Abstract
Background
Lung adenocarcinoma (LUAD) is the main subtype of primary lung cancer and is a leading cause of cancer‐related death worldwide. PIWI‐interacting RNAs (piRNAs) are a type of small non‐coding RNAs that may play crucial roles in cancer progression and serve as biomarkers for tumor detection. This study aimed to explore the expression profiles and diagnostic values of piRNAs in LUAD.
Methods
Small RNA sequencing was performed to investigate tissue piRNA profiles of LUAD. The expression of selected upregulated piRNAs were detected in tissues and serum exosome samples by quantitative real‐time polymerase chain reaction (qRT‐PCR). Serum exosomes were identified by transmission electron microscope, nanoparticle tracking analysis, and western blot analysis. Receiver operating characteristic (ROC) curve was adopted to quantify the diagnostic potentials of piRNAs in LUAD. Finally, a piRNA panel was developed by multivariate logistic regression model.
Results
We identified that 76 piRNAs were overexpressed and 9 piRNAs were underexpressed in LUAD tissues compared with adjacent non‐tumor tissues. Among the top 10 overexpressed piRNAs, 4 piRNAs (piR‐hsa‐26925, piR‐hsa‐5444, piR‐hsa‐30636, and piR‐hsa‐8757) were verified by qRT‐PCR to be significantly upregulated in LUAD tissues. Moreover, piR‐hsa‐26925 and piR‐hsa‐5444 had a significantly higher level in serum exosome samples of LUAD patients than those of healthy controls. We finally established a 2‐piRNA panel composed of piR‐hsa‐26925 and piR‐hsa‐5444, which showed higher diagnostic performance for LUAD with an AUC of 0.833.
Conclusions
Our finding revealed the abnormally expressed piRNAs in LUAD, and serum exosomal piR‐hsa‐26925 and piR‐hsa‐5444 could serve as potential biomarkers for LUAD diagnosis.
Keywords: biomarker, diagnosis, lung adenocarcinoma, PIWI‐interacting RNAs, serum exosome
Serum exosomal piR‐hsa‐26925 and piR‐hsa‐5444 had a significantly higher level in LUAD patients than those of healthy controls. We established a 2‐piRNA panel composed of piR‐hsa‐26925 and piR‐hsa‐5444, which showed higher diagnostic performance for LUAD with an AUC of 0.833. In summary, this study suggested that serum exosomal piRNAs could be used as potential biomarkers for the diagnosis of LUAD.
INTRODUCTION
As one of the most common malignant tumors worldwide, lung cancer is the leading cause of cancer‐related morbidity in 2020, seriously threatening human health.1 According to the morphological characteristics of tumor cells, lung cancer is typically divided into two main types: small cell lung cancer (SCLC) and non‐small cell lung cancer (NSCLC). NSCLC is the deadliest malignancy worldwide, among which lung adenocarcinoma (LUAD) is the most prevalent histopathologic type, with increasing morbidity year by year.2, 3 The tumorigenesis of LUAD is a complex process involving multiple stages and factors.4, 5 Despite recent advances in medical technology, the 5‐year survival rate of LUAD remains low,6, 7 mainly because LUAD lacks noticeable symptoms in the early stage, making it difficult to detect. Therefore, it is urgently necessary to identify potential hallmarks correlated with effective diagnosis of LUAD.
PIWI‐interacting RNAs (piRNAs) are a type of small non‐coding RNAs with a length of ~30 nucleotides (nt) and more than 30 000 members were described in the human genome.8, 9, 10 Investigation into the role of piRNAs was initially limited to germ cells and gonadal cells during early embryonic development.11 Recent reports have shown that piRNAs are expressed in a tissue‐specific manner in a variety of human tissues, and regulate key signaling pathways at the transcriptional or post‐transcriptional level.12 An increasing number of studies have indicated that the abnormal expressions of piRNAs and PIWI proteins in various cancers may serve as new biomarkers and therapeutic targets for tumor diagnosis and treatment.13 For example, Mai et al.14 suggested that piRNA‐54265 can be used as a biomarker for the early detection and clinical monitoring of colorectal cancer. Tan et al.15 revealed that piRNA‐36712 is a novel tumor suppressor and may serve as a prognostic predictor of breast cancer. Furthermore, recent studies have revealed that piRNAs can be stably present in body fluids and detected in exosomes of cancer cells.16, 17 However, the potential clinical significance of serum exosomal piRNAs as biomarkers for the diagnosis of LUAD remain elusive.
To fill this important gap in knowledge, in the present study, we systematically analyzed the expression profiles of piRNAs in five pairs of LUAD tissues and adjacent normal tissues by small RNA sequencing (SRS). Following rigorous bioinformatic analysis and prioritization of aberrantly piRNAs, the top 10 upregulated piRNAs were validated in 14 LUAD tissues and paired normal tissues, four (piR‐hsa‐26925, piR‐hsa‐5444, piR‐hsa‐30636, and piR‐hsa‐8757) of which were significantly upregulated in LUAD tissues. We thereafter, conducted a systematic analysis on the serum exosomal levels of the above‐mentioned four piRNAs and established a panel comprised of 2 piRNAs (piR‐hsa‐26925 and piR‐hsa‐5444) for the diagnosis of LUAD. In summary, all these findings suggested that serum exosomal piRNAs could be used as potential biomarkers for the diagnosis of LUAD.
