Abstract
Currently, therapeutics for COVID‐19 are limited. To overcome this, it is important that we use physiologically relevant models to reproduce the pathology of infection and evaluate the efficacy of antiviral drugs. Models of airway infection, including the use of a human infection challenge model or well‐defined, disease relevant in vitro systems can help determine the key components that perpetuate the severity of the disease. Here, we briefly review the human models that are currently being used in COVID‐19 research and drug development.
Keywords: cilia, differentitated, infection, respiratory, SARS‐CoV‐2
Abstract figure legend Current physiologically relevant models for studying COVID19. Image made using BioRender.com.
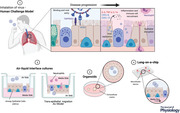
Introduction
Coronavirus disease 2019 (COVID‐19)is an infectious disease caused by a newly emerged severe acute respiratory syndrome (SARS)‐coronavirus‐2 (SARS‐CoV‐2). COVID‐19 presents a diverse clinical spectrum, ranging from an asymptomatic carrier state to patients with life‐threatening multi‐organ failure and death (Huang et al. 2020 a). The greatest risk factor for severe disease is age, with higher morbidity and mortality rates in the elderly population, despite younger people shedding similar levels of virus (Yanez et al. 2020). The overall case fatality rate of COVID‐19 is 2.3%, rising to 14.8% in patients over the age of 80, and 49% among the critically ill (Kang & Jung, 2020).
The human airway epithelium is the first line of defence against inhaled pathogens and, to help prevent damage to the vulnerable tissues, the airways have developed physical and innate molecular barriers. These include polyps, hairs and mucus to trap foreign objects. At the microscopic level, the airway epithelial barrier is composed of differentiated cell types including muco‐secretory goblet cells and ciliated cells (Pack et al. 1981). These cells function together to maintain a healthy homeostasis through the production of secretions that regulate the volume and viscosity of the fluid layer, and motile ciliary beating that coordinates clearance (Empey & Kolls, 2017).
SARS‐CoV‐2 virus enters the airway through the oral and nasal cavities (Gallo et al. 2020). The inhaled virus can evade these initial barriers and infect the respiratory epithelium. SARS‐CoV‐2 is thought to primarily target and infect airway epithelial cells via interaction of the viral spike glycoprotein with the angiotensin converting enzyme 2 (ACE2) host cell receptor (Zhou et al. 2020 b; Hoffmann et al. 2020), although other receptors have been implicated (i.e. NRP1, BSG, TFRC) (Wang et al. 2020; Tang et al. 2020; Cantuti‐Castelvetri et al. 2020; Daly et al. 2020). An impressive body of RNA sequencing data has confirmed that ACE2 is expressed in approximately 1% of epithelial cells (Ziegler et al. 2020) and there is a gradient of ACE2 expression from the upper to the lower airways (Hou et al. 2020). Within the airways, ACE2 protein is thought to be most abundantly found on ciliated cells, nasal goblet cells and alveolar type II cells (Sungnak et al. 2020; Li et al. 2020; Ortiz et al. 2020; Lee et al. 2020 b). There is also evidence that some secretory cell types, notably the ‘secretory3’ cell type (an intermediate position between club or goblet cells and ciliated cells) may be an important viral target as they are ACE2+ and co‐express viral spike priming proteases transmembrane serine protease 2 (TMPRSS2) and/or furin (Sungnak et al. 2020; Lukassen et al. 2020; Ziegler et al. 2020; Hou et al. 2020; Schuler et al. 2021). Histological examination of the lungs of deceased COVID‐19 patients showed it was predominantly the ciliated cells that were infected, whereas MUC5B+ club cells, MUC5AC+ goblet cells and p63+ basal cells were not infected (Hou et al. 2020; Schaefer et al. 2020). Alveolar type 1 (AT1) and 2 (AT2) cells (or AT2 cells that had transitioned to AT1 cells) can also become infected (Hou et al. 2020; Schaefer et al. 2020). Recently, a novel epithelial cell population that is found bridging the secretory and ciliated clusters (referred to as inflammatory epithelial transit cells) was found to yield the highest viral load in airway epithelial cells (Yoshida et al. 2021).
Once infected, the airways have been shown to become inflamed and damaged. Patients with severe COVID‐19 have demonstrated acute diffuse alveolar damage and diffuse inflammatory infiltrates (consisting of interstitial and peribronchial lymphocytes and intra‐alveolar macrophages) (Hou et al. 2020; Schaefer et al. 2020). Compared to moderate cases and controls, patients with critical COVID‐19 disease exhibited epithelial cells with significant expression of chemokine‐ligand encoding genes that promote recruitment of neutrophils, T cells and mast cells (Chua et al. 2020; Lee et al. 2020 c). Severe cases also show greater enrichment for neutrophils, comprising >60% of cellular composition of upper airway samples (Chua et al. 2020). The recruitment of immune cells to sites of epithelial infection is an important early innate defence mechanism, regulated by the secretion of cytokines and chemokines by infected epithelial cells, which can help control inflammation and promote pathogen clearance.
To study the disease mechanisms and evaluate the efficacy of antiviral drugs, it is important that models of COVID‐19 reproduce the physiology and pathology of infection. Here, we briefly review the human models that are currently being used in COVID‐19 research and drug development.
Models of the airway
Human infection challenge model
The human infection challenge model has, for many decades, helped to shorten the timeline of new therapeutic drugs and vaccines, and to understand the role of viruses in disease pathogenesis. Edward Jenner's 1796 iconic challenge experiment demonstrated the protective effects of cowpox against smallpox and launched the field of vaccinology. We now have well‐defined ethical criteria for human‐challenge trials (WHO, 2020) and state‐of‐the‐art facilities that satisfy safety and regulatory requirements, making this a scientifically acceptable and ethically valid method in the modern age. (For a review of ethical considerations, see Miller & Grady, 2001). Previous human infection challenge trials, of several hundred adult subjects, have established this as a safe and effective method in which to study the viral life cycle of many respiratory viruses including influenza virus, respiratory syncytial virus (RSV) and rhinovirus, the latter of which has been shown to reproduce the natural acquired infection (Lambkin‐Williams et al. 2018). A human COVID‐19 infection challenge trial is currently taking place at the Royal Free Hospital, London, infecting 90 healthy adults (aged 18–30) to determine the lowest possible dose of SARS‐CoV‐2 to cause disease (Callaway, 2020).
