ABSTRACT
Remdesivir (RDV; GS-5734) is currently the only FDA-approved antiviral drug for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. The drug is approved for use in adults or children 12 years or older who are hospitalized for the treatment of COVID-19 on the basis of an acceleration of clinical recovery for inpatients with this disease. Unfortunately, the drug must be administered intravenously, restricting its use to those requiring hospitalization for relatively advanced disease. RDV is also unstable in plasma and has a complex activation pathway which may contribute to its highly variable antiviral efficacy in SARS-CoV-2-infected cells. Potent orally bioavailable antiviral drugs for early treatment of SARS-CoV-2 infection are urgently needed, and several, including molnupiravir and PF-07321332, are currently in clinical development. We focused on making simple, orally bioavailable lipid analogs of remdesivir nucleoside (RVn; GS-441524) that are processed to RVn monophosphate, the precursor of the active RVn triphosphate, by a single-step intracellular cleavage. In addition to high oral bioavailability, stability in plasma, and simpler metabolic activation, new oral lipid prodrugs of RVn had submicromolar anti-SARS-CoV-2 activity in a variety of cell types, including Vero E6, Calu-3, Caco-2, human pluripotent stem cell (PSC)-derived lung cells, and Huh7.5 cells. In Syrian hamsters, oral treatment with 1-O-octadecyl-2-O-benzyl-glycero-3-phosphate RVn (ODBG-P-RVn) was well tolerated and achieved therapeutic levels in plasma above the 90% effective concentration (EC90) for SARS-CoV-2. The results suggest further evaluation as an early oral treatment for SARS-CoV-2 infection to minimize severe disease and reduce hospitalizations.
KEYWORDS: SARS-CoV-2, remdesivir, remdesivir nucleoside, antiviral agents, lipid prodrugs, Vero E6 cells, Calu-3 cells, Caco-2 cells, Huh7.5 cells, PSC-derived human lung cells
INTRODUCTION
Over the past 18 years, spillover events have introduced the highly transmissible betacoronavirus strains severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 into the human population (1–3). Although case fatality ratios have varied, each virus induces substantial morbidity and mortality, especially among those >55 years of age and/or those with underlying comorbid medical conditions (4, 5). Although SARS-CoV and MERS-CoV were largely contained by epidemiological interventions, the most recent 2019 outbreak has evolved into a global pandemic responsible for >160 million infections and >3.5 million deaths (6). With >30 million cases and nearly 600,000 deaths at this writing, the United States remains in the center of the pandemic. Intensive and economically disruptive social distancing measures have blunted several viral surges, but this approach is not sustainable, and infection rates have increased each time restrictions have eased (7). Fortunately, several highly effective vaccines have emerged over the past 6 months, and their distribution in the United States and many other high-income countries has had a major impact on SARS-CoV-2-associated morbidity and mortality (8, 9). Despite these highly noteworthy successes in vaccine development, gaps and delays in global vaccine delivery, the emergence of viral variants against which vaccine protection is compromised, and a relatively sizable immunocompromised population who are unable to fully respond to vaccination make it clear that infections will continue and that highly effective antiviral therapy is required.
Remdesivir nucleoside triphosphate (RVn triphosphate) potently inhibits enzymatic activity of the polymerase of every coronavirus tested thus far, including SARS CoV-2 (10–13). The drug has recently been approved by the FDA for the treatment of adults and children aged 12 or older who are hospitalized for COVID-19 (14). This broad activity reflects the relative molecular conservation of the coronavirus RNA-dependent RNA polymerase (RdRp). Remdesivir (RDV) is an aryloxy phosphoramidate triester prodrug that must be converted by a series of reactions to RVn triphosphate, the active antiviral metabolite (Fig. 1). Although RVn triphosphate is an excellent inhibitor of the viral RdRp (11, 15), RDV’s antiviral activity is highly variable in different cell types, which may be due to variable expression of the four enzymes required for conversion to remdesivir nucleoside monophosphate (RVn-P) (16). RDV’s base is a 1′-cyano-substituted adenine C nucleoside (GS-441524, RVn) that is thought to be poorly phosphorylated. To bypass the perceived slow first phosphorylation, the developers relied on an aryloxy phosphoramidate triester prodrug that is converted by a complex series of four reactions to RVn-P that is then efficiently converted to RVn triphosphate, the active metabolite. RDV may be more active in some SARS-CoV-2-infected tissues than in others, a possible reason for its incomplete clinical impact on SARS-CoV-2. A recent report suggests that some tissues express low levels of the four enzymes that activate RDV, and some tissues may be responsible for tissue-specific differences in antiviral activity (13). Yan and Muller have recently published a detailed analysis of the potential weaknesses of remdesivir and suggested that RVn (GS-441524) might be a preferable therapy (16). Remdesivir has beneficial antiviral and clinical effects in animal models of coronavirus infection (17, 18). These effects are primarily demonstrable when administered before or very soon after viral challenge. However, RDV is not highly bioavailable following oral administration and must be administered intravenously, functionally limiting its clinical application to hospitalized patients with relatively advanced disease. It would be clinically useful to have a highly active, orally bioavailable analog of RVn which provides sustained levels of intact antiviral drug in plasma, since RDV persistence in plasma is known to be brief. In monkeys treated with intravenous RDV, the plasma level declined by roughly two log10 2 h after the infusion ended (16, 19). In two patients with COVID-19 treated with intravenous RDV, 1 h after the intravenous infusion stopped, a drop of >90% was observed (20).
FIG 1.
Structures of remdesivir and related intermediates.
Here, we report the synthesis and antiviral evaluation of three novel lipid prodrugs of RVn monophosphate that are active at submicromolar concentrations against SARS-CoV-2 infection in a variety of cell types, including Vero E6, Calu-3, Caco-2, pluripotent stem cell (PSC)-derived human lung cells, and Huh7.5 cells. These compounds are stable in human plasma in contrast to remdesivir and are orally bioavailable as predicted by our prior work with other antivirals of this general design (21, 22). These oral remdesivir nucleoside phosphate prodrugs could allow earlier and more effective treatment at the time of diagnosis of SARS-CoV-2 infection. In addition, one of these prodrugs represents an approach that may be capable of delivering the antiviral to the lung and away from the liver, the site of remdesivir’s dose-limiting toxicity, due to its route through intestinal lymph bypassing the portal vein and the liver (23, 24).
(This article was submitted to an online preprint archive [25].)
RESULTS
Synthesis of RVn monophosphate prodrugs.
We synthesized the hexadecyloxypropyl-, octadecyloxyethyl-, and 1-O-octadecyl-2-O-benzyl-sn-glyceryl esters of RVn 5′-monophosphate. Compounds 5a to 5c were synthesized as shown in Fig. 2. Analyses by nuclear magnetic resonance (NMR), electrospray ionization (ESI) mass spectrometry, and high-performance liquid chromatography (HPLC) were consistent with each structure and demonstrated purities of >95%.
FIG 2.
