Abstract
Considering the worldwide health crisis associated with highly contagious severe respiratory disease of COVID-19 outbreak, the development of multiplexed, simple and rapid diagnostic platforms to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is in high demand. Here, a nucleic acid amplification-free electrochemical biosensor based on four-way junction (4-WJ) hybridization is presented for the detection of SARS-CoV-2. To form a 4-WJ structure, a Universal DNA-Hairpin (UDH) probe is hybridized with two adaptor strands and a SARS-CoV-2 RNA target. One of the adaptor strands is functionalized with a redox mediator that can be detected using an electrochemical biosensor. The biosensor could simultaneously detect 5.0 and 6.8 ag/μL of S and Orf1ab genes, respectively, within 1 h. The biosensor was evaluated with 21 clinical samples (16 positive and 5 negative). The results revealed a satisfactory agreement with qRT-PCR. In conclusion, this biosensor has the potential to be used as an on-site, real-time diagnostic test for COVID-19.
Keywords: COVID-19, Electrochemical biosensor, SARS-CoV-2 RNA, S gene, Orf1ab gene
1. Introduction
The global coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been surging rapidly all over the world. With over 3,937,437 deaths worldwide, by July 2021 (WHO Coronavirus (COVID-19) Dashboard, 2021), the world will be confronting a huge health, social, and economic burden. As a result, identifying and monitoring coronavirus infection is crucial to controlling the transmission of the disease and ultimately saving lives. Several diagnostic methods for COVID-19 disease detection have been developed or are being developed (Broughton et al., 2020; Fozouni et al., 2021; Guo et al., 2020; Hajian et al., 2019; Kim et al., 2021; Wang et al., 2020). Despite the improved availability of COVID-19 diagnostic tests, which has allowed for more extensive monitoring of disease transmission, many diagnostic challenges remain. Currently, the gold standard method for COVID-19 diagnosis is a diagnostic test based on real-time reverse transcription-polymerase chain reaction (RT-PCR) for viral RNA detection. While molecular diagnosis using real-time RT-PCR is able to provide sensitive as well as specific detection of SARS-CoV-2, it takes 4–5 h and necessitates the use of high-priced instruments and reagents, as well as skilled personnel (Torrente-Rodríguez et al., 2020; Yuan et al., 2020). Moreover, RT-PCR tests for SARS-CoV-2 detection have yielded a few false-negative results on confirmed infection patients (Xie et al., 2020; Yang et al., 2020). Besides, low viral concentrations in clinical samples collected from asymptomatic, mildly symptomatic, or convalescent patients may be inadequate for RT-PCR detection (Yuan et al., 2020). Alternatively, several loop-mediated isothermal amplification (LAMP) assays have been developed for rapid detection of COVID-19 in clinical samples (Lamb et al., 2020; Yu et al., 2020). However, LAMP assays can detect only one target gene. Moreover, false-positive results can be produced when homologous sequences are present in the clinical samples (Li et al., 2020). Additionally, LAMP results are normally confirmed by agarose gel electrophoresis and SYBR dyes. Gel electrophoresis is a tiresome and time-consuming technique, as well as the evaluation of color changes with the naked eye, is possibly subject to uncertain results (Li et al., 2020). Conversely, serological tests are rapid and require minimal equipment yet take several days to weeks following disease symptoms onset to mount a detectable antibody response (Zhang et al., 2020). Given the aforementioned challenges, there is an immediate and urgent requirement for rapid, accurate, specific, and cost-effective tests to identify COVID-19 at an early stage and, as a result, mitigate the risks of its spread. Furthermore, COVID-19 disease demands point-of-care testing (POCT) without the need for skilled staff and central laboratories due to its transmissible nature.
Biosensors, as powerful analytical tools in medical diagnostics, have attracted considerable attention to detecting and quantifying numerous targets such as diseases’ biomarkers (Kashefi-Kheyrabadi et al., 2018, 2020, 2021; Lee et al., 2020; Stobiecka et al., 2019). The application of biosensors in diagnostics offers various advantages over other diagnostic procedures, such as high sensitivity, simplicity, rapid response, and cost-effectiveness. Until now, a number of biosensors have been reported for the detection of SARS-CoV-2 (Alafeef et al., 2020; Eissa and Zourob, 2021; Kim et al., 2021; Zhao et al., 2021; Zhu et al., 2020). COVID-19 detection biosensors are classified into two types: immunological diagnostic biosensors and viral nucleic acid-based biosensors. Immunological diagnostic biosensors mainly focus on the detection of antibodies specific to SARS-CoV-2 antigens, developed in patients due to their exposure to the virus or the presence of viral antigens in biofluids (Eissa and Zourob, 2021; Kim et al., 2021; Seo et al., 2020; Torrente-Rodríguez et al., 2020; Zhang et al., 2020). Although immunosensors are rapid and require minimal equipment, it may take several days to weeks for a patient to produce a detectable antibody response after the onset of the symptoms. Thus, an antibody test could not be used to diagnose an individual with an active infection. The human immune system is not involved enough in the early stages of infection, which may lead to false-negative diagnoses. Furthermore, the variety of antibodies to detect various viral antigens is limited.
Alternatively, viral nucleic acid monitoring can be used to reliably diagnose an active COVID-19 infection. A number of nucleic acid-based biosensing systems have been developed for the detection of COVID-19 (Carter et al., 2020; Kim et al., 2021; Moitra et al., 2020; Qiu et al., 2020; Zhao et al., 2021; Zhu et al., 2020). Nucleic acid-based sensors are beneficial because they can detect SARS-CoV-2 in its early stages in a highly sensitive, specific, and rapid manner.
Among the nucleic acid-based biosensors currently available, electrochemical nucleic acid-based sensors possess several advantages, including instantaneous and multiplex analysis as well as ease of use for POC applications (Alafeef et al., 2020; Chaibun et al., 2021; Mahari et al., 2020; Seo et al., 2020; Torrente-Rodríguez et al., 2020). Although nucleic acid-based biosensors have demonstrated outstanding performance in DNA/RNA analysis, they may be limited in some ways by nucleic acid amplification methods, which complicate the operation.
