Abstract
Objective
Non‐small‐cell lung cancer (NSCLC) is one of the most common fatal cancers in the world. Although the treatment of NSCLC has been significantly improved, there is still an unmet need to identify novel targets for developing therapeutic agents and diagnostic/prognostic markers. The aim of this study is explore the role and underlying mechanism of the epithelial splicing regulatory protein (ESRP1) in the development and progression of NSCLC.
Methods
A total of 115 participants, 65 cases of NSCLC, 20 cases of precancerous lesions, and 30 cases of benign lung nodules, were included in this study. The expressions of ESRP1 and related transcription factor Twist in enrolled lung tissues were evaluated by histochemistry and immunohistochemistry assay. The survival analysis and related prognosis factors were evaluated by the Kaplan–Meier curve and Cox regression. In addition, the expression of ESRP1 and epithelial‐mesenchymal transition (EMT)related transcription factor Twist and EMT markers E‐cadherin and N‐cadherin were ascertained by immunohistochemical and immunoblotting assay on A549 lung adenocarcinoma cell lines that were exposed to transforming growth factor β1 (TGFβ1).
Results
Compared with normal lung tissues, the abundance of ESRP1 protein was significantly increased in precancerous lesions and lung cancer. Correlation analysis demonstrated that ESRP1 was an independent prognostic factor in NSCLC. The expression of ESRP1 and Twist was positively correlated in lung tissues (r = 0.285, p < 0.001). In vitro analysis further showed that TGFβ1 could upregulate the expression of EMT transcription factor Twist while downregulating ESRP1.
Conclusions
Our data suggest that the aberrant expression of ESRP1 is an early event in the development of NSCLC. The ESRP1 could serve as a prognostic biomarker for NSCLC, particularly when combined with Twist. The Twist negatively regulated the expression of ESRP1, emphasizing the role of the TGFβ/ESRP1 pathway in the development of NSCLC, which warrants further investigation.
Keywords: biomarker, EMT, ESRP1, non‐small‐cell lung cancer, Twist
Study flowchat: A total 115 samples of lung tissues were inclued in study, which including 30 cases of benign samples, 20 cases of precancerous lesions and 65 cases of NSCLC lesions for further study.

INTRODUCTION
Lung cancer remains the most common cancer‐related cause of death, accounting for about 12% of all cancer deaths.1 In the United States, the case mortality of lung cancer is about 25.3%, first place in causes of cancer death. Among lung cancers, non‐small‐cell lung cancer (NSCLC) is the main subtype, accounting for 80–85% of the total number of lung cancers.2 In the past decade, with the introduction of chemotherapy, targeted therapy, and emerging immunotherapy, as well as the improvement of oncological management of late‐stage lung cancer, the 5‐year survival rate in patients with NSCLC has increased. However, survival remains poor in most patients (~75%) with advanced disease at the time of diagnosis (stage III/IV).3 One of the main causes of death in patients with advanced lung cancer is distant metastasis. It has been reported that the 5‐year relative survival rate is only 19.7% in NSCLC with distant metastasis,4 therefore early diagnosis is necessary to reduce the high mortality in lung cancers. Understanding the biological mechanisms of tumor development and biomarker expression typical for lung cancer is crucial for accurate diagnosis, prognosis, and development of treatment regimens.
During embryonic development, cells can transition between epithelial and mesenchymal states in a highly plastic and dynamic manner. In this context, the expression of adhesion molecules of epithelial cells is altered to adopt the migratory and invasive behavior of mesenchymal cells. The process of cells shifting from the epithelial state to the mesenchymal state is referred to as epithelial‐mesenchymal transition (EMT).5 Functionally, EMT is involved in important processes of embryonic development, wound healing, organ fibrosis, and cancer progression.6, 7 Oncogenically, EMT commonly occurs during cancer progression, including metastasis, cancer stem cell proliferation, and/or acquired immune escape.7 EMT is highly regulated by various signaling pathways and factors. Transforming growth factor β (TGFβ) signaling is one of the best documented pathways and plays in pivotal role in EMT. In addition, the epithelial splicing regulatory proteins (ESRPs) are a family of newly discovered epithelial cell‐specific alternative splicing regulatory proteins which are known to be involved in cell stemness, proliferation, and resistance to chemotherapy and radiotherapy in various cancers.8
Twist is a transcription factor able to regulate cell migration and tissue reorganization during early embryogenesis, which also plays an important role in EMT and tumor metastasis.9, 10 Twist induces the downregulation of E‐cadherin (E‐Ca) and increased expression of mesenchymal markers such as fibronectin, vimentin, smooth muscle actin (α‐SMA), and N‐cadherin (N‐Ca).11 In addition to the level of transcription and epigenetic modification, alternative splicing (AS) of mRNA precursors has become another important EMT regulation mechanism.12 AS regulates EMT by modifying RNA splicing during mRNA maturation, which can extensively affect cell morphology, adhesion function, secretion function, and biological behaviors such as cell migration. The epithelial splicing regulatory protein 1 (ESRP1) is the key determinant protein of alternative splicing. Twist, a transcription factor that induces EMT, binds to the key cis‐acting element (eg. E‐box) of ESRP1 gene promoter to inhibit splicing regulation, and promote and induce EMT to occur in tumors.13 In the views of that the ESRP1 has been used as a biomarker for the identification and a target of treatment of a variety of cancers, however the role of Twist in prognosis of lung cancer and potential target in lung cancer treatment are still poorly understood. In this study, we investigated expression of ESRP1 and EMT transcription factor Twist in NSCLC patient samples and explored its effect on diagnostic/prognostic values in NSCLC.
