Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy characterized by aberrant proliferation of immature thymocytes. Despite an overall survival of 80% in the pediatric setting, 20% of patients with T-ALL ultimately die from relapsed or refractory disease. Therefore, there is an urgent need for novel therapies. Molecular genetic analyses and sequencing studies have led to the identification of recurrent T-ALL genetic drivers. This review summarizes the main genetic drivers and targetable lesions of T-ALL and gives a comprehensive overview of the novel treatments for patients with T-ALL that are currently under clinical investigation or that are emerging from preclinical research.
Significance:
T-ALL is driven by oncogenic transcription factors that act along with secondary acquired mutations. These lesions, together with active signaling pathways, may be targeted by therapeutic agents. Bridging research and clinical practice can accelerate the testing of novel treatments in clinical trials, offering an opportunity for patients with poor outcome.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) arises from the accumulation of genetic lesions during T-cell development in the thymus, resulting in differentiation arrest and aberrant proliferation of immature progenitors. T-ALL accounts for only 10% to 15% of pediatric and up to 25% of adult ALL cases (1), with an overall survival (OS) of 80% in the pediatric setting that has been achieved using a risk-based stratification toward intensive multiagent combination chemotherapeutic protocols (2). OS rates for adult patients with T-ALL are lower than 50% due to higher treatment-related toxicities (1). Patients are assigned to standard-, medium-, or high-risk group based on initial steroid response and minimal residual disease (MRD) after the first two courses of chemotherapy (3, 4). The risk-based therapeutic regimen consists of steroids, microtubule-destabilizing agents (vincristine), alkylating agents (cyclophosphamide), anthracyclines (doxorubicin or daunorubicin), antimetabolites (methotrexate, MTX), nucleoside analogues (6-mercaptopurine, thioguanine, or cytarabine), and hydrolyzing enzymes (l-asparaginase), and in some cases, it is followed by stem cell transplantation. Some of these conventional chemotherapeutics have a lymphoid lineage–specific effect in ALL. In fact, lymphoblasts have low asparagine synthetase activity, and thus, they are very sensitive to exogenous asparagine depletion by l-asparaginase. Moreover, ALL blasts are susceptible to MTX treatment due to a higher accumulation of MTX-polyglutamate metabolites that increases MTX intracellular retention and its antileukemic effect in these cells (5). Risk-based intensification of the therapeutic regimen has greatly improved the survival rate for pediatric (6) and young adult patients treated on pediatric-based protocols (1). Nevertheless, still 1 of 5 pediatric patients with T-ALL dies within 5 years after first diagnosis from relapsed disease and therapy resistance (refractory disease) or from treatment-related mortalities, including toxicity and infections. Therefore, further intensification of the treatment protocol does not seem feasible for high-risk patients (6), and there is an urgent need for implementation of targeted therapies. Furthermore, molecular biomarkers, in addition to MRD detection, could improve the upfront identification of high-risk patients and therefore guide the treatment of these patients with an intensified chemotherapeutic regimen or, whenever available, targeted agents. Unfortunately, such genetic biomarkers are not yet included in the risk stratification of newly diagnosed patients with T-ALL.
The clinical testing of targeted agents in the oncology field has dramatically increased over the last years. Nevertheless, targeted treatment options for patients with T-ALL remain limited. In fact, unlike other leukemias such as chronic myeloid leukemia (CML) and Philadelphia-positive ALL, which are kinase-driven malignancies, the initiating events in T-ALL cause the ectopic expression of transcription factors (type A aberrations) that drive leukemogenesis. However, the additional genetic lesions that are required for full transformation into malignancy (the so-called type B mutations) potentially serve as druggable vulnerabilities. Therefore, the thorough investigation of T-ALL oncogenic molecular pathways and their intricate RNA and protein signaling networks that sustain proliferation and survival can offer opportunities for the implementation of personalized targeted therapies (7). Potential limitations to the use of targeted drugs in pediatric T-ALL include clonal heterogeneity of the disease, resulting in only partial elimination of leukemia cells upon therapy. Therefore, resistant clones may be selected and survive under the selective pressure of treatment (8, 9). Similar resistance mechanisms have already been demonstrated for conventional chemotherapeutics such as the glucocorticoid-selected NR3C1 mutations (10, 11, 12) and the 6-mercaptopurine–selected NT5C2 mutations in chemoresistant relapsed ALL (11, 13). Already in 2017, the Innovative Therapies for Children with Cancer (ITCC) Consortium advised a change in the setup of early-phase pediatric clinical trials in order to accelerate the access of interesting drugs to randomized trials (14). ITCC has proposed to extrapolate data from adult clinical trials as a starting point for first-in-child trial designs. In addition, ITCC suggested the addition of homogeneous expansion cohorts to assess pharmacodynamic and pharmacokinetic parameters for the therapeutic agents tested and to detect early signs of antitumor activities. Furthermore, it has become evident that molecular tumor profiling is needed to study cancer heterogeneity and to understand therapy-induced mutations and the insurgence of relapse (14). Supplementary Table S1 offers an overview of current clinical trials that investigate targeted agents for T-ALL. In the following paragraphs, we summarize the main recurrent T-ALL oncogenic drivers and targetable genetic lesions and highlight the most important preclinical and clinical evidence to implement promising drugs in clinical trials for patients with T-ALL. In particular, we discuss agents that target activated pathways by specific genomic lesions in T-ALL and drugs already approved for cancer treatment that are under clinical investigation for patients with T-ALL. Moreover, we briefly discuss novel therapeutic options for which promising preclinical results were obtained in T-ALL models and that should be taken into consideration for future research. The agents discussed here include modifiers of apoptosis; inhibitors of transcriptional regulators, signal transduction, and the cell cycle; and immunotherapies. Figure 1 offers a visual summary of the relevant targets and therapeutic agents described throughout this review.
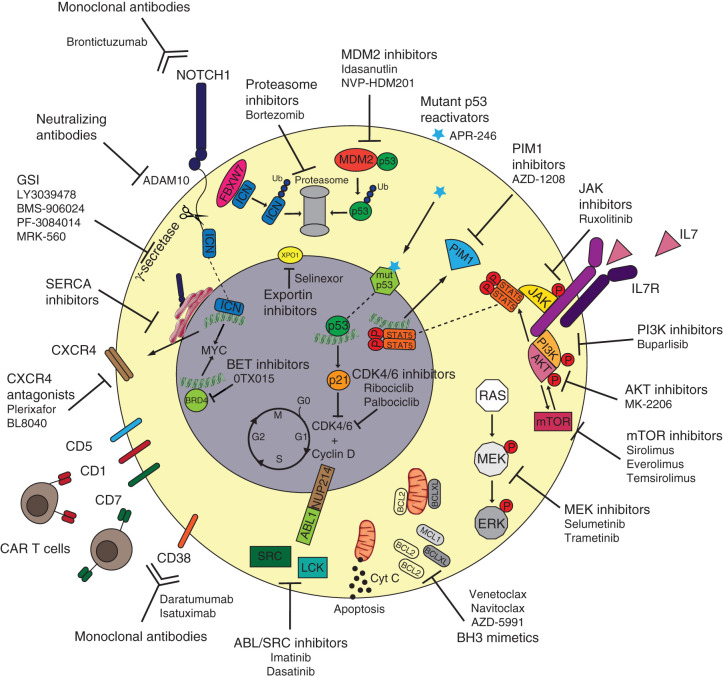
Figure 1.
Targeted therapies to tackle T-ALL vulnerabilities. Oncogenic NOTCH1 signaling can be inhibited via different strategies such as monoclonal antibodies blocking the NOTCH1 receptor itself (brontictuzumab), monoclonal antibodies blocking the ADAM10 metalloprotease that releases extracellular NOTCH1, gamma-secretase inhibitors (GSI) preventing the release of intracellular ICN1, and SERCA inhibitors that block the maturation of NOTCH1 and its localization on the cell surface. Because NOTCH1-mutated T-ALL cases can present higher CXCR4 surface expression, CXCR4 antagonists (plerixafor and BL8040) can be used to tackle NOTCH1-driven T-ALL as well. Immunotherapy approaches for T-ALL include monoclonal antibodies against surface CD38 (daratumumab or isatuximab) as well as CAR T cells directed toward surface CD1, CD5, CD7, and CD38. The increased expression of antiapoptotic BH3 proteins such as BCL2 and BCLXL can be counteracted by the use of BH3 mimetics (venetoclax, navitoclax, and AZD-5991). The oncogenic signaling of ABL1 fusion proteins as well as aberrant activity of Src-family kinases can be inhibited by the tyrosine kinase inhibitors imatinib and dasatinib. The aberrant IL7R signaling cascade can be tackled using multiple targeted agents including JAK inhibitors (ruxolitinib), PIM1 inhibitors (AZD-1208), PI3K inhibitors (buparlisib), AKT inhibitors (MK-2206), mTOR inhibitors (sirolimus, everolimus, or temsirolimus), and MEK inhibitors (selumetinib or trametinib). APR-246 can bind mutant p53 and restore its wild-type, tumor-suppressor function, whereas MDM2 inhibitors (idasanutlin and NVP-HDM201) can prevent wild-type p53 ubiquitination and consequent degradation via the proteasome. Alternatively, tumor-suppressor proteins' degradation can be prevented by proteasome inhibitors (bortezomib). Increased activity of cell-cycle regulators CDK4/6 can be blocked by CDK inhibitors (ribociclib or palbociclib), whereas aberrant transcription induced by BRD4 can be targeted by BET inhibitors (OTX015). Nuclear trafficking of oncogenic mRNA and proteins can be targeted via XPO1 inhibitors (selinexor).