Human in vitro models
In vitro models are essential for the pre‐clinical evaluation of potential therapeutics and can give new insight into the mechanism of infection and viral pathogenesis. Indeed, high‐throughput cell line assays measuring cytopathic effects were instrumental in the discovery and development of anti‐viral drugs including remdesivir (Eastman et al. 2020). Since the detection of SARS‐CoV‐2 in November 2019, several in vitro models have been used to determine replication, infection and cytopathic effects of the virus. The essential characteristics of cell models required for SARS‐CoV‐2 infection are thought to be the expression of ACE2 and, secondarily, spike priming proteases TMPRSS2 and furin. Investigations into the susceptibility of cell lines to SARS‐COV‐2 infections showed that replication was most robust in Calu‐3 (a human lung adenocarcinoma cell line) and Caco2 (a human intestinal epithelial cell line), whilst hepatic Huh7, renal HEK293T and neuronal U251 cells were also able to propagate the virus (Chu et al. 2020). However, the commonly used lung adenocarcinoma cell line A549 (often used to model alveolar type II cells) and the immortalised human bronchial epithelium cell line BEAS2B are poor models for SARS‐CoV‐2 investigation as they are not permissive to infection, unless transformed with ACE2 (Kam et al. 2009; Blanco‐Melo et al. 2020; Chu et al. 2020). This is especially intriguing as their respective areas of origin (alveoli and bronchi) have demonstrated ACE2 expression and infection in vivo (Ziegler et al. 2020; Hou et al. 2020; Salahudeen et al. 2020). Other non‐human primate cell lines that have been historically employed for virology and replicate SARS‐CoV‐2 include kidney cells FRhK4, LLCMK2 and Vero E6 cells (Matsuyama et al. 2020; Chu et al. 2020). Interestingly, the heavily used Vero E6 cell line does not express TMPRSS2 yet produces robust propagation of SARS‐CoV‐2 and is commonly used to generate viral stocks from clinical isolates (Harcourt et al. 2020; Ogando et al. 2020). A biochemical cleavage assay that uses Caco2, HEK293T or Vero E6 cells has been used to show SARS‐CoV‐2 spike protein harbours a distinct four‐amino‐acid insertion at the S1/S2 that can be cleaved by furin‐like, trypsin‐like and cathepsin proteases (Hoffmann et al. 2020; Jaimes et al. 2020). Whilst this is of interest for potential development of protease inhibitors (i.e. camostat) for therapeutics, a caveat here is the cathepsin spike priming pathway may only target the residual protein unprimed by TMPRSS2 and therefore act redundantly within COVID‐19 disease severity and progression (Hoffmann et al. 2020).
It is also crucial to note that whilst passaging SARS‐CoV‐2 in cell lines, and specifically Vero E6 cells, the virus is under strong selection pressure to acquire adaptive mutations in its spike protein gene, due to the lack of relevant protease expression (Klimstra et al. 2020; Ogando et al. 2020; Peacock et al. 2020). This selection pressure can lead to attenuated replication in human bronchial epithelial cells (Pohl et al. 2021). These mutations may be negated by propagating SARS‐CoV‐2 in the serine protease expressing Calu‐3 cell line (Lamers et al. 2021). These are important considerations for drug discovery. For example, a screen of ∼3000 potential anti‐viral drugs using Vero E6 cells showed few hits, but the same library screened in Calu‐3 revealed nine hits, seven of which are currently being used in human trials (Dittmar et al. 2021). Development of SARS‐CoV‐2 pseudoviruses also offers a potential solution for the genetic instability apparent with Vero E6 propagation, with the additional benefit of increased safety, reproducibility and scalability for screening assays (for reviews see Li et al. 2018; Chen & Zhang, 2021). Proof of principle for this has been demonstrated with a HIV‐based lentiviral pseudovirus assay in a HEK293T cell line expressing human ACE2 and TMPRSS2 (Neerukonda et al. 2021). This assay has shown success in screening for neutralizing antibodies for SARS‐CoV‐2 (Neerukonda et al. 2021).
Calu‐3 cells have also been a useful model in determining the function of SARS‐CoV‐2 non‐structural proteins (NSPs). Employing a range of functional analyses including RNA/protein crosslinking, splice reporter assays and surface sensing of translation (SUnSET) assays, NSPs were shown to disrupt mRNA processing. Mechanisms included suppression of global mRNA splicing (NSP16), binding to 18S ribosomal RNA causing global inhibition of mRNA translation (NSP1) and disruption of protein trafficking to the cell membrane (NSP8 and NSP9) (Banerjee et al. 2020).
Whilst easy to obtain, with the generation of reproducible assays, these immortalised cell cultures (using conventional submerged culture techniques) lack appropriate cell polarisation and many other distinguishing properties found in the lung, such as transport proteins, mucus and motile cilia. Fortunately, the relative accessibility to airway epithelium and air–liquid interface (ALI) culture techniques (described below) has allowed the development of a gold standard for in vitro epithelial models using primary airway epithelial cells.
Air–liquid interface cell culture
The ALI culture platform allows a physiological and pragmatic in vitro model that resides at the top of the experimental hierarchy for preclinical airway epithelial model systems (Karp et al. 2002). Here, cells are grown on semipermeable membranes with a basolateral medium supply and apical exposure to air in a well humidified (>95%) environment. This method is commonly used in a 24‐ or 12‐well plate format, but recently groups have demonstrated the application of a miniaturised 96‐well microplate system (Hyang Lee et al. 2020) and robotics for exchange of media that enable higher throughput drug testing (Bluhmki et al. 2020). Exposure to air stimulates the epithelial progenitor cells to differentiate into heterogeneous pseudostratified ciliated, goblet and basal cells, alongside other specialised cell types such as the ionocyte (Montoro et al. 2018). After 28 days, this differentiated epithelium exhibits mucus production and motile cilia, and transcriptionally has been shown to strongly correlate (>96% similarity) with the expression profile of epithelial cells obtained from the original nasal brushings from the same patients (Ghosh et al. 2020).
There are two major advantages of the ALI system. First, it allows for several precise, functional readouts of airway physiology, including the measurement of ciliary beating, mucociliary clearance, current or voltage across the membrane, differential protein secretion, airway surface liquid height measurements, ion transport measurements and wound healing assays (see review articles: Gianotti et al. 2018; Hiemstra et al. 2019). Secondly, airway epithelial cell culture models can be generated from any donor of interest, allowing the modelling of a range of human phenotypes. For example, models can be developed from donors that are diverse for variables associated with COVID‐19 severity, such as age, ethnicity, sex, comorbidities such as diabetes and smoking/vaping and respiratory disorders. All of these have demonstrated distinct phenotypic and/or functional characteristics in ALI culture (Clunes et al. 2012; Bilodeau et al. 2016; Castellani et al. 2018; Carlier et al. 2018; Gaiha et al. 2020; Woodall et al. 2020; Zhu et al. 2021).
Primary ALI cell cultures
Primary airway progenitor cells are the principal choice for the ALI method. These cells can be refined from small tissue biopsies taken from donated lungs, nasal brushings or bronchial brushings by bronchoscopy. The main challenge of working with primary cell culture lies with procurement of the appropriate cells and access to ethically sourced lung/airway tissue. Even when readily available, the process of retrieving these tissues requires specialist and expensive culture methods and carries risk of contamination. There is also a substantial amount of evidence that shows that different culture practices, including different medium preparations, membrane pore size and collagen coating method, and the duration at ALI can vary the characteristics of the epithelial model (Gianotti et al. 2018; Lee et al. 2020 a; Leung et al. 2020).
In traditional airway epithelial cell culture systems, primary cells do not survive for more than a few passages and their characteristics can deteriorate rapidly with age. One solution of many (see review article: Orr & Hynds, 2021) to increase proliferative capacity of the progenitor cells has been to expand the progenitor cells in co‐culture with irradiated/mitotically inactivated fibroblasts prior to conversion to ALI for differentiation (Butler et al. 2016). Another solution has been to introduce anti‐senescent mechanisms such as viral oncogenes or the human polycomb protein BMI‐1 (Fulcher et al. 2009; Munye et al. 2016; Gianotti et al. 2018). The use of induced pluripotent stem cells (iPSCs) is also becoming increasingly popular, as fully differentiated cells such as fibroblasts can be expanded rapidly as iPSCs and then reprogrammed to differentiate into club cells, goblet cells or ciliated cells that self‐assemble into a functional pseudostratified airway epithelium (Hawkins et al. 2021), reproduce characteristics of the proximal and distal airways (Pollard & Pollard, 2018) and are capable of replicating SARS‐CoV‐2 (Huang et al. 2020 b).