MATERIALS AND METHODS
Study design
Ethics statement
Human samples were collected using a protocol approved by the Ethic Committee for the Conduct of Human Research at Ningxia Medical University (NXMU‐2015‐205). Written consent was obtained from every participating individual according to the Ethic Committee for the Conduct of Human Research protocol. All participants signed an informed consent for scientific research of clinical data during hospitalization and provided written informed consent for the publication of the data. The Ethic Committee for the Conduct of Human Research approved the consent procedure for this study (NXMU‐2015‐205).
Subjects and data collection
A total of 115 lung tissue samples were collected in the General Hospital of Ningxia Medical University from 1 January 2015 to 31 January 2018. The participants included 65 cases with NSCLC who underwent pneumonectomy, lobectomy or lung biopsy. In addition, 50 cases of lung nodules at a suspected early stage of NSCLC who underwent segmentectomy, in which 20 cases were confirmed by pathology as precancerous lesions (precancerous group, n = 20) and 30 cases were confirmed by pathology as benign lung lesions (benign group, n = 30), were also enrolled in this study. All tissue samples were collected during the same study period. The diagnosis of NSCLC was according to the criteria of the National Comprehensive Cancer Network (NCCN) guideline.14 Patients aged under 18 and patients with other malignant disease were excluded in this study (Figure 1).
FIGURE 1.
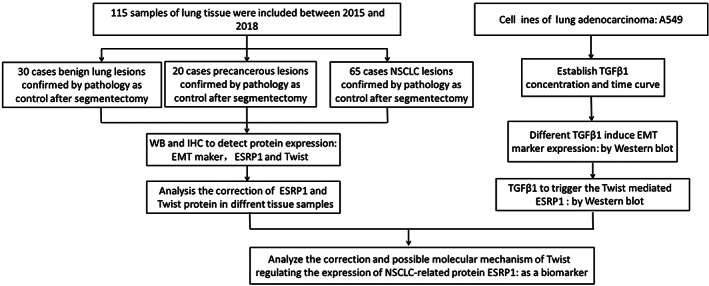
Flowchart of materials included in the study. EMT, epithelial‐mesenchymal transition; ESRP1, epithelial splicing regulatory protein 1; IHC, immunohistochemistry; NSCLC, non‐small‐cell lung cancer; TGFβ1, transforming growth factor β1; WB, Western blot
Data collection and follow‐up
Basic demographic information was collected through medical records, including patient age, tumor diameter, stage determined by the International Lung Cancer Federation, degree of differentiation, lymphatic infiltration, alveolar infiltration, bronchial infiltration, and pathological type. Patient follow‐up was carried out through regular telephone visits in the hospital's outpatient clinic and clinical follow‐up department. The follow‐up ran from cancer diagnosis to the end data of the study (31 January 2020), whichever came first. The overall survival time was defined as the date of surgery to the date of death from any cause or the date of the last follow‐up, whichever came first.
