VAF at day 0 were associated with subsequent response (Extended Data 5). VAF declined within the first week of treatment for 16/17 patients, and the magnitude of decline was significantly associated with ongoing CR by PET/CT at 3 months (Two-sided Wilcoxon rank-sum P=0.008; Figure 4a–b). We applied a threshold of 5-fold reduction in VAF to dichotomize patients evaluable for response. Of the 8 patients with >5-fold molecular response (>5FMR), 6 (75%) had ongoing CR at 3 months. Two patients (ac17 and ac21) with >5FMR at day 7 progressed by the time of their 3 month PET/CT, consistent with the ~16% of patients in the ZUMA-1 pivotal study of axi-cel that had a CR at their 1 month evaluation but progressed prior to their 3 month follow-up4. Notably, these patients had an observable increase in their VAFs at day 14 or day 30 (Extended Data 5). Of the 8 evaluable patients with <5-fold molecular response (<5FMR), none were in CR at their 3-month follow-up (0%). Patients with <5FMR also showed a trend towards reduced CAR T-cell expansion, as measured by qPCR analysis of the CAR transcript from cfDNA (Supplementary Figure 6). The association between early molecular response (EMR) and long-term outcome following CAR T-cell therapy requires prospective validation in a large independent cohort, but supported its use as a relevant metric to evaluate using scRNA-seq.
Figure 4: Association between early molecular response measured by cfDNA sequencing and clinical response measured by PET/CT.
a) Molecular response measured by deep targeted cfDNA sequencing over the first month following infusion. Variant allele fraction (VAF) for each patient (n = 17) are normalized to the infusion day time point (day 0) and lines are colored according to response assessed by PET/CT at their 3-month follow-up or prior disease progression. b) Fold change in molecular disease burden at the day 7 time point relative the day 0 time point is shown for each patient, with bars colored by clinical response determined by PET/CT at their 3 month follow-up. The fold reduction of molecular disease burden was significantly associated with clinical response at 3 months (two-sided Wilcoxon rank-sum P = 0.008). The 16 patients with evaluable response were split into two groups according to whether they achieved >5-fold molecular response (>5FMR) or had <5-fold molecular response (<5FMR).
Comparison of the cellular functional states within products from patients with >5FMR versus <5FMR revealed a striking association between transcriptional profiles of T-cell exhaustion and a poor EMR (Figure 5a). This was particularly significant within the CD8 T-cell compartment. All of the co-inhibitory receptor genes used to classify exhausted CD8 T-cells were expressed in a higher fraction of CD8 T-cells from patients with <5FMR compared to >5FMR (Figure 5b; q-value<0.01). Of these, LAG3 and TIM3 were the most abundantly expressed, and cells co-expressing both LAG3 and TIM3 were highly discriminatory between the <5FMR and >5FMR groups (Figure 5b; q-value<2.38×10−170). Specifically, 36.8% of CD8 T-cells from patients with <5FMR were LAG3+TIM3+, compared to 22.7% from patients with >5FMR (Figure 5c). Frequencies of LAG3+TIM3+ cells were only moderately higher in infusion products of patients with PR/PD compared to patients with CR determined by their 3 month PET/CT (Supplementary Figure 7), and not significantly different between memory CD8 T-cells compared to other CD8 T-cells (Supplementary Figure 7). The frequency of LAG3+TIM3+ CD8 T-cells were also higher in patients with high IPI (3–4) and lower in patients that had received an autologous stem cell transplantation prior to their CAR T-cell therapy (Supplementary Figure 3). Patients with poor responses had moderately reduced TCR clonotypic diversity (Extended Data 6) but higher rates of the CD8 T-cell exhaustion signature were not due to the presence of highly expanded and exhausted clonotypes. Rather, each clonotype from patients with <5FMR had a significantly higher fraction of LAG3+TIM3+ cells compared to patients with >5FMR (Figure 5d, P=4.8×10−9), indicating that T-cells with the exhaustion signature are polyclonal. Further studies to examine the cytotoxicity and proliferative capacity of LAG3+TIM3+ CD8 T-cells are required to validate that this profile is associated with a loss of effector function and/or in vivo expansion of CD8 CAR T-cells, respectively.
Figure 5: Association between CD8 T-cell exhaustion markers and early molecular response.
a) Cell types and functional states associated with >5-fold molecular response at day 7 (>5FMR, blue) or failure to achieve 5-fold molecular response at day 7 (<5FMR, red). Q-values were calculated by a two-sided Fisher exact test with a Benjamini-Hochberg correction. b) The percentage of cells expressing co-inhibitory molecules utilized in the classification of exhausted CD8 T-cells, and the percentage of cells co-expressing LAG3 and TIM3, is shown for cells from patients with >5FMR (turquoise) compared to those from patients with <5FMR (red). c) Scatter plots show the expression of LAG3 and TIM3 in cells from patients who achieved >5FMR (left, blue) compared to those from patients with <5FMR (right, red). Each point represents a single cell and the proportion of cells at each state is labelled on the plot. Expression levels are normalized UMI counts from CapID sequencing, with normalized UMI counts >2 defined as positive expression. d) The percentage of cells from each TCR clonotype (identified from single cell TCR sequencing) co-expressing LAG3 and TIM3 are shown and data are compared between clonotypes within infusion products of patients with >5FMR (above, blue) and those from patients with <5FMR (below, red). The size of each point indicates the clonal fraction of each clonotype within each infusion product. >5FMR; n=72 clonotypes from 8 patients. <5FMR; n=196 clonotypes from 9 patients. Boxes, median +/− the interquartile range; whiskers, 1.5x interquartile range. P value was calculated by two-sided Wilcoxon rank-sum test. e) Transcripts were measured in 38,601 cells from fresh core needle biopsies of 9 rrLBCL tumors by CapID; 5 patients progressing following chemo-immunotherapy or targeted therapy (CAR T naïve) and 4 patients progressing following axi-cel CAR T-cell therapy (post-CAR T). The expression of TIM3 and LAG3 was quantified by CapID within CD8 T-cells in CAR T naïve and post-CAR T tumors, and within CD8 T-cells expressing the CAR transcript in post-CAR T tumors. The fraction of single- and double-positive cells annotated.
We next investigated whether the LAG3+TIM3+ CD8 T-cell signature is important in the setting of CAR T-cell failure. Using CapID+ analysis of fresh core needle biopsies of 9 LBCL tumors, we compared the frequencies of tumor-infiltrating LAG3+TIM3+ CD8 T-cells between 5 tumors from patients that had never received CAR T-cell therapy (CAR T naïve) and 4 patients progressing following axi-cel with PR/PD as best response (Post-CAR T; Figure 5e; Supplementary Table 8). There was no significant difference in the number of preceding lines of therapy between these cohorts. This showed that 34% (1,402/4,114) of CD8 T-cells within tumors progressing from CAR T-cell therapy were LAG3+TIM3+, as compared to 19% (587/3,168) from CAR T naïve tumors progressing from chemo-immunotherapy. CAR+ T-cells were detectable in all tumors from patients progressing following axi-cel, and are the only cells that can be definitively traced to the CAR T-cell infusion product. 51% (295/589) of CAR+ CD8 T-cells within tumors progressing from axi-cel were LAG3+TIM3+ (Figure 5e). Patients failing to achieve a robust EMR therefore have a significantly higher frequency of CD8 T-cell exhaustion signatures characterized by increased proportion of LAG3+TIM3+ cells, which is a signature that is proportionately over-represented among intratumoral CD8 T-cells from patients progressing from axi-cel CAR T-cell therapy.
