Abstract
Nitrate commands genome‐wide gene expression changes that impact metabolism, physiology, plant growth, and development. In an effort to identify new components involved in nitrate responses in plants, we analyze the Arabidopsis thaliana root phosphoproteome in response to nitrate treatments via liquid chromatography coupled to tandem mass spectrometry. 176 phosphoproteins show significant changes at 5 or 20 min after nitrate treatments. Proteins identified by 5 min include signaling components such as kinases or transcription factors. In contrast, by 20 min, proteins identified were associated with transporter activity or hormone metabolism functions, among others. The phosphorylation profile of NITRATE TRANSPORTER 1.1 (NRT1.1) mutant plants was significantly altered as compared to wild‐type plants, confirming its key role in nitrate signaling pathways that involves phosphorylation changes. Integrative bioinformatics analysis highlights auxin transport as an important mechanism modulated by nitrate signaling at the post‐translational level. We validated a new phosphorylation site in PIN2 and provide evidence that it functions in primary and lateral root growth responses to nitrate.
Keywords: auxin, HPLC‐MS/MS, nitrate signaling, nitrogen response, phosphorylation
Subject Categories: Methods & Resources, Plant Biology, Proteomics
Phosphoproteomic analysis in response to nitrate treatment reveals early nitrate‐induced changes in Arabidopsis thaliana root phosphoproteins associated to signaling while late changes are associated to transport and hormone metabolism. Moreover, nitrate promotes the dephosphorylation of PIN2, impacting PIN2 subcellular localization and root development.
Introduction
Nitrogen (N) is the mineral nutrient required in the greatest amounts by plants. N is often scarce in natural and agricultural systems, constituting a major factor limiting plant growth and agricultural yield. During the last 50 years, global demand for synthetic N fertilizers has dramatically increased in response to growing agricultural demand. Depending on soil conditions and plant species, less than 50% of the applied N fertilizer is taken up by crops. Excess N may contaminate aquatic systems (Robertson & Vitousek, 2009) or be released into the atmosphere as N oxide gases (Crutzen et al, 2008; Davidson, 2009), both leading to detrimental effects on the environment and human health.
The relevance of N for plants is exemplified by its effects on leaf growth (Wirén et al, 2000), senescence (Vanacker et al, 2006), root system architecture (Zhang et al, 1999; Vidal et al, 2010), and flowering time (Marín et al, 2010; Gras et al, 2018). Due to its importance, plants have evolved sophisticated mechanisms to adapt to fluctuating N availability. Furthermore, growth and developmental processes can be regulated by varying the amount of N supplied to plants. For instance, exogenous nitrate applications stimulate lateral root elongation, enabling root growth and colonization in nitrate‐rich soil patches (Zhang & Forde, 1998; Gojon et al, 2009). However, high nitrate concentrations reduce primary and lateral root elongation under homogeneous growth conditions (Zhang et al, 2007). Nitrate is the main form of inorganic N for plants in natural and agricultural soils (Crawford & Forde, 2002; Gutiérrez, 2012). Besides its nutritional role, nitrate acts as a signaling molecule that regulates several genes involved in a wide range of biological processes (Gutiérrez et al, 2006; Vidal & Gutiérrez, 2008). With advances in genomic technologies and system approaches, thousands of nitrate‐responsive genes have been identified in Arabidopsis thaliana roots and shoots (Wang et al, 2003; Scheible et al, 2004; Wang et al, 2004; Gutiérrez et al, 2007, 2008; Gaudinier et al, 2018; Varala et al, 2018; Brooks et al, 2019). These N‐responsive genes include nitrate transporters, nitrate reductase (NR) and nitrite reductase (NiR), putative transcription factors, and stress response genes, as well as genes whose products play roles in glycolysis, N metabolism, and hormone pathways. Moreover, nitrate elicits local and systemic signals to synchronize its availability with plant growth and development (Ruffel et al, 2011, 2016; Chen et al, 2016; Ohkubo et al, 2017; Poitout et al, 2018). Although transcriptional responses activated by nitrate have been described in great detail, it is clear that regulation at the post‐translational level is key for N responses (Liu & Tsay, 2003; Liu et al, 2017).
The role of protein phosphorylation in response to nitrate was initially identified in post‐translational modifications in N metabolism. The activity of NR, the enzyme that catalyzes the first step of nitrate reduction, is modulated by protein phosphorylation and then inhibited by 14‐3‐3 protein interaction (Bachmann et al, 1996; Kaiser et al, 2002). Studies in spinach leaves using 32P labeling and kinase assays demonstrated that the regulation of NR by light/dark and photosynthetic activity involves protein phosphorylation (Huber et al, 1992; MacKintosh, 1992). A subsequent study showed that a 14‐3‐3 family protein interacts with and inactivates phosphorylated NR in the presence of covalent ions (Bachmann et al, 1996; Athwal & Huber, 2002). Earlier experiments also indicated that changes in gene expression in response to nitrate treatments require kinase and phosphatase activities. In maize leaves for example, treatments with inhibitors of calmodulin‐dependent protein kinases repress nitrate induction of genes encoding nitrate assimilatory enzymes such as NR, NiR, glutamine synthetase 2 (GS2), and ferredoxin glutamate synthase (Fd‐GOGAT) (Sakakibara et al, 1997). Conversely, inhibition of protein phosphatases blocked the nitrate response of NR, NIR, and GS2 (Sakakibara et al, 1997). In another study, pharmacological inhibitors of serine–threonine protein phosphatase and tyrosine protein kinases repressed the nitrate‐induced accumulation of transcripts for NR and NiR in barley leaves (Sueyoshi et al, 1999). These early experiments suggested that changes in the status of protein phosphorylation were important for the regulation of gene expression in response to nitrate treatments.
The discovery that a kinase protein complex can directly phosphorylate the nitrate transceptor NRT1.1/NPF6.3 demonstrated that phosphorylation plays an important role in nitrate signaling. Under low‐nitrate conditions, NRT1.1/NPF6.3 is phosphorylated in a threonine residue (T101) by CIPK23‐CBL9 complex (CIPK, CLB‐interacting protein kinase; CBL, calcineurin B‐like protein), shifting into a high‐affinity nitrate transporter (Liu & Tsay, 2003; Ho et al, 2009). In contrast, at high‐nitrate levels, NRT1.1/NPF6.3 is dephosphorylated at T101 and turns into a low‐affinity transporter. Experiments with a mutant mimicking the phosphorylated form of the transceptor showed the importance of this phosphorylation for the regulation of gene expression at low nitrate concentrations (Ho et al, 2009). Phosphorylation of NRT1.1/NPF6.3 also appears to play a role in the modulation of auxin transport and repression of lateral root emergence under low‐nitrate conditions (Bouguyon et al, 2015, 2016). Conversely, the dephosphorylated form of NRT1.1/NPF6.3 is critical for upregulation of NITRATE TRANSPORTER 2.1 (NRT2.1) gene expression in response to nitrate (Ho et al, 2009; Bouguyon et al, 2015). Bouguyon et al, (2015) studied different mutant alleles of NRT1.1/NPF6.3 (T101D, T101A, and P492L substitution) and proposed a different NRT1.1/NPF6.3‐dependent signaling mechanism. The short‐term induction of NRT2.1 at high nitrate is negatively affected by T101D substitutions but not by P492L and T101A. Long‐term regulation of NRT2.1 transcripts at high nitrate and repression of lateral root emergence at low nitrate showed an opposite pattern, where signaling was suppressed by both T101A and P492L mutations but not affected by T101 substitution. Another kinase involved in signaling is CIPK8 (Hu et al, 2009). In cipk8 mutants, the rapid induction of genes or primary nitrate response was strongly reduced (40–65% of WT levels), particularly in the low‐affinity phase (Hu et al, 2009). Both CIPK8 and CIPK23 are rapidly induced by nitrate treatments and down‐regulated in the chl1‐5 and chl1‐9 mutants, respectively (Ho et al, 2009; Hu et al, 2009). More recently, the calcium sensor CBL1 and the protein phosphatase 2C (ABA‐insensitive) ABI2 were also identified as components of this signaling pathway, which regulates NRT1.1/NPF6.3 transport and sensing (Léran et al, 2015). The calcium sensor CBL1 also interacts with CIPK23, and this complex was dephosphorylated by ABI2 (Léran et al, 2015).
In higher plants, CBL/CIPK complexes sense and decode Ca2+ signals, triggering specific transduction pathways (reviewed by Kudla et al, 2010). Recent studies have shown that Ca2+ plays a role in nitrate signaling transduction and is important for the primary nitrate response in Arabidopsis roots (Riveras et al, 2015). Calcium is a key secondary messenger that triggers changes in signaling pathways, including changes in phosphorylation levels (Dodds & Rathjen, 2010; Hashimoto & Kudla, 2011). More recently, results published by Liu et al, (2017) have contributed to our understanding of the relationship between Ca2+ signaling and the first layer of transcriptional regulators. They used the luciferase (LUC) reporter gene NIR‐LUC, which exhibits a physiological nitrate response in transgenic Arabidopsis plants, to identify three CPKs (CPK10, CPK30, and CPK32) that activated the NIR‐LUC reporter in an effective and synergistic manner. Additionally, CPK10, CPK30, and CPK32 phosphorylated the transcription factor NIN‐LIKE PROTEIN 7 (NLP7) in a Ca2+‐dependent manner (Liu et al, 2017), suggesting that CPK‐NLP signaling is a key regulator of primary nitrate responses (Liu et al, 2020). All these studies provide evidence that NRT1.1/NPF6.3, calcium, and phosphorylation of target proteins are key elements of a signaling pathway involved in the nitrate response.
Global‐scale proteomic analysis performed in Arabidopsis seedlings, mostly shoot organs, showed that nitrogen starvation and resupply (nitrate or ammonium) modulates protein phosphorylation over a time course of 30 min (Engelsberger & Schulze, 2012). In general, proteins such as receptor kinases and transcription factors change their phosphorylation levels after nitrogen resupply at 5–10 min (fast response). Proteins involved in protein synthesis and degradation, central and hormone metabolism showed changes in their phosphorylation level after 10 min (late response). Another study showed that nitrate deprivation affects both protein abundance and phosphorylation status (Menz et al, 2016). Nitrate deprivation assays revealed that some proteins, mostly involved in transport, contain sites that are dephosphorylated early in the response (Menz et al, 2016).
In this study, we performed quantitative time‐course analyses of the Arabidopsis root phosphoproteome in response to nitrate via liquid chromatography coupled to tandem mass spectrometry detection (HPLC‐MS/MS). We chose to focus on root‐phosphoproteomics profiling in response to nitrate for several reasons: (i) phosphoproteomics and proteomics studies describe phosphorylation levels as more dynamic and mainly independent of protein abundance (Huttlin et al, 2010; Walley et al, 2013, 2016), suggesting that many proteins are regulated by phosphorylation independent of their changes in protein abundance. (ii) Previous global studies of N treatment focused on the proteome and phosphoproteome in Arabidopsis seedlings, which interrogates mostly shoot tissues (Engelsberger & Schulze, 2012; Menz et al, 2016). In order to search for new N‐regulatory factors, our experimental approach focused on Arabidopsis roots because early sensing and responses to N supply occur in the roots. Several studies have shown that HPLC‐MS/MS provides accurate estimates of dynamic phosphorylation levels in vivo (Umezawa et al, 2013; Zhang et al, 2013; Zhang et al, 2014; Lin et al, 2015).
We used HPLC‐MS/MS to identify phosphorylated proteins with differential profiles in response to nitrate treatments at 5 or 20 min. We found that the nature of these phosphorylated proteins differed significantly from those encoded by genes implicated in nitrate via transcriptomic studies. We found different types of phosphoproteins changing at 5 or 20 min after nitrate treatments. Interestingly, the large majority of these changes depend on NRT1.1/NPF6.3. Kinases and transcription factors were over‐represented at 5 min, while proteins involved in protein binding and transporter activity were common by 20 min of nitrate treatments. We found several phosphoproteins involved in auxin transport, including the auxin efflux carriers PIN2 and PIN4. We validated the role of PIN2 and found dephosphorylation of PIN2 to be important for modulation of root system architecture in response to nitrate. Our analysis reveals that the nitrate signaling pathway mediated by NRT1.1/NPF6.3 leads to important changes in protein phosphorylation patterns and proposes new players that participate in the developmental responses to nitrate in plants.
