Abstract
The morphogenesis checkpoint in budding yeast delays cell cycle progression in G2 when the actin cytoskeleton is perturbed, providing time for cells to complete bud formation prior to mitosis. Checkpoint-induced G2 arrest involves the inhibition of the master cell cycle regulatory cyclin-dependent kinase, Cdc28p, by the Wee1 family kinase Swe1p. Results of experiments using a nonphosphorylatable CDC28Y19F allele suggested that the checkpoint stimulated two inhibitory pathways, one that promoted phosphorylation at tyrosine 19 (Y19) and a poorly characterized second pathway that did not require Cdc28p Y19 phosphorylation. We present the results from a genetic screen for checkpoint-defective mutants that led to the repeated isolation of the dominant CDC28E12K allele that is resistant to Swe1p-mediated inhibition. Comparison of this allele with the nonphosphorylatable CDC28Y19F allele suggested that Swe1p is still able to inhibit CDC28Y19F in a phosphorylation-independent manner and that both the Y19 phosphorylation-dependent and -independent checkpoint pathways in fact reflect Swe1p inhibition of Cdc28p. Remarkably, we found that a Swe1p mutant lacking catalytic activity could significantly delay the cell cycle in vivo during a physiological checkpoint response, even when expressed at single copy. The finding that a Wee1 family kinase expressed at physiological levels can inhibit a nonphosphorylatable cyclin-dependent kinase has broad implications for many checkpoint studies using such mutants in other organisms.
Cell cycle progression in eukaryotic cells is orchestrated by cyclin-dependent kinases (Cdks), whose activity is subject to many layers of regulation (for reviews, see references 22, 24, and 26). Cdks (primarily Cdc2 in Schizosaccharomyces pombe and Cdc28p in Saccharomyces cerevisiae) become activated by forming a heterodimeric complex with one of a diverse family of regulatory proteins called cyclins. The activity of cyclin-Cdk complexes can be inhibited by phosphorylation of a tyrosine residue in the Cdk, catalyzed by the Wee1 family of protein kinases. In S. pombe, Cdc2 tyrosine phosphorylation is critical for restraining cyclin-Cdc2 activation until cells reach a threshold size in G2, at which point dephosphorylation of Cdc2, catalyzed by the phosphatase Cdc25, triggers cyclin-Cdc2 activation and consequent entry into mitosis (reviewed in references 8 and 20).
Cdk tyrosine phosphorylation has also been implicated in the regulation of cell cycle progression by checkpoint controls. Studies in fission yeast have implicated Wee1 and Cdc25 in the checkpoint-mediated G2 arrest following DNA damage or a block to DNA replication (6, 10, 11, 28). In other species, phosphorylation-site cdc2 mutants have been used to dissect the contribution of inhibitory Cdc2 phosphorylation to these checkpoints. Nonphosphorylatable cdc2 mutants permitted inappropriate cell cycle progression in some cells exposed to DNA damage or DNA replication inhibitors, indicating that Cdc2 phosphorylation was important for the checkpoint response (5, 16, 39). However, severe DNA damage or high doses of DNA replication inhibitors still arrested the cell cycle in most cells containing the cdc2 mutants. This complex result indicated that checkpoint responses can halt the cell cycle through Cdc2 phosphorylation-independent mechanisms as well as through inhibitory Cdc2 phosphorylation (reviewed in reference 20).
In budding yeast, the DNA damage and DNA replication checkpoints do not require tyrosine phosphorylation of Cdc28p (1, 33, 34). It has been suggested that these checkpoints operate at a different stage of the cell cycle in this organism, halting anaphase onset rather than cyclin-Cdc28p activation (38). In contrast, we have described a morphogenesis checkpoint in budding yeast that acts to delay or block cyclin-Cdc28p activation (21, 23, 30, 31). In yeast mutants that fail to polarize the actin cytoskeleton, nuclear division is delayed. This delay helps cells to recover from cytoskeletal insults and to complete bud formation prior to nuclear division, forestalling the generation of binucleate cells. The cell cycle delay was reduced, though not abolished, in cells containing a nonphosphorylatable mutation of CDC28 (21). This finding indicated that much of the checkpoint response was due to Cdc28p tyrosine phosphorylation, but that as in the studies cited above, a Cdk tyrosine phosphorylation-independent mechanism also played some role.
We report here the isolation of a dominant checkpoint-defective mutation of CDC28 in a screen for morphogenesis checkpoint mutants of budding yeast. This allele of CDC28 was more resistant to the action of the morphogenesis checkpoint than was the nonphosphorylatable CDC28 mutation. This result prompted us to reexamine the different components of the checkpoint response, and we found that both the phosphorylation-dependent and the phosphorylation-independent pathways require the Wee1 family kinase, Swe1p. The phosphorylation-site CDC28 mutant was still partly susceptible to inhibition by Swe1p, both in vitro and in vivo. Furthermore, catalytically inactive Swe1p was still able to inhibit Cdc28p in vitro and to sustain a partial checkpoint response. We conclude that the phosphorylation-independent branch of the morphogenesis checkpoint also reflects inhibition of Cdc28p by Swe1p, which can inhibit the nonphosphorylatable CDC28 mutant. Furthermore, our experiments suggest that catalytically inactive Swe1p can respond to a signal from the morphogenesis checkpoint to inhibit Cdc28p.
MATERIALS AND METHODS
Reagents.
4′,6′-Diamidino-2-phenylindole (DAPI) was purchased from Sigma Chemical Co. (St. Louis, Mo.) and stored as a 1-mg/ml stock solution in H2O at −20°C. Latrunculin B (Lat-B) was purchased from BIOMOL Research Laboratories, Inc. (Plymouth Meeting, Pa.) and stored as a 10 mM stock solution in dimethyl sulfoxide at −20°C. Latrunculin-A (Lat-A), rhodamine-conjugated phalloidin, and Sytox were purchased from Molecular Probes (Eugene, Oreg.). Lat-A was also provided as a generous gift from Phil Crews (University of California, Santa Cruz) and stored as a 20 mM stock solution in dimethyl sulfoxide at −20°C. Rhodamine-conjugated phalloidin was stored as a 200-U/ml stock solution in methanol at −20°C. Mounting medium was made as described elsewhere (27). 5-Fluoroorotic acid (5-FOA) was purchased from Toronto Research Chemicals, Inc. (North York, Ontario, Canada). Anti-Myc antibody 9E10 was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), and anti-phospho(Tyr15)-Cdc2 antibody was purchased from New England Biolabs (Beverly, Mass.).
Yeast strains and plasmid constructions.
The yeast strains used in this study are listed in Table 1. All are in the BF264-15DU background (ade1 his2 leu2-3, 112 trp1-1a ura3Δns). The previously described swe1ΔLEU2 and GAL:SWE1::LEU2 (7), GAP:SWE1::HIS2 (31), GAL:SWE1-myc::URA3 and GAL:MIH1::TRP1 (23), zds1ΔHIS3 (4), GAL:CLB2::LEU2 (34), cdc28-4 (37), zds2ΔURA3, gal:SWE1K473P-myc::URA3, SWE1K473P-myc::TRP1, CDC28Y19F, and CDC28E12K alleles were introduced into the BF264-15DU background by direct transformation.
TABLE 1.