METHODS
Sample collection
A total of 19 pairs of LUAD tissues and matched adjacent non‐tumor tissues were obtained from patients who underwent surgery at Qilu Hospital of Shandong University from December 2020 to May 2021. None of the patients received chemotherapy or radiotherapy before surgery, and all LUAD tissues and paired non‐tumor tissues were confirmed by experienced pathologists (Dr. Chengjun Zhou and Dr. Binxin Guan). These tissues were immediately frozen in liquid nitrogen and then stored at −80°C. Among these samples, five pairs of samples were used for SRS and 14 pairs were used for quantitative real‐time polymerase chain reaction (qRT‐PCR) verification. Serum samples from 70 patients with LUAD and 57 healthy controls were also obtained at Qilu Hospital of Shandong University and frozen at −80°C. The study protocol was approved by the Institutional Review Board of Qilu Hospital of Shandong University, and written informed consents from all participants were obtained before the initiation of the research.
SRS
Five pairs of LUAD tissues and the corresponding adjacent non‐tumor tissues were used for SRS. Total RNA was isolated using miRNeasy Mini Kit (50) (Qiagen) according to the manufacturer's instructions. The concentration and quality of RNA were determined by the Qubit 2.0 fluorometer (Life Technologies) and Nanodrop One spectrophotometer (Thermo Fisher Scientific), respectively. The integrity of RNA was assessed by Agilent 2100 bioanalyzer (Agilent Technologies), and samples with an RNA integrity number (RIN) above 7.0 were used for subsequent sequencing.
Paired‐end libraries were synthesized using the QIAseq miRNA Library Kit (Qiagen) following TruSeq RNA Sample Preparation Guide. The products were then purified and enriched with PCR to establish the final complementary DNA (cDNA) library. The purified libraries were quantified by Qubit 2.0 fluorometer (Life Technologies) and validated by Agilent 2100 bioanalyzer (Agilent Technologies) to confirm the insert size and calculate the mole concentration. Clusters were generated by cBot with the library diluted to 10 pM, and then the SRS was performed by an Illumina Hiseq Xten System (Illumina). The library construction and sequencing were performed by Sinotech Genomics.
For data analysis, reads of each sequencing sample were mapped to the piRbank using CLC genomics_workbench 5.5 to calculate the expression level of each piRNA (count number). The abundance of piRNA was estimated by the transcripts per million (TPM). The differentially expressed piRNAs were selected using the criteria of |log2 (fold change)| >1 and p value <0.05 by edgeR software.
Prediction of piRNA target genes
Possible target genes of the known piRNAs were predicted using the miRanda software. The interaction between piRNAs and the potential target genes were visualized using Cytoscape 3.6.0. To clarify the biological functions of the genes and the involved signaling pathways, Gene Ontology (GO) analysis for biological processes, cellular components, and molecular function was performed. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.ad.jp/kegg) was conducted via “enrichR” package.
Exosome extraction and verification
Exosomes were isolated from serum using ExoQuick Exosome Precipitation Solution (System Biosciences) according to the manufacturer's protocols. Subsequently, morphology observation of exosomes was conducted using a transmission electron microscope (TEM). The concentration, size distribution, and zeta potential of isolated exosomes were detected by nanoparticle tracking analysis (NTA) using a ZetaView (Particle Metrix). TSG101 and CD9 proteins were used as markers to identify exosomes by western blot analysis.
RNA extraction
Total RNA was extracted from tissues using miRNeasy Mini Kit and from serum exosomes using miRNeasy Micro Kit (50) according to the manufacturer's instructions. The quality and concentration of the purified total RNA were detected using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific).
qRT‐PCR
Total RNA was reversely transcribed to synthesize cDNA using PrimeScript RT reagent kit (Takara) on a T100 Thermal Cycler (Bio‐Rad). qRT‐PCR was carried out using SYBR Green Premix Ex Taq reagent kit (Takara) on a CFX96 Real‐Time System (Bio‐Rad) in a 10‐μL reaction system consisting of 1 μL cDNA, 5 μL SYBR Green PCR Master Mix, 0.3 μL miScript Universal Primer (RiboBio), 0.3 μL specific primers for each piRNA (RiboBio), and 3.4 μL RNase‐free water. Briefly, after an initial denaturation step at 95°C for 10 minutes, the amplifications were carried out with 40 cycles at a melting temperature of 95°C for 2 seconds, an annealing temperature of 60°C for 20 seconds, and an extension temperature of 70°C for 10 seconds. The threshold cycle (Ct) was defined as the fractional cycle number, at which the fluorescence passed the fixed threshold. The relative expression level of each piRNA was determined by the 2−ΔΔCt method and normalized against U6.
