Abstract
More than 60% of pharmaceuticals are related to natural products (NPs), chemicals produced by living organisms. Despite this fact, the rate of natural product discovery has slowed over the past few decades. In many cases the rate-limiting step in NP discovery is structural characterization. Here we report the use of microcrystal electron diffraction (MicroED), an emerging cryogenic electron microscopy (CryoEM) method, in combination with genome mining to accelerate NP discovery and structural elucidation. As powerful proofs-of-principle we rapidly determine the structure of a new 2-pyridone NP, Py-469 (1), and revise the structure of fischerin (2), an NP isolated more than 25 years ago, with potent cytotoxicity but hitherto ambiguous structural assignment. This study serves as a powerful demonstration of the synergy of MicroED and synthetic biology in NP discovery, technologies that when taken together will ultimately accelerate the rate at which new drugs are discovered.
Introduction
Natural products (NPs) remain a treasure trove for the development of bioactive molecules. NPs in isolated form and derivatives arising from chemical modification form the basis set for new therapeutics and agrochemicals.1 This role is becoming ever more important as resistance to existing therapeutic compounds is rapidly increasing as pathogenic organisms evolve in response to treatment.2 Ideally, the fast evolution of pathogenic organisms would be met by an equally rapid rate of NP discovery; however, for structurally novel natural products, the rate of discovery is decreasing, and is currently lagging behind the emergence of resistance.3,4 The recent explosion of sequenced microbial genomes has fueled a renaissance in NP discovery, where synthetic biology can be leveraged to produce novel metabolites with unprecedented structural complexity or to rediscover compounds previously isolated, but no longer available for study.5 While these cutting-edge methods in synthetic biology have advanced discovery efforts dramatically, the structural elucidation of the NPs remains a rate-limiting step in NP discovery campaigns.
Such difficulties in structural elucidation can arise from i) the lack of sufficient quantities of material for traditional analytical methods (e.g. nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography); ii) intrinsic physical properties of the NP, such as poor solubility and stability in NMR solvents, etc.;6,7 and iii) limitations of NMR capabilities in determining relative stereochemistry, which is accentuated by analytes with distal stereocenters, especially when interrupted by rigid substructures bearing multiple rotatable bonds.8 X-ray crystallography remains the gold-standard for unambiguous structural determination, including the assignment of stereochemistry. However, X-ray crystallographic analysis of newly-isolated natural products is often thwarted by insufficient quantities to provide crystals large enough for single-crystal diffraction (~0.1 mm3), or poor solid-state properties that preclude the formation of large, pristine crystals even when sufficient material is available. Given these challenges, we envisioned that application of the recently reported CryoEM modality MicroED9,10 could lead to vertical advances in the field of natural product discovery, as MicroED has recently been demonstrated to provide unambiguous structures from sub-micron-sized crystals of chemical compounds that had failed to yield large crystals suitable for X-ray analysis.11,12
Results
Discovery and Structural Characterization of Py-469
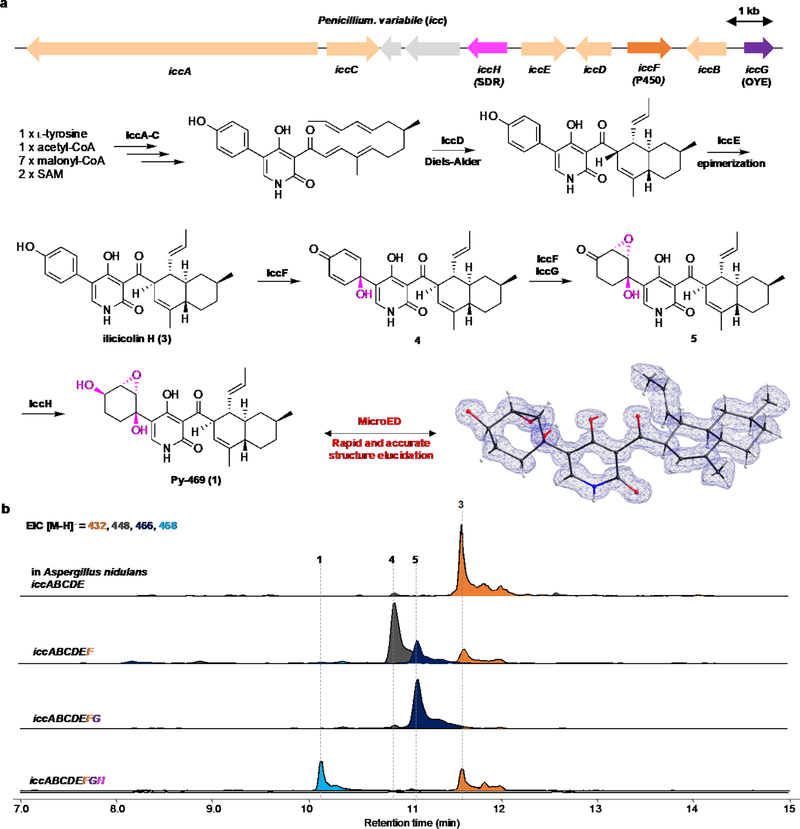
As an entry into the area, we became interested in α-pyridone containing fungal metabolites. These compounds, including ilicicolin H (3, Fig. 1a),13 fischerin (2, Fig. 1a), maximiscin,14 and atpenin A515 are particularly appealing as targets for genome mining16 and MicroED because of their diverse biological activities and structural complexity. Ilicicolin H (3) is a potent inhibitor of eukaryotic respiratory chain and has been thoroughly explored as an antifungal compound through medicinal chemistry.17 To identify structurally more complex, natural variants of ilicicolin H (3), we focused on the icc biosynthetic gene cluster (BGC) from Penicillium variable that has been shown to produce Ilicicolin H (3) (Fig. 1b).18 Compared to other clusters that biosynthesize 3, this BGC contains three additional genes that encode enzymes of biosynthetic relevance, including a P450 (iccF), a short-chain dehydrogenase/reductase (SDR) (iccH) and a flavin dependent oxidoreductase with homology to old-yellow ene-reductase (OYE) (iccG). We reasoned the activities of these three enzymes may further derivatize 3 to impart increased structural complexity to the already densely functionalized ilicicolin H (3).
