ABSTRACT
Infections caused by antimicrobial-resistant bacterial pathogens are fast becoming an important global health issue. Strains of Escherichia coli are common causal agents of urinary tract infection and can carry multiple resistance genes. This includes the gene blaCTX-M-15, which encodes an extended-spectrum beta-lactamase (ESBL). While studying antimicrobial resistance (AMR) in the environment, we isolated several strains of E. coli ST131 downstream of a wastewater treatment plan (WWTP) in a local river. These isolates were surviving in the river sediment, and characterization proved that a multiresistant phenotype was evident. Here, we show that E. coli strain 48 (river isolate ST131) provided a protective effect against a third-generation cephalosporin (cefotaxime) for susceptible E. coli strain 33 (river isolate ST3576) through secretion of a functional ESBL into the growth medium. Furthermore, extracellular ESBL activity was stable for at least 24 h after secretion. Proteomic and molecular genetic analyses identified CTX-M-15 as the major secreted ESBL responsible for the observed protective effect. In contrast to previous studies, outer membrane vesicles (OMVs) were not the route for CTX-M-15 secretion. Indeed, mutation of the type I secretion system led to a significant reduction in the growth of the ESBL-producing strain as well as a significantly reduced ability to confer protective effect. We speculate that CTX-M-15 secretion, mediated through active secretion using molecular machinery, provides a public goods service by facilitating the survival of otherwise susceptible bacteria in the presence of cefotaxime.
KEYWORDS: Escherichia coli, antibiotic resistance genes, beta-lactamases, enzyme secretion, secretion systems
INTRODUCTION
Pathogenic strains of Escherichia coli producing CTX-M β-lactamases have recently emerged worldwide and now present the most common type of extended-spectrum β-lactamase (ESBL) enzymes in Enterobacteriaceae (1–5). Limited treatment is available for patients infected with these E. coli strains, which presents severe challenges to health care (1, 4, 6–9). The global emergence of CTX-M-producing E. coli is driven by the rapid dissemination of the gene blaCTX-M located on highly mobilizable elements, such as plasmids and transposons (2, 10). Over 172 variants of CTX-M have been identified, which cluster into five groups, CTX-M-1, -2, -8, -9, and -25 groups (11). blaCTX-M-15 belongs to group 1 and is the predominant variant in the human population globally, including the United Kingdom (2, 7).
Clinical studies have suggested that secretion of hydrolytic enzymes, such as β-lactamases, irreversibly inactivate antibiotics outside the cell and protect both the producer and otherwise susceptible bacteria in close proximity (12). One proposed mechanism for the secretion of ESBLs is the formation and likely stochastic release of outer membrane vesicles (OMVs), which are common in Gram-negative bacteria (13). This secretory process eliminates the need for bacterial contact or complex molecular architectures at the cell wall-periplasm interface typically required for long-distance dissemination of extracellular proteins (14). The packaging of β-lactamases into OMVs has been demonstrated in Pseudomonas aeruginosa by microscopy and enzymatic studies (15). In addition, the release of OMVs containing various antibiotic-related proteins from a drug-resistant E. coli isolate facilitated the survival of various susceptible bacteria in the presence of β-lactam antibiotics (16). However, in this study, the relative contribution of ESBLs compared to other antibiotic-related proteins, such as bacterial transporter systems, was not conclusively determined. Thus, the mechanism of CTX-M variants, such as CTX-M-15, in providing a protective effect remains uncertain.
In Gram-negative bacteria, secretion of extracellular enzymes is achieved through either a one- or two-step process. In the two-step process, initial translocation across the cytoplasmic membrane to the periplasm is achieved through two main pathways: the twin-arginine (Tat) or the general secretory (Sec) pathway. While the Tat pathway translocates a small number of folded proteins across the cytoplasmic membrane, the majority of proteins are translocated in an unfolded state using the Sec pathway (17). The second translocation event across the outer membrane is coordinated by various specialized export systems classified as type II and type V secretion systems (T2SS and T5SS, respectively). The one-step process, performed by type 1 (T1SS), type III (T3SS), type IV (T4SS), and type VI (T6SS) secretion systems, translocates proteins directly from the cytoplasm to the extracellular milieu, bypassing the periplasmic space (18, 19). The architecture of the T1SS is closely related to the secretion system of the multidrug efflux pumps, called resistance nodulation division (RND), which secretes most antibacterial molecules out of the cells, contributing to antibiotic resistance (20). In contrast, the T2SS, including components called the general secretion pathway (Gsp), ensures the transport of hydrolyzing enzymes and toxins (21–23). Secretion of extracellular enzymes is often thought of as a public goods service, as they can provide an auxiliary function to bacteria otherwise lacking a given phenotype, for example, the degradation of recalcitrant organic phosphorus or carbohydrate polymers (24–27).