Results
Phosphoproteome analysis of Arabidopsis roots in response to nitrate treatments
In an effort to identify new components involved in nitrate responses in plants, we performed large‐scale mass spectrometry‐based phosphoproteome experiments following nitrate treatments in Arabidopsis roots. A. thaliana (L.) Columbia‐0 (Col‐0) seedlings were grown hydroponically, with a full nutrient solution (Murashige and Skoog basal medium without N) containing 1 mM ammonium as the only N source for 14 days (time 0, see experimental procedures). Two‐week‐old plants were treated with 5 mM KNO3 or KCl, as control. We and other laboratories have used this experimental setup in prior studies because it elicits robust primary nitrate responses in Arabidopsis plants (Gutiérrez et al, 2006; Gifford et al, 2008; Vidal et al, 2010; Alvarez et al, 2014; Riveras et al, 2015). A label‐free high‐performance liquid chromatography–tandem mass spectrometry (HPLC‐MS/MS) method was used to identify changes in protein phosphorylation at 5 or 20 min after nitrate treatments. We chose these time points because they have shown to be effective in describing transient and persistent protein phosphorylation responses (Engelsberger & Schulze, 2012; Lin et al, 2015). Moreover, this experimental design allows for comparison with transcriptomics data obtained using the same experimental conditions (Vidal et al, 2010; Alvarez et al, 2014; Riveras et al, 2015). For phosphoproteome analysis, we used a previously validated experimental pipeline (Facette et al, 2013; Walley et al, ,2013, 2016). Briefly, phosphopeptides were enriched using cerium oxide affinity capture and analyzed with a HPLC‐MS/MS instrument (Appendix Fig S1). The spectra were assigned to specific peptide sequences by the MASCOT search engine (FDR < 0.1%). We quantified the relative abundance of each phosphoprotein using average normalized spectral counts (nSPCs) of the total number of spectral‐peptide matches to protein sequences in two–three independent biological replicates for each treatment condition. In total, we identified and measured 6,560 unique phosphopeptides which unambiguously mapped to 2,048 phosphoprotein groups (Dataset EV1). The majority of identified phosphopeptides (82%) were phosphorylated in a single residue (Appendix Fig S2A). The relative distribution of each phosphorylated residue—80% serine, 18% threonine, and 2% tyrosine (Appendix Fig S2B)—was consistent with prior plant phosphoproteomic studies (Lan et al, 2012; Umezawa et al, 2013; Menz et al, 2016). The identified phosphopeptides were mapped and grouped in phosphoprotein groups, where proteins that shared peptides were clustered together. A group leader was assigned to each group, based on having the highest number of peptide identifications; throughout the remainder of the article, “phosphoproteins” is synonymous with “group leaders”. The majority of the identified phosphoproteins present one (50%), two (25%), or three (12%) phosphorylated peptides (Appendix Fig S2C) with similar distributions of phosphorylated residue (69% Ser, 28% Thr, and 3% Tyr; Appendix Fig S2D). We recognized phosphoproteins across several biological process, subcellular compartments, and cellular functions based on the Gene Ontology (GO) classification (Appendix Fig S3). No overrepresented GO categories were observed when comparing against the Arabidopsis genome, showing that our experimental strategy was unbiased with regard to annotated protein functions, subcellular locations, or biological processes and represents an unbiased Arabidopsis proteome sampling.
To identify nitrate‐regulated phosphoproteins in Arabidopsis roots, the phosphoproteomics dataset was filtered by quality following the workflow described in DEP (Zhang et al, 2018) and MSnBase (Gatto & Lilley, 2012) packages using R/Bioconductor (Huber et al, 2015). Reverse hits and phosphoproteins with missing values in several experimental conditions were eliminated (see Materials and Methods for details). We obtained a dataset with 34% of missing values, after which quantile normalization, data imputation using random draws from a Gaussian distribution centered to the minimal value in each experimental condition and a log2 transformation were applied (Database EV2). Then, we performed a two‐way ANOVA statistical analysis (significance: P < 0.05). We considered nitrate treatment (N), time (Ti), and the interaction between nitrate and time (N‐Ti) as the factor for the ANOVA models. We identified 120 (N), 197 (Ti), and 106 (N‐Ti) phosphoproteins that were significantly affected under our experimental conditions. The group whose phosphoprotein levels depend on nitrate (significant P value for N or N‐Ti) were selected for further analysis and their ANOVA model was examined (Fig EV1A). We found 54 phosphoproteins regulated at 5 min after nitrate treatments, 36 of which were induced (Fig 1A, Dataset EV3). We found 145 phosphoproteins differentially regulated at 20 min, 102 of which were induced by the nitrate treatments. The large majority of phosphoproteins were found to be nitrate‐regulated at only one time‐point, indicating that most changes in the phosphoproteome are transient with an early (5 min) and late (20 min) component in response to nitrate treatments (Fig 1A and 1B). Three previous studies characterized the phosphoproteome in response to nitrate depletion (Engelsberger & Schulze, 2012) or nitrate resupply (Menz et al, 2016; Wu et al, 2017) (Fig 1C). Response to nitrate resupply resulted in 15 and 34 common phosphoproteins, while nitrate deprivation in 7 common proteins. We observed an overlap of 4 phosphoproteins between all experiments using the Sungear tool (Poultney et al, 2007), which shows few phosphoproteins common to nitrate studies. These proteins were mostly associated with nitrate metabolism, such as transporters (NRT2.1; ammonium transporter 1.3, AMT1.3) and nitrate reductase 2 (NIA2). The comparison of the phosphoproteins identified here with transcriptome data revealed that 95% of the phosphoproteins are encoded by genes that do not change expression at the mRNA level (Fig EV1B). We compared our results with an integrated analysis of available root microarray data under contrasting nitrate conditions (27 experimental datasets corresponding to 131 arrays (Canales et al, 2014)). This study identified a group of 2,286 nitrate‐responsive genes regulated at transcription levels. Only 10 of these genes were found in our phosphoproteomic dataset.
Figure EV1. Phosphoproteins affected by nitrate response.
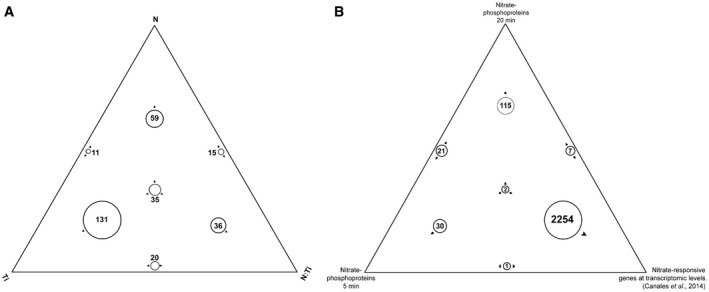
- The list of phosphoproteins with significant factors achieved by two‐way ANOVA was represented by the Sungear tool. The Sungear figure was represented by a triangle that shows the factors of ANOVA model at the vertices [nitrate treatment (N), time (Ti), and the interaction between treatment and genotype (N–Ti)]. The circles inside the triangle represent the phosphoproteins with statistical changes by the different factors (P < 0.05), as indicated by the arrows around the circles. The size of each circle was represented proportional to the number of phosphoproteins associated with that circle. The number of phosphoproteins in each circle is shown inside or next to the corresponding circle.
- The list of encoded genes in response to nitrate at transcriptomic (dataset 27 affymetrix experiment Canales et al, 2014) and phosphoproteomic (our work) levels were compared and represented using the Sungear tool. The triangle shows each dataset at the vertices: transcriptomic dataset, phosphoproteomic data (5 min), and phosphoproteomic data (20 min). The position and the arrows of the circle indicated which vertices or anchor the data belong. The largest circles are on the perimeter that indicate most of the data are associated with only one dataset.
Figure 1. Characterization of the phosphoproteome profile in Arabidopsis roots in response to nitrate.
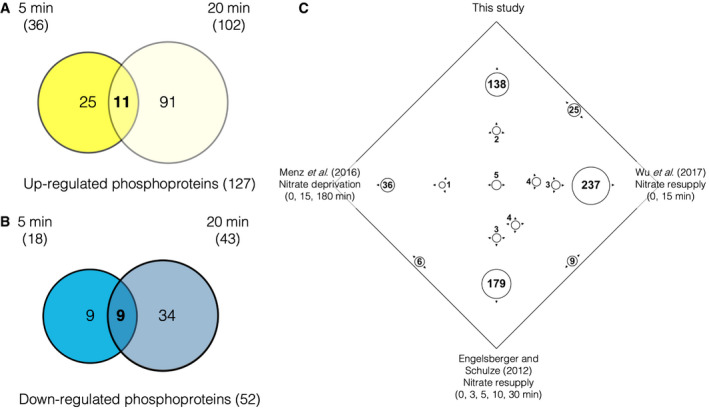
Venn diagrams show the overlap of transient (5 min) and more persistent (20 min) changes in phosphoprotein relative abundance after 5 mM nitrate treatments compared with each control condition (5 mM KCl) in Arabidopsis roots.
- Overlap of phosphoproteins that were “up‐regulated” by nitrate at 5 min (dark yellow) and 20 min (light yellow)
- Overlap of phosphoproteins that were “down‐regulated” by nitrate at 5 min (blue) and 20 min (light blue).
- Identification of common phosphoproteins with published phosphoproteome datasets in response to nitrate deprivation (Menz et al, 2016; Wu et al, 2017) or nitrate resupply (Engelsberger & Schulze, 2012). In the Sungear figure, vertices represent the different phosphoprotein sets identified in different studies. The circles with arrows within the square represent different intersections among the phosphoprotein studies and the size is proportional to the number of proteins in that circle. The position and the arrows of the circle indicated which vertices or data sets the proteins belong to. The largest circles on the perimeter indicate most of the phosphoprotein are associated with only one experiment.
In summary, our phosphoproteome analysis identified new genes coding for phosphoproteins involved in nitrate responses.
Functional enrichment in the phosphoproteome reveals distinctive signaling and regulatory processes occurring in early and late responses to nitrate
To evaluate the biological significance of the phosphoproteome patterns observed in response to nitrate, hierarchical clustering analysis was performed on the phosphoprotein dataset at 5 and 20 min following nitrate treatments in Col‐0 roots (Figs 2 and EV2). In order to identify the most prominent functional categories affected, we searched for overrepresented biological terms in each cluster using the BioMaps program (Katari et al, 2010) and the PANTHER classification system (Mi et al, 2018, 2019) (significance: P < 0.05, corrected by FDR). This analysis highlighted several signaling, regulatory, or metabolic functions differentially associated with early (5 min) and late (20 min) components in response to nitrate treatments. “Nucleic acid binding” category was overrepresented in cluster 6 (Figs 2 and EV2G), containing phosphoproteins that increase their levels at 5 min and do not change at 20 min. This cluster also includes previously undescribed transcription factors (TFs) in the N‐response cascade in diverse transcriptomic studies (Gutiérrez et al, 2008; Gaudinier et al, 2018; Varala et al, 2018; Brooks et al, 2019; Alvarez et al, 2020). In contrast, cluster groups containing mostly phosphoproteins that changed their levels at 20 min in response to nitrate were enriched in the functional categories: “RNA binding” and “protein binding” (clusters 3 and 4, Figs 2 and EV2C and D). We also found increases in phosphoprotein levels in NRT2.1 and AMT1‐3 only at 20 min following nitrate treatment. Increased abundance of the phosphopeptide Ser501, corresponding to NRT2.1, was observed at 20 min after nitrate treatment. This site is localized in a C‐terminal phosphorylation hotspot (phosphorylation site database and predictor Phosphat4.0 (Heazlewood et al, 2007; Durek et al, 2010)) and was also identified in nitrate deprivation experiments with an opposite phosphorylation response (Menz et al, 2016). Also, phosphopeptides for AMT1.3 with phosphorylation at Thr‐464 and Ser487 were identified as “up‐regulated” by nitrate treatments at 20 min. The phosphorylation of both sites inhibits transport function (Lanquar et al, 2009) and was also identified as phosphopeptides in nitrate deprivation (Menz et al, 2016) and resupply (Engelsberger & Schulze, 2012) experiments. We also observed that nitrate strongly increased levels of phosphorylated nitrate reductase NIA2 at the highly conserved and regulatory site Ser534 (Su et al, 1996). These results are consistent with previous studies and suggest overall regulation of N metabolism by phosphorylation of key players by 20 min after nitrate treatments.
Figure 2. Functional analysis of the phosphoproteome profile reveals signaling and regulatory processes occurring in early and late response to nitrate.
