Abstract
The current pandemic caused by a novel coronavirus (SARS‐CoV‐2) has underlined the importance of emerging diseases of zoonotic importance. Along with human beings, several species of wild and pet animals have been demonstrated to be infected by SARS‐CoV‐2, both naturally and experimentally. In addition, with constant emergence of new variants, the species susceptibility might further change which warrants intensified screening efforts. India is a vast and second most populated country, with a habitat of a very diverse range of animal species. In this study we place on record of SARS‐CoV‐2 infections in three captive Asiatic lions. Detailed genomic characterization revealed involvement of Delta mutant (Pango lineage B.1.617.2) of SARS‐CoV‐2 at two different locations. Interestingly, no other feline species enclosed in the zoo/park were found infected. The epidemiological and molecular analysis will contribute to the understanding of the emerging mutants of SARS‐CoV‐2 in wild and domestic animals.
Keywords: COVID‐19, Delta mutant, lions, SARS‐CoV‐2
1. INTRODUCTION
The entire world is currently struggling with the pandemic of Coronavirus Disease 2019 (COVID‐19), caused by newly emerged beta coronavirus, SARS‐CoV‐2 (Gorbalenya et al., 2020). The disease is characterized by development of atypical pneumonia and pyrexia which was later termed by WHO as COVID‐19 (https://www.who.int). Virus has spread to almost all the countries in a very short span, and by June 2021, over 177 million people have been infected and more than 0.38 million succumbed (https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019; accessed 20 June 2021).
The precise origin of SARS‐CoV‐2 is still unknown but the closest ancestor of SARS‐CoV‐2 is bat coronavirus (RaTG13‐2013) which probably entered into humans via one or more intermediate host (Zhou et al., 2020; Andersen et al., 2020). Increasing evidence indicates the newly emerging variants may find a reservoir host other than humans, hence continuous screening of captive and free range wild animals is important.
Several lineages/clades and their sub‐lineages have emerged due to accumulation of mutations during spread in the population. Among several variants of concern (VOC), Delta mutant (Pango lineage B.1.617.2), first identified in the month of October 2020 in India, is currently spreading to other parts of the world, and by June 2021; it has been reported from more than 80 countries (https://www.who.int). India first reported COVID‐19 cases on 15 February 2020, which spread rapidly to the entire country, and about 11.5 million cases were tested positive from February 2020 to March 2021. About 18 million people were tested positive in just 3 months (from April 21 to June 21) with the spread of Delta mutant (https://www.icmr.gov.in/).
India is a vast and second most populated country with diverse flora and fauna. COVID‐19 has badly struck the country infecting several million people. Close association with human beings to domesticated and wild animals in India often resulted in enhanced risk of reverse zoonosis. SARS‐CoV‐2 has been shown to infect different mammalian species including mink, dogs, cats and Panthera species (McAloose et al., 2020; Newman et al., 2020; Oreshkova et al., 2020; Sit et al., 2020). Recent detection of this virus in pet, zoo, wild and farm animals has necessitated the investigation regarding the zoonotic (animal‐to‐human) and reverse zoonotic (human‐to‐animal) transmissibility of SARS‐CoV‐2 with the potential of COVID‐19 pandemic evolving into a panzootic (Mallapaty, 2021). We have been continuously screening wild animals with respiratory signs and dead animals for SARS‐CoV‐2 since the beginning of the pandemic. However, no animal species was found positive in India till April 2021.
In this study, we report natural infection of SARS‐CoV‐2 in captive Asiatic lions (Panthera leo persica) at two different locations in India. Detailed genomic characterization confirmed first animal infection with Delta mutant of SARS‐CoV‐2.
2. MATERIALS AND METHODS
2.1. Clinical investigation
First suspected case was reported on 30 April 2021 from Lion Safari Park, Etawah, Uttar Pradesh, India (26.7680°N, 79.0009°E). Two adult Asiatic lionesses, namely Jennifer, aged 7 years (lion 1) and Gauri, aged 4 years (lion 2) developed symptoms of increased respiratory rate with distress, anorexia, fever (105.6°F & 104.5°F), mild epistaxis and increased water intake. Both the lionesses were housed separately but located in adjacent enclosures. The clinical samples (blood, serum, nasopharyngeal and rectal swabs) from affected and healthy animals were collected and subjected for laboratory investigations.
