Abstract
Muenke syndrome is the leading genetic cause of craniosynostosis and results in a variety of disabling clinical phenotypes. To model the disease and study the pathogenic mechanisms, a human induced pluripotent stem cell (hiPSC) line was generated from a patient diagnosed with Muenke syndrome. Successful reprogramming was validated by morphological features, karyotyping, loss of reprogramming factors, expression of pluripotency markers, mutation analysis and teratoma formation.
1. Resource utility
Muenke syndrome (MS), caused by a p.Pro250Arg (c.749C>G) gain-of-function mutation in the FGFR3 gene (Muenke et al., 1996; Wilkie et al., 2013), is a common form of craniosynostosis. Human induced pluripotent stem cells (hiPSCs) from an MS patient may serve as a disease model to elucidate underlying pathogenetic mechanism(s).
2. Resource details
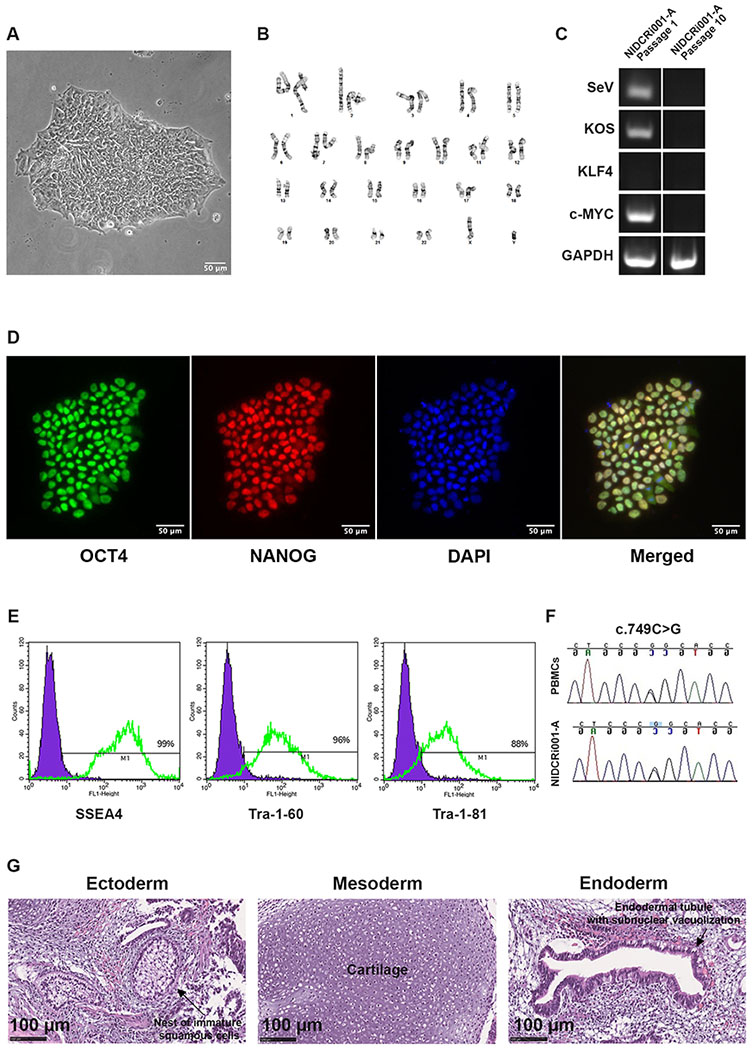
The clinical manifestations of MS patients are highly variable with incomplete penetration, ranging from developmental delay and hearing impairment to increased intracranial pressure often requiring surgery for decompression (Kruszka et al., 2016; Twigg et al., 2009). Through surface plasmon resonance and X-ray crystallography, one study demonstrated that fibroblast growth factor receptor 3 (FGFR3) with the MS mutation has enhanced affinity for atypical ligands, such as FGF2 and FGF9 (Ibrahimi et al., 2004). Loss of ligand specificity may potentially be a unique mechanism that separates MS from other FGFR3-related diseases that tend to severely affect long bone growth. However, a detailed description of the aberrant molecular signalling pathways involved in this disease has not yet been reported. The iPSC line, NIDCRi001-A, was generated from a 7-year-old male patient previously diagnosed with MS. Peripheral blood mononuclear cells (PBMCs) were expanded in StemSpan CD34+ Expansion Supplement (Cat# 02691, STEMCELL Technologies Inc) for 9 days to expand CD34+ cells. Reprogramming into iPSCs was accomplished with Sendai virus containing the factors OCT4, SOX2, KLF4 and c-MYC. The resulting line showed appropriate morphology (Fig. 1A); a normal diploid male karyotype (46, XY) (Fig. 1B); and clearance of reprogramming factors by passage 10, confirmed by RT-PCR compared with passage 1 (Fig 1C). NIDCRi001-A expressed pluripotency markers, evaluated by nuclear staining for OCT3/4 and NANOG and by FACS analysis for SSEA4, Tra-1-60, and Tra-1-81 (Fig. 1D, E). Short Tandem Repeat (STR) analysis confirmed that the iPSCs had an identical genetic background as the donor PBMCs (data available from the authors), and Sanger sequencing showed that the FGFR3 heterozygous missense mutation was present in both cell types (Fig. 1F). Pluripotency was further confirmed by differentiation of hiPSCs into derivatives of all three germ layers through a teratoma assay in immunocompromised mice (Fig. 1G). Mycoplasma testing was negative (Supplementary Data).