Cells with a monocyte-like transcriptional signature are associated with high-grade ICANS.
We aimed to identify features of the CAR T-cell infusion products that were significantly associated with related toxicities. We first evaluated differences between patients who developed high-grade CRS (n=4) compared to those who did not (n=20). Although the number of patients with high-grade CRS was small, we observed a negative association with exhausted CD8 T-cells and a positive association with exhausted CD4 T-cells (Extended Data 7; Supplementary Tables 9 and 10). However, CRS is effectively managed with the use of corticosteroids and IL-6/IL-6R antagonists13, which may have confounded these results.
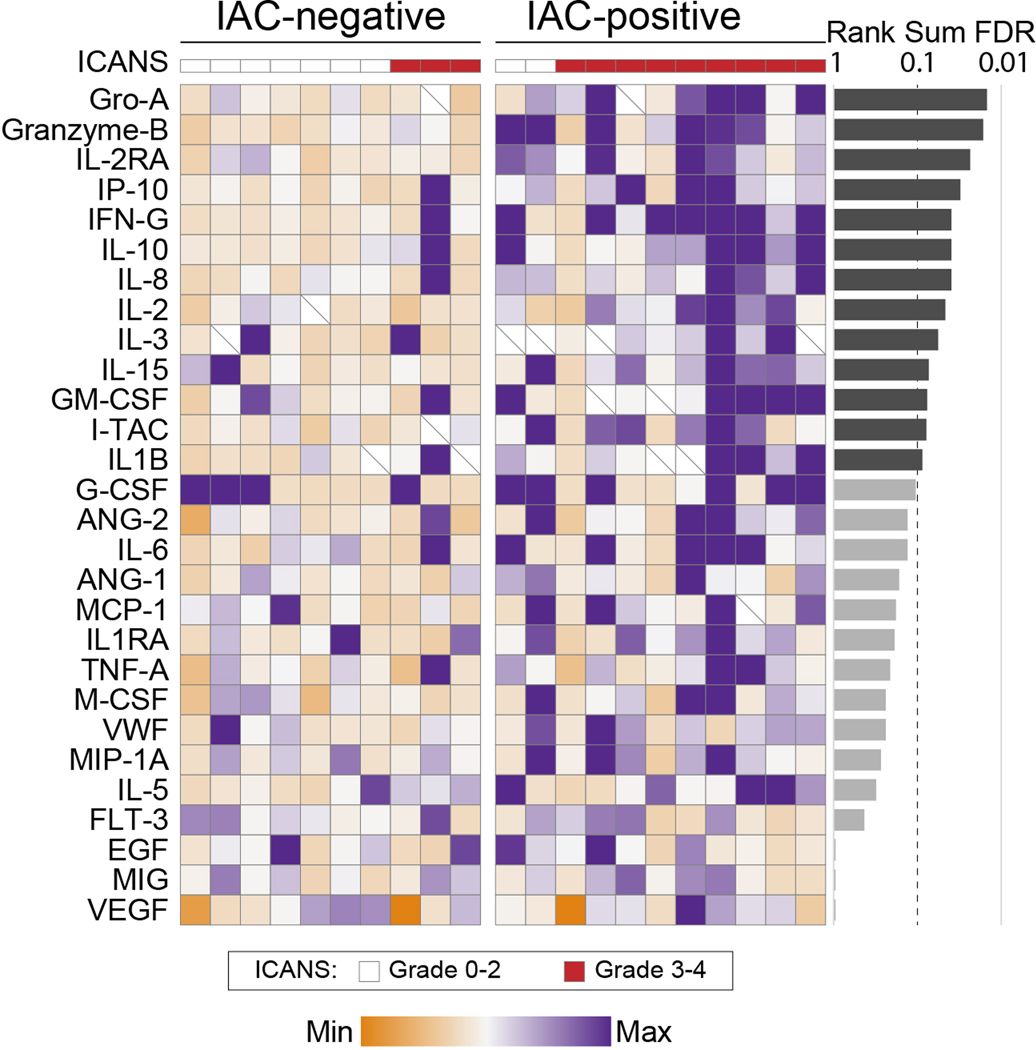
We next evaluated differences between patients that developed grade 3–4 ICANS (ICANSgr3–4, n=12) compared to patients with grade 0–2 ICANS (ICANSgr0–2, n=12). We did not observe significant differences in gene expression within the major CD4 or CD8 T-cell compartments, but patients with ICANSgr3–4 had a significantly lower frequency of CAR+ cells within their infusion products (Extended Data 8). As axi-cel dosing is calculated based upon transduction efficiency to achieve 2×106 CAR-positive cells per kilogram, this association may suggest that higher total cell doses per kilogram contributes to neurological toxicity, but this requires validation. We further evaluated whether ICANSgr3–4 is associated with any unique cluster of cells with distinct transcriptomic features, which identified a small cluster (n=254) that was significantly over-represented in the infusion products of patients who developed ICANSgr3–4 (Figure 6a). These cells were unlikely to be cellular debris or doublets (Supplementary Figure 8), and are henceforth referred to as ICANS-associated cells (IACs). A significantly lower fraction of IACs had detectable CAR expression (10.6% IACs vs 36.2% other cells; two-tailed Wilcoxon rank-sum P<0.001), TCR rearrangements (14.6% IACs vs 92% other cells; two-tailed Wilcoxon rank-sum P<0.001), CD3D expression (22.4% IACs vs 93.3% other cells; two-tailed Wilcoxon rank-sum P<0.001) and CD8B expression (4.3% IACs vs 49.3% other cells; two-tailed Wilcoxon rank-sum P<0.001; Figure 6b–c) compared to cells from other clusters. Expression of CD4 was detected in 49.6% of IACs, compared to 32.9% of other cells, but this can be expressed on both the myeloid and T-lymphoid lineages in response to IL-231,32. We identified 257 genes with significantly higher expression in IACs compared to other cells (Two-sided Wilcoxon rank-sum q-value<0.01, FC>2; Supplementary Table 11), including multiple genes that are typically expressed within the myeloid lineage such as CD68, LYZ, SPI1, LILRB4 and SIRPA (Figure 6b–d). Multiple cytokines and chemokines were among the genes with significantly higher expression in IACs compared to other cells, including IL1B and CXCL8 (IL8) which have been previously implicated in ICANS pathophysiology17,20. These cytokines were higher in the serum of patients with IACs, but this was part of a broad pattern of inflammation17 (Extended Data 9). Using ssGSEA of the IACs signature genes within profiles from purified hematopoietic cell subsets33 we observed that the IACs signature is significantly higher in cells of myeloid lineages compared to lymphoid lineages (p<2.2×10−16), and highest in classical monocytes (Figure 6e). However, IACs do not express genes encoding canonical monocyte markers such as CD14 or CD16 (FCGR3A/B) (Figure 6c). Therefore, despite the similarities of the IACs transcriptional signature to monocytes, the expression of CD3D on 22.4% of IACs and the lack of expression of canonical monocyte cell markers means that we cannot confidently assign these cells to the monocyte lineage.