Second suspected case of an Asiatic lion named Tripur aged 5 years (lion 3) was reported on 7 May 2021 from Nahargarh Biological Park, a part of the Nahargarh Sanctuary, located about 12 km from Jaipur, Rajasthan, India (27.0161°N, 75.8645°E). As in the previous case, samples were collected from all the housed animals (n = 20) for laboratory investigations. Animals were re‐sampled on days 4 and 21 post‐appearance of symptoms.
All the samples were collected and handled by wearing personal protective gear, masks, gloves, etc. The bio‐waste materials generated were suitably discarded as per the standard guidelines. The samples were packed in three layers following the guidelines issued by WHO and transported to bio‐safety level 3 laboratory of Center for Animal Disease Research and Diagnosis (CADRAD), ICAR‐Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh. Zoo authorities were advised to screen all the staff workers immediately for SARS‐CoV‐2 and continue monitoring till the animal recovers.
2.2. Detection of SARS‐CoV‐2
Total RNA was extracted from 160 µl of virus transport medium (VTM) using Qiamp Viral RNA extraction kit (Qiagen, Germany) as per the manufactures’ instructions. Real‐time PCR reaction was assembled as per the recommendations using SuperScript III One‐Step RT‐PCR System with Platinum Taq DNA Polymerase (Invitrogen USA) and primers and probes described in Table S1 in multiplex format. All samples were tested in duplicate in real‐time PCR machine (AriaMx Real time PCR systems, Agilent) with conditions consisted of reverse transcription and enzyme activation at 50°C for 15 min and 95°C for 3 min, respectively, followed by 45 cycles of 95°C for 15 s and 58°C for 30 s. RNA extracted from in‐house cultivated virus (GenBank Accession no. MZ203529) was used as positive control whereas nuclease free water was included as a negative control. Quality of RNA was assessed by using Beta actin primers using SYBR chemistry.
2.3. Amplicon and whole genome sequencing
RNA from all the three lion samples collected on day 1 were reverse transcribed using RevertAid reverse transcriptase enzyme (ThermoFisher, USA) as per the recommendation. The reverse transcription (RT) reaction was incubated at 42°C for 45 min followed by inactivation of RT enzyme at 75°C for 10 min.
Amplicon length of 4251 nucleotides containing the complete gene of Spike protein (3822 nts) was amplified and gel purified in three overlapping segments of size 1057 bp, 981 bp and 2563 bp from the cDNA. Similarly, an amplicon length of 575 bp containing E gene (228 nts) and amplicon of 1499 bp containing N gene (1260 nts) were amplified from template cDNA. Primers were designed (Table S2) for amplification using Wuhan Reference virus sequence (GenBank Reference NC_045512.2)
Gel purified products were subjected to nucleotide sequencing by the Sanger sequencing method using the same primer sets used for amplification. In addition to the primers sets used for amplification, another reverse primer (CVR3) was also used for sequencing of the third segment of S gene.
Full length genome sequencing of one sample from each location was done by Next Generation Sequencing method (Eurofins India Limited). Briefly, library was prepared from the extracted RNA using Eurofins NG Seq ARTIC SARS‐CoV‐2 kit followed by paired end Illumina Truseq library preparation kit. Sequencing was performed on the NextSeq 500 platform of Illumina. The high quality reads of the samples were aligned to the reference genome of NC_045512.2followed by annotation and variant calling.
2.4. Isolation of virus in cell culture
The Vero E6 cells were sub‐cultured in 25 cm2 flasks in Dulbecco modified Eagle's medium (DMEM) medium with 10% foetal bovine serum (FBS), antibiotics and antifungal agents. When the cells grown to 70−80% confluency, the nasal swabs processed and filtered through the 0.2 µm disposable filters at the rate of 0.5 ml per flask. The flasks containing the cells were incubated for 2 h at 37°C with 5% CO2 tension. Cells were washed with DMEM without serum and fresh 5 ml DMEM with 2% FBS was added. The cells were observed for 72 h for the development of any cytopathic effect (CPE). However, there was no development of CPE, hence cell culture supernatant was tested by real‐time PCR assay.
2.5. Serum neutralization test
Serum samples were periodically collected and sero‐conversion was assessed by a virus neutralisation assay as described by (McAloose et al., 2020). Briefly, serum samples were diluted in 1:8 in DMEM maintenance media followed by incubation at 56°C for 45 min for de‐complementation. Samples were further two fold diluted to 1:1028. Each dilution was mixed with 102 TCID50 of in‐house cultivated virus SARS‐CoV‐2 virus (GenBank Accession no. MZ203529) and incubated at 37°C for 1 h. Serum‐virus mixture was then transferred to the wells of 96‐well plates and mixed with 50 µl of Vero cells suspension. Plates were tapped gently and incubated for 72 h at 5% CO2 tension at 37°C. Virus‐induced CPE was used as an indicator of virus infection/replication. Neutralizing antibody titres were expressed as the reciprocal of the highest dilution of serum that completely inhibited CPE. Post‐vaccine human control sera were included as positive control and all samples were tested in triplicate with results averaged. A cell culture control was included in the assays, and the virus working dilution was back‐titrated.
