Abstract
Participate in tumorigenic, oncogenic, and tumor suppressive pathways through gene expression regulation. We aimed to build an immune-related long noncoding RNA (lncRNA) prognostic model to enhance nonsmall cell lung cancer (NSCLC) prognostic prediction.
The original data were collected from the cancer genome atlas database. Perl and R software were used for statistical analysis. The effects of lncRNAs expression on prognosis were analyzed by Gene Expression Profiling Interactive Analysis. Silico functional analysis were performed by DAVID Bioinformatics Resources.
The median risk score as a dividing value separated patients into high- and low-risk groups. These 2 groups had different 5-year survival rates, median survival times, and immune statuses. The 5-lncRNA signature was validated as an independent prognostic factor with high accuracy (area under the receiver operating characteristic = 0.722). Silico functional analysis connected the lncRNAs with immune-related biological processes and pathways in carcinogenesis.
The novel immune-related lncRNA prognostic model had significant clinical implication for enhancing lung adenocarcinoma outcome prediction and guiding the choice of treatment.
Keywords: Kyoto Encyclopedia of Genes and Genomes, long noncoding RNA, nonsmall cell lung cancer, prognostic model, the cancer genome atlas
1. Introduction
Nonsmall cell lung cancer (NSCLC) is one of the most important causes of cancer mortality around the world. NSCLC accounts for 80% to 85% of new cases of lung cancer in North America.[1] Recently, there has been growing progress in therapeutic strategies for NSCLC, but the 5-year survival is less than 15%. Several NSCLCs are found in late stages with low survival rates; nonetheless, patients who were in early stages of NSCLC may have longer survival rates. There are many clinical features or molecular biomarkers used for predicting the prognosis of NSCLC, such as tumor, lymph node, and metastasis staging and other grading systems, but these systems have limitations. For example, we sometimes met patients who diagnosed NSCLC in late stage with a relatively long disease-free survival time, nevertheless, some patients in early stage encountered metastasis fast. Furthermore, the National Comprehensive Cancer Network guidelines advised that adjuvant chemotherapy was uncalled for patient in IA stage, and the side effect may overtake the benefits, but the IB stage patients with poor prognosis risk were required additional adjuvant chemotherapy to increase survival time. It is important to figure out high-risk NSCLC patients with poor prognosis. Therefore, building a new prognostic model gives us a chance to more accurately predict the prognosis of patients and select treatment.
Lung cancer cells escape immune detection and activation by inhibiting pivotal steps in the process of CD8+ T cell response. Previous studies have indicated that the immune escape was a main cause for tumor progression.[2,3] However, the potential prognostic biomarkers related to the interrelationship of NSCLC-immune were still needed to be identified.
Long noncoding RNAs (lncRNAs) are transcripts over 200 nucleotides in length that are not translated into proteins.[4] LncRNAs are involved in tumorigenesis, proliferation, and metastasis.[5–7] In addition, many studies indicate that lncRNAs could not only regulate the innate immune response but are also involved in the adaptive immune response.[8–11] Emerging evidence shows that lncRNAs are related to the progression of NSCLC, and their dysregulation is correlated with prognosis. For example, TMPO-AS1 and C1orf132 could influence the prognosis of lung adenocarcinoma (LUAD) patients by affecting the cell cycle and cell adhesion regulation.[12] These advances in genomics study extremely boosted the understanding of the molecular mechanisms of NSCLC and stressed the clinical needs of newly identified molecular biomarkers in early diagnosis and prognosis prediction to increase the survival of NSCLC patients. However, the functions of most lncRNAs are not clear.
With the continuous development and improvement of genome sequencing, it is possible to use bioinformatics methods to deeply mine sequencing data and identify new biomarkers. Moreover, this method allows for the preliminary screening of potential biomarkers and establishes a foundation for further research. The cancer genome atlas (TCGA) database is a cancer database established in 2005. The molecular signatures database is a widely applied bioinformatic tool with huge numbers of gene sets for biologically regulatory pathways.[13]
In the study, we generated a 5 immune-related lncRNAs prognostic model of LUAD by bioinformatics methods, and the prognostic performance was assessed by Cox regression and Receiver operating characteristic (ROC) curve analysis. The model highly increased prediction accuracy of LUAD prognosis synthesized the variables like age, gender, and stage. We further explore the biological functions of the immune-related lncRNAs to facilitate the mechanism research.
2. Materials and methods
2.1. Data collection and data preprocessing
Firstly, the publicly available RNA-seq data (Fragments Per Kilobase per Million values) of squamous cell carcinoma and adenocarcinoma of lung cohort were downloaded from TCGA website (https://cancergenome.nih.gov/), Level 3 data of gene expression profiles (gdc_download_20200101_125616.017009.tar.gz, metadata.cart.2020-01-01.json and MANIFEST.txt) and relevant clinical information were included. A total of 551 samples (497 LUAD samples and 54 normal samples) were included in the LUAD group and 551 samples (502 lung squamous carcinoma (LUSC) samples and 49 normal samples) were incorporated in the LUSC group. Perl (https://www.perl.org/, v5.30.1, 64-Bit; New York, USA, Adam Kaplan) was used for screening lncRNA expression data and data integration. We merged the data and transformed the gene ID by using Perl software (merge.pl and getmRNA.pl). Patients who survived less than 15 days and patients with missing or no clinical data were excluded. R (https://www.r-project.org/, v3.6.2; New York, USA, Lucent Technologies) and corresponding R packages were also used for data processing and analysis. The research based on public sources data, which contains its ethnic approval. Thus, the study do not need any further ethnic approval.