Figure 6: ICANS-associated cells in CD19 CAR T-cell infusion products.
a) Clusters are shown for 10X transcriptome data of 24 patients in a tSNE plot, colored by the significance of their association with ICANS. The significant cluster of ICANs-associated cells (IACs) is circled. P-values were calculated using a two-sided Wilcoxon rank-sum test. b) Genes that are most highly expressed in cells from the IACs cluster compared to cells from other clusters are shown in a heatmap. Expression of additional T-cell markers, T-cell receptor (TCR) gene rearrangements and the CAR transcript are shown in tracks at the top. c) The expression of representative IACs markers, T-cell markers and canonical monocyte markers are shown for each cluster. The size of bubbles is relative to the percentage of cells within a cluster that express a given gene, and the color is relative to the mean expression of a given gene within each cluster. d) Density plots show the cellular distribution of transcript abundance of IACs marker genes in cells from the IAC cluster compared to cells from other clusters, determined by 10X transcriptome data. e) Single sample gene set enrichment (ssGSEA) values are shown for the IACs signature genes within gene expression data from purified populations of normal hematopoietic cell subsets, with increasing scores representing higher expression of the gene set. Myeloid lineage subsets [left, peach] have a significantly higher expression of genes that are characteristic of the IACs cluster compared to lymphoid lineage cells (Two-sided Wilcoxon rank-sum test, p<2.2×10−16). The highest expression of the IACs signature genes observed in monocytes. N=6 for all cell types except for basophils (n=4), myeloid DCs (n=5), non-classical monocytes (n=5), and memory CD8 T-cells (n=5). Boxes, median +/− the interquartile range; whiskers, 1.5x interquartile range. f) The cellular proportion of IACs determined by measurement of signature genes by CapID the infusion products of 269,164 single cells from 40 patients’ CAR T-cell infusion products (24 patient discovery cohort, 16 patient validation cohort [bold sample ID]) is shown. The samples are grouped according to patients with grade 3–4 ICANS (n=18, orange) compared to grade 0–2 ICANS (n=22, blue). Box, median +/− interquartile range. Whiskers, 1.5x interquartile range. P value was calculated by a two-sided Wilcoxon rank-sum test.
We validated the IACs population within the original cohort plus an additional 16 patients (total n=40; Supplementary Table 1) using an approach that is statistically independent of clustering. Specifically, the IACs signature genes (Supplementary Table 2) were measured by CapID and the signature quantified by scGSVA (Extended Data 10). Of the 254 IACs identified by clustering of transcriptome data, 231 were classified as IACs with high confidence using CapID and scGSVA. An additional 184 IACs were identified among 131,924 cells from the 16-patient validation cohort. IACs were both qualitatively and quantitatively associated with the development of ICANSgr3–4 (Figure 6f). 61.1% (11/18) of patients that developed ICANSg3–4 had detectable IACs within their infusion products, compared with 9.1% (2/22) of patients with ICANSgr0–2 (Two-sided Fisher-exact P=0.0007). IACs consisted of a mean of 0.39% of cells (range 0 – 2.27%) from patients with ICANSgr3–4, compared to 0.003% of cells from patients with ICANSgr0–2 (range 0 – 0.4%; 130-fold higher in ICANSgr3–4; Wilcoxon rank-sum P=0.0002). All patients with grade 4 ICANS (3/3) had detectable IACs, but the difference between patients with grade 3 vs grade 4 was not significant (Supplementary Figure 9). It will be important to investigate the origin and function of IACs in future studies and to validate its potential predictive capacity for ICANSgr3–4 in a large prospective series.
DISCUSSION
Our results suggest that cellular and molecular diversity of infused CAR T-cell products are a major factor contributing to the variability in efficacy and toxicity among LBCL patients treated with CD19 CAR T-cell therapy. Quantifiable phenotypes associated with the infusion product are potentially actionable by enriching desirable, or depleting undesirable, cellular populations or functional states during manufacturing. Furthermore, our understanding of the mechanisms by which discrete cellular populations lead to poor response or high-grade toxicity may identify avenues for therapeutic intervention to improve efficacy and decrease toxicity after CAR T-cell infusion.
We did not observe significant differences in gene expression profiles between cells expressing the CAR transcript compared to those with no detectable CAR expression, presumably because the signaling induced by stimulation during the manufacturing process may overwhelm the effects of any tonic signaling originating from the CAR construct. Comparisons of CAR-transduced and untransduced T-cells will therefore need to be performed separately using cells isolated from patients after infusion, when the transcriptional consequences of tonic or antigen-driven signaling will be more evident. We showed that cells from the infusion products of patients with ongoing CR at 3 months had an enrichment of CD8 memory T-cell phenotypes compared patients with PR/PD. This is in line with a prior study that evaluated CAR T-cells with CD3ζ and 4–1BB signaling domains in the setting of chronic lymphocytic leukemia, in which a memory CD8 phenotype was associated with superior responses10. However, we observed a high expression of CCR7 that is consistent with a central memory phenotype. Manufacturing conditions or rapid expansion protocols can be adapted to favor the generation of central memory CD8 T-cells34,35, which we would expect to be associated with greater efficacy.
Patients with PR/PD had a significant enrichment of cells with a CD8 T-cell exhaustion signature within their infusion products, and this association was most significant in patients failing to achieve a robust EMR at day 7. This is consistent with the kinetics of CAR T-cell expansion, which peaks ~1 week after axi-cel infusion and declines thereafter. Tumor-derived somatic alterations can be readily detected in the cfDNA of DLBCL patients30,36–38 and has been shown to correlate with tumor burden measured by functional imaging36. The kinetics of tumor-derived cfDNA following chemo-immunotherapy may be predictive of patient outcome37,38, but to our knowledge this has not been previously examined in patients treated with CD19 CAR T-cells. Furthermore, no other metrics are available to predict the outcome of LBCL patients treated with CD19 CAR T-cells prior to their first clinical assessment at 1 month post-infusion. In our cohort, the magnitude of EMR was correlated with efficacy assessed at the 3-month follow-up by PET/CT. This association requires prospective validation in a large independent series, but raised the possibility that failure to achieve EMR within the first week may be a biomarker for poor CAR T-cell function. In line with this notion, we observed a significant association between poor EMR and gene expression signatures associated with T-cell exhaustion. Other studies aimed at addressing exhaustion of CAR T-cells have primarily focused on targeting PD-1 signaling39–41. However, PD-1 (PDCD1) was expressed in a very small subset of cells and the most informative exhaustion markers in our cohort were co-expression of LAG3 and TIM3. These co-inhibitory molecules limit T-cell activation and expansion42, and are potentially targetable using blocking antibodies43. Therefore, these results suggest that the use of LAG3 and/or TIM3 blockade following infusion might improve efficacy in patients with poor EMR. The association between T-cell exhaustion signatures in autologous infusion products and adverse patient characteristics also suggests that allogeneic CAR T-cells, which are currently under clinical development, might be more beneficial in some patients. However, this requires further validation as prior studies have not identified any association between patient characteristics and response4.
Finally, we identified a rare monocyte-like cell population within axi-cel infusion products that was significantly associated with the development of high-grade ICANS. The mechanism by which IACs might act in vivo to promote neurotoxicity is unclear, though IL-1 signaling has been recently implicated in the pathogenesis of ICANS20 and the expression of IL1B and other inflammatory cytokines was significantly higher in IACs compared to other cells within the infusion products. The association between this cell population and the development of high-grade ICANS requires validation and characterization by orthogonal approaches to scRNA-seq in a large prospective series, including the evaluation of whether they are present in products from other manufacturers.