Immortalised cell lines
Some immortalised airway epithelial cell lines, such as 16HBE14, and spontaneous cancer cells (i.e. Calu‐3 and H441) (see table in review article: Orr & Hynds, 2021) that are cultured on semipermeable membranes can form tight junctions, secrete mucins, and show differential distribution of plasma membrane transport proteins between the apical membrane and the basolateral membrane epithelium (Castellani et al. 2018), but do not produce ciliated and other important specialised cell types. Calu‐3 cells are a popular epithelial cell line for infection studies as they are known to generate higher transepithelial resistance, produce mucus, and express a diverse set of immune and inflammatory modulators (Grainger et al. 2006; Braakhuis et al. 2020). The HBEC3 series are thought to be the most favourable airway model due to generation of motile cilia (Lodes et al. 2020). To facilitate the study of basal cell biology, a few research groups have successfully immortalised basal cell lines expanding the experimental scope. In one instance, basal cell immortalised non‐smoker 1 (BCi‐NS1) is an immortalised human large airway basal cell line generated via retrovirus‐mediated expression of human telomerase (hTERT) (Walters et al. 2013). The small airway epithelium basal cell line hSABCi‐NS1.1 was generated via a similar methodology (Wang et al. 2019). The development of these basal cell lines with a multipotent differentiation capacity retaining characteristics to the original primary basal cells for over 40 passages is a useful tool for understanding basal cell biology, pathogenesis and related diseases allowing long term experimentation. Notably, expression of genes encoding ACE2, TMPRSS2 and other essential factors necessary for SARS‐CoV‐2 infection has been detected in BCi‐NS1.1 and hSABCi‐NS1.1 cell lines, thus advocating for their suitability in COVID‐19 research (Zhang et al. 2020).
Whilst immortalised cell lines are pragmatic for high throughput assays, especially those required for drug screening, a resounding problem in their use is the inability to represent age phenotypes when age‐related severity is a distinguishing characteristic of COVID‐19.
Primary ALI cultures for SARS‐CoV‐2 research
Primary cells grown using ALI culture have been shown to propagate SARS‐CoV‐2 effectively in epithelial cell types expressing ACE2 (Hou et al. 2020; Robinot et al. 2020; Zhu et al. 2020, 2021; Pohl et al. 2021). Interestingly, the number of ciliated cells in airway epithelial cell cultures did not correlate with susceptibility to infection (Hou et al. 2020). This may be due to the presence of secondary entry receptors (NRP1, BSG, TFRC), whose expression has shown some correlation with actively infected cells in scRNA‐seq studies and cell line assays (Wang et al. 2020; Tang et al. 2020; Cantuti‐Castelvetri et al. 2020; Daly et al. 2020; Yoshida et al. 2021), or changes to epithelial defence mechanisms, including apparent basal cell mobilization (Robinot et al. 2020). ALI cultures have also been shown to support long‐ and short‐term modelling of SARS‐CoV‐2. Short‐term studies are useful to investigate virus replication and cytopathic effects, and there is rapidly emerging data suggesting some prevalent variants of SARS‐CoV‐2 may have altered replication dynamics within human airway epithelial cells, whilst some in vitro passaged isolates may develop mutations that severely decrease infectivity of human airway epithelial cells (Peacock et al. 2020; Liu et al. 2021; Pohl et al. 2021). Long‐term modelling allows us to study the airway epithelium's ability to repair and regenerate (Hao et al. 2020).
Functional readouts from ALI‐specific assays have shown a transient decrease in epithelial barrier function and disruption of tight junctions, though infectious viral particles almost exclusively remain at the apical side of cultures (Fig. 1) (Robinot et al. 2020; Hao et al. 2020; Zhu et al. 2020). SARS‐CoV‐2 infection also led to a rapid loss of the ciliary layer and this resulted in reduction of motile cilia function as measured in a mucociliary clearance assay (Robinot et al. 2020). Furthermore, iPSC‐derived AT2 cultures demonstrated rapid transcriptomic change in SARS‐CoV‐2‐infected cells to an inflammatory phenotype, characterised by an upregulation of nuclear factor κB signalling (Huang et al. 2020 b).
Figure 1. Transmission electron micrographs of SARS‐CoV‐2‐infected ciliated nasal epithelial cells grown in culture at an air–liquid interface.
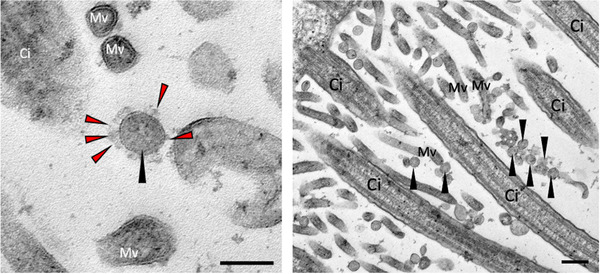
Black arrows indicate SARS‐CoV‐2 viral particles; red arrowheads indicate the viral spike protein. Ci, cilia; Mv, microvilli. Scale bar: 200 nm. Image credit Andreia Pinto.
Studies have shown that ALI cultures display the age‐, sex‐ and disease‐dependent changes in ACE2 mRNA levels that have been observed in lung tissue from different donors (Lukassen et al. 2020). Specifically, the expression of putative SARS‐CoV‐2 receptors was found to be lower in the upper and lower airways in children compared to other age groups, whilst expression of both ACE2 and TMPRSS2 were upregulated in smokers and patients with chronic obstructive pulmonary disease compared with healthy subjects (Saheb Sharif‐Askari et al. 2020). The role of entry factors in determining disease severity remains inconclusive, and other studies suggest that the immune response to SARS‐CoV‐2 may play a more important role (Koch et al. 2021), with elevated production of type I and III interferons, rather than differential receptor expression, suggested as the cause of the lower viral replication in paediatric compared to adult nasal epithelial cultures (Zhu et al. 2021).
Trans‐epithelial migration models
The migration of neutrophils and other polymorphonuclear leukocytes (PMNs) across columnar epithelia is a key component of mucosal defence and inflammation, and it is becoming increasingly evident that COVID‐19 disease severity is associated with a dysregulation of the immune response, including increased neutrophil recruitment (Lee et al. 2020 b). To study this further, a modified ALI model has been developed that facilitates neutrophil transepithelial migration in response to airway infection or other stimuli (for review see Adams et al. 2021). This three‐dimensional in vitro technique differs from the aforementioned ALI model in that airway epithelial cells are cultured on the underside of the membrane insert. The inverted insert is coated with collagen to assist in the epithelial cell attachment. Once fully differentiated (at ALI for 28 days) and infected, neutrophils or PMNs are added to the basolateral (medium) side of the epithelial cells so they can migrate in the physiological direction, from basolateral to apical surface. This model has already revealed important insights into the contribution of neutrophils to airway damage and viral clearance during RSV infection (Deng et al. 2018; Herbert et al. 2020). Additionally, this model was used to study neutrophil transepithelial migration during Pseudomonas aeruginosa infection (Kusek et al. 2014; Yonker et al. 2017). Although this model has not yet been utilised for COVID‐19 research, work is underway in our group to study neutrophil transepithelial migration in response to SARS‐CoV‐2 infection.
Immortalised cell lines have also been applied in co‐culture models. Specifically, Calu‐3 cells have been used as the structural barrier to create co‐culture models with human macrophages, to model inflammatory responses to aerosols (Grainger et al. 2006).
