ABSTRACT
Outer membrane vesicles (OMVs) act as carriers of bacterial products such as plasmids and resistance determinants, including metallo-β-lactamases. The lipidated, membrane-anchored metallo-β-lactamase NDM-1 can be detected in Gram-negative OMVs. The soluble domain of NDM-1 also forms electrostatic interactions with the membrane. Here, we show that these interactions promote its packaging into OMVs produced by Escherichia coli. We report that favorable electrostatic protein-membrane interactions are also at work in the soluble enzyme IMP-1 while being absent in VIM-2. These interactions correlate with an enhanced incorporation of IMP-1 compared to VIM-2 into OMVs. Disruption of these interactions in NDM-1 and IMP-1 impairs their inclusion into vesicles, confirming their role in defining the protein cargo in OMVs. These results also indicate that packaging of metallo-β-lactamases into vesicles in their active form is a common phenomenon that involves cargo selection based on specific molecular interactions.
KEYWORDS: metallo-β-lactamases, outer membrane vesicles, NDM-1, IMP-1, protein-membrane interactions
INTRODUCTION
The rise of carbapenem-resistant bacteria producing metallo-β-lactamases is of great concern since this class of β-lactam antibiotics is often used in severe infections (1, 2). Metallo-β-lactamases (MBLs) represent the largest family of carbapenemases (3–6). Among MBLs, the plasmid-borne NDMs, VIMs, and IMPs are the enzymes with the highest clinical relevance and geographical dissemination (3, 5, 7). This genetic localization has favored their dissemination into different opportunistic and pathogenic bacteria (8–10).
MBLs from the NDM family are lipoproteins anchored to the outer membrane (OM) of Gram-negative bacteria (11, 12). This cellular localization enables packaging of NDM-1 into outer membrane vesicles (OMVs) (12). OMVs are spherical lipid bilayer nanostructures released by all Gram-negative bacteria (13) which are recognized as “public goods,” as they can transport different molecules outside the boundaries of the bacterial cell and share them with neighboring communities of bacteria (14, 15). Ciofu and coworkers reported, for the first time, the presence of β-lactamases into OMVs (16). Later studies in recent years have strengthened this observation, showing that OMVs from different bacteria are able to incorporate serine-β-lactamases, MBLs, and genes coding for these enzymes (16–21). These observations suggest that this phenomenon is common rather than being an exception and supports the hypothesis that these vesicles play a pivotal role in the dissemination of bacterial antibiotic resistance. However, the features that govern the selection of the protein cargo into OMVs are largely unknown.
NDM-1 is packaged into OMVs produced by different bacteria as a fully active β-lactamase, endowing the vesicles with a potent carbapenemase activity (12, 22). NDM-1-loaded vesicles are able to inactivate the antibiotic in the near environment and protect nearby populations of bacteria susceptible to antibiotics (12). NDM-1 is selectively packaged into vesicles by being anchored to the outer membrane (12). However, a soluble variant engineered in the lab (NDM-1 C26A, lacking the lipidated Cys residue) is also present in OMVs (although in smaller amounts) (12). This observation compelled us to ask whether there are other molecular features in addition to membrane anchoring that favor the cargo selection into vesicles. Indeed, the globular domain of NDM-1 was shown to interact with the negatively charged anionic lipids in the bacterial membrane through electrostatic interactions mediated by two Arg residues, Arg45 and Arg52, which are conserved in all known NDM variants (23). Substitution of these two residues to negatively charged Glu disrupts the interaction with the lipid bilayer (23). Here, we show that these interactions are relevant in determining the amount of active NDM-1 present in Escherichia coli vesicles. We also analyzed the protein-membrane interactions of IMP-1 and VIM-2, clinically relevant MBLs that are soluble periplasmic proteins. We report that attractive electrostatic interactions between the membrane and the soluble domains of MBLs favor their packaging into OMVs, as is the case for NDM-1 and IMP-1. In contrast, for VIM-2 the lack of interaction with the membrane results in decreased levels in vesicles. Disruption of the membrane interactions in NDM-1 and IMP-1 induces a reduction in the MBL levels in vesicles, confirming that these are important molecular interactions active in selecting the protein cargo into vesicles.
RESULTS AND DISCUSSION
NDM-1 packaging into OMVs is promoted by specific electrostatic interactions with the membrane.