Future studies should be conducted to validate the potential predictive capacity of day 7 molecular response, and whether rational therapeutic interventions can be made in high-risk patients to improve their outcomes. Furthermore, future studies should determine whether the detection of ICANS-associated cells within infusion products may identify a subset of patients that are at high risk of neurotoxicity and would benefit from prophylactic therapy with agents that target myeloid cell function, such as interleukin 1 receptor antagonists44.
DATA AVAILABILITY
All requests for raw and analyzed data and materials are promptly reviewed by the University of Texas MD Anderson Cancer Center to verify if the request is subject to any intellectual property or confidentiality obligations. Patient-related data not included in the paper may be subject to patient confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. Transcriptome and CapID scRNA-seq datasets are available through the gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accessions GSE150992 and GSE151511, respectively. Raw data used in the generation of Figures 1, 2, 3, 4, 5, and 6, and Extended Data 1 to 10 are available through the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home), accessions EGAD00001006327 and EGAD00001006325.
METHODS
Patient samples
Patients with diffuse large B-cell lymphoma (DLBCL), transformed follicular lymphoma (TFL), primary mediastinal large B-cell lymphoma or high-grade B-cell lymphoma being treated with axicabtagene ciloleucel (axi-cel) anti-CD19 CAR T-cell as standard of care were eligible for this study. Those on active therapy at the time of their apheresis were excluded due to potential confounding effects on the phenotype of CAR T-cell infusion products. Patients provided informed consent for use of their cells, blood samples and the use and disclosure of deidentified health information in research as part of a protocol that was approved by the Institutional Review Board of University of Texas MD Anderson Cancer Center. Following completion of CAR T-cell infusion, the infusion bag was washed with PBS to obtain residual cells, Samples were deidentified and transferred to the laboratory for single-cell RNA sequencing (scRNA-seq). At the 3-month follow-up, 13 patients had progressive disease (PD, 50%), 1 was in partial response (PR, 4%) and 9 were in complete response (CR, 38%) and 1 was not evaluable (NE) due to death from sepsis. Peripheral blood samples from patients were drawn in EDTA vacutainers at the time of a clinically indicated procedure, and processed by centrifugation to obtain plasma. De-identified plasma from 24 healthy controls was obtained from the Gulf Coast Blood Center. Samples were available from patients at the infusion (day 0–1, n=22), and 1 week (6–8 days, n=22), 2 weeks (12–18 days, n=18) and 1 month (25–35 days, n=16) post infusion. Analysis of cfDNA from patient samples and healthy controls was approved by the Institutional Review Board of University of Texas MD Anderson Cancer Center.
Flow cytometry
When sufficient residual cells were available from CAR T-cell infusion products, flow cytometry was performed using Horizon Fixable Viability Stain (BD Biosciences) and the following antibodies; CD134 FITC (ACT35, BD Biosciences), anti-CAR T PE (KIP-1, Kite), ICOS PE-TR (C398.4A, BD Biosciences), CD127 PerCP Cy5.5 (HIL-7R-M21, BD Biosciences), CD69 APC (FN50, BD Biosciences), CD28 APC-H7 (CD28.2, BD Biosciences), CD4 AF700 (RPA-T4, BD Biosciences), 4–1BB BV421 (4B4–1, BD Biosciences), CD14 BV605 (M5E2, BD Biosciences), CD8 BV650 (RPA-T8, BD Biosciences), CD3 BV711 (UCHT1, BD Biosciences), and PD-1 BV786 (EH12.1, BD Biosciences). All antibodies were used at the manufacturer’s recommended concentration. Flow cytometry flow cytometry was performed on a BD Fortessa instrument and the data were analyzed using Cytobank.
Single cell RNA-sequencing
Fresh cells were interrogated using the 10X Chromium platform for simultaneous transcriptome and T-cell receptor (TCR) profiling, with 133,405 cells (range 1,732–8,573 cells per patient) passing our quality control thresholds and taken into subsequent analysis. These cells were sequenced to an average depth of 73,521 reads per cell, yielding an average of 2,472 genes per cell (Supplementary Figure 1). All major TCR clonotypes had both CAR-positive and CAR-negative cells detected, suggesting no integration-site-driven jackpot effects45 in this cohort.
5’ gene expression profiling using 10X.
Single cell RNA-seq was performed using GemCodeTM Single Cell Platform and Chromium Single Cell 5’ Reagent kit (10X Genomics Inc.) according to the manufacturer’s instructions. Briefly, CAR-T cells were resuspended in PBS with 0.04% BSA and gently pipette mix 10–15 times using a wide-bore pipette tip. Cell concentration was determined by cell counting with a hemocytometer and an approximate concentration (between 700–1200 cells/μl) was adjusted to maximize the likelihood of achieving the desired cell recovery target. Samples were at least 85% viable cells confirmed by trypan blue stain prior to capture and always handled on ice when possible. Subsequently, single-cell suspension was mixed with RT master mix and loaded together with barcoded single cell 5’ gel beads and partitioning oil onto Single Cell A Chip to generate GEM (Gel bead-in-Emulsion) using Chromium Controller. Cell lysis and barcoded reverse transcription of RNAs from single cells are finished inside each GEM. Barcoded cDNA product was recovered through post GEM-RT cleanup and PCR amplification. cDNA QC & quantification were determined by High Sensitivity D5000 DNA Screen Tape analysis (Agilent Technologies) and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). 50ng of cDNA was used for 5ʹ Gene Expression (GEX) library construction and each sample was indexed by Chromium i7 Sample Index Kit, which was run on Illumina HiSeq4000 sequencer with 2×100 bp paired reads to achieve a depth of at least 50,000 read pairs per cell.
Single-cell T Cell Receptor (TCR) repertoire sequencing
TCR α/β sequencing was performed with 10× Genomics Single Cell V(D)J Immune Profiling Solution (10×Genomics Inc.). Briefly, full-length V, D, and J-gene segments were amplified from barcoded cDNA using Chromium Single Cell V(D)J Enrichment Kit (Human T Cell) to generate enrichment products. Enriched product was measured by D5000 DNA Screen Tape analysis and Qubit dsDNA HS Assay Kit, and then 50 ng of enrichment TCR product was used for library construction. Single Cell V(D)J enriched libraries were pooled for sequencing on HiSeq4000 to produce paired 2×150 bp reads, taking into account depth requirement (5,000 read pairs per cell) between the pooled libraries.
Hybrid capture sequencing of marker genes of CapID
xGen Lockdown probes for 207 gene (Supplementary Table 2) were designed and synthesized by Integrated DNA Technologies, Inc. (IDT). Hybridization capture of DNA libraries was performed using xGen hybridization and wash kit (IDT Inc.). First, pool 500 ng of each library to multiplex 8–12 libraries in a low-bind tube and add the blocker components, including Human Cot DNA and xGen blocking oligos. Dry down the mixture in a SpeedVac system. Second, thaw all contents of the xGen Hybridization and Wash Kit to room temperature, and prepare the Hybridization Master Mix according to the manufacturer’s protocol. Perform hybridization capture reaction in a thermal cycler with 100 ºC lid at a HYB program (95ºC, 30 sec; 65 ºC, overnight). Third, prepare wash buffers and equilibrate Streptavidin beads at room temperature for a minimum of 30 min before performing the washes. In the thermal cycler with 70 ºC lid, perform bead capture at 65 ºC for 45 min. Every 10–12 min, gently pipette to ensure the sample is fully resuspended. At the end of the 45 min, take the sample off the thermal cycler. Proceed immediately to heated washes and room temperature washes. Finally, Perform post-capture PCR and purify post-capture PCR fragment. We employed 10% of post-capture elute as template to run real-time quantitative PCR to determine the optimal number of PCR cycles for the custom probes panel. The captured library was measured using TapeStation System and Qubit dsDNA HS Assay Kit, and run on Illumina Miseq.
