Graphical abstract
Abstract
The formation of small hybrid aggregates between excipient and drug molecules is one of the mechanisms that contributes to the solubilization of active principles in pharmaceutical formulations. The characterization of the formation, governing interactions and structure of such entities is not trivial since they are highly flexible and dynamic, quickly exchanging molecules from one to another. In the case of cyclodextrins, this mechanism and the formation of inclusion complexes synergistically cooperate to favour the bioavailability of drugs. In a previous study we reported a detailed characterization of the possible formation of inclusion complexes with 1:1 stoichiometry between remdesivir, the only antiviral medication currently approved by the United States Food and Drug Administration for treating COVID-19, and sulphobutylether-β-cyclodextrins. Here we extend our study to assess the role of the spontaneous aggregation in the solubilization of the same drug, by molecular dynamics simulations at different relative concentrations of both compounds. The number of sulphobutylether substitutions in the cyclodextrin structure and two different protonation states of the remdesivir molecule are considered. We aim to shed light in the solubilization mechanism of sulphobutylether-β-cyclodextrins, broadly used as an excipient in many pharmaceutical formulations, in particular in the case of remdesivir as an active compound.
1. Introduction
Cyclodextrins (CDs) are widely used as excipients to solubilize or/and stabilize drugs [1]. It is typically assumed that two different interaction mechanisms contribute to these aims, namely the formation of inclusion complexes by threading the CD hydrophobic cavity and the encapsulation of the drug into CD aggregates [2], [3], [4]. The former mechanism is much more specific than the latter, but both are expected to depend on the structure of the CD to be employed: number or glucopyranoside rings as well as number, type and location of the substitutions in non-native CDs. The probability to form inclusion complexes is typically quantified by means of the so-called association or binding constants which can be estimated by different experimental and computational methods. Isothermal Titration Calorimetry (ITC) is recognized as the gold standard technique for this type of characterizations [5] although any physical observable able to detect changes in the concentration of complexes versus free species can be employed for this aim [6]. Such characterization is much simpler for native than for modified CDs since the latter are always heterogenous mixtures of many different structures with different number and location of substituted groups, leading to a different ability to encapsulate molecular groups in their cavity. Thus, the affinity constants obtained from these methods should necessarily be understood as apparent or average constants representing the ensemble of different CD structures. Computational simulations with atomic resolution can also be employed to characterize the association between molecules [7]. In contrast to the calculations obtained from typical wet-lab experiments, these methods do allow considering specific structures and to provide a full energy profile with energy barriers and wells for the different possible conformations of the molecules forming the supramolecular complex [8], [9]. When CDs are used as excipients, moderate affinity constants are convenient, since a too strong binding would prevent the delivery of the active molecule and a too weak binding would require a large amount of CD to increase the solubility. Actually, CDs with high affinity for specific drugs have been designed to remove them from the target organism in order to cancel their activity when it is not required anymore. A well-known example is Sugammadex, a modified CD used to revert the rocuronium-induced neuromuscular blockade [10].
It is worth to comment that encapsulation of molecules in the cavity of CDs forming inclusion complexes does not necessarily favor the solubility of the drug since the supramolecular structures resulting from this process can exhibit a significant trend to aggregate, eventually forming a precipitate or even leading to exotic structures in aqueous solution [11]. The alternative mechanism exhibited by CDs to increase the solubilization of drugs is the formation of relatively small aggregates, with CD molecules surrounding the drug and thus avoiding its self-aggregation [12]. This mechanism also requires a convenient balance since the formation of too large aggregates could lead to a precipitate while the null interaction between molecules does not contribute to the bioavailability of the drug. This mechanism could also be assessed by different experimental methods such as turbidity, dynamic light scattering, transmission electron microscopy or even nuclear magnetic resonance measurements [11], [12], [13], [14], [15], [16] , but it is less clear how to perform a suitable thermodynamic characterization since the apparent binding would be associated to the interaction between a non-specific number of molecules with no clear internal structure. As in the case of the inclusion complexes, the situation is more difficult for modified CDs due to their heterogeneous composition regarding the number and location of substitutions, which are expected to exhibit different aggregation properties. Despite a substantial amount of experimental data is available on CD aggregation, the mechanisms of CD aggregation and CD complex aggregation has not been fully understood yet [17], [18], [19], [20], especially for sulphobutylether-β-CD (SBE-β-CD), which is currently being used in 13 FDA approved injectables and numerous clinical candidates. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7196545/]
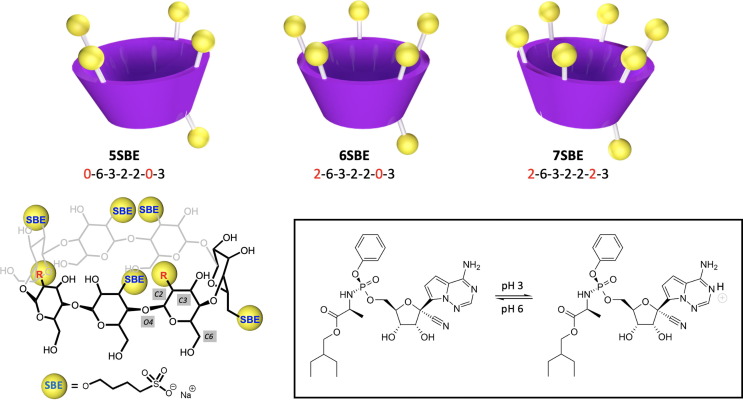