Organoids
Another in vitro model that closely recapitulates lung epithelial function and architecture is three dimensional organoids. These self‐assembling structures can be cultured in a three dimensional extracellular matrix using a variety of progenitor cells including basal cells, airway secretory club cells, epithelial cells, iPSC and crypt stem cells (Lancaster & Knoblich, 2014; Barkauskas et al. 2017). Though there are some limitations with these models, such as the restricted accessibility to the luminal surface, they are quickly becoming valuable in development of personalised therapies (Dekkers et al. 2016; Berkers et al. 2019), to study lung development (Vazquez‐Armendariz et al. 2020) and infection with viruses such as rotavirus, norovirus, enterovirus 71 and human adenovirus (Ramani et al. 2018). Both airway and intestinal organoids have been shown to express high levels of ACE2 and membrane‐bound serine proteases TMPRSS2 and TMPRSS4 enabling cleavage of the SARS‐CoV‐2 spike protein to facilitate viral entry (Zang et al. 2020; Suzuki et al. 2020). Indeed, three simultaneous studies (Zang et al. 2020; Zhou et al. 2020 a; Lamers et al. 2020) used human adipose‐derived stem‐derived intestinal organoids to provide evidence that SARS‐CoV‐2 could establish itself in the gastrointestinal tract, showing that the most common cell type of the intestinal epithelium, the enterocyte, is readily infected and strongly upregulates viral response genes (Lamers et al. 2020). One recent development is the generation of distal lung organoids by embedding cells in extracellular matrices to form cyst‐like organoids with apical‐out polarity (Danahay et al. 2015; Lukassen et al. 2020) to present ACE2 on the exposed external surface (Salahudeen et al. 2020). This polarity allows for the more physiological, non‐invasive apical infection of AT2 and basal cultures with SARS‐CoV‐2, already leading to the identification of club cells as another target population via scRNA‐seq analysis (Salahudeen et al. 2020).
Organoid cultures have also shown potential for studying immune cell–epithelium interactions. The co‐culture of human intestinal stem cell‐derived enteroid monolayers with human monocyte‐derived macrophages have demonstrated communication between the epithelium and macrophages through morphological changes and cytokine production in response to E. coli infections (Noel et al. 2017).
Lung‐on‐a‐chip
A precursor to the lung organoid culture was the ‘lung‐on‐a‐chip’ model. This biological device uses microfluidics technologies and allows communication between epithelial and endothelial cells through a porous and elastic membrane. Here, epithelial cells in the upper chamber are exposed to the ALI, whilst the submerged endothelial cells (grown in the bottom layer) co‐ordinate the uptake of nutrients from the media (Benam et al. 2016 b). The lung‐on‐a‐chip model has been employed successfully in some drug discovery and toxicity studies (Huh et al. 2012; Gkatzis et al. 2018), including chips connected to an instrument that mimics ‘breathing’ (Benam et al. 2016 a; Huang et al. 2021). Recently, the human airway‐on‐a‐chip model has shown utility in testing potential antiviral therapeutics against COVID‐19 using pseudotyped (Si et al. 2021) and wild‐type SARS‐CoV‐2 virus (Thacker et al. 2021; Deinhardt‐Emmer et al. 2021).
Future directions
Ion transport in COVID‐19
Despite the consensus that airway epithelial cell function is key in COVID‐19 pathogenesis, there has been a lack of research on how critical epithelial functions, such as ion transport, are affected by SARS‐CoV‐2 infection (Gentzsch & Rossier, 2020). For example, it has been suggested that SARS‐CoV‐2 could disrupt conserved second messenger signalling cascades via G protein‐coupled receptors, adversely modulating transepithelial transport processes (Hameid et al. 2021). The role of cystic fibrosis transmembrane conductance regulator (CFTR) in COVID‐19 is also undescribed and may be of significance as a key regulator of mucociliary clearance and airway liquid pH, especially since SARS‐CoV‐2 entry into airway epithelial cells is pH‐dependent (Hoffmann et al. 2020; Shang et al. 2020). Investigation here may be valuable as it has been shown in an influenza model that potentiating CFTR expression and function with corrector lumacaftor reverses in vitro down‐regulation of CFTR and ENaC following viral infection, rehydrating the airway surface liquid (Brand et al. 2018), which could aid viral clearance.
Tissue engineering
Advances in whole lung bioengineering using engineered three‐dimensional scaffolds and microenvironments have opened new possibilities for studying lung regeneration and infection ex vivo using acellular human and non‐human derived lung tissue scaffolds. Methods to decellularize whole human lungs, lobes or resected segments from normal and diseased human lungs have been developed using both perfusion and immersion‐based techniques (Lin et al. 2009; Asnaghi et al. 2009; Castellani et al. 2018). These bioreactors, containing for example autologous respiratory epithelial cells and mesenchymal stem cells (BMSCs, then differentiated into chondrocytes), have been used clinically in tracheal transplantation (Macchiarini et al. 2008; Day, 2019) but also may be able to predict oxygen profiles (Asnaghi et al. 2009) following infection. These cellular systems, combined with improved sensitivity in readouts, offer immense potential to study the functional responses to respiratory virus infection using superior, physiologically relevant human models.
Conclusion
Every experimental model of the human airway has its limitations. Whilst cell lines are pragmatic models for reproducible high throughput assays, complex primary cell models, produced from donors from a range of demographics, are the only ones capable of representing the variability of disease severity that has become characteristic of COVID‐19 disease.
Additional information
Competing interests
All authors declare no competing interests.
Author contributions
M.W. contributed to paper conception, manuscript preparation and data collection (Fig. 1). T.M. and K.C. contributed to manuscript preparation. C.M.S. contributed to paper conception, manuscript preparation and graphical abstract. All authors have read and approved the final version of this manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by the National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. T.M. would also like to thank A.G Leventis Foundation for their support and help during her studies. C.M.S is currently a recipient of grants from Animal Free Research UK (AFR19‐20274), BBSRC (BB/V006738/1), GOSH Children's charity (COVID_CSmith_017) and the Wellcome Trust (212516/Z/18/Z).
Funding for Open Access was approved for this article.
Supporting information
Peer Review History
Video abstract and summary
Acknowledgements
All authors want to thank Dr Robert Hynds (University College London and The Francis Crick Institute, London) for feedback on draft manuscripts, and Dr Andreia Pinto (Royal Brompton Hospital, London) for providing transmission electron microscopy images.
Biographies
Claire M. Smith, PhD, is an Associate Professor in the department of Infection, Immunity and Inflammation, GOS Institute of Child Health, University College London. Her research focuses on anti‐viral therapies, mucosal immunity and the interaction between the airway epithelium and neutrophils during respiratory virus infection.

Maximillian Woodall, PhD, is a Research Fellow working alongside C.M.S. His research employs an experimental model of the human airway to investigate age‐dependent factors driving COVID‐19 disease severity.
Tereza Masonou, MSc, is a PhD student working under the supervision of C.M.S. Her research investigates neutrophil transepithelial migration and function during SARS‐CoV2 infection, and how this differs between older adults and children.
Katie M. Case, MSc, is a Research Assistant working alongside M.W. She maintains airway epithelial cells from different age groups and explores baseline differences such as cell motility, cilia beating and protein levels.
Edited by: Ian Forsythe & Frank Powell
The peer review history is available in the Supporting Information section of this article (https://10.1113/JP281499#support‐information‐section).
This is an Editor's Choice article from the 15 September 2021 issue.