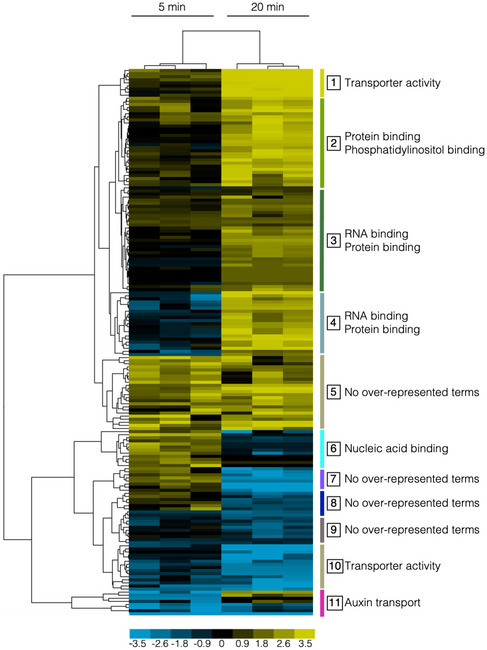
The Figure shows a hierarchical clustering of nitrate phosphoproteins with differential abundance at 5 or 20 min in response to nitrate. Phosphoproteins were clustered using the Euclidean distance method with average linkage. The resulting clusters are shown as a heat map, where vertical bars and numbers to the right of the map denote the group (composed of all terminal nodes in the hierarchical tree) of phosphoprotein (correlation > 0.9) with a selected profile in early (5‐min) or late (20‐min) nitrate response (log 2 fold‐change for each phosphoproteins upon nitrate treatment are referred to KCl‐treated samples). Functional Gene Ontology (GO) categories significantly enriched (hypergeometric test with FDR, P < 0.05) in each cluster are highlighted to the right of the group.
Source data are available online for this figure.
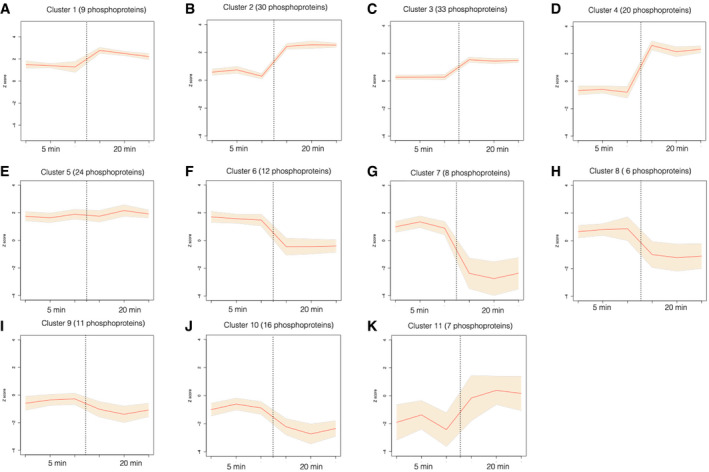
Figure EV2. Hierarchical clustering of nitrate phosphoproteins with differential abundance at 5 or 20 min in response to nitrate treatments.
-
A–KNitrate phosphoprotein abundance patterns for eleven clusters in response to nitrate at 5 or 20 min (correlation distance threshold < 0.9) as described in Figure 2. Graphs show mean values of Z‐scored normalized phosphoproteins levels (orange line) and 95% confidence interval for mean values of each Cluster for the independent experiments (shadow).
Other group of clusters showed enrichment in process involved in nitrate response, with different profiles at 5 min and 20 min. Clusters 1 and 10 (Figs 2 and EV2A and J) were enriched in “transport activity” and showed phosphoproteins with opposite regulation. Clusters 2, 4, and 5 showed phosphoproteins associated with microRNA processing, phosphoinositide, and phosphatidylinositol binding functions. Phosphoinositides can act in signaling pathways and serve as precursors for phospholipase C (PLC)‐mediated signaling. A previous study in our laboratory implicated a PLC activity in the nitrate signaling pathway (Riveras et al, 2015). Our results are consistent with these results and show that PLC2, represented by the phosphopeptide Ser280, was identified as up‐regulated in response to nitrate at 20 min (in Cluster 4). Cluster 11 (Figs 2 and EV2K) is interesting for nitrate responses because it includes components involved in a classical process regulated by nitrate in Arabidopsis roots. Auxin transport is a biological function enriched in this group, which is consistent with auxin pathways being modulated by nitrate (Gutiérrez et al, 2006; Vidal et al, 2013). Several reports indicate that auxin acts as regulator of root system architecture in response to nitrate availability (Vidal et al, 2010, 2013; Ma et al, 2014).
Overall, our dataset captures the dynamic effects of N signaling on phosphoproteome profiles, which implicate a cascade of nonoverlapping processes in early and late responses. The earliest steps in the N phospho‐dynamics were involved in signal transduction and transcription factor activity. In contrast, the later phosphoprotein dataset was enriched in metabolic, transferase, and transport processes (Appendix Table S1). These temporal mechanisms show a transition of phosphorylation dynamics from phosphoproteins essential to signaling networks to proteins associated with biological processes involved in nitrate response.
NRT1.1/AtNPF6.3 is essential for transient phosphoprotein changes in response to nitrate treatments
The only nitrate sensor described to date is the nitrate transporter, NRT1.1/NPF6.3 (Ho et al, 2009; Wang et al, 2009). In order to understand the importance of NRT1.1/NPF6.3 for nitrate‐elicited changes in the phosphoproteome observed, we analyzed the phosphoproteomic profile of roots treated with 5 mM KNO3 or KCl (control) in a nrt1.1‐null background (mutant chl1‐5), using the same experimental conditions described above. 74% of phosphoproteins were detected in both datasets, yet only 4% of the nitrate‐phosphoproteome response observed in wild‐type plants was maintained in the chl1‐5 mutant (Fig 3, Dataset EV4). Moreover, 96% of phosphoprotein levels were altered in the chl1‐5 mutant plants. This result indicates that this gene is important for modulating protein phosphorylation in response to nitrate, in addition to its established role as nitrate transceptor. Intriguingly, we found 32 phosphoproteins that changed levels in response to nitrate at 5 min in the chl1‐5 mutant as compared to wild‐type plants (ANOVA P < 0.05; Appendix Table S3), with 22 induced phosphoproteins. These changes suggest an alternative nitrate‐sensing mechanism that can also impact protein phosphorylation. In the absence of NRT1.1 function, the balance between this alternate signaling mechanisms may be disrupted resulting in anomalous phosphorylation patterns. Examples of phosphoproteins affected in chl1‐5 include signaling pathway components such as brassinosteroid signaling positive regulator 1 (BZR1), signal responsive 1 SR1, and a phosphatidylinositol signaling‐related protein.
Figure 3. Characterization of the phosphoproteome profile in response to nitrate in roots of Arabidopsis (Col‐0) or chl1‐5 mutant plants.
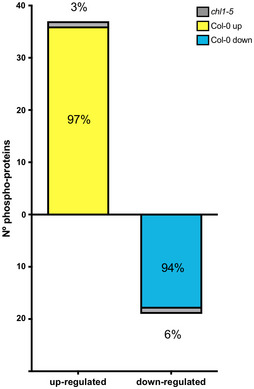
Percentage of nitrate‐phosphoproteome responses at 5 min in wild‐type plants that were maintained in chl1‐5‐mutant plant roots.
Our analysis indicates that NRT1.1/NPF6.3 is critical for maintaining the Arabidopsis phosphoproteome in response to nitrate availability. It also denotes that NRT1.1/NPF6.3 function is required for rapid changes in phosphorylation of key proteins in response to nitrate in Arabidopsis roots. For example, phosphorylated peptides that map to proteins associated with nitrogen metabolism (NRT2.1, AMT1‐3, and NIA2) were identified in chl1‐5 mutant roots, but their levels were not affected in response to nitrate.
Network analysis reveals regulatory subnetworks connected to transcription factors and potential kinases in response to nitrate
To uncover key biological processes modulated by changes in phosphorylation, we performed a multinetwork analysis with our phosphoproteomics data. We generated this network by integrating different levels of information, including protein–protein interactions from BioGRID (Oughtred et al, 2018), predicted protein–DNA interactions of Arabidopsis TFs (DapSeq) (Weirauch et al, 2014; Bartlett et al, 2017), Arabidopsis metabolic pathways (KEGG), and miRNA‐RNA, as described previously (Gutiérrez et al, 2006). We also integrated kinase‐substrate predictions and identified the most significant phosphorylation motifs and their predicted kinase families from our phosphoproteomic datasets using the Motif‐X algorithm (Schwartz & Gygi, 2005) and the PhosPhAt Kinase‐Target interactions database (Zulawski et al, 2012) (Appendix Fig S3). We used the Cytoscape (Shannon et al, 2003) software to visualize the resulting network, wherein genes that encoded each phosphoprotein were represented as nodes linked by edges that signify any of the functional relationships annotated in the various databases indicated above. We generated a network of 196 nodes with 502 interactions (Fig 4). Although the majority of these genes are not regulated by nitrate at the mRNA level, they form a highly interconnected network which includes potential regulatory transcription factors and kinase components. We identified network modules using the community cluster (GLay) algorithm in ClusterMaker tool (Su et al, 2010; Morris et al, 2011), which recognizes network domains with densely connected nodes. They are connected by multiple edges, including protein–protein, protein–DNA, and metabolic interactions (described in legend to Fig 4). This result suggests that the products of these genes form connected biological modules that are coordinately regulated at the (post‐)translational level. This network included several TFs with a high number of regulatory links. The most connected were the Trihelix transcription factor 1 (GTL1), the WRKY DNA‐binding protein 65 (WRKY65), and the RELATED TO VERNALIZATION 1 (RTV1) transcription factors, which had not previously been characterized in the context of nitrate response. Intriguingly, GTL1 regulates root hair growth in Arabidopsis (Shibata et al, 2018), which has recently been described as a biological process modulated by nitrate treatments under the same experimental conditions (Canales et al, 2017). A previous study indicated that WRKY65 interacts at the protein level with the mitogen‐activated protein kinase 10 (MAPK10), which binds with the lateral organ boundaries domain 16 (LBD16), LBD18, and LBD29 transcription factors (Popescu et al, 2008; Feng et al, 2012). These LBDs are inducible by auxin and play a role in the formation of lateral roots. In addition, MAPK10 interacted with other genes involved in the auxin response (Popescu et al, 2008), while LBD29 regulated genes involved in auxin transport, including auxin efflux‐carriers PIN1 and PIN2 (Feng et al, 2012). This evidence suggests that WRKY65 could be involved in nitrate–auxin signaling crosstalk. These three TFs appear to coordinate different subnetworks largely involved in auxin transport and nitrogen metabolism. Consistent with this observation, analysis of over‐represented gene ontology annotations highlights the importance of auxin transport (Fig EV3). Other over‐represented biological functions in our network were mRNA binding and splicing, regulation of translation, and kinase activity (Fig EV3).
Figure 4. Gene network of nitrate‐modulated phosphoproteins.
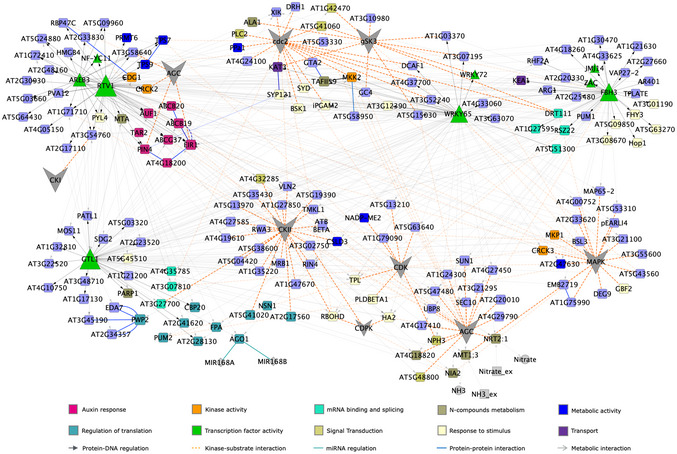
A network for the phosphoproteins (encoded genes) and putative kinases family was represented as nodes with color and shapes assigned according to function (e.g., blue squares: metabolic encoded genes, green triangle: transcription factors or gray hexagon: kinase families). Edges connecting nodes represent functional interactions: transcription factor/TARGET (gray solid arrow), protein–protein (blue solid line), miRNA–RNA (cyan solid line), metabolic (gray solid line with open arrow), or predicted kinase–substrate control (orange dashed line) interactions are coded with indicated colors. Arrowheads in the edges indicate directionality of the interaction. The size of the triangle or hexagon is proportional to the number of targets of the TF or putative kinase family, respectively. The network was grouped by topology using ClusterMaker tool in Cytoscape. The distance between nodes was optimized for visualization purposes.