Figure 1:
3. Materials and Methods
3.1. Reprogramming
PBMCs isolated using lymphocyte separation medium (Lonza) were expanded (9d at 37°C in 5% CO2) in StemSpan SFEM II/CD34+ Expansion Supplement (STEMCELL Technologies Inc) and reprogrammed with CytoTune™-iPS 2.0 Sendai Reprogramming Kit (ThermoFisher Scientific). After 3d, cells were plated on 12-well plates coated with Matrigel® Matrix (Corning) or Vitronectin XF™ (Nucleus Biologics) in complete E8™ medium (ThermoFisher Scientific). Clones were picked (~21d) and expanded in 6-well plates for 10 passages. Subculture (1:6) used 50mM EDTA in calcium-magnesium-free phosphate buffered saline (PBS) for 4-5min every 4-5 days.
3.2. Karyotype analysis
Karyotyping (passage 10) was performed on twenty G-banded metaphase cells at 475 band resolution (Cell Line Genetics).
3.3. Mutation analysis
DNA (100ng/35μL) was PCR-amplified with primers flanking exon 7 of FGFR3 using high fidelity AccuPrime™ Taq DNA Polymerase (ThermoFisher Scientific). Reactions were run in a 96-well thermal cycler: 95°C for 5min, 40 cycles at 95°C for 30sec, 66°C for 30sec, 72°C for 30sec, and a final step at 72°C for 7min. Products were separated on a 1% agarose gel, purified using ExoSAP-IT™ Express (ThermoFisher Scientific) and sequenced (Eurofins Clinical Molecular Testing Services).
3.4. STR analysis
Identity analysis was performed by Cell Line Genetics on PBMCs and the iPSC line using the PowerPlex 16 System (Promega).
3.5. RNA isolation and RT-PCR analysis
Clearance of Sendai virus and reprogramming factors was confirmed by RT-PCR after 10 passages. Total RNA (1×106 cells) was obtained using the RNeasy Mini Kit (Qiagen). cDNA was generated through iScript™ cDNA Synthesis Kit (Bio-Rad) with GAPDH as the control. PCR was performed using GoTaq® Green Master Mix (25μl, Promega) and Proflex PCR System (ThermoFisher Scientific). A three-step PCR for GAPDH was performed: denaturation for 2min at 95°C; 35 cycles of 45sec at 95°C, 45sec at 58°C, and 1min at 72°C; 30sec at 72°C). For Sendai virus genome and reprogramming factors: denaturation for 2min at 95°C; 40 cycles of 30sec at 95°C, 30sec at 55°C, and 30sec at 72°C. cDNA from passage 1 was used as a positive control.