Many pharmacological formulations are based on modified CDs [21], [22]. One of the best known examples in the times of COVID-19 is Veklury® which, even 1 year after the start of the pandemic, is still the only antiviral medication approved by the United States Food and Drug Administration (FDA) for treating this disease (https://www.fda.gov/newsevents/press-announcements/fda-approves-first-treatment-covid-19). The active principle of Veklury® is remdesivir (Fig. 1 ), which is solubilized by sulphobutylether-β-CD (SBE-β-CD, Fig. 1) in this formulation [23]. The mixture between both molecules is preferentially performed at low pH since the solubility of remdesivir, pKa 3.3 significantly increases under this condition. It was observed that the drug remains solubilized upon the addition of NaOH at the proposed concentrations, thus allowing the administration of the formulation at neutral pH into the human body. We have recently reported a detailed thermodynamic and structural characterization on the formation of inclusion complexes between SBE-β-CD (considering different number and location of substitutions) and remdesivir at two different pH values by ITC measurements and molecular dynamics (MD) simulations [24]. Our results indicate that the association constant for the formation of the inclusion complexes is moderate (∼104 M−1) at low pH and even lower (∼102 M−1) at neutral pH. Additionally, our MD simulations indicate that the two molecules can interact as a non-inclusion complex or as an inclusion complex, with similar free energy for both possibilities and with a significant energy barrier preventing the entry of the ligand in the cavity. The inclusion complex was observed just for a specific orientation of the remdesivir in the CD cavity, with the C-nucleoside group of this molecule towards the secondary side of the CD. Furthermore, the association constant was observed to be significantly higher for the structure with more number of substitutions (7SBE), mainly at low pH. This is probably due to the electrostatic attraction between the drug, which is expected to be protonated under those conditions, and the negatively charged CD, which increases with the number of substitutions. Based on ITC measurements, the presence of a variety of species with different affinities or/and interaction mechanisms with remdesivir in the SBE-β-CD distribution was also proposed in the same work. This suggests the existence of more complex stoichiometries or even some kind of aggregation. In view of our previous results, and taking also in account that the typical mass ratio values between SBE-β-CD and remdesivir in the pharmaceutical formulation of this active principle are much higher than those expected for an ideal 1:1 encapsulation, the possible formation of aggregates is now addressed. With this aim, MD simulations of the two molecules at different relative concentrations, for three different structures of SBE-β-CD (5SBE, 6SBE and 7SBE, defined in Fig. 1) and considering again two different protonation states for remdesivir, were performed. The methodology and main results will be presented in what follows.
Fig. 1.
Top: Schematic representation of the employed SBE-β-CDs, 5SBE, 6SBE and 7SBE. The label indicates the counter-clockwise sequence of substitutions, when the CD structure is oriented with the secondary hydroxyl groups pointing out of the plane. The numbers 2, 3 or 6 represent the position of the substitution group in each glucopyranoside ring and 0 indicates that no substitution is present in the corresponding ring. Down: Two-dimensional representation of SBE-β-CD and the equilibrium between neutral and protonated remdesivir. In the CD structure the blue SBE labels represent the groups that are substituted in all the structures studied by MD simulations and the red R label represents the group that is substituted in 6SBE and in 7SBE. The numbers in grey squares represent the position of the carbon atoms C2, C3 and C6 and that of the oxygen O4.
2. Methods
The same three different representative structures of SBE-β-CD with a single substitution in the primary side and 4, 5 or 6 substitutions in the secondary size of the CD employed in our previous work (5SBE, 6SBE and 7SBE, Fig. 1) [24] were selected for the current simulations. For remdesivir, the previously employed neutral and protonated structures were used. The parameterizations of the molecules and the employed general simulation methods were also identical to those of our previous publications [20], [24], [25]. Briefly, GROMACS 2020 package [26], [27], [28] was used to run the simulations in the NPT ensemble at 298 K and 1 bar with the 54a7 parameterization of the GROMOS force field [29] and the SPC water model [30], the v-rescale thermostat [31] and the Parrinello-Rahman barostat [32], the PME algorithm [33], [34] for the long range interactions and a cutoff of 1.2 nm for the short range interactions. The equations of motion were integrated using the leap-frog algorithm [35] with a time step of 2 fs and the bonds were constrained using the SETTLE algorithm [36] for the waters and the LINCS algorithm [37] for the rest of the molecules in the simulation boxes. For each CD structure, 4 simulation boxes were prepared with neutral remdesivir and another 4 with the protonated structure of the drug. Those simulations boxes contained 20 CD units each + ∼31 thousand water molecules and 1, 2, 4 or 8 remdesivir molecules. For each of these systems, the solute molecules were placed at random positions and orientations and recorded the trajectories for a minimum of 500 ns. Additionally simulation boxes with 8 and 20 remdesivir units (+ ∼31 thousand water molecules) using both the neutral and the protonated states of the drug were executed. It is important to note that the total concentrations in our simulations are significantly larger than those employed in the experiments considering just the ratio between the number of molecules. For instance, for the simulation boxes with just one solute molecule, the total concentration is 1.79 mM, while the solubility of remdesivir is 1 order of magnitude lower. However, even when the total concentration is that high, the molecule cannot aggregate nor precipitate because no other solute molecule is reachable in the same simulation box. Thus, these concentrations can be interpreted as local concentrations, focusing on the relative CD to drug concentration more than in the absolute concentration of both compounds. In a solution where the total concentration is relatively low, it is possible to see microscopic regions of relatively high concentration since the molecules are not evenly distributed throughout the container. Thus, a total of 24 simulations boxes with 20 CDs each (35.84 mM) and 1, 2, 4 or 8 remdesivir molecules (1.79, 3.58, 7.17 and 14.34 mM), and another 4 simulation boxes with 8 (14.34 mM) or 20 (35.84 mM) neutral or protonated remdesivir molecules will be prepared. For each of the resulting trajectories the distance between all the CDs and all the drugs, the number of H-bonds between the different types of molecules including the solvent, and the number of clusters of different composition, were determined. These analyses were performed using GROMACS tools [26], [27], [28]. The average distance between each CD and remdesivir molecule as well as the standard deviation of this property using the MDAnalysis package [38], [39] in Python 3.7 were also determined. Finally several representative snapshots of the final configuration were represented using Pymol [40]. Summary sheets containing these analyse were created for all the trajectories and they are included as Supplementary Material (SM).