Figure 1.
Genome mining and MicroED determination of Py-469 (1), a new pyridone natural product. (a) structural diversity of 2-pyridone natural products; (b) the biosynthetic gene clusters that can produce ilicicolin H from Nectoria B-13. The P. variable cluster contains the same five genes (iccA-E) that can produce ilicicolin H (3), and three additional redox enzymes of unassigned function; (c) heterologous expression of the eight genes in the heterologous host A. nidulans led to production of a new compound 1 with the molecular weight of 469. (d) MicroED structure of 1 showed Py-469 is a new natural product.
To test this hypothesis, these three genes, together with the five genes (iccA-E) that produced ilicicolin (3), were expressed in the heterologous biosynthetic host A. nidulans A1145 ΔEMΔST19 (Fig 1c). Comparative mass analysis of the metabolites produced by the host with the three additional genes led to the identification of a new compound (Py-469, 1) with molecular weight of 469, which is 36 mass units higher than that of ilicicolin (3). Metabolite 1 was purified to homogeneity from the host (1 mg/L) and immediately subjected to MicroED for ab initio structural elucidation without additional spectroscopy characterizations. Electron micrographs of the HPLC-purified and lyophilized powder sample revealed crystalline domains that diffracted to sub-atomic resolution. Within hours, two movies collected from a single data collection session were merged to provide a 0.85 Å structure (13.8% R1, refined anisotropically), revealing that metabolite 1 is a derivative of ilicicolin (3) in which the C5’-phenol moiety is modified by these three additional enzymes to form a 2,3-epoxy-syn-1,4-cyclohexane diol (Fig 1d). Importantly, from the MicroED structure the relative stereochemistries of the epoxydiol with respect to the decalin moiety was unambiguously established. Based on previous determination of absolute stereochemistry of 3, the absolute configuration of 1 is also established as shown in Fig 1d. To the best of our knowledge this is a completely new natural product and its increased structural complexity is consistent with the BCG features. The individual functions of three additional icc enzymes in the oxidative dearomatization cascade were also confirmed through additional reconstitution: the P450 IccF catalyzes oxidative dearomatization to 4, followed by IccF-catalyzed epoxidation and IccG-catalyzed ene-reduction to 5, and finishes with IccH-catalyzed reduction to 1 (Extended Data Figure 1b).
NMR spectra of metabolite 1 were collected and as expected the chemical shifts and multiplicities of the pyridone moiety matched to the planar structure of NP 3. However, post facto attempts to assign the relative stereochemistry of the epoxydiol substituents, as well as its stereochemical relationship to the decalin portion, using the NMR data were unsuccessful. Due to molecular topology, such challenges in structural assignment are not suprising for compounds such as 1. Here, distal, stereochemically complex ring systems are linked through freely rotating bonds to a rigid, flat α-pyridone moiety, precluding determination of relative stereochemistry of the two distal fragments. This challenge is well-documented for other structurally related compounds such as apiosporamide (6) (Fig. 2, R=H)20. Apiosporamide (6) is also a 2-pyridone molecule bearing an epoxydiol moiety, and while tentatively assigned by the isolation chemists nearly 30 years ago, it was only unambiguously assigned after a lengthy total synthesis campaign by Williams and coworkers.21
Figure 2.
Rediscovery of fischerin. a, Proposed structure of fischerin (2) and related N-hydroxyapiosporamide (7). b, Alignment of MT domains in PKS-NRPS to identify possible fischerin PKS-NRPS. c, Proposed biosynthetic gene cluster of fischerin (2). d, Heterologous expression of the fin cluster leads to formation and isolation of 2.
Rediscovery and Structural Revision of Fischerin
Encouraged by the success of MicroED in overcoming challenges associated inferring relative stereochemistry from NMR data, we next targeted fischerin (2) for structural elucidation. First isolated more than 25 years ago from Neosartorya fischeri, early studies of fischerin showed that it causes acute peritonitis in mice and has potent cytotoxicity.22 Although the 2-D structure of fischerin (2) was determined upon isolation, the relative stereochemistries between the decalin and the multiply-oxygenated cyclohexane bridged by the 2-pyridone ring have thus far eluded unambiguous assignment, with one attempt reported using computational methods.23 Moreover, unlike apiosporamide (6) and other members of the family, fischerin (2) was hypothesized to possess a rare cis-decalin moiety by the isolation chemists. As an additional obstacle, no other isolation of fischerin (2) had been reported, precluding further structural or biological studies of this compound.