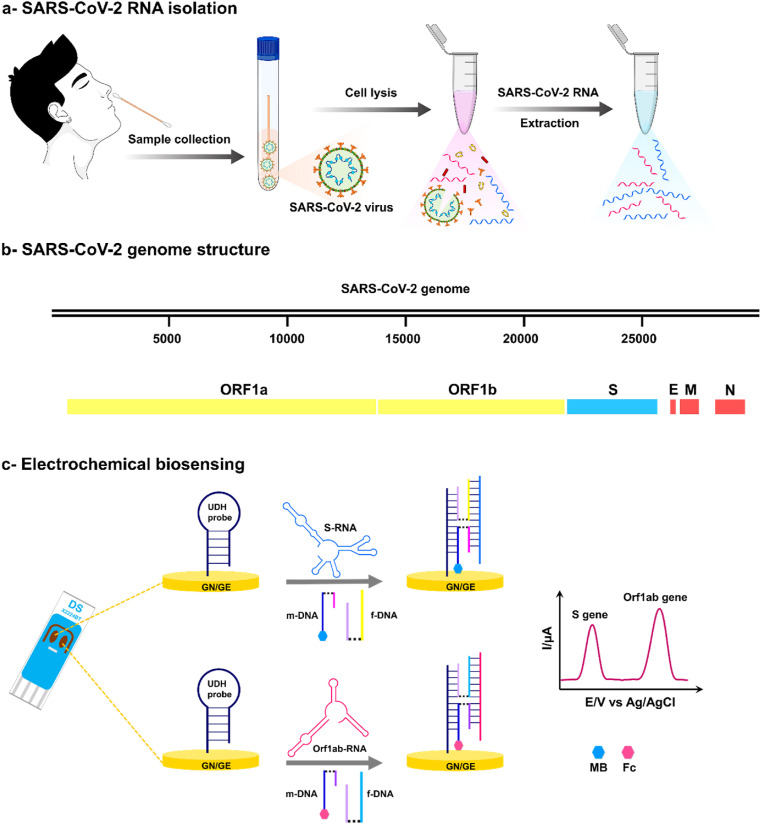
In this study, a nucleic acid amplification-free electrochemical biosensor based on four-way junction (4-WJ) hybridization is developed, which can simultaneously detect SARS-CoV-2 spike (S) and open reading frame (Orf1ab) genes within 1 h (Fig. 1 ). To form a 4-WJ structure, a Universal DNA-Hairpin (UDH) probe is hybridized with an RNA target and two adaptor DNA strands (m and f). One of the adaptor strands (m-strand) is modified with a redox mediator (methylene blue (MB) or ferrocene (Fc)) that can be detected using an electrochemical biosensor. To construct the biosensor, a thiolated UDH probe is immobilized onto a gold nanoneedle structured electrode using sulfur-gold chemistry, and then backfilled with the 6-mercapto-1-hexanol to avoid nonspecific adsorption. The biosensor is then incubated with a mixture solution of two adaptor sequences and the RNA target. In the presence of all biosensor components, the 4-WJ configuration forms, causing the redox mediator-labeled adaptor strand to approach the electrode surface, facilitating the electron transfer, and producing an electrochemical signal. The high selectivity of this strategy distinguishes it. The specificity is attributed to a short m-strand to RNA target hybrid, which allows it to simply bind to the fully complementary target fragment (Gerasimova et al., 2010; Kolpashchikov, 2010, Kolpashchikov, 2006). The biosensor can detect SARS-CoV-2 genes with high sensitivity (2 and 3 copies/μL for the S and Orf1ab genes, respectively) and high specificity even between closely related RNA sequences with a single nucleotide substitution within 1 h. The assay was also evaluated with clinical samples, depicting satisfactory agreement with qRT-PCR results.
Fig. 1.
Schematic of the detection platform. A detection workflow of SARS-CoV-2 RNA sequences from clinical samples using the electrochemical biosensor for detection of the S and Orf1ab genes.
2. Material and methods
2.1. Reagents and materials
Except for the S and Orf1ab RNA sequences, all oligonucleotides used in this study were custom-made by Integrated DNA Technologies (IDT Inc., USA). The oligonucleotide sequences utilized are shown in Table S1 (Supporting Information). Trizma hydrochloride solution (Tris-HCl), tris (2-carboxyethyl) phosphine hydrochloride (TCEP), 6-mercapto-1-hexanol (MCH), sodium chloride (NaCl), sodium hydroxide (NaOH), potassium chloride (KCl), sulfuric acid (H2SO4), acetone, isopropyl alcohol (IPA), ethanol, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES buffer; pH 7.4), potassium ferricyanide (K3[Fe(CN)6]), potassium ferrocyanide (K4[Fe(CN)6]), hexaammineruthenium (iii) chloride (RuHex), and magnesium chloride (MgCl2) were purchased from Sigma-Aldrich (USA). RNase-free phosphate-buffered saline (PBS) solution (10X) pH 7.4, UltraPure™ DNase/RNase-Free Distilled Water (DW), and RNase AWAY solution were provided by Thermo Fisher Scientific (USA). Fivephoton Biochemicals (USA) supplied the ferrocene carboxylic N-hydroxysuccinimide ester (Fc). AmpliScribe™ T7 High Yield Transcription Kit was purchased from Lucigen (USA). TRIzol LS reagent was purchased from Life technology Inc. (USA).
The stock solution of all oligonucleotides was prepared by dissolving its lyophilized powder in UltraPure™ DNase/RNase-Free Distilled Water, and then diluted to the desired concentrations using buffer, prior to use. The immobilization buffer (IB) was prepared using 5 × 10−2 M Tris-HCl and 2.5 × 10−1 M NaCl and adjusted to a pH of 7.4. The hybridization buffer (HB) was prepared using 5 × 10−2 M Tris-HCl, 1 × 10−1 M NaCl, and 5 × 10−2 M MgCl2 and adjusted to a pH 7.4. As a washing buffer (WB), a 1X PBS pH 7.4 solution was used. Metrohm DropSens Inc. (Spain) supplied screen printed gold electrodes (SPGEs) (C220BT and X2224BT).
2.2. Instrumentation
Field emission-scanning electron microscopy (FE-SEM) imaging and Energy Dispersive X-Ray (EDX) analysis were performed with the Carl Zeiss-Sigma instrument (Carl Zeiss, Germany) at an accelerating voltage of 20 kV. Electrochemical experiments were carried out using CHI1030C and CHI-660E (CH Instruments, USA) electrochemical analyzers. For the analysis of clinical samples, the LightCycler® 480 II PCR system (Roche, Basel, Switzerland) was used.