References
- Adams W, Espicha T & Estipona J (2021). Getting your neutrophil: Neutrophil transepithelial migration in the lung. Infect Immun 89, doi: 10.1128/iai.00659-20. [DOI] [PubMed] [Google Scholar]
- Asnaghi MA, Jungebluth P, Raimondi MT, Dickinson SC, Rees LEN, Go T, Cogan TA, Dodson A, Parnigotto PP, Hollander AP, Birchall MA, Conconi MT, Macchiarini P & Mantero S (2009). A double‐chamber rotating bioreactor for the development of tissue‐engineered hollow organs: From concept to clinical trial. Biomaterials 30, 5260–5269. [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees RM, Botten JW, Majumdar D & Guttman M (2020). SARS‐CoV‐2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183, 1325–1339.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H & Hogan BLM (2017). Lung organoids: current uses and future promise. Development 144, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benam KH, Novak R, Nawroth J, Hirano‐Kobayashi M, Ferrante TC, Choe Y, Prantil‐Baun R, Weaver JC, Bahinski A, Parker KK & Ingber DE (2016a). Matched‐comparative modeling of normal and diseased human airway responses using a microengineered breathing lung chip. Cell Syst 3, 456–466.e4. [DOI] [PubMed] [Google Scholar]
- Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, Alves SE, Salmon M, Ferrante TC, Weaver JC, Bahinski A, Hamilton GA & Ingber DE (2016b). Small airway‐on‐a‐chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods 13, 151–157. [DOI] [PubMed] [Google Scholar]
- Berkers G, Van Mourik P, Vonk AM, De Jonge HR, Beekman JM & Van Der CK (2019). Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep 26, 1701–1708.e3. [DOI] [PubMed] [Google Scholar]
- Bilodeau C, Bardou O, Maillé É, Berthiaume Y & Brochiero E (2016). Deleterious impact of hyperglycemia on cystic fibrosis airway ion transport and epithelial repair. J Cyst Fibros 15, 43–51. [DOI] [PubMed] [Google Scholar]
- Blanco‐Melo D, Nilsson‐Payant BE, Liu W‐C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA & Tenoever BR (2020). Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell 181, 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhmki T, Bitzer S, Gindele JA, Schruf E, Kiechle T, Webster M, Schymeinsky J, Ries R, Gantner F, Bischoff D, Garnett J & Heilker R (2020). Development of a miniaturized 96‐Transwell air–liquid interface human small airway epithelial model. Sci Rep 10, 13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis HM, He R, Vandebriel RJ, Gremmer ER, Zwart E, Vermeulen JP, Fokkens P, Boere J, Gosens I & Cassee FR (2020). An air‐liquid interface bronchial epithelial model for realistic, repeated inhalation exposure to airborne particles for toxicity testing. J Vis Exp 2020, 61210. [DOI] [PubMed] [Google Scholar]
- Brand JD, Lazrak A, Trombley JE, Shei RJ, Adewale AT, Tipper JL, Yu Z, Ashtekar AR, Rowe SM, Matalon S & Harrod KS (2018). Influenza‐mediated reduction of lung epithelial ion channel activity leads to dysregulated pulmonary fluid homeostasis. JCI Insight 3, e123467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler CR, Hynds RE, Gowers KHC, Lee DDH, Brown JM, Crowley C, Teixeira VH, Smith CM, Urbani L, Hamilton NJ, Thakrar RM, Booth HL, Birchall MA, De Coppi P, Giangreco A, O'Callaghan C & Janes SM (2016). Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am J Resp Crit Care Med 194, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E (2020). Dozens to be deliberately infected with coronavirus in UK “human challenge” trials. Nature 586, 651–652. [DOI] [PubMed] [Google Scholar]
- Cantuti‐Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T, Levanov L, Szirovicza L, Tobi A, Kallio‐Kokko H, Österlund P, Joensuu M, Meunier FA, Butcher SJ, Winkler MS, Mollenhauer B, Helenius A, Gokce O, Teesalu T, Hepojoki J, Vapalahti O, Stadelmann C, Balistreri G & Simons M (2020). Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science 370, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier F, Detry B, Sibille Y & Pilette C (2018). COPD‐related epithelial inflammation drives EMT in primary epithelial ALI‐cultures. Eur Resp J 52(62), PA4239. [Google Scholar]
- Castellani S, Di Gioia S, di Toma L & Conese M (2018). Human cellular models for the investigation of lung inflammation and mucus production in cystic fibrosis. Anal Cell Pathol 2018, 3839803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M & Zhang XE (2021). Construction and applications of sars‐cov‐2 pseudoviruses: A mini review. Int J Biol Sci 17, 1574–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Chan JFW, Yuen TTT, Shuai H, Yuan S, Wang Y, Hu B, Yip CCY, Tsang JOL, Huang X, Chai Y, Yang D, Hou Y, Chik KKH, Zhang X, Fung AYF, Tsoi HW, Cai JP, Chan WM, Ip JD, Chu AWH, Zhou J, Lung DC, Kok KH, To KKW, Tsang OTY, Chan KH, & Yuen KY (2020). Comparative tropism, replication kinetics, and cell damage profiling of SARS‐CoV‐2 and SARS‐CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID‐19: an observational study. Lancet Microbe 1, e14–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller‐Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander LE & Eils R (2020). COVID‐19 severity correlates with airway epithelium–immune cell interactions identified by single‐cell analysis. Nat Biotechnol 38, 970–979. [DOI] [PubMed] [Google Scholar]
- Clunes LA, Davies CM, Coakley RD, Aleksandrov AA, Henderson AG, Zeman KL, Worthington EN, Gentzsch M, Kreda SM, Cholon D, Bennett WD, Riordan JR, Boucher RC & Tarran R (2012). Cigarette smoke exposure induces CFTR internalization and insolubility, leading to airway surface liquid dehydration. FASEB J 26, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón‐Plágaro C, Shoemark DK, Simón‐Gracia L, Bauer M, Hollandi R, Greber UF, Horvath P, Sessions RB, Helenius A, Hiscox JA, Teesalu T, Matthews DA, Davidson AD, Collins BM, Cullen PJ & Yamauchi Y (2020). Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. Science 370, 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahay H, Pessotti AD, Coote J, Montgomery BE, Xia D, Wilson A, Yang H, Wang Z, Bevan L, Thomas C, Petit S, London A, LeMotte P, Doelemeyer A, Vélez‐Reyes GL, Bernasconi P, Fryer CJ, Edwards M, Capodieci P, Chen A, Hild M & Jaffe AB (2015). Notch2 is required for inflammatory cytokine‐driven goblet cell metaplasia in the lung. Cell Rep 10, 239–252. [DOI] [PubMed] [Google Scholar]
- Day M (2019). Disgraced tracheal transplant surgeon is handed 16 month prison sentence in Italy. BMJ 367, l6676. [DOI] [PubMed] [Google Scholar]
- Deinhardt‐Emmer S, Böttcher S, Häring C, Giebeler L, Henke A, Zell R, Jungwirth J, Jordan PM, Werz O, Hornung F, Brandt C, Marquet M, Mosig AS, Pletz MW, Schacke M, Rödel J, Heller R, Nietzsche S, Löffler B & Ehrhardt C (2021). SARS‐CoV‐2 causes severe epithelial inflammation and barrier dysfunction. J Virol 95(10), 10.1128/jvi.00110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, De Jonge HR, Janssens HM, Bronsveld I, Van De Graaf EA, Nieuwenhuis EES, Houwen RHJ, Vleggaar FP, Escher JC, De Rijke YB, Majoor CJ, Heijerman HGM, De Winter‐De Groot KM, Clevers H, Van Der Ent CK & Beekman JM (2016). Characterizing responses to CFTR‐modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Trans Med 8, 344ra84–344ra84. [DOI] [PubMed] [Google Scholar]