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 5.0 and MedCalc version 19.6. Wilcoxon matched‐pairs tests were used to evaluate the group differences of tissue samples. For serum exosome samples, the expressions of piRNAs between different groups were analyzed by Mann–Whitney tests. The expression levels of key piRNAs were used to construct a multivariate model to distinguish LUAD from healthy controls based on the multivariate logistic regression analysis. The receiver operating characteristic (ROC) curve was adopted to assess the diagnostic potential of piRNAs, and the area under the ROC curve (AUC), sensitivity, and specificity were calculated. Only a two‐sided p value <0.05 was considered statistically significant.
RESULTS
Genome‐wide screening to identify dysregulated piRNAs in LUAD
To identify the most differentially expressed piRNAs, five pairs of LUAD tissues and corresponding adjacent non‐tumor tissues were analyzed by SRS. In general, the classes of small RNAs can be identified by the peak of length distribution, which ranges from 18 to 35 nt. MicroRNAs (miRNAs) are mainly distributed in length of 21 or 22 nt, whereas piRNAs are concentrated in 26–32 nt.18 Consistent with this, Figure 1(a) shows that in the LUAD and paired adjacent non‐tumor tissue libraries, small RNAs were clustered in two ranges by length, 19–24 and 32–34 nt. More SRS data is available in Table S1. Further analysis of the sequencing data showed that 1474 piRNAs were expressed in the tissue samples. A total of 85 differentially expressed piRNAs were identified by the criteria of |log2 (fold change)| >1 and p value <0.05, among which 76 were highly expressed and 9 were lowly expressed in LUAD tissues compared with adjacent non‐tumor tissues (Figure 1(b) and (c)). These data indicated that the above‐mentioned abnormally expressed piRNAs might play a crucial role in the progression of LUAD.
FIGURE 1.
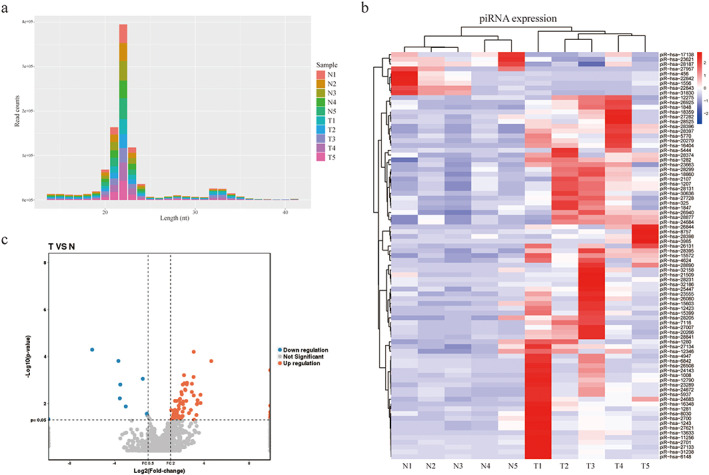
The analysis of dysregulated piRNAs in LUAD patients. (a) Bar graph of small RNA transcripts identified from LUAD patient's tissue samples. (b,c) The heatmap (b) and volcano plots (c) of 85 differently expressed piRNAs in LUAD patients. T, LUAD tissues; N, paired adjacent non‐tumor tissues
Selection of four differentially expressed piRNAs in LUAD tissues
Considering that the multi‐biomarker combination has better diagnostic performance than single candidates,19 this study aims to identify and validate a panel of differentially expressed piRNAs that can distinguish individuals with LUAD from healthy. The top 10 piRNAs were chosen from significantly upregulated piRNAs in LUAD tissues (Table 1, Figure 2(a)). The differential expressions of these piRNAs were verified by qRT‐PCR in tumor tissues and paired adjacent non‐tumor tissue samples from 14 patients with LUAD. The results showed that the expressions of four piRNAs (piR‐hsa‐26925, piR‐hsa‐5444, piR‐hsa‐30636, and piR‐hsa‐8757) were significantly upregulated in LUAD tissues compared with non‐tumor tissues (Figure 2(b)–(e), p < 0.05), which was consistent with the results of SRS. There was no significant difference in the expressions of the other six piRNAs between LUAD tissues and non‐tumor tissues (Figure S1(a)–(f), p > 0.05).
TABLE 1.
Feature of top 10 mostly upregulated piRNAs
| piRNA_ID | log2FC | p value | Regulation |
|---|---|---|---|
| piR‐hsa‐1008 | 4.636623662 | 0.000157193 | UP |
| piR‐hsa‐26925 | 3.680927145 | 0.009539399 | UP |
| piR‐hsa‐28231 | 3.676403107 | 0.004281919 | UP |
| piR‐hsa‐11256 | 3.4113601 | 0.007260661 | UP |
| piR‐hsa‐5444 | 3.388361348 | 0.010085528 | UP |
| piR‐hsa‐30636 | 3.356912918 | 0.010721393 | UP |
| piR‐hsa‐24143 | 3.313713536 | 0.03099702 | UP |
| piR‐hsa‐6842 | 3.100589608 | 0.049094356 | UP |
| piR‐hsa‐8757 | 3.07264821 | 0.004560042 | UP |
| piR‐hsa‐15572 | 3.069194841 | 6.35E‐05 | UP |
FIGURE 2.