3.6. Immunocytochemistry
Cells fixed with 4% paraformaldehyde for 20min at RT and washed 3x with PBS were blocked with 10% normal goat serum (NGS), permeabilized with 0.3% TX-100 for 40min, and washed. Cells were incubated with primary antibodies in 5% NGS/PBS for 1hr, washed, and incubated with secondary antibodies (1hr). After a final wash, nuclei were stained with 0.2μg/ml bisbenzimide in PBS for 10min. An Axiovert 200 (Zeiss) was used for imaging.
3.7. Flow cytometry
Cells released using Accutase™ (STEMCELL Technologies) were passed through a 30μm cell strainer and incubated in 96-well plates with primary antibodies and isotype controls for 30min, followed by incubation with secondary antibodies for another 30min. Analysis was performed using a FACSCalibur™ analyzer, and CellQuest™ Pro software (BD Bioscience).
3.8. Teratoma Assay
iPSCs (1×106), released using 50mM EDTA and suspended in 50% Matrigel, were injected subcutaneously into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (Jackson Laboratory). After 6wk, teratomas were histologically analyzed to identify derivatives of all three germ layers.
3.9. Mycoplasma detection
Mycoplasma (19 species) and Acholeplasma (1 species) were tested for by Cell Line Genetics.
Table 1:
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | normal | Figure 1 panel A |
| Phenotype | Qualitative analysis Immunocytochemistry |
Positive for expression of pluripotency markers: OCT4, NANOG | Figure 1 panel D |
| Quantitative analysis Flow cytometry |
SSEA-4 99% Tra-1-60 96% Tra-1-81 88% |
Figure 1 panel E | |
| Genotype | Karyotype (G-banding) and resolution | 46XY, Resolution 475 |
Figure 1 panel B |
| Identity | STR analysis | 16 loci analyzed, all matching | Available with the authors |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous missense mutation | Figure 1 panel F |
| Microbiology and virology | Mycoplasma | Mycoplasma testing negative | Supplementary |
| Differentiation potential | Teratoma formation | Presence of three embryo germ layers by histology | Figure 1 panel G |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Table 2:
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
|
| |||
| Pluripotency Markers | Mouse anti-TRA-1-60 (IgM) | 1:100 | Santa Cruz Biotechnology cat# sc-21705, RRID: AB_628385 |
| Mouse anti-TRA-1-81 (IgM) | 1:100 | Santa Cruz Biotechnology cat# sc-21706, RRID: AB_628386 | |
| Mouse anti-SSEA4 (IgG3) | 1:100 | Santa Cruz Biotechnology cat# sc-21704, RRID: AB_628289 | |
| Mouse anti-Oct3/4 (IgG2b) | 1:400 | Santa Cruz Biotechnology cat# sc-5279, RRID: RRID: AB_628051 | |
| Rabbit anti-Nanog | 1:200 | Reprocell cat# RCAB004P-F; RRID: RRID: AB_1560380 | |
|
| |||
| Secondary Antibodies | Alexa Fluor 488 goat anti-mouse (IgG) | 1:100 | ThermoFisher Scientific cat# A-11001, RRID: AB_2534069 |
| Alexa Fluor 555 rabbit anti-mouse (IgG) | 1:500 | ThermoFisher Scientific cat# A-21428, RRID: AB_2535849 | |
| Alexa Fluor 488 goat anti-mouse (IgM) | 1:1000 | ThermoFisher Scientific cat# A-21042, RRID: AB_141357 | |
|
| |||
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
|
| |||
| Sendai virus detection | SeV, 181bp | GGATCACTAGGTGATATCGAGC ACCAGACAAGAGTTTAAGAGATATGTATC GenBank Accession #NC_001552.1 |
|
|
| |||
| Transgene detection | KOS, 528bp | ATGCACCGCTACGACGTGAGCGC ACCTTGACAATCCTGATGTGG GenBank Accession #NM_002701.4, NM_003106.2, BC029923.1 |