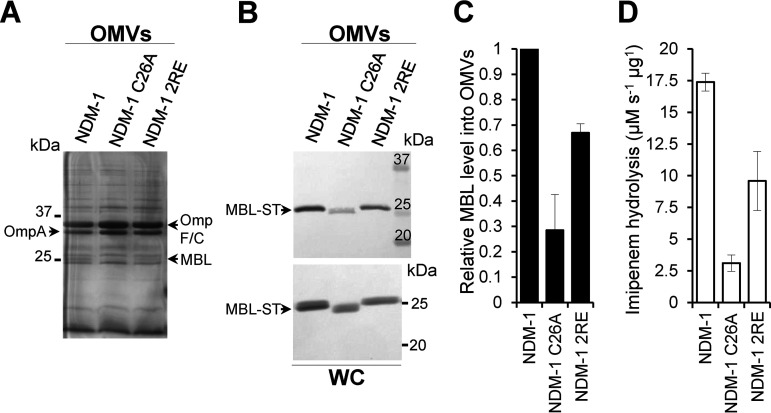
In order to study the contribution of specific interactions between the bacterial membrane and NDM-1 in its packaging in vesicles, we analyzed the amount of active protein in the OMVs produced by E. coli cells expressing (i) the native, lipidated, membrane-bound NDM-1; (ii) a soluble variant in which the lipidation site is removed (NDM-1 C26A); and (iii) a lipidated double mutant in which the two Arg residues (Arg45 and Arg52) are replaced by Glu (NDM-1 2RE). The selected MBLs were fused to a C-terminal Strep-tag sequence (−ST) for immunoblotting quantification. OMVs were purified from the supernatants of cell cultures of E. coli and then analyzed by SDS-PAGE and immunoblotting with anti-ST antibodies. In all cases, the detected levels of MBLs in vesicles are calculated relative to the amount of mature protein in cells, as detailed in Materials and Methods. Fig. 1B and C show that removal of the lipidation site in NDM-1 C26A leads to a 70% decrease in the amount of transported NDM-1, in agreement with previous experiments (12). The NDM-1 2RE mutant, despite preserving the membrane localization, displays decreased packaging in vesicles of almost 35% (Fig. 1B and C). Remarkably, the R45E/R52E mutation did not alter the ability of this MBL to confer resistance against β-lactams in E. coli cells (Table S1 in the supplemental material) nor its carbapenemase activity within vesicles (Fig. 1D), indicating that the catalytic ability of the enzyme remains relatively unaffected by these substitutions in Zn(II)-rich growth media. Considering that these substitutions disrupt the electrostatic interaction of NDM-1 with the membrane, we conclude that interactions between the globular domain of NDM-1 and the inner leaflet of the outer membrane enhance the packaging of this enzyme into vesicles. These interactions may determine the relative amounts of MBL packaged into OMVs.
FIG 1.
The specific interaction between NDM-1, mediated by residues Arg45 and Arg52, and the bacterial membrane promotes its packaging into vesicles. (A) SDS-PAGE of the OMVs purified from E. coli expressing blaNDM-1, blaNDM-1 C26A, or blaNDM-1 2RE. (B) Immunoblotting detection of NDM-1, NDM-1 C26A and NDM-1 2RE (fused to a C-terminal Strep-tag sequence [−ST]) in OMVs and in whole cells (WC) from E. coli strains expressing each MBL. (C) Mature protein levels of NDM-1, NDM-1 C26A, and NDM-1 2RE into OMVs. The plotted values were calculated as described in Materials and Methods. Data correspond to three independent experiments and are shown as the mean value. Error bars represent the standard deviation (SD). (D) Imipenem hydrolysis rate by OMVs purified from E. coli expressing NDM-1, NDM-1 C26A, or NDM-1 2RE. Data correspond to two independent experiments and are shown as the mean value. Error bars represent the SD.
To further evaluate the contribution of the protein-membrane interactions mediated by the two Arg residues in the soluble mutant NDM-1 C26A, we tested the level of incorporation into OMVs of this protein and the variant with the two Arg replaced by Glu (NDM-1 C26A 2RE). We observed that the levels of soluble mutant NDM-1 C26A 2RE in OMVs drop by 27% relative to the soluble mutant NDM-1 C26A, which preserves the bacterial membrane interaction given by the two arginines (Fig. S1). Similar effects were observed for both lipidated NDM-1 and soluble NDM-1 C26A when evaluating the role of disruption of electrostatic interactions in the level of packaging within the vesicles (Fig. 1A to C and Fig. S1). The favorable contribution of these specific interactions in the soluble domain of NDM-1 led us to wonder if these interactions may be present in natural soluble variants of MBLs.