2.6. Phylogenetic analysis
After aligning and trimming the reads, the sequence was searched in the NCBI BLAST tool. Data were analysed in two sets. Initially, representative sequences of each clade with high quality sequences (Data set 1) were downloaded from the GISAID web server. Sequences of each clade were aligned using the Muscle tool (Edgar et al., 2004) and consensus sequences were generated using the cons programme of EMBOSS web explorer. The nucleotide sequences derived from SARS‐CoV‐2 of lion were then aligned with consensus sequences of each clade and with Wuhan‐Hu reference and other SARS related viruses using the MUSCLE tool. In the second set of analysis, representative sequences of each clade were aligned with the SARS‐CoV‐2 sequences infecting lions and aligned using the MUSCLE tool (Data set 2). Phylogenetic tree was constructed using the workspace of https://ngphylogeny.fr web server. Tree was visualised by TreeView X software.
2.7. Haplotype network analysis
In order to validate the results of phylogenetic analysis, haplotype network analysis was conducted using Data set 1. The FASTA format was converted into Nexus format and imported to ProPart software (Leigh and Bryant, 2015). Haplotype network was created using median joining algorithm (Bandelt et al., 1999).
3. RESULTS
3.1. Clinical and laboratory investigations
In case 1, two of 18 lions showed clinical symptoms. Haemato‐biochemical values indicated compromised (secondary bacterial infection, liver and kidney functions) health status of both lionesses. With the appearance of symptoms, animals were isolated and symptomatic treatment was initiated. No other lions showed any clinical signs nor found positive in laboratory investigations. Blood parameters of healthy animals were also in normal range. Infected animals responded to symptomatic treatment with fluid therapy, antibiotics and nutritional supplements. Total leukocyte count, absolute neutrophil count, alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN) and creatinine exhibited an increasing trend in both the lions from 2 to 30 May 2021. These parameters started normalizing from 25 May and came down to normal limits after 5 June. Animals were again screened after 4 days, that is, on 4 May 2021 and found to be infected with SARS‐CoV‐2 (Table S1). Both the animals showed clinical signs up to 30 May 2021 but the animals showed recovery from 25 May with diet intake restored to ∼4.5 kg of buffalo meat daily. Samples were collected on 21 days post‐detection and were found to have a negative SARS‐CoV‐2 genome.
In second case, one out of four lions exhibited mild clinical symptoms and affected lion (Tripur) was immediately isolated and subjected to symptomatic treatment. Samples collected from this animal were found positive whereas rest all the animals were tested negative. Animals were provided with symptomatic and supportive treatment. The mild symptoms quickly disappeared and the infected animal recovered uneventfully. Samples tested after 14 days of infection were found negative for SARS‐CoV‐2 genome. Only Asiatic lions but not any other Panthera species which were housed in the same parks/zoos were found positive for SARS‐CoV‐2 genome.
All the zoo staff members at both the locations were screened for SARS‐CoV‐2 after animals were found positive. Samples were tested by real time PCR by local authorities and everyone was negative for SARS‐CoV‐2. However, the veterinarian who was treating the infected lions in case 1 developed clinical symptoms two days after the detection in lions and found positive in real‐time PCR. He recovered uneventfully after 15 days.
Samples found positive for SARS‐CoV‐2 with primers and probes targeting RdRP, E gene and RT‐PCR were confirmed by Sanger sequencing of S, N and E genes in both the cases.
3.2. Virus isolation
Nasal and oral swabs of two positive animals were attempted by blindly passaging on confluent Vero E6 cells. However, even after 5 blind passages, no CPE appeared in any sample. Also real‐time PCR of cell culture supernatant did not indicate any virus replication. Further attempts are being made to isolate the virus including by RNA transfection method.
3.3. Non SARS‐CoV‐2 pathogens
Blood smears of all the animals were analysed and ruled out for presence of any haemoprotozoan or rickettsial parasites. Total nucleic acid extracted from the swabs samples were screened by real‐time PCR for other viral pathogens such as Canine Distemper virus (Frisk et al., 1997; Bhatt, 2019), Feline panleukopenia virus (Mochizuki et al., 1996), Feline herpes virus (Sykes et al., 2001) and Feline Calicivirus (Sykes et al., 2001), and highly pathogenic Avian influenza (H5 and H7; Spackman et al., 2002).