2.2. Immune-related lncRNA extraction
We downloaded 2 influenced gene sets named “IMMUNE_RESPONSE (M19817)” and “IMMUNE_SYSTEM_PROCESS (M13664)” from the molecular signatures database v7.0 to distinguish immune-related RNAs. Co-expression analysis was conducted to identify immune-related lncRNAs with the thresholds of |cor| > 0.6 and P < .001 by “limma” package in R. A total of 359 immune-related lncRNAs of LUAD were identified.
2.3. Immune-related lncRNA prognostic model construction
First, univariate Cox analysis was utilized to figure out prognostic immune-related lncRNAs. The P values of prognostic immune-related lncRNAs were sorted. Nineteen genes were selected with the threshold of P < .05. Next, we performed multivariate Cox regression analysis to calculate risk score for each patient. The formula was as follows:
where n = the nth gene number. The risk score was assigned using the linear combination of the lncRNA expression weighted by the regression coefficient (coef). The regression coefficient of multivariate Cox analysis was regarded as coef (gene n), on behalf of the contribution of lncRNA(n) for risk score, and expression (gene n) was defined as the expression of the nth prognostic immune-related lncRNA. The lncRNAs for inclusion in the final prognostic model were carefully chosen using forward stepwise selection with Akaike Information Criterion as the stopping rule to optimize the model. The Akaike Information Criterion value for the final model was minimized with the fewest number of lncRNAs. Finally, we identified 5 immune-related lncRNAs for LUAD prognostic model construction. All LUAD patients were separated into high- and low-risk groups refer to the median risk score to construct the prognostic model.
2.4. Statistical analysis
To contrast the survival rate of the high- and low-risk groups, Kaplan–Meier analysis was conducted. The Kaplan–Meier survival analysis was performed by the survfit function in “survival” package in R software. The log-rank test was utilized to value survival difference between the low- and high-risk groups. The correlation between risk factors (clinical characteristics and risk score) and prognosis was evaluated by univariate and multivariate Cox regression analyses. Hazard ratios and 95% confidence intervals (CIs) were estimated by the Cox analysis. The coefficients of risk score and stages were analyzed via logistic regression model. The diagnostic capability of risk score and stages in combination was then calculated by binary logistic regression. ROC curve analysis was performed to investigate the prognostic value of multiple factors in the prognostic model in “survivalROC” package in R software. The association between 5 immune-related lncRNAs expression and overall survival (OS) is evaluated by the Gene Expression Profiling Interactive Analysis (GEPIA) database.[14] The relation between prognostic immune-related lncRNA expression levels and clinical characteristics was examined by Kruskal–Wallis test in “ggpubr” package of R software. To explore the different immune statuses of the 2 risk groups in different gene expression datasets, principle component analysis was conducted by “scatterplot3d” package in R software. P < .05 was regarded as statistically significant.
2.5. Silico functional analysis
To investigate the immune system process and immune response pathways of the immune-related lncRNAs in high-risk groups, functional enrichment analysis was conducted by GSEA (http://www.broadinstitute.org/gsea/index.jsp) was conducted between the high-risk and low-risk groups. Co-expressed lncRNA-mRNA pairs were figured out by computing Pearson correlation coefficients using the paired lncRNA and mRNA expression profiles[15] of 497 LUAD patients. Gene ontology (GO, http://www.geneontology.org/) analysis[16] and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/kegg/pathway.html)[17,18] functional analysis of co-expressed protein-coding genes (PCGs) were implemented to explore the potential function of the 5 lncRNAs signature in LUAD through DAVID Bioinformatics Resources (https://david.ncifcrf.gov/, v6.8). P < .05 was considered statistically significant.