Strains list used in this study
| Strain | Relevant genotype |
|---|---|
| DLY1 | MATa bar1 |
| JMY1208 | MATa zds1ΔHIS3 zds2ΔURA3 GAL:MIH1:TRP1 GAP:SWE1:HIS2 |
| JMY1222 | MATα zds1ΔHIS3 zds2Δura3 GAL:MIH1:TRP1 GAP:SWE1:HIS2 |
| JMY1207 | MATα zds1ΔHIS3 zds2ΔURA3 swe1ΔLEU2 |
| DLY1028 | MATa swe1ΔLEU2 bar1 |
| DLY115 | MATα cdc28-4 |
| JMY1380 | MATa CDC28E12K bar1 |
| JMY1390 | MATa GAL:SWE1:LEU2 CDC28Y19F bar1 |
| JMY1386 | MATa GAL:SWE1:LEU2 CDC28E12K bar1 |
| DLY2626 | MATa GAL:SWE1:LEU2 bar1 |
| JMY1362 | MATa CDC28E12K:URA3:CDC28 bar1 |
| JMY1364 | MATa CDC28Y19F:URA3:CDC28 bar1 |
| JMY1365 | MATa CDC28E12K:URA3:CDC28 swe1ΔLEU2 bar1 |
| JMY1367 | MATa CDC28Y19F:URA3:CDC28 swe1ΔLEU2 bar1 |
| JMY1428 | MATa swe1K473P-12Xmyc:TRP1 bar1 |
| JMY1429 | MATa CDC28E12K:URA3:CDC28 swe1K473P-12Xmyc:TRP1 bar1 |
| DLY101 | MATa cdc28-4 bar1 |
| JMY1455 | MATa cdc28-4 GAL:SWE1-12Xmyc:URA3 bar1 |
| JMY1456 | MATa cdc28-4 GAL:swe1K473P-12Xmyc:URA3 bar1 |
| RSY16 | MATa/MATα GAL:GST-CDC28:LEU2/GAL:CLB2:LEU2 |
| RSY17 | MATa/MATα GAL:GST-CDC28Y19F:LEU2/GAL:CLB2:LEU2 |
The zds2ΔURA3 allele is a deletion of nearly the entire open reading frame (ORF) of ZDS2 (from 31 bases upstream of the start codon to the HindIII site 0.2 kb upstream of the stop codon). The zds2ΔURA3 disruption plasmid, pNC419, was kindly provided by Beverly Errede (University of North Carolina at Chapel Hill). The zds2Δura3 allele in strain JMY1222 was made by plating a zds2ΔURA3 strain onto 5-FOA medium to select for ura3 mutants. PCR amplification of the zds1Δura3 allele from one of these 5-FOA-resistant colonies identified an allele of ura3 likely containing a small deletion. This allele was used in further strain constructions because it was unlikely to revert to a functional URA3.
To create the CDC28Y19F:URA3:CDC28 allele (strains JMY1364 and JMY1367), the XhoI/BamHI DNA fragment from pSF38 (33) carrying CDC28Y19F as well as approximately 340 bp upstream and 790 bp downstream of the CDC28Y19F ORF was ligated into the corresponding sites of pRS306 (32), a URA3 integrating vector. This plasmid, pJM1054, was cut with HindIII to target integration into the CDC28 locus. A similar strategy was used to create the CDC28E12K:URA3:CDC28 allele. The CDC28E12K allele originated from one of the four CDC28E12K mutants isolated in the screen for suppressors of the zds1Δ zds2Δ elongated cell phenotype and was cloned by gap repair as described below. JMY1380, a yeast strain containing only the CDC28E12K allele, was made by plating JMY1362 (CDC28E12K:URA3:CDC28) onto 5-FOA medium to select for cells that had undergone a recombination event between the two alleles of CDC28, looping out the URA3 gene. Those cells having only the CDC28E12K allele were identified by their ability to suppress the elongated cell phenotype when mated to a SWE1-overproducing strain.
To construct and integrate the GAL:GST:CDC28 and GAL:GST:CDC28Y19F alleles (strains RSY16 and RSY17), the glutathione S-transferase (GST) sequence plus the adjacent multiple cloning site from pGEX-KG (14) was amplified by PCR with primers that placed BglII sites at the ends of the PCR product. This PCR product was digested with BglII and ligated into the BamHI site of YIpG2 (12, 34), to place the GST sequence downstream of the GAL1 promoter, creating YIpG2:GST. A 1.8-kb NdeI (start codon)-to-BamHI (3′ of the stop codon) CDC28 fragment from pRD47 (7) and the similar CDC28Y19F NdeI/BamHI fragment from pSF38 were blunt-end ligated into the XhoI site of YIpG2:GST, placing the CDC28 alleles in frame with GST, to create YIpG2:GST-CDC28 and YIpG2:GST-CDC28-YF, respectively. These plasmids were digested with BstEII to integrate the relevant alleles into the yeast LEU2 locus.
All of the plasmids used to express Cdc28p in Fig. 4 (pJM1042 [CDC28], pJM1046 [CDC28E12K], and pAL88 [CDC28Y19F]) were made by cloning XhoI/BamHI fragments containing CDC28 alleles into the corresponding sites of pRS316. CDC28 was isolated from pRD47, CDC28E12K was isolated from a gap-repaired plasmid rescued from a CDC28E12K mutant (see below), and CDC28Y19F came from pSF38. pAL88 was kindly provided by Steve Garrett (University of Medicine and Dentistry of New Jersey [UMDNJ], Newark).
FIG. 4.
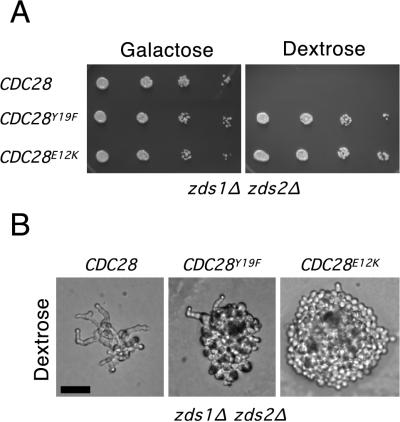
The CDC28E12K allele fully rescues the zds1Δ zds2Δ mutant, while the CDC28Y19F allele only partially rescues cell morphology. (A) Spot assays of a zds1Δ zds2Δ GAL:MIH1 strain (JMY1222) transformed with a centromere plasmid (pRS316) carrying CDC28 or the CDC28E12K or CDC28Y19F alleles, grown on YEPG and YEPD at 30°C for 3 days and 2 days, respectively. Both the CDC28E12K and CDC28Y19F alleles rescue the growth defect in a zds1Δ zds2Δ strain. (B) Colony morphology of the above strains after 1 day of growth on dextrose-containing medium at 30°C. Bar = 20 μm.
To make the catalytically inactive mutant of SWE1, the codon AAA corresponding to lysine at position 473 of Swe1p was mutated to proline CCG by an overlap PCR strategy. Two PCR fragments (up- and downstream from the K473 position) were amplified by using primers that altered K473 to P (actually, through a transcribing error, one of the primers altered the K to P while the other altered the K to R; subsequent sequencing demonstrated that the mutant generated was K473P). Primers were 5′-AAGTATGCAATCCGGGCCATTAAACCAAAC-3′ and 5′-CGATCTCAACTTATGCCATGCG-3′ for the first fragment, and 5′-TGGTTTAATGGCCGGGATTGCATACTTTTTG-3′ and 5′-CGTCTTCTCTAAGTGTTTCCCC-3′ for the second fragment (codon 473 is underlined). The two PCR products were then mixed in an overlap PCR with the outside primers to generate a SWE1 fragment with an internal K473P mutation. This PCR product was swapped into SWE1 residing on pRAS16 (constructed by ligating the XbaI/BamHI GAL:SWE1 fragment from pSWE1-29 [7] into pRS316 [32]) by gap repair of pRAS16 (cut with BstEII to remove the internal BstEII fragment spanning K473) in yeast. The resulting GAL:swe1K473P plasmid (pRAS17) was sequenced to confirm the presence of the K473P mutation and the absence of any other PCR-generated mutations.