The MBL packaging into OMVs depends on the protein-membrane affinity.
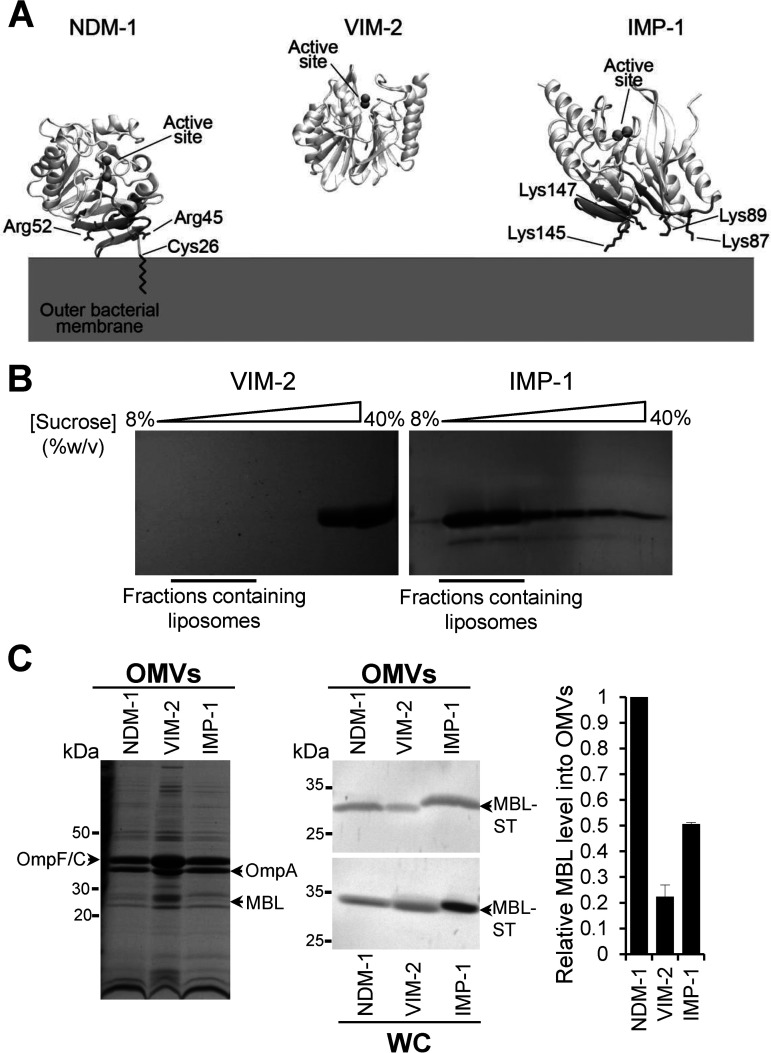
As a result of these observations, we hypothesized that electrostatic interactions of soluble periplasmic MBLs with the bacterial membrane may also favor packaging of these proteins into OMVs, regardless of a lipid anchor. Therefore, we analyzed the affinity of wild-type (WT) NDM-1 and soluble, periplasmic VIM-2 and IMP-1 with the bacterial membrane through coarse-grained (CG) molecular dynamics (MD) simulations (23) (Fig. 2A). The membrane bilayer was modeled mimicking the lipid composition of the bacterial outer membrane, as previously reported (23). IMP-1 was predicted to have a strong interaction with the membrane, as evidenced by the same number of binding events as NDM-1 (Fig. S2A and Fig. 2A). In general, IMP-1 spent a large portion of simulation time in proximity of the bilayer (Fig. S2B). In contrast, VIM-2 did not show significant binding events in any of the MD replicas (Fig. S2A and B and Fig. 2A). In the case of IMP-1, the interaction occurred by means of a specific protein patch spanning from Gly85 to Ser94 and from Thr133 to Pro153. Although this region is different from that previously identified for NDM-1, it also features a cluster of positively charged residues, in particular, four lysine residues (Lys87, Lys89, Lys145, and Lys147) (Fig. 2A).
FIG 2.