Figure EV3. Over‐represented biological processes in the phosphoproteins gene network in response to nitrate (hypergeometric test with FDR, P < 0.05).
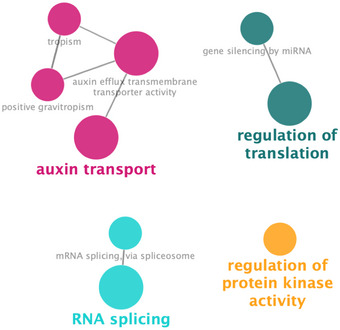
The network was analyzed using BINGO and ClueGO tools in Cytoscape software. Over‐represented gene ontology terms are shown as a node connected by edges based on semantic relationships as defined in the ontology.
Source data are available online for this figure.
Overall, this network analysis highlights a potential role of multiples TFs in linking the N signal and regulatory nitrate responses that show significant enrichment for key functions in signaling pathways and validates the important role auxin plays in the nitrate response of root system architecture.
PIN2 is important for modulation of root system architecture in response to nitrate treatments
Auxin is a key phytohormone in plants, involved in growth and developmental responses. Several reports have shown that auxin mediates root developmental responses to nitrate availability (Gutiérrez et al, 2006; Walch‐Liu et al, 2006; Krouk et al, 2010; Vidal et al, 2010; Mounier et al, 2013; Vidal et al, 2013; Ma et al, 2014). Nitrate can regulate auxin biosynthesis, transport, and accumulation. In response to nitrate, several auxin‐related modules are regulated, including the upregulation of auxin receptor AUXIN SIGNALING F‐BOX 3 (AFB3) and the feedback regulation by miR393 (Vidal et al, 2010, 2013). This auxin signaling component in response to nitrate is implicated in both primary and lateral root growth (Vidal et al, 2010). Other important evidence comes from the analyses of the Arabidopsis transcriptomic response upon nitrate treatments. They show that several genes involved in auxin transport are affected, including auxin efflux carriers PIN1, PIN4, and PIN7 (Gutiérrez et al, 2006; Vidal et al, 2013). Moreover, the nitrate transceptor NRT1.1/NPF6.3 not only senses and transports nitrate but can also transports auxin, this process is modulated by NRT1.1 phosphorylation dynamics (Zhang et al, 2019) and regulates auxin‐localization patterns and lateral root elongation (Krouk et al, 2010). Consistent with these prior observations, auxin transport was conspicuous throughout our entire phosphoproteomic analysis (Figs 2 and 4). PIN phosphorylation has been shown to be essential for auxin transport and distribution (Friml et al, 2004; Michniewicz et al, 2007; Huang et al, 2010; Zhang et al, 2010; Weller et al, 2017; Barbosa et al, 2018). PINs phosphorylation in conserved serine and/or threonine of the central loop controls intracellular trafficking, recycling, and polar membrane localization of PIN proteins (Review by Barbosa et al, 2018). Previous studies indicate that PIN polar localization explains auxin fluxes and distribution patterns, which could mediate differential growth in diverse plant tissue such as roots (Benková et al, 2003; Weller et al, 2017). To validate the relevance of phosphorylation in auxin transport and its connection with nitrate response, we chose the auxin efflux carrier PIN2 (identified in our experimental dataset) due to its potential role in linking nitrate and changes in root system architecture (RSA). We found an uncharacterized PIN2 phosphorylation site (Ser439) at the end of the hydrophilic cytoplasmic loop (C‐loop, Fig 5A). Its phosphopeptide levels decreased by close to 75% in response to nitrate treatments by 5 min (Fig 5B). PIN2 belongs to the PIN‐FORMED protein family of auxin transporters and is the principal component mediating basipetal auxin transport in roots (Luschnig et al, 1998; Müller et al, 1998). This polar auxin transport is essential for root gravitropism (Müller et al, 1998) and lateral root formation (Laskowski et al, 2008). Intriguingly, we detected only one phosphorylated peptide for PIN2 in response to nitrate. Protein sequence alignment indicated that this phosphosite is highly conserved in different plant species representing gymnosperm and mono‐ and dicotyledonous plant lineages of seed plants (Fig 5A). This phosphopeptide has also been described as down‐regulated after auxin treatment but its function remains an open question (Zhang et al, 2013).
Figure 5. Nitrate regulates PIN2 phosphorylation levels.
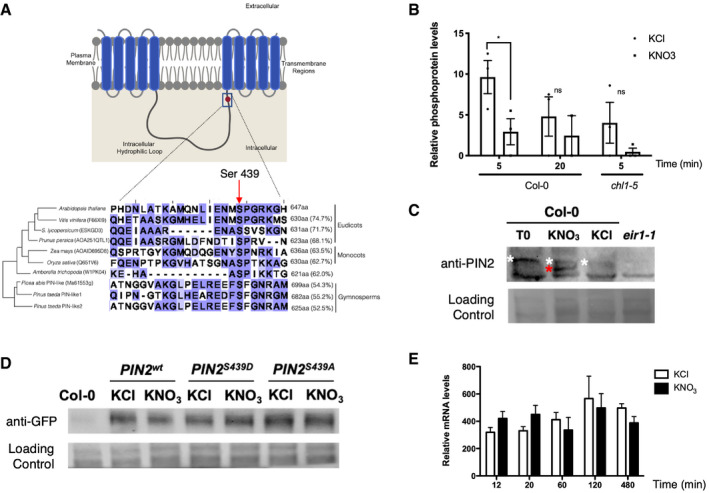
- Schematic representation of the phosphosite (Ser439) identified in PIN2 in our study. PIN2 protein possesses three specific regions: a region with five transmembrane segments (residues 1–163), a central hydrophilic loop extending from residue 164 to 482 and a region with five additional transmembrane segments (residues 483–647). Protein alignment with PIN2 or PIN‐like proteins from different species indicates conservation of the serine residue in eudicot, monocot, and gymnosperm species. The red arrow highlights the S439 in the central loop of PIN2 proteins in different plants. The size of each protein in amino acids and the percentage of identity against the Arabidopsis PIN2 are indicated.
- Levels of PIN2 phosphopeptide in our experiments. Bars represent the mean plus standard error of replicates (2 biological replicates for nitrate treatments at 20 min (Col‐0 roots) and 3 biological replicates for all other experimental conditions). Each independent biological replicate consisted of a pool of 4.500 roots collected from Arabidopsis plants grown independently under the same experimental conditions. The asterisk indicates statistically significant differences in phosphoproteomic analysis (multiple t‐test comparison without testing corrections, assuming same variance, P < 0.05).
- Detection of phosphorylation PIN2 by Phos‐tag Western blotting. Arabidopsis plants (Col‐0) were growth in ammonium as only nitrogen source and treated with 5 mM KNO3 or 5mM KCl as control. Total protein from roots were analyzed in SDS–PAGE using Phos‐tag to detect changes in phosphorylation status. Immunoblotting was performed with PIN2 antibody. Total proteins isolated from eir1.1 roots were used as a negative control. White and red asterisks indicate a slow‐ or fast‐mobility band corresponding to a more or less phosphorylated PIN2, respectively.
- Western blot against PIN2 protein comparing nitrate‐treated (KNO3) and control (KCl) condition in Arabidopsis roots for all genotypes eir1‐1 mutant background was complemented with PIN2::PIN2wt‐GFP (PIN2wt), PIN2::PIN2S439D‐GFP (PIN2S439D phospho‐mimic point mutation), or PIN2::PIN2S439A‐GFP (PIN2S439A, phospho‐null point mutation).
- Time‐course analysis of PIN2 mRNA levels in response to nitrate treatments in Arabidopsis roots. Bars represent the mean plus standard deviation of 3 biological replicates.
Source data are available online for this figure.
As a first step to understand the function of PIN2 phosphorylation in the nitrate response, we performed a Phos‐tag Western blot analysis to confirm the changes in PIN2 phosphorylation after nitrate treatment (Figs 5C and EV4). We detected two, fast and slow mobility (red and white asterisk in Fig 5C, respectively), PIN2‐specific bands indicating the presence of two phospho‐populations in response to nitrate: one less and another more phosphorylated PIN2. In contrast, only one slow‐mobility band corresponding to the more phosphorylated PIN2 subpopulation could be observed at time 0 (ammonium‐supplied roots) or under control conditions (KCl‐treated roots) (white asterisk, Fig 5C). To assess whether these changes in PIN2 phosphorylation status are a result of a change in protein abundance, we analyzed PIN2‐GPF protein levels by Western blot under our experimental conditions. We introgressed the construct PIN2::PIN2‐GFP (PIN2wt‐GFP) into the pin2 loss‐of‐function mutant plant eir1‐1 (Roman et al, 1995). No differences in protein levels were observed in roots treated with nitrate as compared to roots at control condition (KCl) (Figs 5D and EV4). To understand the function of this specific PIN2 phosphosite Ser439, we also analyzed the protein levels in pin2 null mutant complemented with phospho‐null (PIN2::PIN2S439A‐GFP) or phospho‐mimic (PIN2::PIN2S439D‐GFP) versions of PIN2‐GFP. Similarly, PIN2 protein concentrations were similar when our experimental conditions mimicked PIN2 phospho‐modifications (Figs 5D and EV4). Moreover, no regulation at the mRNA level was observed in PIN2 during nitrate responses (Fig 5E). These results indicated that nitrate regulates PIN2 at the post‐translational level, causing dephosphorylation of PIN2 at a specific phosphosite.
Figure EV4. PIN2 protein analysis in response to nitrate.
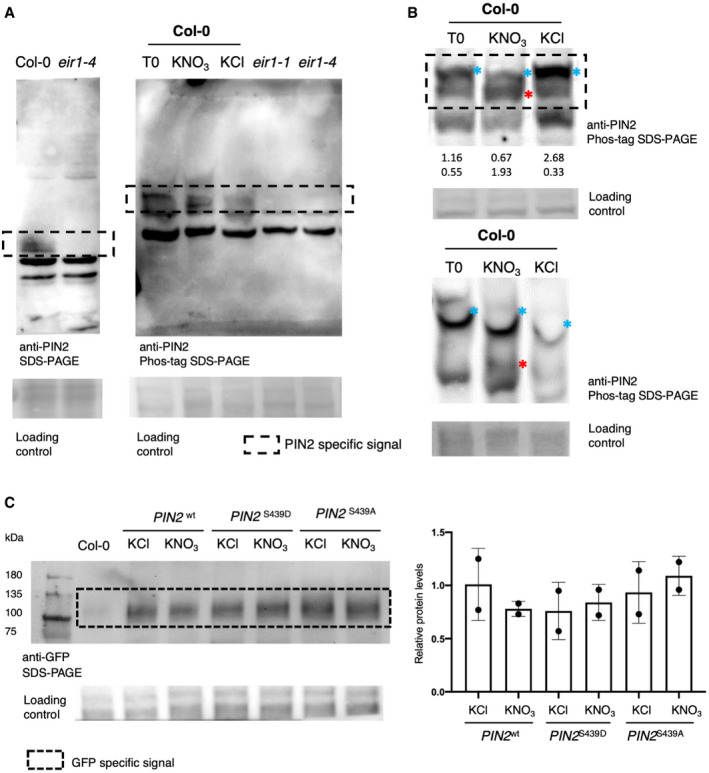
- PIN2 detection by Phos‐tag Western blotting (source data Fig 5C). Arabidopsis plants (Col‐0) were grown in ammonium as only nitrogen source and treated with 5 mM KNO3 or 5mM KCl as control. Total protein from roots was analyzed in SDS–PAGE using Phos‐tag to detect changes in phosphorylation status. Immunoblotting was performed with PIN2 antibody. Total proteins isolated from eir1.1 roots were used as a negative control. PIN2‐specific signal was tested as control in Col‐0 and eir1‐4 mutant by normal Western blot SDS–PAGE.
- Phos‐tag Western blotting analysis to detect PIN2 phosphorylation (two independent biological replicates). Arabidopsis plants (Col‐0) were grown in ammonium as only nitrogen source and treated with 5 mM KNO3 or 5 mM KCl, as a control condition. Total proteins from roots were analyzed in SDS–PAGE using Phos‐tag to detect changes in phosphorylation status. Immunoblotting was performed with PIN2 antibody. Blue and red asterisks indicate a slow‐ or fast‐mobility band corresponding to a more or less phosphorylated PIN2, respectively. The numbers below the blot correspond to relative quantification by densitometry using Image J of these two bands, normalized by loading control.