3. Results & discussion
3.1. Aggregation of remdesivir
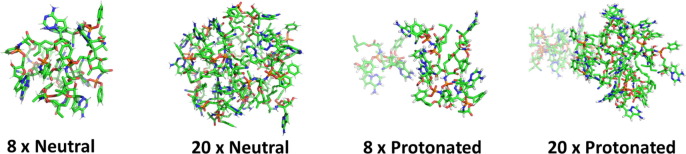
The simulations with remdesivir in the absence of CD quickly formed aggregates. In the case of the neutral species, a single cluster was formed while for the protonated structure two clusters for the trajectories using both 8 and 20 molecules of the drug were obtained (Fig. 2 ). The formation of the clusters was quicker in the simulations with the 20 molecules than in the simulations with 8 molecules. For the simulations with 8 protonated structures, there is a dynamic exchange between clusters, which fluctuate between 2 and 4 aggregates (see SM, page 1). The number of H-bonds between water and the drug molecules do not seem to depend on the charge nor on the local concentration in the simulation box. For all the simulations the number of H-bonds increases during the initial part of the trajectories and then, after 50–100 ns, it reaches a plateau fluctuating around a constant average value close to 1 H-bond per drug molecule. The aggregates resulting from these simulations anticipate the formation of a second phase, thus indicating the lack of solubility of this molecule. The fact that the aggregates are smaller for the charged species is also in line with the experimentally observed fact that remdesivir is significantly more soluble at low pH than at neutral pH, the accumulation of charge preventing the growth of the aggregate in the former case.
Fig. 2.
Structures obtained from the simulation of 8 or 20 neutral or protonated remdesivir molecules, after 200 ns of MD simulations. The number of molecules in the simulation box and the protonation state are indicated.
3.2. Solubilization of remdesivir by SBE-β-CD
In our previous work, the formation of inclusion complexes between 7SBE, 6SBE and 5SBE with neutral and protonated remdesivir was studied. Clearly, the interaction between the CDs and the protonated form of the drug is stronger than with the neutral form. It was also concluded that the affinity constant for the formation of the inclusion complex is lower for the CD structure with lower number of substitutions (5SBE) than for the structures with 6 and 7 SBE groups. The aim of the present work is to evaluate whether or not the protonation state of the drug molecule and the number of substitutions in the CD have any impact in the formation of aggregates. Several relative concentrations of both molecules (CD and drug) were tested in order to see the existence of consistent trends. As explained in the methods section, the analysis was automated for all the obtained trajectories and the corresponding results are shown in the Supplementary Material. Here the main results will be summarized.
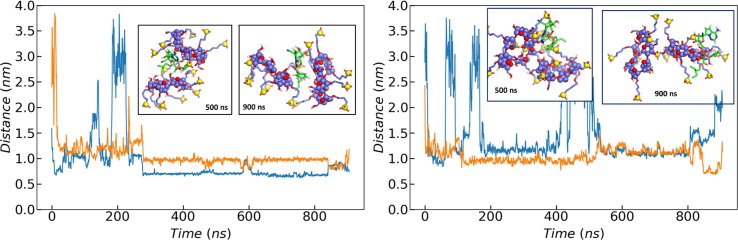
First of all, the spontaneous formation of inclusion complexes with the remdesivir molecule threading the CD cavity was not observed in any of our trajectories. This is not surprising since our previous PMF calculations [24] showed that the free energy corresponding to such arrangements is similar to that of non-inclusion complexes and that both types of structures are separated by an energy barrier [24]. Thus, although the spontaneous formation of inclusion complexes is thermodynamically possible, it is not expected to be dominant. In addition to the simulations described above, several long MD trajectories (∼900 ns) with two 7SBE structures and one neutral remdesivir were computed trying to seek for the spontaneous formation of inclusion complexes (Fig. 2). Again, they were not observed, although the drug appears always interacting with at least one of the CDs present in the solution, with a conformation compatible with that predicted by our previous PMF calculations [24].