Oncogenic Drivers and T-ALL Subtypes
Historically, three main T-ALL differentiation stages were identified based on the expression of cluster of differentiation (CD) markers on the cell surface and were denoted as early/precortical, cortical, and mature in analogy with the thymocytes' developmental stages (15). With the rapid development of (cyto)genetic technologies and next-generation sequencing in the last two decades, it was possible to identify genetic drivers that, in case of T-ALL, are transcription factors that are ectopically activated due to chromosomal rearrangements or deletions (reviewed in ref. 7). Initially using gene expression profiling (16, 17), which has been replaced by the identification of recurrent genomic abnormalities via genome sequencing (18, 19), patients with T-ALL can be clustered in four main subtypes with characteristic oncogenic aberrations, namely early thymocyte progenitor (ETP)/immature-ALL, TLX, TLX1/NKX2.1 (originally denoted as proliferative subgroup), and TAL/LMO. Figure 2 illustrates the main features of each subtype. The ETP-ALL group includes the most immature T-ALL cases (approximately 10% of the total T-ALL cases) that present a gene expression profile similar to hematopoietic stem cells and myeloid progenitors, with a high expression of self-renewal genes including LMO2, LYL1, and HHEX and the antiapoptotic BCL2 (20). The mechanisms for high BCL2 expression in ETP-ALL are still poorly understood—the expression of this antiapoptotic protein could reflect a stem cell–like feature of immature cells, or it could be due to STAT5 activation downstream of recurrent IL7 signaling pathway mutations within this subgroup (21, 22). ETP-ALL cases show increased expression of the transcription factor MEF2C or genetic aberrations of MEF2C-associated transcription regulators such as SPI1, RUNX1, ETV6-NCOA2, and NKX2.5 (16). Interestingly, ETP-ALL blasts have higher mutational loads compared with blasts of other T-ALL subtypes (21, 22). In particular, although NOTCH1-activating mutations and cell-cycle regulators' CDKN2A/2B-inactivating mutations are relatively rare in ETP-ALL, recurrent activating aberrations involve kinase-encoding genes, such as FLT3, NRAS, IL7R, JAK1, and JAK3 (21, 22). In addition, recurrent 5q deletions result in deletion of the NR3C1 locus, encoding for the glucocorticoid receptor (GCR; refs. 22, 23). Interestingly, recent evidence demonstrated that reduced GCR expression can induce steroid resistance in T-ALL (12). Some ETP-ALL cases present genomic aberrations that activate genes of the HOXA locus. Such activating events have been correlated to chemoresistance and inferior outcome in adult ETP-ALL (24).
Figure 2.
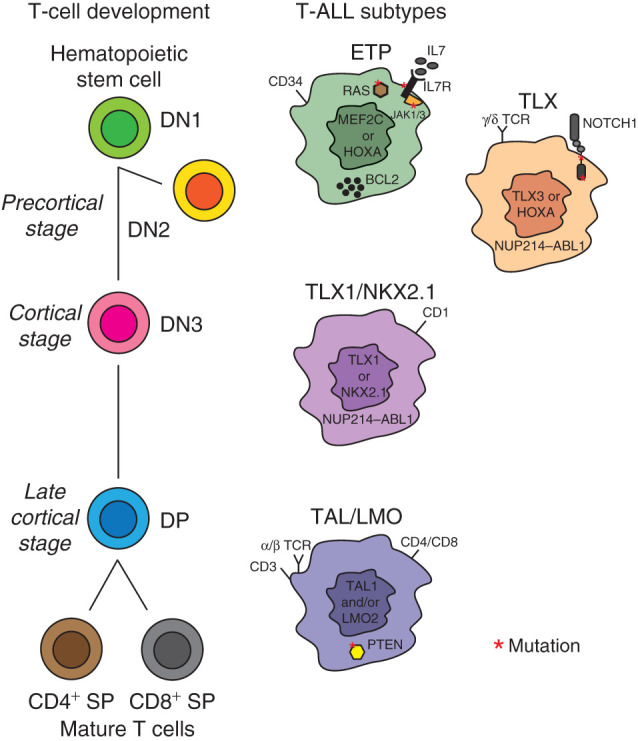
Thymocytes' developmental stages and T-ALL subtypes. The ETP-ALL subtype is driven by aberrant MEF2C or HOXA gene expression, presents frequent mutations in the IL7 signaling cascade, and shows higher BCL2 protein expression. Similarly to hematopoietic progenitors, ETP-ALL blasts express stem cell markers such as CD34. The TLX subgroup, driven by either TLX3- or HOXA-activating events, often presents NOTCH1 mutations and, in some cases, expression of the γ/δ T-cell receptor (TCR), in analogy to the precortical γ/δ T-cell progenitors (DN2 stage). The TLX1/NKX2.1 subgroup is driven by either NKX2.1 or TLX1 aberrations. TLX-rearranged cases can present the oncogenic NUP214–ABL1 fusion. The TAL/LMO subgroup, driven by the expression of the oncogenes TAL1 and LMO2, includes the most mature T-ALL cases. As for late cortical (SP) T-cell progenitors, TAL/LMO blasts express mature T-cell surface markers such as CD4, CD8, CD3, and α/β TCR and often present PTEN mutations.
The TLX group includes immature cases that either lack a functional T-cell receptor (TCR) or present a γ/δ TCR, which is in line with early or γ/δ T-cell lineage development (DN2 stage). A recent study suggests that patients with γ/δ T-ALL have higher MRD levels after induction chemotherapy compared with other T-ALL cases (25). Common genetic lesions within the TLX group include rearrangements of the transcription factor TLX3 (16, 17), mostly as consequence of recurrent TLX3–BCL11B translocations (26). These aberrations result in haploinsufficiency of the tumor suppressor BCL11B (27), which is a crucial regulator of the α/β lineage commitment during differentiation. Moreover, TLX3-rearranged T-ALLs often have NOTCH1-activating mutations (28) and aberrations in epigenetic regulators such as PHF6 and CTCF (18). Similar to various ETP-ALL cases, some TLX patients harbor alternative HOXA-driving events instead of TLX3-activating lesions (16).
The common features of the TLX1/NKX2.1 T-ALL group are genomic rearrangements involving either TLX1 or NKX2.1, CD1 expression, and differentiation arrest at the cortical (DN3-DP) stage of T-cell development. These cases present higher expression of genes involved in cell-cycle regulation and progression, DNA duplication, and spindle assembly (16, 17). T-ALL cases with TLX1 or NKX2.1 aberrations have been associated with excellent treatment outcomes (reviewed in ref. 7).
The TAL/LMO T-ALL subgroup comprises nearly half of all pediatric patients with T-ALL, and it is characterized by ectopic expression of TAL1 (either via translocation or SIL–TAL1 deletion), TAL2, LYL1, LMO1, LMO2, or LMO3 (driven by TCRB or TCRAD rearrangements; refs. 16, 17). Immunophenotypes of TAL/LMO patients mostly resemble late cortical (CD4+ single positive or CD8+ single positive) T-cell development stages. PTEN mutations are most common in this subgroup and have been associated with poor outcome (29). In addition, PIK3R1- or PIK3CG-activating lesions are frequent within this cluster (30, 31). Moreover, TAL1-rearranged cases often have mutations in the ubiquitin-specific protease USP7 that regulates MDM2 and TP53 stability (18).
Current and Novel Possibilities for Targeted Therapy
In the following paragraphs, we will discuss various classes of drugs and biological agents that provide novel strategies for targeted treatment. These are classified as modifiers of apoptosis; inhibitors of transcription regulation, signal transduction, and the cell cycle; and immunotherapies.
Modifiers of Apoptosis
BH3 Mimetics
Encouraged by significant responses of the BCL2 inhibitor venetoclax (ABT-199) in chronic lymphatic leukemia (32), BH3 mimetics became of great interest for the treatment of various hematologic malignancies. The sensitivity toward BH3 mimetics can be determined by BH3 profiling, a functional screening method that determines the “priming of death” state in cells by measuring specific BCL2 family member (e.g., BCL2, BCLXL, and/or MCL1) dependencies (33). BH3 profiling of T-ALL cell lines and patient blasts identified a dependency on BCL2 in ETP-ALL and BCLXL in the remaining subtypes of T-ALL (34). Consequently, immature/ETP-ALL cells are most responsive to venetoclax, whereas other T-ALL subtypes are more sensitive to navitoclax (ABT-263) treatment, respectively (34, 35). The BCL2/BCLW/BCLXL inhibitor navitoclax induces significant cell death in both T-ALL and B-cell precursor (BCP)-ALL patient-derived xenograft (PDX) models (36), but it can induce severe thrombocytopenia in vivo. First reports on pediatric and adult patients with relapsed/refractory T-ALL treated with venetoclax alone or combined with navitoclax showed promising results (37, 38). However, various resistance mechanisms toward venetoclax treatment have been reported in several hematologic malignancies including T-ALL, such as acquired BCL2 mutations, altered mitochondrial fitness, or MCL1 upregulation (36, 39, 40, 41). Combination treatment of venetoclax with other BH3 mimetics or PI3K/AKT/mTOR inhibitors significantly increases cell toxicity and overcomes venetoclax-induced resistance (39, 40). The MCL1 inhibitor S63845 also induces efficient cell death in various T-ALL cell lines as single treatment (39), therefore serving as an interesting alternative to venetoclax, especially given the limited dependency on BCL2 in most patients with T-ALL (34). Measuring BCL2 family dependencies can enable guided application of different BH3 mimetics for individualized medicine. In addition, the mitochondrial priming for apoptosis correlates with clinical responses in ALL and predicts for chemosensitivity, empowering the use of BH3 profiling as a functional screen in pediatric leukemia (42).
Transcriptional Regulator Inhibitors
NOTCH1 Inhibitors
Over 70% of T-ALL cases present NOTCH1-activating mutations (gain-of-function), and up to 25% of patients harbor mutations in the FBXW7 gene (18), which mediates the proteasomal degradation of NOTCH1. Gamma-secretase inhibitors (GSI) have been extensively studied as potential treatment for NOTCH1-activated tumors. Despite promising preclinical results, GSI failed during clinical trials due to insufficient efficacy (even in presence of NOTCH1 mutations) and excessive gastrointestinal toxicity caused by the concomitant inhibition of NOTCH2 in the gut epithelium (reviewed in ref. 43). Preclinical data showed that simultaneous corticosteroid administration can relieve gastrointestinal toxicity and enhance the GSI antitumor activity (44). Current clinical trials are investigating whether combined GSI and dexamethasone administration could be an effective therapeutic approach (NCT02518113 and NCT01363817). As an alternative strategy, Habets and colleagues showed a safe, selective GSI targeting of NOTCH1 signaling in T-ALL using a PSEN1 inhibitor (MRK-560; ref. 45). Although intestinal epithelial cells express both PSEN1 and PSEN2 subunits of the γ-secretase proteolytic complex, T-ALL cells express only PSEN1. In vivo preclinical data showed that γ-secretase inhibition by MRK-560 has antileukemic activity without causing intestinal toxicity in mouse xenografts derived from patients with T-ALL, offering a promising alternative therapeutic approach for NOTCH1-activated T-ALL cases (45). It is fair to question whether, despite high prevalence of NOTCH1 mutations in T-ALL, GSI is a valid strategy to efficiently and safely target this mutant protein and the consequent altered transcriptional program.