The GAL:swe1K473P-myc allele (strain JMY1456) was constructed by first transferring GAL:swe1K473P into the integrating pRS306 vector and then swapping the C terminus and 3′ noncoding fragment of SWE1 (KpnI BamHI) in that plasmid with the corresponding fragment from GAL:SWE1-myc (23), generating pRAS12. pRAS12 was targeted for integration at the URA3 locus by digestion with StuI. To make a SWE1K473P-myc allele transcribed from the SWE1 promoter (strains JMY1428 and JMY1429), the EcoRI/BamHI DNA fragment containing the 3′ end of SWE1K473P-myc and downstream sequence was isolated from pRAS12 and ligated into the corresponding sites of pRS304, a TRP1 integrating vector (32). This plasmid, pJM1050, was digested with BglII, which cuts 5′ of the K473P mutation within the SWE1 ORF, to target integration at the SWE1 locus. Integration creates a full-length SWE1K473P-myc allele under control of the endogenous SWE1 promoter with an adjacent 5′ truncated allele of swe1.
Medium and growth conditions.
Strains were grown in rich medium (YEPD [1% yeast extract, 2% Bacto Peptone, 2% dextrose, 0.01% adenine], YEPS [YEPD with 2% sucrose instead of dextrose], or YEPG [YEPD with 2% galactose instead of dextrose]). Cells transformed with plasmids were grown on synthetic dropout medium (0.67% yeast nitrogen base plus 2% dextrose or galactose, supplemented with amino acids). For α-factor arrest-release experiments, exponentially growing cells (2 × 106 to 5 × 106 cells/ml) in YEPD were incubated with 20 to 25 ng of α-factor per ml for 2 to 3 h, harvested by centrifugation, and resuspended in fresh YEPD to release the α-factor-induced cell cycle block. bar1 strains were used in all the α-factor arrest experiments. To compare the growth of different strains by spot assay, cells were sonicated and counted with a hemacytometer. For each strain, four 2-μl spots were pipetted onto a YEPD or YEPG plate, each spot containing a total of approximately 1,250, 250, 50, or 10 cells. Strains being compared were pipetted onto the same plate and were incubated at 30°C for the indicated times. For growth rate assays in liquid culture, cells were removed from exponentially growing cultures at five time points over a period of 360 min and sonicated. At least 200 cells from each sample were counted on a hemacytometer from which cell density was calculated. The best-fit line (linear regression) was calculated by using KaleidaGraph (Synergy Software, Reading, Pa.).
Screen to identify suppressors of zds1Δ zds2Δ.
The starting strains, JMY1208 (MATa) and JMY1222 (MATα), were first streaked onto YEPG plates; individual colonies containing 5 × 106 to 1 × 107 cells were resuspended in 100 μl of liquid YEPD and then plated onto separate YEPD plates. A total of approximately 109 cells were plated onto YEPD plates to select for suppressors of the zds1Δ zds2Δ mutations. However, this number is conservative because the plated cells divide several times on YEPD before dying. Cell morphology of cells in colonies growing on the YEPD plates was analyzed microscopically, and suppressors with morphologically normal cells were tested for a defect in the morphogenesis checkpoint by using Lat-B as described in Results. Of the eight suppressors isolated that were confirmed to have a morphogenesis checkpoint defect based on the Lat-B assay, four (three from JMY1208 and one from JMY1222) contained the CDC28E12K allele.
Gap repair of CDC28 alleles.
The gap repair plasmid pJM1028 is a pRS315 (32) derivative (LEU2, centromere) containing the CDC28 ORF plus 338 bp upstream and 789 bp downstream. pJM1028 was digested with NdeI (start codon) and HindIII (130 bases upstream of the stop codon), resulting in the deletion of most of the CDC28 ORF. The digested and gel-purified linearized plasmid was transformed into strains isolated from the zds1Δ zds2Δ suppressor screen, allowing repair of the gap within the plasmid by homologous recombination with the genomic copy of CDC28. The gap-repaired plasmids were subsequently rescued and the CDC28 ORFs sequenced.
Fluorescence staining, microscopy, and flow cytometry.
DNA was stained with DAPI, and F-actin was visualized by staining with rhodamine-conjugated phalloidin as reported previously (23). Cells were viewed on an Axioscope (Carl Zeiss, Inc., Thornwood, N.Y.) equipped with epifluorescence and Nomarski optics. Images were captured with a cooled charge-coupled device camera (Princeton Instruments, Princeton, N.J.). Microscopic images of whole yeast microcolonies were similarly captured. Cells were processed for flow cytometry as described previously (31), using 1 μM Sytox to stain the DNA. The percentages of budded and anaphase cells were quantitated for the same samples as processed for flow cytometry.
p18CKS1 precipitation and immunoblotting.
Strains DLY2626 (CDC28 GAL:SWE1), JMY1390 (CDC28Y19F GAL:SWE1), and JMY1386 (CDC28E12K GAL:SWE1) were grown in YEPS and induced to express Swe1p by growth for 4 h after addition of galactose (2%). Cells were then harvested by centrifugation and washed with ice-cold H2O, and the pellets were frozen at −80°C. Yeast lysates were made as described elsewhere (30). Lysates containing 1 mg of total protein were incubated with 50 μl of p18CKS1 beads (see below) in 1 ml of binding buffer containing 50 mM sodium phosphate (pH 7.5), 150 mM sodium chloride, 10% glycerol, 10 mM sodium pyrophosphate, 10 mM EDTA, 10 mM EGTA, 0.1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 2 μg each of aprotinin, leupeptin, and pepstatin A (Sigma) per ml. Affigel-15 beads (Bio-Rad, Hercules, Calif.) were coupled to p18CKS1 (1.5 mg of p18/ml of Affigel-15) according to the manufacturer’s instructions (p18 was a gift from Mark Watson, The Scripps Research Institute, La Jolla, Calif.). Lysates and beads were mixed by gentle rocking for 4 h at 4°C, and the beads were then washed once in binding buffer and once in binding buffer with 0.3 M potassium chloride, following which the beads were drained and resuspended in 2× sample loading buffer (125 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 50% glycerol, 710 mM β-mercaptoethanol, 0.02% bromophenol blue), heated at 95°C for 3 min, diluted twofold with H2O, and loaded on an SDS–15% polyacrylamide gel. Following electrophoresis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.) and stained with anti-phospho-Cdc2 antibody Tyr 15 (used at 1:1,000 dilution according to the manufacturer’s recommendations). The blot was then stripped and reprobed with monoclonal anti-PSTAIRE antibody (used at 1:20,000 dilution of a mouse ascites preparation).
Cdc28p histone H1 kinase assays. (i) Galactose induction.
Yeast strains were grown to a density of 0.5 × 107 to 1 × 107 cells/ml in YEPS either at 30°C for RSY16 (GAL:CDC28-GST GAL:CLB2) and RSY17 (GAL:CDC28Y19F-GST GAL:CLB2) or at 24°C for DLY101 (cdc28-4 control strain), JMY1455 (GAL:SWE1-myc cdc28-4), and JMY1456 (GAL:SWE1K473P-myc cdc28-4). Cells were induced to express GAL-regulated genes by the addition of galactose to a final concentration of 2% followed by a 3-h incubation of the cultures at their growth temperatures, except for Fig. 6B, in which the JMY1455 and JMY1456 cultures were first shifted to 37°C for 3 h, galactose was added, and the cultures were incubated for an additional 3 h at 37°C. After induction, cells were harvested and washed once with ice-cold H2O and the cell pellets were stored at −80°C. cdc28-4 strains were used as the source for Swe1p-Myc because in other experiments (data not shown) we found that Swe1p could associate with a Cdc28p-dependent histone H1 kinase activity: Cdc28-4p does not exhibit in vitro kinase activity (37), and so these strains allowed us to circumvent this complicating factor.
FIG. 6.