2.3. Electrochemical measurements
All electrochemical measurements were performed in a three-electrode system using dual screen-printed electrodes (SPGEs) on the CHI instruments. Cyclic voltammetry (CV) was carried out in a K3[Fe (CN)6] solution (5 × 10−3 M) containing 1 × 10−1 M KCl at a scan rate of 0.1 V s−1. Electrochemical impedance spectroscopy (EIS) experiments were performed in an equimolar K3[Fe (CN)6]/K4[Fe(CN)6] solution (1:1, 5 × 10−3 M) containing 1 × 10−1 M KCl. The Nyquist plots were recorded under a constant potential of 0.21 V and AC potential of 0.005 V over the frequency range (10 kHz–0.1 Hz). The surface density of the UDH probe on the electrode surface was determined using chronocoulometry in the presence of 2 × 10−4 M RuHex solution. Chronocoulometry experiments were performed at a potential step of 0.5 V (+0.1 to −0.4 V) with a pulse width of 0.5 s and a pulse interval of 0.0025 s. Square-wave Voltammetry (SWV) measurements were recorded in HB at a potential range from −0.6 to +0.6 V, frequency of 15 Hz, amplitude of 2.5 × 10−2 V, and step potential of 5 × 10−3 V. All the measurements were performed at least in triplicate at room temperature (RT).
2.4. Fabrication of biosensor platform
Dual SPGEs (Ref. X2224BT) were applied as a substrate for immobilizing the UDH probe. Before analysis, the electrodes were cleaned by rinsing sequentially using acetone, IPA, ethanol, and deionized water for at least 30 s and dried with a flow of nitrogen gas, followed by Oxygen plasma treatment at 100 W for 20 s to produce a hydrophilic surface. They were then activated by potential cycling over the range (−1.2 to 1.2) V at a scan rate of 0.1 V/s in 0.1 M NaOH solution and over the range (0–1.1) V at a scan rate of 0.1 V/s in 0.5 M H2SO4 solution, respectively. The SPGEs were then electrodeposited by gold solutions to create 3D gold nanoneedle structured surface. They were electroplated at −0.4 V for 300 s in a solution of 1.6 × 10−1 M HAuCl4 in 0.5 M H2SO4 and in a solution of 2 × 10−2 M HAuCl4 in 0.5 M HClO4 at −0.4 V for 30 s, respectively. Next, a thiolated UDH probe (1 × 10−6 M) prepared in the IB was activated using TCEP solution (1 × 10−2 M) for 1 h at RT. After that, 3 μL of the UDH probe was immobilized onto the gold nanoneedle structured SPGEs (GN/SPGEs) overnight at 4°C in a humid environment. The as prepared UDH probe modified GN/SPGEs were then thoroughly rinsed using WB and dried with nitrogen gas flow followed by incubation with MCH solution (1 × 10−5 M in 1X PBS pH 7.4) for 10 min to passivate nonspecific adsorptions. The 4-WJ structure was eventually formed by hybridizing the UDH probe with other biosensor components including 2.5 × 10−7 M redox mediator-labeled m-strand, 5 × 10−7 M associated f-strand, and various concentrations of the RNA targets for 30 min. Synthetic COVID-19 RNA oligos (S or Orf1ab gene) or COVID-19 RNA amplicon extracted from clinical samples were used as the RNA target.
2.5. RNA in vitro transcription
The S and Orf1ab RNA sequences were produced using T7 transcription from synthesized DNA oligos (Fig. S6). A T7 promoter was added to each DNA template sequence prior to in vitro transcription with the aid of a PCR primer set (Table S2). The S and Orf1ab gene DNA sequences were then transcribed using AmpliScribe™ High Yield Transcription kit according to the manufacturer’s instruction. Briefly, all reaction components, save the AmpliScribe T7 RNA Polymerase, were first brought to RT. Except for the AmpliScribe T7 RNA Polymerase, the appropriate volume of each reaction component was combined and mixed in the following order: 6.5 μL sterilized Nuclease-Free Water, 1 μg template DNA with appropriate promoter, 2 μL AmpliScribe T7 10X Reaction Buffer, 1.5 μL 1 × 10−1 M of each nucleotides (ATP, CTP, GTP, UTP), 2 μL 1 × 10−1 M DTT, and 0.5 μL RiboGuard RNase Inhibitor. Next, 2 μL AmpliScribe T7 RNA Polymerase was added and mixed. The resulting mixture was then incubated for 3 h at 42 °C with interval inversion of the mixture. After the reaction completed, 5 × 10−2 M EDTA was added to the mixtures to remove magnesium pyrophosphate formed. The mixture was then treated with RNase-Free DNase I to remove DNA template. The RNA products were ultimately purified using TRIzol LS reagent. The purified RNA transcripts were stored at −80 °C for the further use.
2.6. Biosensor characterization
The characterization of the electrochemical biosensor was initiated using synthetic SARS-CoV-2 RNA fragments. The SARS-CoV-2 RNA sequences were produced using T7 transcription from synthesized DNA oligos (previous section). The UDH probe-modified GN/SPGEs were first incubated with the HB solution containing the associated f-strand (5 × 10−7 M), m-strand (2.5 × 10−7 M), and RNA target (5 × 10−8 M). To achieve an outstanding analytical performance, various experimental parameters were first optimized such as the concentration of UDH probe immobilized on the GN/SPGE as well as the time required for the adaptor strands and the RNA target to hybridize with the UDH probe. To optimize the UDH probe concentration, different concentrations of the UDH probe (0.01, 0.1, 0.25, 0.5, 1, 2, and 5 × 10−6 M) were immobilized on the electrode surface while the quantities of the other components remained constant. Following that, the oxidation peak currents of redox markers were measured. Using chronocoulometry, the surface density of the UDH probe at its optimum concentration was then calculated. (Steel, Adam B. et al., 1998).
The hybridization time was appraised at various intervals of 1, 5, 15, 30, 60, 90, and 120 min. The concentration of the UDH probe, as well as the concentration of other components of the biosensor, were kept constant while optimizing the hybridization time.
Experiments with one or more missing components were also carried out as controls. The control samples lacked one or more of the following components: (1) UDH, (2) m-strand, (2) f-strand, and (4) RNA target. Calibration curves were obtained in the presence of both COVID-19 RNA oligos (S, and Orf1ab genes) over a range (1 × 10−16 to 1 × 10−11 M). The specificity of the biosensor was evaluated using synthesized mismatched COVID-19 RNA sequences (1 × 10−12 M), as well as influenza RNA sequences isolated from clinical samples. Finally, the performance of the biosensor was evaluated for the detection of the COVID-19 RNA fragments in clinical samples.