3. Results
3.1. Identification of prognostic immune-related lncRNAs
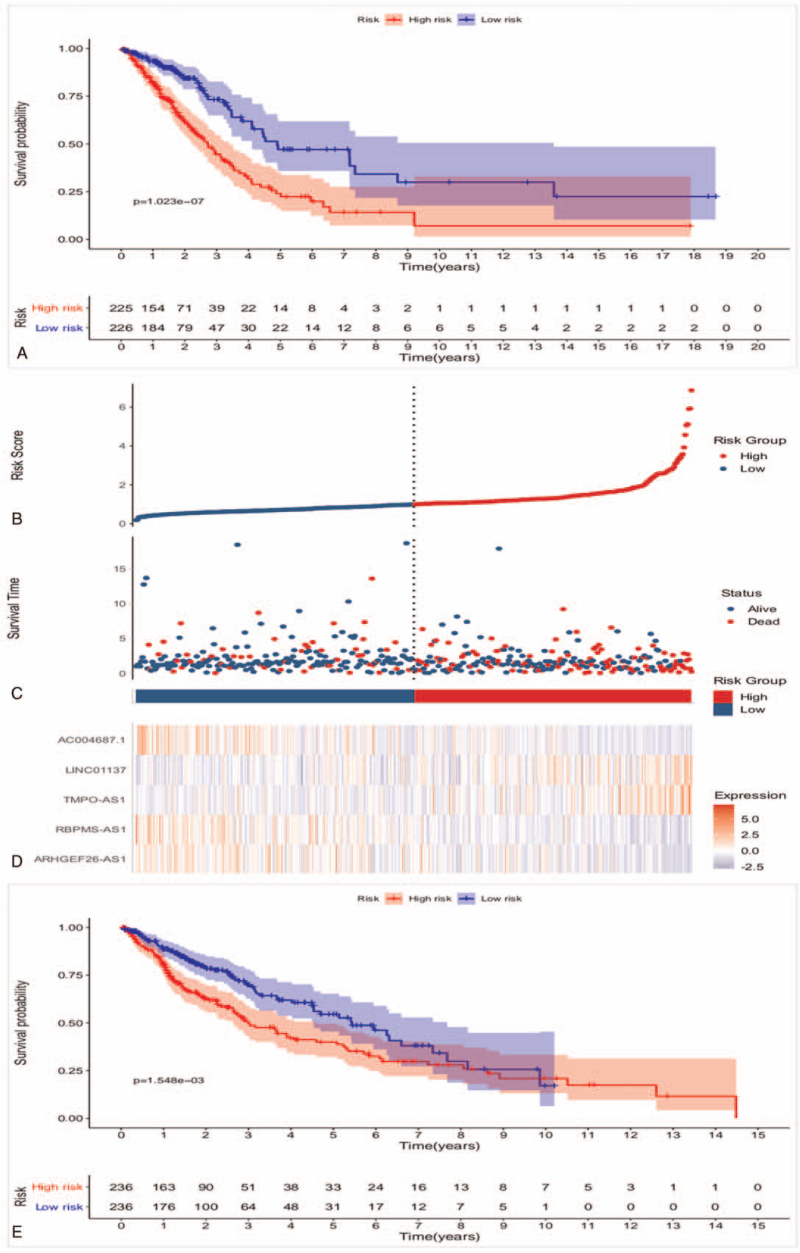
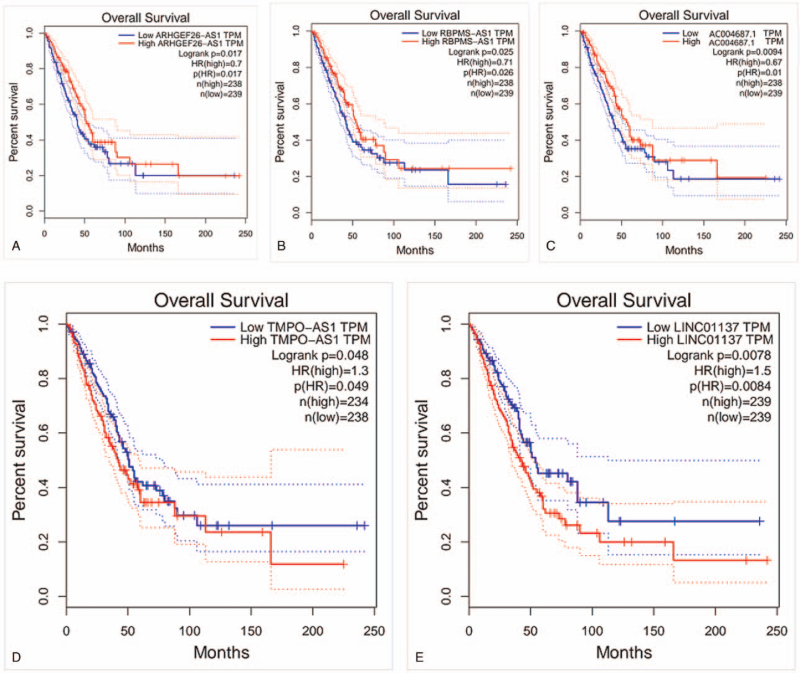
Through univariate Cox regression analysis, 11 prognostic lncRNAs of LUAD were initially selected (P < .01, Table 1), then multivariate Cox regression analysis was carried out to further screen the key prognostic immune-related lncRNAs, 5 lncRNAs were identified finally (Table 1). Based on the coefficients in univariate Cox regression results, the lncRNAs with negative coefficient were regarded as protective lncRNAs. The up-regulation of ARHGEF26-AS1, RBPMS-AS1 and AC004687.1 were related to better OS. Oppositely, up-regulation of risky lncRNAs TMPO-AS1 and LINC01137 with positive coefficient were associated with poor survival. However, only 2 key prognostic immune-related lncRNAs of LUSC were identified through the same method mentioned above (Table 1). As it shown in Figure 1A, all LUAD patients were divided into high- and low-risk groups based on the median risk score. The result of Kaplan–Meier analysis indicated that the 5-year survival rate of the high-risk group was 22.4% (95% CI = 0.149–0.337), which was significantly lower than the 5-year survival rate of the low-risk group (47.1%, 95% CI = 0.359–0.618). Patients in the high-risk group showed worse survival probability compared with the low-risk group (log-rank test P < .001). The median survival time of low-risk group 4.90 years, which was longer than the high-risk group (2.70 years). The 5-lncRNAs were selected to construct a heat map to illustrate the differences in prognostic immune-related lncRNAs expression between high- and low-risk groups (Fig. 1D). The result of heat map indicated that TMPO-AS1 and LINC01137 were overexpressed in high-risk group, whereas ARHGEF26-AS1, RBPMS-AS1 and AC004687.1 had higher expression level in the low-risk group (Fig. 1D). The distribution of risk scores and survival statuses are displayed in Figure 1B and C, respectively. The Kaplan–Meier analysis result of LUSC revealed that survival probability was not significantly different between the high-risk and low-risk group (log-rank test P > .001, Fig. 1E), which indicated that the construction of immune-related lncRNAs prognostic model of LUSC was not successful. Thus, the study only verify the immune-related lncRNAs prognostic model of LUAD in the following research. GEPIA database was used to further verify the role of immune-related lncRNAs in LUAD prognosis, the results showed that higher ARHGEF26−AS1, RBPMS−AS1 and AC004687.1 levels were related to longer OS (log-rank test P < .05, Fig. 2A–C), however patients with high TMPO − AS1 or LINC01137 levels had shortened OS (log-rank test P < .05, Fig. 2D and E). In summary, the 5-lncRNAs could act as potential biomarkers for the LUAD prognosis prediction.
