Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) targets the respiratory and gastric epithelium, causing coronavirus disease 2019 (COVID‐19). Tissue antigen expression variations influence host susceptibility to many infections. This study aimed to investigate the closely linked Lewis (FUT3) and ABO histo‐blood types, including secretor (FUT2) status, to infections with SARS‐CoV‐2 and the corresponding severity of COVID‐19.
Study Design and Methods
Patients (Caucasians, n = 338) were genotyped for ABO, FUT3, and FUT2, and compared to a reference population of blood donors (n = 250,298). The association between blood types and severity of COVID‐19 was addressed by dividing patients into four categories: hospitalized individuals in general wards, patients admitted to the intensive care unit with and without intubation, and deceased patients. Comorbidities were considered in subsequent analyses.
Results
Patients with blood type Lewis (a−b−) or O were significantly less likely to be hospitalized (odds ratio [OR] 0.669, confidence interval [CI] 0.446–0.971, OR 0.710, CI 0.556–0.900, respectively), while type AB was significantly more prevalent in the patient cohort (OR 1.519, CI 1.014–2.203). The proportions of secretors/nonsecretors, and Lewis a+ or Lewis b+ types were consistent between patients and controls. The analyzed blood groups were not associated with the clinical outcome as defined.
Discussion
Blood types Lewis (a−b−) and O were found to be protective factors, whereas the group AB is suggested to be a risk factor for COVID‐19. The antigens investigated may not be prognostic for disease severity, but a role for ABO isoagglutinins in SARS‐CoV‐2 infections is strongly suggested.
Keywords: ABO, COVID‐19, host–pathogen interaction, Lewis, secretor
1. INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) targets the respiratory mucosa, and it can infect and replicate in the gastric and intestinal epithelium, causing coronavirus disease 2019 (COVID‐19).1, 2 This emerging infectious disease is usually characterized by an acute respiratory infection, with symptoms varying from mild to severe, including pneumonia, respiratory failure, coagulopathy, multiple organ failure, and death.3, 4 COVID‐19 is a complex and multifactorial disease, where inherited predispositions together with existing comorbidities and acquired risk factors are likely to influence the severity of the disease.
Differences in blood group antigen expression can increase or decrease host susceptibility to many infections.5 Pathogens including viruses, bacteria, and eukaryotic parasites carry lectins on their surfaces, which can bind to glycan structures such as blood group antigens on the host cell surface as a first step in the infection process. Blood group antigen structures may be involved in the binding of their toxins, facilitating invasion and colonization or evasion of host clearance mechanisms. They can also serve as false receptors, preventing pathogen binding to target tissues. In addition, it was reported that microorganisms can stimulate antibodies against blood group antigens, including ABO and Lewis.5, 6 Molecular mimicry can modify the innate immune response to infections, when directed against enveloped viruses that express blood group‐like carbohydrate antigens.7
The carbohydrate histo‐blood group antigens Lewis and ABO are widely expressed in many tissues, including respiratory and gastric mucosa, endothelium, kidney, and heart.8 The Lewis enzyme (fucosyltransferase 3), encoded by FUT3, is responsible for the last step in the biosynthesis of Lewis antigens.9 The immunodominant glycan structures defining the ABO antigens are synthesized by glycosyltransferases, encoded by the ABO gene.10 The expression of soluble ABO(H) antigens in secretory epithelium is regulated by Fucosyltransferse 2 (Fut 2), encoded by the FUT2 gene.11 The activity of fucosyltransferase 2 (Fut 2) is also required for the production of Lewis b (Le b) antigen, and reflected by the Le (a−b+) phenotype. In contrast, the Le (a+b−) phenotype is found in ABH nonsecretors (18%–22%).8, 11
The naturally occurring anti‐A and anti‐B isoagglutinins have a high titer and avidity, activate complement, and consist of mainly IgM and to a lesser extent IgG. Different titers of these antibodies may be the result of environmental rather than hereditary factors.12, 13, 14
The soluble ABH and Le b antigens, which are found in the respiratory and gastric mucosa of secretors, may influence the mechanisms of SARS‐CoV‐2 invasion to the target tissues. The interaction between ABO, FUT2, and FUT3 impacts the amount of soluble ABH and Lewis antigens present15 and thus may increase the risk for a severe course of the disease.
Lewis antigens were identified as Helicobacter pylori virulence factors, enabling bacterial adherence to and invasion of gastric epithelial cells.16, 17, 18, 19 A relationship between ABO/Lewis phenotype, gastric ulcers, chronic H. pylori infections, and urinary tract infections has been discussed.20, 21, 22, 23, 24 ABO blood types have been reported to affect susceptibility to several infections,25, 26, 27, 28 and an additional role for ABO glycans and glycosyltransferases in inflammatory vascular diseases, cardiovascular diseases, and acute respiratory distress syndrome has been suggested.29, 30, 31, 32, 33, 34, 35, 36, 37
European and American studies support preliminary investigations in China regarding an association between the ABO blood group system and SARS‐CoV‐2 infections.38, 39, 40, 41, 42, 43, 44, 45, 46, 47 The secretor phenotype was suggested to possibly moderate disease progression, especially among type A carriers.47 A contribution of the Lewis blood type to infection with SARS‐CoV‐2 has not yet been studied.
This is the first study investigating the closely linked Lewis and ABO blood group systems and blood group secretor type simultaneously, relative to the severity of COVID‐19 in hospitalized patients. As recently discussed,48 our results strongly implicate a possible role for the ABO isoagglutinins in the process of infection with SARS‐CoV‐2.