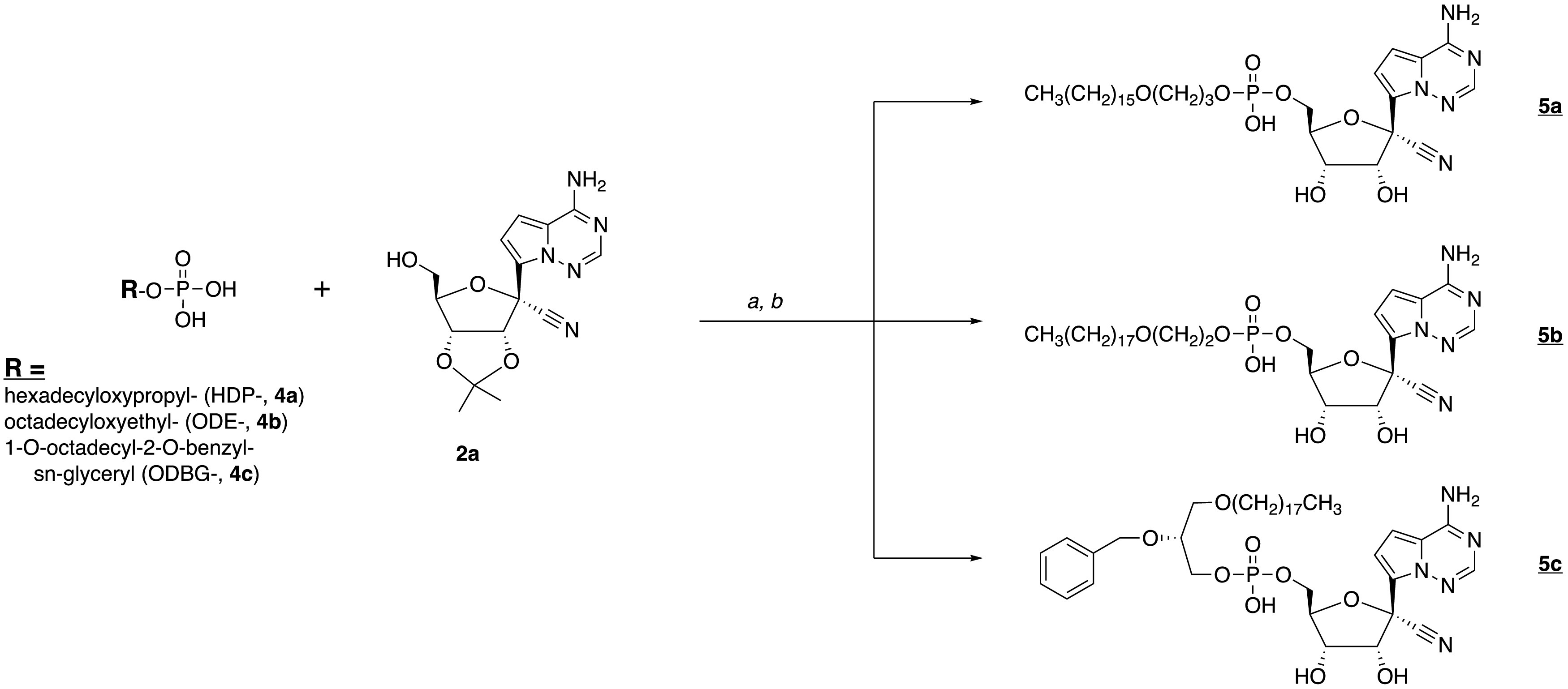
Synthesis of antiviral prodrugs 5a to 5c. a, 2′,3′-isopropylidene RVn (2a), DCC, DMAP, and pyridine, 90°C, 24 to 72 h; b, 37% HCl, THF, 3 to 18 h.
Ex vivo stability of ODBG-P-RVn in human plasma.
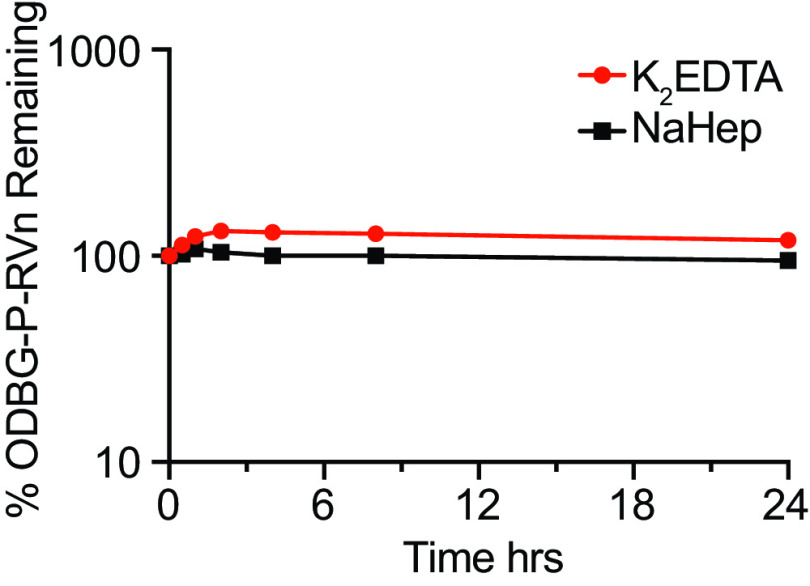
One of the disadvantages of remdesivir is its in vivo and ex vivo instability in plasma. In vivo, it has been reported to persist for less than 2 h after intravenous infusion in rhesus monkeys (13, 16, 19) and in COVID-19 patients (20). Ex vivo, remdesivir is not stable in human plasma, with a reported half-life (t1/2) of 69 min (26). We tested the ex vivo stability of 1-O-octadecyl-2-O-benzyl-glycero-3-phosphate RVn (ODBG-P-RVn) in human plasma with either K2EDTA or sodium heparin (NaHep) as anticoagulant. ODBG-P-RVn was added to human plasma and incubated at 37°C for 24 h. ODBG-P-RVn levels were measured at the indicated times by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 3). ODBG-P-RVn has a t1/2 in human plasma of >24 h versus 69 min for remdesivir.
FIG 3.
ODBG-P-RVn stability in human plasma at 37°C. Plasma was spiked with 2-μg/ml concentrations of ODBG-P-RVn and incubated at 37°C for 24 h. Samples were taken at 0.5, 1, 2, 4, 8, and 24 h and frozen for later analysis by LC-MS/MS.
Lipid RVn monophosphate prodrugs are potent inhibitors of SARS-CoV-2.
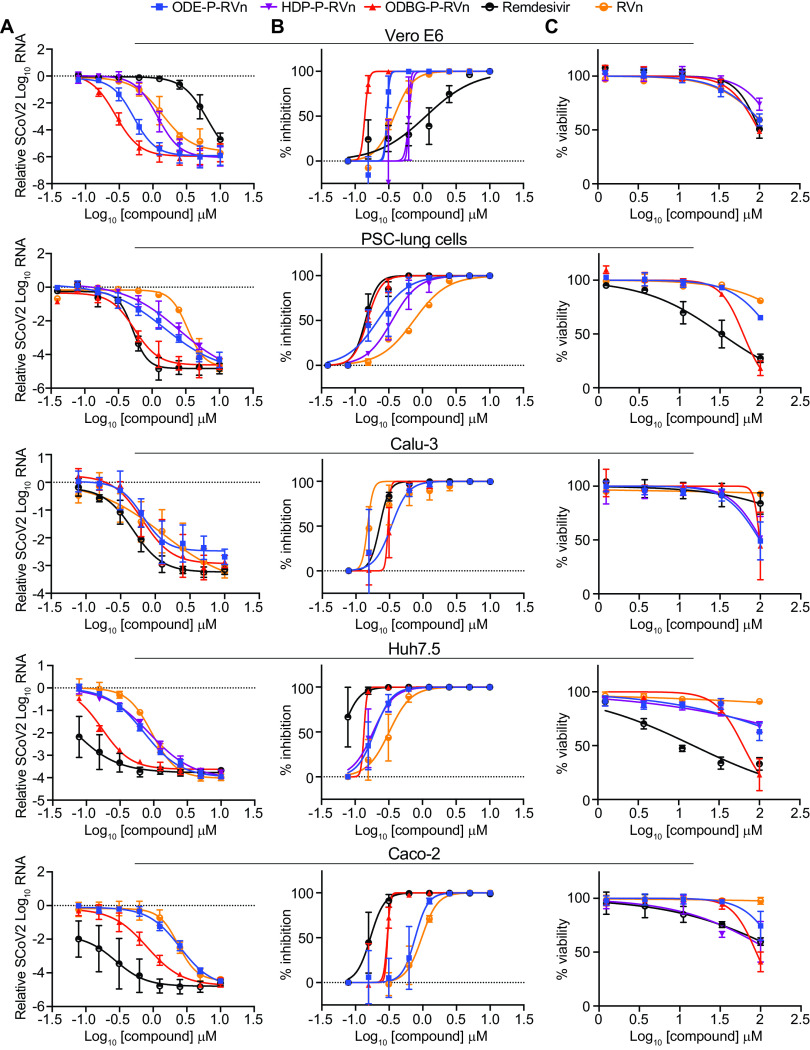
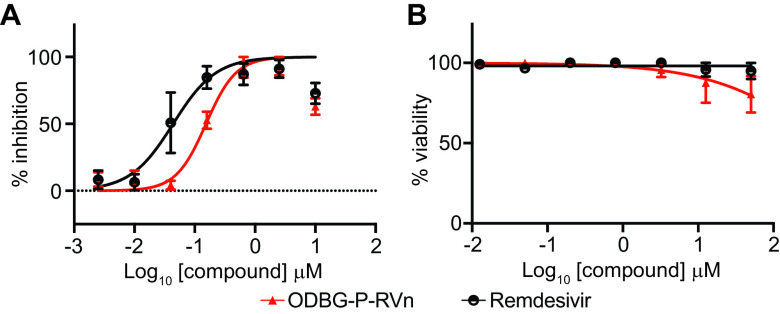
To determine if lipid RVn monophosphate prodrugs inhibit SARS-CoV-2 RNA replication, we performed antiviral assays using a clinical isolate of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) in multiple cell lines: African green monkey kidney cells (Vero E6), human PSC-derived lung (PSC-lung) cells, human lung epithelial cell line Calu-3, human colonic epithelial cell line Caco-2, and human hepatocyte cell line Huh7.5. Each cell line was treated with the above-indicated compounds for 30 min prior to infection and incubated for 48 h postinfection. Intracellular viral RNA was measured by quantitative reverse transcription-PCR (qRT-PCR). In all cell lines, there was a dose-dependent inhibition of viral RNA by ODBG-P-RVn, ODE-P-RVn, octadecyloxyethyl phosphate RVn (HDP-P-RVn), remdesivir (RDV), and remdesivir nucleoside (RVn) (Fig. 4A). In Vero E6 cells, the average half-maximal effective concentration (EC50) and average 90% effective concentration (EC90) of ODBG-P-RVn were 0.14 μM and 0.16 μM, respectively (Fig. 4B and Table 1). The EC50 of ODBG-P-RVn in Vero E6 cells was significantly lower than that of RDV (Table 1). ODE-P-RVn and HDP-P-RVn were also potently antiviral, with EC50 values of 0.3 μM and 0.63 μM, respectively, in Vero E6 cells (Fig. 4B and Table 1). The EC50s of ODBG-P-RVn and ODE-P-RVn were less than 0.35 μM in PSC-lung and Calu-3 cells, both models of human lung infection (Fig. 4B; Table 1). The antiviral activities of ODBG-P-RVn and ODE-P-RVn were significantly better than RVn in PSC-lung cells (Table 1). ODBG-P-RVn, ODE-P-RVn, and HDP-P-RVn demonstrated strong antiviral activity in Huh7.5 cells with EC50s of less than 0.2 μM that were not significantly different from that of RDV or RVn (Fig. 4B and Table 1). In the Caco-2 cell line, the EC50 of ODBG-P-RVn was 0.3 μM, which was significantly lower than that of RVn but similar to that of RDV (Fig. 4B and Table 1). In the same cell line, the EC50 of ODE-P-RVn was 0.77 μM, which was significantly higher than that of RDV (Fig. 4B and Table 1).
