Abstract
The role of airborne particles in the spread of severe acute respiratory syndrome coronavirus type 2 (SARS‐CoV‐2) is well explored. The novel coronavirus can survive in aerosol for extended periods, and its interaction with other viral communities can cause additional virulence and infectivity. This baseline study reports concentrations of SARS‐CoV‐2, other respiratory viruses, and pathogenic bacteria in the indoor air from three major hospitals (Sheikh Jaber, Mubarak Al‐Kabeer, and Al‐Amiri) in Kuwait dealing with coronavirus disease 2019 (COVID‐19) patients. The indoor aerosol samples showed 12–99 copies of SARS‐CoV‐2 per m3 of air. Two non‐SARS‐coronavirus (strain HKU1 and NL63), respiratory syncytial virus (RSV), and human bocavirus, human rhinoviruses, Influenza B (FluB), and human enteroviruses were also detected in COVID‐positive areas of Mubarak Al Kabeer hospital (MKH). Pathogenic bacteria such as Mycoplasma pneumonia, Streptococcus pneumonia and, Haemophilus influenza were also found in the hospital aerosols. Our results suggest that the existing interventions such as social distancing, use of masks, hand hygiene, surface sanitization, and avoidance of crowded indoor spaces are adequate to prevent the spread of SARS‐CoV‐2 in enclosed areas. However, increased ventilation can significantly reduce the concentration of SARS‐CoV‐2 in indoor aerosols. The synergistic or inhibitory effects of other respiratory pathogens in the spread, severity, and complexity of SARS‐CoV‐2 need further investigation.
Keywords: indoor air, pathogenic bacteria, qPCR, respiratory viruses, SARS‐CoV‐2
Practical Implications.
The presence of 12–99 copies of SARS‐CoV‐2 per m3 of indoor hospital air indicates likelihood of infection transmission via aerosol.
The presence of pathogenic bacteria such as Mycoplasma pneumonia, Streptococcus pneumonia, and Haemophilus influenza, along with non‐SARS‐coronavirus (strain HKU1 and NL63), respiratory syncytial virus (RSV), and human bocavirus, human rhinoviruses, Influenza B (FluB), and human enteroviruses within the COVID wards indicates need to increase air exchange.
This study utilized a simplified and safe sampling technique that can be employed even in labs that fall short of biosafety level III compliance.
The RNA‐based culture‐independent approach used in this study quantifies only the viable bacterial cells that have direct health implications in indoor hospital air.
1. INTRODUCTION
Airborne particles of biological origin have a repertoire of atmospheric bacteria, fungal spores, pollen, allergens, and viruses. 1 , 2 These particles are known to be the direct cause of epidemics of various infectious and non‐infectious diseases. The microbes bound to fine and ultrafine particulates settle deep into the respiratory system, reaching in alveolar and tracheobronchial regions and increasing the chance of infective transmission. The World Health Organization (WHO) in January 2020 declared the spread of a new coronavirus, severe acute respiratory syndrome coronavirus type 2 (SARS‐CoV‐2) as a public health emergency of international concern. Since then, the outbreak has claimed about 2.92 million lives worldwide as of 9th April 2021. 3
The transmission of SARS‐CoV‐2 is believed to be via human‐to‐human contact, touching infected surfaces, and inhalation of the exhaled virus in respiratory droplets. 4 , 5 , 6 Recent studies have also provided evidences of aerosol‐mediated spread of SARS‐CoV‐2, 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 mostly from confined spaces in hospitals and quarantine camps. 15 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 Thus, it becomes an issue of preeminent importance to understand the microbial assemblage in indoor aerosols to ensure the safety of healthcare personnel and uninfected people involved in the care of COVID patients. Drawing similarities with the previous SARS epidemic in Hong Kong, the airborne spread was suggested as the primary transmission pathway of the type I coronavirus. 29 The understanding of the presence and concentration of SARS‐CoV‐2 in aerosols becomes challenging, because of the difficulties in sampling virus‐containing aerosols in real‐world settings and problems in their quantification at low concentrations. 4 , 7
Van Doremalen, et al. 2020 30 showed the viability of SARS‐CoV‐1 and SARS‐CoV‐2 in aerosols and on surfaces like plastic, stainless steel, copper, and cardboard. Both SARS‐CoV‐1 and SARS‐CoV‐2 remained viable in the aerosol after 3 h. However, there was a reduction in infectious titer from 104.3 to 103.5 TCID50/L of air for SARS‐CoV‐1, and 103.5 to 102.7 TCID50/L of air for SARS‐CoV‐2. It has been reported that the viral load is high in the respiratory tract of infected persons and can be transmitted through droplets via coughing or sneezing, 31 and it is a widely accepted fact. Still, there is no information reported on whether there are viral loads in the exhaled air that probably adds to indoor air. Besides, in closed environments such as hospital wards, droplets can remain suspended for more than 10 min and cover long distances, potentially maintaining their ability to transmit disease. 32 , 33 , 34 The information on the levels and concentrations of SARS‐CoV‐2 and its spread in indoor scenarios is still in its infancy. To the best of our knowledge, there has been no assessment made so far on SARS‐CoV‐2 concentration in the indoor air of the hospitals in Kuwait.
A recent study has suggested the need for investigations on adsorption, survival, and behavior of the SARS‐CoV‐2 virus within the aerosol community to understand its spread and its interaction with other viral communities 6 that are possibly having a cumulative effect on virulence and infectivity. The clinical presentation of the illness caused by SARS‐CoV‐2 appears to range from mild or asymptomatic to severe and fatal respiratory illness. 35 The clinical manifestation of COVID‐19 fatalities is reported to be heterogeneous. 36 , 37 One of the risk factors is the presence of other co‐infections that require management and treatment. 38 The frequency of respiratory failure and mortality among patients with COVID‐19 and co‐infections is high, 36 similar to influenza, 39 SARS‐CoV‐1, 40 and the Middle East Respiratory Syndrome coronavirus (MERS‐CoV). 41
This baseline study demonstrates the presence of viral and pathogenic bacterial species present in the aerosols, including the SARS‐CoV‐2 in the indoor atmosphere of hospitals dealing with COVID‐19 patients in Kuwait. A custom‐designed sampler was used, the details of which have been described elsewhere, 7 and the culture‐independent quantitative polymerase chain reaction (qPCR) method was employed for identification and quantification.
2. MATERIALS AND METHODS
2.1. Sampling and RNA isolation
This study was conducted to identify and characterize the novel coronavirus, associated respiratory viruses, and pathogenic bacteria in indoor aerosols of three major hospitals in Kuwait. Samples were collected using a specialized sampler developed for this purpose 7 ; it is efficient in capturing the entire aerosol load and safe for use, as it lyses the captured microbes (bacteria, fungi, and viruses) during sampling itself. This sampling device utilizes a variable speed suction pump (Tisch, Environmental International) with a flow controller that allows air to pass through gas wash bottles. Three glass gas wash bottles attached in series were filled with 100 ml of TRIzol® (APB Biosciences) (henceforth mentioned as Trizol). The sampler was deployed and the air was pumped through this sampling setup for 2 hours @ 30 L per min. In total, 3.6 m 3 (3600 L) of air was collected from four, seven, and two locations in Sheikh Jaber Hospital (SJH), Mubarak Al Kabeer hospital (MKH), and Al Amiri hospital (AMH), respectively, in Kuwait (Table 1). With ongoing restrictions imposed due to the COVID‐19 pandemic, prior approval of the Ministry of Health (MOH), Kuwait, was obtained for sample collection. In this study, only the locations approved by the MOH within the hospital premises were covered. Samples were also collected from five areas within the Kuwait Institute for Scientific Research (KISR) to see the aerosol microbial load in a non‐hospital setting. Two ambient air samples from an outdoor site in a residential area were collected as control samples for this study. The standard procedure of RNA isolation from Trizol was followed to purify total RNA. 42 The high‐sensitivity Qubit HS ssRNA kit was used for fluorometric estimation 43 of isolated RNA on a Qubit 4 fluorometer (Thermo Scientific). All these steps were performed under BSL2 cabinets.