2. STUDY DESIGN AND METHODS
2.1. Patients
This retrospective study included 338 Caucasian patients, over the age of 18, admitted to the Department of Internal Medicine at the University Hospital Graz and to Landeskrankenhaus Graz II, with a diagnosis of COVID‐19, between March and May 2020 during the first wave of coronavirus infection in Austria. All study participants tested positive for SARS‐CoV‐2 RNA by real‐time PCR (qPCR). After extraction using the EMAG® platform (bioMérieux S.A., Marcy l'Etoile, France), nucleic acids were amplified using the RIDA® GENE SARS‐CoV‐2 (r‐biopharm, Darmstadt, Germany) with the LightCycler® 480 II (Roche Molecular Diagnostics, Rotkreuz, Switzerland). Additionally, the Cobas® SARS‐CoV‐2 test (Roche Molecular Systems, Branchburg, NJ) was applied on the Cobas® 6800/8800 system (Roche Molecular Diagnostics).49
The diagnosis of COVID‐19 was established based on the national guidelines published by the Austrian Ministry of Health.50 Admission referred to COVID‐19‐related hospitalization, and mortality was defined as all‐cause mortality in SARS‐CoV‐2 infected patients.51, 52
Demographic information and concurrent diagnoses, based on the International Classification of Diseases (ICD‐10), were obtained from the electronic health records of patients.
The study was approved by the local Ethics Committee at the Medical University of Graz (32‐436 ex 19/20).
2.2. Genetic analysis
Nasal‐ and pharyngeal swab specimens of the 338 patients were used for the extraction of human DNA with the Qiamp DNA Micro Kit (Qiagen, GmbH, Germany).
The ABO region, containing exons 6 and 7, was amplified using the primer pair ABO_Ex6_F (5′‐GCCTCTCTCCATGTGCAGTA‐3′),53 and ABO_Ex7_R (5′‐CCTAGGCTTCAGTTACTCAC‐3′). Sanger sequencing was done with ABO_In6_223R (5′‐GCCTCTGGAGAAGGAGCT‐3′) and ABO_Ex7_F1 (5′‐CATCGCTGGGAAGAGGATGAAGTG‐3′) as recently described.54
The single nucleotide polymorphisms (SNPs) c.261delG (rs1556058284), c.297A>G (rs8176720), c.467C>T (rs1053878), c.526C>G (rs7853989), c.646T>A (rs8176740), c.681G>A (rs8176742), c.703G>A (rs8176743), c.771C>T (rs8176745), c.796C>A (rs8176746), c.802G>A (rs41302905), c.803G>C (rs8176747), c.829G>A (rs8176748), c.930G>A (rs8176749), and c.1061delC (rs56392308) were analyzed to discriminate the presence of ABO*A1, A2, B.O1, O.01, O.02, or O.03 alleles according to Olsson et al.55 and the ISBT database.
The FUT2 gene was investigated by restriction fragment length polymorphism analysis using recombinant AvaII (R0153S, New England Biolabs, Frankfurt, Germany) for detecting the c.428G>A (rs601338) inactivating SNP.56 The presence of this variant, which is the most common FUT2 null allele in Caucasians (47%–49.5%),57 was used to determine secretor status. Homozygosity of c.428A results in the nonsecretor phenotype.
The coding region of FUT3 was analyzed using Sanger Sequencing. The detected SNPs were assigned to the corresponding functional or nonfunctional alleles as reported before.58
The Lewis types were inferred from the results of FUT2 and FUT3 genotyping. If at least one functional FUT3 allele was present, secretors were predicted to have the Lewis type Le b+, whereas nonsecretors were predicted to have type Le a+. Homozygous nonfunctional FUT3 genotypes were defined as Le (a−b−), regardless the FUT2 genotype detected. Using this method, it was not possible to predict the Le (a+b+) phenotype as a separate proportion. However, the frequency of the Le (a+b+) phenotype as observed in our blood donors is very low (0.4%), and therefore was considered to be negligible for the purposes of our analysis. The reliability of this approach was previously validated, and confirmed by consistent results for serologic and genetic Lewis typing as reported before.58
2.3. Blood donors
Data about the ABO and Lewis blood groups of Styrian blood donors (n = 250,298) were used to calculate the blood type distributions of healthy controls. Repeat donors' data were excluded. A selection bias due to selective retention of blood group O individuals in our blood donor campaign can be ruled out. Serological ABO typing was performed on the Olympus automated blood group testing system (Olympus PK7300, Beckman Coulter, Hamburg, Germany). Lewis blood groups were determined with standard serologic and gel matrix techniques (MicroTyping System, Bio‐Rad).
The secretor status of randomly selected blood donors (n = 480) was defined by FUT2 genotyping, as described earlier for the patients. Genomic DNA was prepared from peripheral blood leucocytes from the donors' blood by silica‐magnetic particle technology using a DNA purification kit and a biorobot system (EZ1 DSP DNA and EZ1 Advanced XL, respectively, Qiagen GmbH, Germany).
2.4. Statistical analysis
The COVID‐19 cohort was classified into four categories reflecting symptom severity: patients hospitalized in the general ward (1); patients who needed admission to the intensive care unit (ICU) without (2) and with intubation (3); and deceased patients (4). A broader outcome classification that distinguished between surviving and deceased patients was included.
The distribution of blood types in hospitalized COVID‐19 patients was compared to that of Styrian blood donors (n = 250,298 for ABO, n = 7241 for Lewis) by means of chi‐squared tests, along with odds ratios (ORs) with 95% confidence intervals (CI).
The association between blood groups and symptom severity was investigated with chi‐squared tests or by Fisher's exact test as appropriate. Group differences in continuous variables were conducted with Kruskal–Wallis tests. To test the contribution of blood types to patients' outcomes (deceased vs. recovered), a logistic regression, adjusting for potential confounders, was performed. These confounders comprised age, sex, and predictors with p < .02 in the univariable analysis (malignant neoplasm, diabetes mellitus, hypertension, and acute kidney injury/chronic kidney disease). Adjusted ORs with 95% CI were generated. A p value of ≤.05 was considered significant. All statistical analyses were conducted using R version 4.0.2 (https://www.r-project.org).
