ABSTRACT
In vitro MICs and in vivo pharmacodynamics of ceftazidime and cefepime human-simulated regimens (HSR) against modified carbapenem inactivation method (mCIM)-positive Pseudomonas aeruginosa isolates harboring different OXA-10-like subtypes were described. The murine thigh model assessed ceftazidime (2 g every 8 h [q8h] HSR) and cefepime (2 g and 1 g q8h HSR). Phenotypes were similar despite possessing OXA-10-like subtypes with differing spectra. Ceftazidime produced ≥1-log10 killing in all isolates. Cefepime activity was dose dependent and MIC driven. This approach may be useful in assessing the implications of β-lactamase variants.
KEYWORDS: Pseudomonas aeruginosa, carbapenem resistant, cefepime, ceftazidime, in vivo, pharmacodynamics, pharmacokinetics
TEXT
Pseudomonas aeruginosa is notorious for having numerous mechanisms that contribute to phenotypic resistance to first-line antimicrobial agents (1). For example, class D oxacillinases, which can have a variety of amino acid substitutions and produce a range of different hydrolytic spectra that result in various phenotypes (2). Of the OXA-10-like enzymes, some are considered narrow-spectrum β-lactamases (i.e., penicillinases), while others have extended-spectrum activity (i.e., can hydrolyze third- and fourth-generation cephalosporins) (3). For example, OXA-35 typically produces a ceftazidime-susceptible, cefepime-resistant phenotype due to its greater hydrolytic activity against cefepime, while OXA-10 is considered narrow spectrum because of its inability to hydrolyze any extended-spectrum cephalosporins (3, 4). This raises the question of how the various amino acid changes in OXA-10-like variants (i.e., genotypes) translate into phenotypic differences and, most importantly, differing clinical efficacy in vivo.
The availability of specific genotypic data for resistance genes in P. aeruginosa isolates from clinical specimens can provide clinicians with more information than just an antibiogram to consider when selecting antibiotic therapy. However, data that correlate the genotype, the phenotype, and the clinical efficacy are needed. Here, we describe the in vivo activity of ceftazidime and cefepime, administered at doses that produce exposures mimicking human exposures, to assess the potential efficacy and clinical implications of carbapenem-resistant P. aeruginosa isolates harboring OXA-10-like enzymes.
Isolates of P. aeruginosa were collected during the Enhancing Rational Antimicrobials for Carbapenem-resistant P. aeruginosa (ERACE-PA) Global Study, which includes 17 health care centers worldwide (12 international and five in the United States). The three P. aeruginosa isolates assessed in this study were evaluated using broth microdilution per CLSI standards to determine the in vitro activity of ceftazidime, cefepime, ceftazidime-avibactam, ceftolozane-tazobactam, imipenem, and meropenem. Testing was performed in triplicate, and results were interpreted using both CLSI and EUCAST breakpoints (5, 6). Isolates were screened using the modified carbapenem inactivation method (mCIM) and whole-genome sequencing to identify OXA-10-like genotypes (Table 1) (5). Genomic DNA was prepared with the DNeasy blood and tissue kit (Qiagen, USA) from pure cultures grown overnight in Trypticase soy broth (Hardy Diagnostics, USA). DNA was quantitated with the NanoPhotometer system (Implen, Germany). Libraries were prepared with the Nextera XT DNA library kit (Illumina, USA) and were sequenced on the MiSeq platform using reagent kit v3 chemistry (Illumina, USA). Paired-end reads were trimmed and de novo assembled with the CLC Genomics Workbench 21.0.3 tools (Qiagen Bioinformatics, Denmark) according to the manufacturer’s instructions. Acquired resistance genes were identified with the CLC Microbial Genomics Module 21.0 “Find Resistance” tool using the ResFinder database (version 2021-04-06) (Qiagen Bioinformatics) (7). To confirm β-lactamase gene subtypes, the sequences were compared using BLASTn to those published in GenBank nucleotide database and in the β-Lactamase Database (BLDB) (8).
TABLE 1.