TABLE 1.
Sampling details for aerosol collection from three major hospitals, a non‐hospital site, and a control site in Kuwait
| Hospital name/GPS | Sampling point | Code | Remarks |
|---|---|---|---|
| Mubarak Al Kabeer Hospital (MKH) 29.3260° N, 48.0350° E | Waiting Area near Pharmacy | MKHP | Main pharmacy delivering medications with non‐stop human intervention. |
| Main Gate Entrance | MKHE | The common area near outdoor air | |
| Pediatric Casualty | MKHPC | Reception and emergency wards receiving and treating pediatric patients | |
| Laboratory 1 | MKHL1 | Central laboratory receiving patient samples including COVID for testing | |
| Laboratory 2 | MKHL2 | Interior area of main laboratory exclusively dealing with COVID samples | |
| COVID Isolation Area | MKHCO | Common ward dealing with COVID suspects | |
| COVID Ward | MKHCW | Common ward taking care of symptomatic COVID patients | |
| Sheikh Jaber Hospital (SJH) 29.2768oN, 48.0063oE | Cytology Laboratory | SJHCL | Laboratory processing samples for cytological testing |
| COVID Observation Area | SJHCO | Common ward dealing with COVID suspects | |
| COVID Ward | SJHCW | Common ward for symptomatic COVID patients | |
| Virology Laboratory | SJHVL | Laboratory processing samples for viral testing | |
| Amiri Hospital (AMH) 29.3878° N, 47.9875° E | Laboratory Reception Area | AMHLR | Open space outside the virology laboratory receiving samples and dispatching results regularly |
| Virology Laboratory | AMHVL | Laboratory processing samples for COVID testing | |
| Kuwait Institute for Scientific Research (KISR) 29.3369° N, 47.9064° E | The corridor on 1st Floor | KFF1 | East End of Link bridge opposite to stairs frequented by staff |
| The pavement on the First Floor | KFF2 | West End of Link bridge near the stairs and lift frequented by staff | |
| Ground Floor Reception area | KR1 | Attendance reader in the reception area. A large number of employee pass through it | |
| KISR Gate | KR2 | The area near the backside entrance and the card punching machine | |
| KISR Laboratories | KL | Laboratory area with regular laboratory personnel presence | |
| *Outdoor Air (OUT) 29.3135° N, 48.0071° E | Outdoor 1 | OUT1 | Parking lot in Residential Area beside the main street |
| Outdoor 2 | OUT2 | Parking lot in Residential Area beside the main street |
Ambient air samples from this area were collected as controls.
2.2. Heating ventilation and air conditioning in Kuwait's Hospitals
The hospitals in Kuwait are constructed as per the Ministry of Health Kuwait hospital design guidelines. The design parameters ensure protection of outside air intakes; use of a variable‐air‐volume system to minimize the need of air exchange while ensuring that there is no stagnation of air inside the wards. The ventilation is met by central air conditioning system, with filtration and humidification provisions. The fresh air intakes are located at least 7.62 m from the exhaust outlets of ventilating systems, combustion vents (including those serving rooftop air handling equipment), medical‐surgical vacuum systems, plumbing vents, or areas that may collect vehicular exhaust or other noxious fumes; however, this is not applicable to relief air. The air conditioning design parameter also accounts for the prevailing wind directions and proximity to other structures ensuring appropriate clearances. The plumbing vents are at least 3.05 meters above the intake, while the bottom of outdoor air intakes serving central systems shall be 0.91 m above roof level, as most of the central air conditioning systems are located above the roof. The air exchange in the wards used for patient care is 6–10 times per hour. To maintain asepsis control, airflow supply, and exhaust, the air movement is generally controlled to ensure movement of air from “clean” to “less clean” areas. 44
2.3. Live bacterial load in aerosols of indoor hospital environments in Kuwait
The culture‐independent RNA‐based 16sRNA gene amplification was used to estimate the live bacterial load of the collected samples. 45 Total bacterial load was estimated through the quantitative polymerase chain reaction (qPCR). The isolated RNA was converted to complementary DNA (cDNA) using iScript™ Reverse Transcription Supermix (Bio Rad). The reverse transcription was carried out for 20 min at 46°C after priming for 5 min at 20°C. The reverse transcriptase enzyme was inactivated by incubating the reaction mix at 95°C for 1 min. The cDNA (2 µl) was added to 2 × iQTM SYBR® Green supermix (BioRad) along with universal bacterial primers (300 nM Forward 5′‐ CCTACGGGNBGCASCAG‐3′ and Reverse 5′‐GACTACNVGGGTATCTAATCC‐3′ targeting the V3‐V4 region). The total reaction volume was made up to 20 µl with nuclease‐free water and the qPCR was performed on the CFX96TM Deep Well thermal cycler (BioRad). Standard curves for bacteria were based on the extraction of a known quantity of Escherichia coli. The cycling conditions were set for initial denaturation at 95°C (3 mins), followed by 40 rounds of denaturation at 95°C (20 s); annealing and extension 60°C (45 s). The melt curve analysis was carried out at 60°C. The mean Ct values were used for calculating the number of cells per m3 of air (Table S1).
2.4. Multiplex panel qPCR for detection of respiratory viruses and pathogenic bacteria
We performed a multiplex qPCR to detect the presence of sixteen airborne respiratory viruses namely, influenza A (Flu A), influenza B (Flu B), human respiratory syncytial virus (RSV); parainfluenza 1, parainfluenza 2, parainfluenza 3, parainfluenza 4; human adenovirus, metapneumovirus, bocavirus, human rhinovirus, human enterovirus and non‐SARS strains of coronaviruses namely 229E, NL63, OC43, and HKU1. In the same panel, we also tested the hospital aerosols for six pathogenic bacteria such as Legionella pneumophila, Chlamydophila pneumonia, Mycoplasma pneumonia, Haemophilus influenza, Streptococcus pneumonia, and Moraxella catarrhalis associated with respiratory diseases. Respective targets of each organism are given in Table S2. The respiratory panel III kit 46 , 47 from VIASURE (CerTest, Biotec) was used to set up the multiplex qPCR reaction. As per the manufacturer's instruction, the PCR reaction was assembled in a volume of 20 µl by adding 5µl of isolated RNA to each well to the reconstituted master mix. 46 The PCR was performed on the CFX96TM Deep Well (BioRad) thermal cycler. The cycling conditions of the reaction included the steps of reverse transcription (15 min at 45°C) followed by initial denaturation (2 min at 95°C) and then 45 cycles of denaturation (10 s at 95°C), annealing/extension (50 s at 60°C). Fluorogenic data were collected during the extension step through the FAM, ROX, Cy5, and HEX channels as per the respective targets (Table S2). The sample was considered positive if the Ct value was ≤40 after baseline subtraction and fluorescence drift correction. All the wells were checked for amplification of internal controls, positive controls, and non‐amplification in negative controls. All the reactions were done in duplicate and the Ct values were estimated by the CFX MaestroTM software (BioRad).