Single cell RNA-sequencing bioinformatics
Raw sequencing data processing, QC, data filtering, and normalization.
The raw single cell RNA sequencing (scRNA-seq) data were pre-processed (demultiplex cellular barcodes, read alignment, and generation of feature-barcode matrix) using Cell Ranger (10x Genomics, v2.1.1). Detailed QC metrics were generated and evaluated. Genes detected in <3 cells and cells where < 200 genes had nonzero counts were filtered out and excluded from subsequent analysis. Low quality cells where >15% of the read counts derived from the mitochondrial genome were also discarded. In addition, cells with number of detected genes >7,000 were discarded to remove likely doublet or multiplet captures. In this study, all sequencing libraries were constructed using the same version of reagent kits, by the same individual, following the same protocols, on the same 10X Chromium controller, and the libraries were sequenced on the same Illumina HiSeq 4000 platform. The results of principal component analysis (PCA), t-SNE plots and sample-by-cluster (Supplementary Figure 1), flow-cell-by-cluster (Supplementary Figure 10), and bead-lot-by-cluster (Supplementary Figure 10) distribution were carefully reviewed and integrated information revealed minimal batch effects. Seurat46 was applied to the filtered gene-cell matrix to generate the normalized UMI counts as previously described47.
Unsupervised cell clustering and dimensionality reduction.
Seurat46 v3.0.2 was applied to the normalized gene-cell matrix to identify highly variable genes. The elbow plot was generated with the ElbowPlot function of Seurat46 and based on which, the number of significant principal components (PCs) were determined. Different resolution parameters for unsupervised clustering were then examined in order to determine the optimal number of clusters. For this study, the first 20 PCs and 1,437 highly variable genes identified by Seurat46 were used for unsupervised clustering analysis with the resolution set to 0.6, yielding a total of 17 cell clusters. The t-distributed stochastic neighbor embedding (t-SNE) method was used for dimensionality reduction and 2-D visualization of the single cell clusters.
Determination of major cell types and cell states.
The major cell type (CD4 and CD8) was defined by marker gene expression (CD4, CD40LG, TNFRSF4, CD8A, CD8B) by 10X transcriptome and CapID sequencing data with an SNN boosting procedure. The functional state of each single cell (activated, memory, exhausted, regulatory) was determined using a hierarchical classification schema recently described by Sade-Feldman et al.21. All canonical marker genes in this schema were included on the CapID hybrid capture panel for sensitive detection (Supplementary Table 2) and cell type was automatically assigned to each individual cell based on expression status of these genes21. On average, this supervised classification approach led to the unambiguous classification of 72.5% of T cells. CAR-positive T cells were identified by the presence (normalized UMI >0) of CAR-specific sequence contigs48 (FMC63-CD19scFV, GenBank: HM852952.1) in the aligned reads and this approach identified an average of 24% of QC-passed T cells as CAR-positive (range 7.1% - 42.6%). To describe the cell types and states that were defined by each tSNE cluster, we performed a manual review of the differentially expressed genes (DEGs) that were identified for each cell cluster by Seurat46 (Extended Data 1, Supplementary Table 12).
Capture-sequencing based identification plus SNN boosting (CapID+) for cell type classification.
Gene dropouts are common events in scRNA-seq data and represent a challenge for cell type identification. For the remaining unclassified cells (27.5%) in above steps, we evaluated the gene dropout events and applied two strategies to improve cell type identification. First, we employed CapID sequencing data to assign positive and negative expression. Second, we applied an SNN (shared nearest neighbor) boosting approach. Briefly, for each cell cluster, we first examined the diversity (the composition of cell types detected) and dominance (‘purity’, the proportion of cells in the cluster that are representative of a particular cell type) of classified cell types through supervised approach as described above. The cell type diversity of each cluster was evaluated by normalized Shannon entropy, the lower the Shannon entropy value, the lower the diversity and the higher the dominance of a classified cell type within each individual cell cluster. SNN boosting was performed on the high-dominance clusters (dominance score ≥ 0.8 and normalized Shannon entropy ≤ 0.25, the clusters that are dominated by a single cell type) with the cluster size ≥ 50 cells. Cell type was assigned based on the hypothesis that phenotypically similar cells are grouped together, and cell type annotation was updated and used for SNN boosting in the next iteration. The boosting process was iterated on each cell cluster for 100 times (or when all unclassified cells were assessed). On average, this process rescued additional 19.3% of cells in total (70% of unclassified cells) and improved the identification of CAR-positive cell from 24% to 35%.
Inferring cell cycle stage, hierarchical clustering, differentially expressed genes (DEGs), and pathway enrichment analysis.
The cell cycle stage was computationally assigned to each individual cell using the R code implemented in Seurat46 based on expression profiles of the cell cycle-related signature genes, as previously described49. Differentially expressed genes (DEGs) were identified for each cluster using the FindMarkers function of in Seurat R package46 and DEG list was filtered with the following criteria: the gene should expressed in 20% or more cells in the more abundant group; the absolute expression fold change >1.2; and FDR q-value <0.05. Hierarchical clustering was performed for each cell type using the Ward’s minimum variance method. Heat map was then generated using the heatmap function in pheatmap R package for filtered DEGs. For pathway analysis, the curated gene sets were downloaded from the Molecular Signature Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/index.jsp), single-sample gene set enrichment analysis (ssGSEA) was applied and pathway scores were calculated for each cell using the GSVA software package50. The CD8 T-cell memory phenotype was defined by enrichment of a CD8 memory gene signature21 in CD8 subset and the T-cell dysfunction phenotype was defined by enrichment of T-cell dysfunction-related markers51. Pathway enrichment analysis was done with the limma R software package. Significant signaling pathways were identified with FDR q-value < 0.05. Differences in CD27, CCR7, LAG3, and TIM3 (HAVCR2) that were identified by 10X whole-transcriptome data were validated by CapID to increase their sequencing saturation and thereby mitigate transcript dropout. There was no significant difference in the number of reads per cell between clinical response (CR vs PR/PD) or molecular response (>5FMR vs <5FMR) groups, further supporting little or no contribution of transcript dropout to differences observed between these groups (Supplementary Figure 10).
Identification and characterization of the IAC cell cluster.
To identify ICANS associated cell cluster(s) (IAC cluster), we computed the proportion of cells assigned to a given cluster from each sample for the CRES-high and CRES-low/no groups, and applied a two-sided Wilcoxon rank-sum test to determine the significance. P-values were adjusted for multiple testing using the Benjamini-Hochberg method and false discovery rates (FDR q-value) were calculated. The IAC cluster was then identified and characterized. IAC feature genes were identified by performed DEG analysis between the IAC and the rest of other (non-IAC) cell clusters and the top 20 highly variable DEGs were selected as the signature genes of IAC, followed by a manual evaluation and annotation process. To further determine the cell phenotype of IAC, we performed GSEA and calculated GSEA scores for the IAC signature genes using the curated gene expression data matrix from a genome-wide transcriptomic analysis of protein coding genes in human blood cells33. The nonparametric two-sided Wilcoxon rank-sum test was used to determine the significance between the myeloid cell lineages (classical monocyte, intermediate monocyte, non-classical monocyte, myeloid DC, neutrophil, basophil, eosinophil, plasmacytoid DC) and non-myeloid cell types (naive CD4 T-cell, naïve CD8 T-cell, memory CD4 T-cell, memory CD8 T-cell, T-regulatory cell, MAIT T-cell, gdT-cell, NK cell, naïve B-cell, memory B-cell).