- PIN2‐GFP detection by SDS–PAGE Western blotting. Quantification of two independent replicate Western blot experiments against PIN2‐GFP protein comparing nitrate‐treated (KNO3) and control (KCl) condition at 5 min in Arabidopsis roots for all genotypes eir1‐1 mutant background was complemented with PIN2::PIN2wt‐GFP (PIN2wt), PIN2::PIN2S439D‐GFP (PIN2S439D phospho‐mimic point mutation), or PIN2::PIN2S439A‐GFP (PIN2S439A, phospho‐null point mutation). Total proteins isolated from Col‐0 roots were used as a negative control. Bars represent the mean plus standard deviation of 2 biological replicates.
Next, we evaluated the role of PIN2 in root system architecture (RSA) in response to nitrate treatments. We grew wild‐type (Col‐0) and pin2 mutant (eir1.1) plants for 2 weeks on ammonium as sole N source (time 0) and evaluated RSA after nitrate treatments. We measured primary root length 3 days after 5 mM KNO3 or KCl treatments. As expected for this experimental setup, we found that nitrate‐treated wild‐type plants developed shorter primary roots as compared to KCl‐treated plants, consistent with earlier results indicating that nitrate treatments inhibit primary root elongation under these experimental conditions (Vidal et al, 2010) (Fig 6A). However, primary roots of eir1‐1 plants were not significantly inhibited by nitrate treatments as compared to wild‐type plants. We also analyzed the density of lateral roots in response to nitrate treatments. In wild‐type plants, nitrate treatments increased the number of lateral roots (emerged and initiating) as compared with KCl treatments (Fig 6B). In contrast, the lateral root response to nitrate treatment was altered in the eir1‐1 mutant and the density of lateral roots was significantly reduced as compared with wild‐type plants (Fig 6B). These results show that PIN2 plays an important role in modulating RSA in response to nitrate treatments. Accordingly, Ötvös et al, (2021) recently reported that PIN2 plays an important function regulating distinct root growth patterns in response to different nitrogen sources and concentrations.
Figure 6. PIN2 is essential for nitrate regulation of primary root growth and lateral root density.
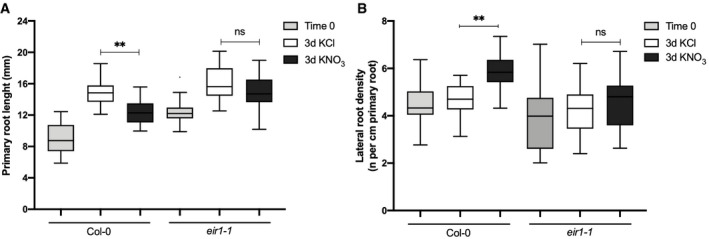
- Primary root length of Col‐0 wild‐type plants or eir1‐1 mutant plants was measured using the ImageJ program 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 10–15 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5× IQR (interquartile range). Outlier data are plotted individually. Asterisk indicates statistically significant difference between analyzed by unpaired, two‐tailed and assuming equal variance t‐test (**P < 0.01).
- Number of initiating and emerging lateral roots of Col‐0 or eir1‐1 mutant plants were counted using light microscopy 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 10–15 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5× IQR (interquartile range). Asterisk indicates statistically significant difference between analyzed by unpaired, two‐tailed and assuming equal variance t‐test (**P < 0.01).
Source data are available online for this figure.
PIN2 dephosphorylation of Ser439 plays a role in modulating the root system architecture in response to nitrate treatments
To understand the function of the PIN2 phosphosite identified in this study, we analyzed the pin2 null mutant eir1‐1 complemented with phospho‐null (S439A) or phospho‐mimic (S439D) versions of PIN2‐GFP. As PIN2 plays an important role in root gravitropism, we evaluated the gravitropic curvature in wild‐type, eir1‐1 mutant plants, and PIN2‐GFP transgenic plants. All seedlings were germinated and grown in MS media in the absence of nitrate for 7 days, and then, they were transferred to agar plates with or without nitrate. Agar plates were rotated 90° and root curvature was measured at 24 h. As expected, pin2 mutant plants showed an agravitropic phenotype. Both PIN2S439D‐GFP and PIN2S439A‐GFP were able to rescue the agravitropic phenotype of eir1‐1 mutant plants. This result indicates that the phosphorylation status of PIN2 at S439 is not relevant for root gravitropic responses in Arabidopsis (Appendix Fig S4). This result also indicates that phosphorylation at S439 does not impair all PIN2 functions.
Next, we evaluated the RSA in response to nitrate treatments in the same phospho‐mimicking (PIN2S439D‐GFP) or phospho‐null (PIN2S439A‐GFP) genotypes and compared it to wild‐type plants. Plants were grown for 2 weeks on ammonium as sole N source and then treated with 5 mM KNO3 or KCl for three days. Complementation of the eir1‐1 mutant with a wild‐type version of PIN2 (PIN2wt ) restored normal RSA responses to nitrate (Fig 7). Eir1‐1 mutant plants complemented with PIN2S439A showed RSA changes similar to wild‐type plants in response to nitrate treatments, albeit slightly less pronounced. Three days after nitrate treatments, primary root growth was inhibited 42% by nitrate in the PIN2S439A‐GFP genotype as compared with nitrate‐treated wild‐type plants (Fig 7A). Similarly, lateral root density in PIN2S439A‐GFP plants increased 47% as compared to wild‐type plants in response to nitrate treatments (Fig 7B). In contrast at the end of the 3‐day treatment, the primary root length and lateral root density did not differ between nitrate or KCl treatment in eir1‐1 mutant plants complemented with PIN2S439D‐GFP. This result, comparable to the response in eir1‐1 roots, indicates that regulation of PIN2 phosphorylation status at S439 is necessary for normal RSA modulation in response to nitrate treatments.
Figure 7. PIN2 dephosphorylation at S439 is important for modulation of primary root growth and lateral root density in response to nitrate treatments.
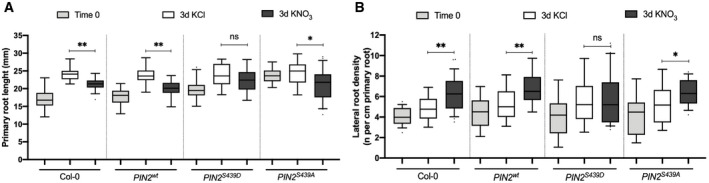
- Primary root length of the different genotypes was measured using the ImageJ program 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 8–10 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5x IQR (interquartile range). Outlier data are plotted individually. Asterisk indicates statistically significant difference between means analyzed by unpaired, two‐tailed and assuming equal variance t‐test (*P < 0.05, **P < 0.01).
- Number of initiating and emerging lateral roots for all genotypes were counted using light microscopy 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 8–10 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5× IQR (interquartile range). Outlier data are plotted individually. Asterisk indicates statistically significant difference between means analyzed by unpaired, two‐tailed and assuming equal variance t‐test (*P < 0.05, **P < 0.01).
Source data are available online for this figure.
PIN2 phosphosite regulates polar plasma membrane localization in response to nitrate
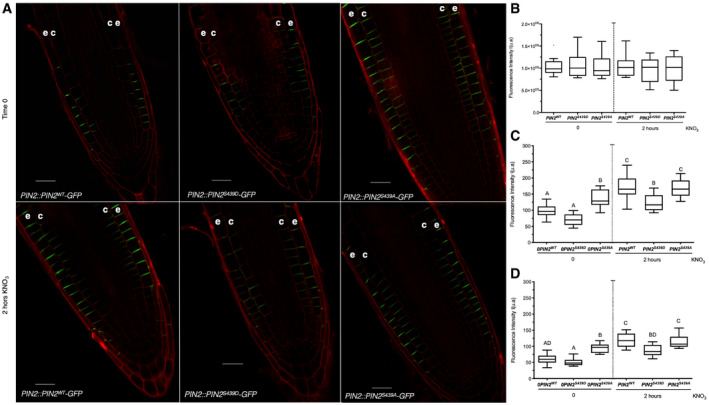
To explore the impact of nitrate‐regulated phosphorylation of PIN2 on cellular localization, we examined the subcellular localization pattern in PIN2‐GFP genotypes with phosphosite substitutions. PIN2WT‐GFP proteins were accumulated at the plasma membrane of epidermal and cortical cells, as previously described (Fig 8A) (Müller et al, 1998). PIN2WT‐GFP fluorescence signal increased in epidermal and cortical cells 2 h after nitrate treatments as compared to roots in control conditions (roots without nitrate treatments, Fig 8A). Interestingly, mutations at S439 altered this pattern. PIN2S439A‐GFP plants showed higher fluorescence in the plasma membrane even without nitrate treatment as compared to PIN2wt‐GFP or PIN2S439D‐GFP (Fig 8A). The total fluorescence in all experimental conditions analyzed here was similar (Fig 8B). In response to nitrate, PIN2WT‐GFP and PIN2S439A‐GFP were accumulated at the plasma membrane of epidermal and cortical cells at comparable levels (Fig 8C and D). On the contrary, PIN2S439D‐GFP plants showed lower levels at epidermal and cortical cells in response to nitrate treatments as compared to PIN2WT‐GFP or PIN2S439A‐GFP (Fig 8C and D).
Figure 8. PIN2 phosphorylation at S439 is important for modulation of protein localization in response to nitrate treatments.
-
AConfocal microscopy using Zeiss Airyscan microscope and Zeiss‐blue3.1 software was used to visualize PIN2 in Arabidopsis roots. “e” denotes epidermis and “c” cortex. Scale bar = 20 µm.
-
B–DTuckey box plots show PIN2‐GFP fluorescence intensity quantification (number of roots analyzed per experimental conditions = 8–10; arbitrary units, a.u.) at the total cell membrane in roots (B), and from epidermal (C) and cortex (D) plasma membrane in the different genotypes. The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5× IQR (interquartile range). Outlier data are plotted individually. Letters denote statistically significant difference between means as determined by one‐way ANOVA followed by Tukey HSD post hoc test (P < 0.05).
Source data are available online for this figure.
These results indicate that PIN2 phosphorylation status at S439 is important for a correct subcellular localization pattern in response to nitrate treatments. Moreover, these results indicate that post‐translational control impinging upon PIN2 localization is required for RSA changes in response to nitrate treatments.
Discussion
A key plant nutrient, N also acts as a signal that regulates a myriad of plant growth and developmental processes. Nitrate, a main N source in natural and agriculture soils, elicits genome‐wide changes in gene expression for thousands of genes involved in various biological functions. Nitrate responses have been characterized in great detail at the transcriptome level. However, post‐translational modifications have not been characterized in detail. In this study, we evaluated phosphoproteomic profiles in Arabidopsis roots in response to nitrate treatments. We focused on characterizing early nitrate‐elicited changes in protein phosphorylation. Protein phosphorylation and dephosphorylation play a central role in modulating protein function in plant signaling pathways involved in a wide range of processes relating to hormones, nutrients, and responses to stress (Lan et al, 2012; Umezawa et al, 2013; Zhang et al, 2013, 2014; Lin et al, 2015; Vu et al, 2016).
Our analysis demonstrated that early and late changes in phosphoprotein levels occur in response to nitrate in roots. Furthermore, we identified candidates for nitrate signaling and biological functions underlying the nitrate response in roots. Interestingly, the majority of proteins and corresponding genes identified in our analysis have not been previously associated with nitrate responses. The phosphoproteomic profile was characteristic of each time‐point, showing dynamic changes in phosphorylation patterns in response to nitrate treatments. Early changes in phosphorylation levels (5 min) mainly affected proteins associated with gene regulation, including transcription factors and splicing process. These results are consistent with rapid and dynamic N responses observed at the mRNA level described in previous studies (Vidal et al, 2013; Gaudinier et al, 2018; Varala et al, 2018; Brooks et al, 2019). Recent studies have identified new transcription factors (Gaudinier et al, 2018; Varala et al, 2018; Brooks et al, 2019) involved in the control of gene expression by nitrate. Interestingly, the majority of the transcription factors that we detected as differentially phosphorylated had not been identified as part of the nitrate response. The phosphoproteomic response to nitrate at 20 min revealed a group of different phosphoproteins, mostly involved in protein binding and transport. In this dataset, we found proteins known to be involved in nitrogen response as differentially phosphorylated, including the high‐affinity nitrate transporter NRT2.1 (Engelsberger & Schulze, 2012; Menz et al, 2016) and AMT1.3 (Lanquar et al, 2009). In our experimental conditions, site T464 in AMT1.3 was phosphorylated. Since phosphorylation of this site inhibits ammonium transport (Lanquar et al, 2009), the increased phosphorylation status at this site may be related to the fact that our experimental conditions focused on nitrate transport in Arabidopsis roots. We also identified phosphoproteins associated with signaling pathways and transcription factors, uncovering regulatory networks linked to transcriptomic changes occurring later in the response to nitrate. Changes in phosphoprotein patterns improve our understanding of signaling mechanisms that connect nitrate transporters/sensors with transcriptomic responses and other biological processes. The small overlap between the phosphoproteomic studies to date highlights the importance of ours and additional future studies to address this important aspect of post‐translational modifications in response to N signals in plants.