FIG 4.
SARS-CoV-2 inhibitory activity replicate experiments. (A and B) Antiviral dose-response curves for three remdesivir analogs, remdesivir (GS-5734), and remdesivir nucleoside (GS-441524) against SARS-CoV-2 infection in multiple cell types. SARS-CoV-2 relative viral RNA reduction (A) and percent inhibition (B) in the indicated cell types. Cells were pretreated with the indicated dose of the indicated drug for 30 min and then infected with SARS-CoV-2 isolate USA-WA1/2020 for 48 h. The relative SARS-CoV-2 spike RNA expression was determined by qRT-PCR. (C) Cytotoxicity in the indicated cells incubated in the presence of the indicated drug at the indicated concentration for 48 h, after which, cell viability was measured by the CellTiter-Glo assay. (A to C) Each antiviral and cytotoxicity dose-response data point indicates the average from 3 independent experiments performed in duplicates, except as indicated in Table 1. Error bars represent the standard errors of the means (SEMs).
TABLE 1.
Antiviral activity, cytotoxicity, and selectivity of the compoundsa
| Compound | EC50 (μM) | EC90 (μM) | CC50 (μM) | Selectivity indexb | P value (EC50 vs RDV, RVn) |
|---|---|---|---|---|---|
| Vero E6 cells | |||||
| RDV | 1.13 | 7.05 | 101 | 89.4 | |
| RVn | 0.38 | 0.77 | >100 | >263 | |
| HDP-P-RVn | 0.63 | 0.73 | >100 | >158 | NS |
| ODE-P-RVn | 0.30 | 0.33 | >100 | >333 | NS |
| ODBG-P-RVn | 0.14 | 0.16 | 97.9 | 699 | 0.010, 0.311 |
| PSC-human lung cells | |||||
| RDV | 0.14 | 0.23 | 32.7c | 234 | |
| RVn | 0.74 | 2.62 | >100c | >135 | |
| HDP-P-RVnc | 0.35 | 0.94 | ND | ||
| ODE-P-RVn | 0.22 | 0.70 | >100c | >454 | 0.791, 0.006 |
| ODBG-P-RVn | 0.15 | 0.26 | 61.5c | 410 | >0.999, 0.002 |
| Calu-3 cells | |||||
| RDV | 0.23 | 0.31 | >100 | >434 | |
| RVn | 0.15 | 0.18 | >100 | >666 | |
| ODE-P-RVn | 0.34 | 0.64 | 98.7 | 290 | NS |
| ODBG-P-RVn | 0.30 | 0.33 | 98.2 | 327 | NS |
| Huh7.5 cells | |||||
| RDV | 0.06 | 0.12 | 15.2 | 253 | |
| RVn | 0.32 | 0.73 | >100 | >312 | |
| HDP-P-RVn | 0.19 | 0.40 | >100 | >526 | NS |
| ODE-P-RVn | 0.19 | 0.37 | >100 | >526 | NS |
| ODBG-P-RVn | 0.14 | 0.15 | 62.9 | 449 | NS |
| Caco-2 cells | |||||
| RDV | 0.17 | 0.28 | >100 | >588 | |
| RVn | 0.96 | 1.75 | >100 | >104 | |
| ODE-P-RVn | 0.77 | 1.25 | >100 | >129 | 0.007, 0.971 |
| ODBG-P-RVn | 0.30 | 0.33 | 88.4 | 295 | 0.968, 0.007 |
RDV, remdesivir (GS-5734); RVn, remdesivir nucleoside (GS-441524); HDP-P, hexadecyloxypropyl-P; ODE-P, octadecyloxyethyl phosphate; ODBG-P, 1-O-octadecyl-2-O-benzyl-glycero-3-P; EC50, half-maximal effective concentration; CC50, 50% cytotoxic concentration, results by CellTiter-Glo; ND, not detected; NS, not significant.
Selectivity index, CC50/EC50; statistical analysis comparing log EC50 values from separate experiments by one-way analysis of variance (ANOVA).
Experiments performed only twice in duplicates (all others were performed three times in duplicates).
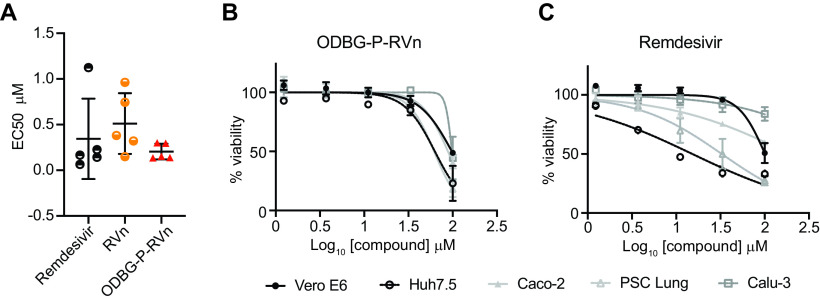
We next measured the cytotoxicity of each compound by incubating each of these cell lines with serial dilutions of each compound from 1.23 μM to 100 μM for 48 h (Fig. 4C). The average 50% cytotoxic concentrations (CC50) for all compounds were greater than 60 μM in all cell lines except for RDV, which had a CC50s of 32.7 μM in PSC-lung cells and 15.2 μM in Huh7.5 cells, a human hepatocyte cell line (Fig. 4C and Table 1). The selectivity index of ODBG-P-RV ranged from 295 to 699 in the five cell types tested (Table 1). The ranges of antiviral activity and cytotoxicity of ODBG-P-RVn (EC50, 0.14 μM to 0.30 μM; CC50, 61.5 μM o 98.2 μM) were more consistent across cell types than those for RDV (EC50, 0.06 μM to 1.13 μM; CC50 15.2 μM to >100 μM) (Fig. 5A to C and Table 1). Collectively, these data demonstrate that lipid RVn monophosphate prodrugs are potent antivirals against SARS-CoV-2 in vitro with low toxicity and excellent selectivity indexes.
FIG 5.
ODBG-P-RVn antiviral activity and cytotoxicity is highly reproducible across cell lines. (A) EC50 values for RDV, RVn, and ODBG-P-RVn in 5 different cell lines derived from lung (human PSC-derived, Calu-3), kidney (Vero E6), colon (Caco-2), or liver (Huh7.5). Cytotoxicity of ODBG-P-RVn (B) or RDV (C) in the indicated cells at the indicated concentration for 48 h, as measured by the CellTiter-Glo assay.
ODBG-P-RVn inhibits the human Alphacoronavirus 229E.