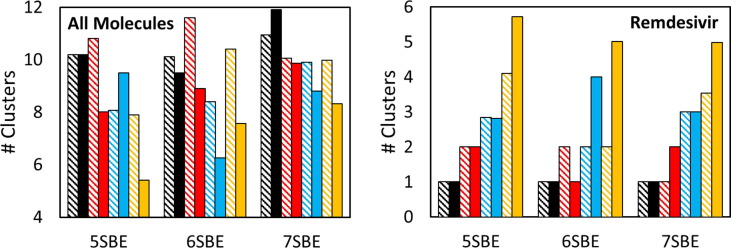
The number of clusters formed during the MD simulations is the most evident quantitative parameter to illustrate the aggregation process. This is a kinetic parameter that evolves during the trajectories. Thus, in order to make an objective comparison between the different simulations, the average number of clusters during the last 100 ns of all the trajectories was computed (Fig. 4 and Table S1). A cluster is defined as a group of molecules in close contact (less than 3.5 Å) with at least one molecule of the same group. Isolated molecules are considered as independent clusters.
Fig. 4.
Average number of clusters formed by remdesivir and CD (left) and just for the remdesivir molecules (right) in the simulations with 20 CD molecules and 1 (black), 2 (red), 4 (blue) or 8 (orange) neutral (lined bars) or protonated (solid bars) remdesivir molecules, determined through the last 100 ns of all the trajectories. The numerical values for these plots, including standard deviations, are in Table S1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The number of initial clusters is typically equivalent to the number of molecules since they were randomly located in the simulation box (see plots in the Supplementary Material) but it quickly decreases in all cases indicating the formation of small aggregates. In general, the presence of remdesivir seems to slightly favor the aggregation of the total molecules but not the self-aggregation of the drug. This could explain the high ratio of excipient employed to dissolve remdesivir in the pharmaceutical formulation [23]. The coincidence of several drug molecules in the same cluster seems to be exceptional, even at the largest concentrations of the drug. Thus, it is evident that the self-aggregation of remdesivir is disrupted by the presence of the CD molecules. When just one drug molecule is present in the simulation box, more than ten clusters were observed for practically all the simulations, indicating that the SBE-β-CD molecules interact with each other but that they do not form large aggregates even at the relatively high concentrations employed in our simulations. The interaction between CD molecules in solution partially explains the significant signal observed in the dilution Isothermal Titration Calorimetry (ITC) experiments performed with this compound [24]. Another important factor is the strong interaction of the CDs with the surrounding water molecules induced by the presence of the SBE groups (see below). The number of total clusters is, in general, lower for the simulations with the protonated structure than for those with the neutral drug (Fig. 4). This is more evident for 6SBE and 7SBE than for 5SBE and for the simulations with the largest concentration of remdesivir, probably due to the electrostatic attraction between the positively charged remdesivir and the negatively charged cyclodextrins under those conditions. Such electrostatic attraction has a significant impact also in the formation of inclusion complexes, as we have already seen in our previous work [24].
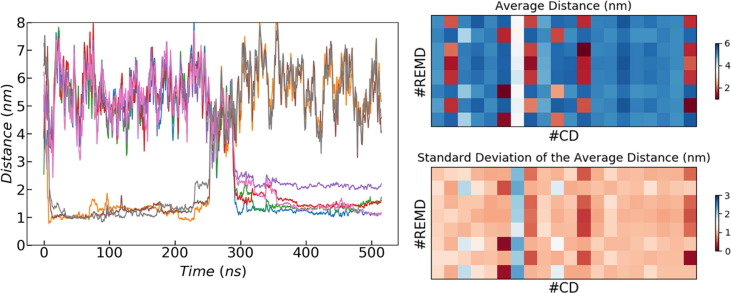
Throughout the simulations, the remdesivir molecules are surrounded by CDs in a dynamic equilibrium. This means that both the drug and the CDs move from one cluster to another. The time scale for these movements, as observed in the different trajectories, is of the order of several hundred ns. As an example, the 20 × 6SBE CDs of the simulation with 8 × neutral REMD molecules are distributed in an average of 10.4 clusters while the drugs are distributed in just 2 clusters (Fig. 4). Along the trajectory it is clearly seen how one of the CDs moves from one these two clusters to the other, spending more or less half of the trajectory interacting with each of them (Fig. 5 ). This type of movements is observed in almost all the simulations, including those shown in Fig. 3, which have only 3 solute molecules (2 CDs and 1 drug) in the simulation boxes.
Fig. 5.
Left: Distance between each of the 8 neutral remdesivir molecules and one of the 6SBE molecules present in the simulation box as a function of time. Right: Average distance between all the CD and remdesivir molecules of the same simulation throughout the whole trajectory (top) together with the corresponding standard deviation (bottom). The CD corresponding to the left plot can be easily identified in the right plots as the #7 columns. In the top right plot it can also be seen that several CDs do not interact with any remdesivir molecule during the trajectory while others remain most of the time interacting with the same drug structure.
Fig. 3.
Distance between the neutral remdesivir molecule (REMD) and the two 7SBE molecules present in the simulation box (each color line corresponds to the distance to a different CD molecule) as a function of time for two independent simulations, together with two snapshots at 500 ns and at 900 ns.