Additional strategies to block aberrant NOTCH1 signaling include monoclonal antibodies (46) or sarco-endoplasmic reticulum Ca2+-ATPase (SERCA) inhibitors that block NOTCH1 protein maturation by preventing its localization on the cell membrane (47). Other approaches to tackle oncogenic NOTCH1 involve the targeting of molecules that are activated upon NOTCH1-induced signaling. For example, it has been reported that GSI-resistant T-ALL cells express lower levels of the antiapoptotic protein MCL1. Because MCL1 can counteract the inhibition of BCL2 and BCLXL, cells with lower MCL1 expression are vulnerable to navitoclax treatment (48). At last, another emerging druggable player within NOTCH1 oncogenic signaling is CXCR4 (CD184), the chemokine receptor for CXCL12 that is released by stromal cells in the thymus. CXCR4 is upregulated in NOTCH1-driven T-ALL and promotes survival and proliferation in the bone marrow niche (reviewed in ref. 49). Therefore, CXCR4 antagonists, which are already largely used in the clinic to promote stem cells' mobilization into the bloodstream, could be repurposed as a therapeutic option for patients with leukemia. In fact, the novel CXCR4 inhibitor BL8040 is now in phase II clinical trial for patients with relapsed T-ALL/lymphoblastic lymphoma (LBL; Supplementary Table S1). Together, these studies show that there is potential for targeting mutant NOTCH1 or its downstream signaling.
BET Inhibitors
Bromodomain (BRD)-containing proteins affect gene transcription via binding to acetylated histones. Their functions include remodeling of the chromatin, modifying histones, and modulating transcription itself (50). The BRD and extraterminal (BET) family of BRD-containing proteins consists of four members: BRD2, BRD3, BRD4, and the testis-specific BRDT. One of the first small molecules developed to selectively inhibit BET proteins was JQ1 (50). In leukemia, BRD4 activity can drive aberrant MYC expression. Because MYC is an important and direct NOTCH1 target gene (51), NOTCH1-mutated T-ALL cases have increased MYC expression. In preclinical T-ALL models, JQ1 competes with BRD4, resulting in reduced MYC expression, decreased cell proliferation, and impaired tumor growth (52). Moreover, JQ1 treatment can synergize with vincristine (53) and with the BCL2 inhibitor venetoclax (54). Interestingly, T-ALL cells that acquire resistance to GSIs remain responsive to BRD4 inhibition by JQ1 (55), indicating that NOTCH1-mutated patients could benefit from BET inhibitor treatment. In addition to MYC, JQ1 also lowers the transcription of another important NOTCH1 target gene, the IL7 receptor (IL7R; ref. 56). Moreover, another BRD4-dependent transcription factor, ETS1, can cooperate with NOTCH1 during leukemogenesis. Because Ets1 deletion sensitizes T-ALL cells to GSI (57), targeting NOTCH1 transcriptional cofactors could offer an alternative strategy to treat NOTCH1-driven T-ALL cases.
Cancer cells often use superenhancer structures to restore and sustain oncogene expression. Guo and colleagues (58) showed that JQ1-resistant leukemic cells can restore MYC expression via enhancer remodeling. However, combined BET and cyclin-dependent kinase (CDK) 7 (transcriptional regulator) inhibition in JQ1-resistant cells effectively abrogates MYC expression. Pharmacologic targeting of CDK7 results in decreased enhancer activity in T-ALL and epigenetic reprogramming, in particular for NOTCH1-related enhancers that are not affected by GSI treatment (59). CDK7 inhibition also effectively disrupts the TAL1 superenhancer (60), highlighting that disruption of oncogenic transcription complexes may be an effective approach for T-ALL treatment when direct targeting of mutant genes, proteins, or pathways is not possible. Therefore, the investigation of the epigenetic state of leukemia cells can provide additional insights to guide the use of targeted treatments. Despite promising results in preclinical models, JQ1 has a very short half-life that limits its applicability as a therapeutic agent in vivo. Nevertheless, several novel BET inhibitors have been developed by multiple companies, and they are currently under investigation in oncology trials, highlighting the great interest in these epigenetic drugs and their potential application (61). Among these novel agents, OTX015 was proven effective in preclinical leukemia models (62).
Signal Transduction Inhibitors
ABL1/Src-Family Kinase Inhibitors
Unlike B-cell ALL (B-ALL) cases, patients with T-ALL with ABL1 fusions are rare (18, 63). The most common ABL1 aberration in T-ALL is the NUP214–ABL1 fusion due to an episomal amplification of the 9q34 region, which was one of the few discovered T-ALL lesions that can be directly targeted by a kinase inhibitor (63). Usually, NUP214–ABL1 rearrangements are particularly present at the subclonal level (64). Novel ZBTB16–ABL1 and ZMIZ1–ABL1 fusions have been identified in rare T-ALL cases (ref. 65 and unpublished observations), resulting in sensitivity toward imatinib and dasatinib treatment in preclinical models (65). Interestingly, in 2017, Frismantas and colleagues identified a subgroup of patients with T-ALL who are highly sensitive to dasatinib treatment in vitro despite the absence of ABL1 aberrations, suggesting a role for the SRC kinase as a putative novel target for therapy (66). Other studies proposed the lymphocytic-specific kinase LCK, which is often highly expressed in T-ALL, as a prime dasatinib target in T-ALL (67, 68). Based on these preclinical data, patients presenting high SRC phosphorylation and/or increased LCK expression could potentially benefit from dasatinib treatment. Therefore, in addition to genomic analyses, further investigation of the phospho-proteome could highlight aberrantly activated proteins (7) that could serve as biomarkers for dasatinib responsiveness when ABL1 abnormalities are not present.
JAK Inhibitors
JAK–STAT pathway activation in T-ALL is mainly observed in the context of IL7-induced signaling or caused by activating mutations in the IL7R gene or in genes encoding downstream effectors (e.g., JAK1, JAK3, or STAT5) that are recurrently found at diagnosis (18, 21, 69). Active JAK–STAT signaling leads to the upregulation of various antiapoptotic and prosurvival proteins including BCL2 and PIM1 and contributes to steroid resistance (21, 70, 71). Ruxolitinib, an FDA-approved JAK1/2 inhibitor for the treatment of myeloproliferative neoplasms (MPN) and graft-versus-host disease (GvHD), blocks JAK–STAT signaling regardless of the presence of mutations (72). In T-ALL, ruxolitinib shows efficacy in IL7-responsive T-ALL and ETP-ALL (69). Ruxolitinib treatment can synergize with dexamethasone treatment to overcome IL7-induced steroid resistance in patients with T-ALL and ETP-ALL. Multiple trials are underway to test the efficacy of ruxolitinib for JAK-mutated T-ALL (Supplementary Table S1) or Philadelphia-like BCP-ALL with CRLF2 rearrangements and/or JAK mutations (NCT2723994, NCT03117751, and NCT02420717) despite the fact that the clinical responses to ruxolitinib in MPNs seem rather limited (73). This indicates that the role of JAK inhibitors should be carefully considered in future treatment regimens of T-ALL.
PIM1 Inhibitors
When exploring alternative treatment options for aberrant JAK–STAT signaling, PIM1 was identified as a direct STAT5 transcriptional target gene that is also upregulated by physiologic IL7-induced signaling (71, 74, 75). Expression of the prosurvival PIM1 kinase is mainly observed in precortical T-ALL, with the highest expression in the TLX and ETP-ALL subtypes (71, 74, 76, 77). This is in agreement with the higher occurrence of activating mutations in the IL7R signaling pathway in these T-ALL subtypes, including JAK1/3 and STAT5B mutations (18, 21, 22, 78). PIM1 inhibition has proven efficacy in T-ALL using in vitro and in vivo models, with an increased effect observed for ETP-ALL blasts (74, 77). Both phospho-STAT5 and PIM1 expression levels can be used as a predictive biomarker for response to JAK inhibitors (74). PIM1 inhibition paradoxically results in enhanced MAPK–ERK signaling and may explain the observed synergy of combined PIM1 and MEK inhibitor treatment (74, 79). In addition, synergistic effects of PIM1 inhibitors in combination with venetoclax or dexamethasone have been observed (77, 80), indicating that PIM1 could be a valuable therapeutic target to counteract unfavorable hallmarks of immature/ETP-ALL cases such as high BCL2 expression and steroid resistance.
PI3K–AKT–mTOR Inhibitors
High PI3K–AKT–mTOR signaling is frequently observed in T-ALL and can be caused by a variety of cellular events, including activating mutations in PI3K or AKT, inactivating lesions in the tumor-suppressor gene PTEN, or posttranslational modification of these molecules (21, 30, 31, 81). PTEN-inactivating events are predominantly observed in patients with T-ALL that belong to the TAL/LMO subtype. PTEN loss is associated with poor prognosis in T-ALL, resulting in higher risk of disease relapse (29, 30, 81, 82). In addition, IL7R signaling mutations that frequently occur in ETP-ALL and TLX subtypes also activate the downstream PI3K–AKT pathway and correlate with steroid resistance and inferior event-free survival (21, 78). Pan-PI3K inhibitors have shown higher efficacy in inhibiting cell growth and survival of T-ALL cell lines compared with inhibitors that target only specific catalytic subunit(s) of PI3K (83). Preclinical in vitro studies demonstrate synergy between PI3K inhibitors and several chemotherapeutic agents, including doxorubicin, nelarabine, and glucocorticoids (21, 84, 85). Moreover, dual PI3K/mTOR inhibitors seem to be even more effective and also synergize with a wide range of chemotherapeutics (85, 86, 87, 88).