3.4. Virus neutralization test
The sequential serum samples collected on 1, 4 and 21 days post‐infection were evaluated by serum neutralization assay. Titre of 64 was observed in lion 1 and 3 on day 1 whereas lion 2 showed titre of 128. Titres remain unchanged on day 4 but increased to 128 in lion 1 and 3 and 256 in case of the lion 2.
3.5. Comparative genomics and phylogenetic analysis
Complete genome sequences of SARS‐CoV‐2 detected in the Asiatic lions of each case (lion 1 and lion 3) were generated directly from the nasal and rectal swabs. The high‐quality reads of the samples were aligned to the reference sequence using BWA MEM (version 0.7.17) (Li and Durbin, 2009). Reads were submitted BioSample database (Accession numbers; SAMN20424376 and SAMN20424377). Consensus sequence of 29903 bp was extracted using Samtools mpileup. Both the sequences genome coverage was more than 99% with the reference sequence. Annotated sequences were submitted to NCBI (lion 1; GenBank accession number MZ413340 and lion 3; GenBank accession number MZ413341). The mpileup utility of Samtools (v 0.1.18) was used to identify SNPs and InDels from the sorted BAM file of the sample (Li et al., 2009). The variants were filtered based on a minimum read depth of 5, quality threshold of 25. Compared to the Wuhan‐Hu‐1 sequence (GenBank accession number NC_045512), lion 1 (MZ413340) contained 32 polymorphisms (SNPs) of which 28 were in the coding regions and 4 were present in intergenic regions (Figure 1 and Table S3). Out of 28, 6 resulted in synonymous mutations. Total of 7 amino acid mutations were found in the spike protein when compared to Wuhan‐Hu‐1 virus.
FIGURE 1.
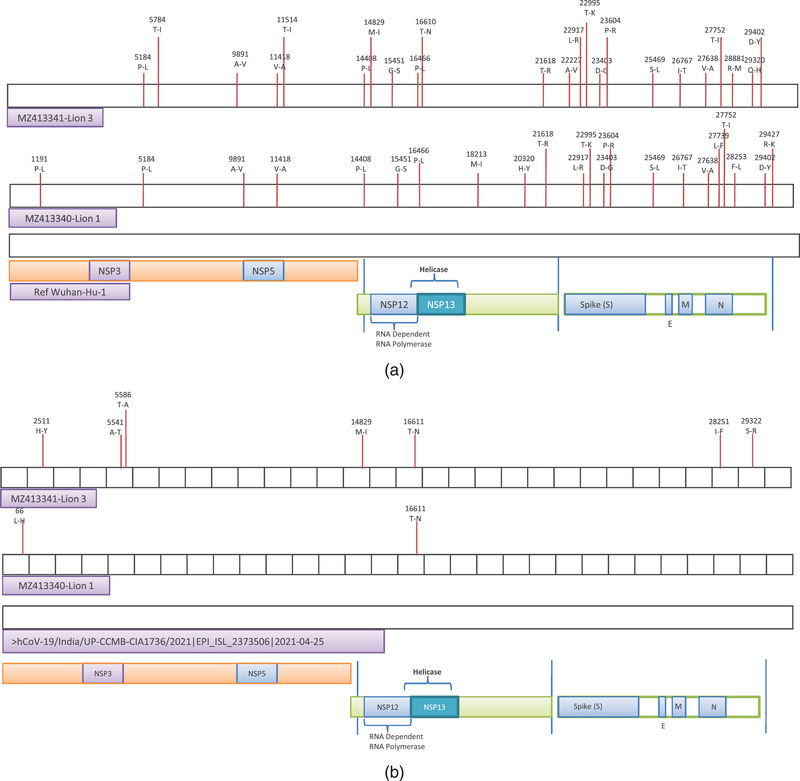
(a) Schematic representation of SARS‐CoV‐2 genome organisation and functional proteins. Amino acid changes in comparison with Wuhan‐Hu‐1 (NC_045512) virus. Non‐synonymous mutations highlighted with red lines. (b) Comparative genomics of Delta mutant virus sequence in comparison with one high quality Delta mutant determined from humans in India. Non‐synonymous mutations resulted in amino acid changes are highlighted with red lines
Lion 3 (MZ413341) contained 28 polymorphisms (SNPs) compared to reference virus genomes of which 25 were present in the coding and 3 in the intergenic regions (Figure 1) and only 2 substitutions resulted in synonymous mutations. Total of 6 amino acid mutations were found in the spike proteins in comparison to the reference virus. There were a difference of 20 nucleotides between the two lions and only one was in the non‐coding region.