To reconstruct the biosynthesis of fischerin (2) without access to the reported strain, we mined a possible BGC from the National Center for Biotechnology Information (NCBI) database. We reasoned a functional fin BGC may be present in fungi other than the original producer, albeit silent and not producing fischerin (2) under culturing conditions. We hypothesized the BGC should resemble that of the related compound N-hydroxyapiosporamide (7, R = OH) (Fig. 2a), which is a trans-decalin NP with C12 methyl substitution. The BGC of N-hydroxyapiosporamide (7, R = OH) consists of a polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) pathway.24 The lack of the C12 methyl substituent in fischerin (2) suggests that the methyltransferase (MT) domain in the corresponding PKS-NRPS should be inactive and may contain a mutated active site. To identify such a PKS-NRPS, we categorized fungal BGCs that are homologous to api and encode the required accessory enzymes, including analogs to the three enzymes that can generate the epoxydiol (Fig. 2 and Extended Data Fig. 2). Sequence scanning of the PKS-NRPS MT domain was then performed on the candidate BGCs, especially at the conserved MT active site GXGTG motif that binds S-adenosyl-methionine (SAM).25 Gratifyingly, we were able to identify one such PKS-NRPS, from a cluster in Aspergillus carbonarius (renamed fin), to contain a MT domain with a mutated and presumably inactivating GXGAG motif (Fig. 2b). This PKS-NRPS is therefore expected to be devoid of MT activity (the MT domain is designated as MT°) and could be involved in biosynthesis of NP 2.
The fin BGC was then completely refactored and expressed in A. nidulans A1145 ΔEMΔST for metabolite production. As shown in Fig. 2d, coexpression of finACDEHIJ produced a new metabolite with the same expected molecular weight of 431 as fischerin (2) with a titer of 5 mg/L. The purified compound was judged to be >98% pure by both LC-MS (Fig. 2d) and NMR analyses (Extended Data Fig. 3 and see Supplementary Figs. S22–S26). Comparison of NMR peaks to those of the published data in the same solvent showed strong overlap of both proton and carbon signals. The purified sample also showed the same negative optical rotation with fischerin ([α]20D −28° (c = 0.10, CHCl3)). Therefore, we are confident that we have correctly identified the fin cluster and rediscovered fischerin (2) (Extended Data Fig. 4).
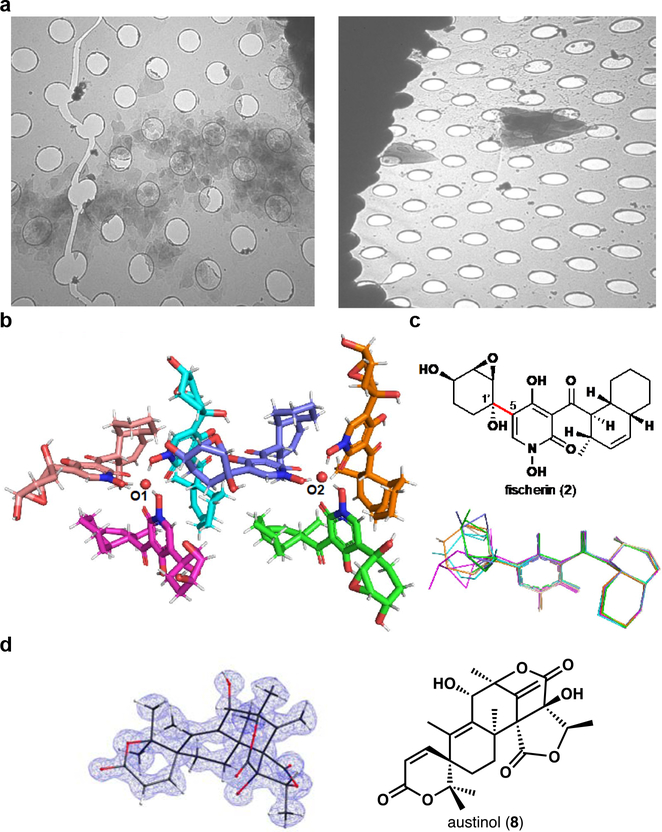
Analogous to the initial isolation study, our attempts to unambiguously establish the relative stereochemistry of fischerin (2) using 2D NMR or X-ray crystallography were unsuccessful.22 We then turned to electron diffraction. While crystallization from various solvents did not yield large single crystals, electron micrographs of pale-yellow particles precipitated from a mixture of acetonitrile and water revealed microcrystals with a distinct morphology. These thin triangular plates were (~0.5–3 μm longest dimension) prone to stacking, thereby hindering the growth of larger crystalline domains and leading to complex intractable diffraction patterns in initial screens (Fig. 3a). Even in cases where we could isolate uniform crystalline domains in selected area diffraction mode, the resolution and completeness of the acquired data were too poor for ab initio structural determination (60% complete and 1.3–1.5 Å). Attempts to merge multiple data sets did not lead to an increase in completeness or resolution, as crystal morphology biased orientation of crystals on the grid; in MicroED studies, where stage rotation is limited, this situation can prevent structural solution.26 Evaluation of hundreds of crystallization conditions and optimization of vitrification conditions,27,28 ultimately allowed for embedding of the microcrystals in a layer of vitreous ice in different orientations,29 increasing the completeness of the data sets.
Figure 3.