For each cell in the CapID data from the discovery (n=24) and validation (n=16) cohort, the IAC gene signature score was calculated as the average gene expression level of IAC signature genes identified in the discovery cohort. IAC scores were first examined across cell clusters, which confirmed the expected significantly higher score in cells originally assigned to the IAC cluster (p < 0.001), as determined by their unique cell barcodes. IACs were therefore classified as cells with IAC signature gene scores > 1.5, which identified a total 440 IACs across 40 patients from the discovery and validation cohorts. The qualitative difference (present vs absent) between patients with grade 0–2 ICANS vs patients with grade 3–4 ICANS was tested using a Fisher’s exact test. The quantitative difference in IAC frequency between patients with grade 0–2 ICANS vs patients with grade 3–4 ICANS was tested using a Wilcoxon rank-sum test.
TCR V(D)J sequence assembly, paired clonotype calling, TCR diversity and clonality analysis and integration with scRNA-seq data.
Cell Ranger v3.0.2 for V(D)J sequence assembly was applied for TCR reconstruction and paired TCR clonotype calling. The CDR3 motif was located and the productivity was determined for each single cell. The clonotype landscape was then assessed and the clonal fraction of each identified clonotype was calculated. The TCR clonotype diversity matrix was calculated using the tcR R package 52. TCR clonality was defined as 1-Peilou’s evenness and was calculated on productive rearrangements as previously described 53. Clonality values approaching 0 indicate a very even distribution of clone frequencies, whereas values approaching 1 indicate an increasingly asymmetric distribution in which a few clones are present at high frequencies. The TCR clonotype data was then integrated with the T-cell phenotype data inferred from single cell gene expression analysis based on the shared cell barcodes.
Statistical analysis
In addition to the bioinformatics approaches described above for scRNA/TCR-seq data analysis, all other statistical analysis was performed using statistical software R v3.5.2. Analysis of differences in immunological features (continuous variables) between patient groups (R vs. NR; CRES-high vs. CRES-low/no) was determined by the nonparametric two-sided Wilcoxon rank-sum test. To control for multiple hypothesis testing, we applied the Benjamini-Hochberg method to correct p values and the false discovery rates (q-values) were calculated. All statistical significance testing was two-sided and results were considered statistically significant at p-value < 0.05. In R v3.5.2, 2.2e-16 is the smallest value that can be accurately calculated. Therefore some highly significant p-values or q-values are represented as p<2.2e-16 or q<2.2e-16, respectively.
Cell-free DNA sequencing
DNA extraction
Cell-free DNA was extracted from 2mL of plasma for each time point using Qiagen Circulating Nucleic Acid kits (Qiagen), and quality assessed using a TapeStation instrument and D1000 high sensitivity tapes (Agilent). Genomic DNA was extracted from peripheral blood mononuclear cells of each patient using Qiagen AllPrep Mini kits (Qiagen) and used as a germline control. DNA was quantified using Qubit high-sensitivity reagents (ThermoFisher).
Library preparation, hybrid capture and sequencing
Libraries were prepared from up to 150ng of cfDNA using Kapa Hyper Prep Kits (Roche) and XGen CS Adapters (IDT). In brief, DNA was end-repaired and a-tailed, then ligated using a 1:10 molar ratio of stubby duplex unique molecular identifiers (UMIs). Index-ligated products were cleaned up with AmPure beads (Beckman Coulter), subjected to up to 8 cycles of PCR with oligos containing unique sequencing adapters, and cleaned up with AmPure beads. Libraries were 12-plexed and captured using a 196kb Nimblegen SeqCap EZ custom reagent (Roche) targeting previously described recurrently mutated regions30. Enriched DNA was amplified with up to 8 cycles of PCR, cleaned up with AmpPure beads and sequenced on a single lane of a HiSeq 4000 instrument using 100bp paired-end reads at the MD Anderson Sequencing and Microarray Facility. Samples were sequenced to an average coverage of 12,427X (min 5,921X, max 21,856X).
Mutation detection and enumeration
The read data was demultiplexed into two read files and one UMI file, all in .fastq format. The read files were first combined to form an unaligned BAM file (uBAM) with UMI information using the FastqToBam tool of fgbio version 0.6.1. An interleaved paired fastq file was generated using fgbio’s SamToFastq tool and was used align to the hg19 genome using BWA-MEM. UMI information was mapped from the uBAM file to the aligned reads using Picard 2.18.9 / 2.9.0’s MergeBamAlignment tool. The alignments with UMI information then undergo several pre-processing steps using GATK version 3.7, including, in that order, indel realignment and two rounds of base quality recalibration. True PCR duplicates were then removed by using Picard’s UmiAwareMarkDuplicatesWithMateCigar tool. Variants were called from the calibration time point (day 0–1) using the intersection of VarScan254 and the GATK Unified Genotyper55. Variants detected within sequencing of PBMC samples from the same patient were treated as germline and filtered. Variants in repetitive elements, the dbSNP data (build 151), the ExAC database of 60,706 healthy subjects56, or identified at variant allele fractions of >0.1% by deep sequencing of 24 healthy controls were filtered. An average of 11 variants were detected per patient (min=0, max=43; Table S1) and 18/23 patients with available calibration samples passed the required threshold of ≥3 variants for subsequent monitoring. These variants were enumerated (supporting reads divided by total reads over the position) in data from each time point using Picard tools.
Definition of early molecular response
The fold change in variant allele fraction was calculated for each variant relative to the calibration time point. The mean of the fold changes for all variants was calculated for every patient at each time point. The day 6/7 fold change in variant allele fraction was tested for association with clinical response by dichotomizing patients into those that achieved complete response vs those with partial response or progressive disease at their 3 month follow-up. The association between quantitative fold change in variant allele fraction and clinical response group was tested using a log rank test. Patients were thereafter split into two groups of equal size with available follow-up data (n=8) using a 5-fold reduction in variant allele fraction as the threshold, referred to as 5-fold molecular response (5FMR).
Extended Data
Extended Data 1:
Heatmap showing the top 50 signature genes of each cluster and putative assignments to cell types according to canonical marker genes.
Extended Data 2: Increased sequencing saturation and marker gene detection rate by CapID.
a) Sub-sampling of reads from a single CAR T infusion product with 600 million reads for whole transcriptome and 20 million reads for CapID, showing the saturation (flattening of curve) for CapID (orange) at ~10 million reads, for 10X whole transcriptome sequencing (blue) at ~400M reads, and the effect of supplementing whole transcriptome data with 10 million reads of CapID data (green). b) Density plots from the entire dataset show the reduced number of cells with UMI counts of zero and increased signal-to-noise ratio for CapID sequencing compared to 10X whole transcriptome sequencing. The 10X whole transcriptome sequencing in this study were performed to an average of 73,521 reads per cell, vastly exceeding the minimum of 20,000 reads per cell recommended by 10X.
Extended Data 3: Correlation of cell frequencies measured by scRNA-seq and flow cytometry.