Phylogenetic analysis consisting of representative sequences of all the clades showed that both the lion samples clustered into common SARS‐CoV‐2 G‐clade (GISAID classification) or Delta variant (B.1.617.2 Pango lineage) (Figure 2). Both viruses showed a close relationship with each other. Both viruses also showed a close relationship with the consensus sequences of Delta mutant virus found in this region in humans at the time of detection (UP‐April‐21).
FIGURE 2.
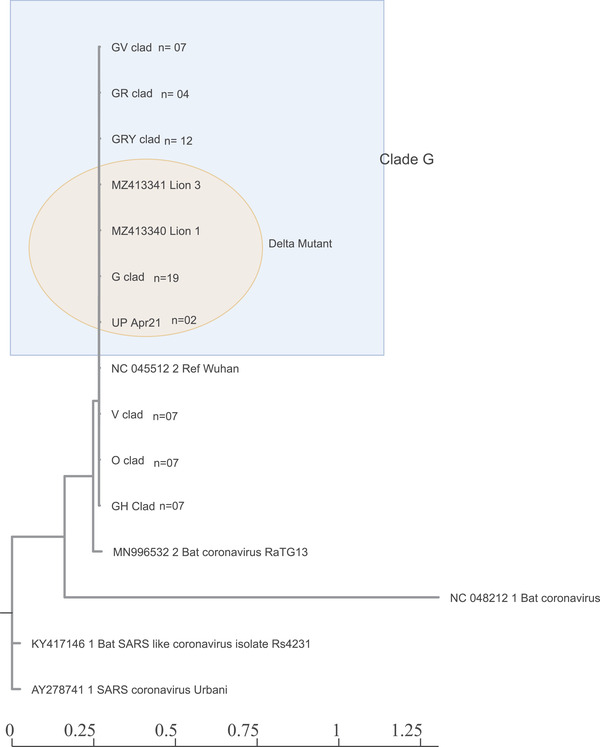
Phylogenetic analysis of Lion's sequences. Whole‐genome phylogeny of lion's sequences with Wuhan‐Hu‐1 reference genome (NC_045512.2) and consensus sequences of other clades available in the GISAID database. Lion's sequences clustered with the G‐clad (B.1.617.2 pango lineage)
The findings were also corroborated through haplotype network analysis of the sequences using the Median‐joining method in PopArt analysis tool (Figure 3). It was evident that both the virus and UP‐April‐21 consensus sequences emerged from Clade G.
FIGURE 3.

Haplotype network analysis of lion's samples with Wuhan‐Hu‐1 reference virus and consensus sequences of other clades. Haplotype analysis shows relatedness and levels of genetic variation between lion's and virus circulating in the nearby areas. Differences are indicated by one‐step edges (lines) between blue dots
In comparison with Delta mutants circulating in human population in the area it was observed that lion 1 showed substitution of only two amino acids, whereas lion 3 showed seven amino acid substitutions of which five were in the ORF1ab region and one each in ORF8 and N gene regions (Figure 1).
4. DISCUSSION
In this report we have identified and characterized first incidence of SARS‐CoV‐2 Delta mutant infection in Asiatic lions of India. Cases of SARS‐CoV‐2 have been reported in other species including Asiatic lions. Although these incidences have largely been considered as spill over infections from human beings, intensive screening of various species is essential in order to identify the potential reservoir host especially in light of emergence of new variants of concern of SARS‐CoV‐2 (McAloose et al., 2020; Sit et al., 2020; Newman et al., 2020; Oreshkova et al., 2020). Montagutelli et al. (2021) recently demonstrated susceptibility of mice to B1.351 (Beta mutant) of SARS‐CoV‐2 which previously was unable to infect mice cells.
The cases reported here are second incidences of infection in Asiatic lions, before that eight animals were found positive in Nehru Zoological Park, Hyderabad of India (https://www.thehindu.com/news/cities/Hyderabad/eight‐asiatic‐lions‐test‐positive‐for‐coronavirus‐in‐hyderabad‐zoo/article34480453.ece); however, detailed genomic analysis is awaited.