Phenotypic and genotypic profiles of three clinical carbapenem-resistant P. aeruginosa isolates harboring OXA-10-like β-lactamases, evaluated in vivo
| Organism | Genotype | MLSTh | MIC (mg/liter) |
|||||
|---|---|---|---|---|---|---|---|---|
| Ceftazidime | Cefepime | Ceftolozane-tazobactam | Ceftazidime-avibactam | Imipenem | Meropenem | |||
| PSA INT-7-65 | blaOXA-56,ablaOXA-50b (OXA-904 by BLAST), blaOXA-392c, blaPAO (blaPDC-117) | 260 | 4d | 32f | 4 | 4 | 16 | 32 |
| PSA INT-9-5 | blaOXA-35,ablaOXA-488,bblaPAO (blaPDC-35) | 235 | 4d | 32f | 2 | 2 | 32 | >64 |
| PSA INT-9-11 | blaOXA-10,ablaOXA-488,bblaPAO (blaPDC-35) | 235 | 8e | 64g | 8 | 4 | 16 | 64 |
blaOXA-10-like family. For PSA INT-7-65, blaOXA-56 could not be distinguished from blaOXA-101.
blaOXA-50-like family. For PSA INT-7-65, blaOXA-50 could not be distinguished from blaOXA-396 or blaOXA-494.
blaOXA-1-like family.
Ceftazidime 2 g q8h 2-h infusion HSR, 100% fT > MIC (9).
Ceftazidime 2 g q8h 2-h infusion HSR, 87% fT > MIC (9).
Cefepime 2 g q8h 2-h infusion HSR, 38% fT > MIC; cefepime 1 g q8h 2-h infusion HSR, 8% fT > MIC (10).
Cefepime 2 g q8h 2-h infusion HSR, 8% fT > MIC; cefepime 1 g q8h 2-h infusion HSR, 0% fT > MIC (10).
MLST, multilocus sequence type.
All animal studies were approved by the local Institutional Animal Care and Use Committee (IACUC) and conducted per the standards of the National Research Council of the National Academy of Sciences. Specific-pathogen-free, female, CD-1 mice (20 to 22 g) were acquired from Charles River Laboratories (Raleigh, NC). Animals were housed and pretreated with cyclophosphamide and uranyl nitrate as previously described (9, 10). Mice were inoculated as previously described, and antibiotic dosing began 2 h postinoculation and continued for 24 h (9, 10). Each treatment group contained three mice allocated to 0-h control (baseline bacterial burden), 24-h control (received normal saline), ceftazidime human-simulated regimen (HSR), cefepime 1 g HSR, or cefepime 2 g HSR. The HSRs of ceftazidime 2 g intravenously (i.v.) every 8 h (9) and cefepime 2 g i.v. every 8 h (10), both given as 2-h infusions, were administered as previously described. Cefepime 1 g i.v. every 8 h was simulated by halving the doses of the cefepime 2 g regimen. Thighs were aseptically harvested at 24 h and serially diluted to enumerate CFU/thigh. Bacterial enumeration was described as the mean plus or minus standard deviation log10 CFU/thigh. Changes in log10 CFU/thigh were calculated using the mean result of each group at 24 h minus the mean result at 0 h result. Achievement of a 24-h decrease of ≥1-log10 CFU/thigh was utilized as a benchmark associated with clinical success and was compared to the cumulative percentage of 24 h of treatment that the unbound drug concentration exceeded the MIC (fT > MIC) exposures for each dosing regimen (9–11).
Modal MICs and genotypes for the test isolates are presented in Table 1. All three isolates were susceptible to ceftazidime but resistant to cefepime, meropenem, and imipenem per CLSI and EUCAST interpretations. All three isolates tested positive by mCIM for carbapenemase activity.