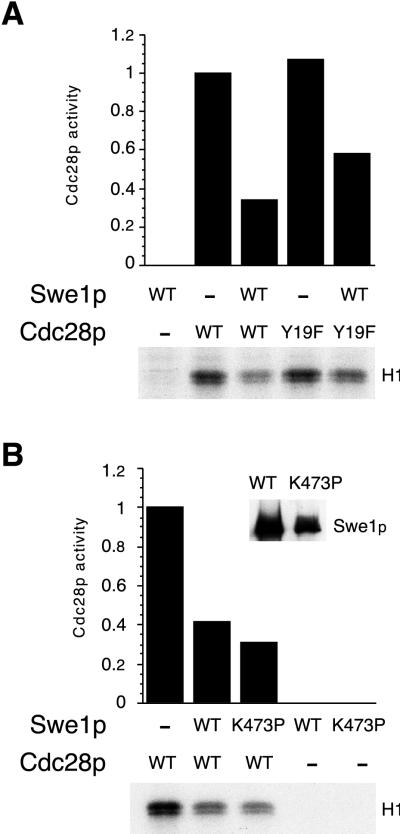
Phosphorylation-independent inhibition of Cdc28p by Swe1p in vitro. (A) Immunoprecipitates containing Swe1p from strains expressing (WT [wild type]) or not expressing (−) GAL:SWE1-myc were mixed as indicated with GST-Cdc28p/Clb2p (WT) or GST-Cdc28pY19F/Clb2p (Y19F) complexes isolated from yeast and subsequently assayed for Cdc28p-dependent histone H1 kinase activity as described in Materials and Methods. In the autoradiograph of radioactive histone H1, histone bands were excised from the gel and the incorporated radioactivity was quantitated and plotted above the lanes. Activity was normalized so that the activity of wild-type Cdc28p in the absence of Swe1p was made equal to 1. Swe1p inhibited wild-type and Y19F Cdc28p to similar extents. (B) Assays were performed as for panel A but with immunoprecipitated Swe1p (WT) or Swe1pK473P (K473P). The inset shows a Western blot of the Myc-tagged Swe1p proteins from the same immunoprecipitates used for the kinase assay. Wild-type and kinase-dead forms of Swe1p inhibited CDC28p to similar extents.
(ii) Lysates and immunoprecipitation.
Yeast lysates were made as described previously (30). To purify GST-Cdc28p and GST-Cdc28pY19F, a 50% glutathione bead slurry (Pharmacia) was added to 500 μg of total protein in 1 ml of Nonidet P-40 (NP-40) lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 1 mM sodium pyrophosphate, 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 2 μg each of aprotinin, pepstatin, and leupeptin per ml) with 10 mM dithiothreitol (DTT). The bead-lysate mixture was gently mixed for 5 min at 20°C and then given three 1-ml washes in NP-40 lysis buffer with 10 mM DTT. To elute the bound protein, the beads were resuspended in 50 μl of a 25 mM glutathione–50 mM Tris (pH 8)–10 mM DTT solution, gently mixed for 5 min at 20°C, and centrifuged, and the supernatant was removed to a new tube. Immediately before use in the kinase assay, the eluted GST-Cdc28p and GST-Cdc28pY19F proteins were diluted 1:1 with reaction buffer (7.5 mM MgCl2, 20 mM Tris [pH 7.5]). To purify Swe1p-Myc and Swe1pK473P-Myc, 5 μl of anti-Myc antibody 9E10 was added to 2.5 mg of total protein in 1 ml of NP-40 lysis buffer and mixed for 1 h at 4°C. After addition of 150 μl of a 50% protein A slurry, the tubes were incubated for 1 h at 4°C with mixing. The protein A beads were then washed three times in NP-40 lysis buffer and resuspended in 100 μl of NP-40 lysis buffer.
(iii) Histone H1 kinase assay.
Swe1p-Myc-bound beads (10 μl) and eluted GST-Cdc28p 1 μl were added to 19 μl of reaction buffer and incubated for 20 min at 20°C (GST-Cdc28p was not added to the kinase assay in Fig. 7A). After addition of 15 μl of a kinase reaction cocktail, the mixture was incubated at 30°C for 30 min. The final kinase reaction mixture contained 7.5 mM MgCl2, 20 mM Tris (pH 7.5), 2 μg of histone H1, 0.1 nM ATP, 10 μCi of [γ-32P]ATP (3,000 Ci/mmol), 10 μl of Swe1p-Myc (or Swe1pK473P-Myc) beads, and 1 μl of GST-Cdc28p (or GST-Cdc28pY19F) in a total volume of 45 μl. The reaction was stopped with the addition of 15 μl of sample loading buffer, and the mixture was heated at 100°C for 5 min. Then 20 μl of each reaction mixture was loaded onto an SDS–11% polyacrylamide gel. After electrophoresis, the gel was stained with Coomassie blue to visualize the histone H1, boiled for 10 min in 5% trichloroacetic acid to remove remaining unincorporated nucleotide, and dried on Whatman 3MM paper. After exposure to film, the stained bands corresponding to histone H1 were cut out of the gel and counted on a scintillation counter. Cdc28p activity in the presence of Swe1p was normalized to the activity in the absence of Swe1p (following subtraction of background counts).
FIG. 7.
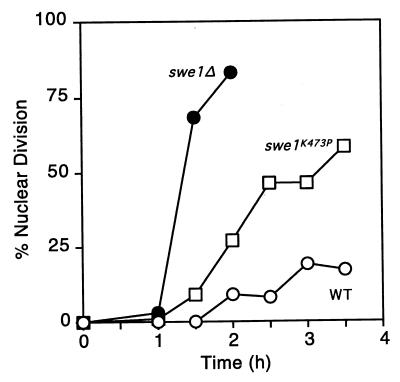
Morphogenesis checkpoint-induced delay of nuclear division in a swe1K473P mutant. Strains were released from α-factor-induced cell cycle arrest into fresh medium containing 50 μM Lat-A. At the indicated times, aliquots were fixed and subsequently stained with DAPI to visualize DNA; 200 cells were scored in each sample. Strains were DLY1 (WT [wild type]), DLY1028 (swe1Δ), and JMY1428 (swe1K473P-myc).
RESULTS
Isolation of morphogenesis checkpoint-defective mutants.
We devised a two-step genetic screen for mutants with a defective morphogenesis checkpoint. The first step depends on the phenotype of zds1 zds2 mutant strains. Bi and Pringle (4) identified ZDS1 in a screen for regulators of Cdc42p and demonstrated that a GST-Zds1p fusion protein was concentrated at presumptive bud sites and at the tip of small buds, suggestive of a role in cell morphogenesis. A homolog of ZDS1, called ZDS2, was identified; the zds1 zds2 mutant strain grows very slowly, is delayed in G2, and displays morphological abnormalities that include elongated buds. The exact role of the ZDS genes remains obscure, particularly since they have been identified independently by multiple investigators in diverse screens (zds stands for “zillion different screens” [cited in reference 4]). However, Wang and Burke (36) reported that the poor growth and morphologic abnormalities of zds1 zds2 mutants could be rescued by expression of the CDC28Y19F allele that cannot be phosphorylated at Y19. They suggested that one primary defect in zds1 zds2 mutants was a mild morphogenesis defect and that this defect triggered the morphogenesis checkpoint to cause a prolonged G2 delay through Cdc28p Y19 phosphorylation (36). In this scenario, the morphogenesis checkpoint was in fact compromising the growth of an otherwise only mildly defective strain.