Screening of various animal species of domesticated, pet and wild animals were intensified with rising in cases of COVID‐19 in India. Two incidences of SARS‐CoV‐2 were confirmed and thoroughly investigated. Case 1 occurred in Lion Safari, Etawah, Uttar Pradesh, which includes multiple safaris (deer, antelopes, leopard and bear) and more importantly lion safari with Asiatic lion Breeding Centre. Currently, 18 Asiatic lions (2 cubs, 8 male and 8 female) are housed of which only two lions were found infected. On day 1, lion 1 showed weak amplification with an average Ct value of 34.0 whereas lion 2 showed average Ct values of 30.7, which could be possible during either very early infection or very late infection stage. Lion 2 developed only mild clinical symptoms and recovered uneventfully, whereas lion 1 developed respiratory distress along with pyrexia and off‐feed. The average Ct value of lion 1 reduced to 24.0 on day 4 whereas the average Ct value of lion 2 increased to 34.41. It was hypothesised that lion 1 was in active infection but not lion 2. Serological analysis also revealed that lion 1 has a titre of 64 whereas lion 2 showed an increased titre of 128, which further increased as infection advanced. However, no other feline species enclosed nearby, were found positive for both RT‐PCR and virus neutralization assay. The infected animals showed mild to moderate clinical symptoms but were negative for the major pathogens. They responded to the palliative treatment and recovered uneventfully.
Case 2 occurred at Nahargarh Biological Park, Jaipur, Rajasthan, India. This park housed several wild animals including lions, tigers, leopards, etc. The lions are housed separately at Lion Safari which is somewhat distant from enclosures of other species. On 7 May one male lion namely Tripur aged 5 years (lion 3) developed mild clinical signs like anorexia and dullness, but not pyrexia. This animal recovered uneventfully.
Since the beginning of the pandemic, no cases in any wild animals were reported/detected in spite of intensive screening. However, with the emergence of Delta mutants of SARS‐CoV‐2, cases at two different locations were confirmed which may be associated with high affection of the particular mutants to these species. Similarly, in Nehru Zoological Park located in Hyderabad India, only Asiatic lions were found positive but not any other feline species. On the other hand, there was a sudden rise in the number of cases in humans in most parts of the country, which increased the associated risk of virus transmission to these animals that might explain the possible infection in these animals.
Till February 2021, the alpha VOC (B.1.1.7) was detected in parts of India but it has been reported that it did not spread to a wide population. However, March 2021 onwards, B.1.617 and its sub‐lineage B.1.617.2 rapidly rise in the population displacing B.1.1.7 and other B.1.617 (Dhar et al., 2021). The group generated complete genome sequences of currently prevailing SARS‐CoV‐2 in humans in the city of Delhi and concluded that B.1.617.2 shows high transmissibility rates by more than 50% when compared to the B.1.1.7 clade in unvaccinated population (Dhar et al., 2021). Both the lions were found to be infected with the Delta mutant. All the zoo handlers were found negative for SARS‐CoV‐2 but the virus detected in lions closely grouped with the viruses circulating in that area. Both the zoos were closed for the public and a clear association for virus transmission could not be established but there could be possibility of virus transmission via in‐direct means (e.g. fomite, food handling/preparation or shared enrichment items). Four tigers and three lions were found positive for the genome of SARS‐CoV‐2 who developed respiratory clinical symptoms at Bronx zoo, New York (McAloose et al., 2020). Tigers and tiger keepers were found to be infected by Clade G virus and infection was closely associated to be transmitted from Tiger keepers, whereas strain infecting the lions were of V clad (McAloose et al., 2020). However, in this report, neither all the Asiatic lions enclosed in both the zoos nor any other feline species were found positive and were also serologically negative. Probably the animals were housed individually in separate enclosures and handled by independent zoo keepers.
The lion's sequences were highly similar to that of Delta mutants circulating among humans in that area as determined by comparative genomics. However, the sequence of lion 1 does not show many changes except substitutions of two amino acids whereas the lion 3 sequence showed several substitutions in comparison to the reference genome (Figure 1). Detailed analysis needs to be done if the substitutions have any phenotypic characters. It has to be emphasised again that lion 1 showed moderate clinical symptoms whereas lion 3 showed only mild clinical symptoms.
The spike protein interacts with the angiotensin‐converting enzyme II (ACE2) (Bibiana et al., 2020). Conservation of amino acids residues that are critical for binding with SARS‐CoV‐2 has been predicated in felines, which might explain the high susceptibility of these species over others (Zhai et al., 2020). However, studies are required to further understanding on differential susceptibility of native wild animals.