To determine if ODBG-P-RVn inhibits other human coronaviruses, we performed antiviral assays in the human lung fibroblast cell line MRC-5 using a clinical isolate of human coronavirus 229E. MRC-5 cells were infected with 229E for 2 h followed by incubation in the presence of various concentrations of RDV, ODBG-P-RVn, or vehicle for 72 h postinfection. Both ODBG-P-RVn and RDV demonstrated a dose-dependent inhibition of the cytopathic effect (CPE). The EC50 values of ODBG-P-RVn and RDV were 0.15 μM and 0.04 μM, respectively, and the EC90s were 0.54 μM and 0.26 μM, respectively (Fig. 6A). The CC50s for ODBG-P-RVn and RDV were greater than 50 μM in MRC-5 cells (Fig. 6B). Together with the antiviral data for SARS-CoV-2, this demonstrates that ODBG-P-RVn has antiviral activity against two genetically distinct human-pathogenic coronaviruses.
FIG 6.
ODBG-P-RVn inhibits the human Alphacoronavirus 229E. (A) Antiviral dose-response curves for remdesivir (GS-5734) and ODBG-P-RVn against the human coronavirus 229E in MRC-5 cells. Cells were infected with 229E for 2 h followed by treatment with the indicated dose of the indicated drug for 72 h. The relative CPE was determined by measuring cell viability using an MTT assay. (B) Cytotoxicity in MRC-5 cells incubated in the presence of the indicated drug at the indicated concentration for 72 h, after which cell viability was measured by the MTT assay. Data points indicate the averages from 3 independent experiments performed in duplicates. Error bars represent the standard errors of the means (SEMs).
Orally administered ODBG-P-RVn achieves therapeutic plasma levels in Syrian hamsters.
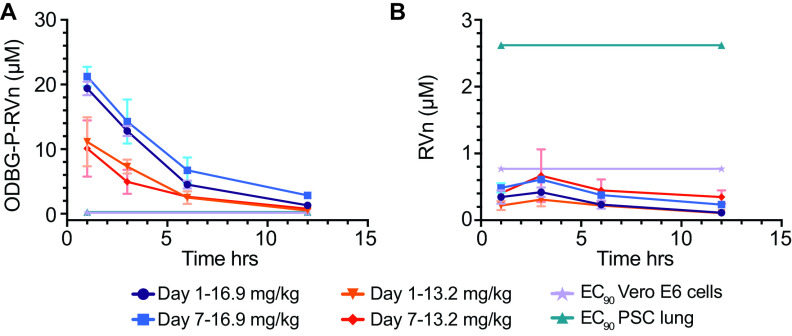
ODBG-P-RVn administered to Syrian hamsters by oral gavage every 12 h for 7 days was well tolerated, and no adverse clinical signs were noted. Peak plasma levels of ODBG-P-RVn were noted at 1 h and fell by 50% in approximately 5 h (Fig. 7A). Plasma curves were generally similar at days 1 and 7, except at 16.9 mg/kg body weight, the 7-day values were slightly higher than the levels at day 1. At 12 h, ODBG-P-RVn levels were higher than the EC90 for ODBG-P-RVn in all cell lines studied, including Vero E6 cells and PSC-lung cells on both days 1 and 7 (Fig. 7A). Levels of RVn, the nucleoside metabolite of ODBG-P-RVn, peaked at 3 h after administration and declined thereafter (Fig. 7B). Plasma levels of RVn were less than the EC90 for RVn in both PSC-lung cells and Vero E6 cells (Fig. 7B). The observed low levels of RVn suggest that antiviral activity attributable to this metabolite will be minimal and are also consistent with finding of ODBG-P-RVn ex vivo stability in human plasma (Fig. 3). Collectively, these results suggest that ODBG-P-RVn will be effective in suppressing viral replication in a variety of tissue types in vivo.
FIG 7.
Seven-day oral pharmacokinetics in Syrian hamsters. Syrian hamsters were given vehicle or ODBG-P-RVn by oral gavage every 12 h for 7 days. Groups of 3 animals received vehicle or drug at doses of 16.9 and 13.2 mg/kg body weight. Animals were weighed daily and monitored for clinical signs. Plasma samples were obtained at 1, 3, 6, and 12 h on day 1 and day 7 and frozen for analysis of ODBG-P-RVn (A) and RVn (B) by LC-MS/MS. The EC90 values in Vero E6 and PSC-lung cells for ODBG-P-RVn (A) and RVn (B) are shown.
DISCUSSION
RDV is a prodrug designed to bypass the first phosphorylation of the remdesivir nucleoside (RVn), which may be rate limiting in the synthesis of RVn triphosphate, the active metabolite. This occurs by the successive action of carboxylesterases, cathepsin A, and phosphoamidases (16, 27). However, this approach does not appear to provide any benefit in Vero E6 cells, a monkey kidney cell line, as shown by Pruijssers et al. (28) and by our results showing the antiviral activity of RVn is greater than that of RDV. Other perceived disadvantages of RDV include a lack of oral bioavailability, a difficult synthesis, instability in plasma, inadequate delivery to lung, and hepatotoxicity (13, 16). In patients with COVID-19 and in the Syrian hamster model of SARS-CoV-2 disease, while high viral loads are notably present in the nasal turbinate, trachea, and lung, as the infection proceeds, many other tissues also become infected, including the intestine, heart, liver, spleen, kidney, brain, lymph nodes, and vascular endothelium (29–33). However, RDV antiviral activity varies widely in lung and kidney cell lines, with EC50 values of 1.65 μM in Vero E6 cells, 0.28 μM in Calu-3 2B4 cells, and 0.010 μM in human alveolar epithelial (HAE) cells, a 165-fold difference (28). We found similar results in Vero E6 (EC50, 1.13 μM) and Calu-3 (0.23 μM) cells. It has been suggested that this is due to variable amounts of the enzymes which convert RDV to RVn (13, 16). The antiviral activity of ODBG-P-RVn was consistently high in all five cell types we tested (Fig. 5A).
We chose to design prodrugs of RVn which could provide oral bioavailability, because an effective oral drug would allow for much earlier treatment of persons diagnosed with SARS-CoV-2 infection, when active viral replication is believed to be the key driver of the subsequent course of the illness. We accomplished this by constructing liponucleotides of RVn resembling lysophospholipids that are normally highly absorbed intact in the gastrointestinal (GI) tract (34, 35). Liponucleotides of this type are not metabolized in plasma and gain rapid entry to the cell, often exhibiting greatly increased antiviral activity (36–39). In contrast to the activation of RDV, which requires four transformations, intracellular kinase bypass with ODBG-P-RVn generates the RVn monophosphate directly when the lipid ester moiety is cleaved in a single reaction catalyzed by acid phospholipase C (40, 41) or acid sphingomyelinase (sphingomyelin phosphodiesterase I) (K. Sandhoff and K. Y. Hostetler, unpublished). ODBG-P-RVn is likely to deliver relatively more drug to the lung and less to the liver, as shown previously in lethal mousepox infection, because of its apparent route to circulation via intestinal lymph rather that the portal vein (23, 24). Finally, the synthesis of these lipid prodrugs is much simpler than that of RDV and is readily scalable.
The limitations of RDV, including a lack of oral bioavailability, a difficult synthesis, instability in plasma with rapid conversion to RVn, a less active metabolite, inadequate delivery to lung, and dose-limiting hepatotoxicity, provide opportunities to improve its clinical utility. As reported here, we synthesized three lipid prodrugs of RVn which were highly active in five cell types infected with SARS-CoV-2. The most active compound, ODBG-P-RVn, is 8 times more active than RDV in Vero E6 cells, and the activity is equivalent to that of RDV in four other cell types. The cytotoxicity of ODBG-P-RVn is generally lower than that of RDV, is more consistent across 5 cell lines, and is not selectively higher in Huh7.5 cells, a hepatocyte cell line (Fig. 5). Selectivity indexes are excellent and range from 295 to 699 in the five cell lines studied. ODBG-P-RVn achieved therapeutic levels in Syrian hamsters by twice daily oral gavage and was well tolerated over a 7-day period of administration. Collectively, our data support the development of lipid prodrugs of RVn as potent oral antivirals that could be used orally early in the course of COVID-19 to prevent serious disease requiring hospitalization.
MATERIALS AND METHODS
Chemistry.