2.7. Detection of SARS-CoV-2 from clinical samples
To verify the performance of the biosensor, RNA samples extracted from 21 anonymized respiratory clinical were used as the RNA target in the 4-WJ platform and detected using the electrochemical biosensor. Samkwang Medical Laboratories (Seoul, South Korea) provided the SARS-CoV-2 positive samples in the form of purified RNA under IRB code number S-IRB-2020-029-09-17. They were ready-to-use samples that did not require pre-treatment before use. Furthermore, isolated RNA samples were subjected to RT-qPCR analysis to be compared with the electrochemical results. The RT-qPCR analysis was carried out for Orflab gene primers and probe sets as well as S gene primers and probe sets (Park et al., 2020; Zhen and Berry, 2020) using TaqPath™ 1-Step RT-qPCR Master Mix (ThermoFisher Scientific, Applied Biosystems™, USA), according to the manufacturer’s protocol. The RT-qPCR system was set at 25°C for 2 min, 50°C for 15 min, 95°C for 2 min, followed by 45 cycles at 95°C for 15 s and 55°C–60°C (S gene – Orflab gene) for 30 s.
To assess the method’s functionality, a total of 21 respiratory clinical samples were used. The 16 SARS-CoV-2-positive clinical samples yielded positive results, while the 5 SARS-CoV-2-negative clinical samples produced negative results. A few specimens containing RNA sequences from other respiratory diseases, such as Influenza A and B, were also examined to assess biosensor specificity.
3. Results and discussion
3.1. The biosensing assay principle
In the current research, an electrochemical biosensor was developed to simultaneously detect S and Orf1ab gene fragments of SARS-CoV-2 RNA using the 4-WJ hybridization platform (Fig. 1). A Universal DNA-Hairpin (UDH) probe is immobilized onto a gold nanoneedle structured SPGE (GN/SPGE) via sulfur-gold chemistry, then hybridized to the two adaptor strands (m and f) and an RNA target to form the biosensor's skeleton. Both m- and f-adaptor strands contain fragments complementary to the UDH (UDH binding arm) and the SARS-CoV-2 RNA (analyte binding arm). The analyte binding arm and the UDH binding arm are linked through a triethylene glycol spacer. One of the adaptor strands (m-strand) was labeled with a redox marker (MB for detection of S gene and Fc for detection of Orf1ab gene, respectively).
In the presence of the biosensor components (i.e., RNA target, m- and f-strands), the UDH probe switches from a hairpin to a straight strand, and the adaptor strands are attached to both the UDH and the RNA target. The redox markers approach the electrode surface due to the creation of the 4-WJ structure. The electron transfer between the redox probes and the electrode surface is facilitated by this phenomenon, resulting in a detectable electrochemical signal. It should be noted that the f- and m-strand sequences were designed to identify SARS-CoV-2 RNA fragments specifically (Table S1).
The m strand's analyte binding arm is 10-mer long and only hybridizes to the completely matched RNA target fragment. The biosensor's high specificity is ensured by the m-strand feature, which allows it to distinguish even single-base substitutions in RNA target sequences. The f-strand with a longer target-binding arm, alternatively, effectively unfolds the secondary structure of the RNA target (Kolpashchikov, 2006; Labib et al., 2015).
3.2. The impact of biosensor surface morphology on detection efficiency
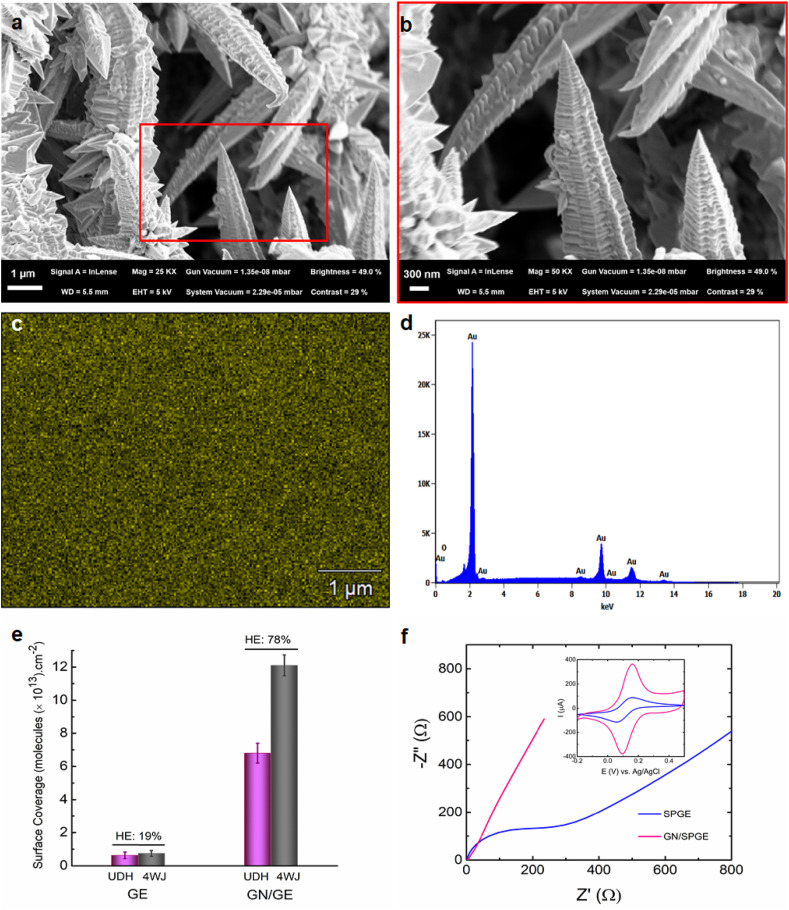
Analysis of clinical samples using the nucleic acid-based biosensors has often encountered significant obstacles, such as poor sensitivity and low hybridization process efficiencies on the surface of bulk electrodes. The use of gold nanoneedle structured electrodes offered a way to address the aforementioned issues by either increasing the surface area of the electrodes, thereby increasing the capture sites, or enhancing the accessibility of the probes immobilized on the nanoneedles surface, resulting in more efficient and rapid analyte binding (Bin et al., 2010). Here, 3D gold nanoneedles were constructed on the electrode surface with the aid of electrodeposition. The morphological structure of the gold nanoneedles was examined using FE-SEM. As shown in Fig. 2 a, the morphology features needle structures. The needles are wrinkled heavily, depicting a pattern of peaks and valleys running the length of them. Fig. 2b displays the nanoneedles at different magnification. The average distance between peaks ranges from 50 to 120 nm. Fig. 2c and d shows the distribution of gold on the electrode as well as its EDX analysis spectrum. Based on elemental mapping, the gold element clearly dominates, with a percentage of Au atoms of 99.3%. The development of a 3D hierarchical nanotextured electrode considerably increased the surface coverage of UDH probe molecules on the electrode surface as well as the hybridization process efficiency to form a 4-WJ structure (Fig. 2e). The UDH probe surface density and hybridization efficiency were quantified using a chronocoulometric assay based on electrostatic adsorption of RuHex on the UDH probe-modified electrodes (Steel, Adam B et al., 1998). As shown in Fig. S1a, the observed charge for the UDH-modified GN/SPGE differs significantly from that of the smoothed electrode (SPGE). The UDH probe density on the GN/SPGE ((6.8 ± 0.87) × 1013 molecules/cm2) enhanced 10-fold compared to that on the SPGE ((6.2 ± 0.68) × 1012 molecules/cm2). This growth could be attributed to the presence of fine nanoneedles, which increase the deflection angle between the probe molecules, allowing them to form more compact and uniform monolayers. As a result of the increased deflection angle, RNA target molecules have better access to the UDH probes, resulting in increased hybridization efficiency (78% and 19% for GN/SPGE and SPGE, respectively) Fig. 2e and S1b.