Possible role of domestic and wild animals in SARS‐CoV‐2 epidemiology is now being sorted out. It has been speculated that the receptor binding domain (RBD) region of the spike protein of SARS‐CoV‐2 may bind to ACE2 protein receptors of different species, giving them a higher likelihood of SARS‐CoV‐2 crossing the species barrier (Zhai et al., 2020; Bibiana et al., 2020). Thus, many animals could be infected with SARS‐CoV‐2 and serve as reservoir hosts in the spread of the virus. Although very few reports are available describing the natural infection of animals, with the constant emergence of new variants of concern, there is always the possibility this virus finds a new host species for transmission (Frutos et al., 2021). This warrants further intensification of screening various species which act as potential reservoirs and sources of infection to both humans and animals.
CONFLICT OF INTEREST
Authors disclose no conflict of interest with anyone
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
We are thankful to all the zoo staff and central zoo authority of India for collection of samples and data. We are also thankful to ICAR, NASF and DST‐SERB for providing funds and infrastructure to take up the work. We are also highly thankful to Dr R K Singh and Dr V K Gupta, ex‐director and ex Joint Director CADRAD, respectively for their contributions in SARS‐CoV‐2 screening in the institute.
Karikalan, M. , Chander, V. , Mahajan, S. , Deol, P. , Agrawal, R. K. , Nandi, S. , Rai, S. K. , Mathur, A. , Pawde, A. , Singh, K. P. , & Sharma, G. K. (2021). Natural infection of Delta mutant of SARS‐CoV‐2 in Asiatic Lions of India. Transboundary and Emerging Diseases, 00, 1–9. 10.1111/tbed.14290
M. Karikalan, V. Chander and S. Mahajan contributed equally.
DATA AVAILABILITY STATEMENT
Annotated complete genome sequences are available in NCBI (GenBank accession numbers MZ413340 and MZ413341). Raw sequencing data are available at NCBI BioSample (Accession numbers; SAMN20424376 and SAMN20424377). Data sets 1 and 2 are available from the corresponding author upon reasonable request.
REFERENCES
- Andersen, K. G. , Rambaut, A. , Lipkin, W. I. , Holmes, E. C. , & Garry, R. F. (2020). The proximal origin of SARS‐CoV‐2. Nature Medicine, 26, 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt, H. J. , Forster, P. , & Röhl, A. (1999). Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48. 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- Bhatt, M. , Rajak, K. K. , Chakravarti, S. , Yadav, A. K. , Kumar, A. , Gupta, V. , & Singh, R. K. (2019). Phylogenetic analysis of haemagglutinin gene deciphering a new genetically distinct lineage of canine distemper virus circulating among domestic dogs in India. Transboundary and Emerging Diseases, 66, 1252–1267. 10.1111/tbed.13142 [DOI] [PubMed] [Google Scholar]
- Bibiana, S. O. F. , Vargas‐Pinilla, P. , Amorim, C. E. G. , Sortica, V. A. , & Bortolini, M. C. (2020). ACE2 diversity in placental mammals reveals the evolutionary strategy of SARS‐CoV‐2. Genetics and Molecular Biology, 43(2), e20200104. 10.1590/1678-4685-gmb-2020-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam, B. S. , Vargas‐Pinilla, P. , Amorim, C. E. G. , Sortica, V. A. , & Bortolini, M. C. (2020). ACE2 diversity in placental mammals reveals the evolutionary strategy of SARS‐CoV‐2. Genetics and molecular biology, 43. 10.1128/JCM.37.11.3634-3643.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisk, A. L. , Konig, M. , Moritz, A. , & Baumgartner, W. (1999). Detection of canine distemper virus nucleoprotein RNA by reverse transcription‐PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. Journal of clinical microbiology, 37, 3634–3643. 10.1128/JCM.37.11.3634-3643.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutos, R. , Gavotte, L. , Serra‐Cobo, J. , Chen, T. , & Devaux, C. (2021). COVID‐19 and emerging infectious diseases: The society is still unprepared for the next pandemic. Environmental Research, 111676. 10.1016/j.envres.2021.111676. Epub ahead of print. PMID: 34252435; PMCID: PMC8268624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E. , Baker, S. C. , Baric, R. S. , de Groot, R. J. , Drosten, C. , Gulyaeva, A. A. , & Ziebuhr, J. (2020). Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome‐related coronavirus: Classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nature Microbiology, 5, 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J. W. , & Bryant, D. (2015). popart: Full‐feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110–1116. 10.1111/2041-210X.12410 [DOI] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , & Durbin, R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352. Epub 2009 Jun 8. PMID: 19505943; PMCID: PMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty, S. (2021). The search for animals harbouring coronavirus and why it matters. Nature, 591, 26–28. 10.1038/d41586-021-00531-z [DOI] [PubMed] [Google Scholar]