3. RESULTS
3.1. Characteristics of the COVID‐19 patients
Of the 338 individuals, more females (n = 187) were hospitalized than males (n = 151). The females (median age of 80.0 [IQR = 69.0–88.0]) were significantly older than the males (median age of 74.0 [IQR = 64.5–80.0]), p < .001. A higher proportion of males (70.4%, n = 19) than females (29.6%, n = 8), p = .004, underwent treatment in the ICU. Furthermore, males had higher rates of malignant neoplasm (12.6%, n = 19) than females (5.9%, n = 11), p = .031, diabetes mellitus (males: 20.5%, n = 31; females: 12.3%, n = 23), p = .040, and gastrointestinal diseases (males: 19.9%, n = 30; females: 9.6%, n = 18), p = .007. The mortality rate among male patients was 26.5% (n = 40), versus 20.3% (n = 38) of female patients, which was not significantly different, p = .181.
In Table 1, the demographics and comorbidities of the patients in relation to the four categories reflecting symptom severity are outlined.
TABLE 1.
Demographics and patient comorbidities presented by symptom severity
| Total (N = 338) | General ward (N = 233) | ICU no intubation (N = 10) | ICU with intubation (N = 17) | Deceased (N = 78) | p | |
|---|---|---|---|---|---|---|
| Sex | .008a | |||||
| Male | 151 (44.7%) | 92 (39.5%) | 8 (80.0%) | 11 (64.7%) | 40 (51.3%) | |
| Female | 187 (55.3%) | 141 (60.5%) | 2 (20.0%) | 6 (35.3%) | 38 (48.7%) | |
| Age (years) | <.001b | |||||
| Median | 77.0 | 76.0 | 60.0 | 67.0 | 82.5 | |
| Min–Max | 23.0–100.0 | 23.0–99.0 | 36.0–77.0 | 51.0–79.0 | 53.0–100.0 | |
| Hospitalization (days) | <.001b | |||||
| Median | 11.0 | 12.0 | 11.0 | 28.0 | 8.5 | |
| Min–Max | 1.0–133.0 | 1.0–133.0 | 6.0–30.0 | 11.0–100.0 | 1.0–90.0 | |
| Concurrent conditions | ||||||
| COPD | 27 (8.0%) | 17 (7.3%) | 3 (30.0%) | 1 (5.9%) | 6 (7.7%) | .123a |
| Malignant neoplasm | 30 (8.9%) | 23 (9.9%) | 0 (0.0%) | 3 (17.6%) | 4 (5.1%) | .254a |
| AKI/CKD | 70 (20.7%) | 42 (18.0%) | 0 (0.0%) | 2 (11.8%) | 26 (33.3%) | .009a |
| Diabetes mellitus | 54 (16.0%) | 32 (13.7%) | 1 (10.0%) | 2 (11.8%) | 19 (24.4%) | .161a |
| Coronary artery disease | 14 (4.1%) | 7 (3.0%) | 1 (10.0%) | 2 (11.8%) | 4 (5.1%) | .128a |
| Deep vein thrombosis | 6 (1.8%) | 4 (1.7%) | 0 (0.0%) | 1 (5.9%) | 1 (1.3%) | .498a |
| Gastrointestinal diseases | 48 (14.2%) | 33 (14.2%) | 1 (10.0%) | 5 (29.4%) | 9 (11.5%) | .289a |
| Hypertensive diseases | 173 (51.2%) | 127 (54.5%) | 4 (40.0%) | 8 (47.1%) | 34 (43.6%) | .324a |
| Infectious and parasitic diseases (excluding Coronavirus) | 12 (3.6%) | 7 (3.0%) | 2 (20.0%) | 0 (0.0%) | 3 (3.8%) | .109a |
Note: Demographic data and underlying chronic diseases of the patients, in relation to the 4 categories of symptom severity.
Abbreviations: AKI, acute kidney injury; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; ICU, intensive care unit.
The association of symptom severity with demographic and clinical information was tested by means of Fisher's exact tests for categorical variables.
The association of symptom severity with demographic and clinical information was tested by means of Kruskal–Wallis rank sum tests for continuous variables. The proportion of males/females, age, days of hospitalization, as well as the proportion of patients with acute kidney injury/chronic kidney disease, differed significantly among the four groups.
3.2. Lewis and ABO types and frequencies of secretors/nonsecretors
Of the pool of 338 hospitalized patients, DNA quality issues prevented the successful analysis of two ABO and two FUT3 genotypes, leaving a sample size of 336 patients for ABO‐ and Lewis‐related analyses. The data set for secretor status was complete (n = 338).
Table 2 indicates the proportions of the Lewis and ABO blood types, as well as the frequencies of secretors/nonsecretors in the reference sample of blood donors, as compared to the cohort of COVID‐19 patients. The frequencies of the different blood types observed in the blood donors are largely in line with data reported among Europeans or Caucasian individuals.8, 59
TABLE 2.
Frequencies of blood types in healthy blood donors versus hospitalized patients and their associations with COVID‐19
| Blood type | Blood donors in % (N) | COVID‐19 patients in % (N) | p | OR | 95% CI |
|---|---|---|---|---|---|
| O | 37.4 (93,579) | 29.8 (100) | .008a | 0.710 | 0.556–0.900 |
| A | 42.7 (106,861) | 44.9 (151) | 1.095 | 0.878–1.366 | |
| B | 13.7 (34,164) | 16.1 (54) | 1.211 | 0.888–1.626 | |
| AB | 6.3 (15,694) | 9.2 (31) | 1.519 | 1.014–2.203 | |
| Secretor | 82.3 (395) | 84.0 (284) | .404a | 1.132 | 0.767–1.679 |
| Nonsecretor | 17.7 (85) | 16.0 (54) | |||
| Le a+ | 14.8 (1070) | 14.6 (49) | .083a | 0.985 | 0.707–1.347 |
| Le b+ | 71.6 (5186) | 75.9 (255) | 1.247 | 0.962–1.630 | |
| Le (a−b−) | 13.6 (985) | 9.5 (32) | 0.669 | 0.446–0.971 |
Note: The proportions of the ABO and Lewis blood types, as well as the frequencies of secretors and nonsecretors in COVID‐19 patients are compared against the population of blood donors. The odds ratios of having a specific blood type against the other blood types in patients are reported. Secretors were compared to Nonsecretors. Le a+ were compared to Non‐Le a+, Le b+ were compared to Non‐Le b+, and Le (a−b−) were tested versus Le a+ and Le b+ types.