Structure of fischerin (2). a, Electron micrograph of fischerin (2) microcrystalline aggregates (left) and at maximum stage tilt (ca. 60°, right); holes are 1 μm wide in diameter. b, Asymmetric unit showing two trimers. c, Proposed relative stereochemistry (top) and overlay of six fischerin (2) molecules showing various degrees of epoxydiol ring rotation (hydrogens omitted for clarity) (bottom). d, Structural determination of minor impurity austinol.
Of over 200 movies collected, four were merged to provide a 1.05 Å ab initio solution (R1 = 13.8%, refined anisotropically) (Fig. 3b, Supplementary Table S5 and Supplementary Fig. S3). The asymmetric unit revealed six unique fischerin (2) residues, each with varying degrees of rotation about the carbon-carbon bond connecting the epoxydiol ring system to the pyridone moiety (C1’–C5, Fig. 2c). The lack of symmetry and the large size of the unit cell further explain the difficulties in obtaining high quality data, and the evident facile rotation about the C1’–C5 bond points to potential reasons for challenging assignment of relative stereochemistry with NOE studies (Fig. 3b).22 Fischerin (2) residues in the asymmetric unit are arranged into two distinct trimers around a common central atom (O1 and O2, Fig. 3b). The identity of the central atom of each trimer was not apparent from initial refinement efforts. Given the challenge of accurately discriminating elements in MicroED experiments, we devised an atom substitution test similar to the approach described by Kato and co-workers to attempt to assign this atom.30 Minimization of R1 and wR2 values when refining the structure using the electron scattering factors of various atoms against measured structure factors and phases from the initial SHELXD output allowed for convergence to an oxygen atom based on residuals. This test suggested that the central atoms at each trimer are disordered water molecules (Extended Data Fig. 5).
From the refined crystal structure, we unambiguously assigned the relative stereochemistry of the substituents on the oxygenated cyclohexyl ring, bearing trans hydroxyl groups with the epoxy moiety trans to the tertiary C1’ –OH group (e.g. 2, Figure 3c). Interestingly, the relative stereochemistry of the epoxycyclohexane diol group in 2 differs from that of 1, indicating varied stereospecificity of the responsible enzymes. We also validated the predicted stereochemistry of the rare cis-decalin system (Fig. 3c). Importantly, the relative stereochemistry of these two stereochemically complex functionalities from the ab initio structural solution differs from Amini’s calculated structure derived from reported NMR chemical shifts.23 This discordance demonstrates the challenges of using NMR shift calculations to predict stereochemistry on fluxional groups and also highlights the importance of experimental validation of computed structures.
Detection of Trace Impurity Austinol
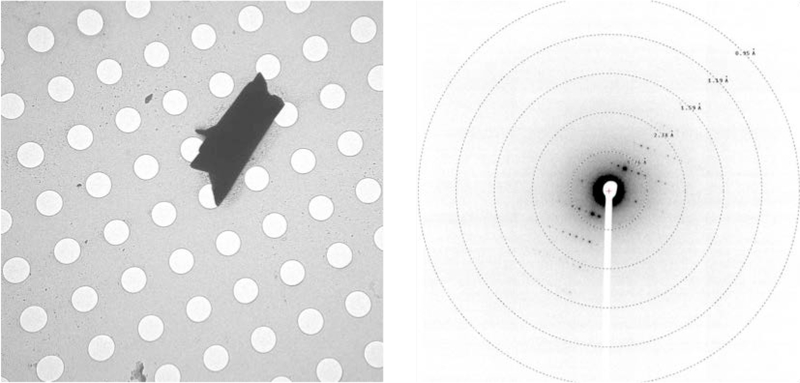
During our crystallographic studies of fischerin, we noticed small amounts of a highly crystalline impurity, present in sufficient quantities to be detectable by electron microscopy but insufficiently abundant in solution to be detectable by 1H NMR (Extended Data Fig. 3). From two such microcrystals, we collected a high-completeness data set with reflections beyond 1.00 Å (R1 value of 14.7%, see Supplementary Table S5), leading to an ab initio structure of the complex polycyclic natural product austinol (8) (Fig. 3d), an endogenous A. nidulans meroterpenoid NP that is co-purified with 2 in trace amounts.31 Intrigued by the ability to obtain structural information from a trace impurity, we proceeded to determine the limit of detection for austinol (8) in MicroED experiments. To our delight, we observed that deposition of a 1.5 ng/μL solution of austinol (8) in acetonitrile and water on a TEM grid followed by slow evaporation led to formation of microcrystals (Extended Data Fig. 6). These crystals were of sufficient quality to obtain a high resolution structure, demonstrating the impressive sensitivity of electron crystallography to trace quantities of analytes crystallized directly on a TEM grid. Moreover, the ability to identify NPs from a mixture further highlights the applicability of MicroED in NP discovery and characterization, particularly for compounds produced in scarce amounts that may be overlooked by other techniques. These results showcase the exciting possibility of serendipitous discoveries in the field of NP isolation, where biological extracts could often contain minor NP impurities that may not be detectable using other established analytical methods.
Discussion
We report the discovery of Py-469 (1) and rediscovery of fischerin (2) using genome mining approaches. For new NP 1, MicroED was used for rapid and complete ab initio structural determination. For 2, we report the first crystal structure of this NP that has eluded full structural characterization for decades, establishing for the first time the relative stereochemistry of the epoxydiol moiety and the stereochemical relationship of these functionalities. We also report the structure of a co-metabolite impurity, austinol (8), that was present in amounts below the detection limit in our initial 1H NMR experiments, and we demonstrate the exquisite sensitivity of MicroED by obtaining structural information from merely 3 ng of material. These results demonstrate the synergy of synthetic biology and MicroED in NP discovery and highlight the importance of developing novel characterization techniques that can complement and overcome limitations of the current state-of-the-art. Taken together, our findings provide a powerful approach toward discovery and structural determination of novel and elusive NPs of ever increasing structural complexity.