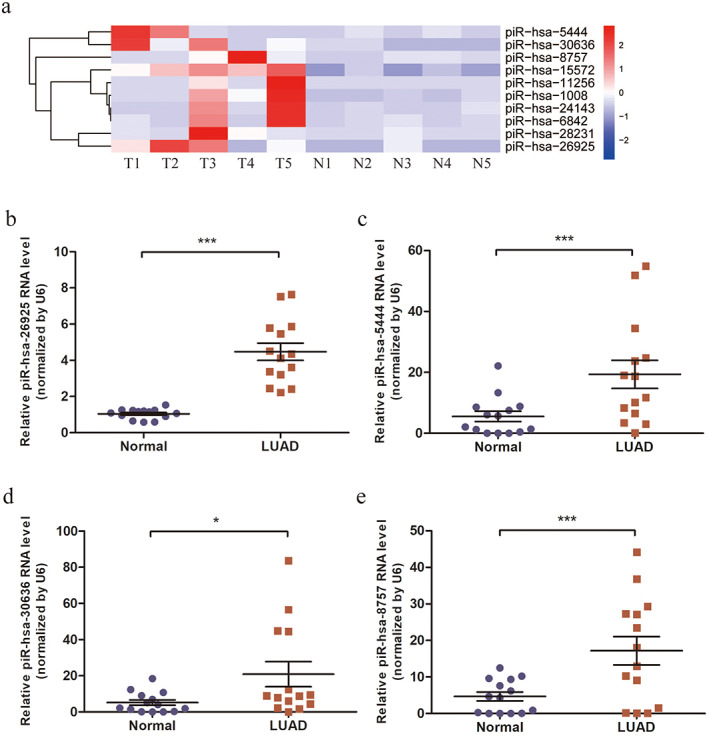
Screening and verification of candidate upregulated piRNAs in tissues. (a) Heatmap of top 10 candidate piRNAs with statistical significance. T, LUAD tissues; N, paired adjacent non‐tumor tissues. (b–e) Relative expressions of piR‐hsa‐26925 (b), piR‐hsa‐5444 (c), piR‐hsa‐30636 (d) and piR‐hsa‐8757 (e) in LUAD patients. *p < 0.05, ***p < 0.001
To predict the possible functions of the four candidates, gene‐set enrichment analysis was performed, showing 283 target mRNAs in the interaction network (Figure S2(a)). GO analysis was also performed, and the results revealed significant enrichment of multiple target genes in vesicle‐mediated transport, intracellular signal transduction, cell junction, and purine nucleotide‐binding (Figure S2(b)). KEGG pathway analysis showed that differential genes were enriched in the Wnt signaling pathway, pathways in cancer, and NSCLC (Figure S2(c)). These data indicated that the candidate piRNAs investigated in our study might regulate several downstream genes involved in the progression of LUAD.
Characterization of serum exosomes
Exosomes were isolated from the serum of LUAD patients and healthy controls. The TEM results showed that the exosomes obtained from serum were 40–150 nm in size with saucer‐shaped morphology (Figure 3(a)). The exosomes were also confirmed by NTA, with data showing that the average diameter of isolated exosomes was ~100 nm (Figure 3(b)). Western blot analysis further indicated the presence of exosomal markers TSG101 and CD9 (Figure 3(c)). Taken together, these results suggested that exosomes were successfully isolated from serum samples.
FIGURE 3.
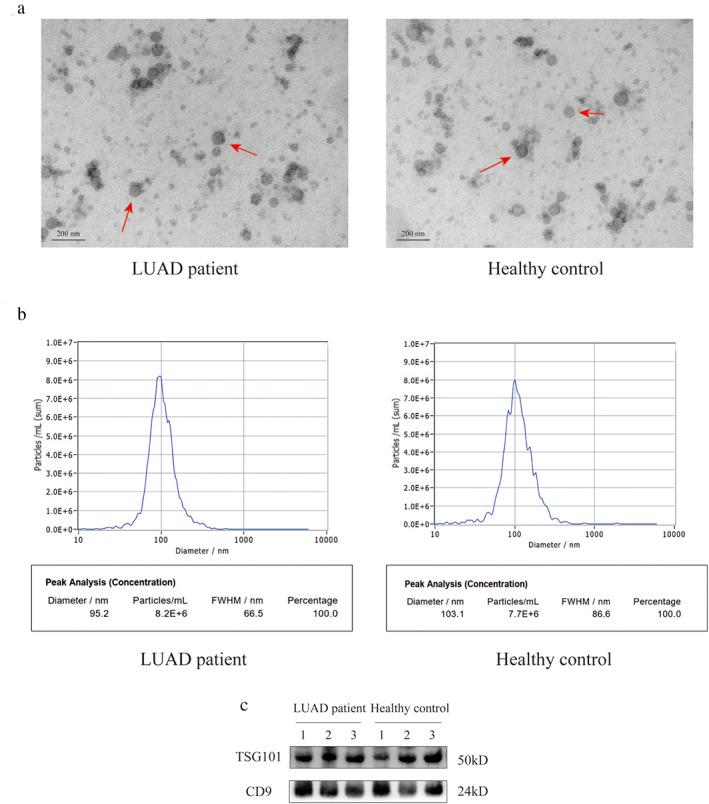
Characterization of serum exosomes. (a) TEM images of exosomes isolated and detected from serum samples (scale bar, 200 nm). (b) the NTA showed the size distribution and the number of serum exosomes. (c) Western blot analysis of exosomal markers, including TSG101 and CD9
Validation of exosomal piRNAs in serum samples
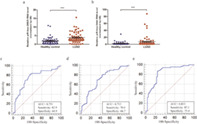
To support the translation of our findings from LUAD tissues into clinical practice, we extracted exosomal piRNAs in serum samples from 70 LUAD patients and 57 healthy controls, and explored the potential clinical value of exosomal piRNAs for LUAD diagnosis. The differential expressions of four candidate piRNAs in serum exosomes from patients with LUAD and healthy controls were evaluated by qRT‐PCR. Data analysis showed that piR‐hsa‐26925 and piR‐hsa‐5444 were significantly upregulated in LUAD patients compared to healthy controls (Figure 4(a) and (b), p < 0.001)), whereas there was no