In addition to the description of the general behavior of these simulations and to the difference between the simulations with the protonated and the neutral forms of the drug, it is interesting to observe how the number of substitutions of the CD affect the aggregation of remdesivir. In general, it is seen that the number of total clusters tends to increase with the number of substitutions, suggesting a better solubilization. The differences are more clear for the largest concentrations of the drug where the number of clusters computed for all the solute molecules is 7.9 and 5.4 for the neutral and protonated forms of remdesivir with 5SBE and 9.98 and 8.32, respectively, with 7SBE (Table S1). However, even for these cases, the drug molecules are distributed in a larger number of clusters for the simulations with 5SBE (4.1 and 5.72) than for the simulations with 7SBE (3.54 and 4.99) (Fig. 4 and Table S1). This is likely due to the higher negative charge of the CD molecules with larger number of substitutions, that may favor the interaction with the drug and, at the same time, prevent the aggregation of the CDs.
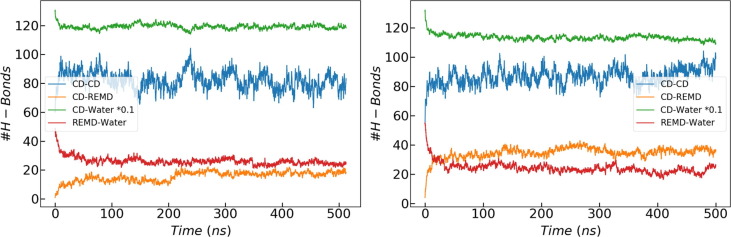
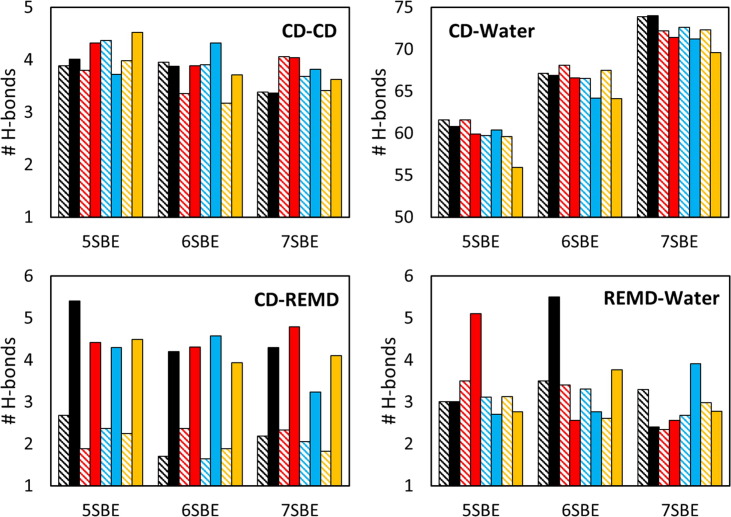
Regarding the number of hydrogen bonds (H-bonds) between different molecules, the kinetic pattern is similar for all the simulations: the number of H-bonds between remdesivir and CD increases with time while it decreases between the drug and water (Fig. 6 ). A similar behavior takes place for the H-bonds between CDs and between CD and water: the former increases while the latter decreases. These values stabilize along the simulation and they seem to oscillate around a constant value during the last 100 ns. The average values for this last segment of the trajectories were computed for all the simulations (Fig. 7 and Table S2). The most significant difference is that the protonated drug forms almost twice as many H-bonds with the CDs than the neutral structure while the number of H-bonds between the remdesivir and water does not seem to depend significantly on the protonation state of the drug. This clearly indicates a stronger interaction between the SBE-β-CD and the protonated remdesivir than with the uncharged structure, in agreement with our previous experimental and computational results [24]. Actually, the number of H-bonds between the protonated drug and the CDs is higher than between the same structure and the solvent while the opposite happens with the neutral form of remdesivir (Fig. 6, Fig. 7). It is also clearly seen that the number of H-bonds between CDs and water significantly increases with the number of substituted groups in the CD. This is due to the large number of H-bonds involved in each SBE group, as shown in Fig. 8 , which induce a highly ordered solvation shell. As anticipated above, this is expected to contribute to the important power signal obtained from ITC dilution experiments of SBE-β-CD in aqueous solution [24]. It is worth to remark that such signal should strongly depend on the number of substitutions in the different CD structures consisting the sample. Thus, the distribution of different species in the of SBE-β-CD formulation is expected to seriously affect the structure and general properties of the solvent.
Fig. 6.
Number of H-bonds between different types of molecules, as indicated in the legends, for the simulations with 20 × 5SBE molecules and 8 × neutral (left) or protonated (right) remdesivir molecules.
Fig. 7.
Average number of H-bonds between different groups of molecules (see labels in the plots), determined through the last 100 ns of all the trajectories. The plots containing the CD-CD and CD-Water H-bonds (top row) are normalized by CD molecule, while those containing the CD-REMD and REMD-Water H-bonds (bottom row) are normalized by remdesivir molecule. The plots correspond to simulations with 20 CD molecules and 1 (black), 2 (red), 4 (blue) or 8 (orange) neutral (lined bars) or protonated (solid bars) remdesivir molecules, determined through the last 100 ns of all the trajectories. The numerical values for these plots, including standard deviations, are shown in Table S2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Representative structures of 7SBE (left) and 5SBE (right) after 500 ns of MD simulations, showing the water molecules at less than 3 Å from any atom of the CDs.
