LETTER
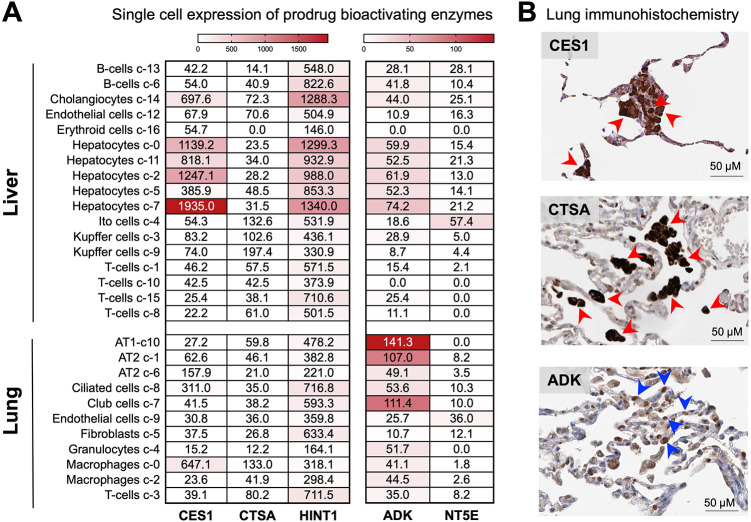
A recently published article by Li et al. titled, “Key Metabolic Enzymes in Remdesivir Activation in Lung Cells,” validates the canonical McGuigan enzymes (CES1, CTSA, and HINT1) involved in remdesivir (RDV)’s bioactivation using bioinformatic and biochemical approaches (1). Yet in what appears to be a dismissal of the narrative portrayed by their own data, the authors’ discussion does not account for cell type-specific differences in the lung and tissue-wide levels. Despite the omission in their discussion, the authors’ data clearly support that key McGuigan-bioactivating enzymes are expressed at exceptionally high levels in the liver (Li et al., in their Fig. 5B) (1). Nevertheless, we commend the authors for supporting the assertion we made over a year ago (2) on the uniquely high liver expression of CES1/CTSA/HINT1 as a culprit behind the dose-limiting hepatotoxicity of remdesivir (RDV) observed in healthy volunteers (3, 4). Paradoxically, while the authors use single-cell RNA sequencing (scRNAseq) data to evaluate the expression of McGuigan prodrug-activating enzymes in the lung, their discussion of these data ignores the cell type-specific nuance apparent in their figures and instead views their lung as a single homogenous entity. The authors’ oversight of these cell type-specific differences prompts the inaccurate conclusion that CES1, CTSA, and HINT1 are “highly expressed in the human lung” and that such enzymes are “highly expressed in cell types that are involved in SARS-CoV-2 infection and replication.” It is well-established that type II alveolar (AT2) cells are the primary site of SARS-CoV-2 infection (5, 6). Close inspection of the same scRNAseq data from the Human Protein Atlas (HPA) shows that the high expression of CES1/CTSA/HINT1 to which the authors refer is largely restricted to alveolar macrophages (Fig. 1A), which are not the main cell types involved in “SARS-CoV-2 infection and replication”; these scRNAseq data are further corroborated by immunohistochemistry staining of primary lung tissue, which shows intense staining for the relevant enzymes in macrophages but not AT2 cells (Fig. 1B, red arrows). According to HPA data, the modest expression of McGuigan prodrug enzymes in AT2 cells is vastly overshadowed by that measured in hepatocytes (Fig. 1). Viewed in the context of RDV’s low 50% cytotoxic concentration (CC50) value in primary human hepatocytes (7) and the inability to up-dose RDV to achieve more obvious reductions in SARS-CoV-2 viral titers in the clinic due to liver-related dose-limiting toxicities become self-evident (3, 8). The authors’ oversight of the AT2 versus macrophage difference and omission of the tissue-wide relevance of their findings in their discussion is thus misleading and offers an incomplete portrayal of the broader in vivo significance of their data.
FIG 1.
Single-cell RNA sequencing and immunohistological data unambiguously show high expression of remdesivir-bioactivating enzymes in the liver. (A) Heatmap of HPA scRNAseq expression data for RDV (CES1/CTSA/HINT1)- and GS-441524 (ADK)-bioactivating enzymes in the lung placed in context of the liver. Individual values per cell type are expressed in protein-coding transcripts per million. (B) Immunohistochemistry staining of normal primary human lung tissue obtained from HPA. High lung expression of CES1 and CTSA is unique to alveolar macrophages (red arrows) rather than AT2 cells. In contrast, ADK expression is largely present in AT2 cells (blue arrows). Note: HINT1 expression data are not shown due to lack of staining in primary lung tissue. Antibodies include CES1, HPAO12023; CTSA, CAB024930; and ADK, HPA038409. Patient IDs include CES1, 1678; CTSA, 2268; and ADK, 2268.
Footnotes
For the author reply, see https://doi.org/10.1128/AAC.01394-21.
REFERENCES
- 1.Li R, Liclican A, Xu Y, Pitts J, Niu C, Zhang J, Kim C, Zhao X, Soohoo D, Babusis D, Yue Q, Ma B, Murray BP, Subramanian R, Xie X, Zou J, Bilello JP, Li L, Schultz BE, Sakowicz R, Smith BJ, Shi P-Y, Murakami E, Feng JY. 2021. Key metabolic enzymes involved in remdesivir activation in human lung cells. Antimicrob Agents Chemother 65:e00602-21. 10.1128/aac.00602-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan VC, Muller FL. 2020. Advantages of the parent nucleoside GS-441524 over remdesivir for Covid-19 treatment. ACS Med Chem Lett 11:1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humeniuk R, Mathias A, Cao H, Osinusi A, Shen G, Chng E, Ling J, Vu A, German P. 2020. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin Transl Sci 13:896–906. 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan VC, Muller FL. 2021. Remdesivir for COVID-19: why not dose higher? Antimicrob Agents Chemother 65:e2713-20. 10.1128/AAC.02713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer I-M, Padera RF, Solomon IH, Kanjilal S, Hammer MM, Hornick JL, Sholl LM. 2020. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol 33:2104–2114. 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speranza E, Williamson BN, Feldmann F, Sturdevant GL, Pérez-Pérez L, Meade-White K, Smith BJ, Lovaglio J, Martens C, Munster VJ, Okumura A, Shaia C, Feldmann H, Best SM, De Wit E. 2021. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med 13:eabe8146. 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Barauskas O, Kim C, Babusis D, Murakami E, Kornyeyev D, Lee G, Stepan G, Perron M, Bannister R, Schultz BE, Sakowicz R, Porter D, Cihlar T, Feng JY. 2020. Off-target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrob Agents Chemother 65:e02237-20. 10.1128/AAC.02237-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. 2020. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395:1569–1578. 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]