- Deng Y, Herbert JA, Smith CM & Smyth RL (2018). An in vitro transepithelial migration assay to evaluate the role of neutrophils in Respiratory Syncytial Virus (RSV) induced epithelial damage. Sci Rep 8, 6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar M, Lee JS, Whig K, Segrist E, Li M, Jurado K, Samby K, Ramage H, Schultz D & Cherry S (2021). Drug repurposing screens reveal FDA approved drugs active against SARS‐Cov‐2. Cell Rep, 35 108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S & Hall MD (2020). Remdesivir: A review of its discovery and development leading to emergency use authorization for treatment of COVID‐19. ACS Cent Sci 6, 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good M, Kolls JK & Empey, KM (2017). Neonatal pulmonary host defense. In Polin RA, Abman SH, Rowitch DH, Benitz WE & Fox WW (eds.), Fetal and Neonatal Physiology, pp. 1262–1293. 5, Elsevier. [Google Scholar]
- Fulcher ML, Gabriel SE, Olsen JC, Tatreau JR, Gentzsch M, Livanos E, Saavedra MT, Salmon P & Randell SH (2009). Novel human bronchial epithelial cell lines for cystic fibrosis research. Am J Physiol Lung Cell Mol Physiol 296, L82–L91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiha SM, Cheng J & Halpern‐Felsher B (2020). Association between youth smoking, electronic cigarette use, and COVID‐19. J Adolesc Health 67, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo O, Locatello LG, Mazzoni A, Novelli L & Annunziato F (2020). The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS‐CoV‐2 infection. Mucosal Immunol 14, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M & Rossier BC (2020). A pathophysiological model for COVID‐19: Critical importance of transepithelial sodium transport upon airway infection. Function 1, zqaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Park B, Bhowmik D, Nishida K, Lauver M, Putcha N, Gao P, Ramanathan M, Hansel N, Biswal S & Sidhaye VK (2020). Strong correlation between air‐liquid interface cultures and in vivo transcriptomics of nasal brush biopsy. Am J Physiol Lung Cell Mol Physiol 318, L1056–L1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianotti A, Delpiano L & Caci E (2018). In vitro methods for the development and analysis of human primary airway epithelia. Front Pharmacol 9, 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkatzis K, Taghizadeh S, Huh D, Stainier DYR & Bellusci S (2018). Use of three‐dimensional organoids and lung‐on‐a‐chip methods to study lung development, regeneration and disease. Eur Resp J 52, 1800876. [DOI] [PubMed] [Google Scholar]
- Grainger CI, Greenwell LL, Lockley DJ, Martin GP & Forbes B (2006). Culture of Calu‐3 cells at the air interface provides a representative model of the airway epithelial barrier. Pharm Res 23, 1482–1490. [DOI] [PubMed] [Google Scholar]
- Hameid RA, Cormet‐Boyaka E, Kuebler WM, Uddin M & Berdiev BK (2021). SARS‐CoV‐2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am J Physiol Lung Cell Mol Physiol 320, L430–L435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Ning K, Kuz CA, Vorhies K, Yan Z & Qiu J (2020). Long‐term modeling of SARS‐CoV‐2 infection of in vitro cultured polarized human airway epithelium. MBio 11, e02852‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD & Thornburg NJ (2020). Isolation and characterization of SARS‐CoV‐2 from the first US COVID‐19 patient. bioRxiv, 2020.03.02.972935. [Google Scholar]
- Hawkins FJ, Suzuki S, Beermann ML, Barillà C, Wang R, Villacorta‐Martin C, Berical A, Jean JC, Suer JL, Matte T, Simone‐Roach C, Tang Y, Schlaeger TM, Crane AM, Matthias N, Huang SXL, Randell SH, Wu J, Spence JR, Carraro G, Stripp BR, Rab A, Sorsher EJ, Horani A, Brody SL, Davis BR & Kotton DN (2021). Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell 28, 79–95.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert JA, Deng Y, Hardelid P, Robinson E, Ren L, Moulding D, Smyth RL & Smith CM (2020). β2 integrin LFA1 mediates airway damage following neutrophil trans‐epithelial migration during RSV infection. Eur Resp J 56, 1902216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemstra PS, Tetley TD & Janes SM (2019). Airway and alveolar epithelial cells in culture. Eur Resp J 54, 1900742. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine‐Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C & Pöhlmann S (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH 3rd, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schäfer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi‐Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC & Baric RS (2020). SARS‐CoV‐2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J & Cao B (2020a). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Liu T, Liao J, Maharjan S, Xie X, Pérez M, Anaya I, Wang S, Mayer AT, Kang Z, Kong W, Mainardi VL, Garciamendez‐Mijares CE, Martínez GG, Moretti M, Zhang W, Gu Z, Ghaemmaghami AM & Zhang YS (2021). Reversed‐engineered human alveolar lung‐on‐a‐chip model. Proc Natl Acad Sci U S A 118, e2016146118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hume AJ, Abo KM, Wilson AA, Mühlberger E & Kotton Correspondence DN (2020b). SARS‐CoV‐2 Infection of pluripotent stem cell‐derived human lung alveolar type 2 cells elicits a rapid epithelial‐intrinsic inflammatory response. Stem Cell 27, 962–973.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA & Ingber DE (2012). A human disease model of drug toxicity‐induced pulmonary edema in a lung‐on‐a‐chip microdevice. Sci Trans Med 4, 159ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyang Lee DD, Cardinale D, Nigro E, Butler CR, Rutman A, Fassad MR, Hirst RA, Moulding D, Agrotis A, Forsythe E, Peckham D, Robson E, Smith CM, Somavarapu S, Beales PL, Hart SL, Janes SM, Mitchison HM, Ketteler R, Hynds RE & O'Callaghan C (2020). High‐content screening for rare respiratory diseases: readthrough therapy in primary ciliary dyskinesia. bioRxiv, 2020.02.28.959189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Millet JK & Whittaker GR (2020). Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel s1/s2 site. iScience 23, 101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Okumura Y, Kido H, Ng LFP, Bruzzone R & Altmeyer R (2009). Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One 4, e7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ & Jung SI (2020). Age‐related morbidity and mortality among patients with COVID‐19. Infect Chemother 52, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J & Welsh MJ (2002). An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol 188, 115–137. [DOI] [PubMed] [Google Scholar]
- Klimstra WB, Tilston‐Lunel NL, Nambulli S, Boslett J, McMillen CM, Gilliland T, Dunn MD, Sun C, Wheeler SE, Wells A, Hartman AL, McElroy AK, Reed DS, Rennick LJ & Paul Duprex W (2020). SARS‐CoV‐2 growth, furin‐cleavage‐site adaptation and neutralization using serum from acutely infected hospitalized COVID‐19 patients. J Gen Virol 101, 1156–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Prigge AD, Anekalla KR, Shukla A, Chi Do‐Umehara H, Setar L, Chavez J, Abdala‐Valencia H, Politanska Y, Markov NS, Hahn GR, Heald‐Sargent T, Nelson Sanchez‐Pinto L, Muller WJ, Misharin A V, Ridge KM & Coates BM (2021). Immune response to SARS‐CoV‐2 in the nasal mucosa in children and adults. medRxiv, 2021.01.26.21250269. [Google Scholar]