Abbreviations: CI, confidence interval; N, number of individuals typed for a specific ABO type in the cohort of blood donors and the patients; OR, odds ratio.
p‐Values refer to chi‐squared tests.
No significant differences were found in the proportion of secretors/nonsecretors and Lewis types between the patients and the blood donors. However, the Le (a−b−) type was present more frequently in the blood donors than in the patients (13.6% vs. 9.5%). The distribution of ABO blood types in the COVID‐19 patients differed significantly from the distribution observed in the blood donors (p = .008).
Additionally, the cooperative interaction of ABO blood type with secretor status was analyzed by comparing the proportion of secretors versus nonsecretors in each ABO group in patients versus blood donors. No significant differences in the distributions were found (A secretor/A nonsecretor, p = .312; B secretor/B nonsecretor, p = .999; AB secretor/AB nonsecretor, p = .996; O secretor/O nonsecretor, p = .816).
3.3. Association of blood types with COVID‐19
The odds ratio of patients in the COVID‐19 study group with Lewis type Le (a−b−) was significantly lower, than those with other Lewis types (Le a+ and Le b+), as shown in Table 2. The blood type O was detected significantly less often in the COVID‐19 patients, as compared to the other ABO types (A, B, AB), and the odds ratio of patients with blood type AB was significantly higher.
The proportions of blood types did not differ significantly in the four categories of hospitalized patients (Table 3). Both univariable and multivariable analyses revealed no significant contribution of the blood types Lewis, ABO, or secretor/nonsecretor status to the outcome “deceased” versus “recovered” (Table 4).
TABLE 3.
Blood types in relation to the severity of COVID‐19
| Blood type | General ward (N = 233) | ICU without intubation (N = 10) | ICU with intubation (N = 17) | Deceased (N = 78) | p |
|---|---|---|---|---|---|
| ABO | .925a | ||||
| O | 68 (29.4%) | 5 (50.0%) | 5 (29.4%) | 22 (28.2%) | |
| A | 105 (45.5%) | 3 (30.0%) | 6 (35.3%) | 37 (47.4%) | |
| B | 37 (16.0%) | 1 (10.0%) | 4 (23.5%) | 12 (15.4%) | |
| AB | 21 (9.1%) | 1 (10.0%) | 2 (11.8%) | 7 (9.0%) | |
| Secretor status | .801a | ||||
| Secretor | 192 (82.4%) | 9 (90.0%) | 15 (88.2%) | 68 (87.2%) | |
| Nonsecretor | 41 (17.6%) | 1 (10.0%) | 2 (11.8%) | 10 (12.8%) | |
| Lewis | .917a | ||||
| Le a+ | 36 (15.6%) | 1 (10.0%) | 2 (11.8%) | 10 (12.8%) | |
| Le b+ | 170 (73.6%) | 8 (80.0%) | 14 (82.4%) | 63 (80.8%) | |
| Le (a−b−) | 25 (10.8%) | 1 (10.0%) | 1 (5.9%) | 5 (6.4%) |
Note: The investigated blood types are indicated in relation to the four hospitalized patient categories reflecting symptom severity.
Abbreviation: ICU, Intensive care unit.
Fisher's exact test.
TABLE 4.
Blood types and the risk for the outcomes “deceased” or “recovered”
| Blood type | Deceased (N = 78) | Recovered (N = 260) | OR univariable | CI | p | AOR multivarable | 95% CI | p |
|---|---|---|---|---|---|---|---|---|
| A | 37 (47.4%) | 114 (44.2%) | Reference | |||||
| AB | 7 (9.0%) | 24 (9.3%) | 0.89 | 0.33–2.14 | .805 | 1.12 | 0.36–3.20 | .840 |
| B | 12 (15.4%) | 42 (16.3%) | 0.87 | 0.40–1.79 | .719 | 0.77 | 0.32–1.81 | .562 |
| O | 22 (28.2%) | 78 (30.2%) | 0.86 | 0.47–1.56 | .627 | 1.28 | 0.64–2.54 | .486 |
| Secretor | 68 (87.2%) | 216 (83.1%) | Reference | |||||
| Nonsecretor | 10 (12.8%) | 44 (16.9%) | 0.75 | 0.34–1.53 | .453 | 0.71 | 0.29–1.6 | .421 |
| Le b+ | 63 (80.8%) | 192 (74.4%) | Reference | |||||
| Le a+ | 10 (12.8%) | 39 (15.1%) | 0.80 | 0.36–1.65 | .565 | 0.73 | 0.30–1.66 | .469 |
| Le (a−b−) | 5 (6.4%) | 27 (10.5%) | 0.56 | 0.19–1.41 | .260 | 0.64 | 0.20–1.74 | .408 |
Note: The ABO types AB, B, and O were tested compared to blood group A. Nonsecretors were compared to secretors, and patients with the Le (a−b−) and Le a+ types were compared to individuals positive for Le b. Multivariable analyses were conducted, adjusting for sex, age, and concurrent diagnoses (cancer, diabetes mellitus, hypertension, and acute kidney injury/chronic kidney disease).
Abbreviations: AOR, adjusted odds ratio; CI, confidence interval; OR, odds ratio.
4. DISCUSSION
This study investigated the contribution of Lewis, ABO, and secretor type to COVID‐19 outcomes in hospitalized Caucasian patients.
Consistent with the sex ratio of people infected with SARS‐CoV‐2 as reported by the Austrian Federal Office for Safety in Health Care,60 more females than males needed inpatient treatment in this study. This may be explained by the higher percentage of females in the 60‐and‐over age group in the Austrian population. However, more males than females were admitted to the ICU, which can be explained by the comorbidities from which predominantly the males suffered. Consistent with the clinical characteristics of COVID‐19 cohorts investigated in Italy, Spain, and the USA, the patients predominantly suffered from comorbidities such as hypertension (51.2%) and diabetes mellitus (16.0%). There were higher rates of AKI/CKD (20.7%), whereas CAD was present less frequently (4.1%), compared to recently reported studies.43, 44, 61, 62, 63, 64 The deceased patients (23.01%) were older (median age = 82.5), than the survivors, regardless of sex, and had a higher incidence of AKI/CKD (33.3%) and diabetes mellitus (24%).