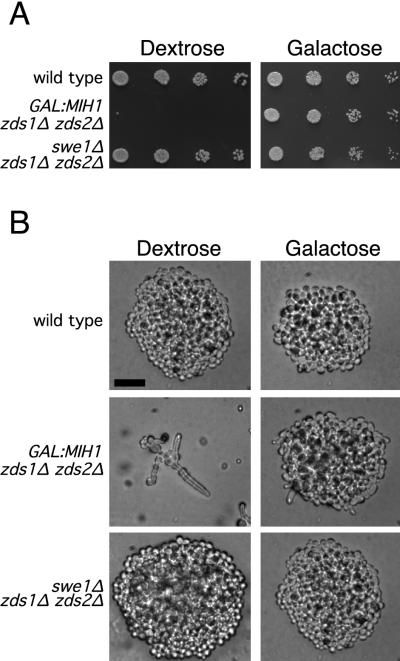
In agreement with Wang and Burke’s observations, we found that either deletion of SWE1 (the S. cerevisiae Wee1 homolog [7]) or overexpression of MIH1 (the S. cerevisiae Cdc25 homolog [29]) suppressed the slow growth and morphologic abnormalities of zds1 zds2 mutants in our strain background (Fig. 1). We reasoned that if the zds1 zds2 strain was in fact triggering the morphogenesis checkpoint, a simple selection for suppressors of the zds1 zds2 growth defect should yield checkpoint mutants. To perform this selection, we made a zds1 zds2 GAL::MIH1 strain in which the MIH1 gene was replaced by a copy of MIH1 transcribed from the regulatable GAL1 promoter. This strain overexpressed MIH1 and rescued the zds1 zds2 phenotype on galactose-containing medium but did not produce functional MIH1 on dextrose-containing medium, where the cells showed a severe growth defect (Fig. 1). In addition, an extra copy of SWE1, expressed from a heterologous constitutive promoter (31), was integrated at HIS2 in order to avoid the repeated isolation of swe1 mutants. Individual colonies from MATa (JMY1208) or MATα (JMY1222) strains growing on galactose-containing medium were plated onto dextrose-containing medium to select for spontaneous suppressors of the zds1 zds2 growth defect. Only one suppressor was picked from any one starting colony to ensure that independent mutants would be analyzed. Suppressors were selected that displayed an apparently complete rescue of the morphologic abnormality as well as the slow-growth phenotype of the starting strain.
FIG. 1.
Rescue of zds1Δ zds2Δ strains by overexpression of MIH1 or deletion of SWE1. (A) Spot assays of wild-type (DLY1), zds1Δ zds2Δ GAL:MIH1 (JMY1208), and zds1Δ zds2Δ swe1Δ (JMY1207) on YEPD (dextrose) and YEPG (galactose) after 2 days of growth at 30°C. Both the overexpression of MIH1 and deletion of SWE1 rescue the growth defect of a zds1Δ zds2Δ mutant. (B) Colony morphology of the above strains after 1 day of growth on dextrose or galactose-containing media at 30°C. Bar = 20 μm.
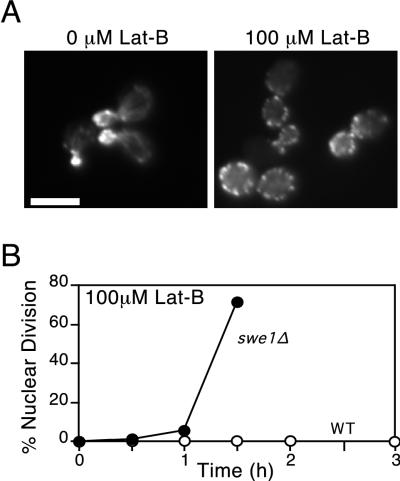
To determine which of the suppressors conferred a morphogenesis checkpoint defect, we performed a secondary screen using the antiactin drug Lat-B (2). Previous experiments had shown that treatment of cells with Lat-A caused a rapid depolymerization of the actin cytoskeleton, blocked bud formation, and triggered a checkpoint-mediated G2 arrest (2, 23). We found that the actin perturbation caused by similar concentrations of Lat-B was less severe but nevertheless sufficient to block bud formation and trigger Swe1p-dependent G2 arrest (Fig. 2). Given the considerably lower cost of Lat-B than Lat-A, we used Lat-B for mutant screening (similar results were subsequently obtained with Lat-A [see Fig. 5]). Cells were grown in dextrose-containing medium to mid-log phase, and Lat-B was added to 0.1 mM (final concentration). Following a 3-h incubation, the cells were fixed and stained with DAPI to visualize the DNA. Checkpoint-defective mutants were scored as those in which >30% of unbudded cells underwent nuclear division to become binucleate during the 3-h incubation, indicating a failure of the checkpoint-induced G2 arrest. For comparison, wild type cells displayed <5% binucleate cells while swe1Δ mutants displayed >50% binucleate cells in this assay.
FIG. 2.
Cell cycle arrest in Lat-B-treated cells. (A) F-actin organization in wild-type (DLY1) cells exposed to 0 and 100 μM Lat-B. Actin was visualized by staining cells with rhodamine-conjugated phalloidin. In the cells treated with 100 μM Lat-B, cables are absent and the actin patches are depolarized. Bar = 5 μm. (B) Kinetics of nuclear division for wild-type (WT; DLY1) and swe1Δ (DLY1028) cells after release from α-factor-induced G1 arrest into fresh medium containing 100 μM Lat-B. All cultures were grown in YEPD at 30°C.
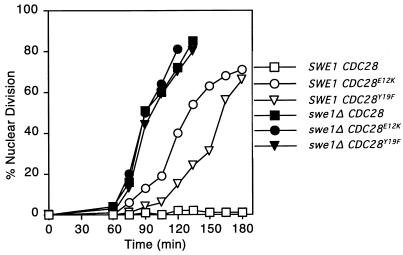
FIG. 5.
Morphogenesis checkpoint-induced delay of nuclear division in CDC28Y19F and CDC28E12K mutants. Strains were released from α-factor-induced cell cycle arrest into fresh medium containing 50 μM Lat-A. At the indicated times, aliquots were fixed and subsequently stained with DAPI to visualize DNA; 200 cells were scored in each sample. Strains used were DLY1 (SWE1 CDC28), JMY1362 (SWE1 CDC28E12K), JMY1364 (SWE1 CDC28Y19F), DLY1028 (swe1Δ CDC28), JMY1365 (swe1Δ CDC28E12K), and JMY1367 (swe1Δ CDC28Y19F).
We isolated eight independent mutants that suppressed both the growth defect and the morphologic abnormalities of the starting strains and displayed severe checkpoint defects in the secondary screen. These mutants were crossed to a wild-type strain of the opposite mating type, and the resulting diploids were tested for checkpoint defects by using Lat-B as described above. Four mutants were recessive, while four showed a dominant checkpoint defect. Sporulation and dissection revealed 2:2 segregation of the checkpoint defect in all cases (in >6 tetrads each), suggesting that single mutations were responsible for the phenotype. One of the recessive mutants was transformed with a low-copy-number genomic library and screened for restoration of the zds1 zds2 slow-growth phenotype. Two complementing plasmids were isolated from ∼3,000 transformants, and PCR analysis revealed that both contained SWE1. The SWE1 plasmids were able to complement both the zds1 zds2 suppression and the checkpoint defect of all four independent recessive mutants, and segregation analysis showed that at least one of the mutants was tightly linked to SWE1. We conclude that the recessive mutants are all likely to contain mutations in both of the copies of SWE1 present in the original strain.
The dominant checkpoint-defective mutants all contain a single mutation in CDC28.
Since Swe1p is known to inhibit cyclin-Cdc28p complexes through phosphorylation of Cdc28p Y19, it seemed possible that the dominant mutants we obtained contained alterations (e.g., at the Y19 position) in the CDC28 gene. One of the four independent dominant mutants was crossed to each of the others and to a strain containing a cdc28-4 temperature-sensitive allele. The resulting diploids were sporulated, and tetrads were dissected to determine whether the mutants were in linked genes. No wild-type recombinants were obtained in 22 tetrads from these crosses, suggesting that all four strains contained mutations in linked genes, possibly all in the CDC28 gene. To test this, we rescued the CDC28 gene from the genome of each mutant strain (as well as a wild-type parent strain) by using gap repair (see Materials and Methods). The plasmid-borne CDC28 alleles from the mutant strains were all able to suppress the poor growth and morphologic abnormalities of the parent zds1 zds2 strain and conferred a morphogenesis checkpoint defect (see Fig. 4 and 5), confirming that CDC28 had been mutated in each of the strains.