significant difference in piR‐hsa‐30636 and piR‐hsa‐8757 (Figure S3(a) and (b), p > 0.05). The corresponding AUC values of ROC curves were 0.751 and 0.713, respectively. The sensitivity and specificity were shown in Figure 4(c) and Figure 4(d), respectively. We thereafter, explored the correlation between serum exosomal piR‐hsa‐26925 and piR‐hsa‐5444 expression and the clinicopathological characteristics of LUAD patients. The results of Table S2 showed that there was not any association between the expression of these piRNAs and patients' age, sex, tumor size, (tumor, nodes, and metastases [TNM] stage), or lymph node metastasis (p > 0.05).
FIGURE 4.
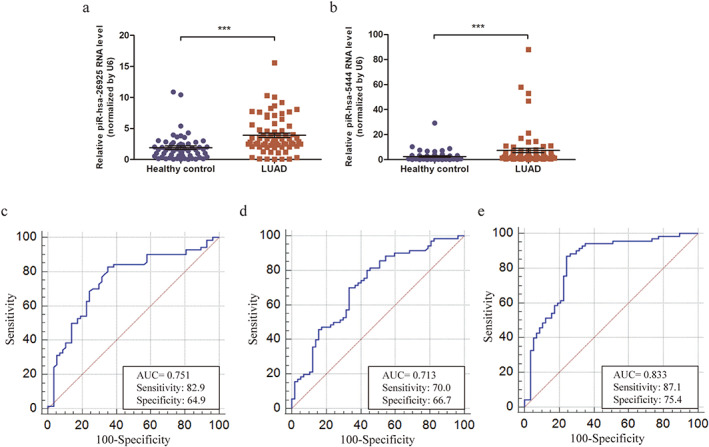
The diagnostic values of piRNAs in serum exosomes. (a,b) Relative expressions of piR‐hsa‐26925 (a) and piR‐hsa‐5444 (b) in the serum exosome samples from LUAD patients and healthy controls. (c,d). ROC curve for piR‐hsa‐26925 (c) and piR‐hsa‐5444 (d) in serum exosome samples. (e) ROC curve for the 2‐piRNA panel in serum exosome samples. ***p < 0.001
To further improve the diagnostic accuracy, we established a multivariate logistic regression model based on the expressions of piR‐hsa‐26925 and piR‐hsa‐5444. The predictive probability of LUAD from the 2‐piRNA panel was calculated using the following formula: Logit (P) = 0.2809–0.07054* piR‐hsa‐26925‐0.01127* piR‐hsa‐5444. The performance of the 2‐piRNA panel in the diagnosis of LUAD was evaluated, and the results of ROC curve showed that the AUC of this panel was 0.833 (95% confidence interval [CI] = 0.756–0.893, sensitivity = 87.1% and specificity = 75.4%, Figure 4(e)). Compared with individual piRNAs, the 2‐piRNA panel showed higher diagnostic performance, suggesting that it was a potential diagnostic biomarker for LUAD.
Stability of identified piRNAs in serum exosomes
We next attempted to investigate the stability of the identified piRNAs in serum exosomes, given that this is another essential prerequisite for ideal biomarkers application. Four exosome samples were subjected to harsh conditions, including incubation at room temperature for 0, 4, 8, 12, and 24 hour (Figure 5(a) and (d)), incubation at −80°C for 0, 1, 2, and 3 months (Figure 5(b) and (e)), and multiple freeze–thaw cycles (0, 2, 4 and 8) (Figure 5(c) and (f)). Total RNA was extracted, and the Ct values of piRNAs were detected by qRT‐PCR. Data showed that there was no obvious change in expressions of the exosomal piRNAs under the above‐mentioned conditions. Collectively, the results indicated that exosomal piRNAs were sufficiently stable in serum exosomes to serve as promising biomarkers for the diagnosis of LUAD.
FIGURE 5.