Our results revealed a significantly different frequency of ABO blood types in the patients, compared to the frequencies observed in a healthy control group. Proportions of the Lewis blood types and frequencies of secretors and nonsecretors did not differ significantly in our COVID‐19 sample group, as compared to the healthy population. None of our analyses predicted a certain blood type to be a risk factor for a severe course of the disease.
4.1. Association between the Lewis type and COVID‐19
The Le (a−b−) type appeared to be at least a mitigating factor against hospitalization with COVID‐19. It may therefore be speculated that the presence of certain fucosylated glycosphingolipids in Le (a+b−) and Le (a−b+) phenotypes may also play a role in the progression of this infectious disease. Mucins, secreted by epithelial cells, are highly glycosylated proteins, and are suggested to be substrates for active Fut 3, which is expressed in secretory mucosa. The composition of bound and mobile mucins, including the presence or absence of a certain type of glycan, may influence the binding affinities or the persistence of infectious organisms in secretory tissues.65, 66
The frequency of possessing an inactive Lewis gene responsible for the Le (a−b−) blood type is much higher among African Americans than among Europeans and Caucasian Americans (22%–29% vs. 4%–11%). However, the Le (a+b+) phenotype is very rare (0% to <1%) in European and Caucasian Americans, it has an incidence of 6%–25% and 27% in Taiwan Chinese and Hong Kong Chinese, respectively.8, 67, 68 A possible impact of the Lewis carbohydrate structures may merit further investigation, especially with regard to their prevalence in African Americans and Chinese.
This study confirms previous evidence reporting a protective effect of ABO blood group O, and a higher risk for COVID‐19 associated with blood type AB.43, 47, 69, 70 Our data are different from studies also reporting risk associations for the blood types A38, 40, 42, 45 or B.43 As well, the ABO types were not associated with higher odds of suffering from more severe COVID‐19 symptoms in our study.45, 47, 70 Our data are in line with previous results, regarding disease severity, confirming no association between the ABO type and the risk of intubation or of a fatal outcome of COVID‐19 infection among hospitalized individuals.43, 71 Likewise, the ABO blood type of critically ill patients with COVID‐19 was not related to 28‐day mortality in another study performed.44 Our results do not confirm the protective effect of nonsecretor phenotype as observed by Valenti et al.47
Divergent observations regarding associations of ABO blood types may be explained by different blood group distributions observed in different geographic regions or ethnicities.8, 72, 73
Recently, adhesion of the SARS‐CoV‐2 receptor binding domain to A antigen on a solid phase glycan microarray was demonstrated in vitro, indicating a possible contribution of ABO(H) antigens to the infection.74 The lower incidence of individuals with ABO blood type O, accompanied by a higher incidence of individuals with blood type AB in our cohort of patients, suggests that the variable susceptibility to the infection with SARS‐CoV‐2 could also be related to an interference caused by circulating ABO antibodies (Figure 1). The lack of any significant differences regarding the interactions of the ABO and secretor types in the cohort of patients compared to the blood donors, strengthens our theory that emphasis should not be put on the ABH antigenic structures, but rather on the isoagglutinins.
FIGURE 1.
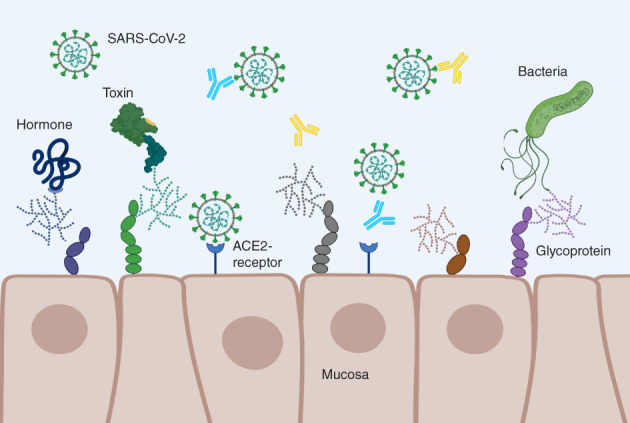
Schematic illustration of mucosa cells with glycosylated structures and receptor–ligand interactions. Hormones, antibodies, as well as viruses and bacteria or their toxins, interact with oligosaccharide epitopes (glycoproteins) that serve as receptors or co‐receptors. The suggested interference of ABO antibodies with SARS‐CoV‐2 targeting of host cells via angiotensin‐converting enzyme 2 (ACE‐2) receptor is indicated. Made with Biorender.com
SARS‐CoV‐2 is an enveloped virus. It targets host cells via interaction of the viral adhesion glycoprotein, SARS‐CoV spike (S) protein, with the angiotensin‐converting enzyme 2 (ACE‐2), which serves as its cellular receptor.75, 76 The S protein consists of a complex glycan structure, which was reported to be capable of supporting ABH epitopes in an in vitro study investigating infections with SARS‐CoV in 2008.77 The S protein, experimentally modified to express A antigen, was effectively blocked by high titer (>1:256) monoclonal anti‐A and human anti‐A. Inhibitory effects of anti‐B were not investigated in their experiments. As the virus targets respiratory and gastrointestinal mucosa, it was suggested that human isolates express ABH antigens on the S protein and host envelope glycosphingolipids by utilizing the host's enzymes and post‐translational glycosylation machineries.77, 78, 79 Thus, the ability of ABO antibodies to decrease the risk of initial infection may depend on ABO incompatibility between the infected and exposed individuals. However, the presence of ABO antibodies may at least delay the spread of the virus among people, particularly when an individual is exposed to aerosols with only a low viral load. The rates of infection with SARS‐CoV‐2 throughout the population may be further influenced by the titer of ABO isoagglutinins and the incidence of blood group O in the affected population or region.