Immunohistochemical analysis
Hematoxylin and eosin staining was used to confirm the main pathological diagnosis. An immunohistochemistry (IHC) test was performed to evaluate the abundance of E‐Ca, vimentin, α‐SMA, ESRP1 and Twist proteins. The primary antibodies to E‐Ca (Abcam, ab76055, dilution 1:200), vimentin (ab57595, dilution 1:250; Abcam), α‐SMA (ab5694, dilution 1:250; Abcam), ESRP1 (ab57595, dilution 1:50; Abcam), and Twist (ab183105, dilution 1:250; Abcam) were incubated on 4 μm section samples at room temperature for 1.5 h. Two pathologists who were unaware of the results independently assessed protein expression according to a defined assessment system. For each lung tissue sample, two blocks were evaluated for each case and the average score was applied. By reassessing and recording samples, the difference in the scores of the two pathologists was resolved. We used the IHC scoring scale for E‐Ca, vimentin and α‐SMA according to the previously published method and scored the total score by a combination of intensity and percentage of positive cells15 (Supporting Information Table S2). Twist and ESRP1 are nuclear‐located proteins and are expressed in almost every cell. The intensities of ESRP1 and Twist staining were graded on a scale from 0 to 3 (0 for no staining, 1 for weak, 2 for moderate, and 3 for strong immunoreactivity). The percentage immunoreactivity was scored on a scale from 0 to 3 (0 for no positive cells, 1 for <30% of cells being positive, 2 for 30% to 60% of cells being positive, and 3 for >60% of cells being positive). The two scores were multiplied to obtain a composite expression score. The final expression level was classified as negative (−) (score of 0), weakly positive (+) (score of 1 to 4), or strongly positive (++) (score of 6 to 9). E‐Ca expression was considered positive (+) if >90% of tumor cells exhibited a staining pattern similar to that in normal epithelial cells. Vimentin and α‐SMA expression was classified as positive (+) when >10% tumor cells were stained.
Protein extraction and Western blotting analysis
Cells were dissociated with 1 ml of trypsin‐ethylenediamine tetraacetic acid (EDTA) solution (Beyotime) and 500 μl of Radioimmunoprecipitation assay (RIPA)lysis buffer per 100 pack cell volume (PCV) was used to lyse the cells and extract the proteins. Western blotting was performed to evaluate the expression of the ESRP1 and Twist proteins. The primary antibodies to E‐Ca (ab76055, dilution 1:1000; Abcam), N‐Ca (ab18203, dilution 1:1000; Abcam), ESRP1 (ab57595, dilution 1:1000; Abcam), Twist (ab32020, dilution 1:1000; Abcam), and Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) (dilution 1:2000) were incubated overnight at 4°C. Enhanced chemiluminescence(ECL, Advansta) was employed to develop a blot of the immunological reaction of the protein of interest for visualization. The intensity and area of the blot were analyzed using ImageJ software for semiquantification of the expression of proteins.
Statistical analysis
The chi‐square test was used to evaluate the correlation between IHC expression of ESRP1 or Twist and the development of NSCLC. Bivariate correlation analysis (Spearman coefficient) was used to evaluate the relationship between ESRP1 or Twist expression. The Kaplan–Meier curve and log‐rank test were performed to compare the overall survival of patients. To determine independent prognostic factors, a multivariate analysis was performed using the Cox proportional hazards model. In vitro results were given as mean ± standard error of mean (SEM) from no less three independent experiments using Student's t‐test. Statistical analysis was performed in SPSS 20.0 (SPSS Inc.). All statistical analyses were two‐way and p < 0.05 was considered statistically significant.
RESULTS
The expression of EMT‐related proteins was significantly different between lungs of patients with NSCLC and precancer or benign tumors
To explore the expression of EMT‐related proteins in different lung samples, the expression of the EMT stromal phenotypic marker proteins vimentin, α‐SMA, N‐Ca, and E‐Ca were evaluated by Western blotting assay in 65 cases of NSCLC patients, 20 cases of precancerous tumor, and 30 cases of benign tumor (Figure 2(b)). The Western blotting results show that among the 65 NSCLC samples, the EMT marker E‐Ca was significantly reduced compared to that of the benign and precancerous tissues (p < 0.001; Figure 2(b),(c)). In addition, vimentin and α‐SMA began to show high expression in the precancerous lesions (p < 0.01), and expression in NSCLC tissue was significantly higher than that in benign tissue (p < 0.05; Figure 2(b),(c)). IHC assay results exhibited similar trends. In the lung cancer group, the EMT‐related protein E‐Ca was positively expressed in 17% (11/65) of NSCLC tissues and as high as 70% (21/30) in the benign group (Supporting Information Table S1). However, the expression of vimentin was higher in the NSCLC group than in the benign group, but no statistically significant difference between the NSCLC group and the precancerous group was detected (Figure 2(a) and Supporting Information Table S1).
FIGURE 2.