Table 1.
LncRNAs of NSCLC identified from TCGA.
| Category | ID | HR | HR.95L | HR.95H | P value | coef |
| LncRNA of LUAD | ELN-AS1 | 0.811 | 0.692 | 0.949 | .009 | – |
| AC090948.1 | 0.533 | 0.338 | 0.841 | .007 | – | |
| ARHGEF26-AS1 | 0.614 | 0.438 | 0.862 | .005 | – | |
| RBPMS-AS1 | 0.711 | 0.572 | 0.885 | .002 | – | |
| AC026369.3 | 0.678 | 0.516 | 0.891 | .005 | – | |
| TMPO-AS1 | 1.742 | 1.278 | 2.373 | <.001 | – | |
| AC135050.6 | 0.68 | 0.539 | 0.859 | .001 | – | |
| LINC01137 | 1.335 | 1.081 | 1.65 | .007 | – | |
| GAS6-AS1 | 0.654 | 0.485 | 0.884 | .006 | – | |
| AC004687.1 | 0.731 | 0.59 | 0.906 | .004 | – | |
| AC090559.1 | 0.697 | 0.534 | 0.908 | .007 | – | |
| Key lncRNA of LUAD | ARHGEF26-AS1 | 0.6701 | 0.4768 | 0.94171 | .0021 | −0.4004 |
| RBPMS-AS1 | 0.7947 | 0.6381 | 0.98964 | .004 | −0.2299 | |
| TMPO-AS1 | 1.5397 | 1.1232 | 2.11072 | .0073 | 0.4316 | |
| LINC01137 | 1.475 | 1.2045 | 1.80613 | .0002 | 0.3886 | |
| AC004687.1 | 0.7429 | 0.5992 | 0.92096 | .0067 | −0.2972 | |
| LncRNA of LUSC | AC007098.1 | 0.737 | 0.569 | 0.955 | .021 | – |
| AP002026.1 | 0.671 | 0.464 | 0.97 | .034 | – | |
| AP001830.1 | 0.668 | 0.47 | 0.949 | .024 | – | |
| AC020765.2 | 0.671 | 0.48 | 0.937 | .019 | – | |
| TMEM44-AS1 | 0.836 | 0.712 | 0.981 | .029 | – | |
| Key lncRNA of LUSC | AC007098.1 | 0.77287 | 0.59579 | 1.00258 | .0209 | −0.258 |
| AC020765.2 | 0.67093 | 0.48017 | 0.93747 | .0194 | −0.339 |
coef = regression coefficient, HR = hazard ratio, LUAD = lung adenocarcinoma, LUSC = lung squamous carcinoma, TCGA = the cancer genome atlas.
Figure 1.
Immune-related lncRNA prognostic model construction. (A) Kaplan–Meier analysis of survival probability of LUAD according to the median of risk score. (B) Heat map of the 5 immune-related lncRNAs. (C) Distribution of 5 immune-related lncRNAs-based risk score. (D) Distribution of 5 immune-related lncRNAs-based risk score for survival status. (E) Kaplan–Meier analysis of survival probability of LUSC according to the median of risk score. lncRNAs = long noncoding RNAs, LUAD = lung adenocarcinoma, LUSC = lung squamous carcinoma.
Figure 2.
The association between 5 immune-related lncRNAs expression and OS determined with the GEPIA database. (A) ARHGEF26-AS1. (B) RBPMS-AS1. (C) AC004687.1. (D) TMPO-AS1. (E) LINC01137. lncRNAs = long noncoding RNAs, OS = overall survival.