NRT1.1/NPF6.3 is the main nitrate sensor described to date (Liu & Tsay, 2003; Ho et al, 2009). Interestingly, we observed important differences in the phosphoproteome of chl1‐5 mutant as compared to wild‐type plants in response to nitrate treatments. These results emphasize that NRT1.1/NPF6.3, calcium, and kinases/phosphatases make up the canonical nitrate signaling pathway of Arabidopsis roots. Intriguingly, chl1‐5 mutant plants show differences in phosphoprotein levels as compared to wild‐type plants. The small number of proteins identified in the absence of NRT1.1 function suggests an alternative nitrate signaling pathway exists. Alternative nitrate sensor candidates exist, such as NRT2.1, which acts as a repressor of lateral root initiation in response to nitrate, a role shown to be independent of nitrate uptake (Little et al, 2005).
Recent evidence showed that NRT1.1/NPF6.3 and PLC activity are required for nitrate‐induced increases in cytoplasmic Ca2+ levels (Riveras et al, 2015). Our results indicated that PLC2 phosphoprotein levels increased in response to nitrate. Nitrate signaling pathways also involve CBL–CIPK complex and CPK–NLP regulatory network (Liu et al, 2020). Furthermore, we identified the molecular function “kinases” as overrepresented in our network analyses and potential phosphoprotein kinases that change its abundance in response to nitrate. The levels of two phosphoproteins involved in MAPK cascades (MKP1 and MKK2) were regulated by nitrate under our experimental conditions. A previous study identified a MAPK kinase cascade under nitrate resupply conditions, where MKP1 was also found phosphorylated under low‐affinity nitrate uptake (Wu et al, 2017). Consistent with these results, we found the overrepresented the MAPK motif in nitrate‐regulated phosphopeptides, suggesting a role of MAPKs in response to N availability.
The transcriptomic nitrate response was not mirrored at phosphoproteomic levels. This lack of correlation between mRNA and protein or phosphoprotein levels has been documented in other nitrogen‐phosphoproteomic studies (Engelsberger & Schulze, 2012; Menz et al, 2016), plant responses to different stimulus (Walley et al, 2013, 2016), and responses in other organisms (Vogel & Marcotte, 2012). Furthermore, post‐translational regulation does not always require a change in gene expression or de novo protein synthesis. Post‐translational control could be faster, allowing rapid adaptation to environmental changes (Zhang et al, 2015). Interestingly, the genes coding for nitrate‐modulated phosphoproteins identified in this study are highly co‐expressed across many different experimental conditions but not regulated by nitrate treatments (Obayashi et al, 2014). This finding suggests that this group of genes is functionally related and regulated at the mRNA level in response to several endogenous or exogenous cues. In the context of nitrate responses, the products of these genes are regulated at the post‐translational level, uncovering a new layer of control that enables signal crosstalk and fine‐tuning. The analysis of mRNA levels in response to nitrate under a number of experimental conditions (27 experimental datasets corresponding to 131 array, Canales et al, 2014) revealed that most of the phosphoprotein encoding genes regulated by nitrate exhibit high mRNA levels. Specifically, 63% have greater than average mRNA levels. Only 10% of them display low mRNA levels. This finding suggests the genes coding for phosphoproteins that we identified are expressed and susceptible to regulation by phosphorylation. Modulation at the phosphoprotein level would be a regulatory layer independent of the nitrate‐mediated changes in mRNA levels. Our results also highlight the need for integrated analysis with a multi‐omics approach to decipher plant responses to environmental cues.
Moreover, our phosphoproteomic analysis was performed assuming most proteins were not differentially expressed, and the most significant missing values were produced because they are below or around the detection limit. Missing values are expected in phosphoproteomics experiments using MS/MS methodology mainly due to sensitivity issues and biological factors, including a low abundance and transitory nature of phosphorylated proteins (Zhang et al, 2018). Two approaches to deal with missing values are removing proteins that have insufficient samples for analysis (i) or using an imputing method for the missing values (ii). The first approach involved perform the analyses on a limited part of the dataset and unnecessarily excluding data. Thus, replace the missing values with a selected strategy avoids apparent mathematic problems (infinite fold‐change values) or excludes a large part of the data. We selected the MinProp method explicitly designed for the low‐abundant nature of these absences and impute the left‐censored data appropriately. The main limitation of this approach is that missing values can also occur throughout the entire range of values and left‐censored methods had been described as less effective with this type of missing values (Lazar et al, 2016). Thus, imputing below a detection limit may inappropriately take values too small and influence statistical analysis.
It is now clear that auxin plays a central role in the plant root response to changes in nitrate availability. Nitrate regulates primary root growth, lateral root initiation, and elongation. Auxin in turn is key during root development (Laskowski et al, 2008), particularly in initiation and growth of lateral roots (Benková et al, 2003). Several reports show that auxin signaling, biosynthesis, transport, and accumulation are affected during nitrate responses (Krouk et al, 2010; Ma et al, 2014), and transcriptomic analyses demonstrate that genes involved in auxin response are controlled by nitrate (Gutiérrez et al, 2006; Vidal et al, 2010, 2013). The main nitrate transporter NRT1.1/NPF6.3 can also transport auxin (Krouk et al, 2010; Mounier et al, 2013; Zhang et al, 2019). A recent study also showed that NRT1.1/NPF6.3 negatively regulates the TAR2 auxin‐biosynthetic gene and the LAX3 auxin‐influx transport gene at low nitrate concentrations, repressing lateral root development (Maghiaoui et al, 2020). These results suggest that an interplay between nitrate signaling and auxin transport occurs at different levels (Krouk et al, 2011; Krouk, 2016; Vega et al, 2019). Consistent with these findings, we found that the molecular function “auxin transport” was overrepresented in our cluster and network analyses. We showed that dephosphorylation of PIN2 in a novel phosphosite is a part of a regulatory mechanism for RSA responses triggered by nitrate. The phosphorylation/dephosphorylation of PIN proteins at specific sites (serine or threonine) located in their higher loops has been shown to play important roles in modulating PIN functions (Michniewicz et al, 2007; Dhonukshe et al, 2010; Huang et al, 2010; Weller et al, 2017), including trafficking (Ganguly et al, 2014). We showed that phosphorylation/dephosphorylation of PIN2 at S439 is important for PIN2 plasma membrane localization in epidermal and cortical cells in response to nitrate. Previous studies have shown that changes in PIN2 membrane localization and polarity interfere with PIN2 function in auxin transport with an impact on RSA during alkaline stress (Xu et al, 2012) or low phosphate (Kumar et al, 2015). In addition, recent studies showed that kinase cascade modules 3′‐PHOSPHOINOSITIDE‐DEPENDENT PROTEIN KINASE 1 (PDK1)‐D6 PROTEIN KINASEs (D6PK) and PDK1‐AGC1 kinase PROTEIN KINASE ASSOCIATED WITH BRX (PAX) regulate auxin distribution through PIN phosphorylation (Tan et al, 2020; Xiao & Offringa, 2020). The phosphorylation of PIN affects PIN‐mediated auxin transport, which controls plant growth (Tan et al, 2020) and other developmental processes (Xiao & Offringa, 2020). These results suggest that PIN phosphorylation is part of a regulatory switch that strictly controls the directional transport of auxin and subsequent growth or developmental processes.
Our results suggest a model (Fig 9) in which nitrate promotes dephosphorylation of PIN2, which then impacts localization and auxin transport. Modulation of PIN2 function could affect growth of primary and lateral roots for optimal nutrient uptake. Beyond this new regulatory mechanism involving PIN2 protein, our phosphoproteomics results identify novel proteins, which may be interesting targets for future studies or biotechnological developments for improved nitrogen use efficiency or crop yield.
Figure 9. Schematic model of the role of PIN2 and phosphorylation in root nitrate responses.
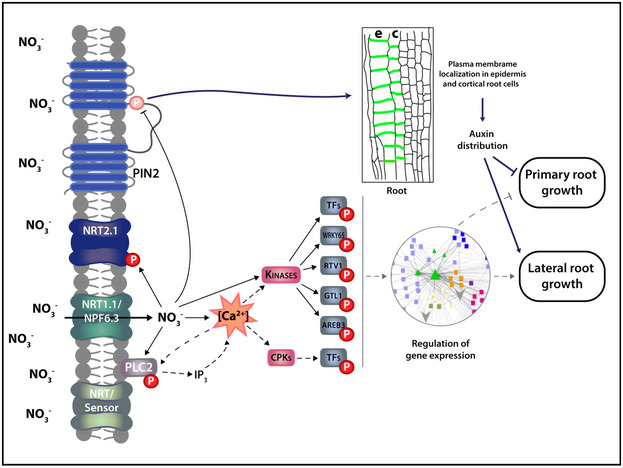
Nitrate treatments increase cytosolic calcium levels which interact with calcium‐binding proteins such as kinases in the nitrate signaling pathway. Kinases phosphorylate protein targets such as transcription factors that can mediate changes in gene expression in response to nitrate treatments. Nitrate can also cause dephosphorylation of specific proteins. We showed nitrate treatments promote PIN2 dephosphorylation at S439 which impact membrane localization and polarity of the PIN2 protein. This post‐translational regulatory mechanism is important for modulation of primary root growth and lateral root density in response to nitrate treatments. Arrows and bar‐headed solid lines represent activation or inhibition in response to nitrate, respectively. Dashed arrows indicate proposed connections. A working model (depicted in synopsis image) was constructed with the BioRender.com software.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. Columbia‐0 accession plants were used as wild‐type genotype in all experiments. The transgenic lines PIN2::PIN2wt‐GFP, PIN2::PIN2S439D‐GFP and PIN2::PIN2S439A‐GFP were introduced into eir1‐1 background (pin2 null mutant plants), using strategies described previously. PIN2 lines were generated by Gybson assembly as described (Gibson et al, 2009) using pGREEN backbone vector (Hellens et al, 2000). PIN2::PIN2wt‐GFP line was generated by insertion of mGFP5 into PIN2 coding sequence at nucleotide 1,215 from ATG (between Thr405 and Arg406) (Luschnig et al, 1998; Xu & Scheres, 2005). For PIN2 promoter, 2,178 bp upstream of the start codon was used. For the generations of PIN2::PIN2S439D‐GFP and PIN2::PIN2S439A‐GFP lines, Serine 439 was replaced by aspartate (PIN2S439D ) or alanine (PIN2S439A ) by site‐directed mutagenesis using Gibson Assembly.
Seeds were sterilized using 50% chlorine solution for 7 min and washed with sterile distilled water three times. Then, 1,500 Arabidopsis seedlings were placed into a hydroponic system (Phytatrays) with MS‐modified basal salt media without N (Phytotechnology Laboratories, M531) supplemented with 1 mM ammonium as the only N source. Plants were grown under long‐day photoperiods (16 h light/8 h dark and a temperature of 22°C) for 14 days using a plant growth incubator (Percival Scientific, Inc.). At day 15, plants were treated with 5 mM KNO3 or 5 mM KCl for different time periods as indicated, using a protocol described previously (Vidal et al, 2010). For the phenotypic study of root response to nitrate, plants were grown as described above and were treated with 5 mM KNO3 or 5 mM KCl for 3 days.
Root architecture analysis
For root phenotyping, plants were scanned in plates using an Epson Perfection V700 Photo scanner, and root length was measured using Fiji (v1.52). Initiating and emerging lateral roots were analyzed using DIC optics on a Nikon Eclipse 80i microscope, as described (Vidal et al, 2010). The data were statistically analyzed in the GraphPad Prism 8.0 Program.