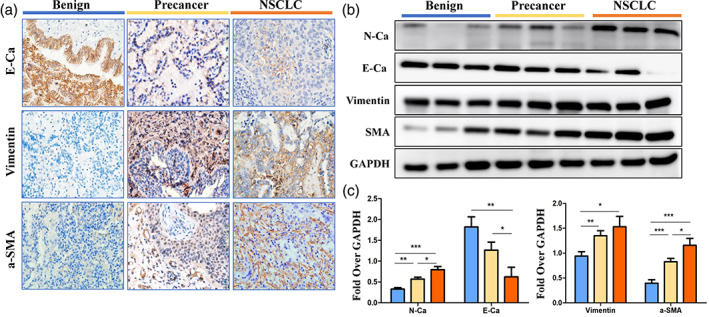
An elevated expression of epithelial‐mesenchymal transition (EMT) markers in lung tissues of benign, precancerous, and non‐small‐cell lung cancer (NSCLC) individuals. (a) Representative image immunohistochemical staining of E‐cadherin, vimentin, and smooth muscle actin (α‐SMA) in lung tissues of the benign (n = 30), precancerous lesion (n = 20), and NSCLC (n = 65) groups. (b) Protein expression of N‐cadherin (N‐Ca), E‐cadherin (E‐Ca), vimentin, and smooth muscle actin (α‐SMA) was measured by Western blotting in the lung tissues of the precancerous lesion (n = 20), NSCLC (n = 65) and benign (n = 30) groups. (c) Densitometric analysis of Western‐blot bands in b. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. GAPDH, Glyceraldehyde‐3‐phosphate dehydrogenase
Aberrant expression of ESRP1 and Twist in NSCLC and precancerous lesions
Twist is considered to be an important transcription factor for splicing protein activity, which may link to the ESRP1. Therefore, we tested the expression of Twist and ESRP1 in the same batch of samples (n = 115). Compared with the benign group, Twist and ESRP1 were highly expressed in the NSCLC group (Figure 3(a),(b)) and in the precancerous group (p < 0.05; Figure 3(b),(c)). The expression of ESRP1 and Twist was not significant between the NSCLC group and the precancerous group (p > 0.05; Figure 3(b),(c)).
FIGURE 3.
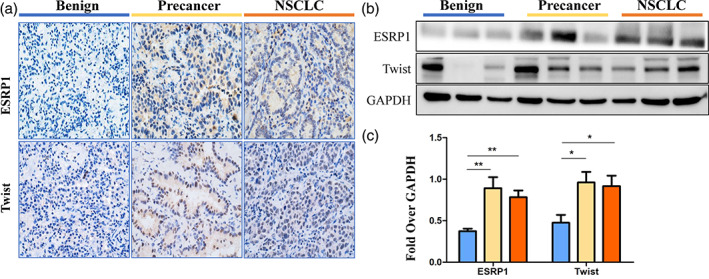
An elevated expression of markers in lung tissue samples of the benign, precancerous lesion, and non‐small‐cell lung cancer (NSCLC) groups. (a) Representative image immunohistochemical staining of epithelial splicing regulatory protein 1 (ESRP1) and Twist in the lungs of the precancerous lesion (n = 20), NSCLC (n = 65), and benign (n = 30) groups. (b) Protein expression of ESRP1 and Twist was measured by Western blotting assay in the lungs of the precancerous lesion (n = 20), NSCLC (n = 65), and benign (n = 30) groups. (c) Densitometric analysis of Western‐blot bands in b. Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. GAPDH, Glyceraldehyde‐3‐phosphate dehydrogenase
Correlation between ESRP1 expression and clinicopathological factors in patients with NSCLC
To explore the clinical characteristics of NSCLC patients for relationships with ESRP1, the correlation between ESRP1 expression and clinicopathological factors was examined (n = 115; Table 1). The results show that low expression of ESRP1 was correlated to poor differentiation (p = 0.01), tumor stage (p = 0.042), and distant metastasis (p = 0.012). In addition, no difference in ESRP1 protein expression was determined by age (≤60 years old compared with >60 years old, p = 0.565), gender (p = 0.215), and type of NSCLC (p = 0.115) (Tables 1 and 2).
TABLE 1.