3.2. The prognostic value of the risk score and clinical characteristics correlation
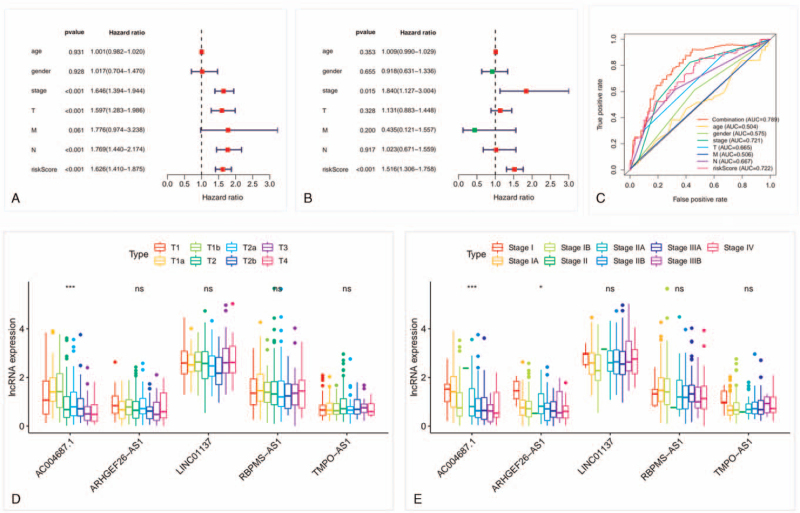
Univariate Cox regression analysis was conducted to value the prognostic performance of the clinical characteristics and risk score. The risk score, tumor size, N stage, and stage were identified as prognostic risk factors (Fig. 3A). Multivariate Cox regression analysis further validated the risk score as an independent prognostic factor (P < .001, Fig. 3B). The predicting performance of the 5-lncRNAs model was calculated by the area under the ROC curve (AUC). The ROC curve had a risk score AUC area of 0.722.
Figure 3.
Risk score as an independent prognostic factor and correlation between 5 lncRNAs and clinical factors. (A) Univariate Cox regression analysis of the prognostic value of risk score. (B) Multivariate Cox regression analysis of risk score prognostic value. (C) The ROC analysis of the performance value for LUAD prognostic prediction. (D) Five immune-related lncRNAs and tumor size. (E) Five immune-related lncRNAs and stage. ∗P < .1; ∗∗∗P < .001. lncRNAs = long noncoding RNAs, LUAD = lung adenocarcinoma, ns = no significance, ROC = receiver operating characteristic.
To further improve the prognostic value, the risk score and stage were combined to predict the prognosis, and the combination AUC was increased to 0.789 (Fig. 3C). These results indicated that the risk score and the combination had better prognostic value than other prognostic factors. Kruskal–Wallis test was conducted to explore the relationship between the 5-lncRNAs and clinical characteristics. As shown in Figure 3D and E, the higher AC004687.1 expression was obviously correlated with smaller tumor size and earlier stage (P < .001).
3.3. Low- and high-risk groups showed different immune statuses
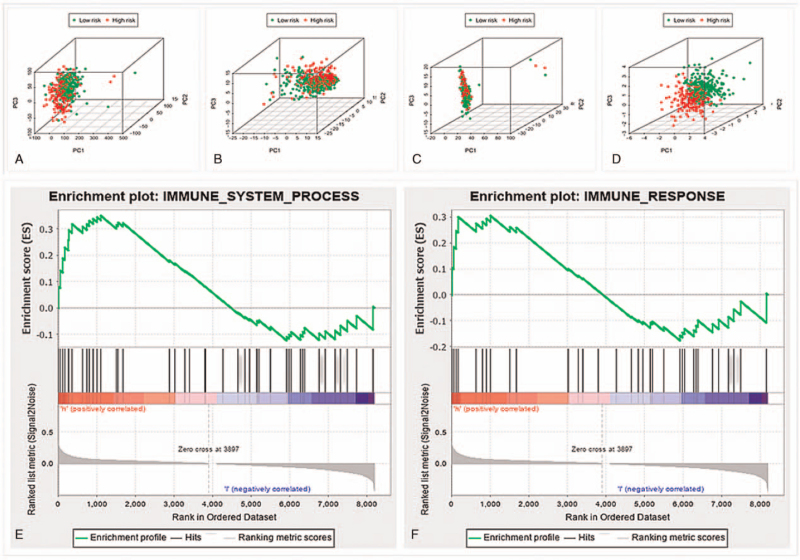
Principle component analysis was utilized to compare the difference of immune status between the low- and high-risk groups in different gene datasets (Fig. 4A–D). The results of all gene datasets indicated that the low- and high-risk groups were randomly located (Fig. 4A). Prognostic immune-related lncRNAs perfectly stratified the low- and high-risk groups (Fig. 4D). Furthermore, results of functional annotation performed by GSEA demonstrated that differentially expressed genes between the 2 groups were enriched in immune system process and immune response pathways (Fig. 4E and F). To sum up, the low- and high-risk groups showed totally different immune statuses.
Figure 4.
The difference of immune status between low-and high-risk groups. Principal Component Analysis of all gene (A), immune gene (B), all immune lncRNAs (C), and risk immune lncRNAs (D) data set. The high-risk group showed significant enrichment of immune phenotype in immune system process (E) and immune response pathways (F). lncRNAs = long noncoding RNAs.