Fig. 2.
FE-SEM images of (a) 3D gold nanoneedle structures, (b) Magnified image from the boxed region in (a), (c) EDX image of 3D gold nanoneedles, (d) EDX spectrum of 3D gold nanoneedles, (e) The surface coverage of the UDH probe molecules and significantly improved hybridization efficiency of the 4-WJ structure using a chronocoulometric assay, (f) Nyquist plots obtained for GN/SPGE (pink) and SPGE (blue); the corresponding cyclic voltammograms of GN/SPGE and SPGE were represented as insets to the Nyquist plots. CV was carried out in a K3[Fe (CN)6] solution (5 × 10−3 M) containing 1 × 10−1 M KCl at a scan rate of 0.1 V s−1. Nyquist plots were recorded in an equimolar K3[Fe(CN)6]/K4[Fe(CN)6] solution (1:1, 5 × 10−3 M) containing 1 × 10−1 M KCl at a constant potential of 0.21 V and AC potential of 0.005 V over the frequency range of 10 kHz to 0.1 Hz. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, the electrochemical activity of the GN/SPGE was determined and compared to that of the SPGE using CV and EIS techniques (Fig. 2f). The Nyquist plots represent charge transfer resistance of 335 Ω for SPGE while barely a semicircle can be observed for the GN/SPGE, indicating that the gold nanoneedle structured electrode has higher electron conductivity. Similarly, the cyclic voltammograms show significantly higher faradic currents for the GN/SPGE compared to the SPGE. Besides, the real surface area of both electrodes was calculated using CV (Li et al., 2007). As it was represented in Fig. S2, the real surface area of the GN/SPGE was 12 folds greater than that of the SPGE.
3.3. Biosensor response
The detection performance of the biosensor was initially evaluated using two different types of synthetic SARS-CoV-2 RNA targets including a 177-mer (S gene) and a 159-mer (Orf1ab gene) RNA fragments. Following that, the feasibility of the sensing platform for multiplexed quantification of the SARS-CoV-2 RNA sequences in clinical samples was investigated.
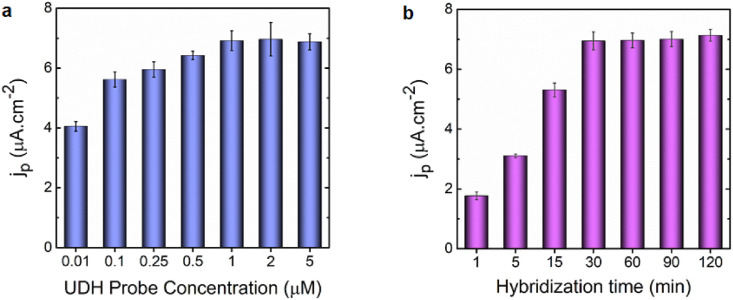
The optimum concentration of the UDH probe immobilized on the GN/SPGE was quantified using the square-wave voltammetry (SWV). The square-wave voltammograms demonstrated that the immobilization of the UDH probe rose with increasing concentration up to 1.0 × 10−6 M (Fig. 3 a), and this optimum concentration was thus applied for further experiments. By increasing the concentration above 1.0 × 10−6 M, the UDH probe immobilization on the electrode surface reached a plateau, and the electrochemical signal leveled out.
Fig. 3.
Biosensor response (a) at various concentrations of the UDH probe (0.01, 0.1, 0.25, 0.5, 1, 2, and 5) × 10−6 M and fixed concentrations of m- and f- and RNA target strands with hybridization time of 30 min. (b) at varied hybridization times (1, 5, 15, 30, 60, 90 and 120) minutes and fixed concentrations of UDH probe, m- and f-, and RNA target strands. All measurements were taken in triplicate, and the error bars represent standard deviations for independent measurements (n = 3).
The time required for the hybridization process on the UDH probe modified electrode was then optimized. As can be seen in Fig. 3b, after incubating the adaptor strands along with RNA targets with the UDH probe modified electrode, the electrochemical signal could be measured nearly on the spot (within 1 min). Increasing the hybridization period to 30 min, however, resulted in a dramatic increase in the target-induced response (a 10-fold increase over the blank signal). However, the signal practically stabilized at a longer hybridization time than 30 min; as a result, the time of 30 min was chosen for subsequent experiments. In the following phase, the control experiments were performed to validate the response signal in the absence of RNA target and/or other elements of the 4-WJ structure.
3.4. Analytical performances of the biosensor
SWV generally offers the best choice of all pulse methods for practical analysis, since it provides background suppression with the same efficacy as differential pulse voltammetry (DPV), higher sensitivity than DPV, much faster scanning times, and applicability to a broader variety of electrode material and systems (Bard and Faulkner, 2001). The biosensing platform's detection limit and sensitivity can be greatly enhanced by combining the best aspects of the SWV method (i.e., background suppression and high detection sensitivity) with the nanoscale roughness of a gold nanoneedles electrode.
In the current study, a multiplexed detection of SARS-CoV-2 RNA sequences, which are the S and Orf1ab genes, was performed using an electrochemical biosensor. For laboratory confirmation of cases by nucleic acid testing in areas where COVID-19 virus transmission is uncertain, the World Health Organization (WHO) recommends that the test be positive for at least two separate targets on the COVID-19 virus genome (World Health, 2020). Furthermore, since the SARS-CoV-2 RNA virus has a high mutation rate (Pachetti et al., 2020), to minimize the risk of false-negatives due to mutation in the target genes, it is advised that a SARS-CoV-2 diagnostic test include at least two targets.