Correlation between ESRP1 expression and clinicopathologic characteristics of NSCLC
| Variable | ESRP1 expression | X2 | p value | ||
|---|---|---|---|---|---|
| Total | −/+ | ++ | |||
| Age year | |||||
| <60 | 30 | 15 (23%) | 15 (23%) | 0.332 | 0.565 |
| ≥60 | 35 | 15 (23%) | 20 (31%) | ||
| Sex | |||||
| Male | 42 | 17 (26%) | 25 (38%) | 1.540 | 0.215 |
| Female | 23 | 13 (20%) | 10 (16%) | ||
| Smoking History | |||||
| Yes | 34 | 10 (16%) | 24 (37%) | 8.041 | 0.005 |
| No | 31 | 20 (30%) | 11 (17%) | ||
| Pathologic category | |||||
| Squamous cell carcinoma | 35 | 13 (20%) | 22 (34%) | 2.478 | 0.115 |
| Adenocarcinoma | 30 | 17 (26%) | 13 (20%) | ||
| Differentiation | |||||
| Poorly differentiated | 35 | 11 (17%) | 24 (37%) | 6.616 | 0.010 |
| Moderately‐well differentiated | 30 | 19 (29%) | 11 (17%) | ||
| TNM Stage | |||||
| T1 + T2 | 45 | 17 (26%) | 28 (43%) | 4.129 | 0.042 |
| T3 + T4 | 20 | 13 (20%) | 7 (11%) | ||
| Metastasis | |||||
| M0 | 57 | 23 (35%) | 34 (52%) | 6.275 | 0.012 |
| M1 | 8 | 7 (11%) | 1 (2%) | ||
| Lymph metastasis | |||||
| N0 | 40 | 21 (32%) | 19 (29%) | 1.685 | 0.194 |
| N1 + N2 | 25 | 9 (14%) | 16 (25%) | ||
p values <0.05 was considered significant.
TABLE 2.
Univariate and multivariate Cox proportional hazard regression analyses for overall survival of NSCLC
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age, ≥60/<60 | 0.742 (0.321–1.719) | 0.487 | 0.511 (0.166–1.570) | 0.241 |
| Sex, male/female | 1.425 (0.633–3.209) | 0.392 | 1.130 (0.232–5.515) | 0.880 |
| Tumor location, left upper/left lower/right upper/right lower | 1.077 (0.342–3.390) | 0.767 | 2.300 (0.567–9.329) | 0.073 |
| Smoking, yes/no | 2.013 (0.877–4.619) | 0.099 | 1.384 (0.207–9.273) | 0.737 |
| Differentiation, poorly/moderately well | 4.704 (1.742–1.706) | 0.002 | 5.905 (1.506–23.149) | 0.011 |
| TNM stage, T1 + T2/T3 + T4 | 1.372 (0.532–3.536) | 0.513 | 1.421 (0.323–6.251) | 0.642 |
| Metastasis, M0/M1 | 0.046 (0.000–881.637) | 0.540 | 0.000 (0.000–0.000) | 0.998 |
| Lymph metastasis, N0/N1 + N2 | 2.023 (0.874–4.682) | 0.100 | 1.834 (0.598–5.621) | 0.289 |
| ESRP1, −+/++ | 17.501 (4.041–75.787) | 0.000 | 20.559 (3.938–107.336) | 0.000 |
p values <0.05 was considered significant.
Prognostic value of ESRP1 and Twist in NSCLC patients
The prognostic value of ESRP1 expression was further confirmed in the 65 cases of NSCLC group. The median Overall Survival (OS) was not reached. Survival analysis showed that high expression of ESRP1 was significantly related to better prognosis of NSCLC (p = 0.026; Figure 4(a)). Multivariate analysis showed that ESRP1 is an independent prognostic factor for NSCLC patients (p = 0.008; Table 3). Twist binds to the E‐boxs of ESRP1 promoter to inhibit splicing regulation and promote EMT in the progression of tumors.16 Both ESRP1 and Twist were highly expressed, accounting for 37.39% (37/85) of all patients (Table 3). Interestingly, even in the subgroup of high expression of Twist, the expression level of ESRP1 could improve the OS of patients (p = 0.039; Figure 4(b)).
FIGURE 4.
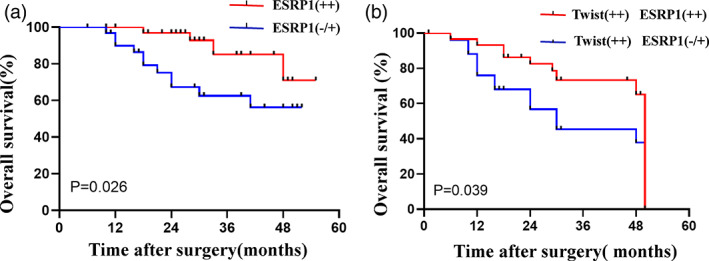
Correlation between epithelial splicing regulatory protein 1 (ESRP1) expression and survival in non‐small‐cell lung cancer (NSCLC) patients. (a) Kaplan–Meier curves displaying the overall survival of NSCLC patients (n = 65) with high ESRP1 expression versus negative or low ESRP1 expression. (b) Kaplan–Meier curves displaying the overall survival of participants with high expression of Twist and different ESRP1 expression in the NSCLC group (n = 47)
TABLE 3.