|
|
| |||
| Transgene detection | KLF4, 410bp | TTCCTGCATGCCAGAGGAGCCC AATGTATCGAAGGTGCTCAA GenBank Accession #BC029923.1 |
|
|
| |||
| Transgene detection | c-MYC, 532bp | TAACTGACTAGCAGGCTTGTCG TCCACATACAGTCCTGGATGATGATG GenBank Accession #K02276.1 |
|
|
| |||
| House-keeping gene | GAPDH, 816bp | AGCCGCATCTTCTTTTGCGTC TCATATTTGGCAGGTTTTTCT GenBank Accession #NG_007073.2 |
|
|
| |||
| Mutation sequencing | FGFR3 (c.749C>G), 191bp | CGGCAGTGGCGGTGGTGGTGA GACCCAAATCCTCACGCAACC GenBank Accession #NG_012632.1 |
|
Resource Table:
| Unique stem cell line identifier | NIDCRi001-A |
| Alternative name(s) of stem cell line | MS-iPSC-1a |
| Institution | National Institutes of Health, Bethesda, USA |
| Contact information of distributor | Pamela G. Robey; probey@dir.nidcr.nih.gov Fahad Kidwai; fahad.kidwai@nih.gov |
| Type of cell line | hiPSC |
| Origin | human |
| Additional origin info | Age: 7 years Sex: Male Ethnicity: unknown |
| Cell Source | PBMCs |
| Clonality | Clonal |
| Method of reprogramming | Sendai Virus |
| Genetic Modification | Yes |
| Type of Modification | Sporadic mutation |
| Associated disease | Muenke syndrome |
| Gene/locus | Gene: FGFR3 Locus: 4p16.3 Mutation: heterozygous p.Pro250Arg (c.749C>G) |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | November 25th, 2019 |
| Cell line repository/bank | https://hpscreg.eu/browse/provider/954 |
| Ethical approval | NIH Combined Neuroscience Institutional Review Board (IRB) (Protocol 16-D-0040) |
Acknowledgements
This research was made possible through the NIH Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, Genentech, the American Association for Dental Research, the Colgate-Palmolive Company, and other private donors (to BWHM), from the Division of Intramural Research, National Institute of Dental and Craniofacial Research (ZIA DE000380 to DA, FKK, JSL and PGR) and from the Division of Intramural Research, National Human Genome Research Institutes (to AFM), both of which are part of the Intramural Research Program, National Institutes of Health, Department of Health and Human Services.
Footnotes
Declaration of competing interest
The authors have no conflicts of interest.
Appendix: Supplementary Materials
1. Mycoplasma test analysis certificate
2. Karyotyping analysis certificate
References
- 1.Muenke M, Gripp KW, McDonald-McGinn M et al. A unique point mutation in the fibroblast growth factor receptor 3 gene (FGFR3) defines a new craniosynostosis syndrome. Am. J. Hum. Genet, 60 (3) 1997, pp. 555–564 [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkie AO, Byren JC, Hurst JA et al. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics, 126 (2) 2010, pp. e391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruszka P, Addissie YA, Yarnell CM et al. Muenke syndrome: An international multicenter natural history study. Am. J. Med. Genet. A 170A (4) 2016, pp. 918–929 [DOI] [PubMed] [Google Scholar]
- 4.Twigg SR, Healy C, Babbs C et al. Skeletal analysis of the Fgfr3(P244R) mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev Dyn. 238 (2) 2009, pp. 331–342 [DOI] [PubMed] [Google Scholar]
- 5.Ibrahimi OA, Zhang F, Eliseenkova AV et al. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum Mol Genet. 13 (1) 2003, pp. 69–78 [DOI] [PubMed] [Google Scholar]