2.5. Detection and quantification of SARS‐CoV‐2 in indoor aerosols
The qPCR was conducted on aerosol samples to detect and quantify the SARS‐CoV‐2 on the isolated RNA samples. The VIASURE SARS‐CoV‐ Real‐Time PCR Detection Kit‐CE‐IVD (Certest‐Biotec) based on two targets that are ORF1ab and N genes were used for this purpose. 4 The PCR reaction was assembled as per the kit instructions. A reaction volume of 20 µl was assembled by adding 5 µl of isolated RNA to the rehydrated master mix. 48 Internal control was added to each well of the reaction mix to rule out PCR inhibition. A known concentration of non‐infectious synthetic construct of cDNA comprising the N gene and ORF1ab genes of SARS‐CoV‐2 was used as a positive control. Serial dilutions of the positive control were amplified along with the unknown samples. The PCR reaction mix was loaded on the QuantStudio 5 Real‐Time PCR System (Applied Biosystems). The amplification profile was set for reverse transcription (15 min at 45°C), initial denaturation (2 min at 95°C) and then 45 cycles of denaturation (10 s at 95°C), and annealing/extension (50 s at 60°C). Fluorogenic data were collected during the extension steps as per the manufactures’ instructions. The Ct values of the positive samples were converted to copies per cubic meter of air employing the QuantStudio Design and Analysis software (Table S3). Samples exhibiting Ct above 40 were excluded from the analysis.
3. RESULTS
3.1. Total live bacterial load in indoor aerosols
The culture‐independent RNA‐based bacterial quantification is recently being used for the detection of metabolically active bacterial cell counts. The qPCR results demonstrated the presence of live bacterial cells in the indoor aerosols collected from all the locations of the hospital environment with wide variations noticed at each sampling point within each site (Figure 1). At MKH, the cell counts ranged from a minimum of 5.50E + 02 (MKHP) per m3 of air to a maximum of ca.1.00E + 05 cells per m3 of air (MKHL1). In the same hospital, the live cell counts at the COVID handling areas of MKHCW, MKHCWI, and MKHE were at almost comparable levels (~1.00E + 03 cells per m3 of air). At MKHL2 (central laboratory) and MKHPC (pediatric causality), 9.50E+04 and 2.00E+03 cells per m3 of air were recorded, respectively. At the main hospital of SJH dealing with the maximum number of COVID patients daily, the cell counts varied from the lowest 4.60E+02 cells per m3 of air in the cytology lab (SJHCL) to the highest 1.90E+03 cells per m3 of air in the COVID isolation area (SJHCO). Almost similar levels of 7.00E+02 (SJHVL) and 9.50E+02 (SJHCW) of viable bacterial cells per m3 of air were recorded at the virology lab and COVID ward of SJH. AMH had higher counts at the Lab reception (AMHLR −1.80E+04 cells per m3) as compared to inside the virology laboratory (AMHVL −7.50E+02 cells per m3). The non‐hospital site, that is, KISR, also has live bacterial cells in the range of 6.11E+02 (KFF2) to 1.98E+03 (KFF1) cells per m3 of air. Whereas, the areas near the attendance machine (KR1, KR2) and the laboratories (KL) at KISR have almost comparable levels of 1.11E+03, 1.06E+03, and 1.73E+03 cells per m3 of air respectively. The average bacterial counts in the indoor hospital air at MKH were 1.68E+04 cells per m3 of air, followed by 9.37 E+03 cells per m3 of air at AMH and 1.00E+03 cells per m3 of air at SJH. The average numbers of live cells in the indoor aerosols at KISR (non‐hospitalized setting) were comparable to SJH and considerably less than the MKH and AMH. Live bacterial cell concentration in the outdoor air was much higher than indoor, that is, 8.19 E+04 and 4.31E+04 cells per m3 of the air at site OUT1 and OUT2, respectively. The average live bacterial counts (6.24E+04 cells per m3 of air) at OUT were 3.7, 6.7, 62.3, and 47.9 folds higher than MKH, AMH, SJH, and KISR, respectively. The reason for this lower bacterial concentration in indoor is attributed to the fact that all buildings in Kuwait are air‐conditioned and the sites investigated use central air conditioning plants and air handling units are equipped with HEPA filters.
FIGURE 1.
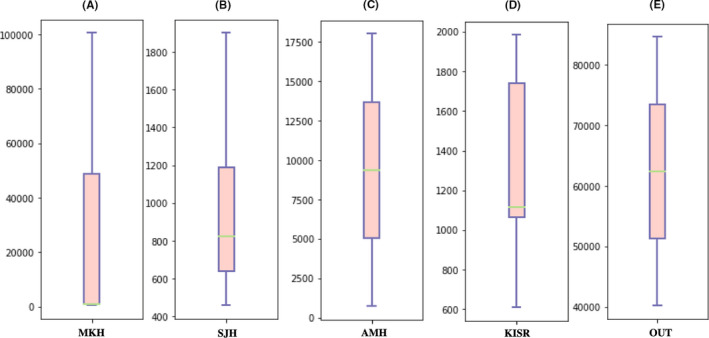
Box whisker plots representing the live bacterial cell numbers per m3 of air from (A) Mubarak Al‐Kabeer Hospital (MKH); (B) Sheikh Jaber Hospital (SJH); (C) Al‐Amiri Hospital (AMH); (D) Kuwait Institute for Scientific Research (KISR) and (E) Outdoor aerosol (control sample). RNA‐based cell counts (copies per m3 of air) obtained through quantitative polymerase chain reaction (qPCR) are plotted on the Y‐axis
3.2. Respiratory viruses and pathogenic bacteria as detected by multiplex panel qPCR
The respiratory panel PCR revealed some common (frequently detected at most sampling locations) and unique (not so frequently detected at most sampling locations) viruses in the ambient indoor air of the three hospitals dealing with COVID‐19 patients in Kuwait. The corresponding Ct values of each virus are given in Table S2. Of the sixteen types of viruses tested, rhinovirus was omnipresent in hospitalized (MKH, SJH, and AMH), non‐hospitalized (KISR), and outdoor samples (OUT). In MKH, the rhinovirus was found at MKHP, MKHPC, MKHE, that is, 3 of 7 sampling locations, in SJH at SJHVL, SJHCL, and SJHCW, that is, 3 of 4 sampled sites, while in AMH, it was detected at AMHLR (Table 2). Out of the five sampled locations, rhinovirus was detected at a single location in KISR, whereas it was present in both the samples collected outdoor. Another common respiratory virus was Flu B with the occurrence at all the four sampled locations at SJH (SJHCW, SJHCO, SJHVL, and SJHCL). However, four of the site seven sites at MKH (MKHP, MKHCO, MKHE, and MKHL1), a single location at both AMH (AMHLR), and KISR (KFF2) were positive for Flu B. In addition to the above, human enterovirus was another common virus detected at three sites in MKH (MKHCW, MKHPC, and MKHCO), two in SJH (SJHCW, SJHVL), and one in KISR (KL).
TABLE 2.