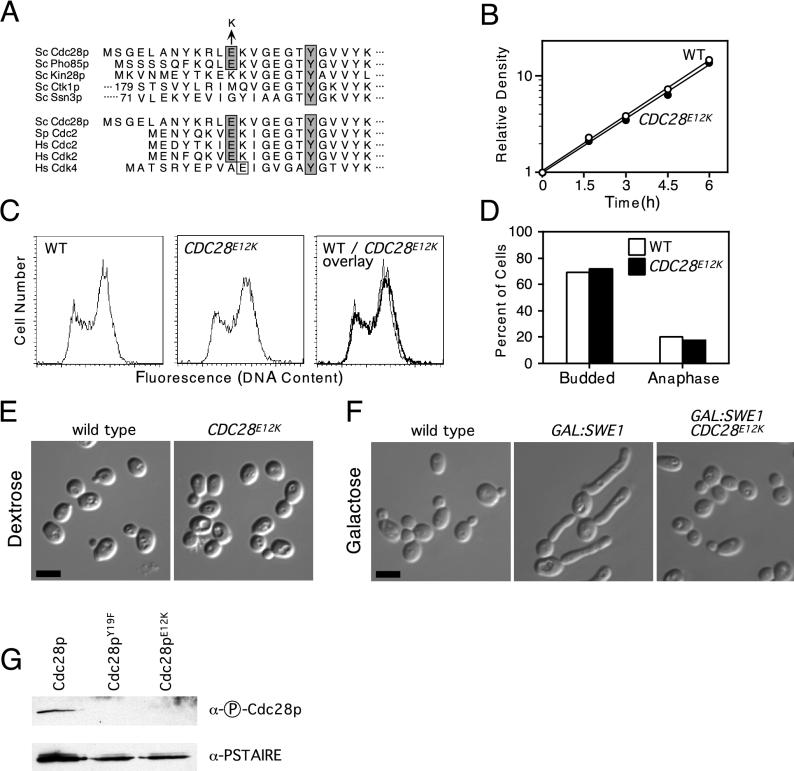
The entire ORF of the gap-repaired CDC28 genes from the parent strain and each mutant were sequenced. The parent (wild-type) CDC28 sequence displayed a 100% match to the CDC28 sequence in the Saccharomyces genome database, but each mutant displayed a single G-to-A nucleotide change from the wild type. Remarkably, all four independent mutants had altered the same residue, causing a change from glutamic acid to lysine at position 12 (Fig. 3A). This residue is highly conserved among Cdks known to trigger entry into mitosis in different organisms but is not conserved between different yeast Cdks (Fig. 3A; see also Discussion).
FIG. 3.
A CDC28E12K mutant has a normal growth profile but is resistant to SWE1 overexpression. (A) Sequence homology of the amino terminus of S. cerevisiae (Sc) Cdc28p with the other Cdks in S. cerevisiae and with S. pombe and human (Hs) Cdks. The conserved tyrosine (Y) residue phosphorylated by Wee1 family kinases and the glutamic acid (E) residue mutated to lysine (K) in the CDC28 mutants isolated from the zds1Δ zds2Δ suppression screen are shaded. S. cerevisiae Kin28p, Ctk1p, and Ssn3p and human Cdk4 do not have a glutamic acid at this position; however, the human Cdk4 does have an adjacent glutamic acid residue that is boxed but not shaded. Ctk1p and Ssn3p have large amino-terminal regions, and the sequence shown begins at residue 179 for Ctk1p and 71 for Ssn3p. (B) Growth rates of exponentially proliferating wild-type and CDC28E12K strains. (C) Flow cytometric analysis of the DNA content of exponentially proliferating wild-type (WT) and CDC28E12K cells. (D) Percentage of budded and anaphase cells, calculated from microscopic examination of the same cells stained for flow cytometric analysis in panel C. (E) Cell morphology of exponentially growing wild-type and CDC28E12K cells. Bar = 5 μm. (F) Cell morphology of wild-type, GAL:SWE1, and GAL:SWE1 CDC28E12K strains 4 h after addition of galactose to cells growing in YEPS medium. Bar = 5 μm. The strains used were DLY1 (wild type), JMY1380 (CDC28E12K), DLY2626 (GAL:SWE1), and JMY1386 (CDC28E12K GAL:SWE1). (G) Tyrosine phosphorylation of Cdc28p alleles upon overexpression of Swe1p. GAL:SWE1 strains containing wild-type (DLY2626), Y19F (JMY1390), and E12K (JMY1386) alleles of Cdc28p were induced to overexpress Swe1p by growth for 4 h after addition of galactose to cells growing in YEPS medium. p18Cks1 beads were then used to precipitate the different Cdc28p proteins, which were immunoblotted with anti-phospho-Cdc2 antibody to detect tyrosine-phosphorylated Cdc28p and anti-PSATIRE antibodies to detect total Cdc28p. All cultures were grown at 30°C and, unless otherwise indicated, were grown in YEPD.
CDC28E12K does not affect cell cycle progression during the unperturbed cell cycle.
To analyze the phenotype of the CDC28E12K mutant in more detail, we constructed a strain in which the wild-type CDC28 gene, at its normal locus, was replaced with the E12K allele (see Materials and Methods). This strain allowed us to determine whether the allele conferred any recessive cell cycle phenotype. Cells containing CDC28E12K grew at the same rate as wild-type cells (Fig. 3B), displayed similar cell cycle profiles by flow cytometry (Fig. 3C), and exhibited a normal cell morphology (Fig. 3E). In addition, asynchronous populations from these strains displayed similar percentages of budded and anaphase cells (Fig. 3D). Thus, the E12K allele has no detectable effect on the unperturbed cell cycle of budding yeast.
CDC28E12K renders cells resistant to Swe1p overexpression.
Both the morphogenesis checkpoint defect exhibited by CDC28E12K and its lack of a cell cycle defect in the unperturbed cycle are reminiscent of the CDC28Y19F mutant that cannot be phosphorylated by Swe1p. If the E12K allele is simply insensitive to inhibition by Swe1p, then strains containing this allele should be resistant to the G2 arrest caused by Swe1p overexpression. To test this, strains containing GAL-SWE1 and either wild-type or E12K CDC28 were induced to express Swe1p by addition of galactose to the medium. While the cells containing wild-type CDC28 formed elongated buds characteristic of G2 arrest on galactose, the cells containing the E12K allele were resistant to the GAL-SWE1 and proliferated with normal cell morphology (Fig. 3F). In principle, this resistance might arise in two ways: Swe1p may be unable to phosphorylate Cdc28pE12K, or Cdc28pE12K might retain activity even following phosphorylation on Y19. To distinguish between these possibilities, we examined whether Cdc28pE12K became phosphorylated at Y19 following overexpression of Swe1p (Fig. 3G). We used an anti-phospho-Cdc2/Cdc28p antibody to immunoblot Cdc28p proteins isolated from Swe1p-overexpressing strains by using p18CKS1 beads (see Materials and Methods). This reagent detected wild-type Cdc28p but not Cdc28pY19F, confirming its specificity for Y19-phosphorylated Cdc28p (Fig. 3G). Cdc28pE12K was not detected by the anti-phospho-Cdc2/Cdc28p antibody, although the protein was present, as evidenced by immunoblotting the same membrane with anti-PSTAIRE antibody (Fig. 3G). Thus, the E12K substitution confers resistance to Swe1p by reducing or eliminating Y19 phosphorylation.
Comparison of the CDC28Y19F and CDC28E12K mutants.
The repeated independent isolation of the CDC28E12K mutation and the failure to isolate a CDC28Y19F mutation (which would require a single A-to-T change) seemed surprising. To determine whether this was a chance result or whether there might be phenotypic differences between the E12K and Y19F alleles, we transformed these alleles on a low-copy-number plasmid into the strains used for the genetic screen. In the zds1 zds2 strain (JMY1222), both alleles were able to suppress the slow-growth defect, but only the CDC28E12K allele effectively suppressed the morphologic abnormalities (Fig. 4). zds1 zds2 mutants containing the CDC28Y19F allele continued to display abnormal morphologies including elongated buds, although to a lesser degree than the parent strain (Fig. 4). This result explains our failure to isolate such alleles by using our stringent primary screen criteria.