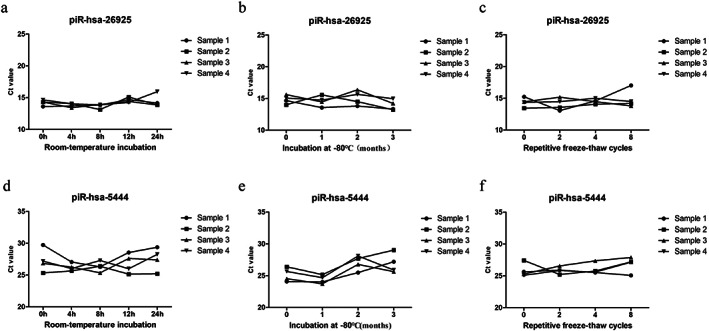
The stability of serum exosomal piRNAs. Serum exosome samples were subjected to incubation at room temperature, repeated freeze–thaw cycles, and incubation at −80°C, and qRT‐PCR was used to detect the expressions of piR‐has‐26925 (a–c) and piR‐has‐5444 (d–f)
DISCUSSION
Regardless of advances in the treatment of LUAD, the outcome of these patients remains poor.20, 21 The main reason lies in the lack of effective screening methods for LUAD, leading to the majority of patients diagnosed at an advanced stage. Serological biomarkers are considered to be one of the most promising diagnostic methods for tumors. However, markers for LUAD, such as CEA, is of limited use because of the lack of sufficient diagnostic sensitivity and specificity.22 Hence, it is urgently necessary to identify new biomarkers for the diagnosis of LUAD.
With the advanced gene sequencing technologies, non‐coding RNAs have emerged as novel genomic regulators in molecular biology. Numerous studies have shown that non‐coding RNAs, especially small RNAs, are involved in the pathogenesis of many diseases and emerge as new tumor markers for clinical diagnosis. In terms of small RNAs, previous studies mainly focused on miRNAs, some of which have been validated in lung cancer, including hsa‐miR‐205‐5p, hsa‐miR‐564, hsa‐miR‐1260b, and hsa‐miR‐337.23, 24, 25, 26 Moreover, Da et al. have confirmed that miR‐1205 can promote the growth of LUAD cells and act as a potential non‐invasive biomarker and therapeutic target for LUAD.27 These studies further suggest that small RNAs can participate in the development of LUAD and have potential as biomarkers.
It is worth noting that piRNAs have emerged as another novel class of small non‐coding RNAs with more than 30 000 members in the human genome, which is much greater than that of miRNAs.9, 10 Emerging studies suggest that piRNAs and PIWI proteins are also involved in tumorigenesis.13 For instance, piR‐021285 is associated with the methylation of cancer‐related genes in breast cancer,28 and PIWIL1‐interacting RNA piR‐017061 inhibits pancreatic cancer by regulating EFNA5.29 More importantly, piRNAs in body fluids are more resistant to oxidation and degradation because of the 2′‐O‐methyl modification at their 3′‐ends,8, 30, 31 indicating that they are more suitable as biomarkers for tumor detection. Five differentially expressed serum piRNAs, including piR‐001311, piR‐004153, piR‐017723, piR‐017724, and piR‐020365, have been reported and proposed as potential non‐invasive diagnostic and prognostic biomarkers in colorectal cancer.14, 16 Moreover, piR‐13643 and piR‐21238 have been identified, which may be promising diagnostic biomarkers for the accurate detection of papillary thyroid carcinoma.32 However, the profile and potential as biomarkers of piRNAs in lung cancer remain largely undetermined.
In the present study, we mapped the differentially expressed piRNAs between LUAD tissues and paired adjacent non‐tumor tissues by high‐throughput sequencing, and found 85 dysregulated piRNAs. After further screening the top 10 significantly upregulated piRNAs in tissue samples by qRT‐PCR, we verified that four (piR‐hsa‐26925, piR‐hsa‐5444, piR‐hsa‐30636, and piR‐hsa‐8757) of them were differentially expressed in LUAD, which was consistent with SRS data. None of these four piRNAs has been reported in previous studies, and further prediction of their target genes indicated that they might regulate several downstream genes, providing clues for investigating their functions and mechanisms involved in LUAD.