The effects of the first-generation allosteric mTOR inhibitors rapamycin (sirolimus) and rapalogs RAD001 (everolimus) and CCI-779 (temsirolimus) have been largely investigated in T-ALL (86, 89, 90). These inhibitors target only mTORC1 and can paradoxically activate AKT via PI3K/mTORC2 in some cell types (reviewed in ref. 91). Second-generation ATP-competitive dual mTORC1/mTORC2 inhibitors are more efficient in inducing apoptosis in T-ALL blasts because they also interfere with more downstream PI3K–AKT–mTOR signaling effectors, including a strong inhibition of 4EBP1 phosphorylation (92). The stronger cytotoxic effects and broad PI3K–AKT pathway regulation of dual inhibitors (e.g., PI3K/mTOR and mTORC1/mTORC2 inhibitors) compared with PI3K- or mTORC1-only inhibitors provide evidence that dual inhibitors are more suitable for future clinical trials (91, 93).
Alternatively, the oncogenic signaling of the PI3K–AKT–mTOR axis can also be targeted by direct AKT inhibition. The allosteric AKT inhibitor MK-2206 inhibits AKT and impairs downstream activation of mTORC1, mTORC2, GSK3, and FOXO in various T-ALL cell lines (94). In addition, MK-2206 synergizes with steroids in primary samples of patients with T-ALL (21, 94). ATP-competitive AKT inhibitors like AZD5363 also demonstrate cytotoxic effect against T-ALL cells in vitro (95).
MEK Inhibitors
The presence of mutations in N- and K-RAS genes at diagnosis, which strongly activate the MAPK–ERK signaling, predicts for inferior outcome in both patients with BCP- and T-ALL (82, 96, 97, 98). In addition, a high prevalence of these mutations in patients with ALL is found at relapse (10). Although not significantly enriched in relapsed T-ALL, the presence of RAS mutations in relapsed pediatric patients with T-ALL predicts for extremely poor outcome (99). MAPK–ERK-activating mutations, which may be selected under the pressure of treatment, can contribute to steroid resistance (21, 78, 100). MEK inhibitors induce cell death in RAS-mutant cells and synergize with glucocorticoids in primary T-ALL patient cells and in vivo BCP-ALL models (21, 97, 101, 102). These findings led to the ongoing SeluDex trial that combines the MEK inhibitor selumetinib with dexamethasone for the treatment of relapsed adult and pediatric patients with BCP- and T-ALL (NCT03705507; Supplementary Table S1). As IL7R and JAK1 signaling mutations strongly activate downstream MEK–ERK signaling, in addition to the JAK-STAT and PI3K-AKT pathways, and strongly provoke steroid resistance in T-ALL (21), patients having such IL7R signaling mutations should also become eligible for selumetinib treatment.
Cell-Cycle Inhibitors
CDK Inhibitors
More than 70% of T-ALL cases downregulate CDKN2A/B (18), negative regulators of CDK4/6, either via recurrent gene deletions, sporadic mutations, or promoter hypermethylation (103). Therefore, the CDK4/6 inhibitors palbociclib and ribociclib could be potential therapeutic options for patients with T-ALL. Palbociclib induces cell-cycle arrest in T-ALL cells and can suppress leukemia progression in animal models (104). Moreover, another preclinical study proved that the CDK4/6 inhibitor ribociclib can act synergistically with glucocorticoids and mTOR inhibitors in both T-ALL cell lines and murine models (90). Current phase I clinical trials for relapsed/refractory pediatric ALL (Supplementary Table S1) are investigating the tolerability of the combination of ribociclib with everolimus and dexamethasone (NCT03740334) or the addition of palbociclib to the standard reinduction chemotherapeutic regimen (NCT03792256). Other aberrations involving cell-cycle regulators include overexpression of the NOTCH1 target Cyclin D3 and CDK6 (18, 19, 21, 65, 99). Moreover, deletions of CDKN1B (p27KIP1), which is a negative regulator of the Cyclin E–CDK2 complex, have been reported in about 13% of patients with T-ALL (18). Therefore, inhibitors targeting CDK2 might be of interest for the treatment of T-ALL as well. In 2017, Moharram and colleagues reported the efficacy of the CDK1/2/5/9 inhibitor dinaciclib in preclinical T-ALL models (105). Despite the promising results, a clinical trial had already shown only transient effect of dinaciclib treatment for adult patients with leukemia (106).
Nelarabine
Active cell cycle may increase the sensitivity to nucleoside analogue treatment. Nelarabine is a purine nucleoside analogue that inhibits DNA synthesis and shows higher efficacy in T-ALL compared with other malignancies. Whether this is an exclusive T-ALL effect still remains debatable. Nevertheless, T-lymphoblasts show higher accumulation of nelarabine-active metabolite ara-G with consequent increased cytotoxicity compared with other hematopoietic cells (107), making T-ALL cells more susceptible to this treatment. At the moment, it is the only novel drug approved for the treatment of relapsed T-ALL/LBL cases. As a single agent for relapsed or refractory T-ALL in children and young adults, nelarabine had a response rate of over 50% (108). In adults, these response rates were somewhat lower (36% achieved complete remission), but they still provided encouraging results for relapsed cases by inducing clinical remissions that facilitated access to stem cell transplantation (109). However, nelarabine treatment can have significant neurologic side effects depending on other central nervous system (CNS)–directed therapy, in particular in children older than 10 years of age (110). The results of nelarabine safety and efficacy trials in patients with T-ALL/lymphoma highlight considerable single-agent activity in the relapse setting that facilitates disease control. Moreover, nelarabine can be combined with other drugs with nonoverlapping toxicities. The Children's Oncology Group recently published the results of a randomized phase III trial investigating the addition of nelarabine to the chemotherapeutic treatment for newly diagnosed pediatric and young adult patients with T-ALL. The increased disease free-survival rate as well as the decreased CNS relapse incidence without excessive toxicity support the inclusion of nelarabine into frontline therapy for pediatric T-ALL, especially for high-risk cases (111).
Drugs Targeting Mutant p53
Mutations that inactivate p53 are rare in patients with T-ALL at diagnosis (1%–6%) but show an increased incidence at relapse and correlate with poor prognosis (18, 99). A recent study showed that p53-mutant subclones that were detected at first relapse can give rise to clonal p53 mutations detectable in post–stem cell transplantation relapses. Furthermore, in these patients, p53 mutations correlated with an extremely short time to relapse (112). Various reactivators of mutant p53 that induce restoration of the wild-type conformation are in preclinical investigation (113). Interestingly, leukemic blasts from a patient with T-ALL who relapsed after stem cell transplantation showed sensitivity ex vivo to the p53 reactivator APR-246 (112). APR-246 has already shown promising results for p53-mutant patients affected by other hematologic malignancies (NCT00900614) and could be a suitable option for patients with T-ALL who relapse after stem cell transplantation and present with p53 mutations.
Drugs Targeting Wild-Type p53
The p53 signaling pathway can be impaired despite the presence of wild-type p53 by overexpression of physiologic p53 inhibitors such as MDM2 or MDM4. In fact, p53 activity can be restored by targeting the E3 ubiquitin ligase MDM2. The MDM2 antagonist idasanutlin disrupts the MDM2–p53 interaction and prevents p53 degradation. Currently, idasanutlin has reached phase I/II clinical trial investigation for pediatric ALL (NCT04029688). Furthermore, another MDM2 inhibitor, NVP-HDM201, is currently being investigated in a phase I/II clinical trial for wild-type p53 tumors, including relapsed ALL (NCT02143635). Lastly, the MDM2/MDM4 stapled peptide ALRN-6924 has reached clinical investigation in pediatric patients with relapsed ALL (NCT03654716).
Immunotherapies
Antibody-Based Therapy
Monoclonal antibodies can be applied in immunotherapies and have entered various trials for T-cell lymphoma (reviewed and summarized in ref. 114). Surprisingly, only a few have been considered in the treatment of ALL, such as anti-CD38 antibodies. CD38 is a transmembrane receptor that is expressed on subsets of myeloid, lymphoid, and some nonhematologic cells. The anti-CD38 monoclonal antibody daratumumab was initially developed for multiple myeloma and was approved by the FDA in 2015 and the European Medicines Agency in 2016 as a single agent for patients with relapsed/refractory multiple myeloma. CD38 is also a promising target for T-ALL as it is robustly and consistently expressed on T-ALL and ETP-ALL blasts at diagnosis, during chemotherapy treatment, and at relapse (115). Moreover, daratumumab displayed great efficacy in 14 of 15 PDX models in NSG mice (115). Of note, the cytotoxic efficacy of daratumumab in NSG mice—that do not have B, T, and natural killer cells, and complement factors—seems therefore independent of T-cell–mediated or complement-dependent cytotoxicity. CD38 expression on regulatory B and T cells as well as on myeloid suppressor cells results in their depletion by daratumumab, which could boost antitumor responses (116). Clinical trials will reveal whether daratumumab has an even higher efficacy than that observed in NSG mice, as both T-cell–mediated toxicity and repression of regulatory cells will be active in patients with T-ALL. Recently, daratumumab was successfully administered for compassionate use to 3 patients with CD38-positive ALL who experienced multiple relapses, with 1 patient who relapsed after an allogeneic stem cell transplantation (117). Two patients had T-ALL, whereas the third had a CD19/CD22-negative pre–B-ALL, and all three achieved an MRD-negative remission after daratumumab treatment. Trials combining daratumumab treatment with standard chemotherapy for pediatric and young adult patients with ALL are in phase II (NCT03384654; EudraCT 2017-003377-34). Another anti-CD38 monoclonal antibody that is under clinical investigation is isatuximab. An isatuximab trial for adult patients with T-ALL in the United States was closed prematurely due to lack of response, whereas the NCT03860844 trial for pediatric patients with refractory/relapsed acute leukemia is still ongoing.