In this study, our aim was to further investigate β-lactamase resistance in E. coli ST131 isolated from a UK river system downstream of a waste water treatment plant (WWTP) to establish the mechanism of enzyme secretion (28–33). We report that this strain provided a protective effect to a susceptible E. coli river isolate against cefotaxime. Further investigations demonstrated that CTX-M-15 was secreted and a role for T1SS was established.
RESULTS
Phenotypic and genotypic testing of the two isolates.
The resistance profile of two environmental E. coli strains, strains 33 and 48, isolated from a UK river system against five antibiotics were determined. Strain 33 showed no phenotypic resistance to any of the antibiotics tested; in contrast, strain 48 was resistant to three of the five tested antibiotics (see Table S4 in the supplemental material). Whole-genome sequencing of strains 33 and 48 revealed the presence of three β-lactamase genes of clinical relevance in strain 48 only, blaTEM-1, blaOXA-1, and blaCTX-M-15 with CTX-M, the main type of ESBLs. PCR and sequencing confirmed the presence of the three β-lactamase genes in strain 48.
Strain 48 constitutively expresses a secreted β-lactamase.
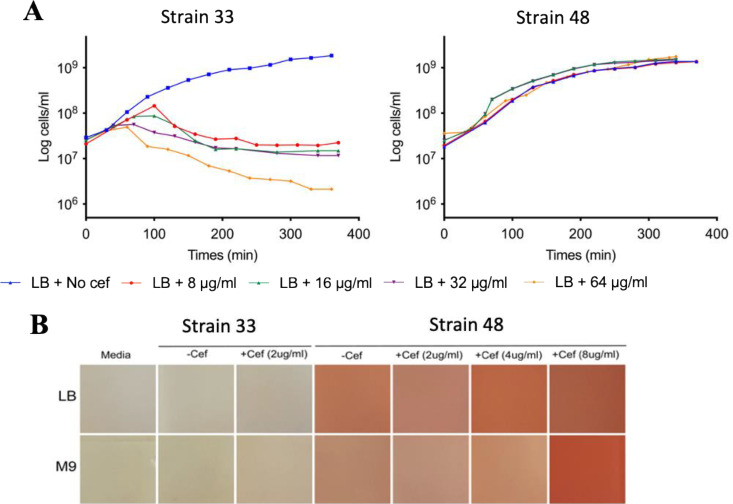
The two E. coli river isolates, 33 and 48, were challenged with cefotaxime (8 μg/ml, 16 μg/ml, 32 μg/ml, and 64 μg/ml); growth of strain 33 was inhibited by all cefotaxime concentrations, while strain 48 was completely resistant (Fig. 1A). Previous genomic analyses revealed strain 48 is predicted to possess three ESBLs, encoded by blaTEM-1, blaOXA-1, and blaCTX-M-15. Biochemical analyses revealed that strain 48 possessed extracellular β-lactamase activity in either the presence or absence of cefotaxime (Fig. 1B), although the greatest activity was observed during growth on the highest concentration of this antibiotic. As expected, E. coli strain 33, which was sensitive to the β-lactams, showed no secreted β-lactamase activity (Fig. 1B).
FIG 1.
Secretion of β-lactamases by strain 48. (A) Growth of E. coli strains 33 and 48 cultivated in LB medium in various cefotaxime (cef) concentrations, 8 μg/ml (in red), 16 μg/ml (in green), 32 μg/ml (in purple), and 64 μg/ml (in yellow). (B) Nitrocefin assay with various concentrations of cefotaxime (2 μg/ml, 4 μg/ml, and 8 μg/ml) in the presence of E. coli strain 33 or strain 48.
CTX-M-15 is responsible for conferring cefotaxime resistance.
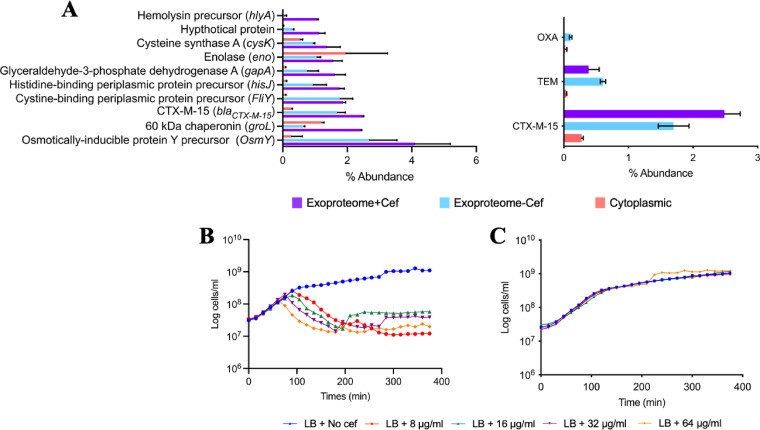
To identify which of the three ESBLs was responsible for β-lactamase activity in strain 48, the proteome of this bacterium, partitioned into cellular (CP) and extracellular (XP) fractions, was analyzed. Cells were grown in either the presence (8 μg/ml) or absence of cefotaxime. Both TEM and CTX-M-15 β-lactamases were identified in the CP and XP; however, the relative abundance of these in the CP was ∼10-fold lower than their relative abundance in the XP (Fig. 2A). Exoproteomics identified 1,845 proteins across all treatments, the majority of which represented a long tail of very-low-abundance proteins (<0.1%). Notably, CTX-M-15 was the third most abundant protein in the XP of strain 48, in either the presence (1.7%) or absence (2.48%) of cefotaxime (Fig. 2A). The abundance of TEM in the XP was lower (absence, 0.38%; presence 0.6%). Constitutive expression of CTX-M-15 was confirmed by reverse transcription-quantitative PCR (RT-qPCR), which showed no significant difference in blaCTX-M-15 transcription in either the presence or absence of antibiotic (see Fig. S1 in the supplemental material).