To determine whether the alleles differed in the ability to sustain a checkpoint response, cells containing a single copy of each CDC28 allele integrated in tandem with the wild-type CDC28 at the CDC28 chromosomal locus were synchronized in G1 with α-factor and released into fresh medium containing the actin-depolymerizing drug Lat-A (Fig. 5). At the high concentrations used (50 μM), actin was largely depolymerized and all cells failed to bud (23). Consistent with previous findings, wild-type cells arrested the cell cycle and failed to undergo nuclear division. In contrast, cells containing the mutant CDC28E12K allele underwent nuclear division at around 2 h, while cells containing the CDC28Y19F allele underwent nuclear division at around 2.5 h (Fig. 5). Thus, the CDC28E12K allele was more potent than the CDC28Y19F allele both in rescuing zds1 zds2 cells and in abolishing the morphogenesis checkpoint. Presumably its potency in these assays explains why we repeatedly isolated the same allele from independent cell populations in our screen.
Cells lacking Swe1p have a more severe checkpoint defect than cells containing the checkpoint-defective CDC28 alleles.
In previous studies, we had interpreted the residual checkpoint-induced G2 delay in cells containing the CDC28Y19F allele to indicate the existence of a second pathway capable of inhibiting Cdc28p in response to morphogenesis insults (21). The reduced G2 delay in cells containing the CDC28E12K allele might therefore indicate that this mutant was resistant both to the Swe1p-mediated phosphorylation pathway and the putative second pathway. To address this issue further, we compared the kinetics of nuclear division in cells lacking Swe1p versus cells containing nonphosphorylatable Cdc28p (Fig. 5). Surprisingly, swe1Δ cells underwent nuclear division even earlier than the cells containing the CDC28E12K allele (Fig. 5). In addition, the swe1Δ cells underwent nuclear division at the same time regardless of which CDC28 allele was present (Fig. 5). This finding suggests that both the E12K and Y19F forms of Cdc28p are still sensitive (to a reduced extent) to inhibition by Swe1p: Cdc28pE12K is more resistant to inhibition by Swe1p than Cdc28pY19F.
Phosphorylation-independent inhibition of Cdc28p by Swe1p.
Booher and colleagues have reported that Swe1p can inhibit Cdc28pY19F in vitro, in a manner that is sensitive to dilution (7). We confirmed the observation that Swe1p could inhibit Cdc28pY19F as well as wild-type Cdc28p (Fig. 6A). In this experiment, GST-Cdc28p/Clb2p complexes were isolated from yeast cells as described in Materials and Methods and mixed with Swe1p immunoprecipitates in the absence of ATP to allow binding. Histone H1 and [γ-32P]ATP were then added, and histone H1 kinase activity was monitored. Under these conditions, Swe1p inhibited wild-type and Y19F Cdc28p to similar extents (Fig. 6A).
The simplest interpretation of these data would be that Swe1p can inhibit Cdc28p by direct binding, with no requirement for phosphorylation. In principle, however, the inhibition could also be mediated by some other phosphorylation catalyzed by Swe1p, either on Cdc28p itself or on some associated protein. To distinguish between these possibilities, we generated a swe1 mutant containing a K473P change in subdomain II of the catalytic domain. This highly conserved lysine residue has been demonstrated to be critical for catalytic activity in many protein kinases (15), and the nonconservative substitution of a proline at that position should eliminate Swe1p catalytic activity. In mixing experiments similar to those described above, we found that Swe1pK473P was capable of inhibiting Cdc28p histone H1 kinase activity to the same extent as wild-type Swe1p (Fig. 6B). Thus, Swe1p catalytic activity is not required for Cdc28p inhibition, and it seems likely that direct binding of Cdc28p by Swe1p can inhibit Cdc28p activity in a phosphorylation-independent manner.
Endogenous levels of Swe1pK473P provide a partial checkpoint response.
The inhibition of Cdc28p by catalytically inactive Swe1p in vitro suggested that catalytically inert Swe1p might be able to function similarly in vivo and provide a partial checkpoint response. To test this, we generated a strain in which Myc-tagged Swe1pK473P was expressed at the SWE1 genomic locus from its own promoter (see Materials and Methods). Cells containing wild-type Swe1p, catalytically inactive Swe1pK473P, or no Swe1p were then synchronized, and their response to actin perturbation by Lat-A was monitored (Fig. 7). As expected, cells containing wild-type Swe1p underwent a G2 arrest, while cells lacking Swe1p did not. Cells containing Swe1pK473P displayed an intermediate response, delaying but not blocking nuclear division (Fig. 7). This demonstrates that noncatalytic inhibition of Cdc28p by Swe1p can play a role during a physiological checkpoint response.
DISCUSSION
Genetic screen for morphogenesis checkpoint mutants.
We report the results of a two-part screen for spontaneous mutants that abrogate the morphogenesis checkpoint in budding yeast. The first step was a selection for suppressors of the zds1 zds2 slow-growth phenotype, predicated on the assumption that the cause of slow growth in these mutants was an inappropriately strong checkpoint arrest. Consistent with this assumption (first suggested by Wang and Burke [36]), we showed that the slow growth and abnormal morphology of these mutants were suppressed by deletion of SWE1 or overexpression of MIH1. The second step was a direct screen for mutants that failed to arrest the cell cycle following perturbation of the actin cytoskeleton. This screen gave rise to two classes of mutants that are expected to arise at very low frequency; the first class inactivated both of the copies of SWE1 present in the starting strain, while the second class consisted of a single dominant checkpoint-defective allele of CDC28.
The fact that such rare types of mutants were isolated repeatedly and that no simple recessive checkpoint-deficient mutants were isolated suggests that our starting assumption may have been wrong. These results are most easily accommodated by postulating that the hyperactivation of Cdc28p tyrosine phosphorylation that leads to the slow growth of the zds1 zds2 mutants arises through a pathway separate from that which causes Cdc28p tyrosine phosphorylation during a checkpoint response. If this is true, then in order to pass both parts of our screen, a mutant would have to abolish two separate pathways causing Swe1p-mediated Cdc28p inhibition, and this would occur only for mutants that actually altered Swe1p or Cdc28p. An alternative possibility is that zds1 zds2 mutants do in fact trigger the checkpoint, but that we could not isolate mutants in upstream components of this pathway because of functional redundancy.
CDC28E12K is resistant to inhibition by Swe1p.
A further surprise stemming from our screen was that all four independent dominant mutations in CDC28 caused the same amino acid change, E12 to K. Phenotypic analysis demonstrated that a strain containing this mutant grew normally under nonperturbing conditions but was largely unable to delay the cell cycle in response to the morphogenesis checkpoint. In addition, the E12K mutant rendered cells resistant to overexpression of Swe1p, which arrests cells containing the wild-type Cdc28p in G2. Finally, we found that Cdc28pE12K was not phosphorylated at Y19 upon overexpression of Swe1p.
Interestingly, a screen for fission yeast cdc2 mutants defective in the DNA replication checkpoint identified a mutant that altered the homologous residue of Cdc2, E8, to V (3). In contrast to the CDC28E12K phenotype reported here, the cdc2E8V mutant phenotype was less severe than that of cells containing the nonphosphorylatable cdc2Y15F mutant, and no cdc2E8K mutants were isolated even though that screen was specifically focused on cdc2 (3). This is probably because tyrosine phosphorylation of Cdc2 is crucial for proliferation in fission yeast even in the absence of a checkpoint (13), so that a cdc2 mutant as severe as CDC28E12K would be lethal in that organism. The fact that mutation of homologous residues in CDC28 (budding yeast) and cdc2 (fission yeast) affect cell cycle arrest by the morphogenesis checkpoint in budding yeast and by the DNA replication checkpoint in fission yeast is consistent with the fact that in both cases, arrest of cell cycle progression occurs through enhanced tyrosine phosphorylation of cdc2 (see the introduction). This strengthens the conclusion that these alleles are resistant to inhibition by tyrosine phosphorylation.