NDM-1, IMP-1, and VIM-2 MBLs have different affinities for the bacterial membrane that impact on the protein cargo into vesicles. (A) Identified surfaces of interaction between NDM-1, VIM-2, and IMP-1 and the bacterial outer membrane. The positively charged residues are in sticks. VIM-2 did not interact with the membrane in any of the MD replicas and, therefore, does not present an interaction patch. (B) SDS-PAGE analysis of sucrose gradient fractions from liposome flotation assays of VIM-2 and IMP-1. The flotation assays were carried out using liposomes made with an outer membrane composition from E. coli. (C) SDS-PAGE analysis (left) and immunoblot analysis (middle) of OMVs and whole cells (WC) from E. coli expressing NDM-1, VIM-2, or IMP-1, after induction with 20 μM IPTG. (C, Right) Comparison between levels of NDM-1, VIM-2, and IMP-1 into OMVs. The plotted values, relativized to the NDM-1 values, were obtained as described in Materials and Methods. Data correspond to two independent experiments and are shown as the mean value. Error bars represent the standard deviation (SD).
To experimentally test these predictions, we studied the interaction of IMP-1 with the membrane in vitro by means of liposome flotation assays (23), incubating soluble IMP-1 with liposomes mimicking the lipid composition of the bacterial outer membrane (Fig. 2B). Samples were loaded at the bottom of a sucrose gradient and were then ultracentrifuged. In these experiments, liposomes float toward lower concentrations of sucrose, and free proteins remain at the bottom of the gradient. SDS-PAGE analysis of these samples showed that IMP-1 colocalizes with liposomes, i.e., IMP-1 is strongly associated with the membrane (Fig. 2B). VIM-2, instead, was largely found in the fractions corresponding to unbound proteins (Fig. 2B). Next, we sought to evaluate if the interaction of IMP-1 with membranes favors its transport into vesicles. We compared the levels of IMP-1, NDM-1, and VIM-2 transported into vesicles from E. coli cells expressing these enzymes. Despite being a soluble protein, the amount of IMP-1 in OMVs is 50% compared to the levels of the membrane-bound NDM-1 (Fig. 2C). This is significant compared to other soluble MBLs, such as VIM-2 (Fig. 2C) or the soluble variant NDM-1 C26A itself. The protein levels of VIM-2 and NDM-1 C26A in vesicles were less than half of those observed for IMP-1.
The interaction of the Lys-rich patch in IMP-1 contributes to its packaging in OMVs.
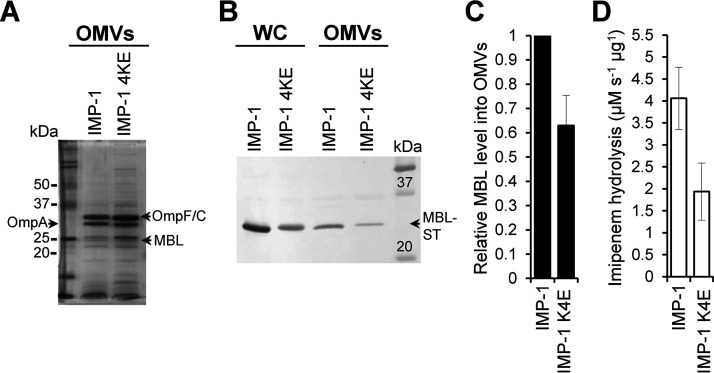
The results obtained heretofore indicate that electrostatic interactions leading to membrane association are responsible for increasing the packaging of MBLs into OMVs, regardless of the presence of a lipid anchor. To test this hypothesis, we decided to disrupt the interactions of IMP-1 with the membrane by replacing the 4 Lys residues (Lys87, Lys89, Lys145, and Lys147) by Glu residues. CG-MD simulations of this IMP-1 mutant (IMP-1 4KE) predict a significant drop in the number of binding events compared to WT IMP-1 (3 out of 5 replicas did not show any binding; Fig. S2A) and, in general, a lower affinity toward the membrane, as highlighted by the significantly smaller amount of simulation time spent by the enzyme in the proximity of the bilayer (Fig. S2B). Then, we evaluated the amount of IMP-1 and IMP-1 4KE mutant in vesicles from E. coli cells expressing these MBLs. Figure 3A to C shows that the amount of IMP-1 4KE in vesicles experiences a decay of almost 35% with respect to WT IMP-1, confirming the role of these specific electrostatic interactions in promoting the IMP-1 cargo into OMVs. Notably, the resistance profile (Table S1) and the carbapenemase activity (Fig. 3D) displayed by IMP-1 and the IMP-1 4KE mutant were preserved in E. coli cells and within the vesicles, indicating that these mutations did not alter the folding nor the metal binding capability of the enzyme. Remarkably, the Lys residues at positions 89 and 147 are fully conserved among the 80 allelic variants in the IMP family (24). Lys145 is replaced by an Asn in only 2 allelic variants, while Lys87 is conserved in 65 variants (Fig. S3). This high degree of conservation predicts that members of the IMP family may possess structural features that favor interactions with the bacterial membrane and consequently be packaged into vesicles.