Methods
Material, fungal strains and culture conditions
Aspergillus carbonarius NRRL346 was obtained from Agricultural Research Service Culture Collection (NRRL). A. carbonarius and Penicillium variabile were maintained on PDA (potato dextrose agar, BD) for 3 days for sporulation or in liquid PDB medium (PDA medium without agar) for isolation of genomic DNA. A. nidulans was maintained on Czapek-Dox (CD) agar for sporulation or on CD–ST agar for gene overexpression, compound production and RNA extraction (http://www.fgsc.net/).
General DNA manipulation technique
All DNA manipulation technique was performed as described previously.32 E. coli TOP10 was used for cloning, following standard recombinant DNA techniques as described. DNA restriction enzymes were used as recommended by the manufacturer (New England Biolabs, NEB). PCR was performed using Phusion High-Fidelity DNA Polymerase (NEB). The gene-specific primers are listed in Supplementary Table S3. PCR products were confirmed by DNA sequencing. In vivo homologous recombination using Saccharomyces cerevisiae was used for the construction of the A. nidulans overexpression plasmids.
Preparation of overexpression plasmids for A. nidulans.
Full length finD was amplified by PCR with three sets of primers of pMO30001-f1/r1, pMO30001-f2/r2, pMO30001-f3/r3 using the genomic DNA of Aspergillus carbonarius NRRL346 as the template. The three overlapping DNA fragments and NotI/PacI-digested pYTU1 expression vector were transformed into yeast to generate pMO30001 by yeast homologous recombination. The genes of finC and finA were individually amplified with primer pairs of pMO30004-f1/r1 and pMO30004-f3/r3 using genomic DNA of A. carbonarius as the template. The glaA promoter was amplified with primer pairs of pMO30004-f2/r2 using pYTU as the template. The three overlapping DNA fragments were ligated into expression vector pYTR1 in the sites of BamHI/PacI to create plasmid pMO30004 (finC and finA). The genes of finJ and finI were amplified with the primer pairs of pMO30009-f1/r1 and pMO30009-f3/r3 using genomic DNA of A. carbonarius, respectively. The gpdA promoter was amplified with primer pairs of pMO30009-f2/r2 using pYTR as the template. Each amplified fragment was ligated into expression vector pYTP in the sites of BamHI/HindIII to create plasmid pMO30009 (finJ and finI). The gene of finH was amplified with primer pairs of pMO300010-f2/r2 using genomic DNA of A. carbonarius as the template. The amyB promoter was amplified with primer pairs of pMO30010-f1/r1 using pYTP as the template. The two overlapping DNA fragments were ligated into NotI-digested pMO30004 to create plasmid pMO30010 (finC, finA, and finH). The gene of finE was amplified with primer pairs of pMO300011-f2/r2 using genomic DNA of A. carbonarius as the template. The glaA promoter was amplified with primer pairs of pMO30011-f1/r1 using pYTU as the template. The two overlapping DNA fragments were ligated into SwaI-digested pMO30009 to create plasmid pMO30011 (finJ, finI, and finE). These plasmids are shown in Supplementary Figure S1.
Heterologous expression in A. nidulans
A. nidulans A1145ΔEMΔST33 was initially grown on oatmeal agar plates containing 10 mM uracil, 0.5 μg/mL pyridoxine HCl and 2.5 μg/mL riboflavin at 30 °C for 5 days. Fresh spores of A. nidulans were inoculated into 45 mL liquid CD media (1 L: 10 g glucose, 50 mL 20 × nitrate salts, 1 mL trace elements, pH 6.5) in 125 mL Erlenmeyer flask and germinated at 30 °C and 250 rpm for approximately 16 h. For the preparation of 20 × Nitrate salts, 120 g NaNO3, 10.4 g KCl, 10.4 g MgSO4·7H2O, 30.4 g KH2PO4 were dissolved in 1 L double distilled water. The 100 mL trace elements with pH 6.5 contains 2.20 g ZnSO4·7H2O, 1.10 g H3BO3, 0.50 g, MnCl2· 4H2O, 0.16 g FeSO4·7H2O, 0.16 g CoCl2·5H2O, 0.16 g CuSO4·5H2O, and 0.11 g (NH4)6Mo7O24 ·4H2O. Mycelia were harvested by centrifugation at 4000 g for 10 min, and washed with 10 mL Osmotic buffer (1.2 M MgSO4, 10 mM sodium phosphate, pH 5.8). Then the mycelia were transferred into 10 mL of Osmotic buffer containing 100 mg lysing enzymes from Trichoderma and 60 mg Yatalase in a 125 mL flask. The flask was kept in shaker at 80 rpm for 4 h at 30 °C. Cells were collected in a 30 mL Corex tube and overlaid gently by 10 mL of Trapping buffer (0.6 M sorbitol, 0.1 M Tris-HCI, pH 7.0). After centrifugation at 3,500 rpm for 15 min at 4 °C, protoplasts were collected in the interface of the two buffers. The protoplasts were then transferred to a sterile 50 mL falcon tube and washed by 10 mL STC buffer (1.2 M sorbitol, 10 mM CaCl2, 10 mM Tris-HCI, pH 7.5). The protoplasts were resuspended in 1 mL STC buffer for transformation. Then, the plasmids (see Fig. S1) were added to 60 μL protoplast suspension and the mixture incubated 60 min on ice. Then 600 μL of PEG solution at pH 7.5 (60% PEG, 50 mM CaCl2 and 50 mM Tris-HCl) was added to the protoplast mixture, and the mixture was incubated at room temperature for additional 20 min. The mixture was spread on the regeneration dropout solid medium (CD solid medium with 1.2 M sorbitol and appropriate supplements) and incubated at 30 °C for 2 to 3 days. The transformants were grown on CD-ST (1 L: 20 g starch, 20 g casamino acids, 50 mL 20 × nitrate salts, 1 mL trace elements, 15 g agar) agar for 3 to 4 days before extraction.