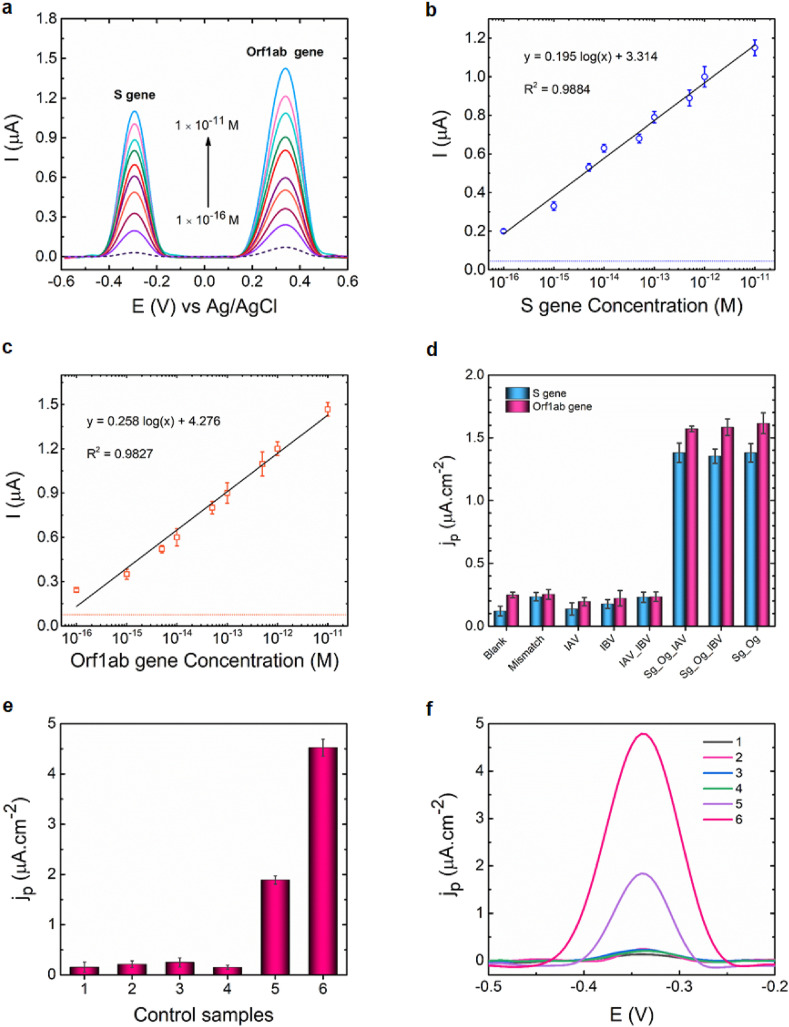
Fig. 4a depicts the SWVs in the presence of various concentrations of SARS-CoV-2 RNA target sequences. The electro-oxidation of MB at −0.29 V and Fc at +0.3 V produced two perfectly distinct peaks, which were attributed to the identification of the S and the Orf1ab genes, respectively. As can be observed, the oxidation currents of MB and Fc increased progressively with rising S and Orf1ab genes concentrations from 1 × 10−16 to 1 × 10−11 M, owing to the hybridization of more RNA target strands with the corresponding UDH probe molecules immobilized on the sensing surfaces. The slight difference in current could be attributed to the different structures of the redox probes. Because redox labels have different structures, their configurations to the electrode differ. As for Fc, it was confined close to the electrode surface, resulting in a higher electrochemical signal, whereas MB was kept relatively far from the electrode surface, causing steric hindrance to electron transfer between the MB and the electrode, resulting in lower current.
Fig. 4.
Biosensor performance. (a) The SWVs represent the increase in the current signal as the concentrations of the S and Orf1ab genes increased. The blank signal is represented by a dashed line. Sensitivity assay for (b) S gene and (c) Orf1ab gene demonstrates a positive correlation in current response to the concentration for both viral genes. The blank signal is represented by dashed lines. (d) The specificity of the biosensor with S (blue bar) and Orf1ab (pink bar) genes, examined using 1 × 10−12 M perfect complementary targets (Sg and Og), 1 × 10−12 M mismatched RNA targets (Mismatch), and non-complementary targets (IAV, IBV) extracted from clinical samples. Biosensor response (e) in the presence of (1) m-, f-strands and S gene (absence of UDH), (2) UDH probe, f strand, and S gene (absence of m strand), (3) UDH probe, m strand, and S gene (absence of f strand), (4) UDH probe, m- and f-strands (absence of target), and UDH probe, m- and f-strands and S gene at (5) 1 × 10−15 M and (6) 1 × 10−13 M. The fixed concentrations of UDH probe, m- and f-strands and RNA target are 1 × 10−6 M, 2.5 × 10−7, 5.0 × 10−7, and 5 × 10−8 M, respectively. (f) The SWVs associated with the control assay, which was represented in Fig. 4e. SWV measurements were recorded in HB at a potential range from −0.6 to +0.6 V, frequency of 15 Hz, amplitude of 2.5 × 10−2 V, and step potential of 5 × 10−3 V. All current responses are represented as mean values ± SD (n = 4). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As can be seen in Fig. 4b and c, the developed biosensor demonstrated superior analytical performance for detection of both SARS-CoV-2 RNA sequences at concentrations ranging from 1 × 10−16 to 1 × 10−11 M. The limits of detection (LODs) for S and Orf1ab genes were determined 5.0 and 6.8 ag/μL, respectively.
There are also a few reported assays targeting S and Orf1ab genes separately using different techniques (Chaibun et al., 2021; Hajian et al., 2019; Patchsung et al., 2020; Yan et al., 2020) (Table S3). Furthermore, other diagnostic tests have been reported to target other commonly used gene fragments (i.e., RdRp, E, and N) for detecting SARS-CoV-2 (Broughton et al., 2020; Qiu et al., 2020; Seo et al., 2020; Zhu et al., 2020). Among the recently reported methods, CRISPR-based assays have piqued the interest of scientists. Although this technology has impressive features such as high sensitivity, specificity, low LOD and rapid response, it requires nucleic acids amplification, costly equipment and highly skilled personnel (Ackerman et al., 2020; Broughton et al., 2020; Huang, Z. et al., 2020; Patchsung et al., 2020; Qiu et al., 2020; Zhang et al., 2021), limiting its application in resource-constrained areas. The reverse transcription loop-mediated isothermal amplification method is another widely reported method (RT-LAMP) (Huang, W.E. et al., 2020; Yan et al., 2020). However, while this technique has a short reaction time and high specificity, it suffers from carry-over contamination, which results in false positive results. Several simple and sensitive protocols for detecting a single target have also been reported (Jiao et al., 2020; Tian et al., 2020; Yao et al., 2021; Zhao et al., 2021). Given that nucleic acid testing must be positive for at least two different targets on the COVID-19 virus genome for laboratory confirmation of cases, single detection procedures increase the possibility of false-negative results. Another recently published study developed an electrochemical biosensor for detecting N and S genes (Chaibun et al., 2021). Despite the biosensor's great sensitivity to its targets, it requires rolling circle amplification (RCA) before detection, that in turn complicates the detection procedure and lengthens the assay time. Furthermore, N and S genes could be detected separately using the biosensor. In contrast to previously described assays, our assay can be performed utilizing a nucleic acid amplification-free strategy in a single step with minimal reagents and can simultaneously detect its RNA targets. Moreover, due to multiplexed detection of SARS-CoV-2 RNA targets, the probability of false-negatives is reduced. Furthermore, the LOD for both the S and Orf1ab genes is satisfactorily comparable to test kits and current biosensors (Table S3).