Numerous studies have shown that exosomes from cancer cells may exhibit the characteristics of the original tumor, and the nucleic acids and proteins contained in tumor‐specific exosomes may serve as more stable tumor diagnostic biomarkers because of lipid bilayer protection compared with cell‐free RNA.33, 34, 35, 36 Previous studies have shown that the expression levels of RNAs in exosomes remain unchanged even on ribonuclease (RNase) A treatment or incubation with strong acid–base treatment.37, 38 A great deal of attention has been paid to the key roles of lung cancer cell‐derived exosomes in the development of lung cancer and early detection/diagnosis. The exosome proteomics of NSCLC has identified enriched proteins, such as EGFR, GRB2, and Src, for the early detection of lung cancer.39 Another report has shown that serum exosomal miR‐1290 can be a potential diagnostic and prognostic biomarker for LUAD.40 However, the presence of piRNAs in serum exosomes as diagnostic markers for LUAD has not been studied. In the present study, we extracted piRNAs in serum exosomes from 70 patients with LUAD and 57 healthy controls. The expressions of four candidate piRNAs (piR‐hsa‐26925, piR‐hsa‐5444, piR‐hsa‐30636, and piR‐hsa‐8757) were assessed by qRT‐PCR. The results showed that the expression of piR‐hsa‐26925 and piR‐hsa‐5444 were significantly higher in patients with LUAD than in healthy controls. The corresponding AUC of ROC curves was 0.751 and 0.713, respectively. To further investigate their combined diagnostic value, we established a multivariate logistic regression model based on the exosomal piR‐hsa‐26925 and piR‐hsa‐5444. Compared with single piRNAs, the predictive capability of the 2‐piRNA panel was higher with an AUC of 0.833 (95% CI = 0.756–0.893, sensitivity = 87.1% and specificity = 75.4%). In addition, a previous study has shown that the expression of piRNA in serum samples is stable.16 Consistent with this study, we found that repeated freeze–thawing, long‐term incubation at room temperature or −80°C did not affect the expressions of piR‐hsa‐26925 and piR‐hsa‐5444 in serum exosomes of patients with LUAD. These data supported the potential of serum exosomal piRNAs as promising diagnostic markers for LUAD.
Collectively, our study revealed the abnormally expressed piRNAs in LUAD, and serum exosomal piR‐hsa‐26925 and piR‐hsa‐5444 could serve as potential biomarkers for the diagnosis of LUAD. However, there are some limitations in this study. First, the number of subjects included in the study was limited. Second, it was uncertain whether serum exosomal piR‐hsa‐26925 and piR‐hsa‐5444 were only specific for LUAD. Therefore, the potential applications of our findings should be validated in larger independent samples.
CONFLICT OF INTEREST
No conflict of interest has been declared by the authors.
Supporting information
Figure S1 Relative expressions of piR‐hsa‐1008 (a), piR‐hsa‐28231 (b), piR‐hsa‐11256 (c), piR‐hsa‐24143 (d), piR‐hsa‐6842 (e), and piR‐hsa‐15572 (f) in tissues of LUAD patients. ns, not significant.
Figure S2 (a) The interaction network regulated by the four upregulated piRNAs. (b) GO classification for biological process, cellular component, and molecular function of the targets of four upregulated piRNAs. (c) KEGG analysis of the targets of four upregulated piRNAs.
Figure S3 Relative expressions of piR‐hsa‐30636 (a) and piR‐hsa‐8757 (b) in serum exosomes. ns, not significant.
Table S1 Small RNA sequence data in LUAD tissues and corresponding adjacent non‐tumor tissues
Table S2 Correlation between serum exosomal piRNAs and clinicopathological characteristics of patients with LUAD [median (interquartile range)]
ACKNOWLEDGMENTS
This research was supported by grant from the Natural Science Foundation of Shandong Province, China (ZR2020MH008) and Young Taishan Scholar Program of Shandong Province.
Li J, Wang N, Zhang F, Jin S, Dong Y, Dong X, et al. PIWI‐interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac Cancer. 2021;12:2468–2477. 10.1111/1759-7714.14094
Funding information Natural Science Foundation of Shandong Province, Grant/Award Number: ZR2020MH008; Young Taishan Scholar Program of Shandong Province
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446–54. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Adjei AA. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev. 2015;34:169–71. [DOI] [PubMed] [Google Scholar]
- 6.Duma N, Santana‐Davila R, Molina JR. Non‐small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623–40. [DOI] [PubMed] [Google Scholar]
- 7.Seijo LM, Peled N, Ajona D, Boeri M, Field JK, Sozzi G, et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14:343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD. PIWI‐interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- 9.Weng W, Li H, Goel A. Piwi‐interacting RNAs (piRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. 2019;1871:160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinasco‐Sandoval T, Moreira FC, Vidal AF, Pinto P, Ribeiro‐Dos‐Santos AM, Cruz RLS, et al. Global analyses of expressed Piwi‐interacting RNAs in gastric cancer. Int J Mol Sci. 2020;21(20):7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas‐Rios P, Simonelig M. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development. 2018;145(17):dev161786. [DOI] [PubMed] [Google Scholar]