FIG 3.
The specific interaction between IMP-1, mediated by Lys87, Lys89, Lys145, and Lys147, and the bacterial membrane promotes its packaging into vesicles. (A) SDS-PAGE analysis of OMVs from E. coli expressing IMP-1 and IMP-1 4KE mutant (IMP-1 K87E/K89E/K145E/K147E). (B) Immunoblot analysis of OMVs and whole cells (WC) from E. coli expressing IMP-1 or IMP-1 4KE. (C) Comparison between levels of WT IMP-1 and IMP-1 4KE into OMVs. The plotted values, relativized to the IMP-1 value, were obtained as described in Materials and Methods. Data correspond to three independent experiments and are shown as the mean value. Error bars represent the standard deviation (SD). (D) Imipenem hydrolysis rate by OMVs purified from E. coli expressing IMP-1 or IMP-1 4KE. Data correspond to two independent experiments and are shown as the mean value. Error bars represent SD.
Concluding remarks.
OMVs have been reported to carry MBLs in an active form, contributing to resistance by hydrolyzing antibiotics in situ and by protecting bacterial populations otherwise susceptible to antibiotics (12, 25). Elucidation of the molecular features that contribute to the packaging of their active forms is essential to understanding the role of OMVs in antibiotic resistance. The current results reveal a direct correlation between attractive electrostatic interactions of MBLs with the bacterial outer membrane and the protein levels packaged in OMVs in an active form. In this work, we demonstrate the role of positively charged regions in promoting the incorporation of MBL into vesicles. Since the inner membrane is also negatively charged, attractive interactions could also take place with this lipid bilayer. Regardless of this fact, altering the positive patches results in a net change in the amount of the protein cargo both in NDM-1 and IMP-1, suggesting that the transient nature of this interaction increases the concentration of MBLs close to the membranes, favoring inclusion into OMVs.
The features identified here add to the already reported effect of membrane anchoring in NDM-1 in favoring inclusion into vesicles and to recent findings in Bacteroides spp. reporting the impact of the efficient packaging of lipoproteins for OMVs sorting (26). Overall, this reveals that there are multiple molecular features synergistically operative in selecting the OMV cargo related to antimicrobial resistance.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and construction of MBL mutants.
Escherichia coli DH5α was used for expression of the pMBLe-Gmr vector as an empty vector and included the following genes coding for the different MBLs: NDM-1, VIM-2 and IMP-1 and their mutants, NDM-1 C26A, NDM-1 R45E/R52E (NDM-1 2RE), NDM-1 C26A R45E/R52E (NDM-1 C26A 2RE), and IMP-1 K87E/K89E/K145E/K147E (IMP-1 4KE). All variants were cloned in the full-length version into pMBLe, fused to a C-terminal Strep-tag II sequence and under the control of a β-isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pTac promoter, as previously described (12, 22). Cells were routinely grown aerobically at 37°C in lysogeny broth (LB) or on LB agar plates except when indicated. Gentamicin was used when necessary at 20 μg/ml. The pMBLe-NDM-1 2RE-ST was obtained from plasmid pMBLe-NDM-1-ST through site-directed mutagenesis by plasmid amplification using primers NDM-1 2RE forward (5′-GAAACTGGCGACCAAGAGTTTGGCGATCTGGTTTTCGAGCAGCTCGCACCGAATG-3′) and NDM-1 2RE reverse (5′-CATTCGGTGCGAGCTGCTCGAAAACCAGATCGCCAAACTCTTGGTCGCCAGTTTC-3′). The same oligonucleotides were used for the construction of pMBLe-NDM-1 C26A 2RE-ST from plasmid pMBLe-NDM-1 C26A. Plasmid pMBLe-IMP-1 K87EK89E-ST was obtained by directed mutagenesis by plasmid amplification on plasmid pMBLe-IMP-1-ST, using primers IMP-1 K87EK89E forward (5′-GGTTTGTGGAGCGTGGCTATGAAATAGAAGGCAGCATTTCCTCTC-3′) and IMP-1 K87EK89E reverse (5′-GAGAGGAAATGCTGCCTTCTATTTCATAGCCACGCTCCACAAACC-3′). Then, using the latter plasmid as a template, the pMBLe-IMP-1 4KE-ST was obtained, using primers K145EK147E forward (5′-GGAGTTAACTATTGGCTAGTTGAAAATGAAATTGAAGTTTTTTATCCAGG-3′) and K145EK147E reverse (5′-CCTGGATAAAAAACTTCAATTTCATTTTCAACTAGCCAATAGTTAACTCC-3′).