Positive sites for SARS‐CoV‐2, common respiratory viruses, and bacterial pathogen in the indoor aerosols of hospitals dealing with COVID‐19 patients in Kuwait
| Sampling Location | RV | Flu B | EV | SARS‐CoV−2 | HI |
|---|---|---|---|---|---|
| MKH | 3 (7) | 4 (7) | 3 (7) | 3 (7) | 2 (7) |
| SJH | 3 (4) | 4 (4) | 2 (4) | 2 (4) | 4 (4) |
| AMH | 1 (2) | 1 (2) | 0 (2) | 0 (2) | 1 (2) |
| KISR | 1 (5) | 1 (5) | 1 (5) | 0 (5) | 0 (5) |
| OUT(C*) | 2 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) |
Number of positive sites for occurrence of viruses and pathogens and total number of sites sampled present in bracket at each location.
Abbreviations: EV, Enteroviruses; Flu B, Influenza B virus; HI, Haemophilus influenza; RV, Rhinoviruses; SARS‐CoV‐2, Severe acute respiratory syndrome coronavirus type 2.
In addition to these common viruses, few unique viruses were detected in the indoor aerosols of MKH. The bocavirus was only found in the MKHCW along with the non‐SARS coronavirus (CoV‐NL63). At MKHP, non‐SARS coronavirus (CoV‐HKU1), respiratory syncytial virus (RSV), and pathogenic bacteria were also found along with SARS‐CoV‐2. None of these unique viruses were detected in SJH, AMH, KISR, and the outdoor air. In all the samples the FluA, parainfluenza 1, parainfluenza 2, parainfluenza 3, parainfluenza 4, adenovirus, metapneumovirus, coronavirus 229E, and coronavirus OC43 were not detected. The limit of detection (LOD) of the kit used was ten copies. To have a more comprehensive representation of airborne viruses, high‐throughput methods based on next‐generation sequencing should be applied.
Our qualitative estimations also confirmed the presence of the bacterial pathogen H. influenzae at all the sites in SJH (SJHVL, SJHCL, SJHCO, and SJHCW), two sites in MKH (MKHCO and MKHL1), and a single site in AMH (AMHLR). Two other bacterial pathogens were unique to MKH. M. pneumonia was detected at MKHP and S. pneumonia at MKHP, MKHPC, MKHCW, and MKHE. Only two locations, MKHP and MKHCW, were positive for the SARS‐CoV‐2. None of the collected samples showed the presence of L. pneumophila, C. pneumonia, and M. catarrhallis.
The highest microbial diversity was observed in MKH, with the presence of multiple viruses such as FluB, rhinovirus, enterovirus, SARS, and non‐SARS coronaviruses (HKU1 and NL63), as well as bacterial pathogens such as H. influenza, S. pneumonia, and M. pneumonia (Figure 2). This was followed by SJH and AMH in terms of microbial variability. The indoor air within a non‐hospitalized setting (KISR) is cleaner with few common respiratory viruses in low frequencies. The outdoor air (OUT) was free from pathogenic forms with the exception of rhinovirus.
FIGURE 2.
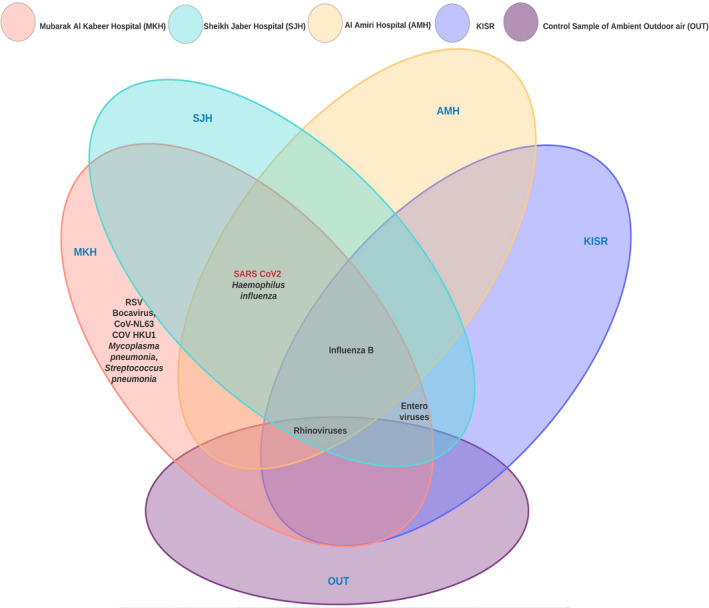
Common and unique respiratory viruses including SARS‐CoV‐2 and pathogenic bacteria detected by the qPCR in three major hospitals of Kuwait (MKH, SJH &AMH). KISR represents samples collected from a non‐hospitalized setting and OUT signifies aerosols in ambient air from a residential area used as a control in the present study
3.3. Detection and quantification of SARS‐CoV‐2 in indoor aerosols
The indoor aerosols showed positive amplification of N‐gene of the SARS‐CoV‐2 at three, two, and one location at MKH, SJH, and AMH, respectively (Table 2). In contrast, negligible amplification of the ORF1ab gene was obtained in all the aerosol samples tested for SARS‐CoV‐2. Hence, these samples were quantified based on the Ct values obtained for the N‐gene only. The SARS‐CoV‐2 concentrations in indoor aerosol varied between <10 and 100 copies per m3 of air. Maximum copies of ca. 100 copies per m3 of air were discovered at MKHCW (main ward taking care of COVID patients), followed by 43 copies per m3 of air at AMHLR (virology lab reception area receiving samples for testing and dispatching results), 35 copies per m3 of air at SJHCO (isolation area before COVID testing), 18 copies per m3 of air at MKHP (waiting area near the pharmacy close to the entrance of the main laboratory), 13 copies per m3 of air at MKHL1 (main lab receiving COVID samples for testing), and 12 copies per m3 of air SJHCW (main ward treating COVID patients) as shown in Figure 3. The variation in the copy numbers of SARS‐CoV‐2 is attributed to the policies and procedures followed in each hospital and possibly the number of patients handled during the time sampling was done. The average concentration of SARS‐CoV‐2 in MKH and SJH was ~45.00 and 23.56 copies per m3 of air, respectively. The copy numbers were relatively higher in COVID wards of MKH and COVID isolation areas in SJH, posing a question about their source in these locations. SARS‐CoV‐2 was not detected in the aerosols from non‐hospital samples of KISR and outdoor air (control).
FIGURE 3.
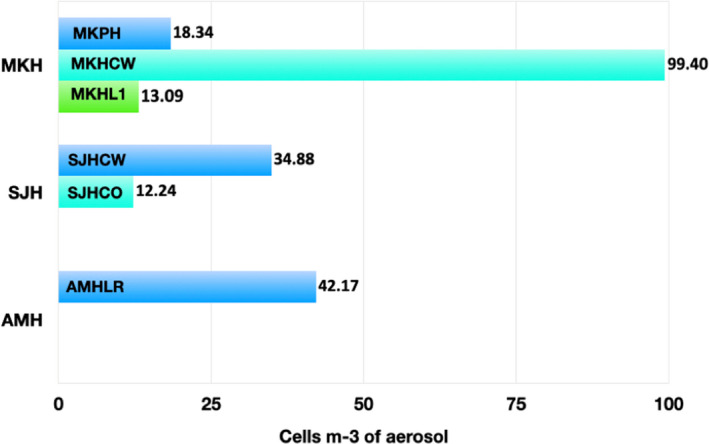
Concentration of SARS‐CoV‐2 in the indoor air of Mubarak Al‐Kabeer hospital (MKH‐sub‐locations MKHP, MKHCW and MKHL1), Sheikh Jaber Hospital (SJH‐ sublocations SJHCW and SJHCO), and Al‐Amiri hospitals (AMH‐sublocation AMHLR) of Kuwait. Quantitative estimations are based on Ct values obtained through the qPCR
4. DISCUSSION
Nosocomial aerosols have made news highlights, especially the notion that SARS‐CoV‐2 also follows an airborne transmission. The reality is that COVID‐19 is just the latest episode of a century‐long attack on the human race by zoonotic respiratory viruses and a much longer assault by several respiratory bacterial pathogens. 49 , 50 The co‐inhalation of respiratory viruses and bacterial pathogens has been hypothesized to influence the respiratory stress induced by the SARS‐CoV‐2 virus. 6 Hospital‐acquired infections cause one of the most severe complications in COVID patients admitted in ICU and among the immunosuppressed people. 6 The results from this study provide an insight into the viral and bacterial load in the indoor aerosols in hospitals in Kuwait serving to combat COVID‐19.