- Kusek ME, Pazos MA, Pirzai W & Hurley BP (2014). In vitro coculture assay to assess pathogen induced neutrophil trans‐epithelial migration. J Vis Exp e50823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambkin‐Williams R, Noulin N, Mann A, Catchpole A & Gilbert AS (2018). The human viral challenge model: Accelerating the evaluation of respiratory antivirals, vaccines and novel diagnostics. Respir Res 19, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, van Schayck JP, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL & Clevers H (2020). SARS‐CoV‐2 productively infects human gut enterocytes. Science 369, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Mykytyn AZ, Breugem TI, Wang Y, Wu DC, Riesebosch S, van den Doel PB, Schipper D, Bestebroer T & Haagmans BL (2021). Human airway cells prevent SARS‐CoV‐2 multibasic cleavage site cell culture adaptation. bioRxiv, 2021.01.22.427802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA & Knoblich JA (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345, 1247125–1247125. [DOI] [PubMed] [Google Scholar]
- Lee DDH, Petris A, Hynds RE & O'Callaghan C (2020a). Ciliated epithelial cell differentiation at air‐liquid interface using commercially available culture media. In Turksen K (ed.), Methods in Molecular Biology, pp. 275–291. Humana Press Inc., New York. https://link.springer.com/protocol/10.1007/7651_2019_269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IT, Nakayama T, Wu CT, Goltsev Y, Jiang S, Gall PA, Liao CK, Shih LC, Schürch CM, McIlwain DR, Chu P, Borchard NA, Zarabanda D, Dholakia SS, Yang A, Kim D, Chen H, Kanie T, Lin CD, Tsai MH, Phillips KM, Kim R, Overdevest JB, Tyler MA, Yan CH, Lin CF, Lin YT, Bau DT, Tsay GJ, Patel ZM, Tsou YA, Tzankov A, Matter MS, Tai CJ, Yeh TH, Hwang PH, Nolan GP, Nayak JV & Jackson PK (2020b). ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun 11, 5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Channappanavar R & Kanneganti TD (2020c). Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol 41, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, Wadsworth SJ, Yang SJ & Dorscheid DR (2020). Structural and functional variations in human bronchial epithelial cells cultured in air liquid interface using different growth media. Am J Physiol Cell Mol Physiol 318, L1063–L1073. [DOI] [PubMed] [Google Scholar]
- Li MY, Li L, Zhang Y & Wang XS (2020). Expression of the SARS‐CoV‐2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liu Q, Huang W, Li X & Wang Y (2018). Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol 28, e1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Hsu S, Huang CE, Cheng WT & Su JM (2009). A scaffold‐bioreactor system for a tissue‐engineered trachea. Biomaterials 30, 4117–4126. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, Ku Z, An Z, Scharton D, Schindewolf C, Menachery VD, Shi P‐Y & Weaver SC (2021). The N501Y spike substitution enhances SARS‐CoV‐2 transmission. bioRxiv, 2021.03.08.434499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes N, Seidensticker K, Perniss A, Nietzer S, Oberwinkler H, May T, Walles T, Hebestreit H, Hackenberg S & Steinke M (2020). Investigation on ciliary functionality of different airway epithelial cell lines in three‐dimensional cell culture. Tissue Eng A 26, 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C & Eils R (2020). SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39, e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto PP, Dickinson SC, Hollander AP, Mantero S, Conconi MT & Birchall MA (2008). Clinical transplantation of a tissue‐engineered airway. Lancet 372, 2023–2030. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T & Takeda M (2020). Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐ expressing cells. Proc Natl Acad Sci U S A 117, 7001–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FG & Grady C (2001). The ethical challenge of infection‐inducing challenge experiments. Clin Infect Dis 33, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Montoro DT, et al. (2018). A revised airway epithelial hierarchy includes CFTR‐expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munye MM, Shoemark A, Hirst RA, Delhove JM, Sharp T V., McKay TR, O'Callaghan C, Baines DL, Howe SJ & Hart SL (2016). BMI‐1 extends proliferative potential of human bronchial epithelial cells while retaining their mucociliary differentiation capacity. Am J Physiol Lung Cell Mol Physiol 312, L258–L267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerukonda SN, Vassell R, Herrup R, Liu S, Wang T, Takeda K, Yang Y, Lin TL, Wang W & Weiss CD (2021). Establishment of a well‐characterized SARSCoV‐2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One 16, e0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF & Zachos NC (2017). A primary human macrophage‐enteroid co‐culture model to investigate mucosal gut physiology and host‐pathogen interactions. Sci Rep 7, 45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando NS, Dalebout TJ, Zevenhoven‐Dobbe JC, Limpens RW, van der Meer Y, Caly L, Druce J, de Vries JJC, Kikkert M, Bárcena M, Sidorov I & Snijder EJ (2020). SARS‐coronavirus‐2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol 101, 925–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr JC & Hynds RE (2021). Stem cell‐derived respiratory epithelial cell cultures as human disease models. Am J Respir Cell Mol Biol 64, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz ME, Thurman A, Pezzulo AA, Leidinger MR, Klesney‐Tait JA, Karp PH, Tan P, Wohlford‐Lenane C, McCray PB & Meyerholz DK (2020). Heterogeneous expression of the SARS‐Coronavirus‐2 receptor ACE2 in the human respiratory tract. EBioMedicine 60, 102976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack RJ, Al‐Ugaily LH & Morris G (1981). The cells of the tracheobronchial epithelium of the mouse: a quantitative light and electron microscope study. J Anat 132, 71–84. [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Jonathan C, Brown JC, Sanchez‐David RY, Braga L, Williamson MK, Hassard JA, Staller E, Hanley B, Osborn M, Giacca M, Davidson AD, Matthews DA & Barclay WS (2020). The furin cleavage site of SARS‐CoV‐2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv, 2020.09.30.318311. [Google Scholar]
- Pohl MO, Busnadiego I, Kufner V, Glas I, Karakus U, Schmutz S, Zaheri M, Abela I, Trkola A, Huber M, Stertz S & Hale BG (2021). SARS‐CoV‐2 variants reveal features critical for replication in primary human cells. PLoS Biol 19, e3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard BS & Pollard HB (2018). Induced pluripotent stem cells for treating cystic fibrosis: State of the science. Pediatr Pulmonol 53, S12–S29. [DOI] [PubMed] [Google Scholar]