Liposome preparation.
Pure lyophilized phospholipids (1-palmitoyl-2-oleoyl phosphatidylethanolamine [PE], tetraoleoyl cardiolipin [CDL], and 1-palmitoyl-2-oleoyl-phosphatidylglycerol [PG]) were purchased from Avanti Polar Lipids. The lipids were dissolved in chloroform, and, after mixing the required proportions of each pure lipid, the lipid mixtures were dried under a nitrogen atmosphere and then kept under vacuum for 2 h. The dried lipid films were hydrated with 50 mM HEPES, pH 7, and heated at 65°C for 1 h with periodic vortexing. Lipid suspensions were frozen in liquid nitrogen and then thawed at 65°C for a total of 5 cycles and, afterward, were passed through a 400-nm polycarbonate filter using an Avanti mini-extruder apparatus (Avanti Polar Lipids) at 65°C, with >20 passes through the device.
Liposome flotation assay.
Samples containing 55 μM purified protein (VIM-2 or IMP-1) in 50 mM HEPES at pH 7 were incubated with liposomes for 30 min at room temperature. Sucrose was added to 40% (wt/vol), the samples were loaded in an ultracentrifuge tube, and sucrose 25% (wt/vol) and sucrose 8% (wt/vol) (both buffered with 50 mM HEPES, pH 7) were layered on top, forming a discontinuous sucrose gradient. Afterward, samples were ultracentrifuged for 1 h at 4°C and 125,000 × g in a Beckman SW Ti90 rotor, and fractions along the gradient were analyzed by 14% SDS-PAGE gels to assess the final distribution of the MBL protein.
MIC determinations.
Ceftazidime (CAZ) and imipenem (IMI) MIC determinations for E. coli DH5α cells expressing the different MBLs, their mutants, or carrying the pMBLe empty vector (EV) lacking any MBL gene (as a control) were carried out in LB following the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) protocols (27).
Purification of OMVs and quantification of MBLs levels into OMVs.
Three hundred milliliters of LB broth were inoculated with 3 ml of saturated E. coli pMBLe-bla culture, grown at 37°C up to an optical density at 600 nm (OD600) of 0.4, and induced with 20 μM IPTG, and growth continued overnight with agitation. The cells were harvested, and the supernatant was filtered through a 0.45-μm membrane (Millipore). Ammonium sulfate was added to the filtrate at a concentration of 55% (wt/vol), followed by overnight incubation with stirring at 4°C. Precipitated material was separated by centrifugation at 12,800 × g for 10 min, resuspended in 10 mM HEPES, 200 mM NaCl at pH 7.4, and dialyzed overnight against >100 volumes of the same buffer. Next, samples were filtered through a 0.45-μm membrane, layered over an equal volume of 50% (wt/vol) sucrose solution, and ultracentrifuged at 150,000 × g for 1 h and 4°C. Pellets containing the OMVs were washed once with 10 mM HEPES, 200 mM NaCl at pH 7.4, and stored at −80°C until use.
OMVs were quantified by total protein dosage with the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). OMVs were then analyzed by SDS-PAGE and immunoblotting. To determine the levels of NDM-1, NDM-1 C26A, NDM-1 2RE, NDM-1 C26A 2RE, VIM-2, IMP-1, and IMP-1 4KE into OMVs, the mature protein band intensities in the whole cells (WC) and in the OMVs, from E. coli expressing each MBL, were quantified from polyvinylidene difluoride (PVDF) membranes with the software ImageJ (28). In addition, the band intensity of the main outer membrane protein (OmpF/C) in the OMVs preparations was quantified from SDS-PAGE gels. The quantity of each MBL in the OMVs (from immunoblots) was normalized to the amount of OmpF/C (from SDS-PAGE) in the same sample, and then this value was divided by the quantity of each MBL in whole cells (from immunoblot). Finally, the values plotted in Fig. 1C, Fig. 2C, Fig. S1C in the supplemental material, and Fig. 3C correspond to the relativization to NDM-1 (Fig. 1C and Fig. 2C), NDM-1 C26A (Fig. S1C), or IMP-1 (Fig. 3C) value, as we show in the following equation:
where MBLsOMVs represent relative level of MBLs into OMVs, MBL_WC is the intensity of the MBL band in whole cells, and MBL_OMV is the intensity of the MBL band in OMVs normalized to the Omp band in the same sample.