Infectious bacteria‐laden aerosols are generated in hospitals by COVID‐19 and other patients through breathing, talking, coughing, and sneezing. 49 According to a risk assessment study, hospital staff, and people having frequent encounters with healthcare facilities, a risk ratio (RR) of 2.5 was established for acquiring viral or bacterial infections. 51 , 52 In the current investigation, we have used the RNA‐based qPCR approach to quantify viable bacterial cells in aerosol samples. The technique has been applied in determining live bacterial cells in water 45 and biofilms. 53 We report an average of 1.68E+04 (MKH), 9.37E+03 (AMH), and 1.00E+03 (SJH) bacterial cells per m3 of air collected from three hospitals in Kuwait. Microbial cell counts (collective copy numbers based on 16s RNA gene for bacteria) ranging from 4.91E+06 to 9.01E+06 cells per m2 were reported in the air samples (collected through passive sedimentation) from selected areas of a hospital in Kandy, Sri Lanka. 54 Similarly, Perkins and his team estimated 3.4E+04 bacterial cells per m3 in shower aerosols of a Stem cell transplant unit. 55 However, these assumptions were DNA‐based that took into account both the live and dead microbes. The hospital aerosol captured through Anderson impactors has between 0.8 and 3.8 colony‐forming units (CFU) m3 56 , 57 of cultivable forms only. Other than this, fluorescence dye 58 and adenosine triphosphate (ATP)‐ 59 based approaches have also been used to estimate live microbial cells. The latter was employed in one of our previous studies in which we reported the microbial equivalents in aerosols collected from the indoor air of non‐hospitalized settings (Kuwait) as 3.0E+04 cells per m3 of air. 7
The highest number of live bacterial cells were recorded in outdoor air, but none was pathogenic. Among the viral communities only, rhinovirus (Average Ct‐23.16) was found in outdoor air. This was very perplexing to find rhinovirus in outdoor air, even in higher concentration than indoor air. However, in congruence with our findings, a study from Virginia, United States, also found bacteria‐like particles (BLP) and virus‐like particles (VLP) 1.6 and 2.6 times higher in outdoors air as compared to indoors. 60 Similarly, rhinoviruses were detected in outdoor air of a healthcare facility in Portugal in late autumn, whereas the indoor samples were negative. 61 The presence of rhinoviruses in aerosols influenced by meteorological factors and air quality has been reported previously. 61 , 62 , 63 Rhinoviruses were detected in exhaled air, 64 , 65 , 66 which probably explains its presence in indoor hospital settings. The absence of other seasonal viruses such as FluB in the outdoor air is attributed to the global phenomenon of social distancing and reduced human contact as a consequence of the restrictions imposed during the pandemic. 15 , 67 , 68
The susceptibility of acquiring an infectious agent largely depends on the type of pathogenic organisms and their dose from inhalation. Based on the infectious status of a person, the bio‐aerosols are proven to contain bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, Methicillin‐resistant Staphylococcus aureus (MRSA), Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia, Haemophilus influenzae, Legionella pneumophila, Mycoplasma pneumonia, Chlamydia pneumonia, and Mycobacterium tuberculosis. 64 Current investigation revealed the presence of three pathogenic bacterial species, viz. H. influenzae, S. pneumonia, and M. pneumonia, in indoor air of MKH. H. influenza was also found in SJH and AMH indoor air. This study provides the qualitative estimation of these pathogenic bacteria. However, direct comparison with other indoor air microbial studies will be equivocal, as the sampling technique and assessment methodology vary widely. Further studies are required for quantitative assessment.
There is a large body of evidence that COVID patients in the acute phase of respiratory infections can disseminate large numbers of virus‐laden aerosols. 15 , 18 , 19 In the present study, the presence of SARS‐CoV‐2 in the indoor aerosol of hospitals corroborates the above statement. Our observations are consistent with the findings of others where SARS‐CoV‐2 was detected in indoor hospital air 4 , 69 , 70 , 71 , 72 and contrary to results of a few 73 , 74 who have reported the absence of SARS‐CoV‐2 in indoor hospital air. Rhinoviruses, enterovirus, and Flu B virus were detected in the indoor aerosol samples from Kuwait's hospitals, similar to findings reported for bio‐aerosols generated in hospitals elsewhere. 75 , 76 Spatial diversity in terms of unique and common viruses in indoor aerosols is known 77 and these variations are often attributed to the cleanliness and hygiene procedures followed at a particular hospital. We believe in Kuwait's hospitals, the presence of rhinoviruses, enterovirus and Flu B is probably linked to the patient load and the efficacy of air purification systems installed, 78 rather than indoor hygiene, as a common disinfection procedure is followed across the hospitals in Kuwait. In the present study, SJH is a dedicated hospital to treat infectious diseases including COVID‐19, whereas MKH and AMH are makeshift types with specialized units created during the pandemic. In congruence with other studies, aerosols from non‐hospitalized settings and control samples were free from pathogenic microbes. 50 , 51 , 52 This further strengthens the fact that besides hygiene, indoor air in hospitals needs to be closely monitored. The international community has exerted concerted efforts to improve indoor air quality to address airborne SARA‐CoV‐2 issue. The federation of European HVAC Associations has set guidelines to operate HVAC and other building services to prevent the spread of SARS‐CoV‐2 in workplaces. 79 The WHO has laid down a roadmap to improve and ensure good indoor air quality in the context of COVID‐19. 80 The American Society of Heating, Refrigerating and Air‐Conditioning Engineers also developed the position document on infectious aerosols that also addresses the issue to minimize the risk. 81
The reports on co‐infection of SARS‐CoV‐2 with non‐SARS‐CoV‐2 strains and other respiratory viruses such as rhinovirus, enterovirus, and RSV have been published. 77 , 82 The exhaled air from symptomatic and asymptomatic carriers, hospital staff, and visitors can contribute synergistically to the dissemination of COVID‐19. 83 The hospital‐acquired infections further cause one of the most serious complications in COVID‐19 patients admitted in ICU and with compromised immunity. 84 In view of this, the presence of multiple viruses (rhinovirus, FluB, RSV, bocavirus, enterovirus, CoV‐NL63, and CoV‐HKU1) and pathogenic bacteria (S. pneumonia, H. influenza, and M. pneumonia) along with SARS‐CoV‐2 at COVID positive areas might pose additional risk to COVID patients. Although as per the MOH hospital design parameters the air borne infection isolation room should be of single occupancy to avoid infection spread via droplet associated with coughing and inhalation, these room have controlled ventilation, negative air pressure of >2.5 Pa/0.01 inch water gauge in relation to the corridor. These are also equipped with HEPA filters at inlet with 99.97% efficiency. Unfortunately, Kuwait's hospital was not designed to accommodate so many infectious patients, requiring single‐bedded, negative pressure isolation rooms. Our results indicate that possible interactions among these microbial communities do play a role in defining the infectivity of a particular organism. Interactions of SARS‐CoV‐2 with these respiratory viruses, therefore, need to be quantified, possibly using whole transcriptomic studies.