- Ramani S, Crawford SE, Blutt SE & Estes MK (2018). Human organoid cultures: transformative new tools for human virus studies. Curr Opin Virol 29, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N, Levallois S, Larrous F, Fernandes J, Gellenoncourt S, Rigaud S, Gorgette O, Thouvenot C, Trébeau C, Mallet A, Duménil G, Gobaa S, Etournay R, Lledo PM, Lecuit M, Bourhy H, Duffy D, Michel V, Schwartz O & Chakrabarti LA (2020). SARS‐CoV‐2 infection damages airway motile cilia and impairs mucociliary clearance. bioRxiv, 2020.10.06.328369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheb Sharif‐Askari N, Saheb Sharif‐Askari F, Alabed M, Temsah MH, Al Heialy S, Hamid Q & Halwani R (2020). Airways Expression of SARS‐CoV‐2 Receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev 18, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, Flynn RA, Margalef‐Català M, Santos AJM, Ju J, Batish A, Usui T, Zheng GXY, Edwards CE, Wagar LE, Luca V, Anchang B, Nagendran M, Nguyen K, Hart DJ, Terry JM, Belgrader P, Ziraldo SB, Mikkelsen TS, Harbury PB, Glenn JS, Garcia KC, Davis MM, Baric RS, Sabatti C, Amieva MR, Blish CA, Desai TJ & Kuo CJ (2020). Progenitor identification and SARS‐CoV‐2 infection in human distal lung organoids. Nature 588, 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer IM, Padera RF, Solomon IH, Kanjilal S, Hammer MM, Hornick JL & Sholl LM (2020). In situ detection of SARS‐CoV‐2 in lungs and airways of patients with COVID‐19. Mod Pathol 33, 2104–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler BA, Habermann AC, Plosa EJ, Taylor CJ, Jetter C, Negretti NM, Kapp ME, Benjamin JT, Gulleman P, Nichols DS, Braunstein LZ, Hackett A, Koval M, Guttentag SH, Blackwell TS, Webber SA, Banovich NE, Kropski JA & Sucre JMS (2021). Age‐determined expression of priming protease TMPRSS2 and localization of SARS‐CoV‐2 in lung epithelium. J Clin Invest 131, e140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A & Li F (2020). Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A 117, 11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, Moller R, Hoagland D, Oishi K, Horiuchi S, Uhl S, Blanco‐Melo D, Albrecht RA, Liu WC, Jordan T, Nilsson‐Payant WE, Golynker I, Frere J, Logue J, Haupt R, McGrath M, Weston S, Zhang T, Plebani R, Soong M, Nurani A, Kim SM, Zhu DY, Benam KH, Goyal G, Gilpin SE, Prantil‐Baun R, Gygi SP, Powers RK, Carlson KE, Frieman M, tenOever BR & Ingber DE (2021). A human‐airway‐on‐a‐chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng, doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W, Huang N, Bécavin C, Berg M & Network HLB (2020). SARS‐CoV‐2 entry genes are most highly expressed in nasal goblet and ciliated cells within human airways. arXiv 26, 681–687. [Google Scholar]
- Suzuki T, Itoh Y, Sakai Y, Saito A, Okuzaki D, Okuzaki D, Minami S, Kobayashi T, Yamamoto T, Okamoto T & Takayama K (2020). Generation of human bronchial organoids for SARS‐CoV‐2 research. bioRxiv, 2020.05.25.115600. [Google Scholar]
- Tang X, Yang M, Duan Z, Liao Z, Liu L, Cheng R, Fang M, Wang G, Liu H, Xu J, Kamau PM, Zhang Z, Yang L, Zhao X, Peng X & Lai R (2020). Transferrin receptor is another receptor for SARS‐CoV‐2 entry. bioRxiv, 2020.10.23.350348. [Google Scholar]
- Thacker VV, Sharma K, Dhar N, Mancini G‐F, Sordet‐Dessimoz J & McKinney JD (2021). Rapid endotheliitis and vascular damage characterize SARS‐CoV‐2 infection in a human lung‐on‐chip model. EMBO reports, 22(6), 10.15252/embr.202152744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Armendariz AI, Heiner M, Agha EE, Salwig I, Hoek A, Hessler MC, Shalashova I, Shrestha A, Carraro G, Mengel JP, Günther A, Morty RE, Vadász I, Schwemmle M, Kummer W, Hain T, Goesmann A, Bellusci S, Seeger W, Braun T & Herold S (2020). Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J 39, e103476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MS, Gomi K, Ashbridge B, Moore MAS, Arbelaez V, Heldrich J, Sen DB, Rafii S, Staudt MR & Crystal RG (2013). Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res 14, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Lou HH, Salit J, Leopold PL, Driscoll S, Schymeinsky J, Quast K, Visvanathan S, Fine JS, Thomas MJ & Crystal RG (2019). Characterization of an immortalized human small airway basal stem/progenitor cell line with airway region‐specific differentiation capacity. Respir Res 20, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Chen W, Zhou YS, Lian JQ, Zhang Z, Du P, Gong L, Zhang Y, Cui HY, Geng JJ, Wang B, Sun XX, Wang CF, Yang X, Lin P, Deng YQ, Wei D, Yang XM, Zhu YM, Zhang K, Zheng ZH, Miao JL, Guo T, Shi Y, Zhang J, Fu L, Wang QY, Bian H, Zhu P & Chen ZN (2020). SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv, 2020.03.14.988345. [Google Scholar]
- WHO (2020). Key criteria for the ethical acceptability of COVID‐19 human challenge studies. https://www.who.int/blueprint/priority‐diseases/key‐action/novel‐coronavirus‐landscape‐ncov.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodall M, Jacob J, Kalsi KK, Schroeder V, Davis E, Kenyon B, Khan I, Garnett JP, Tarran R & Baines DL (2020). E‐cigarette constituents propylene glycol and vegetable glycerin decrease glucose uptake and its metabolism in airway epithelial cells in vitro. Am J Physiol Lung Cell Mol Physiol 319, L957–L967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez ND, Weiss NS, Romand JA & Treggiari MM (2020). COVID‐19 mortality risk for older men and women. BMC Public Health 20, 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonker LM, Mou H, Chu KK, Pazos MA, Leung H, Cui D, Ryu J, Hibbler RM, Eaton AD, Ford TN, Falck JR, Kinane TB, Tearney GJ, Rajagopal J & Hurley BP (2017). Development of a primary human co‐culture model of inflamed airway mucosa. Sci Rep 7, 8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Worlock KB, Huang N, Lindeboom RGH, Butler CR, Kumasaka N, Conde CD, Mamanova L, Bolt L, Richardson L, Polanski K, Madissoon E, Barnes JL, Allen‐Hyttinen J, Kilich E, Jones BC, de Wilton A, Wilbrey‐Clark A, Sungnak W, Pett JP, Prigmore E, Yung H, Mehta P, Saleh A, Saigal A, Chu V, Cohen JM, Cane C, Iordanidou A, Shibuya S, Reuschl AK, Argento AC, Wunderink RG, Smith SB, Poor TA, Gao CA, Dematte JE, NU SCRIPT Study Investigators , Reynolds G, Haniffa M, Bowyer GS, Coates M, Clatworthy MR, Calero‐Nieto FJ, Göttgens B, O'Callaghan C, Sebire NJ, Jolly C, de Coppi P, Smith CM, Misharin AV, Janes SM, Teichmann SA, Nikolic MZ & Meyer KB (2021). The local and systemic response to SARS‐CoV‐2 infection in children and adults. Fernando J Calero‐Nieto 11, 3. [Google Scholar]
- Zang R, Castro MFG, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ & Ding S (2020). TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol 5, eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rostami MR, Leopold PL, Mezey JG, O SL, Strulovici‐Barel Y & Crystal RG (2020). Expression of the SARS‐CoV‐2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med 202, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai JP, Yip CCY, Chan IHY, Wong KKY, Tsang OTY, Chan KH, Chan JFW, To KKW, Chen H & Yuen KY (2020a). Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med 26, 1077–1083. [DOI] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF & Shi ZL (2020b). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, Deng Y, Mao L, Su C, Qiang G, Jiang T, Zhao J, Wu G, Song J & Tan W (2020). Morphogenesis and cytopathic effect of SARS‐CoV‐2 infection in human airway epithelial cells. Nat Commun 11, 3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Chew KY, Karawita AC, Yamamoto A, Labzin LL, Yarlagadda T, Khromykh AA, Stocks CJ, Xia Y, Kollmann TR, Martino D, Kicic A, Bielefeldt‐Ohmann H, Bowen AC, Sly PD, Spann KM & Short KR (2021). Pediatric nasal epithelial cells are less permissive to SARS‐CoV‐2 replication compared to adult cells. bioRxiv, 2021.03.08.434300. [Google Scholar]
- Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK & Ordovas‐Montanes J; HCA Lung Biological Network (2020). SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peer Review History
Video abstract and summary