Correlation between ESRP1 and Twist in lung tissues of patients with precancerous lesions and NSCLC group
| ESRP1 expression, n (%) | ||||||
|---|---|---|---|---|---|---|
| −/+ | ++ | Total | r | p value | ||
| Twist expression | −/+ | 18 (23.48) | 9 (19.13) | 27 | 0.285 | 0.008 |
| ++ | 21 (20) | 37 (37.39) | 58 | |||
| Total | 39 | 46 | 85 | |||
Note: Spearman correlation analysis in ESRP1and Twist between precancerous and NSCLC groups.
p values <0.05 was considered significant.
Twist‐mediated ESRP1 expression in lung adenocarcinoma cell lines triggered by TGFβ1
To further explore the underlying mechanism of Twist in the regulation of ESRP1, lung adenocarcinoma cell line A549 cells were exposed to TGFβ1 and EMT markers were analyzed (Figure 5). Western blotting was used to confirm the influence of TGFβ1 stimulation on the EMT effect of cells. Under different TGFβ1 concentrations, the expression of EMT markers in the A549 cells increased significantly (Figure 5(b),(c)). The results showed a dynamic induction of Twist and ESRP1 by TGFβ1 at doses of 1–10 ng/ml. In addition, 1–10 ng/ml of TGFβ1 could induced Twist expression for the first 48 h of stimulation, which inhibited ESRP1 expression for 72 h (Figure 5(d)). Interestingly, 5 ng/ml of TGFβ1 could significantly increase the level of Twist in A549 cells at 24 h, but the expression waned to a normal level at 48 and 72 h and bounced again at 96 h. In contrast, the expression of ESRP1 in A549 cells was increased at 48 h and elevated again at 96 h in the presence of 5 ng/ml of TGFβ1 (Figure 5(e)).
FIGURE 5.
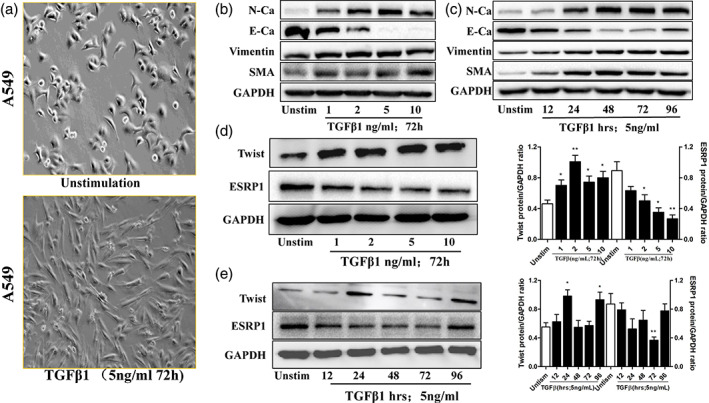
Effects of transforming growth factor β1 (TGFβ1) on the expression of epithelial‐mesenchymal transition markers, Twist, and epithelial splicing regulatory protein 1 (ESRP1) in A549 cells. (a) Morphological examination showed the spindle‐like morphology changes of A549 cells induced by TGFβ (5 ng/ml, 72 h). (b) Cells were incubated with TGFβ1 (72 h) for the indicated concentrations (0–10 ng/ml) and the protein expressions of N‐cadherin (N‐Ca), E‐cadherin (E‐Ca), vimentin, and smooth muscle actin (α‐SMA) were determined by Western blot. (c) Cells were incubated with TGFβ1 (5 ng/ml) for the indicated times (0–96 h) and the protein expressions of N‐Ca, E‐Ca, vimentin, and smooth muscle actin (α‐SMA) were determined by Western blotting assay. (d) Cells were stimulated with various concentrations of TGFβ1 (0–10 ng/ml) for 72 h and the protein expressions of Twist and ESRP1 were determined by Western blotting assay. (e) Cells were stimulated with TGFβ1 (5 ng/ml) for various times (0–96 h) and the protein expressions of Twist and ESRP1 were determined by Western blotting assay. Data was represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 compared to unstimulated control. GAPDH, Glyceraldehyde‐3‐phosphate dehydrogenase
DISCUSSION
The incidence of NSCLC has increased significantly in recent years,1, 2 therefore it is vital to discover the driving factors of the development of NSCLC and identify new potential disease markers. This study showed that the elevated expression of ESRP1 in lung lesions mainly starts in the precancerous phase and remains highly expressed in NSCLC lungs. The high expression of ESRP1 was an independent predictor of poor survival in NSCLC patients, especially when it was highly expressed together with Twist. In addition, it was found that Twist and EMT were positively correlated with the stages of NSCLC. In vitro studies have confirmed that induction of EMT is often accompanied by elevated ESRP1 expression. These findings indicate that ESRP1 may be a clinical predictor and prognostic factor for NSCLC, and it seems to be more sensitive than its transcriptional regulator Twist molecule (data no shown).