For the comprehension of viral pathogenicity, quantification of viral particles to induce an infective response upon inhalation is critical. These values still need to be predicted for SARS‐CoV‐2 72 and the total viral load of SARS‐CoV‐2 ranged between 0 and 100 copies per m3 of air in the indoor hospital air in Kuwait. The concentration was 3‐fold higher in the COVID ward of MKH (~100 copies per m3 of air) as compared to SJH (35 copies per m3 of air). This result is consistent with the findings of Liu et al. 4 that reported the concentrations of airborne SARS‐CoV‐2 virus in the range of 0–42 copies per m3 of air, attributable to the implementation of rigorous sanitization procedures in particular hospitals. 85 The minimum infective dose for SARS‐CoV‐2 via inhalation of droplets was estimated to be 300 particles using computational simulations of the nasopharynx. 86 Other studies on transgenic mice, 87 cynomolgus macaques (Macaca fascicularis), 88 and African green monkeys, 88 , 89 suggested that the minimum tissue culture infectious doses (TCID50) needed to become infected by the aerosol route were 9.0E+02, 4.1E+04, 5.4E+04, and 4.28E+06, respectively. The concentrations found in the indoor aerosol of Kuwait's hospitals were far below the infectious dose reported elsewhere. However, the presence of SARS‐CoV‐2 at these low concentrations within the aerosols will lead to inhalation by individuals in hospital (healthcare professionals and non‐COVID patients) who might become susceptible to infection. It poses a question that requires further investigation, that is, will such a low level of exposure lead to the development of antibodies against COVID‐19 in these individuals?
5. CONCLUSIONS
It is evident from our findings that common respiratory viruses exist in the indoor air of hospitalized settings in a higher frequency as compared to the non‐hospitalized settings and outdoor air. The presence of pathogenic bacteria in hospital settings, especially in COVID‐positive areas, will require further investigations at functional and quantitative levels to confirm their role as a part of the SARS‐CoV‐2 infectome. Rhinovirus was the most prevalent virus in both indoor and outdoor aerosols in Kuwait during the sampling period (Sep‐Nov. 2020). FluB and enteroviruses were found in indoor air only. SARS‐CoV‐2, H. influenzae were unique to indoor aerosols of hospitals only. COVID‐positive areas within the hospital premises depict more diversity as indicated by the presence of viruses and pathogenic bacteria.
The origin of the SARS‐CoV‐2 in the indoor hospital air can be exhalation from an infected person, respiratory shedding from an infected person; inadequate ventilation and air exchange could have resulted in a build‐up of these concentrations. Existing interventions such as social distancing, masks, hand hygiene, surface sanitization, and avoidance of crowded indoor spaces are reasonable prevention measures for spreading SARS‐CoV‐2 in enclosed areas. Increased ventilation can limit the build‐up of SARS‐CoV‐2 concentration in indoor aerosols and shall be a useful corrective measure.
CONFLICT OF INTEREST
The authors explicitly state that there was no conflict of interest.
AUTHOR CONTRIBUTIONS
Nazima Habibi (NH): Conceptualization, methodology, writing—original draft preparation, writing review and editing; Saif Uddin (SU): Conceptualization, sampling, writing—original draft preparation, writing review and editing, data curation; Fadila Al Salameen (FS): Review and editing; Sami Alamad (SA): Sampling and data generation; Vinod Kumar (VK): Methodology; M. Al Otaibi (MO): Data generation; N. Abdul Razzack (NAR) and A. Shajan: Methodology, data generation; F. Shirshikar (FS): sampling.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/ina.12871.
Supporting information
Table S1
Table S2
Table S3
ACKNOWLEDGMENTS
We thank Kuwait Foundation for Advancement of Sciences (KFAS; Grant No. CORONA PROP 86) and Kuwait Institute for Scientific Research (KISR; Grant No. FB157C) for funding this research. Thanks are due to Ms. Farahana Zakir for her participation in experiments. We are thankful to Dr. Scott W. Fowler for editing the manuscript.
Habibi N, Uddin S, Salameen FA, et al. SARS‐CoV‐2, other respiratory viruses and bacteria in aerosols: Report from Kuwait's hospitals. Indoor Air. 2021;31:1815–1825. 10.1111/ina.12871
DATA AVAILABILITY STATEMENT
The entire dataset used in this study is available as supplementary data files S1, S2, and S3 to this submission.
REFERENCES
- 1. Al Salameen F, Habibi N, Uddin S, et al. Characterization and Identification of Micro‐ Organisms Associated with Airborne Dust in Kuwait Final report (EM075C) Kuwait Institute for Scientific Research. 2020; Final Report KISR.
- 2. Al Salameen F, Habibi N, Uddin S, et al. Spatio‐temporal variations in bacterial and fungal community associated with dust aerosol in Kuwait. PLoS ONE. 2020;15(11):e0241283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anon . https://www.worldometers.info/coronavirus/. 2021. Accessed 24 February, 2021
- 4. Liu Y, Ning Z, Chen Y, et al. Aerodynamic analysis of SARS‐CoV‐2 in two Wuhan hospitals. Nature. 2020;582(7813):557‐560. [DOI] [PubMed] [Google Scholar]
- 5. Moore SC, Penrice‐Randal R, Alruwaili M, et al. Amplicon based MinION sequencing of SARS‐CoV‐2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID‐19. medRxiv. 2003;2020(2020):2005. [Google Scholar]
- 6. Qu G, Li X, Hu L, Jiang G. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID‐19). Environ Sci Technol. 2020;54(7):3730‐3732. [DOI] [PubMed] [Google Scholar]
- 7. Habibi N, Behbehani M, Uddin S, Al Salamin F, Shajan A, Zakir F. A safe and effective sample collection method for assessment of SARSCoV2 in aerosol samples. In: Ramanathan AL, Chidambaram S, Jonathan MP, Munoz‐Arriola F, Prasanna MV, Kumar P. ed. Environmental resilience and transformation in times of COVID‐19. Elsevier; 2021. [Google Scholar]