MBL detection and β-lactamase activity measurements.
MBL amounts were determined by SDS-PAGE followed by Western blotting with Strep-tag II monoclonal antibodies (at a 1:1,000 dilution from 200 μg/ml solution) (Novagen) and immunoglobulin G-alkaline phosphatase conjugates (at a 1:3,000 dilution). Briefly, the samples were mixed with loading buffer and heated to denature the peptide structure. SDS-PAGE (14%) was used for separation of the sample components and subsequently transferred onto a PVDF membrane (GE). β-lactamase activity into OMVs purified from E. coli expressing the different MBLs and their variants was measured in a Jasco V-670 spectrophotometer at 30°C in 10 mM HEPES and 200 mM NaCl at pH 7.4 in 0.1-cm cuvettes using 400 μM imipenem as a substrate. Imipenem hydrolysis was monitored at 300 nm (Δε300 = −9,000 M−1 cm−1).
CG-MD simulations.
In CG-MD simulations, the enzymes were located at a 40-Å distance from the bilayer in order to avoid early protein/membrane encounters that might bias the subsequent interactions. The Martini 2.2p (polarizable) force field was used for all the simulations. The atomistic three-dimensional structures of NDM-1, VIM-2, and IMP-1 were taken from the Protein Data Bank (PDB codes 5ZGE [29] for NDM-1, 1KO3 [30] for VIM-2, and 5EV6 [31] for IMP-1) and turned into a coarse-grained model with the Martinize tool. All lipid bilayers were generated with the Insane tool of Martini (32) with a lipid composition that mimics the bacterial outer membrane (91% PEs, 6% CDLs, and 3% PGs, in agreement with lipidomics analyses present in literature [33, 34]). For what concerns the acyl chains, we used the most common motifs for each lipid type, which are one 16:0 and one 18:1 (1-palmitoyl-2-oleoyl) for PEs and PGs, respectively, and four 18:1 for CDLs. All the systems were solvated with the polarizable Martini water model (35) and ionized with 150 mM NaCl. Each enzyme was simulated in 5 distinct replicas. Each replica had a simulation time of 2 μs. Frames for the analysis were collected every 750 ps. A membrane binding event is considered to have occurred when the protein settles at a <3-Å distance from the membrane within the first μs, and this distance remains constant for the rest of the simulation (i.e., no detachment occurs). For each system, the equilibration procedure was run as follows: first, the system went through 5,000 steps of minimization using steepest descent; second, it was equilibrated with 5 ns of MD in the NVT ensemble using Particle Mesh Ewald (PME) for the electrostatic contributions and velocity rescale algorithm for temperature coupling at 310 K. The production phase was conducted in the NPT ensemble using a Parrinello-Rahman semi-isotropic coupling algorithm (36) to maintain the pressure constant at 1 bar. The two zinc ions in the catalytic site were represented as one single Martini bead of Qa type, connected to the 6 coordinating residues through harmonic potentials. Such particle was charged with +2e; that is the total net charge of the catalytic site.
ACKNOWLEDGMENTS
We thank Marina Avecilla (IBR) for excellent technical assistance.
This work was supported by a MinCyT (SUIZ/17/10) and a Swiss National Science Foundation grant (514106) to A.J.V. and M.D.P., a grant from ANPCyT (PICT-2016-1657) to A.J.V., and an NIAID grant (2R01AI100560-06A1) to A.J.V. and R.A.B. C.L. is the recipient of a postdoctoral fellowship from CONICET, and L.J.G. and A.J.V. are staff members from CONICET.