Correlations are shown for 16 patients that had sufficient cells for flow cytometry (a), compared to the fractions measured using traditional 10X data (b) or CapID+ (c). All comparisons showed a significant correlation with Pearson’s correlation 2-tailed P-value < 0.001.
Extended Data 4: Volcano plots of differentially expressed genes between CAR-positive and CAR-negative CD8 and CD4 T-cells.
Q-values were calculated with a two-sided Wilcoxon rank sum test with Bonferonni correction.
Extended Data 5: Variant allele fractions of somatic variants detected by cfDNA sequencing.
a-b) Comparison of the average VAF of mutations at day 0 (a) and the number of calibrated mutations (b) between clinical response groups. P values were calculated by a two-sided Student’s t-test. c) Raw variant allele frequencies for each patient are shown for >5FMR (above) and <5FMR (below) groups. Lines are colored by clinical response as in figure 4a. The grey dashed line represents the 5-fold reduction threshold for each patient.
Extended Data 6: T-cell clonotypic diversity in patients grouped by clinical and molecular response.
a) The frequency of the top 10 clonotypes for each patient among all cells (above), CD4 T-cells (middle) and CD8 T-cells (below). Box, median +/− interquartile range. Whiskers, minimum and maximum. P-values calculated by a two-sided Wilcoxon rank sum test with Benjamini-Hochberg correction. b) Shannon’s clonality score for patients grouped by clinical or molecular response, shown for all cells (above), CD4 T-cells (middle) and CD8 T-cells (below). CR, n=9. PR/PD, n=14. >5FMR, n=8. <5FMR, n=9. Box, median +/− interquartile range. Whiskers, minimum and maximum. P-values calculated by a two-sided Wilcoxon rank sum test with Benjamini-Hochberg correction.
Extended Data 7: Analysis of association between molecular features of CAR T-cell infusion products and the development of high-grade cytokine release syndrome (CRS).
a) Comparison of functional states between patients with grade 0–2 and grade 3–4 identified reduced frequencies of exhausted CD8 T-cells and increased frequencies of exhausted CD4 T-cells to be associated with the development of high-grade CRS. Q-values were calculated by a two-sided Fisher exact test with a Benjamini-Hochberg correction. b-c) Heatmaps show differentially expressed genes between CD4 T-cells (b) and CD8 T-cells. (c) from the infusion products of patients with grade 0–2 CRS versus those that developed grade 3–4 CRS. The CD69 gene shows higher expression and the CCL3 and CLL4 genes show lower expression on both CD4 and CD8 T-cells from the CAR T-cell infusion products of patients that developed high grade CRS. All differentially expressed genes are shown in Supplementary Tables 10 and 11.
Extended Data 8: Percentage of CAR-positive cells in patients with grade 0–2 vs grade 3–4 ICANS.
Grade 0–2 ICANS, n=22. Grade 3–4 ICANS, n=18. Box, median +/− interquartile range. Whiskers, minimum and maximum. P-values calculated by a two-sided Wilcoxon rank-sum test.
Extended Data 9: Cytokine levels in serum between patients with IACs and those without IACs.
Significance level was tested with Mann-Whitney U test. FDR q-value was calculated for multiple testing correction.
Extended Data 10: Quantification of the ICANS-associated cells (IACs) signature by scGSVA in CAR T-cell infusion products.
a) A stringent threshold was set to ensure high confidence classification of IACs by scGSVA analysis of the 109 signature genes measured by CapID, as shown for the cells that were originally identified as IACs by unsupervised clustering of 10X whole-transcriptome data in figure 4a. b) The distribution of scGSVA scores shows a clear difference between infusion products from patients with grade 0–2 ICANS vs patients with grade 3–4 ICANS. The threshold for classification of cells as IACs (scGSVA score >1.5) is shown.
Supplementary Material
Acknowledgements:
This work was supported by the Schweitzer Family Fund (JRW, RED, MRG), the U.T. MD Anderson Cancer Center B-cell Lymphoma Moonshot (SSN and MRG), an NCI cancer center support grant to the University of Texas MD Anderson Cancer Center (P30 CA016672), and the IRG start-up research funds provided to LW and MRG by U.T. MD Anderson Cancer Center. HY is supported by a Fellow award from the Leukemia and Lymphoma Society. MRG is supported by a Scholar award from the Leukemia and Lymphoma Society.
Competing Interests:
B.C. reports advisory board membership for Advanced Accelerator Application and Clovis Oncology. N.F. reports advisory board membership for Roche, BMS, Gilead, and Novartis, research funding from Roche, BMS, Novartis and BostonGene, and employment from BostonGene. H.C.L. reports consulting fees from Adaptive Biotechnologies, Amgen, Celgene, GlaxoSmithKline, Janssen, Sanofi and Takeda, and research funding from Amgen, Celgene, Daiichi Sankyo, GlaxoSmithKline, Janssen, Regeneron and Takeda. L.J.N. reports personal fees and research fees from Celgene, Genentech, Juno, Merck, and TG Therapeutics, and personal fees from Bayer, Novartis, and Spectrum pharmaceuticals. J.W. reports advisory board membership for Kite/Gilead, Juno/Celgene/BMS, Novartis, Genentech, Janssen, Amgen, Astra Zeneca, Curis and Morphosys, and research funding from Kite/Gilead, June/Celgene/BMS, Novartis, Genentech, Janssen, Astra Zeneca, 47, Unum, Curis and Morphosys. S.S.N. has received research support from Kite/Gilead, Cellectis, Poseida, Merck, Acerta, Karus, BMS, Unum Therapeutics, Allogene, and Precision Biosciences; served as consultant and advisory board member for Kite/Gilead, Celgene, Novartis, Unum Therapeutics, Pfizer, Merck, Precision Biosciences, Cell Medica, Incyte, Allogene, Calibr, and Legend Biotech; has royalty income from Takeda Pharmaceuticals; has patents related to cell therapy. M.R.G. reports consulting for VeraStem Oncology and stock ownership interest in KDAc Therapeutics.