- 8. Hsih W‐H, Cheng M‐Y, Ho M‐W, et al. Featuring COVID‐19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan. J Microbiol Immunol Infect. 2020;53:459‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jayaweera M, Perera H, Gunawardana B, Manatunge J. Transmission of COVID‐19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ Res. 2020;188:109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kennedy M, Lee SJ, Epstein M. Modeling aerosol transmission of SARS‐CoV‐2 in multi‐room facility. J Loss Prev Process Ind. 2020;69:104336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang R, Li Y, Zhang AL, Wang Y, Molina MJ. Identifying airborne transmission as the dominant route for the spread of COVID‐19. Proc Natl Acad Sci USA. 2020;117(26):14857‐14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. Ten scientific reasons in support of airborne transmission of SARS‐CoV‐2. Lancet. 2021;397(10285):1603‐1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heneghan CJ, Spencer EA, Brassey J, et al. SARS‐CoV‐2 and the role of airborne transmission: a systematic review. F1000Research. 2021;10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morawska L, Allen J, Bahnfleth W, et al. A paradigm shift to combat indoor respiratory infection. Science. 2021;372(6543):689‐691. [DOI] [PubMed] [Google Scholar]
- 15. Tang S, Mao Y, Jones RM, et al. Aerosol transmission of SARS‐CoV‐2? Evidence, prevention and control. Environ Int. 2020;144:106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang JW, Marr LC, Li Y, Dancer SJ. Covid‐19 has redefined airborne transmission. BMJ. 2021;373(913):1‐2. [DOI] [PubMed] [Google Scholar]
- 17. Lane MA, Brownsword EA, Morgan JS, et al. Bioaerosol sampling of a ventilated patient with COVID‐19. Am J Infect Control. 2020;48(12):1540‐1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma J, Qi X, Chen H, et al. Coronavirus disease 2019 patients in earlier stages exhaled millions of severe acute respiratory syndrome coronavirus 2 per hour. Clin Infect Dis. 2020;72(10):e652‐e654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nissen K, Krambrich J, Akaberi D, et al. Long‐distance airborne dispersal of SARS‐CoV‐2 in COVID‐19 wards. Sci Rep. 2020;10(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaabna K, Doraiswamy S, Mamtani R, Cheema S. Facemask use in community settings to prevent respiratory infection transmission: A rapid review and meta‐analysis. Int J Infect Dis. 2021;104:198‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davido B, Gautier S, Riom I, et al. The first wave of COVID‐19 in hospital staff members of a tertiary care hospital in the greater Paris area: a surveillance and risk factors study. Int J Infect Dis. 2021;105:172‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorny AW, Bagdasarian N, Koh AHK, et al. SARS‐CoV‐2 in migrant worker dormitories: geospatial epidemiology supporting outbreak management. Int J Infect Dis. 2021;103:389‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hwang SE, Chang JH, Oh B, Heo J. Possible aerosol transmission of COVID‐19 associated with an outbreak in an apartment in Seoul, South Korea, 2020. Int J Infect Dis. 2021;104:73‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lednicky JA, Lauzardo M, Fan ZH, et al. Viable SARS‐CoV‐2 in the air of a hospital room with COVID‐19 patients. Int J Infect Dis. 2020;100:476‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. López JH, Romo ÁS, Molina DC, et al. Detection of Sars‐Cov‐2 in the air of two hospitals in Hermosillo, Sonora, México, utilizing a low‐cost environmental monitoring system. Int J Infect Dis. 2021;102:478‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan L, Ma B, Lai X, et al. Air and surface contamination by SARS‐CoV‐2 virus in a tertiary hospital in Wuhan, China. Int J Infect Dis. 2020;99:3‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y, Zeng Y, Chen C. Presence of SARS‐CoV‐2 RNA in isolation ward environment 28 days after exposure. Int J Infect Dis. 2020;97:258‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nissen K, Krambrich J, Akaberi D, et al. Long‐distance airborne dispersal of SARS‐CoV‐2 in COVID‐19 wards. Sci Rep. 2020;10(1):19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu IT, Qiu H, Tse LA, Wong TW. Severe acute respiratory syndrome beyond Amoy Gardens: completing the incomplete legacy. Clin Infect Dis. 2014;58(5):683‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Doremalen N , Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382(16):1564‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. J Autoimmun. 2020;109:102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID‐19. JAMA. 2020;323(18):1837‐1838. [DOI] [PubMed] [Google Scholar]
- 33. Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) from a symptomatic patient. JAMA. 2020;323(16):1610‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stadnytskyi V, Bax CE, Bax A, Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS‐CoV‐2 transmission. Proc Natl Acad Sci USA. 2020;117(22):11875‐11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guan W‐J, Liang W‐H, Zhao Y, et al. Comorbidity and its impact on 1590 patients with Covid‐19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Hu J, Zhang Z, et al. Efficacy of hydroxychloroquine in patients with COVID‐19: results of a randomized clinical trial. medRxiv. 2003;2020(2020):2022. [Google Scholar]
- 37. Nkhata SG, Ngoma TN, Chilenga PM. SARS‐CoV 2 (Covid‐19) heterogeneous mortality rates across countries may be partly explained by life expectancy, calorie intake, and prevalence of diabetes. Human Ecology. 2020;48(5):633‐638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siegel JD, Rhinehart E, Jackson M, Chiarello L. Guideline for isolation precautions: preventing transmission of infectious agents in health care settings. Am J Infect Control. 2007;35(10, Supplement 2):S65‐S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martinez L, Cheng W, Wang X, et al. A risk classification model to predict mortality among laboratory‐confirmed avian influenza A H7N9 patients: a population‐based observational cohort study. J Infect Dis. 2019;220(11):1780‐1789. [DOI] [PubMed] [Google Scholar]
- 40. Booth T, Kournikakis B, Bastien N, et al. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J Infect Dis. 2005;191:1472‐1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rivers CM, Majumder MS, Lofgren ET. Risks of death and severe disease in patients with middle east respiratory syndrome coronavirus, 2012–2015. Am J Epidemiol. 2016;184(6):460‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rio DC, Ares M, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010;2010(6):pdbprot5439. [DOI] [PubMed] [Google Scholar]
- 43. Simbolo M, Gottardi M, Corbo V, et al. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS ONE. 2013;8(6):e62692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anon . Guidelines for general ward design. Task force group for designs and constructions of health care facilities, infection control directorate, Ministry of Health, State of Kuwait. 2008:p 29.