C.L., A.P., G.B., R.A.B., L.J.G., M.D.P., and A.J.V. conceived the project, designed the experiments, and edited the manuscript. C.L. performed the construction of NDM-1 R2E and IMP-1 4KE mutants, MIC determinations, OMVs purification, MBL detection, and determination of β-lactamase into OMVs. G.B. performed the liposome preparation and the liposome flotation assay. C.L. and L.J.G. helped to carry out the IMP-1 liposome flotation assay. A.P. performed the computational experiments. C.L. and A.P. designed the figures. C.L., L.J.G., and A.J.V. wrote the manuscript. All authors discussed the results and commented on the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev 18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meini MR, Llarrull LI, Vila AJ. 2015. Overcoming differences: the catalytic mechanism of metallo-β-lactamases. FEBS Lett 589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Codjoe FS, Donkor ES. 2017. Carbapenem resistance: a review. Med Sci 6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju LC, Cheng Z, Fast W, Bonomo RA, Crowder MW. 2018. The continuing challenge of metallo-β-lactamase inhibition: mechanism matters. Trends Pharmacol Sci 39:635–647. doi: 10.1016/j.tips.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palacios AR, Rossi MA, Mahler GS, Vila AJ. 1975. Metallo‐β‐lactamase inhibitors inspired on snapshots from the catalytic mechanism. Biomolecules 10:854. doi: 10.3390/biom10060854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd SE, Livermore DM, Hooper DC, Hope WW. 1975. Metallo-β-lactamases: structure, function, epidemiology, treatment options, and the development pipeline. Biochem Pharmacol 24:e00397-20. doi: 10.1128/AAC.00397-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 9.Mojica F, Bonomo R, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halat DH, Moubareck CA. 2020. The current burden of carbapenemases: review of significant properties and dissemination among Gram-negative bacteria. Antibiotics 9:186. doi: 10.3390/antibiotics9040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat Chem Biol 12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnington KE, Kuehn MJ. 2014. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruana JC, Walper SA. 2020. Bacterial membrane vesicles as mediators of microbe – microbe and microbe – host community interactions. Front Microbiol 11:432. doi: 10.3389/fmicb.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Høiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Clarke AJ, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 180:5478–5483. doi: 10.1128/JB.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao YT, Kuo SC, Chiang MH, Lee YT, Sung WC, Chen YH, Chen TL, Fung CP. 2015. Acinetobacter baumannii extracellular OXA-58 is primarily and selectively released via outer membrane vesicles after Sec-dependent periplasmic translocation. Antimicrob Agents Chemother 59:7346–7354. doi: 10.1128/AAC.01343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee S, Mondal A, Mitra S, Basu S. 2017. Acinetobacter baumannii transfers the blaNDM-1gene via outer membrane vesicles. J Antimicrob Chemother 72:2201–2207. doi: 10.1093/jac/dkx131. [DOI] [PubMed] [Google Scholar]
- 21.Schaar V, Nordström T, Mörgelin M, Riesbeck K. 2011. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother 55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López C, Ayala JA, Bonomo RA, González LJ, Vila AJ. 2019. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat Commun 10:3617. doi: 10.1038/s41467-019-11615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prunotto A, Bahr G, González LJ, Vila AJ, Dal Peraro M. 2020. Molecular bases of the membrane association mechanism potentiating antibiotic resistance by New Delhi metallo-β-lactamase 1. ACS Infect Dis 6:2719–2731. doi: 10.1021/acsinfecdis.0c00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)–structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devos S, Stremersch S, Raemdonck K, Braeckmans K, Devreese B. 2016. Intra-and interspecies effects of outer membrane vesicles from Stenotrophomonas maltophilia on beta-lactam resistance. Antimicrob Agents Chemother 60:2516–2518. doi: 10.1128/AAC.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valguarnera E, Scott NE, Azimzadeh P, Feldman MF. 1975. Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides. Biochem Pharmacol 24:e00559-18. doi: 10.1128/mSphere.00559-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Ma G, Zhu Y, Zeng L, Ahmad A, Wang C, Pang B, Fang H, Zhao L, Hao Q. 1975. Active-site conformational fluctuations promote the enzymatic activity of NDM-1. Biochem Pharmacol 24:e01579-18. doi: 10.1128/AAC.01579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J Mol Biol 375:604–611. doi: 10.1016/j.jmb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Hinchliffe P, González MM, Mojica MF, González JM, Castillo V, Saiz C, Kosmopoulou M, Tooke CL, Llarrull LI, Mahler G, Bonomo RA, Vila AJ, Spencer J. 2016. Cross-class metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc Natl Acad Sci USA 113:E3745–E3754. doi: 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP, Marrink SJ. 2015. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 33.Lugtenberg EJJ, Peters R. 1976. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim Biophys Acta 441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 34.Morein S, Andersson AS, Rilfors L, Lindblom G. 1996. Wild-type Escherichia coli cells regulate the membrane lipid composition in a “window” between gel and non-lamellar structures. J Biol Chem 271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- 35.Yesylevskyy SO, Schäfer LV, Sengupta D, Marrink SJ. 2010. Polarizable water model for the coarse-grained Martini force field. PLoS Comput Biol 6:e1000810. doi: 10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parrinello M, Rahman A. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table and figures. Download AAC.00507-21-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)