To conclude, our findings suggest that Lewis antigens contribute to infection with SARS‐CoV‐2. Furthermore, we confirm, ABO blood group O, with obligatory anti‐A and anti‐B antibodies present, to be protective against COVID‐19. The blood type AB, lacking those isoagglutinins, is suggested to increase the risk of infection. There are therefore strong indications that ABO antibodies affect susceptibility to COVID‐19, and should be the subject of further research. Sophisticated in vitro studies investigating the mechanism of virus–host cell interactions should be included.
4.2. Limitations of the study
Some limitations of the present study should be kept in mind. Our sample size was restricted to patients hospitalized with COVID‐19. Future studies should take into account the whole cohort of individuals tested positive for SARS‐CoV‐2 to investigate the relationship between the disease and blood group types. The sample size of 338 patients is relatively small compared to other studies on this topic.43, 44, 45, 47, 70 As this is a retrospective observational study, we cannot rule out the fact that unmeasured confounding factors may have influenced the outcome.
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
ACKNOWLEDGMENTS
Anja Stoisser, Annika Lampl, Lisa Rohrhofer, and Marie‐Therese Frisch are gratefully acknowledged for their technical assistance. We thank Wolfgang Helmberg for extracting blood group data from our donor database.
The study was supported by research funding from the City of Graz. Web‐based Resources: Federal Ministry, Republic of Austria: Social Affairs, Health Care, and Consumer Protection. New Coronavirus (COVID‐19): https://www.sozialministerium.at/en/Coronavirus/New‐coronavirus‐(COVID‐19).html. ISBT database: http://www.isbtweb.org/fileadmin/user_upload/Working_parties/WP_on_Red_Cell_Immunogenetics_and/001_ABO_Alleles_v1.2.pdf.
Matzhold EM, Berghold A, Bemelmans MKB, et al. Lewis and ABO histo‐blood types and the secretor status of patients hospitalized with COVID‐19 implicate a role for ABO antibodies in susceptibility to infection with SARS‐CoV‐2. Transfusion. 2021;61:2736–2745. 10.1111/trf.16567
Funding information City of Graz
REFERENCES
- 1.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma C, Cong Y, Zhang H. COVID‐19 and the digestive system. Am J Gastroenterol. 2020;115:1003–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu R, Yi S, Zhang J, Lv Z, Zhu C, Zhang Y. Viral load dynamics in sputum and nasopharyngeal swab in patients with COVID‐19. J Dent Res. 2020;99:1239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centeres for Disease Control and Prevention. Coronavirus (COVID‐19) [monograph on the internet]. https://www.cdc.gov/coronavirus/2019-ncov/index.html. Accessed 28 Sept 2020.
- 5.Cooling L. Blood groups in infection and host susceptibility. Clin Microbiol Rev. 2015;28:801–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel‐Johnson J, Leitman S, Klein H, Alter H, Lee‐Stroka A, Scheinberg P, et al. Probiotic‐associated high‐titer anti‐B in a group A platelet donor as a cause of severe hemolytic transfusion reactions. Transfusion. 2009;49:1845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur CM, Patel SR, Mener A, Kamili NA, Fasano RM, Meyer E, et al. Innate immunity against molecular mimicry: examining galectin‐mediated antimicrobial activity. Bioessays. 2015;37:1327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels G. Human blood groups. 3rd ed.Oxford: Wiley‐Blackwell; 2013. [Google Scholar]
- 9.Kukowska‐Latallo JF, Larsen RD, Nair RP, Lowe JB. A cloned human cDNA determines expression of a mouse stage‐specific embryonic antigen and the Lewis blood group alpha(1,3/1,4)fucosyltransferase. Genes Dev. 1990;4:1288–303. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo‐blood group ABO system. Nature. 1990;345:229–33. [DOI] [PubMed] [Google Scholar]
- 11.Clausen H, Hakomori S. ABH and related histo‐blood group antigens; immunochemical differences in carrier isotypes and their distribution. Vox Sang. 1989;56:1–20. [DOI] [PubMed] [Google Scholar]
- 12.Redman M, Malde R, Contreras M. Comparison of IgM and IgG anti‐A and anti‐B levels in Asian, Caucasian and Negro donors in the North West Thames Region. Vox Sang. 1990;59:89–91. [DOI] [PubMed] [Google Scholar]
- 13.Mazda T, Yabe R, NaThalang O, Thammavong T, Tadokoro K. Differences in ABO antibody levels among blood donors: a comparison between past and present Japanese, Laotian, and Thai populations. Immunohematology. 2007;23:38–41. [PubMed] [Google Scholar]
- 14.Xu Y, Lee JG, Yan JJ, Ryu JH, Xu S, Yang J. Human B1 cells are the main blood group A‐specific B cells that have a moderate correlation with anti‐A antibody titer. Ann Lab Med. 2020;40:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achermann FJ, Julmy F, Gilliver LG, Carrel TP, Nydegger UE. Soluble type A substance in fresh‐frozen plasma as a function of ABO and secretor genotypes and Lewis phenotype. Transfus Apher Sci. 2005;32:255–62. [DOI] [PubMed] [Google Scholar]
- 16.Lozniewski A, Haristoy X, Rasko DA, Hatier R, Plénat F, Taylor DE, et al. Influence of Lewis antigen expression by Helicobacter pylori on bacterial internalization by gastric epithelial cells. Infect Immun. 2003;71:2902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boren T, Normark S, Falk P. Helicobacter pylori: molecular basis for host recognition and bacterial adherence. Trends Microbiol. 1994;2:221–8. [DOI] [PubMed] [Google Scholar]
- 18.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–5. [DOI] [PubMed] [Google Scholar]
- 19.Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, et al. Helicobacter pylori virulence factors‐mechanisms of bacterial pathogenicity in the gastric microenvironment. Cell. 2020;10(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stowell CP, Stowell SR. Biologic roles of the ABH and Lewis histo‐blood group antigens part I: infection and immunity. Vox Sang. 2019;114:426–42. [DOI] [PubMed] [Google Scholar]
- 21.Lindstrom K, Breimer ME, Jovall PA, Lanne B, Pimlott W, Samuelsson BE. Non‐acid glycosphingolipid expression in plasma of an A1 Le(a‐b+) secretor human individual: identification of an ALeb heptaglycosylceramide as major blood group component. J Biochem. 1992;111:337–45. [DOI] [PubMed] [Google Scholar]