All reagents were of commercial quality and used without further purification unless indicated otherwise. Chromatographic purification was performed using the flash method with silica gel 60 (EMD Chemicals, Inc.; 230 to 400 mesh). 1H, 13C, and 31P nuclear magnetic resonance (NMR) spectra were recorded on either a Varian VX-500 or a Varian HG-400 spectrometer and are reported in units of parts per million relative to internal tetramethylsilane at 0.00 ppm. Electrospray ionization mass spectra (ESI-MS) were recorded on a Finnigan LCQDECA mass spectrometer at the small-molecule facility in the Department of Chemistry at University of California, San Diego. Purity of the target compounds was characterized by high-performance liquid chromatography (HPLC) using a Beckman Coulter System Gold chromatography system. The analytical column was Phenomenex Synergi Polar-RP (4.6 by 150 mm) equipped with a SecurityGuard protection column. Mobile phase A was 95% water-5% methanol, and mobile phase B was 95% methanol-5% water. At a flow rate of 0.8 ml/min, gradient elution was as follows: 10% B (0 to 3 min), 10% to 95% B (3 to 20 min), 95% B (20 to 25 min), and 95% to 10% B (25 to 34 min). Compounds were detected by UV light absorption at 274 nm. Purity of compounds was also assessed by thin-layer chromatography (TLC) using Analtech silica gel-GF (250 μm) plates and the following solvent system: CHCl3-methanol [MeOH]-concentrated NH4OH-H2O (70:30:3:3 [vol/vol/vol/vol]). TLC results were visualized with UV light, Phospray (Supelco, Bellefonte, PA, USA), and charring at 400°C.
Compounds.
Remdesivir (GS-5734) and remdesivir nucleoside (GS-441524) were purchased from AA Blocks (San Diego, CA) and Mason-Chem (Palo Alto, CA), respectively. 1-O-Octadecyl-2-O-benzyl-sn-glycerol was obtained from Bachem (Torrance, CA).
Synthesis of HDP-P-RVn: compound 5a {(2R,3S,4R,5R)-5-(4-aminopyrrolo[2,1-f][1, 2, 4]triazin-7-yl)-5-cyano-3,4-dihydroxytetrahydrofuran-2-yl}methyl [3-(hexadecyloxy)propyl] hydrogen phosphate.
N,N-Dicyclohexylcarbodiimide (DCC; 619 mg, 3 mmol) was added to a mixture of compound 2a (300 mg, 0.91 mmol, prepared as described by Warren et al. [19]), HDP phosphate (compound 4a; 414 mg, 1.10 mmol, prepared as described by Kim et al. [42]), and 4-dimethylaminopyridine (DMAP; 122 mg, 1.0 mmol) in 25 ml of dry pyridine, and then the mixture was heated to 90°C and stirred for 24 h. Pyridine was then evaporated, and the residue was purified by flash column chromatography on silica gel 60. Gradient elution (CH2Cl2-methanol, 10% to 20%) afforded 423 mg (67% yield) of 2′,3′-isopropylidene derivative of compound 5a. 1H NMR: (500 MHz, chloroform-d) δ 8.42 (s, 1H), 7.98 (s, 1H), 7.70 (s, 2H), 6.22 (d, J = 6.0 Hz, 1H), 5.68 (d, J = 6.2 Hz, 1H), 5.15 (d, J = 1.0 Hz, 1H), 4.70 (dd, J = 3.8, 0.9 Hz, 1H), 4.48 to 4.42 (m, 1H), 4.26 (ddd, J = 11.2, 8.5, 2.6 Hz, 1H), 4.15 (ddd, J = 11.1, 8.5, 2.6 Hz, 1H), 4.02 (dt, J = 8.5, 6.3 Hz, 2H), 3.49 (t, J = 6.1 Hz, 2H), 3.40 (t, J = 6.1 Hz, 2H), 1.95 (p, J = 6.2 Hz, 2H), 1.54 (tt, J = 7.4, 6.1 Hz, 2H), 1.31 (s, 3H), 1.32 to 1.24 (m, 26H), 0.94 to 0.85 (m, 3H); ESI MS, 691.6 [M − H]−.
Concentrated HCl (0.1 ml) in tetrahydrofuran (THF) was added to a stirred solution of 2′,3′-isopropylidene-5a (100 mg, 0.14 mmol) in THF (10 ml) at room temperature. The mixture was stirred for 3 h, and then sodium bicarbonate (50 mg) and water (2 ml) were added. After stirring an additional 15 min, the solvents were evaporated, and cold water (10 ml) was added to the residue. The solid product was collected by vacuum filtration and dried under vacuum to yield compound 5a (79 mg, 87% yield) as an off-white solid. 1H NMR: (500 MHz, CDCl3-methanol-d4) δ ppm 1H NMR (500 MHz, chloroform-d) δ 8.42 (s, 1H), 7.98 (s, 1H), 7.70 (s, 1H), 6.22 (d, J = 6.0 Hz, 1H), 5.70 (d, J = 6.0 Hz, 1H), 5.12 (d, J = 4.2 Hz, 1H), 4.55 (ddd, J = 5.5, 2.7, 0.9 Hz, 1H), 4.40 (dtd, J = 6.8, 2.6, 0.8 Hz, 1H), 4.33 to 4.27 (m, 2H), 4.25 (ddd, J = 11.1, 8.4, 2.6 Hz, 1H), 4.16 (ddd, J = 11.3, 8.5, 2.6 Hz, 1H), 4.02 (dt, J = 8.5, 6.3 Hz, 2H), 3.49 (t, J = 6.1 Hz, 2H), 3.40 (t, J = 6.1 Hz, 2H), 1.95 (p, J = 6.2 Hz, 2H), 1.59 to 1.50 (m, 1H), 1.34 to 1.24 (m, 23H), 0.94 to 0.85 (m, 3H); ESI MS, 652.39 [M − H]−; purity by HPLC, 99.7%.
Synthesis of ODE-P-RVn: compound 5b {(2R,3S,4R,5R)-5-(4-aminopyrrolo[2,1-f][1, 2, 4]triazin-7-yl)-5-cyano-3,4-dihydroxytetrahydrofuran-2-yl}methyl [2-(octadecyloxy)ethyl] hydrogen phosphate.
N,N-Dicyclohexylcarbodiimide (DCC; 0.3 g, 1.4 mmol) was added to a mixture of compound 2a (0.23 g, 0.7 mmol), ODE phosphate (compound 4b; 0.27 g, 0.68 mmol), and 4-dimethylaminopyridine (DMAP; 0.07 g, 0.6 mmol) in 10 ml of dry pyridine, and then the mixture was heated to 90°C and stirred for 3 days. Pyridine was then evaporated, and the residue was purified by flash column chromatography on silica gel 60. Gradient elution (CH2Cl2-methanol, 10% to 20%) afforded 0.22 g (45% yield) of 2′,3′-isopropylidene-5b. Concentrated HCl (0.3 ml) was added slowly to a stirred solution of 2′,3′-isopropylidene-5b (0.2 g, 0.28 mmol) in tetrahydrofuran (2 ml) at 0°C. The mixture was allowed to warm to room temperature overnight and then was diluted with water (2 ml) and adjusted to pH 8 by adding saturated sodium bicarbonate. The product was extracted with chloroform (3 × 30 ml), and the organic layer was concentrated under reduced pressure. The residue was purified by flash chromatography on silica gel. Elution with 20% MeOH-CH2Cl2 gave 0.10 g (55% yield) of compound 5b. 1H NMR: (400 MHz, CDCl3-methanol-d4) δ ppm 7.89 (s, 1 H), 6.94 (d, J = 4.65 Hz, 1H), 6.89 (d, J = 4.65 Hz, 1H), 4.40 (d, J = 4.65 Hz, 2H), 4.21 to 4.28 (m, 1H), 4.12 to 4.20 (m, 1H), 4.04 to 4.12 (m, 1H), 3.91 (d, J = 4.89 Hz, 2H), 3.46 to 3.57 (m, 2H), 3.42 (td, J = 6.85, 1.96 Hz, 2H), 3.34 (dt, J = 3.18, 1.59 Hz, 2H), 1.53 (d, J = 6.85 Hz, 2H), 1.20 to 1.37 (m, 30H), 0.89 (t, J = 6.97 Hz, 3H); ESI MS, 666.43 [M − H]−; purity by HPLC, 98.4%.
Synthesis of ODBG-P-RVn: compound 5c {(2R,3S,4R,5R)-5-(4-aminopyrrolo[2,1-f][1, 2, 4]triazin-7-yl)-5-cyano-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(R)-2-(benzyloxy)-3-(octadecyloxy)propyl] hydrogen phosphate.