Protein extraction, phosphopeptide enrichment, and mass spectrometry analysis
Proteins were isolated from 1 g of frozen tissue per sample in each experimental condition (2 biological replicates for nitrate treatments at 20 min (Col‐0 roots) and 3 biological replicates for all other experimental conditions). Each biological replicate consisted of a pool of 4.500 roots collected from Arabidopsis plants grown in independent experiments under the same experimental conditions. Sample preparation and protein extraction were performed using previously described methods (Walley et al, 2013, 2016). Phosphopeptide enrichment was performed using 1% (w/v) colloidal CeO2 into an acidified peptide solution at 1:10 (w/w). Mass spectrometry (MS) and peptide identification were based on protocols described previously (Facette et al, 2013). Briefly, the generated spectra were analyzed on LTQ Velos linear ion trap tandem mass spectrometer (Thermo Electron) and phosphorylation sites were identified into a specific amino acid within a peptide by using the variable modification localization score in Agilent Spectrum Mill software (Chalkley & Clauser, 2012). Proteins were grouped based on their shared, common phophopeptides using principles of parsimony to address redundancy in proteins. Proteins classified within the same group share the same subset of phosphopeptides (Dataset EV5 and EV6). Phosphorotein levels were quantified using spectral counting, as described previously (Walley et al, 2013, 2016). MS data were normalized using the total number of spectral counts (nSPC) for each MS run. Expressed phosphoproteins were defined by at least one SPC, after the application of quality score cutoff in MS analysis, in minimum two of the tree biological replicates. Phosphoproteins with reverse protein sequence hit against the Arabidopsis genome or quantified in only one experimental condition were also eliminated, following the proteomic analysis pipeline described in DEP (Zhang et al, 2018) and MSnBase (Gatto & Lilley, 2012) packages in R/Bioconductor (Huber et al, 2015). To identify nitrate‐regulated phosphoproteins in Arabidopsis roots, raw data were log‐transformed, quantile normalized, and the missing values were replaced by random draws from a Gaussian distribution centered to minimal value in the sample processing each experimental conditions using DEP in RStudio (https://rstudio.com) and MEV (http://mev.tm4.org/) software. This imputation method was selected based on the fact missing values in our dataset were mainly on the left tail of the value distribution. The group of proteins with missing values showed low‐intensity values (median value 2.36 in base log2) as compared with proteins without missing values (median value 3.89 in base log2). Two‐way analysis of variance (significance: P < 0.05) was performed using RStudio. For ANOVA, we used a model with abundance Y of a given phosphoprotein i calculated as Yi = β0 + β1N + β2Ti + β3N‐Ti + ɛ, where β0 is the global mean, and where β1, β2, and β3 are the effects of the nitrate treatment, the time and the interaction between these two factors (N–Ti), respectively. The variable ɛ corresponds to the unexplained variance. We selected the group of phosphoproteins significantly affected by N or N–Ti (P < 0.05, Dataset EV3), and we organized them based on changes at 5 or 20 min in response to nitrate. Encoded genes for phosphoproteins showing a similar pattern were analyzed and visualized using the average linkage hierarchical clustering performed in Cluster 2.11 software, as described (Eisen et al, 1998).
Gene network analysis
Arabidopsis encoded genes from our phosphoproteomics data were used. The gene network was generated by integrating different information, including protein–protein interactions from BioGRID (Oughtred et al, 2018), predicted protein–DNA interactions of Arabidopsis TFs (DapSeq) (Weirauch et al, 2014; Bartlett et al, 2017), Arabidopsis metabolic pathways (KEGG), and miRNA‐RNA, as described previously (Gutiérrez et al, 2006). This analysis also included predicted regulatory connections between phosphoproteins that we detected and kinase families. A kinase‐substrate analysis, identifying the most significant phosphorylation motifs and their predicted kinases was performed using the Motif‐X algorithm (Schwartz & Gygi, 2005) and PhosPhAt Kinase‐Target interactions database (Zulawski et al, 2012, 2014). The resulting network was visualized using CYTOSCAPE software (Shannon et al, 2003). Cluster and Gene Ontology analysis into the network were achieved with ClusterMaker (Morris et al, 2011), ClueGO (Bindea et al, 2009), and Bingo (Maere et al, 2005) tools for biological networks in cytoscape.
Phos‐Tag PAGE and Western immunoblotting
Affinity‐based SDS–PAGE identification of phosphorylated PIN2 isoforms was performed based on the protocols by Komis et al, (2014) and the protocol given by the Phos‐tag manufacturer (FUJIFILM Waco Chemicals) with slight modifications. Arabidopsis seedlings were grown on modified MS plates with ammonium as the only N source for 7 days. Next, seedlings (n ≥ 40) were transferred to 5 mM nitrate amended agar plates and were incubated for 6 h in light. Root samples were collected and homogenized in liquid N2 and then extracted with extraction buffer—50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X‐100, 10 µM MG‐132, and 0.1 mM PMSF—supplemented with protease and phosphatase inhibitor cocktails (Roche). Buffer volumes were adjusted to fresh weight (100 µl/100 mg tissues). Homogenized samples were centrifuged (4⁰C, 15 min, 19,098 g), and the supernatant was aliquoted (50 µg protein/20–25 µl) and incubated at 45°C for 5 min in the presence of SDS loading buffer. Next, samples were loaded onto an acrylamide, Bis‐Tris/HCl gel containing 50 µmol/l Phos‐tagTM (AAL‐107) pendant and Zn2+ as cation. Electrophoresis was run at 15 mA/gel for 5–6 h or until the proteins are nicely separated (usually until the 25 kDa prestained protein marker just exit the gel assembly). Next, gels were incubated for 30 min in transfer buffer containing 10 mM EDTA and were blotted to PVDF membranes using Tris/Glycine transfer buffer (25 mM Tris, 192 mM Glycine, 5% methanol). After blocking with 5% milk in TBST, the membrane was probed with α‐PIN2 antibody (1:1,000) for 2 h (RT) followed by α‐rabbit IgG‐HRP (1:15,000) (Amersham) for 1 h (RT). In Western blot analysis, the membrane was probed with monoclonal GFP‐antibody Living Colors® (1:5,000) for 2 h (RT) followed by α‐mouse IgG‐HRP (1:10,000) for 1 h (RT). After treating the membrane with Supersignal West Femto western chemiluminescent HRP substrate (Thermo Fisher Scientific), luminescent signals were detected using a liquid nitrogen‐cooled charge‐coupled device camera (Bio‐Rad). Digital images were analyzed, and signals were quantified using the Fiji software.
Confocal microscopy and image analysis
eir1‐1 mutants complemented with PIN2::PIN2wt‐GFP, PIN2::PIN2S439A‐GFP, PIN2::PIN2S439D‐GFP were grown in modified MS plates with 1 mM ammonium as the only N source. 2‐week‐old plants were treated with 5 mM nitrate or without treatment (control conditions) for 2 h. Roots were stained with propidium iodide (PI) and mounted on a slice for microscopic analysis. Images were acquired with Zeiss LSM880 confocal microscope with Airyscan equipped with a 40×Plan‐Apochromat water immersion objective. Fluorescence signals for GFP (excitation 488 nm, emission 507 nm) and PI (excitation 536 nm, emission 617 nm) were detected.
For image quantification (PIN2‐GFP fluorescence intensity measurements), maximum intensity projections of confocal pictures were used in epidermis and cortex cells. Images were handled and analyzed with Zeiss blue (v3.1) and Fiji (v1.52) software. The experiment was performed with 3 biological replicates and 2‐3 roots per experimental conditions were analyzed in each replicate.
Author contributions
AV performed most experiments with help from IF, KO, RA, ZS, and JO. ZS conducted phosphoproteomic experiments. SB supervised phosphoproteomic experiment. JO and IF conducted phospho‐mimicking plant experiments and helped with their analysis. KO, RA, and AV performed phos‐tag PAGE and Western immunoblotting experiment and analysis. EB supervised immunoblotting experiments and analysis. AV and RAG designed research. AV and RAG wrote the article. All authors were involved in the review of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Source Data for Expanded View/Appendix
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgments
This work was supported by ANID—Millennium Science Initiative Program—ICN17_022, Fondo de Desarrollo de Areas Prioritarias (FONDAP) Center for Genome Regulation (15090007), ANID—Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) 1180759 (to RAG) and 1171631 (to AV). We would like to thank Unidad de Microscopía Avanzada UC (UMA UC).
EMBO reports (2021) 22: e51813.
Data availability
Phosphoproteomic datasets (raw spectra) had been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository with the MassIVE ID MSV000086215 (http://massive.ucsd.edu/MSV000086215).
References
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras‐López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay Let al (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 13 [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Schinke A‐L, Brooks MD, Pasquino A, Leonelli L, Varala K, Safi A, Krouk G, Krapp A, Coruzzi GM (2020) Transient genome‐wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen‐response cascade. Nat Commun 11: 1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal GS, Huber SC (2002) Divalent cations and polyamines bind to loop 8 of 14‐3‐3 proteins, modulating their interaction with phosphorylated nitrate reductase. Plant J 29: 119–129 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Liao P‐C, Gage DA, Huber SC (1996) The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14‐3‐3 protein. Febs Lett 387: 127–131 [DOI] [PubMed] [Google Scholar]
- Barbosa ICR, Hammes UZ, Schwechheimer C (2018) Activation and polarity control of PIN‐FORMED auxin transporters by phosphorylation. Trends Plant Sci 23: 523–538 [DOI] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR (2017) Mapping genome‐wide transcription‐factor binding sites using DAP‐seq. Nat Protoc 12: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W‐H, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová Eet al (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015 [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Perrine‐Walker F, Pervent M, Rochette J, Cuesta C, Benkova E, Martinière A, Bach L, Krouk G, Gojon Aet al (2016) Nitrate controls root development through post‐transcriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol 172: 1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MD, Cirrone J, Pasquino AV, Alvarez JM, Swift J, Mittal S, Juang C‐L, Varala K, Gutiérrez RA, Krouk Get al (2019) Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome‐wide interactions. Nat Commun 10: 1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J, Contreras‐López O, Álvarez JM, Gutiérrez RA (2017) Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana . Plant J 92: 305–316 [DOI] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA (2014) Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley RJ, Clauser KR (2012) Modification site localization scoring: strategies and performance. Mol Cell Proteom Mcp 11: 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yao Q, Gao X, Jiang C, Harberd NP, Fu X (2016) Shoot‐to‐root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr Biol 26: 640–646 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1: e0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutzen PJ, Mosier AR, Smith KA, Winiwarter W (2008) N2O release from agro‐biofuel production negates global warming reduction by replacing fossil fuels. Atmos Chem Phys 8: 389–395 [Google Scholar]
- Davidson EA (2009) The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nat Geosci 2: 659–662 [Google Scholar]
- Dhonukshe P, Huang F, Galvan‐Ampudia CS, Mähönen AP, Kleine‐Vehn J, Xu J, Quint Ab, Prasad K, Friml J, Scheres Bet al (2010) Plasma membrane‐bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome‐wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen‐starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facette MR, Shen Z, Bjornsdottir FR, Briggs SP, Smith LG (2013) Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 25: 2798–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhu J, Du X, Cui X (2012) Effects of three auxin‐inducible LBD members on lateral root formation in Arabidopsis thaliana . Planta 236: 1227–1237 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg Get al (2004) A PINOID‐dependent binary switch in apical‐basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Park M, Kesawat MS, Cho H‐T (2014) Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN‐FORMED proteins. Plant Cell 26: 1570–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto L, Lilley K (2012) MSnbase ‐ an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28: 288–289 [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Rodriguez‐Medina J, Zhang L, Olson A, Liseron‐Monfils C, Bågman A‐M, Foret J, Abbitt S, Tang M, Li Bet al (2018) Transcriptional regulation of nitrogen‐associated metabolism and growth. Nature 563: 259–264 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R‐Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutiérrez RA, Coruzzi GM, Birnbaum KD (2008) Cell‐specific nitrogen responses mediate developmental plasticity. Proc National Acad Sci 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian J‐C (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12: 328–338 [DOI] [PubMed] [Google Scholar]