3.5. Evaluation of the biosensor for in vitro diagnosis
COVID-19 and influenza are both infectious respiratory diseases with symptoms that are almost identical. As a result, a reliable diagnostic test capable of distinguishing COVID-19 RNA from influenza RNA is in high demand to prevent misdiagnosis. To evaluate the accuracy of the biosensor response, the RNA fragments isolated from Influenza A (IAV) and Influenza B (IBV) clinical samples were examined as non-complementary targets. Furthermore, the biosensor was challenged with 1 × 10−12 M synthetic mismatched COVID-19 target sequences (mismatched S and Orf1ab genes). As can be seen in Fig. 4d and Fig. S3, the electrochemical signals for all non-complementary targets (IAV, IBV, and IAV + IBV) were less than or equivalent to blank signal + 3SD (blank signal) and differed significantly from complementary targets for both S and Orf1ab genes. Additionally, the biosensor’s capability to distinguish between perfectly matched and single nucleotide substitution targets reveals its high specificity. As a result, even in the presence of other viruses' genes that cause similar respiratory symptoms, this platform may be useful for detecting SARS-CoV-2. As illustrated in Fig. 4e and f, the absence of any component required to create the 4-WJ complex resulted in an insignificant signal, whereas the presence of all sequences resulted in a remarkable signal.
The biosensor stability was also investigated. As shown in Fig. S4, it could be plainly seen that the biosensor was adequately stable for 2 weeks. The biosensor's reproducibility for both viral genes was also investigated. Across the entire calibration curve, the relative standard deviation (RSD) values for both the S and Orf1ab genes were less than or equal to 10%. For example, the intra-day reproducibility for concentrations of 1 × 10−15 and 1 × 10−13 M S gene was 7.7% and 3.6%, respectively, while the inter-day reproducibility was 8.4% and 7.0%. The intra-day reproducibility for concentrations of 1 × 10−15 and 1 × 10−13 M Orf1ab gene was 9.9% and 8.7%, respectively, while the inter-day reproducibility was 9.9% and 9.7%, as shown in Fig. S5.
3.6. Biosensor validation for COVID-19 diagnosis in clinical samples
The clinical diagnostics feasibility of the developed biosensor was evaluated using analyzing the RNA samples isolated from nasopharyngeal swab specimens of patients.
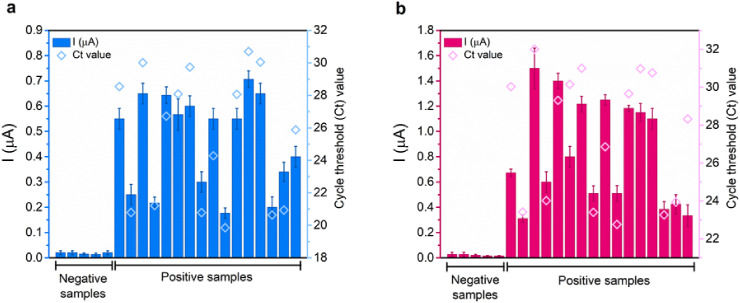
The RNA sequences isolated from clinical specimens were employed as target RNAs in 4-WJ structure for electrochemical detection. In total, 21 clinical samples were assessed, including 5 SARS-CoV-2-negative and 16 SARS-CoV-2-positive samples. The SARS-CoV-2-positive samples generated positive results, while the SARS-CoV-2-negative samples yielded negative results. The electrochemical biosensor evidently distinguished between positive and negative samples (Fig. 5 ). According to previous investigations, the viral load of SARS-CoV-2 in various types of respiratory specimens collected from infected individuals at different stages of infection ranged from 641 copies. mL−1 to 1.34 × 1011 copies. mL−1 (Pan et al., 2020). As a result, the LOD of our biosensor for both viral genes (S and Orf1ab genes), which is 5.0 and 6.8 ag/μL, respectively, is lower than the viral load in clinical samples. This demonstrates that our biosensor satisfies the sensitivity criteria and could be utilized to detect SARS-CoV-2 RNA fragments in the early stages of the disease when the viral load is still low. In addition, the results obtained from the electrochemical biosensor were compared with the RT-PCR results as a standard method, demonstrating a good correlation between the two methods. The correlation study justifies the biosensor’s application as an alternative for COVID-19 detection assays.
Fig. 5.
The detection of (a) S gene and (b) Orf1ab gene in 21 clinical specimens using SWV and RT-PCR. The samples were collected using nasopharyngeal swabs. The electrochemical signals are depicted in bar charts for the S gene (blue bar chart) and the Orf1ab gene (wine bar chart). The RT-PCR Ct values are represented in blue hollow diamond (S gene) and wine hollow diamond (Orf1ab gene). All electrochemical responses are represented as mean values ± SD (n = 3), and RT-PCR Ct values are from single measurements. SWV measurements were recorded in HB at a potential range from −0.6 to +0.6 V, frequency of 15 Hz, amplitude of 2.5 × 10−2 V, and step potential of 5 × 10−3 V. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
In the current study, the development of a nanoneedle structured electrochemical biosensor based on 4-WJ hybridization strategy was presented for the highly sensitive and specific detection of SARS-CoV-2. The S and Orf1ab genes were detected in both synthetic and clinical samples thanks to signal amplification capability provided by nanotextured electrodes and high sensitivity of the 4-WJ based electrochemical detection method. This approach has the following advantages: (i) multiplexed detection that avoids the generation of false-negative results; (ii) high specificity and ability to differentiate between closely related RNA target sequences down to single nucleotide substitution; (iii) a single step procedure and short assay period; (iv) low LOD that satisfies sensitivity requirement and could potentially be used to detect SARS-CoV-2 RNA targets in the early stages of the disease while the viral genes load is low.