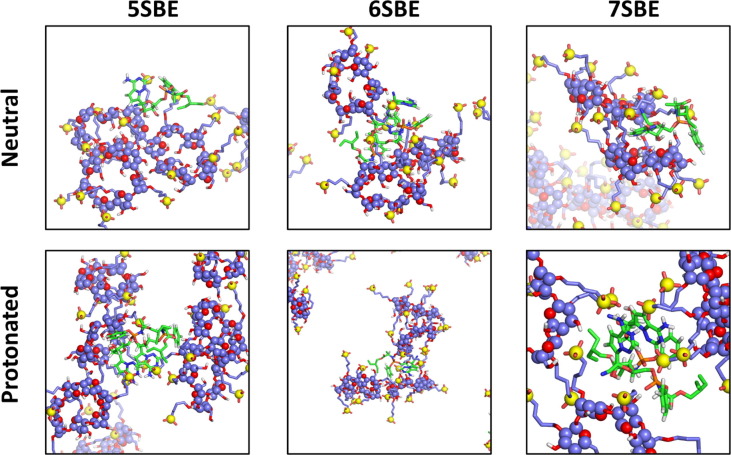
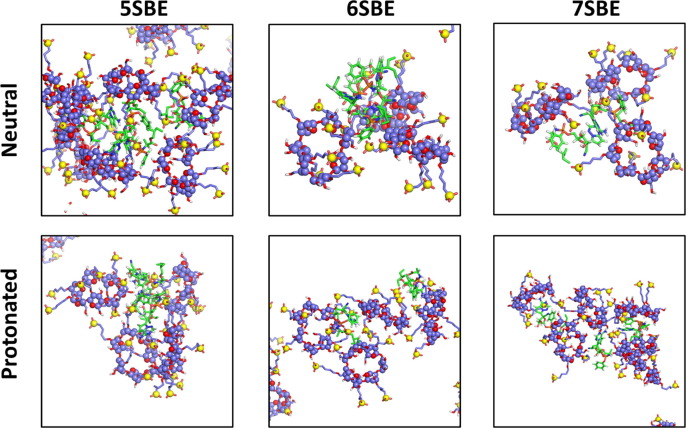
The most typical aggregates in our simulations are formed by one or two molecules of the drug and approximately three CD molecules. As mentioned above, the number of protonated remdesivir molecules in these aggregates at high concentration is, in general, lower than the number of neutral remdesivir within the same cluster, probably due to the repulsive electrostatic interaction between the charged drugs. Some representative structures are shown in Fig. 9, Fig. 10 .
Fig. 9.
Representative structures for small aggregates of SBE-β-CD with neutral or protonated remdesivir. The snapshots were obtained from the frame at 500 ns of the trajectories with 20 × CDs and 4 molecules of the drug.
Fig. 10.
Representative structures for small aggregates of SBE-β-CD with neutral or protonated remdesivir. The snapshots were obtained from the frame at 500 ns of the trajectories with 20 × CDs and 8 molecules of the drug.
4. Conclusions
Using MD simulations, the possible role of SBE-β-CDs aggregates in the solubilization of remdesivir was studied. Both the effect of the number and location of the different SBE substitutions in the CDs present in the excipient (5SBE, 6SBE and 7SBE) and the protonation state of the drug were addressed. It has been illustrated how both the neutral and the protonated remdesivir molecules spontaneously aggregate in aqueous solution. The aggregates of the protonated structure are smaller than those for the neutral form of the drug, due to the electrostatic repulsion that takes place for the charged molecule. The addition of SBE-β-CD to the solution clearly prevents the self-aggregation of remdesivir in both protonation states, even at very high drug concentrations. The solubilization mechanism seems to be the encapsulation of the remdesivir molecule in small clusters formed by a few (2–4) CDs. All our simulations exhibit a dynamic equilibrium between clusters, with molecules exchanging from one to another in a time scale of several hundred ns. The spontaneous formation of inclusion complexes was not observed in any of the simulations but the interaction between CDs and remdesivir was through the peripheral groups of the cyclic CD molecule. The number of substitutions and the protonation state of the drug significantly affect the interactions between molecules and thus their aggregation. The protonated form of the drug seems to favor the cross-aggregation in the presence of SBE-β-CD, as can be seen from the number of total clusters for the charged molecule, typically lower than that for the neutral structure. This results from the electrostatic interaction between the charged form of the drug and SBE-β-CD. The interaction between protonated remdesivir and CD is significantly stronger than that with the neutral structure, showing approximately double amount of CD-drug H-bonds in the former case. Regarding the structure of the aggregates, the pattern observed in our simulations consists of one or two molecules of the drug and approximately three CD molecules. The number of protonated remdesivir molecules within the same cluster at high concentration is, in general, lower than the number of neutral remdesivir under the same conditions. On the other hand, it is seen that SBE-β-CDs notably increase the structure of the solvent, due to the presence of a larger number of H-donors in the SBE groups. Thus, this effect is proportional to the number of substitutions in the CD structure and seriously impact in the self-aggregation properties as well as in interaction with the drug. Increasing the number of substitutions in the CD leads to a lower aggregation of the excipient, while the self-aggregation of the drug slightly increases. This behavior is clearer for the protonated form of the drug than for the neutral structure.