- 22.Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins LC, de Oliveira Corvelo TC, Oti HT, do Socorro Pompeu Loiola R, Aguiar DCF, dos Santos Barile KA, et al. ABH and Lewis antigen distributions in blood, saliva and gastric mucosa and H pylori infection in gastric ulcer patients. World J Gastroenterol. 2006;12:1120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheinfeld J, Schaeffer AJ, Cordon‐Cardo C, Rogatko A, Fair WR. Association of the Lewis blood‐group phenotype with recurrent urinary tract infections in women. N Engl J Med. 1989;320:773–7. [DOI] [PubMed] [Google Scholar]
- 25.Rowe JA, Handel IG, Thera MA, Deans A‐M, Lyke KE, Koné A, et al. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci U S A. 2007;104:17471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo‐blood group type. J Infect Dis. 2002;185:1335–7. [DOI] [PubMed] [Google Scholar]
- 27.Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën‐Clouet N. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J Infect Dis. 2005;192:1071–7. [DOI] [PubMed] [Google Scholar]
- 28.Batool Z, Durrani SH, Tariq S. Association of Abo and Rh blood group types to hepatitis B, hepatitis C, Hiv and syphilis infection, a five Year' experience in healthy blood donors in a tertiary care hospital. J Ayub Med Coll Abbottabad. 2017;29:90–2. [PubMed] [Google Scholar]
- 29.He M, Wolpin B, Rexrode K, Manson JE, Rimm E, Hu FB, et al. ABO blood group and risk of coronary heart disease in two prospective cohort studies. Arterioscler Thromb Vasc Biol. 2012;32:2314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung CM, Wang RY, Chen JW, Fann CSJ, Leu H‐B, Ho H‐Y, et al. A genome‐wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J. 2010;10:537–44. [DOI] [PubMed] [Google Scholar]
- 31.Whincup PH, Danesh J, Walker M, Lennon L, Thomson A, Appleby P, et al. von Willebrand factor and coronary heart disease: prospective study and meta‐analysis. Eur Heart J. 2002;23:1764–70. [DOI] [PubMed] [Google Scholar]
- 32.Pare G, Chasman DI, Kellogg M, Zee RYL, Rifai N, Badola S, et al. Novel association of ABO histo‐blood group antigen with soluble ICAM‐1: results of a genome‐wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, et al. Large‐scale genomic studies reveal central role of ABO in sP‐selectin and sICAM‐1 levels. Hum Mol Genet. 2010;19:1863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, et al. Genetic variants in ABO blood group region, plasma soluble E‐selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19:1856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reilly JP, Meyer NJ, Shashaty MGS, Feng R, Lanken PN, Gallop R, et al. ABO blood type A is associated with increased risk of ARDS in whites following both major trauma and severe sepsis. Chest. 2014;145:753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezoagli E, Gatti S, Villa S, Villa G, Muttini S, Rossi F, et al. ABO blood types and major outcomes in patients with acute hypoxaemic respiratory failure: a multicenter retrospective cohort study. PLoS One. 2018;13:e0206403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiggins KL, Smith NL, Glazer NL, Rosendaal FR, Heckbert SR, Psaty BM, et al. ABO genotype and risk of thrombotic events and hemorrhagic stroke. J Thromb Haemost. 2009;7:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Yang Y, Huang H, Li D, Gu D, Lu X, et al. Relationship between the ABO blood group and the COVID‐19 susceptibility. Clin Infect Dis. 2020;ciaa1150. 10.1093/cid/ciaa1150. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan Q, Zhang W, Li B, Li D‐J, Zhang J, Zhao F. Association between ABO blood group system and COVID‐19 susceptibility in Wuhan. Front Cell Infect Microbiol. 2020;10:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Feng Z, Li P, Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID‐19. Clin Chim Acta. 2020;509:220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnkob MB, Pottegard A, Stovring H, Haunstrup TM, Homburg K, Larsen R, et al. Reduced prevalence of SARS‐CoV‐2 infection in ABO blood group O. Blood Adv. 2020;4:4990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe Covid‐19 with respiratory failure. N Engl J Med. 2020;383(16):1522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latz CA, DeCarlo C, Boitano L, Png CYM, Patell R, Conrad MF, et al. Blood type and outcomes in patients with COVID‐19. Ann Hematol. 2020;99:2113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leaf RK, Al‐Samkari H, Brenner SK, Gupta S, Leaf DE. ABO phenotype and death in critically ill patients with COVID‐19. Br J Haematol. 2020;190:e204–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muniz‐Diaz E, Llopis J, Parra R, Roig I, Ferrer G, Grifols J, et al. Relationship between the ABO blood group and COVID‐19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfus. 2021;19:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pairo‐Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID‐19. Nature. 2021;591:92–8. [DOI] [PubMed] [Google Scholar]
- 47.Valenti L, Villa S, Baselli G, Temporiti R, Bandera A, Scudeller L, et al. Association of ABO blood group and secretor phenotype with severe COVID‐19. Transfusion. 2020;60:3067–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Focosi D. Anti‐A isohaemagglutinin titres and SARS‐CoV‐2 neutralization: implications for children and convalescent plasma selection. Br J Haematol. 2020;190:e148–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poljak M, Korva M, Knap Gasper N, Komloš KF, Sagadin M, Uršič T, et al. Clinical evaluation of the cobas SARS‐CoV‐2 test and a diagnostic platform switch during 48 hours in the midst of the COVID‐19 pandemic. J Clin Microbiol. 2020;58(6):e00599–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Federal Ministry Republic of Austria SA, Health Care and Consumer Protection . New Coronavirus (COVID‐19) [monograph on the internet]. https://www.sozialministerium.at/en/Coronavirus/New-coronavirus-(COVID-19).html. Accessed 03 April 2020.