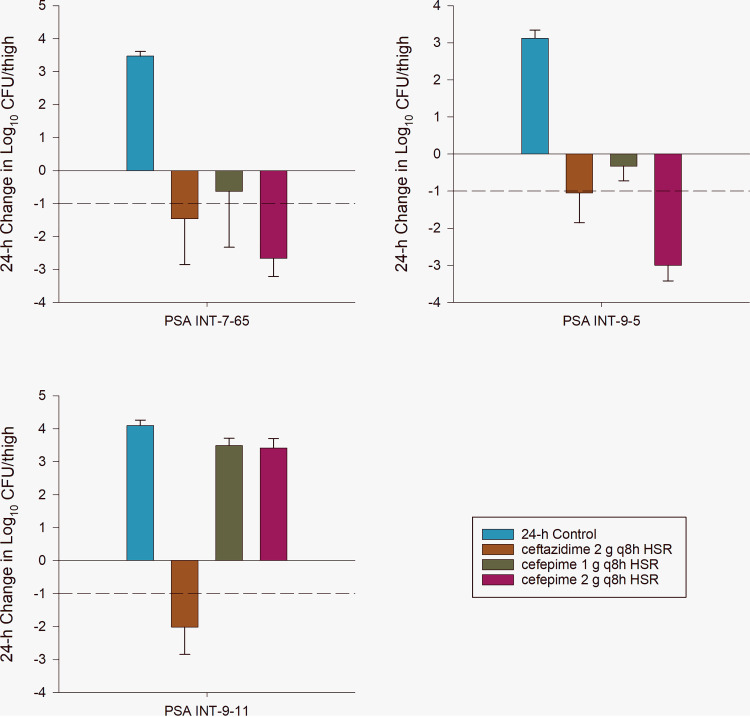
The average baseline bacterial burden at 0 h was 5.43 ± 0.24 log10 CFU/thigh. Untreated controls had a 24-h mean increase in bacterial burden in log10 CFU/thigh of 3.56 ± 0.45. The mean changes in log10 CFU/thigh for each isolate and dosing regimen are presented in Fig. 1. Ceftazidime produced significant bacterial killing of all isolates, which reached a ≥1-log10 reduction in bacterial burden. The cefepime 2 g i.v. every 8 h regimen produced significant killing in two of the three isolates, achieving a ≥1-log10 bacterial reduction in CFU/thigh. Conversely, the cefepime 1 g regimen failed to kill any of the isolates, indicating a cefepime dose-response relationship.
FIG 1.
In vivo change in log10 bacterial burden from 0 h in animals treated with sham control, ceftazidime 2 g every 8 h (q8h) by 2-h infusion human-simulated regimen (HSR), cefepime 1 g q8h by 2-h infusion HSR, or cefepime 2 g q8h by 2-h infusion HSR in the murine thigh infection model.
In vivo, the ability of ceftazidime and cefepime HSRs to kill the three OXA-10-like containing bacterial isolates was consistent with the pharmacodynamic exposures and MICs of the two drugs. The dose-response relationships noted for the two cefepime dosing regimens are consistent with the elevated cefepime MICs observed, as the 1-g regimen (8% fT > MIC32 mg/liter) failed to meet the pharmacodynamic target for bacterial kill for cephalosporin agents (i.e., 50 to 70% fT > MIC32 mg/liter) while the 2 g every 8 h regimen as a 2-h infusion achieved ∼40% fT > MIC32 mg/liter. It should be noted that when evaluating fT > MIC against a doubling dilution MIC scale, the actual MIC of the antibiotic may fall between dilution steps. In this case, the MICs fell between 16 mg/liter and 32 mg/liter, which may translate to bacterial killing, as the fT > MIC will be somewhere between 38 and 59% with the cefepime 2 g dose (8 to 38% fT > MIC for cefepime 1 g HSR) (10, 12). This was not the case for the isolate for which the cefepime MIC was 64 mg/liter, as exhibited by in vivo bacterial growth with the cefepime 2 g HSR in PSA INT-9-11, where the fT > MIC fell to ∼10%. However, the ceftazidime regimen reached 100% fT > MIC4 mg/liter and 87% fT > MIC8 mg/liter where bactericidal activity was expected. Thus, the in vivo findings demonstrate that the exposure-response relationship was unaltered by OXA-10-like enzymes and the other mechanisms expressed by each isolate.
Our data indicate that categorization of OXA β-lactamases as narrow spectrum (i.e., OXA-10 and OXA-56) versus extended spectrum (i.e., OXA-35) may obscure similarities within the OXA-10-like subclass (3). Indeed, despite subtle differences in the reported hydrolytic spectrum of the identified OXA-10-like subtypes determined by whole-genome sequencing, the isolates produced similar in vitro and in vivo response for the three clinical P. aeruginosa isolates. Similarly to what Aubert and colleagues noted during their study of OXA-35-containing P. aeruginosa, our isolates had lower ceftazidime MICs than cefepime MICs (4). However, a cefepime-nonsusceptible phenotype is not consistent with the “narrow spectrum” categorization previously given to OXA-10 and OXA-56, which are highly related enzymes (3). Indeed, there is between 92.6 and 99.8% amino acid conservation within the subclass, with multiple subtypes categorized as being “extended spectrum,” defined as the ability to hydrolyze at least one third- or fourth-generation cephalosporin (3). The phenotypic profiles in the present study show overall similarity despite three different subclasses being identified. Importantly, all three isolates were also carbapenem resistant and mCIM positive. Antunes and colleagues previously described increases in carbapenem MICs that were outside the susceptible range when oxacillinase genes that were not traditionally considered carbapenemases were placed into a wild-type Acinetobacter strain, in contrast to no increase in MICs when the gene was inserted into a wild-type Escherichia coli strain (13). These findings suggest that the background resistance mechanisms, including efflux and/or porin alterations, contribute to the carbapenem-resistant phenotype. This is also notable because all three isolates tested positive by mCIM for carbapenemase activity; however, microbiologic activity was still noted for antipseudomonal cephalosporins in vivo.