In summary, our results suggest two antagonistic consequences of increasing the number of substitutions in the SBE-β-CDs: it favors the interaction with the drug, mainly in its protonated state, whereas it exhibits a slightly weaker ability to disrupt the self-aggregation of remdesivir. In general, it seems that, in order to solubilize remdesivir, a lower concentration of the excipient could be employed if the distribution is shifted towards a larger number of substitutions. This conclusion cannot be extrapolated for other drugs since the nature of the interactions can be totally different in each case.
CRediT authorship contribution statement
Ángel Piñeiro: Conceptualization, Methodology, Investigation, Writing – review & editing. James Pipkin: Conceptualization, Writing – review & editing. Vince Antle: Conceptualization, Writing – review & editing. Rebeca Garcia-Fandino: Conceptualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
R.G.-F thanks to the Spanish Agencia Estatal de Investigación (AEI) and the ERDF (RTI2018-098795-A-I00) and for a “Ramón y Cajal” contract (RYC-2016-20335), to Xunta de Galicia (ED431F 2020/05) and Centro singular de investigación de Galicia accreditation 2019-2022, ED431G 2019/03) and the European Union (European Regional Development Fund - ERDF). Á. P. thanks to the Ministerio de Ciencia e Innovación (PID2019-111327 GB-I00). We thank the “Centro de Supercómputo de Galicia” (CESGA) for computing time as well as for their exceptional technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molliq.2021.117588.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cal K., Centkowska K. Use of cyclodextrins in topical formulations: Practical aspects. Eur. J. Pharm. Biopharm. 2008;68:467–478. doi: 10.1016/j.ejpb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Waleczek K.J., Cabral Marques H.M., Hempel B., Schmidt P.C. Phase solubility studies of pure (-)-α-bisabolol and camomile essential oil with β-cyclodextrin. Eur. J. Pharm. Biopharm. 2003;55:247–251. doi: 10.1016/S0939-6411(02)00166-2. [DOI] [PubMed] [Google Scholar]
- 3.Rawat S., Jain S.K. Solubility enhancement of celecoxib using β-cyclodextrin inclusion complexes. Eur. J. Pharm. Biopharm. 2004;57:263–267. doi: 10.1016/j.ejpb.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Fülöp Z., Kurkov S.V., Nielsen T.T., Larsen K.L., Loftsson T. Self-assembly of cyclodextrins: Formation of cyclodextrin polymer based nanoparticles. J. Drug Deliv. Sci. Technol. 2012;22:215–221. doi: 10.1016/S1773-2247(12)50032-8. [DOI] [Google Scholar]
- 5.M. Bastos, ed., Biocalorimetry: Foundations and Contemporary Approaches, 1st Editio, CRC Press, Taylor & Francis Group, Boca Raton, 2016. https://doi.org/10.1201/b20161.
- 6.Piñeiro Á., Muñoz E., Sabín J., Costas M., Bastos M., Velázquez-Campoy A., Garrido P.F., Dumas P., Ennifar E., García-Río L., Rial J., Pérez D., Fraga P., Rodríguez A., Cotelo C. AFFINImeter: A software to analyze molecular recognition processes from experimental data. Anal. Biochem. 2019;577:117–134. doi: 10.1016/j.ab.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 7.Slochower D.R., Henriksen N.M., Wang L.-P., Chodera J.D., Mobley D.L., Gilson M.K. Binding Thermodynamics of Host-Guest Systems with SMIRNOFF99Frosst 1.0.5 from the Open Force Field Initiative. J. Chem. Theory Comput. 2019;15:6225–6242. doi: 10.1021/acs.jctc.9b00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. Markthaler, S. Jakobtorweihen, N. Hansen, Lessons Learned from the Calculation of One-Dimensional Potentials of Mean Force [Article v1.0], Living J. Comput. Mol. Sci. 1 (2019) 11073. https://doi.org/10.33011/livecoms.1.2.11073.
- 9.Comer J., Gumbart J.C., Hénin J., Lelièvre T., Pohorille A., Chipot C. The Adaptive Biasing Force Method: Everything You Always Wanted To Know but Were Afraid To Ask. J. Phys. Chem. B. 2014;119:1129–1151. doi: 10.1021/jp506633n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nag K., Singh D., Shetti A., Kumar H., Sivashanmugam T., Parthasarathy S. Sugammadex: A revolutionary drug in neuromuscular pharmacology. Anesth. Essays Res. 2013;7:302–306. doi: 10.4103/0259-1162.123211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., Yan Y., Huang J., Petukhov A.V., Kroon-Batenburg L.M.J., Drechsler M., Zhou C., Tu M., Granick S., Jiang L. Giant capsids from lattice self-assembly of cyclodextrin complexes. Nat. Commun. 2017;8:15856. doi: 10.1038/ncomms15856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B. Gidwani, A. Vyas, A Comprehensive Review on Cyclodextrin-Based Carriers for Delivery of Chemotherapeutic Cytotoxic Anticancer Drugs, Biomed Res. Int. 2015 (2015), article ID Article ID 198268. https://doi.org/10.1155/2015/198268. [DOI] [PMC free article] [PubMed]
- 13.Saokham P., Sá Couto A., Ryzhakov A., Loftsson T. The self-assemble of natural cyclodextrins in aqueous solutions: Application of miniature permeation studies for critical aggregation concentration (cac) determinations. Int. J. Pharm. 2016;505:187–193. doi: 10.1016/j.ijpharm.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 14.Kurkov S.V., Loftsson T. Cyclodextrins. Int. J. Pharm. 2013;453:167–180. doi: 10.1016/j.ijpharm.2012.06.055. [DOI] [PubMed] [Google Scholar]
- 15.Stappaerts J., Do Thi T., Dominguez-Vega E., Somsen G.W., Van den Mooter G., Augustijns P. The impact of guest compounds on cyclodextrin aggregation behavior: A series of structurally related parabens. Int. J. Pharm. 2017;529:442–450. doi: 10.1016/j.ijpharm.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Zaman H., Bright A.G., Adams K., Goodall D.M., Forbes R.T. Characterisation of aggregates of cyclodextrin-drug complexes using Taylor Dispersion Analysis. Int. J. Pharm. 2017;522:98–109. doi: 10.1016/j.ijpharm.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 17.T.F.G.G. Cova, S.M.A. Cruz, A.J.M. Valente, P.E. Abreu, J.M.C. Marques, A.A.C.C. Pais, Aggregation of Cyclodextrins: Fundamental Issues and Applications, in: P. Arora, N. Dhingra (Eds.), Cyclodext. - A Versatile Ingred., IntechOpen, 2018. https://doi.org/10.5772/intechopen.73532.