Preclinical evidences suggest that TCR-expressing T-ALL blasts can be targeted by anti-CD3 antibodies. In fact, the activation of persistent TCR signaling induced by antibodies engaging CD3 leads to cell death in vitro and in vivo (118), suggesting a novel targeted therapeutic option for T-ALL cases that present TCR expression.
Cellular Therapy
Genetically engineered autologous chimeric antigen receptor T (CAR T) cells have been used successfully as therapy for various malignancies including relapsed ALL. An extensive review recently addressed the challenges and potential solutions for the use of CAR T cells in T-cell malignancies and lists all currently ongoing trials (119). Initially, the challenge to harvest sufficient mature T cells from patients with T-cell malignancies without any lymphoblast contamination hampered the development of CAR T cells against T-ALL/LBL. Most of the CAR T therapies developed so far are dependent on harvesting sufficient autologous and healthy T cells from a single patient. The production of allogenic CAR T cells would eliminate this challenge by using genetically modified T cells from a healthy donor (reviewed in ref. 120). In addition, the fratricide effect—the paradigm that CAR T cells share the same surface markers with their malignant T-cell targets—would rapidly self-extinguish the CAR T cells. After the first approval of the anti-CD19 CAR T for the treatment of pediatric patients with relapsed B-ALL, many different surface proteins have been investigated for the development of novel CAR T therapies directed toward T-cell malignancies, including CD5, CD7, CD1, and CD38. One of the advantages of anti-CD5 CAR T cells is the rapid internalization of CD5 from their cell surface, resulting in a limited and transient fratricide effect (121). Nevertheless, the internalization of CD5 can happen on blasts as well, offering an escape mechanism for leukemia cells that needs to be taken into account. Currently, a phase I anti-CD5 CAR T-cell trial is ongoing for patients with CD5-positive T-ALL or T-cell lymphoma (NCT03081910). As CD5 is expressed on most T-ALL subtypes, while it is absent or expressed at low levels on ETP-ALL cells, there is need for additional CAR T cells that can target ETP-ALL as well. CD7 is a promising target on T lymphoblasts but is also highly expressed on effector T cells. To minimize the fratricide effect, the CRISPR-Cas9 gene editing technology has been used to remove the endogenous CD7 gene from these CAR T cells (122). A clinical trial using these modified anti-CD7 CAR T cells for treating CD7-positive T-ALL/LBL has been designed (NCT03690011). However, because CD7 is expressed on all thymocytes and T cells, patients receiving CD7 CAR T-cell treatment risk a lifelong T-cell depletion and immunodeficiency that might impair a broad use in the clinic. In order to avoid such side effects and to regulate the activity of these cellular therapies, some CARs have been designed to express an inducible suicide gene (e.g., caspase 9) that can be selectively activated upon administration of a small molecule (reviewed in ref. 123). As an alternative strategy to target CD7, second-generation, fratricide-resistant anti-CD7 CAR T cells have been developed using T cells from healthy donors (UCART7; ref. 124). These CAR T cells have been genetically altered not only to be CD7 deficient but also to lack the TCRAD gene to eliminate the risk for an allogenic CAR T-cell–mediated GvHD. Of note, such an allogenic product can be immediately available for treatment of multiple patients as an “off-the-shelf” product. Promising results on the use of another allogeneic anti-CD7 CAR T-cell treatment were presented at the American Association for Cancer Research virtual meeting in April 2020. Wang and colleagues reported the preliminary exciting data on the efficacy of a single infusion of TruUCAR GC027 (Gracell Biotechnologies) after 6 days of lymphodepleting chemotherapy in 5 adult patients with refractory/relapsed T-ALL enrolled in a phase I clinical trial in China (ChiCTR1900025311). Four patients achieved complete response at day 28 with manageable cytokine release syndrome and absence of neurotoxicities and GvHD, whereas 1 patient who had received the lowest CAR T dose relapsed. Three of 4 patients remained in complete remission at day 161 of follow-up. Future evaluations will investigate the duration of the remissions induced by this treatment (125).
CD1a is another promising target for refractory or relapsed cortical T-ALL (126). Moreover, CD1a is expressed only during the proliferative phase of thymocyte development and not on immature progenitor cells or mature T cells, limiting the risk of complete immunodeficiency after treatment. Recently, the development of fratricide-resistant anti-CD1a CAR T cells for the treatment of CD1a-positive T-ALL has been reported (126). However, because patients with CD1-positive cortical T-ALL have been associated with excellent outcomes, it is not known what percentage of patients with relapsed T-ALL will express CD1 and thus benefit from such a CAR T therapy.
As discussed in the previous section, CD38 is widely expressed on T lymphoblasts, thus the development of anti-CD38 CAR T has also been pursued (127). Recently, the treatment of a relapsed adult patient with B-ALL was reported with the occurrence of serious side effects including cytokine release syndrome and damage to lung and liver tissues that also express the CD38 antigen (128). Therefore, caution and accurate target choices are warranted to extend the repertoire of safe and effective CAR T-cell treatments.
Other Promising Targeted Treatments in Development
Oncology drug development is constantly growing, and several potential novel candidates have recently been put into the spotlight. New, potentially promising compounds that should be kept in consideration for upcoming studies will be discussed below.
OBI-3424 is a first-in-class targeted treatment for liquid and solid tumors that overexpress the Aldo-Keto Reductase 1 c3 (AKR1C3) enzyme such as castrate-resistant prostate cancer and hepatocellular carcinoma. AKR1C3 is also expressed in T-ALL, with the exclusion of TLX1/3-rearranged cases (129). OBI-3424 is a prodrug that releases a potent DNA-alkylating component upon intracellular reduction by AKR1C3. This agent has shown promising cytotoxic activity in T-ALL cell lines and PDXs that express AKR1C3 (129). In September 2017, OBI-3424 received FDA orphan drug designation for AKR1C3-expressing tumors, including ALL, and it is currently being investigated in a phase I/II clinical trial for solid tumors (NCT03592264).
Selinexor (KPT-330) is a selective inhibitor of Exportin-1 (XPO1) that has recently been approved in combination with dexamethasone for the treatment of refractory/relapsed multiple myeloma. XPO1 is the key player in nuclear export of receptors (e.g., NR3C1), tumor-suppressor proteins (e.g., p53 and pRB) but also oncogenic mRNAs transcribed from MDM2, BCL2, and MYC, which will be retained in the nucleus upon XPO1 inhibition. Selinexor treatment is currently being investigated in a phase I clinical trial for relapsed pediatric acute leukemia (NCT02091245). Furthermore, the second-generation XPO1 inhibitor eltanexor (KPT-8602) can induce cytotoxicity and apoptosis in ALL models and can enhance the efficacy of dexamethasone treatment (130).
Histone deacetylases (HDAC) are key enzymes in chromatin remodeling and epigenetic gene regulation. HDACs are frequently overexpressed in cancer, including T-ALL. Samples of patients with T-ALL demonstrate higher HDAC1 and HDAC4 but lower HDAC5 levels compared with B-ALL (131). The pan-HDAC inhibitor panobinostat has shown antileukemic activity in T-ALL preclinical models (132), and it is under clinical investigation for relapsed acute leukemia (Supplementary Table S1). The same applies for vorinostat, which is already approved for the treatment of refractory/relapsed cutaneous T-cell lymphoma.
Additional epigenetic regulators that can be pharmacologically targeted are DNA methyltransferases. DNA methyltransferase inhibitors decitabine and azacitidine induce chromatin hypomethylation with a consequent alteration in gene transcription. They have been approved for the treatment of myelodysplastic syndromes and are currently being investigated in early-phase clinical trials for pediatric patients with ALL (Supplementary Table S1). In 2016, Lu and colleagues showed that decitabine pretreatment enhanced chemosensitivity of preclinical models of ETP-ALL (133). One year later, the successful treatment of a relapsed adult patient with ETP-ALL with decitabine was reported (134), therefore offering a promising opportunity for salvage therapy of ETP-ALL cases.
An alternative way to target oncogenic signaling pathways is by tackling protein stability or degradation. Cancer cells become addicted to the rapid elimination of tumor-suppressor proteins or may require higher protein turnover to sustain their metabolism. Therefore, processes involved in protein degradation can provide leukemia-specific vulnerabilities that can be effectively targeted. Bortezomib, a first-in-class proteasome inhibitor, is approved for the treatment of refractory multiple myeloma. It inhibits the 26S subunit of the proteasome, impairing protein degradation that results in cell-cycle arrest and eventually apoptosis. A recent report of the Children's Oncology Group highlights the safety of bortezomib during reinduction chemotherapy for pediatric relapsed ALL and provided encouraging results for T-ALL, with an increase in patients achieving complete remission (135). Another way of altering protein stability and activity is through inhibition of the Nedd8-activating enzyme (NAE). NAE is an ubiquitin-like protein that regulates the activity and the protein–protein interactions of NF-kB and cullins, which are essential cell-cycle regulators (136). Preclinical data showed that the NAE inhibitor pevonedistat (MLN4924) can induce cell-cycle arrest and apoptosis in T-ALL models (136). Both bortezomib and pevonedistat are currently under clinical investigation for patients with ALL (Supplementary Table S1).
Aurora kinases (AURK) are mitotic regulators often overexpressed in cancer, including pediatric ALL (137). The AURKA inhibitor alisertib (MLN8237) had shown promising results for both ALL and lymphoma cells in vitro (138). Unfortunately, a phase II clinical trial from the Children's Oncology Group reported objective response after alisertib single-agent treatment in less than 5% of the pediatric patients with recurrent/refractory advanced solid tumor or acute leukemia (139). Recent evidence elucidates a role for AURKB in inhibiting proteasomal degradation of MYC, thus stabilizing this oncogenic protein (140). In vitro treatment of T-ALL cells with the AURKB inhibitor barasertib (AZD1152) leads to reduced MYC protein levels (140) and enhanced cell death (140, 141). Furthermore, AZD1152 can act in synergy with vincristine (140).