N,N-Dicyclohexylcarbodiimide (DCC; 310 mg, 1.5 mmol) was added to a mixture of compound 2a (300 mg, 0.91 mmol), ODBG phosphate (compound 4c; 515 mg, 1.0 mmol), and 4-dimethylaminopyridine (DMAP; 122 mg, 1.0 mmol) in 25 ml of dry pyridine, and then the mixture was heated to 90°C and stirred for 24 h. Pyridine was then evaporated, and the residue was purified by flash column chromatography on silica gel 60. Gradient elution (CH2Cl2-methanol, 10% to 20%) afforded 210 mg (28% yield) of compound 2′,3′-isopropylidene-5c; ESI MS, 826.58 [M−H]−. Concentrated HCl (0.1 ml) in tetrahydrofuran (THF) was added to a stirred solution of 2′,3′-isopropylidene-5c (210 mg, 0.25 mmol) in THF (10 ml) at room temperature. The mixture was stirred for 3 h, and then sodium bicarbonate (50 mg) and water (2 ml) were added. After stirring an additional 15 min, the solvents were evaporated, and cold water (10 ml) was added to the residue. The solid product was collected by vacuum filtration and dried under vacuum to yield compound 5c (71 mg, 36% yield) as an off-white solid. 1H NMR: (500 MHz, DMSO-d6) δ 7.89 (s, 1H), 7.79 (s, 1H), 7.33 to 7.24 (m, 4H), 7.22 (ddd, J = 8.7, 5.3, 2.5 Hz, 1H), 6.88 (d, J = 4.5 Hz, 1H), 6.80 (d, J = 4.6 Hz, 1H), 6.14 (d, J = 5.2 Hz, 1H), 5.91 (s, 1H), 4.55 (q, J = 12.1, 12.1, 12.1 Hz, 3H), 4.10 (dt, J = 6.7, 4.3, 4.3 Hz, 1H), 3.93 (t, J = 5.9, 5.9 Hz, 1H), 3.79 (dddd, J = 28.2, 12.1, 7.9, 4.4 Hz, 2H), 3.62 (tdd, J = 10.9, 10.9, 8.2, 5.1 Hz, 4H), 3.43 (dd, J = 10.6, 3.5 Hz, 2H), 1.43 (p, J = 6.6, 6.6, 6.6, 6.6 Hz, 2H), 1.21 (d, J = 8.3 Hz, 30H), 0.83 (t, J = 7.0, 7.0 Hz, 3H); ESI MS, 786.48 [M−H]−; purity by HPLC, 97.6%.
Cells.
Vero E6, Caco-2, and Calu-3 cell lines were obtained from ATCC. Huh7.5 cells were obtained from Apath LLC. Calu-3 and Caco-2 cells were propagated in minimal essential medium (MEM) (Corning) with 10% fetal bovine serum (FBS) and penicillin-streptomycin (Gibco). Vero E6 and Huh7.5 cells were propagated in Dulbecco’s minimal essential medium (DMEM; Corning) with 10% FBS and penicillin-streptomycin (Gibco). For human PSC-lung cell generation, human lung organoids were generated as previously described (43). H9 embryonic stem cells (WiCell) were cultured under feeder-free conditions upon Matrigel (Corning number 354230)-coated plates in mTeSR medium (STEMCELL Technologies number 85850). Medium was changed daily, and stem cells were passaged using enzyme-free dissociation reagent ReLeSR (STEMCELL Technologies number 05872). Cultures were maintained in an undifferentiated state, in a 5% CO2 incubator at 37°C. For proximal lung organoid generation, human PSCs were dissociated into single cells and then seeded on Matrigel-coated plates (BD Biosciences) at a density of 5.3 × 104 cells/cm2 in definitive endoderm (DE) induction medium (RPMI 1640 medium, 2% B27 supplement, 1% HEPES, 1% GlutaMAX, 50 U/ml penicillin-streptomycin) supplemented with 100 ng/ml human activin A (R&D Systems), 5 μM CHIR99021 (Stemgent), and 10 μM ROCK inhibitor, Y-27632 (R&D Systems) on day 1. On days 2 and 3, cells were cultured in DE induction medium with only 100 ng/ml human activin A. Anterior foregut endoderm (AFE) was generated by supplementing serum-free basal medium (3 parts Iscove’s modified Dulbecco’s medium [IMDM] to 1 part F12, B27 plus N2 supplements, 50 U/ml penicillin-streptomycin, 0.25% bovine serum albumin [BSA], 0.05 mg/ml l-ascorbic acid, and 0.4 mM monothioglycerol) with 10 μM SB431542 (R&D Systems) and 2 μM dorsomorphin (Stemgent) on days 4 to 6. On day 7, AFE medium was changed to lung progenitor cell (LPC) induction medium, containing serum-free basal medium supplemented with 10 ng/ml human recombinant BMP4 (R&D Systems), 0.1 μM all-trans retinoic acid (Sigma-Aldrich), and 3 μM CHIR99021. Medium was changed every other day for 9 to 11 days. To generate three-dimensional (3D) human proximal lung organoids, we modified a previously published protocol (44). LPCs were dissociated in Accutase for 10 min and resuspended in Matrigel in a 12-well, 0.4-μm-pore-size Transwell (Corning) culture insert at 5.0 × 104 cells/200 μl of Matrigel. Cells were cultured in proximal lung organoid maturation medium using serum-free basal medium supplemented with 250 ng/ml basic fibroblast growth factor (FGF2), 100 ng/ml recombinant human fibroblast growth factor 10 (rhFGF10), 50 nM dexamethasone (Dex), 100 μM 8-bromoadenosine 3′,5′-cyclic monophosphate sodium salt (Br-cAMP), 100 μM 3-isobutyl-1-methylxanthine (IBMX), and 10 μM ROCK inhibitor (Y-27632). Proximal lung organoid medium was changed every other day for 3 weeks. Human PSC-derived lung organoids were dissociated into single cells and seeded at 20,000 cells per well of a Matrigel-coated 96-well plate 1 day before transfection. Transwells containing the proximal organoids in Matrigel were incubated in 2 U/ml dispase for 30 min at 37°C. Cold phosphate-buffered saline (PBS) was added to the mixture and then centrifuged at 400 × g for 5 min. Supernatant was carefully removed and resuspended in 2 to 3 ml of TrypLE Express (Gibco number 12605010) for 20 min at 37°C. The reaction was quenched with 2% FBS in DMEM/F12 and then centrifuged at 400 × g for 5 min. The supernatant was aspirated, and the cell pellet was resuspended in 1 ml of quenching medium supplemented with 10 μM ROCK inhibitor (Y-27632). A cell count was performed, and the respective volumes of cells were transferred into a reagent reservoir trough, resuspended in proximal lung organoid maturation medium, and plated via multichannel pipette into 96-well plates at 100 μl per well as monolayers.
SARS-CoV-2 infection.
SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources) was propagated and infectious units were quantified by plaque assay by using Vero E6 (ATCC) cells. Approximately 12,000 cells from each cell line were seeded per well in a 96-well plate. Vero E6 and Huh7.5 cells were seeded approximately 24 h prior to treatment/infection. Calu-3 and Caco-2 cells were seeded approximately 48 h prior to treatment/infection. Human PSC-lung cell infections and cytotoxicity experiments were performed when cells reached 100% confluence. Compounds or controls were added at the concentrations indicated in the figures and text 30 min prior to infection, followed by the addition of SARS-CoV-2 at a multiplicity of infection equal to 0.01. After incubation for 48 h at 37°C and 5% CO2, cells were washed twice with PBS and lysed in 200 μl TRIzol (Thermo Fisher). All work with SARS-CoV-2 was conducted under biosafety level 3 conditions at the University of California San Diego with approval from the Institutional Biosafety Committee.
Human coronavirus 229E infection.
Human coronavirus 229E (ATCC) was propagated and infectious units were quantified by 50% tissue culture infective dose (TCID50) using MRC-5 cells. For antiviral testing, approximately 104 MRC-5 cells were seeded per well in Eagle’s minimal essential medium (EMEM) (10% fetal calf serum [FCS]) at 37°C in a 96-well plate overnight. Medium from each well was removed, and cells were infected with 100 TCID50 virus in 100 μl medium for 2 h. Cells were washed one time with medium, and then compounds or controls were added at the concentrations indicated in the text and figures. After 3 days, CPE was observed under a microscope and quantified using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) cell proliferation assay kit (Abcam) and read on an ELx800 universal microplate reader (BioTek Instruments, Inc.). The percent inhibition was calculated as (Atv − Acv)/(Acd − Acv) × 100, where Atv indicates the absorbance of the test compounds with virus-infected cells, and Acv and Acd indicate the absorbance of the virus control and the absorbance of the cell control, respectively. The average half-maximal effective concentration (EC50) was defined as the concentration which achieved 50% inhibition of virus-induced cytopathic effects.
RNA extraction, cDNA synthesis, and qPCR.