References:
- 1.Locke FL et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol 20, 31–42, doi: 10.1016/S1470-2045(18)30864-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuster SJ et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. The New England journal of medicine 380, 45–56, doi: 10.1056/NEJMoa1804980 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Shah NN & Fry TJ Mechanisms of resistance to CAR T cell therapy. Nature reviews. Clinical oncology 16, 372–385, doi: 10.1038/s41571-019-0184-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neelapu SS et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. The New England journal of medicine 377, 2531–2544, doi: 10.1056/NEJMoa1707447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller KT et al. Clinical Pharmacology of Tisagenlecleucel in B-cell Acute Lymphoblastic Leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research 24, 6175–6184, doi: 10.1158/1078-0432.CCR-18-0758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green MR & Neelapu SS Not So FASt: Tumor Cells Resisting Death Drive CAR T-cell Dysfunction. Cancer discovery 10, 492–494, doi: 10.1158/2159-8290.CD-20-0037 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Das RK, Vernau L, Grupp SA & Barrett DM Naive T-cell Deficits at Diagnosis and after Chemotherapy Impair Cell Therapy Potential in Pediatric Cancers. Cancer discovery 9, 492–499, doi: 10.1158/2159-8290.CD-18-1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finney OC et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. The Journal of clinical investigation 130, doi: 10.1172/JCI125423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommermeyer D. et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia 30, 492–500, doi: 10.1038/leu.2015.247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraietta JA et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nature medicine 24, 563–571, doi: 10.1038/s41591-018-0010-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi J. et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 132, 804–814, doi: 10.1182/blood-2018-01-828343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee DW et al. ASTCT Consensus Grading for Cytokine Release Syndrome and Neurologic Toxicity Associated with Immune Effector Cells. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 25, 625–638, doi: 10.1016/j.bbmt.2018.12.758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neelapu SS et al. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nature reviews. Clinical oncology 15, 47–62, doi: 10.1038/nrclinonc.2017.148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teachey DT et al. Identification of Predictive Biomarkers for Cytokine Release Syndrome after Chimeric Antigen Receptor T-cell Therapy for Acute Lymphoblastic Leukemia. Cancer discovery 6, 664–679, doi: 10.1158/2159-8290.CD-16-0040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay KA et al. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor-modified T-cell therapy. Blood 130, 2295–2306, doi: 10.1182/blood-2017-06-793141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le RQ et al. FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 23, 943–947, doi: 10.1634/theoncologist.2018-0028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santomasso BD et al. Clinical and Biological Correlates of Neurotoxicity Associated with CAR T-cell Therapy in Patients with B-cell Acute Lymphoblastic Leukemia. Cancer discovery 8, 958–971, doi: 10.1158/2159-8290.CD-17-1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taraseviciute A. et al. Chimeric Antigen Receptor T Cell-Mediated Neurotoxicity in Nonhuman Primates. Cancer discovery 8, 750–763, doi: 10.1158/2159-8290.CD-17-1368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gust J. et al. Endothelial Activation and Blood-Brain Barrier Disruption in Neurotoxicity after Adoptive Immunotherapy with CD19 CAR-T Cells. Cancer discovery 7, 1404–1419, doi: 10.1158/2159-8290.CD-17-0698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norelli M. et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nature medicine 24, 739–748, doi: 10.1038/s41591-018-0036-4 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Sade-Feldman M. et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013e1020, doi: 10.1016/j.cell.2018.10.038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn SD et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nature immunology 10, 29–37, doi: 10.1038/ni.1679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doering TA et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144, doi: 10.1016/j.immuni.2012.08.021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quigley M. et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nature medicine 16, 1147–1151, doi: 10.1038/nm.2232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nature immunology 20, 326–336, doi: 10.1038/s41590-019-0312-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Man K. et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141e1125, doi: 10.1016/j.immuni.2017.11.021 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Li H. et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789e718, doi: 10.1016/j.cell.2018.11.043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldrath AW, Luckey CJ, Park R, Benoist C & Mathis D The molecular program induced in T cells undergoing homeostatic proliferation. Proceedings of the National Academy of Sciences of the United States of America 101, 16885–16890, doi: 10.1073/pnas.0407417101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josefsson SE et al. TIGIT and PD-1 Mark Intratumoral T Cells with Reduced Effector Function in B-cell Non-Hodgkin Lymphoma. Cancer Immunol Res 7, 355–362, doi: 10.1158/2326-6066.CIR-18-0351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer F. et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Science translational medicine 8, 364ra155, doi: 10.1126/scitranslmed.aai8545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B, Sharron M, Montaner LJ, Weissman D & Doms RW Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proceedings of the National Academy of Sciences of the United States of America 96, 5215–5220, doi: 10.1073/pnas.96.9.5215 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazazi F, Mathijs JM, Foley P & Cunningham AL Variations in CD4 expression by human monocytes and macrophages and their relationships to infection with the human immunodeficiency virus. J Gen Virol 70 ( Pt 10), 2661–2672, doi: 10.1099/0022-1317-70-10-2661 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Uhlen M. et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366, doi: 10.1126/science.aax9198 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Gargett T, Truong N, Ebert LM, Yu W & Brown MP Optimization of manufacturing conditions for chimeric antigen receptor T cells to favor cells with a central memory phenotype. Cytotherapy 21, 593–602, doi: 10.1016/j.jcyt.2019.03.003 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Wang J. et al. Histone Deacetylase Inhibitors and IL21 Cooperate to Reprogram Human Effector CD8(+) T Cells to Memory T Cells. Cancer Immunol Res, doi: 10.1158/2326-6066.CIR-19-0619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz DM et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 125, 3679–3687, doi: 10.1182/blood-2015-03-635169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roschewski M. et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 16, 541–549, doi: 10.1016/S1470-2045(15)70106-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurtz DM et al. Circulating Tumor DNA Measurements As Early Outcome Predictors in Diffuse Large B-Cell Lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36, 2845–2853, doi: 10.1200/JCO.2018.78.5246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherkassky L. et al. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. The Journal of clinical investigation 126, 3130–3144, doi: 10.1172/JCI83092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gargett T. et al. GD2-specific CAR T Cells Undergo Potent Activation and Deletion Following Antigen Encounter but can be Protected From Activation-induced Cell Death by PD-1 Blockade. Mol Ther 24, 1135–1149, doi: 10.1038/mt.2016.63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong EA et al. PD-1 blockade modulates chimeric antigen receptor (CAR)-modified T cells: refueling the CAR. Blood 129, 1039–1041, doi: 10.1182/blood-2016-09-738245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumeister SH, Freeman GJ, Dranoff G & Sharpe AH Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol 34, 539–573, doi: 10.1146/annurev-immunol-032414-112049 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Burugu S, Dancsok AR & Nielsen TO Emerging targets in cancer immunotherapy. Seminars in cancer biology 52, 39–52, doi: 10.1016/j.semcancer.2017.10.001 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Strati P. et al. Clinical efficacy of anakinra to mitigate CAR T-cell therapy-associated toxicity in large B-cell lymphoma. Blood Adv 4, 3123–3127, doi: 10.1182/bloodadvances.2020002328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 45.Fraietta JA et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312, doi: 10.1038/s41586-018-0178-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savas P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 24, 986–993, doi: 10.1038/s41591-018-0078-7 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kochenderfer JN et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother 32, 689–702, doi: 10.1097/CJI.0b013e3181ac6138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jerby-Arnon L. et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984–997e924, doi: 10.1016/j.cell.2018.09.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanzelmann S, Castelo R & Guinney J GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7, doi: 10.1186/1471-2105-14-7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Leun AM, Thommen DS & Schumacher TN CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer 20, 218–232, doi: 10.1038/s41568-019-0235-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nazarov VI et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics 16, 175, doi: 10.1186/s12859-015-0613-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuben A. et al. TCR Repertoire Intratumor Heterogeneity in Localized Lung Adenocarcinomas: An Association with Predicted Neoantigen Heterogeneity and Postsurgical Recurrence. Cancer Discov 7, 1088–1097, doi: 10.1158/2159-8290.CD-17-0256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research 22, 568–576, doi: 10.1101/gr.129684.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research 20, 1297–1303, doi: 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lek M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291, doi: 10.1038/nature19057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All requests for raw and analyzed data and materials are promptly reviewed by the University of Texas MD Anderson Cancer Center to verify if the request is subject to any intellectual property or confidentiality obligations. Patient-related data not included in the paper may be subject to patient confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. Transcriptome and CapID scRNA-seq datasets are available through the gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accessions GSE150992 and GSE151511, respectively. Raw data used in the generation of Figures 1, 2, 3, 4, 5, and 6, and Extended Data 1 to 10 are available through the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home), accessions EGAD00001006327 and EGAD00001006325.