- 51.Skok K, Stelzl E, Trauner M, Kessler HH, Lax SF. Post‐mortem viral dynamics and tropism in COVID‐19 patients in correlation with organ damage. Virchows Arch. 2021;478(2):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lax SF, Skok K, Trauner M, all coauthors . Pulmonary arterial thrombosis as an important complication of COVID‐19 pulmonary disease: letter to the editor. Virchows Arch. 2020;477:467–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferri G, Pelotti S. Multiplex ABO genotyping by minisequencing. Methods Mol Biol. 2009;496:51–8. [DOI] [PubMed] [Google Scholar]
- 54.Matzhold EM, Drexler C, Wagner A, Bernecker C, Pessentheiner A, Bogner‐Strauß JG, et al. A 24‐base pair deletion in the ABO gene causes a hereditary splice site defect: a novel mechanism underlying ABO blood group O. Transfusion. 2020;60:1564–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olsson ML, Chester MA. Polymorphism and recombination events at the ABO locus: a major challenge for genomic ABO blood grouping strategies. Transfus Med. 2001;11:295–313. [DOI] [PubMed] [Google Scholar]
- 56.Serpa J, Mendes N, Reis CA, Santos Silva LF, Almeida R, Le Pendu J, et al. Two new FUT2 (fucosyltransferase 2 gene) missense polymorphisms, 739G–>A and 839T–>C, are partly responsible for non‐secretor status in a Caucasian population from Northern Portugal. Biochem J. 2004;383:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Koda Y, Soejima M, Pang H, Schlaphoff T, du Toit ED, et al. Extensive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum Genet. 1998;103:204–10. [DOI] [PubMed] [Google Scholar]
- 58.Matzhold EM, Helmberg W, Wagner T, Drexler C, Ulrich S, Winkler A, et al. Identification of 14 new alleles at the fucosyltransferase 1, 2, and 3 loci in Styrian blood donors, Austria. Transfusion. 2009;49:2097–108. [DOI] [PubMed] [Google Scholar]
- 59.Reinhold Eckstein RZ. Immunhhämatologie und klinische Transfusionsmedizin. 6th ed.; München: Elsevier GmbH, Urban & Fischer; 2010. [Google Scholar]
- 60.Norbert Handra AC, Puchhammer C, Popovic N, Ali Chakeri ZE‐K, Kanitz E, Maritschnik S, et al. SARS‐CoV2‐Infektion: Täglicher Lagebericht für Österreich. Wien: AGES AIS; 2021. [Google Scholar]
- 61.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, et al. Clinical characteristics of Covid‐19 in New York City. N Engl J Med. 2020;382:2372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colaneri M, Sacchi P, Zuccaro V, Biscarini S, Sachs M, Roda S, et al. Clinical characteristics of coronavirus disease (COVID‐19) early findings from a teaching hospital in Pavia, North Italy, 21 to 28 February 2020. Euro Surveill. 2020;25(16):2000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casas‐Rojo JM, Anton‐Santos JM, Millan‐Nunez‐Cortes J, Lumbreras‐Bermejo C, Ramos‐Rincón JM, Roy‐Vallejo E, et al. Clinical characteristics of patients hospitalized with COVID‐19 in Spain: results from the SEMI‐COVID‐19 Registry. Rev Clin Esp. 2020;220:480–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borobia AM, Carcas AJ, Arnalich F, Álvarez‐Sala R, Monserrat‐Villatoro J, Quintana M, et al. A cohort of patients with COVID‐19 in a major teaching hospital in Europe. J Clin Med. 2020;9(6):1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host‐microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stowell SR, Stowell CP. Biologic roles of the ABH and Lewis histo‐blood group antigens part II: thrombosis, cardiovascular disease and metabolism. Vox Sang. 2019;114:535–52. [DOI] [PubMed] [Google Scholar]
- 67.Molthan L. Lewis phenotypes of American Caucasians, American Negroes and their children. Vox Sang. 1980;39:327–30. [DOI] [PubMed] [Google Scholar]
- 68.Miller EB, Rosenfield RE, Vogel P, Haber G, Gibbel N. The Lewis blood factors in American Negroes. Am J Phys Anthropol. 1954;12:427–43. [DOI] [PubMed] [Google Scholar]
- 69.Abdollahi A, Mahmoudi‐Aliabadi M, Mehrtash V, Jafarzadeh B, Salehi M. The novel coronavirus SARS‐CoV‐2 vulnerability association with ABO/Rh blood types. Iran J Pathol. 2020;15:156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zietz M, Zucker J, Tatonetti NP. Assocciations between blood types and COVID‐19 infection, intubation, and death. Nat Commun. 2020;11(1):5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dzik S, Eliason K, Morris EB, Kaufman RM, North CM. COVID‐19 and ABO blood groups. Transfusion. 2020;60:1883–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu J, Zhang S, Wang Q, Shen H, Zhang Y, Liu M. Frequencies and ethnic distribution of ABO and RhD blood groups in China: a population‐based cross‐sectional study. BMJ Open. 2017;7:e018476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garratty G, Glynn SA, McEntire R, Retrovirus Epidemiology Donor S. ABO and Rh(D) phenotype frequencies of different racial/ethnic groups in the United States. Transfusion. 2004;44:703–6. [DOI] [PubMed] [Google Scholar]
- 74.Wu SC, Arthur CM, Wang J, Verkerke H, Josephson CD, Kalman D, et al. The SARS‐CoV‐2 receptor‐binding domain preferentially recognizes blood group A. Blood Adv. 2021;5:1305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science. 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guillon P, Clement M, Sebille V, Rivain J‐G, Chou C‐F, Ruvoën‐Clouet N, et al. Inhibition of the interaction between the SARS‐CoV spike protein and its cellular receptor by anti‐histo‐blood group antibodies. Glycobiology. 2008;18:1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS‐coronavirus replication/transcription complexes are membrane‐protected and need a host factor for activity in vitro . PLoS Pathog. 2008;4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knoops K, Kikkert M, Worm SH, Zevenhoven‐Dobbe JC, van der Meer Y, Koster AJ, et al. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]