RNA was purified from TRIzol lysates using Direct-zol RNA microprep kits (Zymo Research) according to manufacturer’s recommendations that included DNase treatment. RNA was converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad), and qPCR was performed using iTaq universal SYBR green supermix (Bio-Rad) and an ABI 7300 real-time PCR system. cDNA was amplified using the following primers: RPLP0 F, GTGTTCGACAATGGCAGCAT; RPLP0 R, GACACCCTCCAGGAAGCGA; SARS-CoV-2 spike F, CCTACTAAATTAAATGATCTCTGCTTTACT; SARS-CoV-2 spike R, CAAGCTATAACGCAGCCTGTA. Relative expression of SARS-CoV-2 spike RNA was calculated by the comparative threshold cycle (ΔΔCT) method by first normalizing to the housekeeping gene RPLP0 and then comparing to SARS-CoV-2-infected Vero E6 cells that were untreated (reference control). Curves were fit using the nonlinear regression minus log(inhibitor) versus response (four parameter) model using Prism 9. To calculate effective concentrations (EC50 and EC90 values), qRT-PCR values were normalized to percent inhibition and curves were fit using the nonlinear regression minus log(agonist) versus response (four parameter) model with the bottom and top constrained to 0 and 100, respectively, using Prism 9.
Cell viability assay.
Cell types were seeded as per SARS-CoV-2 infection studies in opaque-walled 96-well cell culture plates or as per 229E infection studies in clear 96-well cell culture plates and incubated overnight. Compounds or controls were added at the concentrations indicated in the text and figures. For SARS-CoV-2-related studies, cells were incubated for 48.5 h at 37°C and 5% CO2; then, equal volumes of CellTiter-Glo reagent (catalog number G7570; Promega, Madison, WI) were added and mixed, and luminescence was recorded on a Veritas microplate luminometer (Turner BioSystems) according to the manufacturer’s recommendations. For 229E-related studies, cells were incubated for 72 h at 37°C and 5% CO2; then, supernatants were removed and 50 μl of serum-free medium and 50 μl of MTT reagent (Abcam ab211091) were added to each well and incubated for 3 h at 37°C. Absorbance was measured on an ELx800 universal microplate reader (BioTek Instruments, Inc.) according to the manufacturer’s recommendations. Percent viability was calculated compared to that of untreated controls, and CC50 values were calculated using Prism 9.
Ex vivo stability in human plasma.
Pooled mixed-gender human plasma samples were obtained from BioIVT (Westbury, NY). ODBG-P-RVn was incubated in human plasma with K2EDTA or sodium heparin as the anticoagulant at a final concentration of 2.00 μg/ml. After incubation at 37°C, duplicate samples of the test article incubations were taken at 0 (preincubation), 0.5, 1, 2, 4, 8, and 24 h and immediately frozen at −70°C. The extracts were prepared by a solid-phase extraction using a Waters Sep Pak tC18 25 mg SPE plate and analyzed as described below.
Analytical methods.
(i) ODBG-P-RVn.
Hamster plasma samples (10 μl) containing ODBG-P-RVn and K2EDTA as the anticoagulant were added to polypropylene tubes containing water (100 μl), internal standard solution (10 μl of 1,000 ng/ml of ODE-P-RVn in acetonitrile [ACN]-dimethylformamide [DMF], 1:1 [vol/vol]), and 10 μl of ACN-DMF (1:1 [vol/vol]). The solutions were mixed and then acidified with phosphoric acid (85% [wt/vol water], 1:19 [vol/vol]; 10 μl) and mixed; then, the solutions were diluted with 200 μl of isopropyl alcohol, mixed, and then diluted with 500 μl of water and mixed. The samples were extracted with a Sep-Pak tC18 96-well solid-phase extraction plate (25 mg; Waters, Milford, MA). Extraction occurred under positive pressure conditions using nitrogen. Samples were washed serially with 1 ml of water-acetonitrile-formic acid (475:25:0.5 [vol/vol/vol]) and 0.4 ml of water-acetonitrile-formic acid (350:150:0.5 [vol/vol/vol]) before being serially eluted with 100 μl and 150 μl of water and acetonitrile-isopropyl alcohol (1:1 [vol/vol])-formic acid-ammonium formate-citric acid solution, 2% [wt/vol] (15:85:0.1:0.1:0.1 [vol/vol/vol/wt/vol]). The citric acid solution was prepared as water-citric acid monohydrate (20:0.4 [vol/wt]). After elution, 100 μl of water was added to each sample. The ODBG-P-RVn extracts were analyzed using an Agilent 1200 HPLC system (Agilent, Santa Clara, CA) coupled to an API5500 mass analyzer (SCIEX, Foster City, CA). Analytes were chromatographically separated using a Dacapo DX-C18 MF column (100 by 2 mm, 2.5 μm; Imtakt USA, Portland, OR) using a mobile phase system consisting of mobile phase A (water-formic acid-[water-ammonium formate-citric acid; 25:5:0.5 {vol/wt/wt}], 1,000:1:1 [vol/vol/vol]) and mobile phase B (acetonitrile-isopropyl alcohol-formic acid-[water-ammonium formate-citric acid; 25:5:0.5 {vol/wt/wt}], 800:200:1:1 [vol/vol/vol/vol]). The total analytical run time was 4.5 min. The mobile phase was nebulized using heated nitrogen in a Turbo-V source/interface set to electrospray positive ionization mode. The ionized compounds were detected using multiple-reaction monitoring with transitions m/z 788.4 > 229 (V2043) and 668.4 > 467.2 (V2041). This method is applicable for measuring ODBG-P-RVn concentrations ranging from 6.25 to 3,000 ng/ml using 10.0 μl of plasma for extraction. The peak areas of ODBG-P-RVn and RVn were acquired using Analyst v. 1.6.2 (SCIEX, Framingham, MA). The calibration curve was obtained by fitting the peak area ratios of the analyte/internal standard (IS) and the standard concentrations to a linear equation with 1/x2 weighting, using Analyst. The equation of the calibration curve was then used to interpolate the concentrations of the analyte in the samples using their peak area ratios. The peak areas used for the calculations were not rounded.
(ii) RVn (GS-441524).
Hamster plasma samples (20 μl) containing GS-441524 and K2EDTA as the anticoagulant were added to Eppendorf LoBind microcentrifuge tubes containing acetonitrile (300 μl) and water-acetonitrile (2:8 [vol/vol]; 60 μl). The solutions were mixed and centrifuged at 16,000 × g for 5 min. The supernatant (300 μl) was then filtered through an Ostro protein precipitation and phospholipid removal plate (25 mg; Waters, Milford, MA). Filtration occurred under positive pressure conditions using nitrogen. Collected filtered samples were capped, mixed, and stored at 10°C pending analysis. The GS-441524 extracts were analyzed using an Acquity ultraperformance liquid chromatography (UPLC) system (Waters) coupled to a G2-S quadrupole time of flight (QTOF) mass analyzer (Waters). Analytes were chromatographically separated using a Unison-UK Amino HT column (100 by 2 mm, 3 μm; Imtakt USA, Portland, OR) using a mobile phase system consisting of mobile phase A (0.008% ammonium hydroxide, 0.012% acetic acid in water [vol/vol/vol]) and mobile phase B (0.008% ammonium hydroxide, 0.012% acetic acid in acetonitrile [vol/vol/vol]). The total analytical run time was 12.5 min. The mobile phase was nebulized using heated nitrogen in a Z-spray source/interface set to electrospray positive ionization mode. The ionized compounds were detected using TOF MS scan monitoring in sensitivity mode scanning from 50.0 to 700 m/z. This method is applicable for measuring GS-441524 concentrations ranging from 1.00 to 1,000 ng/ml using 20.0 μl of plasma for extraction. The peak areas of GS-441524 were acquired using MassLynx V4.2 (Waters). The calibration curve was obtained by fitting the peak area ratios of the analyte and the standard concentrations to a linear equation with 1/x2 weighting using MassLynx. The equation of the calibration curve was then used to interpolate the concentrations of the analyte in the samples using their peak areas. The peak areas used for the calculations were not rounded.
ACKNOWLEDGMENTS
This research was supported by NIAID grant R01 AI131424 and the San Diego Center for AIDS Research as well as an NIH grant (K08 AI130381) and Career Award for Medical Scientists from the Burroughs Welcome Fund to A.F.C., and by CIRM (DISC2COVID19-12022) to S.L.L.
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52281.
Footnotes
Supplemental material is available online only.
Contributor Information
Robert T. Schooley, Email: rschooley@health.ucsd.edu.
Karl Y. Hostetler, Email: khostetler@health.ucsd.edu.
REFERENCES
- 1.Zhong NS, Zheng BJ, Li YM, Poon Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris JSM, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362:1353–1358. 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, Xiong Y, Cheng Z, Gao S, Liang K, Luo M, Chen T, Song S, Ma Z, Chen X, Zheng R, Cao Q, Wang F, Zhang Y. 16March2020. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Center for Systems Science and Engineering of the Johns Hopkins University. 2020. COVID-19 dashboard. https://coronavirus.jhu.edu/map.html. Accessed 1 June 2021.