Analysis of metabolites and isolation of Py-469 (1) and fischerin (2)
For small scale analysis, the transformants of A. nidulans were grown for 3 to 4 days on 25 mL CD-ST agar. Then, a chunk of agar with fungal mycelia was scraped and vortexed with 500 μL of acetone for 30 minutes. The organic phase was dried by speed vacuum and dissolved in methanol for analysis. LC-MS analyses were performed on a Shimadzu 2020 EV LC-MS (Kinetex 1.7 μm C18 100 Å, LC Column 100 × 2.1 mm) using positive- and negative-mode electrospray ionization with a linear gradient of 5–95% acetonitrile MeCN–H2O with 0.5% formic acid in 15 min followed by 95% MeCN for 3 min with a flow rate of 0.3 ml/min.
For isolation of compounds, each transformant of A. nidulans which produce 1 or 2 was grown for 96 h on 4 L CD-ST agar and then solid culture was cut into small pieces and was extracted with acetone. The organic layer was evaporated and the remaining water phase was extracted with 2 × 1 L ethyl acetate. After evaporation of the organic phase, the crude extracts were separated by silica gel chromatography. Fractions containing desired compound were combined and used for further purification by HPLC with a COSMOSIL column (Nacalai Tesque Inc., C18 MS-II, 5 μm, 10 × 250 mm, flowrate 4 mL/min) to yield 4.0 mg of 1 (1.0 mg/L) and 20 mg of 2 (5.0 mg/L), respectively. For elucidation of 1 and 2, 1D and 2D NMR spectra were obtained on Bruker AV500 spectrometer at the UCLA Molecular Instrumentation Center. High resolution mass spectra were obtained from an Agilent Technologies 6545 Accurate Mass QTOF LC/MS at the UCLA Molecular Instrumentation Center.
Recrystallization
Dried, purified fischerin (2) sample (1.0 mg, 2.3 μmol) was suspended in 0.1 mL of tetrahydrofuran (THF), and H2O was added until the solution became clear and homogeneous, changing color to light yellow. Crystals were grown at room temperature by slow evaporation in a dram vial.
0.8 mg (1.7 μmol) of sample containing austinol (8) was dissolved in 0.8 mL of 1:1 MeCN/H2O solution, then precipitated at ambient temperature under a stream of air.
Serial Dilution
0.4 mg (0.9 μmol) of vacuum-dried sample containing austinol (8) was redissolved in a 2:1 MeCN/H2O solution at 333 μM concentration. The sample was then diluted every 100-fold in H2O, vortexed, and centrifuged before the subsequent dilution.
Electron diffraction data acquisition and processing
Recrystallized fischerin (2) microcrystals were diluted to optimize crystal density and deposited onto a Quantifoil holey-carbon EM grid as a 2 μL drop before blotting and plunge freezing in liquid ethane using Vitrobot Mark IV (FEI). The vitrified grid was then transferred to the transmission electron microscope (TEM) on Gatan 626 cryo-holder, maintaining cryogenic temperature (100 K) throughout data acquisition.
Py-469 (1) powder was deposited onto a continuous carbon EM grid as previously described.5 The grid was then transferred to the TEM on a single-tilt sample holder for ambient temperature data acquisition.
2 μL of recrystallized austinol (8) sample was aliquotted from the top of the solution to account for crystal density before deposition onto a continuous carbon EM grid. 2 μL of serial dilution sample was obtained from the bottom of the solution and deposited onto a Quantifoil holey-carbon EM grid. In both cases, solvent was wicked away manually, and the dried grid was transferred to the TEM for ambient temperature data acquisition on a single-tilt sample holder.
All diffraction data was collected using TEM User Interface on FEI Tecnai F200C electron microscope with an operating voltage of 200 keV, corresponding to a wavelength of 0.025 Å. During data acquisition, the crystal of interest was isolated using a selected area aperture and continuously rotated at a rate of −0.3° s−1 over a tilt range of 50–100°. Continuous rotation diffraction data was recorded on TEM Imaging & Analysis (TIA) using rolling shutter mode with a Ceta-D CMOS 4k × 4k camera, integrating at a rate of 3 s per frame and binning by 2 to produce final images of 2k × 2k.34
Diffraction movies saved as SER files were converted to SMV format using ser2smv software as described previously.35 Frames were indexed and integrated in XDS.36 Data from two to four crystals were scaled and merged together using XSCALE37 to produce the final data sets for 1, 2 and 8 (Supplementary Table S5). Finally, intensities were converted to SHELX format using XDSCONV.35
Structure determination and refinement
Structures of 1 and 2 were solved ab initio using direct methods in SHELXD.38 Structure of 8 was also solved ab initio using SHELXT.39 All structures were refined with SHELXL40 in ShelXle.41 Non-hydrogen atoms were refined anisotropically, and hydrogen atoms were placed using the riding model. Residual density corresponding to solvent molecules was observed during refinement; however, solvent disorder hindered unambiguous identification, and oxygen atoms were placed in lieu of water molecules. Overall data quality and refinement statistics are reported in Supplementary Table S5. Structure overlays were illustrated using Pymol.42 Data from atom substitution tests were visualized using Python43.