3.4. Silico functional analysis
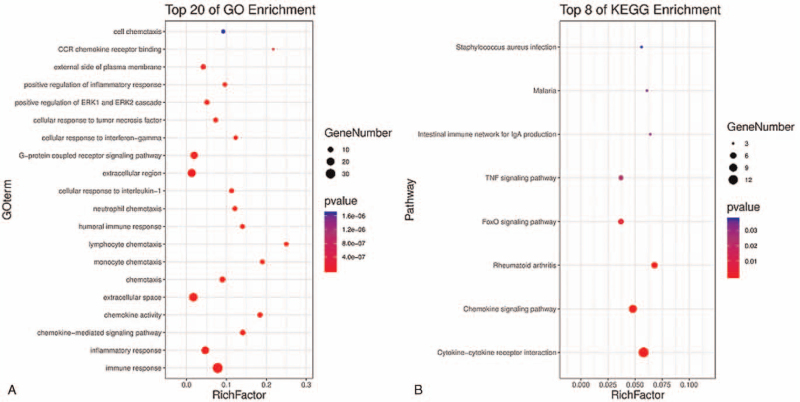
In order to explore functional roles behind the 5-lncRNAs in LUAD biology, we performed in silico analysis for lncRNA function through functional enrichment analysis. The 5-lncRNAs and mRNAs co-expression pairs were obtained by calculating the Pearson correlation coefficient. We screened out 749 PCGs significantly correlated with at least one of lncRNAs (Pearson correlation coefficient > 0.5 and P < .01). GO and KEGG functional enrichment analysis results showed that the 749 PCGs assembled most obviously in 20 GO functional categories (Fig. 5A) and 8 KEGG pathways (P < .05 after Benjamini adjustment) (Fig. 5B). GO enrichment analysis revealed that the PCGs were involved in immune-related processes and pathways such as immune response, inflammatory response, chemokine-mediated signaling pathway evidently (Fig. 5A). The KEGG result suggested that the PCGs were significantly related to cytokine-cytokine receptor interaction, chemokine, FoxO and TNF signaling pathway (Fig. 5B). These results showed that the variation in the 5-lncRNAs expression might influence the immune-related LUAD functional processes and pathways.
Figure 5.
GO and KEGG pathway functional enrichment analysis. (A) GO enrichment results. (B) KEGG pathway enrichment results.[48] GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
4. Discussion
In the past few years, although the appearance of numerous new therapies for NSCLC, the advanced NSCLC patients prognosis still remained poor. The prognostic models of NSCLC development have been a research hotspot because the prognosis of heterogenous NSCLC were varied. LUAD is prone to metastases widely at an early stage and poor prognosis, with the average 5-year survival rate less than 15%.[19] Thus, an investigation into the prognostic models construction of LUAD remains a hot spot to develop novel effective LUAD prognosis prediction. As for lncRNA, recent researches validated its significant position in the progression of NSCLC through different molecular pathways.[20,21] For example, the lncRNA Gm15290 directly interacts with the tumor suppressor miR-615-5p to accelerate NSCLC cell proliferation and invasion[22] and lncRNA AFAP1-AS1 was confirmed to be critical in the progression of NSCLC by epigenetically repressing p21.[23] However, only a single lncRNA was evaluated in previous studies, and we believe that multiple lncRNAs are required for prognosis prediction. Base of the background mentioned above, the study built an immune-related lncRNAs prognostic model for LUAD and screened out 5 immune-related lncRNAs significantly associated with LUAD prognosis. Among the 5 lncRNAs, lncRNA AC004687.1 was firstly reported as an immune-related prognostic lncRNA with significant correlation with stage and tumor size. Combining the previous studied,[24,25] plasma or serum AC004687.1 detection may have the potential to diagnose LUAD at early stage in the future. In addition, the model was proved to be accurate and promising for predicting prognosis, which could assist to make treatment decision in clinical practice and promote new thought of potential prognostic biomarkers development for LUAD patients.
Other scientific studies have undertaken similar investigations to build prognostic models for NSCLC. Compared to other related research, the study firstly proved that the 5-lncRNAs signature as an independent prognostic factor with the AUC of 0.722. In addition, the combination of stage and risk score enhanced the prediction value with the AUC of 0.789. Miao et al[26] reported a lncRNA prognostic model related to tumor immune status regulation in elderly NSCLC patients, the model was similar to ours, the AUC of their model was lower than our immune-related lncRNAs prognostic model (0.669 vs 0.722). Furthermore, compared with the models confined to advanced NSCLC patients or the limited clinical trials data,[27–30] our model was more applicable to a broad crowd. Since the gene detection grow more and more popular, it is more convenient to get the gene detection results without doing the follow-up and imaging examination, which makes our prediction model more available for clinical use. Online lung cancer database is comprehensive, extensively used, specialized and has strong expandability. For example, the support vector machine (SVM) is a traditional machine learning model used to predict the prognosis of cancer and Bayesian network (BN) models could reason under indeterminacy and manage missing data better. Jochems et al[31] validated a survival prediction model for NSCLC patients using distributed learning by BN structure, with many variables included in the BN model, but compared with our model, the AUC of our immune-related lncRNAs prognostic model was higher (0.722 vs 0.67). According to the result reported by Jayasurya et al,[32] the AUC of SVM model and BN model were 0.69 and 0.70, respectively in the Toronto set, which were lower than our immune-related lncRNAs prognostic model. Besides, the BN and SVM models require the integrity of the input variables to increase the accuracy of prediction, which is difficult to achieve in clinical practice. To our knowledge, we are the first to value the immune-related lncRNAs prognostic model of LUAD at any tumor, lymph node, and metastasis stage, ages and regardless of the treatment history with high prediction value. The model is close to clinical practice and suitable to be translated to clinical practice in the near future.