Conclusions
The outcome for children diagnosed with T-ALL has dramatically improved in the last decades. Nevertheless, therapy resistance, disease relapse, treatment-related death, and long-term detrimental side effects for cancer survivors remain serious issues to be solved. In addition, the lack of predictive biomarkers at diagnosis remains an unmet need for patients with T-ALL. In this review, we presented an overview of the current state of drug development and ongoing clinical trials that are of interest for the T-ALL field, integrating preclinical evidence and clinical data. Several molecular tumor profiling protocols have been initiated in Europe (e.g., MOSCATO-01, iTHER, and ESMART; ref. 142) to identify actionable lesions for targeted treatment in specific subgroups of patients. This highlights the importance of bridging preclinical research with clinical practice to accelerate the use of promising novel drugs in effective new treatment combinations for patients with T-ALL.
Authors' Disclosures
No disclosures were reported.
Supplementary Material
Acknowledgments
This study was supported by Dutch Cancer Society (KWF Kankerbestrijding, grant number KWF2016_10355, to V. Cordo') and Stichting Kinderen Kankervrij (grant number KiKa219, to J.C.G. van der Zwet; grant number KiKa295, to K. Canté-Barrett).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Blood Cancer Discov 2021;2:19–31
References
- 1. Patel AA, Thomas J, Rojek AE, Stock W. Biology and treatment paradigms in T cell acute lymphoblastic leukemia in older adolescents and adults. Curr Treat Options Oncol 2020;21:57. [DOI] [PubMed] [Google Scholar]
- 2. Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med 2006;354:166–78. [DOI] [PubMed] [Google Scholar]
- 3. Pieters R, de Groot-Kruseman H, Van der Velden V, Fiocco M, van den Berg H, de Bont E, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol 2016;34:2591–601. [DOI] [PubMed] [Google Scholar]
- 4. Pui CH, Pei D, Coustan-Smith E, Jeha S, Cheng C, Bowman WP, et al. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol 2015;16:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masson E, Relling MV, Synold TW, Liu Q, Schuetz JD, Sandlund JT, et al. Accumulation of methotrexate polyglutamates in lymphoblasts is a determinant of antileukemic effects in vivo. A rationale for high-dose methotrexate. J Clin Invest 1996;97:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeha S, Pei D, Choi J, Cheng C, Sandlund JT, Coustan-Smith E, et al. Improved CNS control of childhood acute lymphoblastic leukemia without cranial irradiation: St Jude Total Therapy Study 16. J Clin Oncol 2019;37:3377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Zwet JCG, Cordo V, Cante-Barrett K, Meijerink JPP. Multi-omic approaches to improve outcome for T-cell acute lymphoblastic leukemia patients. Adv Biol Regul 2019;74:100647. [DOI] [PubMed] [Google Scholar]
- 8. Waanders E, Gu Z, Dobson SM, Antic Z, Crawford JC, Ma X, et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov 2020;1:96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobson SM, Garcia-Prat L, Vanner RJ, Wintersinger J, Waanders E, Gu Z, et al. Relapse-fated latent diagnosis subclones in acute B lineage leukemia are drug tolerant and possess distinct metabolic programs. Cancer Discov 2020;10:568–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B, Brady SW, Ma X, Shen S, Zhang Y, Li Y, et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020;135:41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wandler AM, Huang BJ, Craig JW, Hayes K, Yan H, Meyer LK, et al. Loss of glucocorticoid receptor expression mediates in vivo dexamethasone resistance in T-cell acute lymphoblastic leukemia. Leukemia 2020;34:2025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013;19:368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moreno L, Pearson ADJ, Paoletti X, Jimenez I, Geoerger B, Kearns PR, et al. Early phase clinical trials of anticancer agents in children and adolescents - an ITCC perspective. Nat Rev Clin Oncol 2017;14:497–507. [DOI] [PubMed] [Google Scholar]
- 15. Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia 1995;9:1783–6. [PubMed] [Google Scholar]
- 16. Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 2011;19:484–97. [DOI] [PubMed] [Google Scholar]
- 17. Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 2002;1:75–87. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49:1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet 2017;49:1274–81. [DOI] [PubMed] [Google Scholar]
- 20. Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009;10:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Buijs-Gladdines JG, Cante-Barrett K, Stubbs AP, Vroegindeweij EM, Smits WK, et al. IL-7 receptor mutations and steroid resistance in pediatric T cell acute lymphoblastic leukemia: a Genome Sequencing Study. PLoS Med 2016;13:e1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. La Starza R, Barba G, Demeyer S, Pierini V, Di Giacomo D, Gianfelici V, et al. Deletions of the long arm of chromosome 5 define subgroups of T-cell acute lymphoblastic leukemia. Haematologica 2016;101:951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bond J, Marchand T, Touzart A, Cieslak A, Trinquand A, Sutton L, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica 2016;101:732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pui CH, Pei D, Cheng C, Tomchuck SL, Evans SN, Inaba H, et al. Treatment response and outcome of children with T-cell acute lymphoblastic leukemia expressing the gamma-delta T-cell receptor. Oncoimmunology 2019;8:1599637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su XY, Della-Valle V, Andre-Schmutz I, Lemercier C, Radford-Weiss I, Ballerini P, et al. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t(5;14)(q35;q32). Blood 2006;108:4198–201. [DOI] [PubMed] [Google Scholar]
- 27. Gutierrez A, Kentsis A, Sanda T, Holmfeldt L, Chen SC, Zhang J, et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 2011;118:4169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia 2010;24:2014–22. [DOI] [PubMed] [Google Scholar]
- 29. Paganin M, Grillo MF, Silvestri D, Scapinello G, Buldini B, Cazzaniga G, et al. The presence of mutated and deleted PTEN is associated with an increased risk of relapse in childhood T cell acute lymphoblastic leukaemia treated with AIEOP-BFM ALL protocols. Br J Haematol 2018;182:705–11. [DOI] [PubMed] [Google Scholar]
- 30. Zuurbier L, Petricoin EF, 3rd, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica 2012;97:1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood 2009;114:647–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med 2016;374:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butterworth M, Pettitt A, Varadarajan S, Cohen GM. BH3 profiling and a toolkit of BH3-mimetic drugs predict anti-apoptotic dependence of cancer cells. Br J Cancer 2016;114:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chonghaile TN, Roderick JE, Glenfield C, Ryan J, Sallan SE, Silverman LB, et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov 2014;4:1074–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peirs S, Matthijssens F, Goossens S, Van de Walle I, Ruggero K, de Bock CE, et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood 2014;124:3738–47. [DOI] [PubMed] [Google Scholar]
- 36. Suryani S, Carol H, Chonghaile TN, Frismantas V, Sarmah C, High L, et al. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clin Cancer Res 2014;20:4520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lacayo NJ, Pullarkat VA, Stock W, Jabbour E, Bajel A, Rubnitz J, et al. Safety and efficacy of venetoclax in combination with navitoclax in adult and pediatric relapsed/refractory acute lymphoblastic leukemia and lymphoblastic lymphoma. Blood 2019;134:285. [Google Scholar]
- 38. La Starza R, Cambò B, Pierini A, Bornhauser B, Montanaro A, Bourquin JP, et al. Venetoclax and bortezomib in relapsed/refractory early T-cell precursor acute lymphoblastic leukemia. JCO Precis Oncol 2019;3:PO.19.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z, He S, Look AT. The MCL1-specific inhibitor S63845 acts synergistically with venetoclax/ABT-199 to induce apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia 2019;33:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choudhary GS, Al-Harbi S, Mazumder S, Hill BT, Smith MR, Bodo J, et al. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis 2015;6:e1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen X, Glytsou C, Zhou H, Narang S, Reyna DE, Lopez A, et al. Targeting mitochondrial structure sensitizes acute myeloid leukemia to venetoclax treatment. Cancer Discov 2019;9:890–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 2011;334:1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol Ther 2014;141:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Samon JB, Castillo-Martin M, Hadler M, Ambesi-Impiobato A, Paietta E, Racevskis J, et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther 2012;11:1565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Habets RA, de Bock CE, Serneels L, Lodewijckx I, Verbeke D, Nittner D, et al. Safe targeting of T cell acute lymphoblastic leukemia by pathology-specific NOTCH inhibition. Sci Transl Med 2019;11:eaau6246. [DOI] [PubMed] [Google Scholar]
- 46. Agnusdei V, Minuzzo S, Frasson C, Grassi A, Axelrod F, Satyal S, et al. Therapeutic antibody targeting of Notch1 in T-acute lymphoblastic leukemia xenografts. Leukemia 2014;28:278–88. [DOI] [PubMed] [Google Scholar]
- 47. Marchesini M, Gherli A, Montanaro A, Patrizi L, Sorrentino C, Pagliaro L, et al. Blockade of oncogenic NOTCH1 with the SERCA inhibitor CAD204520 in T cell acute lymphoblastic leukemia. Cell Chem Biol 2020;27:678–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dastur A, Choi A, Costa C, Yin X, Williams A, McClanaghan J, et al. NOTCH1 represses MCL-1 levels in GSI-resistant T-ALL, making them susceptible to ABT-263. Clin Cancer Res 2019;25:312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsaouli G, Ferretti E, Bellavia D, Vacca A, Felli MP. Notch/CXCR4 partnership in acute lymphoblastic leukemia progression. J Immunol Res 2019;2019:5601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 2006;103:18261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood 2014;123:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Loosveld M, Castellano R, Gon S, Goubard A, Crouzet T, Pouyet L, et al. Therapeutic targeting of c-Myc in T-cell acute lymphoblastic leukemia, T-ALL. Oncotarget 2014;5:3168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peirs S, Frismantas V, Matthijssens F, Van Loocke W, Pieters T, Vandamme N, et al. Targeting BET proteins improves the therapeutic efficacy of BCL-2 inhibition in T-cell acute lymphoblastic leukemia. Leukemia 2017;31:2037–47. [DOI] [PubMed] [Google Scholar]