- 18.Z. Bikádi, R. Kurdi, S. Balogh, J. Szemán, E. Hazai, Aggregation of Cyclodextrins as an Important Factor to Determine Their Complexation Behavior, Chem. Biodivers. 3 (2006) 1266–1278. https://doi.org/https://doi.org/10.1002/cbdv.200690129. [DOI] [PubMed]
- 19.Zhu X., Wu G., Chen D. Molecular dynamics simulation of cyclodextrin aggregation and extraction of Anthracene from non-aqueous liquid phase. J. Hazard. Mater. 2016;320:169–175. doi: 10.1016/j.jhazmat.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Mixcoha E., Campos-Terán J., Piñeiro Á. Surface Adsorption and Bulk Aggregation of Cyclodextrins by Computational Molecular Dynamics Simulations as a Function of Temperature: α-CD vs β-CD. J. Phys. Chem. B. 2014;118:6999–7011. doi: 10.1021/jp412533b. [DOI] [PubMed] [Google Scholar]
- 21.Loftsson T. Cyclodextrins in Parenteral Formulations. J. Pharm. Sci. 2021;110:654–664. doi: 10.1016/j.xphs.2020.10.026. [DOI] [PubMed] [Google Scholar]
- 22.C. Muankaew, T. Loftsson, Cyclodextrin-Based Formulations: A Non-Invasive Platform for Targeted Drug Delivery, Basic Clin. Pharmacol. Toxicol. 122 (2018) 46–55. https://doi.org/https://doi.org/10.1111/bcpt.12917. [DOI] [PubMed]
- 23.N. Larson, COMPOSITIONS COMPRISING AN RNA POLYMERASE INHIBITOR AND CYCLODEXTRIN FOR TREATING VIRAL INFECTIONS, US 2019 / 0083525 A1, 2019.
- 24.Á. Piñeiro, J. Pipkin, V. Antle, R. Garcia-Fandino, Remdesivir interactions with sulphobutylether-beta-cyclodextrin: A case study using selected substitution patterns, J. Mol. Liquids, (2021), in press, https://doi.org/10.1016/j.molliq.2021.117157. [DOI] [PMC free article] [PubMed]
- 25.Garrido P.F., Calvelo M., Garcia-Fandiño R., Piñeiro Á. Rings. Hexagons, Petals, and Dipolar Moment Sink-Sources: The Fanciful Behavior of Water around Cyclodextrin Complexes, Biomolecules. 2020;10:431. doi: 10.3390/biom10030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 27.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 28.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 29.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 30.Berendsen H.J.C., Postma J.P., van Gunsteren W.F., Hermans J. In: Intermol. Pullman B., editor. Forces; Dordrecht: 1981. Interaction Models for Water in Relation to Protein HydrationIn; pp. 331–342. [Google Scholar]
- 31.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:14101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 32.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 33.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 34.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 35.Hockney R.W., Eastwood J.W. Computer Simulation Using Particles. CRC Press. 1988 doi: 10.1201/9780367806934. [DOI] [Google Scholar]
- 36.Miyamoto S., Kollman P.A. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 37.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 38.N. Michaud-Agrawal, E.J. Denning, T.B. Woolf, O. Beckstein, MDAnalysis: A toolkit for the analysis of molecular dynamics simulations, J. Comput. Chem. 32 (2011) 2319–2327. https://doi.org/https://doi.org/10.1002/jcc.21787. [DOI] [PMC free article] [PubMed]
- 39.R.J. Gowers, M. Linke, J. Barnoud, T.J.E. Reddy, M.N. Melo, S.L. Seyler, J. Domański, D.L. Dotson, S. Buchoux, I.M. Kenney, O. Beckstein, MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations, Proceedings 15th Python Science Conference, (2016), 98–105. https://doi.org/10.25080/Majora-629e541a-00e.
- 40.Schrodinger L.L.C. The PyMOL Molecular Graphics System. Version. 2015;1:8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.