In the last few years, increasing cognitions on the relationship between the immune system and tumor cells has improved the growth of immunotherapy, which was targeted to fight cancer by promoting the own immune response of patients. As for localised NSCLC (stages I–III), adding immunotherapy (excluding checkpoint inhibitors) to conventional curative surgery or radiotherapy showed no obvious survival benefit.[33] The anti-PD-1 or anti-PD-L1 showed superiority over docetaxel in the treatment of pretreated NSCLC patients, besides, anti-PD-1 emerged more benefits than anti-PD-L1.[34] Going further, a meta-analysis gave a moderate evidence that the addition of immune checkpoint inhibitors to chemotherapy may improve both OS compared with chemotherapy alone.[35] Immunotherapies based on dendritic cells are new choices for cancer treatment, and further research need to prove the potential of checkpoint PD-1 inhibitor and dendritic cell therapy combination in NSCLC treatment.[36] Immune-related signaling pathways play significant roles in several physiologically and pathologically processes,[37–39] and affect the therapeutic response of malignant tumors.[40,41] Especially, several studies have revealed the functional roles of immune-related lncRNAs in human cancers.[42,43] LncRNAs were authenticated to take crucial role in the immune response regulation of tumorigenesis, the PD-1/PD-L1 pathway was also included.[44] Moreover, immune- and tumor microenvironment related genes have already emerged recently, which could act as tumor markers for diagnosis or immunotherapy targets.[45] On account of some specific immunological characteristics, immune-related lncRNA contributes to distinguish cancer subtypes. As reported by Zhang et al,[46] immune-related lncRNA played possible role in evaluating the response to immune checkpoint blockade immunotherapy. In lung cancers, Li et al[47] first reported the lncRNAs that can regulate immune-related pathways in LUAD or LUSC. In the study, we divided LUAD patients based on the expression of these lncRNAs and discovered that the immune statuses of the 2 groups were different, which suggests that immune status may influence the prognosis of NSCLC. LncRNA–PCGs co-expression analysis was conducted to evaluate the function of related lncRNAs. Through the enrichment analysis, we found that the immune-related lncRNAs were involved in immune response, inflammatory response, chemokine-mediated signaling pathway, FoxO, and TNF signaling pathways. The identified 5 prognostic lncRNAs may regulate immune function either directly or indirectly. Therefore, the 5 immune-related lncRNAs might be potential therapeutic targets. In summary, lncRNAs may contribute to LUAD tumorigenesis by interacting with biological pathways related to the immune response and show the great value for deeper explore the potential molecular mechanism.
There are some limitations in our study. The performance of prognostic model can be affected by the different datasets and variables definition. No dataset include adequate number of lncRNA data to check our results. Not only that, there is a lack of strict experimental demonstration in the study. In the near future, in order to build a more stable and reliable prognostic model for clinical application, we wish to collect more follow-up data, explore the mechanism at the molecular level, consider more factors that may affect prognosis, such as clinic pathological characteristics, miRNAs, and mRNAs.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
YJH and CJH performed data analysis work. YJH wrote the manuscript and designed the study. All authors read and approved the final manuscript.
Methodology: Yajie Huang.
Software: Changjie Huang.
Writing – original draft: Yajie Huang.
Writing – review & editing: Yajie Huang.
Footnotes
Abbreviations: BN = Bayesian network, coef = regression coefficient, CI = confidence interval, GEPIA = Gene Expression Profiling Interactive Analysis, GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes, LncRNAs = long noncoding RNAs, LUAD = lung adenocarcinoma, LUSC = lung squamous carcinoma, NSCLC = nonsmall cell lung cancer, OS = overall survival, PCG = protein-coding gene, ROC = receiver operating characteristic, SVM = support vector machine, TCGA = the cancer genome atlas.
How to cite this article: Huang Yj, Huang Cj. Construction of a 5 immune-related lncRNA-based prognostic model of NSCLC via bioinformatics. Medicine. 2021;100:37(e27222).
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analysed during the current study are publicly available on the cancer genome atlas and MSigDB web resources.
References
- [1].Siegel R, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:07–30. [DOI] [PubMed] [Google Scholar]
- [2].Garrido F, Algarra I. MHC antigens and tumor escape from immune surveillance. Adv Cancer Res 2001;83:117–58. [DOI] [PubMed] [Google Scholar]
- [3].Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology 2007;121:01–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].De Rosa M, Pace U, Rega D. Genetics, diagnosis and management of colorectal cancer (review). Oncol Rep 2015;34:1087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014;65:1140–51. [DOI] [PubMed] [Google Scholar]
- [6].He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother 2017;95:331–8. [DOI] [PubMed] [Google Scholar]
- [7].Gupta RA, Shah N, Wang KC. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gomez JA, Wapinsk OL, Yang YW. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 2013;152:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jiang M, Zhang S, Yang Z, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 2018;173:906–19. [DOI] [PubMed] [Google Scholar]