- 55. Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 2014;46:364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 2012;120:2843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCarter AC, Gatta GD, Melnick A, Kim E, Sha C, Wang Q, et al. Combinatorial ETS1-dependent control of oncogenic NOTCH1 enhancers in T-cell leukemia. Blood Cancer Discov 2020;1:178–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guo L, Li J, Zeng H, Guzman AG, Li T, Lee M, et al. A combination strategy targeting enhancer plasticity exerts synergistic lethality against BETi-resistant leukemia cells. Nat Commun 2020;11:740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kloetgen A, Thandapani P, Ntziachristos P, Ghebrechristos Y, Nomikou S, Lazaris C, et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat Genet 2020;52:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014;511:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cochran AG, Conery AR, Sims RJ, 3rd. Bromodomains: a new target class for drug development. Nat Rev Drug Discov 2019;18:609–28. [DOI] [PubMed] [Google Scholar]
- 62. Astorgues-Xerri L, Vazquez R, Odore E, Rezai K, Kahatt C, Mackenzie S, et al. Insights into the cellular pharmacological properties of the BET-inhibitor OTX015/MK-8628 (birabresib), alone and in combination, in leukemia models. Leuk Lymphoma 2019;60:3067–70. [DOI] [PubMed] [Google Scholar]
- 63. Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet 2004;36:1084–9. [DOI] [PubMed] [Google Scholar]
- 64. Graux C, Stevens-Kroef M, Lafage M, Dastugue N, Harrison CJ, Mugneret F, et al. Heterogeneous patterns of amplification of the NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Leukemia 2009;23:125–33. [DOI] [PubMed] [Google Scholar]
- 65. Chen B, Jiang L, Zhong ML, Li JF, Li BS, Peng LJ, et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2018;115:373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Frismantas V, Dobay MP, Rinaldi A, Tchinda J, Dunn SH, Kunz J, et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood 2017;129:e26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Serafin V, Capuzzo G, Milani G, Minuzzo SA, Pinazza M, Bortolozzi R, et al. Glucocorticoid resistance is reverted by LCK inhibition in pediatric T-cell acute lymphoblastic leukemia. Blood 2017;130:2750–61. [DOI] [PubMed] [Google Scholar]
- 68. Shi Y, Beckett MC, Blair HJ, Tirtakusuma R, Nakjang S, Enshaei A, et al. Phase II-like murine trial identifies synergy between dexamethasone and dasatinib in T-cell acute lymphoblastic leukemia. Haematologica 2020. Mar 5 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maude SL, Dolai S, Delgado-Martin C, Vincent T, Robbins A, Selvanathan A, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood 2015;125:1759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Delgado-Martin C, Meyer LK, Huang BJ, Shimano KA, Zinter MS, Nguyen JV, et al. JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias. Leukemia 2017;31:2568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. de Bock CE, Demeyer S, Degryse S, Verbeke D, Sweron B, Gielen O, et al. HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov 2018;8:616–31. [DOI] [PubMed] [Google Scholar]
- 72. Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010;363:1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Greenfield G, McPherson S, Mills K, McMullin MF. The ruxolitinib effect: understanding how molecular pathogenesis and epigenetic dysregulation impact therapeutic efficacy in myeloproliferative neoplasms. J Transl Med 2018;16:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Padi SKR, Luevano LA, An N, Pandey R, Singh N, Song JH, et al. Targeting the PIM protein kinases for the treatment of a T-cell acute lymphoblastic leukemia subset. Oncotarget 2017;8:30199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ribeiro D, Melao A, van Boxtel R, Santos CI, Silva A, Silva MC, et al. STAT5 is essential for IL-7-mediated viability, growth, and proliferation of T-cell acute lymphoblastic leukemia cells. Blood Adv 2018;2:2199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. La Starza R, Messina M, Gianfelici V, Pierini V, Matteucci C, Pierini T, et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia 2018;32:1807–10. [DOI] [PubMed] [Google Scholar]
- 77. De Smedt R, Peirs S, Morscio J, Matthijssens F, Roels J, Reunes L, et al. Pre-clinical evaluation of second generation PIM inhibitors for the treatment of T-cell acute lymphoblastic leukemia and lymphoma. Haematologica 2019;104:e17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cante-Barrett K, Spijkers-Hagelstein JA, Buijs-Gladdines JG, Uitdehaag JC, Smits WK, van der Zwet J, et al. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016;30:1832–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin YW, Beharry ZM, Hill EG, Song JH, Wang W, Xia Z, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood 2010;115:824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. De Smedt R, Morscio J, Reunes L, Roels J, Bardelli V, Lintermans B, et al. Targeting cytokine- and therapy-induced PIM1 activation in preclinical models of T-cell acute lymphoblastic leukemia and lymphoma. Blood 2020;135:1685–95. [DOI] [PubMed] [Google Scholar]
- 81. Mendes RD, Sarmento LM, Cante-Barrett K, Zuurbier L, Buijs-Gladdines JG, Povoa V, et al. PTEN microdeletions in T-cell acute lymphoblastic leukemia are caused by illegitimate RAG-mediated recombination events. Blood 2014;124:567–78. [DOI] [PubMed] [Google Scholar]
- 82. Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengline E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol 2013;31:4333–42. [DOI] [PubMed] [Google Scholar]
- 83. Lonetti A, Cappellini A, Sparta AM, Chiarini F, Buontempo F, Evangelisti C, et al. PI3K pan-inhibition impairs more efficiently proliferation and survival of T-cell acute lymphoblastic leukemia cell lines when compared to isoform-selective PI3K inhibitors. Oncotarget 2015;6:10399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lonetti A, Antunes IL, Chiarini F, Orsini E, Buontempo F, Ricci F, et al. Activity of the pan-class I phosphoinositide 3-kinase inhibitor NVP-BKM120 in T-cell acute lymphoblastic leukemia. Leukemia 2014;28:1196–206. [DOI] [PubMed] [Google Scholar]
- 85. Lonetti A, Cappellini A, Bertaina A, Locatelli F, Pession A, Buontempo F, et al. Improving nelarabine efficacy in T cell acute lymphoblastic leukemia by targeting aberrant PI3K/AKT/mTOR signaling pathway. J Hematol Oncol 2016;9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chiarini F, Fala F, Tazzari PL, Ricci F, Astolfi A, Pession A, et al. Dual inhibition of class IA phosphatidylinositol 3-kinase and mammalian target of rapamycin as a new therapeutic option for T-cell acute lymphoblastic leukemia. Cancer Res 2009;69:3520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hall CP, Reynolds CP, Kang MH. Modulation of glucocorticoid resistance in pediatric T-cell acute lymphoblastic leukemia by increasing BIM expression with the PI3K/mTOR inhibitor BEZ235. Clin Cancer Res 2016;22:621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gazi M, Moharram SA, Marhall A, Kazi JU. The dual specificity PI3K/mTOR inhibitor PKI-587 displays efficacy against T-cell acute lymphoblastic leukemia (T-ALL). Cancer Lett 2017;392:9–16. [DOI] [PubMed] [Google Scholar]
- 89. Batista A, Barata JT, Raderschall E, Sallan SE, Carlesso N, Nadler LM, et al. Targeting of active mTOR inhibits primary leukemia T cells and synergizes with cytotoxic drugs and signaling inhibitors. Exp Hematol 2011;39:457–72. [DOI] [PubMed] [Google Scholar]
- 90. Pikman Y, Alexe G, Roti G, Conway AS, Furman A, Lee ES, et al. Synergistic drug combinations with a CDK4/6 inhibitor in T-cell acute lymphoblastic leukemia. Clin Cancer Res 2017;23:1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov 2011;10:868–80. [DOI] [PubMed] [Google Scholar]
- 92. Yun S, Vincelette ND, Knorr KL, Almada LL, Schneider PA, Peterson KL, et al. 4EBP1/c-MYC/PUMA and NF-kappaB/EGR1/BIM pathways underlie cytotoxicity of mTOR dual inhibitors in malignant lymphoid cells. Blood 2016;127:2711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Martelli AM, Chiarini F, Evangelisti C, Cappellini A, Buontempo F, Bressanin D, et al. Two hits are better than one: targeting both phosphatidylinositol 3-kinase and mammalian target of rapamycin as a therapeutic strategy for acute leukemia treatment. Oncotarget 2012;3:371–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Simioni C, Neri LM, Tabellini G, Ricci F, Bressanin D, Chiarini F, et al. Cytotoxic activity of the novel Akt inhibitor, MK-2206, in T-cell acute lymphoblastic leukemia. Leukemia 2012;26:2336–42. [DOI] [PubMed] [Google Scholar]