Conservation of the E12 residue among Cdks.
A comparison of the amino-terminal sequences in different Cdks revealed that the E12 residue was not highly conserved in general but was present in all Cdks known to be subject to inhibition by Wee1 family kinases (Cdc28p, Cdc2, and Cdk2 [Fig. 3A]). Given our data, we suggest that this is a key residue for recognition by Wee1 family enzymes. Interestingly, among the five budding yeast Cdks only Pho85p shares this residue with Cdc28p (Fig. 3A). This raises the possibility that Pho85p might also be a Swe1p target and suggests that the other yeast Cdks (Kin28p, Ctk1p, and Ssn3p/Ume5p/Srb10p) are not Swe1p targets.
Swe1p inhibits the nonphosphorylatable Cdc28pY19F.
Based on previous findings with strains containing the CDC28Y19F allele, we concluded that in addition to cell cycle arrest mediated by phosphorylation of Y19 in Cdc28p, the morphogenesis checkpoint induced a second phosphorylation-independent arrest pathway (21). The data presented in this report suggest that this second pathway is not distinct, but rather is also mediated by Swe1p, which can still inhibit the Y19F form of Cdc28p. Further, we found that a mutant Swe1p predicted to lack kinase activity was still able to inhibit Cdc28p. Therefore, a single pathway involving Swe1p-mediated Cdc28p inhibition fully accounts for the cell cycle delay (or arrest) in response to the morphogenesis checkpoint.
Recently, Swe1p was found to be required for haploid invasive growth of S. cerevisiae (9). In that study, the effects of deleting SWE1 were found to be more severe than those associated with the CDC28Y19F allele, leading the authors to speculate that additional targets of Swe1p might be involved. However, given our findings we suggest that the relative ineffectiveness of the CDC28Y19F allele might simply reflect residual inhibition of the encoded mutant protein by Swe1p.
Catalytically inactive Swe1p responds to the morphogenesis checkpoint.
Remarkably, we found that Swe1p lacking catalytic activity, when expressed at single copy, could significantly delay the cell cycle in vivo during a physiological checkpoint response. In contrast, even GAL-mediated overexpression of the catalytically inactive Swe1p did not significantly delay the cell cycle in cells not exposed to an actin perturbation (data not shown). This suggests that the checkpoint must enhance Swe1p effectiveness (even for the catalytically inert mutant), and not simply Swe1p abundance. Thus, the recently described stabilization of Swe1p in response to checkpoint insults (30) cannot fully account for increased Swe1p activity during a checkpoint response.
Implications for other systems.
Our findings have far-reaching implications for many studies in other systems that have utilized nonphosphorylatable Cdc2 mutants on the assumption that they completely bypass the action of Wee1-related kinases. In Aspergillus, cells expressing a nonphosphorylatable Cdc2 mutant failed to delay mitosis when incubated in low doses of hydroxyurea but were still able to arrest in response to high doses of hydroxyurea, suggesting that the checkpoint induced a separate pathway (possibly involving BimE [39]). In Xenopus, addition of cyclin B complexed with a nonphosphorylatable Cdc2 mutant to egg extracts promoted entry into mitosis of interphase extracts, but not checkpoint-arrested extracts (18), suggesting that the DNA replication checkpoint induced a separate pathway (possibly involving a Cdc2 inhibitor [18, 19]). In human cells, inhibitory Cdc2 phosphorylation was important for the DNA damage and DNA replication checkpoints (5, 16), but cells containing a nonphosphorylatable Cdc2 mutant could still delay mitosis when these checkpoints were maximally induced (16), suggesting that these checkpoints induced a separate pathway (possibly preventing the nuclear localization of cyclin-Cdc2 complexes [16, 17, 35]). In all of these cases, our findings suggest an alternative possible interpretation in which a single pathway involving Wee1 like kinases may have inhibited the nonphosphorylatable Cdc2 mutant.
Conclusion.
We have isolated a novel morphogenesis checkpoint-defective allele of CDC28 in budding yeast. Analyses using this mutant and a nonphosphorylatable CDC28 mutant show that Swe1p can inhibit Cdc28p without phosphorylating Cdc28p, under physiological conditions. We conclude that a single Swe1p-dependent pathway mediates arrest by this checkpoint, contrary to our previous conclusion (21) indicating a bifurcated pathway. Similar bifurcated pathways (reviewed in reference 20), proposed to account for the action of DNA replication and DNA damage checkpoints in other systems, must now be reevaluated.
ACKNOWLEDGMENTS
We thank Steve Garrett, Erfei Bi, John Pringle, and Beverly Errede for plasmids, Phil Crews for a gift of Lat-A, Mark Watson for a gift of p18CKS1, Haifeng Yang for the suggestion to use the anti-phospho-Cdc2 antibody, and Lynn Martinek and Mike Cook from the Duke Cancer Center Flow Cytometry Shared Resource for help with the flow cytometry. We thank Elizabeth Choi for assistance with the screen, Sally Kornbluth for critical reading of the manuscript, and members of the Lew and Pringle labs for stimulating interactions.
J.N.M. was supported by NIH postdoctoral fellowship GM18455. This work was supported by Public Health Service grant GM53050 and by funds from the Searle Scholars Program/The Chicago Community Trust to D.J.L.
REFERENCES
- 1.Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- 2.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basi G, Enoch T. Identification of residues in fission yeast and human p34cdc2 required for S-M checkpoint control. Genetics. 1996;144:1413–1424. doi: 10.1093/genetics/144.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasina A, Paegle E S, McGowan C H. The role of inhibitory phosphorylation of cdc2 following DNA replication block and radiation-induced DNA damage in human cells. Mol Biol Cell. 1997;8:1013–1023. doi: 10.1091/mbc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 7.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunphy W G. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 9.Edgington N P, Blacketer M J, Bierwagen T A, Myers A M. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 11.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 12.Ghiara J B, Richardson H E, Sugimoto K, Henze M, Lew D J, Wittenberg C, Reed S I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- 13.Gould K, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 14.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S K, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 16.Jin P, Gu Y, Morgan D O. Role of inhibitory CDC2 phosphorylation in radiation induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin P, Hardy S, Morgan D O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai A, Dunphy W G. Control of the Cdc2/cyclin B complex in Xenopus egg extracts arrested at a G2/M checkpoint with DNA synthesis inhibitors. Mol Biol Cell. 1995;6:199–213. doi: 10.1091/mbc.6.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee T H, Kirschner M W. An inhibitor of p34cdc2/cyclin B that regulates the G2/M transition in Xenopus extracts. Proc Natl Acad Sci USA. 1996;93:352–356. doi: 10.1073/pnas.93.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lew D J, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 21.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 607–695. [Google Scholar]
- 23.McMillan J N, Sia R A L, Lew D J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 25.Mueller P R, Coleman T R, Kumagai A, Dunphy W G. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 26.Pines J. Cyclins and cyclin-dependent kinases: theme and variations. Adv Cancer Res. 1995;66:181–212. doi: 10.1016/s0065-230x(08)60254-7. [DOI] [PubMed] [Google Scholar]
- 27.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 28.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 29.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeast. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 30.Sia R A L, Bardes E S G, Lew D J. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorger P K, Murray A W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34CDC28. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- 34.Stueland C S, Lew D J, Cismowski M J, Reed S I. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Burke D J. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of Saccharomyces cerevisiae: cell cycle periodicity, regulation by mating pheromone and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye X S, Fincher R R, Tang A, O’Donnell K, Osmani S A. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 1996;15:3599–3610. [PMC free article] [PubMed] [Google Scholar]