Extended Data
Extended Data Figure 1. Biosynthesis and structure elucidation of Py-469 (1).
(a)The biosynthetic gene cluster and proposed biosynthetic pathway of 1. (b) Product profiles from heterologous expression of iccE, F anf G together with iccA-E in A. nidulans. Expression of iccF with iccA-E in A. nidulans led to the formation of new compounds 4 and 5. The structures of 5 was determined by comparing the LC-MS retention time with the synthesized authentic standard (see Supplementary Figure S5). Further addition of ene-reductase IccG increased the formation of 5 with decreasing the formation of 4. These results suggest that IccF catalyzes the C1’-hydroxylation of the distal phenol ring of 1 and the subsequent epoxidation on the resultant dienone moiety of 4. Then, an endogenous ene-reductase in A. nidulans or IccG catalyzes the ene reduction of the epoxy enone moiety to form 5. Lastly, IccH catalyzes the ketoreduction to give Py-469 (1).
Extended Data Figure 2.
Biosynthetic gene clusters that are homologous to those of fischerin (2) and N-hydroxyapiosporamide (7); and multiple sequence alignment of SAM-binding motif of cis-MT domain in PKS-NRPSs.
Shown here are the putative biosynthetic gene clusters of 2 and 7 and their homologous biosynthetic gene clusters found in NCBI database. SAM binding motif is shown in an alignment with those from FinD and ApiD homologs. Previous study reported that active cis-MT domains contain conserved EXGXGTG sequence as a SAM binding motif.23 Based on this, we hypothesized that the cis-MT domains in PKS-NRPSs which do not have this conserved this motif are inactive, and the biosynthetic gene clusters which contain the PKS-NRPSs could be responsible for formation of 2. For example, the cis-MT domains from FinD (Aspergillus carbonarius), CG_v00450 (Colletotrichum fructicola Nara gc5), and BO_621233 (Aspergillus sclerotioniger) do not contain this conserved EXGXGTG motif as the threonine residues are mutated to alanines. As shown in Fig. 2d, the biosynthetic gene cluster, which contains FinD, is indeed responsible for formation of 2.
Extended Data Figure 3.
1H NMR spectra of 2 in CDCl3, 500 MHz for 1H NMR.
Extended Data Figure 4.
Proposed biosynthetic pathway of 2. Based on the reported proposed biosynthetic pathway of other 2-pyridone alkaloids such as leporins and ilicicolin H (3) (see Extended Data Fig. 1a), we proposed the biosynthetic pathway of 2. FinD (PKS-NRPS) and the partnering FinC (ER) form the tetramic acid intermediate. A P450 FinE catalyzes the oxidative ring-expansion reaction of the tetramic acid to the 2-pyridone compound. Then, a Diels-Alderase likely catalyze the Diels-Alder reaction to form the energetically disfavored cis-decalin ring, since the previous study24 showed that nonenzymatic Diels-Alder reaction of the analog of 2-pyridone compound in water only led to formation of the trans-decalin compound. Further redox modification by FinA, FinE, and FinH forms 2.
Extended Data Figure 5.
Atom substitution test for fischerin with (top) and without (bottom) electron scattering factors.
Extended Data Figure 6.
Electron microgram of austinol crystal and its diffraction pattern from 3 ng of sample. Holes are 1 μm wide in diameter.
Supplementary Material
Acknowledgements:
The authors thank Michael R. Sawaya (UCLA-DOE Institute) for assistance in crystallography with data processing and refinement. This research used resources at the UCLA-DOE Institute’s X-ray Crystallography Core Facility, which is supported by the U.S. Department of Energy (DE-FC02-02ER63421). J.A.R. acknowledges support from STROBE, an NSF Science and Technology Center through Grant DMR-1548924, DOE Grant DE-FC02-02ER63421 and NIH-NIGMS Grant R35 GM128867. J.A.R. is supported as a Pew Scholar and a Beckman Young Investigator. Y.T. acknowledges support from the NIH (1R01AI141481). We also thank the David and Lucile Packard Foundation (Fellowship to H.M.N.; J.A.R. and Y.T.) and Bristol Myers Squibb (Unrestricted Grant in Synthetic Organic Chemistry to H.M.N) for generous support.
Footnotes
Competing Interests Statement:
The authors declare no competing financial interests.
Data availability
Crystallographic information files (CIF) for compounds 1, 2 and 8 containing atomic coordinates and structure factors have been deposited at the Cambridge Crystallographic Data Center (Deposition numbers 2038723, 2020516 and 2020510, respectively). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/.
Source data for Extended Data Figure 5 has been provided in Supplementary Table S6.