- 95. Lynch JT, McEwen R, Crafter C, McDermott U, Garnett MJ, Barry ST, et al. Identification of differential PI3K pathway target dependencies in T-cell acute lymphoblastic leukemia through a large cancer cell panel screen. Oncotarget 2016;7:22128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014;124:3420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jerchel IS, Hoogkamer AQ, Aries IM, Steeghs EMP, Boer JM, Besselink NJM, et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia 2018;32:931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Driessen EM, van Roon EH, Spijkers-Hagelstein JA, Schneider P, de Lorenzo P, Valsecchi MG, et al. Frequencies and prognostic impact of RAS mutations in MLL-rearranged acute lymphoblastic leukemia in infants. Haematologica 2013;98:937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Richter-Pechanska P, Kunz JB, Hof J, Zimmermann M, Rausch T, Bandapalli OR, et al. Identification of a genetically defined ultra-high-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J 2017;7:e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Aries IM, van den Dungen RE, Koudijs MJ, Cuppen E, Voest E, Molenaar JJ, et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica 2015;100:e132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Matheson EC, Thomas H, Case M, Blair H, Jackson RK, Masic D, et al. Glucocorticoids and selumetinib are highly synergistic in RAS pathway-mutated childhood acute lymphoblastic leukemia through upregulation of BIM. Haematologica 2019;104:1804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kerstjens M, Driessen EM, Willekes M, Pinhancos SS, Schneider P, Pieters R, et al. MEK inhibition is a promising therapeutic strategy for MLL-rearranged infant acute lymphoblastic leukemia patients carrying RAS mutations. Oncotarget 2017;8:14835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jang W, Park J, Kwon A, Choi H, Kim J, Lee GD, et al. CDKN2B downregulation and other genetic characteristics in T-acute lymphoblastic leukemia. Exp Mol Med 2019;51:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell 2012;22:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moharram SA, Shah K, Khanum F, Marhall A, Gazi M, Kazi JU. Efficacy of the CDK inhibitor dinaciclib in vitro and in vivo in T-cell acute lymphoblastic leukemia. Cancer Lett 2017;405:73–8. [DOI] [PubMed] [Google Scholar]
- 106. Gojo I, Sadowska M, Walker A, Feldman EJ, Iyer SP, Baer MR, et al. Clinical and laboratory studies of the novel cyclin-dependent kinase inhibitor dinaciclib (SCH 727965) in acute leukemias. Cancer Chemother Pharmacol 2013;72:897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kurtzberg J, Ernst TJ, Keating MJ, Gandhi V, Hodge JP, Kisor DF, et al. Phase I study of 506U78 administered on a consecutive 5-day schedule in children and adults with refractory hematologic malignancies. J Clin Oncol 2005;23:3396–403. [DOI] [PubMed] [Google Scholar]
- 108. Berg SL, Blaney SM, Devidas M, Lampkin TA, Murgo A, Bernstein M, et al. Phase II study of nelarabine (compound 506U78) in children and young adults with refractory T-cell malignancies: a report from the Children's Oncology Group. J Clin Oncol 2005;23:3376–82. [DOI] [PubMed] [Google Scholar]
- 109. Gokbuget N, Basara N, Baurmann H, Beck J, Bruggemann M, Diedrich H, et al. High single-drug activity of nelarabine in relapsed T-lymphoblastic leukemia/lymphoma offers curative option with subsequent stem cell transplantation. Blood 2011;118:3504–11. [DOI] [PubMed] [Google Scholar]
- 110. Malone A, Smith OP. Nelarabine toxicity in children and adolescents with relapsed/refractory T-ALL/T-LBL: can we avoid throwing the baby out with the bathwater? Br J Haematol 2017;179:179–81. [DOI] [PubMed] [Google Scholar]
- 111. Dunsmore KP, Winter SS, Devidas M, Wood BL, Esiashvili N, Chen Z, et al. Children's Oncology Group AALL0434: a phase III randomized clinical trial testing nelarabine in newly diagnosed T-cell acute lymphoblastic leukemia. J Clin Oncol 2020;38:3282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hoell JI, Ginzel S, Kuhlen M, Kloetgen A, Gombert M, Fischer U, et al. Pediatric ALL relapses after allo-SCT show high individuality, clonal dynamics, selective pressure, and druggable targets. Blood Adv 2019;3:3143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018;18:89–102. [DOI] [PubMed] [Google Scholar]
- 114. Ghione P, Moskowitz AJ, De Paola NEK, Horwitz SM, Ruella M. Novel immunotherapies for T cell lymphoma and leukemia. Current Hematol Malig Rep 2018;13:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bride KL, Vincent TL, Im SY, Aplenc R, Barrett DM, Carroll WL, et al. Preclinical efficacy of daratumumab in T-cell acute lymphoblastic leukemia. Blood 2018;131:995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Krejcik J, Casneuf T, Nijhof IS, Verbist B, Bald J, Plesner T, et al. Daratumumab depletes CD38+ immune regulatory cells, promotes T-cell expansion, and skews T-cell repertoire in multiple myeloma. Blood 2016;128:384–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ofran Y, Ringelstein-Harlev S, Slouzkey I, Zuckerman T, Yehudai-Ofir D, Henig I, et al. Daratumumab for eradication of minimal residual disease in high-risk advanced relapse of T-cell/CD19/CD22-negative acute lymphoblastic leukemia. Leukemia 2020;34:293–5. [DOI] [PubMed] [Google Scholar]
- 118. Trinquand A, Dos Santos NR, Tran Quang C, Rocchetti F, Zaniboni B, Belhocine M, et al. Triggering the TCR developmental checkpoint activates a therapeutically targetable tumor suppressive pathway in T-cell leukemia. Cancer Discov 2016;6:972–85. [DOI] [PubMed] [Google Scholar]
- 119. Fleischer LC, Spencer HT, Raikar SS. Targeting T cell malignancies using CAR-based immunotherapy: challenges and potential solutions. J Hematol Oncol 2019;12:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat Rev Drug Discov 2020;19:185–99. [DOI] [PubMed] [Google Scholar]
- 121. Mamonkin M, Rouce RH, Tashiro H, Brenner MK. A T-cell-directed chimeric antigen receptor for the selective treatment of T-cell malignancies. Blood 2015;126:983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gomes-Silva D, Srinivasan M, Sharma S, Lee CM, Wagner DL, Davis TH, et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood 2017;130:285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Brandt LJB, Barnkob MB, Michaels YS, Heiselberg J, Barington T. Emerging approaches for regulation and control of CAR T cells: a mini review. Front Immunol 2020;11:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cooper ML, Choi J, Staser K, Ritchey JK, Devenport JM, Eckardt K, et al. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia 2018;32:1970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang X, Li S, Gao L, Yuan Z, Wu K, Liu L, et al. Clinical safety and efficacy study of TruUCAR™ GC027: the first-in-human, universal CAR-T therapy for adult relapsed/refractory T-cell acute lymphoblastic leukemia (r/r T-ALL) [abstract]. : Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27–28 and Jun 22–24. Philadelphia (PA): AACR; Cancer Res 2020;80(16 Suppl):Abstract nr CT052. [Google Scholar]
- 126. Sanchez-Martinez D, Baroni ML, Gutierrez-Aguera F, Roca-Ho H, Blanch-Lombarte O, Gonzalez-Garcia S, et al. Fratricide-resistant CD1a-specific CAR T cells for the treatment of cortical T-cell acute lymphoblastic leukemia. Blood 2019;133:2291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gao Z, Tong C, Wang Y, Chen D, Wu Z, Han W. Blocking CD38-driven fratricide among T cells enables effective antitumor activity by CD38-specific chimeric antigen receptor T cells. J Genet Genomics 2019;46:367–77. [DOI] [PubMed] [Google Scholar]
- 128. Guo Y, Feng K, Tong C, Jia H, Liu Y, Wang Y, et al. Efficiency and side effects of anti-CD38 CAR T cells in an adult patient with relapsed B-ALL after failure of bi-specific CD19/CD22 CAR T cell treatment. Cell Mol Immunol 2020;17:430–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Evans K, Duan J, Pritchard T, Jones CD, McDermott L, Gu Z, et al. OBI-3424, a novel AKR1C3-activated prodrug, exhibits potent efficacy against preclinical models of T-ALL. Clin Cancer Res 2019;25:4493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Verbeke D, Demeyer S, Prieto C, de Bock CE, De Bie J, Gielen O, et al. The XPO1 inhibitor KPT-8602 synergizes with dexamethasone in acute lymphoblastic leukemia. Clin Cancer Res 2020;26:5747–58. [DOI] [PubMed] [Google Scholar]
- 131. Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, et al. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol 2010;150:665–73. [DOI] [PubMed] [Google Scholar]
- 132. Waibel M, Vervoort SJ, Kong IY, Heinzel S, Ramsbottom KM, Martin BP, et al. Epigenetic targeting of Notch1-driven transcription using the HDACi panobinostat is a potential therapy against T-cell acute lymphoblastic leukemia. Leukemia 2018;32:237–41. [DOI] [PubMed] [Google Scholar]
- 133. Lu BY, Thanawala SU, Zochowski KC, Burke MJ, Carroll WL, Bhatla T. Decitabine enhances chemosensitivity of early T-cell precursor-acute lymphoblastic leukemia cell lines and patient-derived samples. Leuk Lymphoma 2016;57:1938–41. [DOI] [PubMed] [Google Scholar]
- 134. El Chaer F, Holtzman N, Binder E, Porter NC, Singh ZN, Koka M, et al. Durable remission with salvage decitabine and donor lymphocyte infusion (DLI) for relapsed early T-cell precursor ALL. Bone Marrow Transplant 2017;52:1583–4. [DOI] [PubMed] [Google Scholar]
- 135. Horton TM, Whitlock JA, Lu X, O'Brien MM, Borowitz MJ, Devidas M, et al. Bortezomib reinduction chemotherapy in high-risk ALL in first relapse: a report from the Children's Oncology Group. Br J Haematol 2019;186:274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Han K, Wang Q, Cao H, Qiu G, Cao J, Li X, et al. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget 2016;7:23812–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hartsink-Segers SA, Zwaan CM, Exalto C, Luijendijk MW, Calvert VS, Petricoin EF, et al. Aurora kinases in childhood acute leukemia: the promise of aurora B as therapeutic target. Leukemia 2013;27:560–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Park SI, Lin CP, Ren N, Angus SP, Dittmer DP, Foote M, et al. Inhibition of aurora A kinase in combination with chemotherapy induces synthetic lethality and overcomes chemoresistance in myc-overexpressing lymphoma. Targeted Oncol 2019;14:563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mosse YP, Fox E, Teachey DT, Reid JM, Safgren SL, Carol H, et al. A phase II study of alisertib in children with recurrent/refractory solid tumors or leukemia: Children's Oncology Group Phase I and Pilot Consortium (ADVL0921). Clin Cancer Res 2019;25:3229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jiang J, Wang J, Yue M, Cai X, Wang T, Wu C, et al. Direct phosphorylation and stabilization of MYC by aurora B kinase promote T-cell leukemogenesis. Cancer Cell 2020;37:200–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Goto H, Yoshino Y, Ito M, Nagai J, Kumamoto T, Inukai T, et al. Aurora B kinase as a therapeutic target in acute lymphoblastic leukemia. Cancer Chemother Pharmacol 2020;85:773–83. [DOI] [PubMed] [Google Scholar]
- 142. Harttrampf AC, Lacroix L, Deloger M, Deschamps F, Puget S, Auger N, et al. Molecular Screening for Cancer Treatment Optimization (MOSCATO-01) in pediatric patients: a single-institutional prospective molecular stratification trial. Clin Cancer Res 2017;23:6101–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.