- Gras D, Vidal E, Undurraga S, Riveras E, Moreno S, Dominguez‐Figueroa J, Alabadi D, Blázquez M, Medina J, Gutiérrez R (2018) SMZ/SNZ and gibberellin signaling are required for nitrate‐elicited delay of flowering time in Arabidopsis thaliana . J Exp Bot 69: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336: 1673–1675 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Gifford ML, Poultney C, Wang R, Shasha DE, Coruzzi GM, Crawford NM (2007) Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot 58: 2359–2367 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM (2006) Qualitative network models and genome‐wide expression data define carbon/nitrogen‐responsive molecular machines in Arabidopsis . Genome Biol 8: R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez Ra, Stokes Tl, Thum K, Xu X, Obertello M, Katari Ms, Tanurdzic M, Dean A, Nero Dc, McClung Cret al (2008) Systems approach identifies an organic nitrogen‐responsive gene network that is regulated by the master clock control gene CCA1. Proc National Acad Sci 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Kudla J (2011) Calcium decoding mechanisms in plants. Biochimie 93: 2054–2059 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX (2007) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant‐specific phosphorylation site predictor. Nucleic Acids Res 36: D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ho C‐H, Lin S‐H, Hu H‐C, Tsay Y‐F (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hu H‐C, Wang Y‐Y, Tsay Y‐F (2009) AtCIPK8, a CBL‐interacting protein kinase, regulates the low‐affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galván‐Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JL, Huber SC, Campbell WH, Redinbaugh MG (1992) Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch Biochem Biophys 296: 58–65 [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke Tet al (2015) Orchestrating high‐throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP (2010) A tissue‐specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Weiner H, Kandlbinder A, Tsai C, Rockel P, Sonoda M, Planchet E (2002) Modulation of nitrate reductase: some new insights, an unusual case and a potentially important side reaction. J Exp Bot 53: 875–882 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DEet al (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Takáč T, Bekešová S, Vadovič P, Samaj J (2014) Affinity‐based SDS PAGE identification of phosphorylated Arabidopsis MAPKs and substrates by acrylamide pendant Phos‐TagTM . Methods Mol Biology Clifton N J 1171: 47–63 [DOI] [PubMed] [Google Scholar]
- Krouk G (2016) Hormones and nitrate: a two‐way connection. Plant Mol Biol 91: 599–606 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine‐Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung Ket al (2010) Nitrate‐regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Pandya‐Kumar N, Dam A, Haor H, Mayzlish‐Gati E, Belausov E, Wininger S, Abu‐Abied M, McErlean CSP, Bromhead LJet al (2015) Arabidopsis response to low‐phosphate conditions includes active changes in actin filaments and PIN2 polarization and is dependent on strigolactone signalling. J Exp Bot 66: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W (2012) Quantitative phosphoproteome profiling of iron‐deficient Arabidopsis roots. Plant Physiol 159: 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB (2009) Feedback inhibition of ammonium uptake by a phospho‐dependent allosteric mechanism in Arabidopsis . Plant Cell 21: 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CAT, Hogeweg P, Marée AFM, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. Plos Biol 6: e307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar C, Gatto L, Ferro M, Bruley C, Burger T (2016) Accounting for the multiple natures of missing values in label‐free quantitative proteomics data sets to compare imputation strategies. J Proteome Res 15: 1116–1125 [DOI] [PubMed] [Google Scholar]
- Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé‐Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8: ra43 [DOI] [PubMed] [Google Scholar]
- Lin L‐L, Hsu C‐L, Hu C‐W, Ko S‐Y, Hsieh H‐L, Huang H‐C, Juan H‐F (2015) Integrating phosphoproteomics and bioinformatics to study brassinosteroid‐regulated phosphorylation dynamics in Arabidopsis . BMC Genom 16: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel‐Vedele F, Krapp A, Malamy JE (2005) The putative high‐affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K‐H, Diener A, Lin Z, Liu C, Sheen J (2020) Primary nitrate responses mediated by calcium signalling and diverse protein phosphorylation. J Exp Bot 71: 4428–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K‐H, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa Set al (2017) Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Tsay Y‐F (2003) Switching between the two action modes of the dual‐affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root‐specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana . Gene Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y (2014) Auxin biosynthetic gene TAR2 is involved in low nitrogen‐mediated reprogramming of root architecture in Arabidopsis . Plant J 78: 70–79 [DOI] [PubMed] [Google Scholar]
- MacKintosh C (1992) Regulation of spinach‐leaf nitrate reductase by reversible phosphorylation. Biochimica Et Biophysica Acta Bba ‐ Mol Cell Res 1137: 121–126 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Maghiaoui A, Bouguyon E, Cuesta C, Perrine‐Walker F, Alcon C, Krouk G, Benkova E, Nacry P, Gojon A, Bach L (2020) The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J Exp Bot 71: 4480–4494 [DOI] [PubMed] [Google Scholar]
- Marín IC, Loef I, Bartetzko L, Searle I, Coupland G, Stitt M, Osuna D (2010) Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 233: 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz J, Li Z, Schulze WX, Ludewig U (2016) Early nitrogen‐deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho‐) proteome levels between nitrate and ammonium nutrition. Plant J 88: 717–734 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2018) PANTHER version 14: more genomes, a new PANTHER GO‐slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD (2019) Protocol update for large‐scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 14: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang Fet al (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE (2011) clusterMaker: a multi‐algorithm clustering plugin for Cytoscape. BMC Bioinform 12: 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2013) Auxin‐mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37: 162–174 [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Okamura Y, Ito S, Tadaka S, Aoki Y, Shirota M, Kinoshita K (2014) ATTED‐II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol 55: e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa‐Ohnishi M, Matsubayashi Y (2017) Shoot‐to‐root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat Plants 3: 1–6 [DOI] [PubMed] [Google Scholar]
- Ötvös K, Marconi M, Vega A, O’Brien J, Johnson A, Abualia R, Antonielli L, Montesinos JC, Zhang Y, Tan Set al (2021) Modulation of plant root growth by nitrogen source‐defined regulation of polar auxin transport. EMBO J 40: e106862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R, Stark C, Breitkreutz B‐J, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam Ret al (2018) The BioGRID interaction database: 2019 update. Nucleic Acids Res 47: D529–D541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout A, Ruffel S, Novák O, Lacombe B, Crabos A, Krouk G, Petřík I (2018) Responses to systemic nitrogen signaling in Arabidopsis roots involve trans ‐Zeatin in shoots. Plant Cell 30: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh‐Kumar SP (2008) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Gene Dev 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultney CS, Gutiérrez RA, Katari MS, Gifford ML, Paley WB, Coruzzi GM, Shasha DE (2007) Sungear: interactive visualization and functional analysis of genomic datasets. Bioinformatics 23: 259–261 [DOI] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA (2015) The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis . Plant Physiol 169: 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GP, Vitousek PM (2009) Nitrogen in agriculture: balancing the cost of an essential resource. Ann Rev Environ Resour 34: 97–125 [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR (1995) Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate‐cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Poitout A, Krouk G, Coruzzi GM, Lacombe B (2016) Long‐distance nitrate signaling displays cytokinin dependent and independent branches. J Integr Plant Biol 58: 226–229 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T (1997) Partial characterization of the signaling pathway for the nitrate‐dependent expression of genes for nitrogen‐assimilatory enzymes using detached maize leaves. Plant Cell Physiol 38: 837–843 [Google Scholar]
- Scheible W‐R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios‐Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome‐wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large‐scale data sets. Nat Biotechnol 23: 1391–1398 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Breuer C, Kawamura A, Clark NM, Rymen B, Braidwood L, Morohashi K, Busch W, Benfey PN, Sozzani Ret al (2018) GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6‐LIKE 4 in Arabidopsis . Development 145: dev159707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G, Kuchinsky A, Morris JH, States DJ, Meng F (2010) GLay: community structure analysis of biological networks. Bioinformatics 26: 3135–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Huber SC, Crawford NM (1996) Identification in vitro of a post‐translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell 8: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi K, Mitsuyama T, Sugimoto T, Kleinhofs A, Warner RL, Yoshikiyo O (1999) Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Sci Plant Nutr 45: 1015–1019 [Google Scholar]
- Tan S, Zhang X, Kong W, Yang X‐L, Molnár G, Vondráková Z, Filepová R, Petrášek J, Friml J, Xue H‐W (2020) The lipid code‐dependent phosphoswitch PDK1–D6PK activates PIN‐mediated auxin efflux in Arabidopsis . Nat Plants 6: 556–569 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana . Sci Signal 6: rs8 [DOI] [PubMed] [Google Scholar]
- Vanacker H, Sandalio L, Jiménez A, Palma JM, Corpas FJ, Meseguer V, Gómez M, Sevilla F, Leterrier M, Foyer CHet al (2006) Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. J Exp Bot 57: 1735–1745 [DOI] [PubMed] [Google Scholar]
- Varala K, Marshall‐Colón A, Cirrone J, Brooks MD, Pasquino AV, Léran S, Mittal S, Rock TM, Edwards MB, Kim GJet al (2018) Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc National Acad Sci USA 115: 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega A, O’Brien JA, Gutiérrez RA (2019) Nitrate and hormonal signaling crosstalk for plant growth and development. Curr Opin Plant Biol 52: 155–163 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate‐responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana . Proc National Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis . Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras‐Lopez O, Gutierrez RA (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc National Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13: 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LD, Stes E, Bel MV, Nelissen H, Maddelein D, Inzé D, Coppens F, Martens L, Gevaert K, Smet ID (2016) Up‐to‐date workflow for plant (Phospho)proteomics identifies differential drought‐responsive phosphorylation events in maize leaves. J Proteome Res 15: 4304–4317 [DOI] [PubMed] [Google Scholar]
- Walch‐Liu P, Liu L‐H, Remans T, Tester M, Forde BG (2006) Evidence that l ‐Glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana . Plant Cell Physiol 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JRet al (2016) Integration of omic networks in a developmental atlas of maize. Science 353: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Shen Z, Sartor R, Wu KJ, Osborn J, Smith LG, Briggs SP (2013) Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc National Acad Sci USA 110: E4808–E4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1, 000 rapidly responding genes and new linkages to glucose, trehalose‐6‐phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase‐null mutant of Arabidopsis . Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M, Yang A, Albu M, Cote AG, Montenegro‐Montero A, Drewe P, Najafabadi H, Lambert S, Mann I, Cook Ket al (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller B, Zourelidou M, Frank L, Barbosa ICR, Fastner A, Richter S, Jürgens G, Hammes UZ, Schwechheimer C (2017) Dynamic PIN‐FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux‐dependent growth. Proc National Acad Sci USA 114: E887–E896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Lauter FR, Ninnemann O, Gillissen B, Walch‐Liu P, Engels C, Jost W, Frommer WB (2000) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J 21: 167–175 [DOI] [PubMed] [Google Scholar]
- Wu XN, Xi L, Pertl‐Obermeyer H, Li Z, Chu L‐C, Schulze WX (2017) Highly efficient single‐step enrichment of low abundance phosphopeptides from plant membrane preparations. Front Plant Sci 8: 1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Offringa R (2020) PDK1 regulates auxin transport and Arabidopsis vascular development through AGC1 kinase PAX. Nat Plants 6: 544–555 [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP‐RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Baluška F, Ding G, Shi W, Ye N, Zhang J (2012) PIN2 is required for the adaptation of Arabidopsis roots to alkaline stress by modulating proton secretion. J Exp Bot 63: 6105–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient‐induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG, Kay SA, Pruneda‐Paz JL, Davani A, Crawford NM (1999) Dual pathways for regulation of root branching by nitrate. Proc National Acad Sci USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana . J Exp Bot 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou H, Berke L, Heck AJR, Mohammed S, Scheres B, Menke FLH (2013) Quantitative phosphoproteomics after auxin‐stimulated lateral root induction identifies an SNX1 protein phosphorylation site required for growth. Mol Cell Proteomics 12: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencík A, Rolcík J, Friml J (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc National Acad Sci USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lv D, Ge P, Bian Y, Chen G, Zhu G, Li X, Yan Y (2014) Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J Proteomics 109: 290–308 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bhattacharya S, Pi J, Clewell RA, Carmichael PL, Andersen ME (2015) Adaptive posttranslational control in cellular stress response pathways and its relationship to toxicity testing and safety assessment. Toxicol Sci 147: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cui Y, Yu M, Su B, Gong W, Baluška F, Komis G, Samaj J, Shan X, Lin J (2019) Phosphorylation‐mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol 181: 480–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M (2018) Proteome‐wide identification of ubiquitin interactions using UbIA‐MS. Nat Protoc 13: 530–550 [DOI] [PubMed] [Google Scholar]
- Zulawski M, Braginets R, Schulze WX (2012) PhosPhAt goes kinases–searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res 41: D1176–D1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulawski M, Schulze G, Braginets R, Hartmann S, Schulze WX (2014) The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genom 15: 548 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Source Data for Expanded View/Appendix
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Data Availability Statement
Phosphoproteomic datasets (raw spectra) had been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository with the MassIVE ID MSV000086215 (http://massive.ucsd.edu/MSV000086215).