- 7.To KK, Chan WM, Ip JD, Chu AW, Tam AR, Liu R, Wu AK, Lung KC, Tsang OT, Lau DP, To WK, Kwan MY, Yau YS, Ng AC, Yip CC, Chan KH, Tse H, Hung IF, Yuen KY. 2020. Unique SARS-CoV-2 clusters causing a large COVID-19 outbreak in Hong Kong. Clin Infect Dis 73:137–142. 10.1093/cid/ciaa1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, COVE Study Group. 2021. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, FrenckRW, Jr, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group. 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. 2020. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 295:6785–6797. 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan VC, Muller FL. 2020. Advantages of the parent nucleoside GS-441524 over remdesivir for Covid-19 treatment. ACS Med Chem Lett 11:1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh MD, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group Members. 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383:1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. 2020. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem 295:4773–4779. 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan V, Muller F. 29August2020. Comprehensive summary supporting clinical investigation of GS-441524 for Covid-19 treatment. OSFPreprints 10.31219/osf.io/mnhxu. [DOI] [Google Scholar]
- 17.de Wit E, Feldmann F, Cronin J, Jordan R, Okumura A, Thomas T, Scott D, Cihlar T, Feldmann H. 2020. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci USA 117:6771–6776. 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson BN, Feldmann F, Schwarz B, Meade-White K, Porter DP, Schulz J, van Doremalen N, Leighton I, Yinda CK, Pérez-Pérez L, Okumura A, Lovaglio J, Hanley PW, Saturday G, Bosio CM, Anzick S, Barbian K, Cihlar T, Martens C, Scott DP, Munster VJ, de Wit E. 2020. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 585:273–276. 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, et al. 2016. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531:381–385. 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tempestilli M, Caputi P, Avataneo V, Notari S, Forini O, Scorzolini L, Marchioni L, Ascoli Bartoli T, Castilletti C, Lalle E, Capobianchi MR, Nicastri E, D'Avolio A, Ippolito G, Agrati C, COVID 19 INMI Study Group. 2020. Pharmacokinetics of remdesivir and GS-441524 in two critically ill patients who recovered from COVID-19. J Antimicrob Chemother 75:2977–2980. 10.1093/jac/dkaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hostetler KY, Beadle JR, Kini GD, Gardner MF, Wright KN, Wu TH, Korba BA. 1997. Enhanced oral absorption and antiviral activity of 1-O-octadecyl-sn-glycero-3-phospho-acyclovir and related compounds in hepatitis B virus infection, in vitro. Biochem Pharmacol 53:1815–1822. 10.1016/S0006-2952(97)82446-X. [DOI] [PubMed] [Google Scholar]
- 22.Hostetler KY, Beadle JR, Hornbuckle WE, Bellezza CA, Tochkov IA, Cote PJ, Gerin JL, Korba BE, Tennant BC. 2000. Antiviral activities of oral 1-O-hexadecylpropanediol-3-phosphoacyclovir and acyclovir in woodchucks with chronic woodchuck hepatitis virus infection. Antimicrob Agents Chemother 44:1964–1969. 10.1128/AAC.44.7.1964-1969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buller RM, Owens G, Schriewer J, Melman L, Beadle JR, Hostetler KY. 2004. Efficacy of oral active ether lipid analogs of cidofovir in a lethal mousepox model. Virology 318:474–481. 10.1016/j.virol.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Hostetler KY, Beadle JR, Trahan J, Aldern KA, Owens G, Schriewer J, Melman L, Buller RM. 2007. Oral 1-O-octadecyl-2-O-benzyl-sn-glycero-3-cidofovir targets the lung and is effective against a lethal respiratory challenge with ectromelia virus in mice. Antiviral Res 73:212–218. 10.1016/j.antiviral.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schooley RT, Carlin AF, Beadle JR, Valiaeva N, Zhang X-Q, Clark AE, McMillan RE, Leibel SL, McVicar RN, Xie J, Garretson AF, Smith VI, Murphy J, Hostetler KY. 7June2021. Rethinking remdesivir: synthesis, antiviral activity and pharmacokinetics of oral lipid prodrugs. BioRxiv 10.1101/2020.08.26.269159. [DOI] [PMC free article] [PubMed]
- 26.Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, Neville S, Carra E, Lew W, Ross B, Wang Q, Wolfe L, Jordan R, Soloveva V, Knox J, Perry J, Perron M, Stray KM, Barauskas O, Feng JY, Xu Y, Lee G, Rheingold AL, Ray AS, Bannister R, Strickley R, Swaminathan S, Lee WA, Bavari S, Cihlar T, Lo MK, Warren TK, Mackman RL. 2017. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem 60:1648–1661. 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 27.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, Hall MD. 2020. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci 6:672–683. 10.1021/acscentsci.0c00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, DinnonKH, III, Yount BL, Agostini ML, Stevens LJ, Chappell JD, Lu X, Hughes TM, Gully K, Martinez DR, Brown AJ, Graham RL, Perry JK, Du Pont V, Pitts J, Ma B, Babusis D, Murakami E, Feng JY, Bilello JP, Porter DP, Cihlar T, Baric RS, Denison MR, Sheahan TP. 2020. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 32:107940. 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen Z, Chen H, To KK, Yuen KY. 2020. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 71:2428–2446. 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schröder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. 2020. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 383:590–592. 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D. 2020. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med 383:120–128. 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. 2020. Extrapulmonary manifestations of COVID-19. Nat Med 26:1017–1032. 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anonymous. 2020. Progress report on the coronavirus pandemic. Nature 584:325. 10.1038/d41586-020-02414-1. [DOI] [PubMed] [Google Scholar]
- 34.Scow RO, Stein Y, Stein O. 1967. Incorporation of dietary lecithin and lysolecithin into lymph chylomicrons in the rat. J Biol Chem 242:4919–4924. 10.1016/S0021-9258(18)99456-1. [DOI] [PubMed] [Google Scholar]
- 35.Borgström B. 1974. Fat digestion and absorption. Biomembranes 4B:555–620. 10.1007/978-1-4684-3336-4_1. [DOI] [PubMed] [Google Scholar]
- 36.Hostetler KY. 2009. Alkoxyalkyl prodrugs of acyclic nucleoside phosphonates enhance oral antiviral activity and reduce toxicity: current state of the art. Antiviral Res 82:A84–A98. 10.1016/j.antiviral.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hostetler KY. 2010. Synthesis and early development of hexadecyloxypropyl-cidofovir: an oral antipoxvirus nucleoside phosphonate. Viruses 2:2213–2225. 10.3390/v2102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kern ER, Collins DJ, Wan WB, Beadle JR, Hostetler KY, Quenelle DC. 2004. Oral treatment of murine cytomegalovirus infections with ether lipid esters of cidofovir. Antimicrob Agents Chemother 48:3516–3522. 10.1128/AAC.48.9.3516-3522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartline CB, Gustin KM, Wan WB, Ciesla SL, Beadle JR, Hostetler KY, Kern ER. 2005. Ether lipid-ester prodrugs of acyclic nucleoside phosphonates: activity against adenovirus replication in vitro. J Infect Dis 191:396–399. 10.1086/426831. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzawa Y, Hostetler KY. 1980. Properties of phospholipase C isolated from rat liver lysosomes. J Biol Chem 255:646–652. 10.1016/S0021-9258(19)86225-7. [DOI] [PubMed] [Google Scholar]
- 41.Hostetler KY, Hall LB. 1980. Phospholipase C activity of rat tissues. Biochem Biophys Res Commun 96:388–393. 10.1016/0006-291X(80)91227-9. [DOI] [PubMed] [Google Scholar]
- 42.Kim JS, Beadle JR, Freeman WR, Hostetler KY, Hartmann K, Valiaeva N, Kozak I, Conner L, Trahan J, Aldern KA, Cheng L. 2012. A novel cytarabine crystalline lipid prodrug: hexadecyloxypropyl cytarabine 3′,5′-cyclic monophosphate for proliferative vitreoretinopathy. Mol Vis 18:1907–1917. [PMC free article] [PubMed] [Google Scholar]
- 43.Leibel SL, McVicar RN, Winquist AM, Niles WD, Snyder EY. 2020. Snyder EY Generation of complete multi-cell type lung organoids from human embryonic and patient-specific induced pluripotent stem cells for infectious disease modeling and therapeutics validation. Curr Protoc Stem Cell Biol 54:e118. 10.1002/cpsc.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, Kotton DN. 2017. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell 20:844–857. 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1. Download AAC.01155-21-s0001.pdf, PDF file, 496 KB (495.4KB, pdf)