EMT is an important process in tumor malignancy and is also a driver of tumor invasion and metastasis.7, 14 A very important feature of EMT is the lack of expression of E‐Ca. Its biological significance lies in the loss of mutual anchoring adhesion between tumor cells, obtaining a mesenchymal phenotype and showing strong invasion and metastasis ability. The experimental results of this study evidenced that EMT was involved in the occurrence and development of NSCLC. TGFβ signaling plays a key role in controlling embryogenic development, inflammation, and tissue repair, as well as in maintaining adult tissue homeostasis.17 It elicits a broad range of context‐dependent cellular responses and, consequently, alterations in TGFβ signaling have been implicated in many diseases, including cancer.15 Our data also showed that TGFβ1 induced EMT in A549 cells in a time‐ and dose‐dependent manner.
ESRP1 is the key determinant of epithelial splicing regulation and is mainly involved in the regulation of alternative splicing of EMT‐related genes. It regulates alternative splicing during the EMT process.12 We found that ESRP1 was associated with the malignant progression of NSCLC, as indicated by the gradual increase in expression with the progression of cancerogeneisis, i.e. benign, precancerous lesions and NSCLC. Interestingly, the expression of ESRP1 was negatively correlated to tumor differentiation, and local and distant invasion in NSCLC, suggesting that expression of ESRP1 might be negatively correlated with the malignancy of cancer. These data indicate that the EMT process seems to be an early event in the development and malignant transformation of NSCLC, during the process of EMT, there are accompanied with ESRP1 activities triggered the splicing regulator, inhibition of EMT and the process of tumorigenesis.
Previous study showed that Twist, a transcription factor that induces EMT, and E‐box of ESRP1 promoter were found to inhibit splicing regulation and induce EMT in tumors. Increasing evidence shows that Twist is related to the downregulation of ESRP1 in the process of TGF‐induced EMT.16 Mechanistical studies further demonstrated that Twist induced and regulated the process of EMT by inhibiting the activity of the Cadherin‐1 (CDH1) promoter and the expression of E‐Ca protein.11 Our in vitro study also showed that the TGFβ1‐activated Twist in A549 was accompanied by downregulated expression of ESRP1. This implies that the transcription factor Twist downregulated the expression of ESRP1 during the EMT process induced by TGFβ1. These results imply that Twist may bind to the ESRP1 promoter and inhibit its transcriptional activity. The specific molecular mechanism of the related transcription factors Snail and Twist inhibiting ESRP1 has not yet been fully elucidated.
In addition, the present study showed that high expression of ESRP1 is an independent predictor of better survival in NSCLC. Patients with high expression of ESRP1 have a better survival rate than patients with lower ESRP1. In addition, we observe slightly different but not significantly from the correlation between high and low Twist expression groups in NSCLC survivals, additional studies are required to provide a further evidence of it. Indeed, ESRP1 has been linked to the differentiation degree and overall survival rate of a variety of tumors.18, 19 For instance, the study of ESRP involvement in tumor EMT showed that ESRP regulated the fibroblast growth factor (FGFR) genes FGFR‐IIIb (epithelial phenotype) and epidermal growth factor receptor (EGFR)‐IIIc (stromal phenotype) through alternative splicing.20 With the expression of these subtypes, silencing ESRP promoted the transformation of cells from an epithelial phenotype to a mesenchymal phenotype. Overexpression of ESRP1 exhibited an opposite effect that promoted tumor malignancies in certain cancers.21 These studies indicate that ESRP1 may be a prognostic indicator of these cancers with high expression of ESRP1.
The advantages of the study include rigorous clinical data collection, and long‐term systematic and prospective follow‐up analysis of the underlying molecular mechanisms. In vitro experiments using well‐known lung adenocarcinoma cell lines have been carefully designed, and each discovery has been verified through repeated repetitions, demonstrating the reliability of the results. However, there are some limitations that deserve attention. Although we recruited all patients with NSCLC during the study period, this is a single hospital‐based study with a limited sample size, which may hinder its promotion. Therefore, it is necessary to conduct further prospective studies in different populations to evaluate the clinical application of ESRP1. How Twist interacts with ESRP1 and regulates ESRP1, and how ESRP1 promotes the malignant phenotype of NSCLC are under investigation.