In conclusion, in vivo bactericidal activity was noted for ceftazidime HSRs that is consistent with the MICs of three clinical carbapenem-resistant P. aeruginosa isolates harboring OXA-10-like enzymes. Responses to two cefepime dosing regimens were similarly reflective of the MICs of the isolates, suggesting that the elevated in vitro cefepime MICs had in vivo consequence. These data are hypothesis generating, and a similar in vivo approach may be useful in future scenarios to evaluate the translational implications of β-lactamase variants in their responses to clinically relevant antibiotic exposures.
ACKNOWLEDGMENTS
We acknowledge all members of the ERACE-PA Global Study Group: Elif Aktas, Wadha Alfouzan, Carey-Ann Burnham, Rafael Canton, Yehuda Carmeli, Marco Falcone, Lori Bourassa, Carlos Kiffer, Anna Marchese, Octavio Martinez, Michael Satlin, Harald Seifert, Abrar K. Thabit, Maria Virginia Villegas, Julia Wille, Thais Teles Freitas Rezende, Zuhal Cekin, Gulsah Malkocoglu, Desirèe Gijón, Layla Abdullah Tarakmeh, Dalya M. Attallah, Giusy Tiseo, Alessandro Leonildi, Cesira Giordano, Simona Barnini, Francesco Menichetti, Vincenzo Di Pilato, Giulia Codda, Antonio Vena, Daniele Roberto Giacobbe, Lars Westblade, Armando Cardona, Lauren Curtis, Ferric Fang, Gina Thomson, and Ken Thomson. We thank the staff from the Center for Anti-Infective Research and Development for their assistance in the conduct of this study.
This study was internally funded by the Center for Anti-Infective Research and Development.
We have no conflicts of interest relative to this work.
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans BA, Amyes SG. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon EJ, Jeong SH. 2021. Class D β-lactamases. J Antimicrob Chemother 76:836–864. doi: 10.1093/jac/dkaa513. [DOI] [PubMed] [Google Scholar]
- 4.Aubert D, Poirel L, Ali AB, Goldstein FW, Nordmann P. 2001. OXA-35 is an OXA-10-related beta-lactamase from Pseudomonas aeruginosa. J Antimicrob Chemother 48:717–721. doi: 10.1093/jac/48.5.717. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed.CLSI document M100.CLSI, Wayne, PA. [Google Scholar]
- 6.The European Committee on Antimicrobial Susceptibility Testing. 2021. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. http://www.eucast.org.
- 7.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)—structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacVane SH, Crandon JL, Nichols WW, Nicolau DP. 2014. In vivo efficacy of humanized exposures of ceftazidime-avibactam in comparison with ceftazidime against contemporary Enterobacteriaceae isolates. Antimicrob Agents Chemother 58:6913–6919. doi: 10.1128/AAC.03267-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdelraouf K, Almarzoky Abuhussain S, Nicolau DP. 2020. In vivo pharmacodynamics of new-generation β-lactamase inhibitor taniborbactam (formerly VNRX-5133) in combination with cefepime against serine-β-lactamase-producing Gram-negative bacteria. J Antimicrob Chemother 75:3601–3610. doi: 10.1093/jac/dkaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulitta JB, Hope WW, Eakin AE, Guina T, Tam VH, Louie A, Drusano GL, Hoover JL. 2019. Generating robust and informative nonclinical in vitro and in vivo bacterial infection model efficacy data to support translation to humans. Antimicrob Agents Chemother 63:e02307-18. doi: 10.1128/AAC.02307-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong CT, Tessier PR, Li C, Nightingale CH, Nicolau DP. 2007. Comparative in vivo efficacy of meropenem, imipenem, and cefepime against Pseudomonas aeruginosa expressing MexA-MexB-OprM efflux pumps. Diagn Microbiol Infect Dis 57:153–161. doi: 10.1016/j.diagmicrobio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]