- McAloose, D. , Laverack, M. , Wang, L. , Killian, M. L. , Caserta, L. C. , Yuan, F. , Mitchell, P. K. , Queen, K. , Mauldin, M. R. , Cronk, B. D. , Bartlett, S. L. , Sykes, J. M. , Zec, S. , Stokol, T. , Ingerman, K. , Delaney, M. A. , Fredrickson, R. , Ivančić, M. , Jenkins‐Moore, M. , Mozingo, K. , Franzen, K. , Bergeson, N. H. , Goodman, L. , Wang, H. , Fang, Y. , Olmstead, C. , McCann, C. , Thomas, P. , Goodrich, E. , Elvinger, F. , Smith, D. C. , Tong, S. , Slavinski, S. , Calle, P. P. , Terio, K. , Torchetti, M. K. , & Diel, D. G. . (2020). From people to Panthera: Natural SARS‐CoV‐2 infection in tigers and lions at the Bronx Zoo. MBio, 11, e02220–20. 10.1128/mBio.02220-20. PMID: 33051368; PMCID: PMC7554670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, M. , Horiuchi, M. , Hiragi, H. , San Gabriel, M. C. , Yasuda, N. , & Uno, T. (1996). Isolation of canine parvovirus from a cat manifesting clinical signs of feline panleukopenia. Journal of Clinical Microbiology, 34, 2101–2105. 10.1128/jcm.34.9.2101-2105.1996. PMID: 8862565; PMCID: PMC229197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli, X. , Prot, M. , Levillayer, L. , Salazar, E. B. , Jouvion, G. , Conquet, L. , & Simon‐Loriere, E. (2021). The B1. 351 and P. 1 variants extend SARS‐CoV‐2 host range to mice. BioRxiv. 10.1101/2021.03.18.436013 [DOI] [Google Scholar]
- Newman, A. , Smith, D. , Ghai, R. R. , Wallace, R. M. , Torchetti, M. K. , Loiacono, C. , & Behravesh, C. B. (2020). First reported cases of SARS‐CoV‐2 infection in companion animals—New York, March–April 2020. Morbidity and Mortality Weekly Report, 69, 710. 10.15585/mmwr.mm6923e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova, N. , Molenaar, R. J. , Vreman, S. , Harders, F. , Munnink, B. B. O. , Honing, H.‐. D. R. W. , & Stegeman, A. (2020). SARS‐CoV‐2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance, 25, 2001005. 10.2807/1560-7917.ES.2020.25.23.2001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, T. H. , Brackman, C. J. , Ip, S. M. , Tam, K. W. , Law, P. Y. , To, E. M. , & Peiris, M. (2020). Infection of dogs with SARS‐CoV‐2. Nature, 586, 776–778. 10.1038/s41586-020-2334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman, E. , Senne, D. A. , Myers, T. J. , Bulaga, L. L. , Garber, L. P. , Perdue, M. L. , & Suarez, D. L. (2002). Development of a real‐time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of clinical microbiology, 40, 3256–3260. 10.1128/jcm.40.9.3256-3260.2002. PMID: 12202562; PMCID: PMC130722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes, J. E. , Allen, J. L. , Studdert, V. P. , & Browning, G. F. (2001). Detection of feline calicivirus, feline herpesvirus 1 and Chlamydia psittaci mucosal swabs by multiplex RT‐PCR/PCR. Veterinary Microbiology, 81, 95–108. 10.1016/s0378-1135(01)00340-6 [DOI] [PubMed] [Google Scholar]
- Zhai, X. , Sun, J. , Yan, Z. , Zhang, J. , Zhao, J. , Zhao, Z. , & Su, S. (2020). Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. Journal of Virology, 94, e00831–20. 10.1128/JVI.00831-20. PMID: 32404529; PMCID: PMC7375388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. Z. L. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , Wang, X. , Zheng, X. S. , Zhao, K. , Chen, Q. J. , Deng, F. , Liu, L. L. , Yan, B. , Zhan, F. X. , Wang, Y. Y. , Xiao, G. F. , Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Annotated complete genome sequences are available in NCBI (GenBank accession numbers MZ413340 and MZ413341). Raw sequencing data are available at NCBI BioSample (Accession numbers; SAMN20424376 and SAMN20424377). Data sets 1 and 2 are available from the corresponding author upon reasonable request.