References
- 1.Newman DJ & Cragg GM Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Perfect JR The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fair RJ & Tor Y Antibiotics and Bacterial Resistance in the 21st Century: Perspectives in Medicinal Chemistry (2014) doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pye CR, Bertin MJ, Lokey RS, Gerwick WH & Linington RG Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 114, 5601–5606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisch KM et al. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J. Am. Chem. Soc. 133, 16635–16641 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Nicolaou KC & Snyder SA Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem. Int. Ed. 44, 1012–1044 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Maier ME Structural revisions of natural products by total synthesis. Nat. Prod. Rep. 26, 1105–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Bifulco G, Dambruoso P, Gomez-Paloma L & Riccio R Determination of relative configuration in organic compounds by NMR spectroscopy and computational methods. Chem. Rev. 107, 3744–3779 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Jones CG, et al. The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Cent. Sci. 4, 1587–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruene T, et al. Rapid structure determination of microcrystalline molecular compounds using electron diffraction Angew. Chem. Int. Ed. 57, 16313–16317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CJ, et al. Characterization of reactive organometallic species via MicroED. ACS Cent. Sci. 5, 1507–1513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayakawa S, Minato H & Katagiri K The Ilicicolins, antibiotics from Cylindrocladium ilicicola. J. Antibiot. 24, 653–654 (1971). [DOI] [PubMed] [Google Scholar]
- 14.Du L et al. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity. Angew. Chem. Int. Ed. 53, 804–809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyadera H et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc Natl Acad Sci U S A. 100, 473–477 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziemert N, Alanjary M & Weber T The evolution of genome mining in microbes – a review. Nat. Prod. Rep. 33, 988–1005 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Singh SB et al. Antifungal spectrum, in vivo efficacy, and structure–activity relationship of ilicicolin H. ACS Med. Chem. Lett. 3, 814–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z et al. Enzyme-catalyzed inverse-electron demand Diels–Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 141, 5659–5663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu N et al. Identification and Heterologous production of a benzoyl-primed tricarboxylic acid polyketide intermediate from the zaragozic acid A biosynthetic pathway. Org. Lett. 19, 3560–3563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfatafta AA, Gloer JB, Scott JA & Malloch D Apiosporamide, a new antifungal agent from the Coprophilous fungus Apiospora montagnei. J. Nat. Prod. 57, 1696–1702 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Williams DR, Kammler DC, Donnell AF & Goundry WRF Total synthesis of (+)-apiosporamide: Assignment of relative and absolute configuration. Angew. Chem. Int. Ed. 44, 6715–6718 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto H, Ikeda M, Yamamoto K & Yamazaki M Structure of fischerin, a new toxic metabolite from an ascomycete, Neosartorya fischeri var. fischeri. J. Nat. Prod. 56, 1268–1275 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Amini SK Assignment of the absolute configuration of fischerin by computed nmr chemical shifts. J. Struct. Chem. 56, 1334–1341 (2015). [Google Scholar]
- 24.Ugai T, Minami A, Gomi K & Oikawa H Genome mining approach for harnessing the cryptic gene cluster in Alternaria solani: production of PKS–NRPS hybrid metabolite, didymellamide B. Tetrahedron Letters 57, 2793–2796 (2016). [Google Scholar]
- 25.Skiba MA et al. Domain organization and active site architecture of a polyketide synthase C-methyltransferase. ACS Chem. Biol. 11, 3319–3327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nannenga BL MicroED methodology and development. Struct. Dyn. 7, 014304. 10.1063/1.5128226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Cruz MJ, et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat. Methods 14, 399–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Natesh R in Structural Bioinformatics: Applications in Preclinical Drug Discovery Process (ed Mohan CG) 375–400 (Springer Nature; Switzerland AG, 2019). [Google Scholar]
- 30.Kato K, et al. A vault ribonucleoprotein particle exhibiting 39-fold dihedral symmetry. Acta Cryst D64, 525–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda Y & Abe, Ikuro. Biosynthesis of fungal meroterpenoids. Nat. Prod. Rep. 33, 26–53 (2016). [DOI] [PubMed] [Google Scholar]
Methods References:
- 32.Ohashi M et al. SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu N et al. Identification and Heterologous production of a benzoyl-primed tricarboxylic acid polyketide intermediate from the zaragozic acid A biosynthetic pathway. Org. Lett. 19, 3560–3563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nannenga BL, Shi D, Leslie AGW & Gonen T High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods. 11, 927–930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattne J, et al. MicroED data collection and processing. Acta Cryst A71, 353–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W Xds. Acta Cryst D66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W Integration, scaling, space-group assignment and post-refinement. Acta Cryst D66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheldrick GM A short history of SHELX. Acta Cryst A64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Sheldrick GM SHELXT – Integrated space-group and crystal-structure determination. Acta Cryst A71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheldrick GM Crystal structure refinement with SHELXL. Acta Cryst C71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hübschle CB, Sheldrick GM & Dittrich B ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Cryst. 44, 1281–1284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delano W The PyMOL Molecular Graphics System (Schrödinger LLC; ). http://www.pymol.org. [Google Scholar]
- 43.Van Rossum G & Drake FL Python 3 Reference Manual (CreateSpace, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic information files (CIF) for compounds 1, 2 and 8 containing atomic coordinates and structure factors have been deposited at the Cambridge Crystallographic Data Center (Deposition numbers 2038723, 2020516 and 2020510, respectively). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/.
Source data for Extended Data Figure 5 has been provided in Supplementary Table S6.