- [11].Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol 2014;35:408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peng F, Wang R, Zhang Y, et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer 2017;16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhou M, Zhang Z, Zhao H, Bao S, Sun J. A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer 2018;18:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kanehisa M. Goto S: KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res 2019;47:D590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin JJ, Cardarella S, Lydon CA, et al. Five-year survival in EGFR-mutant metastatic lung adenocarcinoma treated with EGFR-TKIs. J Thorac Oncol 2016;11:556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kong X, Zhao Y, Li X, Tao Z, Hou M, Ma H. Overexpression of HIF-2α-dependent NEAT1 promotes the progression of non-small cell lung cancer through miR-101-3p/SOX9/Wnt/β-catenin signal pathway. Cell Physiol Biochem 2019;52:368–438. [DOI] [PubMed] [Google Scholar]
- [21].Lu Z, Li Y, Che##Y, et al. The TGFβ-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett 2018;432:156–68. [DOI] [PubMed] [Google Scholar]
- [22].Dong Y, Huo X, Sun R, Liu Z, Huang M, Yang S. LncRNA Gm15290 promotes cell proliferation and invasion in non-small cell lung cancer through directly interacting with and suppressing the tumor suppressor miR-615-5p. Biosci Rep 2017;38:BSR20181150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer 2018;17:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol 2015;8:12936–42. [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang D, Liu X, Wei B, Qiao G, Jiang T, Chen Z. Plasma lncRNA GAS8-AS1 as a potential biomarker of papillary thyroid carcinoma in Chinese patients. Int J Endocrinol 2017;2017:2645904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Miao R, Ge C, Zhang X, et al. Combined eight-long noncoding RNA signature: a new risk score predicting prognosis in elderly non-small cell lung cancer patients. Aging (Albany NY) 2019;11:467–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mo H, Hao X, Liu Y, et al. A prognostic model for platinum-doublet as second-line chemotherapy in advanced non-small-cell lung cancer patients. Cancer Med 2016;5:1116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hoang T, Dahlberg SE, Sandler AB, et al. Prognostic models to predict survival in non-small-cell lung cancer patients treated with first-line paclitaxel and carboplatin with or without bevacizumab. J Thorac Oncol 2012;7:1361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Motzer RJ, Bacik J, Murphy BA, Russo P, Mazumdar M. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 2002;20:289–96. [DOI] [PubMed] [Google Scholar]
- [30].Voss MH, Reising A, Cheng YZ, et al. Genomically annotated risk model for advanced renal-cell carcinoma: a retrospective cohort study. Lancet Oncol 2018;19:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jochems A, Deist TM, El Naqa I, et al. Developing and validating a survival prediction model for NSCLC patients through distributed learning across 3 countries. Int J Radiat Oncol Biol Phys 2017;99:344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jayasurya K, Fung G, Yu S, et al. Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys 2010;37:1401–7. [DOI] [PubMed] [Google Scholar]
- [33].Zhu J, Li R, Tiselius E, et al. Immunotherapy (excluding checkpoint inhibitors) for stage I to III non-small cell lung cancer treated with surgery or radiotherapy with curative intent. Cochrane Database Syst Rev 2017;12:CD011300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tartarone A, Roviello G, Lerose R, Roudi R, Aieta M, Zoppoli P. Anti-PD-1 versus anti-PD-L1 therapy in patients with pretreated advanced non-small-cell lung cancer: a meta-analysis. Future Oncol 2019;15:2423–33. [DOI] [PubMed] [Google Scholar]
- [35].Petrelli F, Ferrara R, Signorelli D, et al. Immune checkpoint inhibitors and chemotherapy in first-line NSCLC: a meta-analysis. Immunotherapy 2021;13:621–31. [DOI] [PubMed] [Google Scholar]
- [36].Mohsenzadegan M, Peng RW, Roudi R. Dendritic cell/cytokine-induced killer cell-based immunotherapy in lung cancer: what we know and future landscape. J Cell Physiol 2020;235:74–86. [DOI] [PubMed] [Google Scholar]
- [37].Gonzalo S, Coll-Bonfill N. Genomic instability and innate immune responses to self-DNA in progeria. GeroScience 2019;41:255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Walker EM, Slisarenko N, Gerrets GL, et al. Inflammaging phenotype in rhesus macaques is associated with a decline in epithelial barrier-protective functions and increased pro-inflammatory function in CD161-expressing cells. GeroScience 2019;41:739–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kavanagh K, Hsu FC, Davis AT, Kritchevsky SB, Rejeski WJ, Kim S. Biomarkers of leaky gut are related to inflammation and reduced physical function in older adults with cardiometabolic disease and mobility limitations. GeroScience 2019;41:923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Seebacher NA, Stacy AE, Porter GM, Merlot AM. Clinical development of targeted and immune based anti-cancer therapies. J Exp Clin Cancer Res 2019;38:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kruger S, Ilmer M, Kobold S, et al. Advances in cancer immunotherapy 2019 – latest trends. J Exp Clin Cancer Res 2019;38:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hu Q, Ye Y, Chan LC, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 2019;20:835–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lin H, Jiang M, Liu L, et al. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol 2019;20:812–23. [DOI] [PubMed] [Google Scholar]
- [44].Ding L, Lu S, Li Y. Regulation of PD-1/PD-L1 pathway in cancer by noncoding RNAs. Pathol Oncol Res 2020;26:651–63. [DOI] [PubMed] [Google Scholar]
- [45].Fathi Z, Syn NL, Zhou JG, Roudi R. Molecular epidemiology of lung cancer in Iran: implications for drug development and cancer prevention. J Hum Genet 2018;63:783–94. [DOI] [PubMed] [Google Scholar]
- [46].Zhang Y, Zhang L, Xu Y, Wu X, Zhou Y, Mo J. Immune-related long noncoding RNA signature for predicting survival and immune checkpoint blockade in hepatocellular carcinoma. J Cell Physiol 2020;235:9304–16. [DOI] [PubMed] [Google Scholar]
- [47].Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun 2020;11:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2010;38:D355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]