- 45. Li R, Tun HM, Jahan M, et al. Comparison of DNA‐, PMA‐, and RNA‐based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci Rep. 2017;7(1):5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sydenham TV, Bek‐Thomsen M, Andersen SD, et al. Comparative evaluation of the CerTest VIASURE flu A, B & RSV real time RT‐PCR detection kit on the BD MAX system versus a routine in‐house assay for detection of influenza A and B virus during the 2016/17 influenza season. J Clin Virol. 2018;99–100:35‐37. [DOI] [PubMed] [Google Scholar]
- 47. Kukalo O. Molecular‐genetic technologies and their places in the ethyological diagnosis of the infectious combination of bronchial asthma. Global Journal of Clinical Virology. 2019;4(1):001‐007. [Google Scholar]
- 48. Freire‐Paspuel B, Vega‐Mariño P, Velez A, Cruz M, Perez F, Garcia‐Bereguiain MA. Analytical and clinical comparison of Viasure (CerTest Biotec) and 2019‐nCoV CDC (IDT) RT‐qPCR kits for SARS‐CoV2 diagnosis. Virology. 2021;553:154‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumar PS, Subramanian K. Demystifying the mist: sources of microbial bioload in dental aerosols. J Periodontol. 2020;91(9):1113‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kunwar A, Tamrakar S, Poudel S, Sharma S, Parajuli P. Bacteriological assessment of the indoor air of different hospitals of Kathmandu District. Int J Microbiol. 2019;2019:5320807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. MacIntyre R, Dwyer D, Seale H, et al. High risk procedures and respiratory infections in hospital health care workers‐quantifying the risk. Int J Infect Dis. 2012;16:e379. [Google Scholar]
- 52. Macintyre CR, Seale H, Yang P, et al. Quantifying the risk of respiratory infection in healthcare workers performing high‐risk procedures. Epidemiol Infect. 2014;142(9):1802‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Magalhães AP, França Â, Pereira MO, Cerca N. RNA‐based qPCR as a tool to quantify and to characterize dual‐species biofilms. Sci Rep. 2019;9(1):13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sivagnanasundaram P, Amarasekara RW, Madegedara RM, Ekanayake A, Magana‐Arachchi DN. Assessment of airborne bacterial and fungal communities in selected areas of teaching hospital, Kandy, Sri Lanka. Biomed Res Int. 2019;12:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Perkins SD, Mayfield J, Fraser V, Angenent LT. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl Environ Microbiol. 2009;75(16):5363‐5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zemouri C, de Soet H , Crielaard W, Laheij A. A scoping review on bio‐aerosols in healthcare and the dental environment. PLoS ONE. 2017;12(5):e0178007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zemouri C, Volgenant CMC, Buijs MJ, et al. Dental aerosols: microbial composition and spatial distribution. J Oral Microbiol. 2020;12(1):1762040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z, Yomo D, Gradinaru C. Choosing the right fluorophore for single‐molecule fluorescence studies in a lipid environment. Biochim Biophys Acta. 2017;1859(7):1242‐1253. [DOI] [PubMed] [Google Scholar]
- 59. Aycicek H, Oguz U, Karci K. Comparison of results of ATP bioluminescence and traditional hygiene swabbing methods for the determination of surface cleanliness at a hospital kitchen. Int J Hyg Environ Health. 2006;2009:203‐206. [DOI] [PubMed] [Google Scholar]
- 60. Prussin AJ II, Garcia EB, Marr LC. Total virus and bacteria concentrations in indoor and outdoor air. Environ Sci Technol Lett. 2015;2(4):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodrigues AF, Santos AM, Ferreira AM, Marino R, Barreira ME, Cabeda JM. Year‐long rhinovirus infection is influenced by atmospheric conditions, outdoor air virus presence, and immune system‐related genetic polymorphisms. Food Environ Virol. 2019;11(4):340‐349. [DOI] [PubMed] [Google Scholar]
- 62. Price RHM, Graham C, Ramalingam S. Association between viral seasonality and meteorological factors. Sci Rep. 2019;9(1):929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Griffin DW, Garrison VH, Herman JR, Shinn EA. African desert dust in the Caribbean atmosphere: microbiology and public health. Aerobiologia. 2001;17(3):203‐213. [Google Scholar]
- 64. Zheng Y, Chen H, Yao M, Li X. Bacterial pathogens were detected from human exhaled breath using a novel protocol. J Aerosol Sci. 2018;117:224‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huynh KN, Oliver BG, Stelzer S, Rawlinson WD, Tovey ER. A new method for sampling and detection of exhaled respiratory virus aerosols. Clin Infect Dis. 2008;46(1):93‐95. [DOI] [PubMed] [Google Scholar]
- 66. Dick EC, Jennings LC, Mink KA, Wartgow CD, Inhorn SL. Aerosol transmission of rhinovirus colds. J Infect Dis. 1987;156(3):442‐448. [DOI] [PubMed] [Google Scholar]
- 67. Tang JW, Bialasiewicz S, Dwyer DE, et al. Where have all the viruses gone? Disappearance of seasonal respiratory viruses during the COVID‐19 pandemic. J Med Virol. 2021;93(7):4099‐4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sullivan SG, Carlson S, Cheng AC, et al. Where has all the influenza gone? The impact of COVID‐19 on the circulation of influenza and other respiratory viruses, Australia, March to September 2020. Eurosurveillance. 2020;25(47):2001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Razzini K, Castrica M, Menchetti L, et al. SARS‐CoV‐2 RNA detection in the air and on surfaces in the COVID‐19 ward of a hospital in Milan, Italy. Sci Total Environ. 2020;742:140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Santarpia JL, Rivera DN, Herrera V, et al. Aerosol and Surface Transmission Potential of SARS‐CoV‐2. medRxiv. 2003a;2020(2020):2023. [Google Scholar]
- 71. Santarpia JL, Rivera DN, Herrera VL, et al. Aerosol and surface contamination of SARS‐CoV‐2 observed in quarantine and isolation care. Sci Rep. 2020;10(1):12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Girolamo P. Assessment of the potential role of atmospheric particulate pollution and airborne transmission in intensifying the first wave pandemic impact of SARS‐CoV‐2/COVID‐19 in Northern Italy. Bull of Atmos Sci & Technol. 2020;1(3):515‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Colaneri M, Seminari E, Piralla A, et al. Lack of SARS‐CoV‐2 RNA environmental contamination in a tertiary referral hospital for infectious diseases in Northern Italy. J Hosp Infect. 2020;105(3):474‐476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Faridi S, Niazi S, Sadeghi K, et al. A field indoor air measurement of SARS‐CoV‐2 in the patient rooms of the largest hospital in Iran. Sci Total Environ. 2020;725:138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. Lancet Infect Dis. 2007;7(4):257‐265. [DOI] [PubMed] [Google Scholar]
- 76. Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface. 2009;6(suppl_6):S783‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karimzadeh S, Bhopal R, Nguyen Tien H. Review of infective dose, routes of transmission and outcome of COVID‐19 caused by the SARS‐COV‐2: Comparison with other respiratory viruses. Epidemiol Infect. 2021;149:E96. 10.1017/S0950268821000790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anonymous . Guidelines for General Ward Design. 2008. http://www.icdkwt.com/pdf/policiesandguidelines/DesignandConstruction/GuidelinesforGeneralWardDesign‐2008.pdf
- 79. Anon . REHVA COVID‐19 guidance document. Federation of European Heating. Ventillation and Air Conditioning Associations. 2020;17p.
- 80. WHO . Roadmap to improve and ensure good indoor ventilation in the context of COVID‐19. Geneva: World Health Organization. 2021;25p. [Google Scholar]
- 81. Stewart EJ, Schoen LJ, Mead K, et al. ASHRAE Position Document on Infectious Aerosols. American Society of Heating. Georgia, Atlanta, USA. Refrigerating and Air‐Conditioning Engineers, Inc; 2020:20p. [Google Scholar]
- 82. Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co‐infection between SARS‐CoV‐2 and other respiratory pathogens. JAMA. 2020;323(20):2085‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID‐19) infections. Int J Infect Dis. 2020;93:284‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Corse T, Dayan L, Kersten S, Battaglia F, Terlecky SR, Han Z. Clinical outcomes of COVID‐19 patients with pre‐existing, compromised immune systems: a review of case reports. Int J Med Sci. 2020;17(18):2974‐2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li Y. Hypothesis: SARS‐CoV‐2 transmission is predominated by the short‐range airborne route and exacerbated by poor ventilation. Indoor Air. 2021. 10.1111/ina.12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Basu S. Exposure to a COVID‐19 carrier: transmission trends in respiratory tract and estimation of infectious dose. medRxiv. 2007;2020(2020):2027. [Google Scholar]
- 87. Bar‐On YM, Flamholz A, Phillips R, Milo R. SARS‐CoV‐2 (COVID‐19) by the numbers. eLife. 2020;9:e57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Johnston SC, Ricks KM, Jay A, et al. Development of a coronavirus disease 2019 nonhuman primate model using airborne exposure. PLoS ONE. 2021;16(2):e0246366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cross RW, Agans KN, Prasad AN, et al. Intranasal exposure of African green monkeys to SARS‐CoV‐2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol J. 2020;17(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Data Availability Statement
The entire dataset used in this study is available as supplementary data files S1, S2, and S3 to this submission.