Collectively, ESRP1 expression in precancerous lesions and NSCLC tissues was significantly increased compared with normal lung tissues and high expression of ESRP1 associated with better outcome in NSCLC patients suggesting that it is an independent prognostic factor, especially when used in combination with Twist. The two molecules ESRP1 and Twist are positively correlated. Importantly, an aberrant ESRP1 was observed in an early‐stage lung cancer, thus it may be a potential biomarker for identification and prognosis of NSCLC. In addition, the TGF‐induced EMT and accompaied by decreased expression of ESRP1 and elevated expression of Twist, implied that Twist as transcriptional repressors and decreased expression of ESRP1. This finding indicates that Twist may be a potential target by suppressing ESRP1 for NSCLC treatment, but the underlying mechanism needs further study.
CONFLICT OF INTEREST
The authors report that there is no conflict of interest.
Supporting information
Supporting Information Table S1 Immunohistochemistry interpretation results
Supporting Information Table S2 Detailed information for the immunohistochemistry scoring scale
ACKNOWLEDGMENTS
We acknowledge all participants for their agreement to use their clinical pathological data in this study. This study was supported by a grant from the National Natural Science Foundation of China (Nos. 81760004), the Project of the Development Center for Medical Science and Technology, National Health Commission of the PRC(W2017ZWS17), grants from the Key Research and Development Program of Ningxia Hui Autonomous (2018BEG03035), and a grant from the Natural Science Foundation of Ningxia (NO.2020AAC02001;2021AAC03300).
Cui J, Ren P, Li Y, Ma Y, Wang J, Lin C, et al. ESRP1 as a prognostic factor of non‐small‐cell lung cancer is related to the EMT transcription factor of Twist. Thorac Cancer. 2021;12:2449–2457. 10.1111/1759-7714.14088
Jieda Cui, Peng Ren, and Yan Li contributed equally to this work.
Funding information A grant from Natural Science Foundation of Ningxia, Grant/Award Numbers: NO.2020AAC02001, NO.2021AAC03300; grants from Key research and development program of Ningxia hui autonomous, Grant/Award Number: 2018BEG03035; National Natural Science Foundation of China, Grant/Award Number: Nos. 81760004; Project of Development Center for Medical Science and Technology, National Health Commission of the PRC, Grant/Award Number: W2017ZWS17
Contributor Information
Shaohua Ma, Email: chenjuan7419@163.com.
Juan Chen, Email: doctor_msh@bjmu.edu.cn.
REFERENCES
- 1.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, et al. Global surveillance of cancer survival 1995‐2009: analysis of individual data for 25,676,887 patients from 279 population‐based registries in 67 countries (CONCORD‐2). Lancet. 2015;385(9972):977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3.Walters S, Maringe C, Coleman MP, Peake MD, Butler J, Young N, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population‐based study, 2004‐2007. Thorax. 2013;68(6):551–64. [DOI] [PubMed] [Google Scholar]
- 4.Moller H, Henson K, Luchtenborg M, Broggio J, Charman J, Coupland VH, et al. Short‐term breast cancer survival in relation to ethnicity, stage, grade and receptor status: national cohort study in England. Br J Cancer. 2016;115(11):1408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial‐mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42. [DOI] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial‐mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R, Weinberg RA. The basics of epithelial‐mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6(5):546–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang MH, Wu KJ. TWIST activation by hypoxia inducible factor‐1 (HIF‐1): implications in metastasis and development. Cell Cycle. 2008;7(14):2090–6. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, et al. SET8 promotes epithelial‐mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31(1):110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–39. [DOI] [PubMed] [Google Scholar]
- 12.Warzecha CC, Carstens RP. Complex changes in alternative pre‐mRNA splicing play a central role in the epithelial‐to‐mesenchymal transition (EMT). Semin Cancer Biol. 2012;22(5–6):417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. [DOI] [PubMed] [Google Scholar]
- 14.Scheel C, Weinberg RA. Phenotypic plasticity and epithelial‐mesenchymal transitions in cancer and normal stem cells? Int J Cancer. 2011;129(10):2310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Qi L, Qu T, Liu C, Cao L, Huang Q, et al. Epithelial splicing regulatory protein 1 inhibits the invasion and metastasis of lung adenocarcinoma. Am J Pathol. 2018;188(8):1882–94. [DOI] [PubMed] [Google Scholar]
- 16.de Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Lamouille S, Derynck R. TGF‐beta‐induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT‐driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial‐mesenchymal transition. J Biol Chem. 2012;287(43):36435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Fernandez SV, Goodwin S, Russo PA, Russo IH, Sutter TR, et al. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta‐estradiol. Cancer Res. 2007;67(23):11147–57. [DOI] [PubMed] [Google Scholar]
- 21.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial‐to‐mesenchymal transition in lung cancer. J Clin Invest. 2016;126(9):3219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table S1 Immunohistochemistry interpretation results
Supporting Information Table S2 Detailed information for the immunohistochemistry scoring scale