CRediT authorship contribution statement
Leila Kashefi-Kheyrabadi: Conceptualization, Methodology, Validation, Visualization, Formal analysis, Writing – review & editing. Huynh Vu Nguyen: Methodology, Validation, Visualization, Formal analysis, Writing – review & editing. Anna Go: Methodology. Changyoon Baek: Methodology. Nayoon Jang: Methodology, Validation. Jiwon Michelle Lee: Visualization. Nam-Hyuk Cho: Validation. Junhong Min: Validation, Writing – review & editing. Min-Ho Lee: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by a grant from the Korea government’s National Research Foundation (NRF) (No.2020R1A5A1018052) and another funding from the Ministry of Trade, Industry and Energy (Grant no. 20008763).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113649.
Appendix A. Supporting Information
Supplementary data associated with this 1 article can be found in the online version.
Appendix B. Supplementary data
The following is the Supplementary data to this article:
References
- Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., Ye S.H., Boehm C.K., Kosoko-Thoroddsen T.-S.F., Kehe J., Nguyen T.G., Carter A., Kulesa A., Barnes J.R., Dugan V.G., Hung D.T., Blainey P.C., Sabeti P.C. Nature. 2020;582(7811):277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Dighe K., Moitra P., Pan D. ACS Nano. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard A.J., Faulkner L.R. Electroactive Layers and Modified Electrodes. John Wiley & Sons, Inc.; New York: 2001. Electrochemical methods: fundamentals and applications; pp. 580–632. [Google Scholar]
- Bin X., Sargent E.H., Kelley S.O. Anal. Chem. 2010;82(14):5928–5931. doi: 10.1021/ac101164n. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Nat. Commun. 2021;12(1):802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S., Zourob M. Anal. Chem. 2021;93(3):1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.-L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.-Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova Y.V., Hayson A., Ballantyne J., Kolpashchikov D.M. Chembiochem. 2010;11(12):1762–1768. doi: 10.1002/cbic.201000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. Cell Discov. 2020;6(1):34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian R., Balderston S., Tran T., deBoer T., Etienne J., Sandhu M., Wauford N.A., Chung J.-Y., Nokes J., Athaiya M., Paredes J., Peytavi R., Goldsmith B., Murthy N., Conboy I.M., Aran K. Nat. Biomed. Eng. 2019;3(6):427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. Biosens. Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi-Kheyrabadi L., Kim J., Chakravarty S., Park S., Gwak H., Kim S.-I., Mohammadniaei M., Lee M.-H., Hyun K.-A., Jung H.-I. Biosens. Bioelectron. 2020;169:112622. doi: 10.1016/j.bios.2020.112622. [DOI] [PubMed] [Google Scholar]
- Kashefi-Kheyrabadi L., Kim J., Gwak H., Hyun K.-A., Bae N.H., Lee S.J., Jung H.-I. Biosens. Bioelectron. 2018;117:457–463. doi: 10.1016/j.bios.2018.06.036. [DOI] [PubMed] [Google Scholar]
- Kashefi-Kheyrabadi L., Koyappayil A., Kim T., Cheon Y.-P., Lee M.-H. Bioelectrochemistry. 2021;137:107674. doi: 10.1016/j.bioelechem.2020.107674. [DOI] [PubMed] [Google Scholar]
- Kim K., Kashefi-Kheyrabadi L., Joung Y., Kim K., Dang H., Chavan S.G., Lee M.-H., Choo J. Sens. Actuators B Chem. 2021;329:129214. doi: 10.1016/j.snb.2020.129214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov D.M. J. Am. Chem. Soc. 2006;128(32):10625–10628. doi: 10.1021/ja0628093. [DOI] [PubMed] [Google Scholar]
- Kolpashchikov D.M. Chem. Rev. 2010;110(8):4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- Labib M., Khan N., Berezovski M.V. Anal. Chem. 2015;87(2):1395–1403. doi: 10.1021/ac504331c. [DOI] [PubMed] [Google Scholar]
- Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. PloS One. 2020;15(6) doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Yoon T., Yang H.-s., Cha S., Cheon Y.-P., Kashefi-Kheyrabadi L., Jung H.-I. Lab Chip. 2020;20(2):320–331. doi: 10.1039/c9lc01024f. [DOI] [PubMed] [Google Scholar]
- Li S., Jiang W., Huang J., Liu Y., Ren L., Zhuang L., Zheng Q., Wang M., Yang R., Zeng Y., Wang Y. Eur. Respir. J. 2020;56(6):2002060. doi: 10.1183/13993003.02060-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Song Y.-Y., Yang C., Xia X.-H. Electrochem. Commun. 2007;9(5):981–988. [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. BioRxiv. 2020 doi: 10.1101/2020.04.24.059204. [DOI] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. J. Transl. Med. 2020;18(1):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Won J., Choi B.Y., Lee C.J. Exp. Mol. Med. 2020;52(6):963–977. doi: 10.1038/s12276-020-0452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Nat. Biomed. Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Steel A.B., Herne T.M., Tarlov M.J. Anal. Chem. 1998;70(22):4670–4677. doi: 10.1021/ac980037q. [DOI] [PubMed] [Google Scholar]
- Stobiecka M., Ratajczak K., Jakiela S. Biosens. Bioelectron. 2019;137:58–71. doi: 10.1016/j.bios.2019.04.060. [DOI] [PubMed] [Google Scholar]
- Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3(6):1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. J. Am. Med. Assoc. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2020. Laboratory Testing for Coronavirus Disease (COVID-19) in Suspected Human Cases: Interim Guidance, 19 March 2020. [Google Scholar]
- World Health Organization WHO Coronavirus (COVID-19) Dashboard. 2021 https://covid19.who.int/ URL. [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J.J.M. medRxiv. 2020 doi: 10.1101/2020.02.11.20021493. [DOI] [Google Scholar]
- Yao B., Zhang J., Fan Z., Ding Y., Zhou B., Yang R., Zhao J., Zhang K. ACS Appl. Mater. Interfaces. 2021;13(17):19816–19824. doi: 10.1021/acsami.1c04453. [DOI] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yang C., He Q., Chen J., Yu D., Li J., Zhai S., Qin Z., Du K., Chu Z., Qin P. ACS Infect. Dis. 2020;6(8):1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.S., Pan J., Li F., Zhu M., Xu M., Zhu H., Yu Y., Su G. Anal. Chem. 2021;93(8):4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Sens. Actuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Berry G.J. J. Mol. Diagn. 2020;22(12):1367–1372. doi: 10.1016/j.jmoldx.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.