- 12.Chalbatani GM, Dana H, Memari F, Gharagozlou E, Ashjaei S, Kheirandish P, et al. Biological function and molecular mechanism of piRNA in cancer. Pract Lab Med. 2019;13:e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, et al. PIWI‐interacting RNA‐54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8:5213–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, et al. PIWI‐interacting RNA‐36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu A, Wang W, Yang Y, Zhang X, Dong Y, Zheng G, et al. A serum piRNA signature as promising non‐invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res. 2019;11:3703–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Mooij T, Peterson TE, Evans J, McCutcheon B, Parney IF. Short non‐coding RNA sequencing of glioblastoma extracellular vesicles. J Neurooncol. 2020;146:253–63. [DOI] [PubMed] [Google Scholar]
- 18.Williams Z, Morozov P, Mihailovic A, Lin C, Puvvula PK, Juranek S, et al. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015;13:854–63. [DOI] [PubMed] [Google Scholar]
- 19.Muinao T, Deka Boruah HP, Pal M. Multi‐biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon. 2019;5:e02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier‐Valette C, Tessonnier L, et al. Hyperprogressive disease in patients with advanced non‐small cell lung cancer treated with PD‐1/PD‐L1 inhibitors or with single‐agent chemotherapy. JAMA Oncol. 2018;4:1543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CK, Cho HJ, Choi YD, Oh IJ, Kim YC. A phase II trial of Osimertinib in the second‐line treatment of non‐small cell lung cancer with the EGFR T790M mutation, detected from circulating tumor DNA: LiquidLung‐O‐cohort 2. Cancer Res Treat. 2019;51:777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wadowska K, Bil‐Lula I, Trembecki L, Sliwinska‐Mosson M. Genetic markers in lung cancer diagnosis: a review. Int J Mol Sci. 2020;21(13):4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang M, Zhang P, Hu G, Xiao Z, Xu F, Zhong T, et al. Relative expressions of miR‐205‐5p, miR‐205‐3p, and miR‐21 in tissues and serum of non‐small cell lung cancer patients. Mol Cell Biochem. 2013;383:67–75. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, Jia L, Guo Q, Ren H, Hu D, Zhou X, et al. MiR‐564 functions as a tumor suppressor in human lung cancer by targeting ZIC3. Biochem Biophys Res Commun. 2015;467:690–6. [DOI] [PubMed] [Google Scholar]
- 25.Xu L, Li L, Li J, Li H, Shen Q, Ping J, et al. Overexpression of miR‐1260b in non‐small cell lung cancer is associated with lymph node metastasis. Aging Dis. 2015;6:478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Gong WH, Li Y, Zhang HY, Zhang CX. Hsa‐miR‐337 inhibits non‐small cell lung cancer cell invasion and migration by targeting TCF7. Eur Rev Med Pharmacol Sci. 2020;24:7568. [DOI] [PubMed] [Google Scholar]
- 27.Dai B, Kong DL, Tian J, Liu TW, Zhou H, Wang ZF. microRNA‐1205 promotes cell growth by targeting APC2 in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2019;23:1125–33. [DOI] [PubMed] [Google Scholar]
- 28.Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI‐interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J, Xing S, Shen B‐Y, Chen H‐T, Sun B, Wang Z‐T, et al. PIWIL1 interacting RNA piR‐017061 inhibits pancreatic cancer growth via regulating EFNA5. Hum Cell. 2021;34:550–63. [DOI] [PubMed] [Google Scholar]
- 30.Gainetdinov I, Colpan C, Arif A, Cecchini K, Zamore PD. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in Most animals. Mol Cell. 2018;71:775–90. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim SL, Qu ZP, Kortschak RD, Lawrence DM, Geoghegan J, Hempfling AL, et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the Spermatogenic program in the mouse. PLoS Genet. 2015;11:e1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Z, Ji G, Huang R, Chen H, Gao Y, Wang W, et al. PIWI‐interacting RNAs piR‐13643 and piR‐21238 are promising diagnostic biomarkers of papillary thyroid carcinoma. Aging (Albany NY). 2020;12:9292–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Liu W, Wang J, Zhu L, Wang Z, Peng Y. Exosomal noncoding RNAs in colorectal cancer. Cancer Lett. 2020;493:228–35. [DOI] [PubMed] [Google Scholar]
- 34.Seimiya T, Otsuka M, Iwata T, Shibata C, Tanaka E, Suzuki T, et al. Emerging roles of Exosomal circular RNAs in cancer. Front Cell Dev Biol. 2020;8:568366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Shen Z. Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Med. 2020;9:6909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020;476:13–22. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, et al. Evaluation of serum exosomal LncRNA‐based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23:1396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Du LT, Wang YS, Gao SY, Li J, Li PL, et al. Development of a novel serum Exosomal MicroRNA Nomogram for the preoperative prediction of lymph node metastasis in esophageal squamous cell carcinoma. Front Oncol. 2020;10:573501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer‐derived exosomes. J Proteomics. 2016;133:161–9. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Wei J, Zhang W, Xie M, Wang X, Xu J. Serum Exosomal miR‐1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther. 2020;13:7809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Relative expressions of piR‐hsa‐1008 (a), piR‐hsa‐28231 (b), piR‐hsa‐11256 (c), piR‐hsa‐24143 (d), piR‐hsa‐6842 (e), and piR‐hsa‐15572 (f) in tissues of LUAD patients. ns, not significant.
Figure S2 (a) The interaction network regulated by the four upregulated piRNAs. (b) GO classification for biological process, cellular component, and molecular function of the targets of four upregulated piRNAs. (c) KEGG analysis of the targets of four upregulated piRNAs.
Figure S3 Relative expressions of piR‐hsa‐30636 (a) and piR‐hsa‐8757 (b) in serum exosomes. ns, not significant.
Table S1 Small RNA sequence data in LUAD tissues and corresponding adjacent non‐tumor tissues
Table S2 Correlation between serum exosomal piRNAs and clinicopathological characteristics of patients with LUAD [median (interquartile range)]