FIG 2.
Identification of CTX-M-15 as the major secreted ESBL. (A) Top 10 most abundant proteins found in the exoproteome of strain 48 in the presence of cefotaxime. Protein abundance was evaluated by tandem mass spectrometry spectral counts, which correlated linearly with the protein abundance. Error bars indicate means from three replicates. Abundance of the three ESBLs proteins is shown. (B and C) Growth of JM109 empty plasmid (in blue) (B) and JM109 CTX-M-15 (in blue) (C) cultivated in LB medium in various cefotaxime concentrations, 8 μg/ml (in red), 16 μg/ml (in green), 32 μg/ml (in purple), and 64 μg/ml (in yellow).
To confirm if CTX-M-15 was responsible for conferring cefotaxime resistance, blaCTX-M-15, blaTEM, and blaOXA from strain 48 were separately cloned into the expression vector pGEM-T. Plasmids were mobilized into a susceptible host, the commercial strain E. coli JM109, resulting in the strains JM109-OXA, JM109-TEM, and JM109-CTX-M-15. An empty vector control was also mobilized into JM109, creating the strain JM109-pGEM-T (Fig. 2B). Only JM109-CTX-M-15 grew in the presence of cefotaxime, confirming blaCTX-M-15 was essential for resistance to cefotaxime (Fig. 2C).
CTX-M-15 secretion provides protection to susceptible cells.
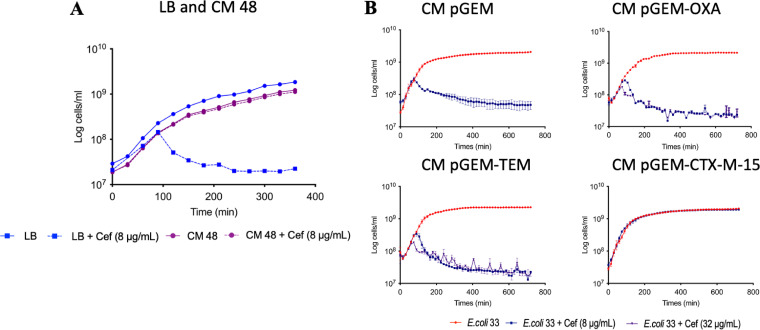
Strain 33 was susceptible to cefotaxime, so to determine if secreted CTX-M-15 from strain 48 could complement a susceptible strain, strain 33 was grown in both the presence and absence of cefotaxime in a conditioned medium (CM) used for growing strain 48. Strain 33 grew in CM in the presence of cefotaxime, demonstrating that strain 48 secreted sufficient quantities of CTX-M-15 into the medium to degrade the antibiotic and prevent inhibition of the otherwise susceptible strain 33 (Fig. 3A).
FIG 3.
Protective effect of conditioned medium obtained from strain 48 growth chambers. (A) Growth of E. coli strain 33 (in blue) cultivated in fresh LB medium compared to growth in CM of strain 48 (in purple). (B) Growth of strain 33 in CM of pGEM, CM of pGEM-OXA, CM of pGEM-TEM, CM of pGEM-CTX-M-15 in absence (in red), and presence of 8 μg/ml (in blue) and 32 μg/ml (in purple) cefotaxime.
The protection given by the secreted CTX-M-15 of strain 48 was confirmed with the engineered JM109 strains. Only CM from JM109-CTX-M-15 facilitated the growth of strain 33, while CM from TEM- and OXA-producing strains did not (Fig. 3B). Finally, we tested the stability of secreted CTX-M-15 by storing CM from JM109-CTX-M-15 at 4°C for 24 h, 48 h, and 72 h prior to inoculation with strain 33. Again, strain 33 grew in the presence of cefotaxime (Fig. S2). Together, this demonstrated that CTX-M-15 is functionally stable after secretion outside the cell and can provide protection to otherwise susceptible bacterial strains.
The T1SS is involved in the secretion of CTX-M-15.
To identify a mechanism of secretion for this CTX-M-15, we first investigated the role of OMVs, which have previously been reported to express ESBL activity (16). To remove OMVs from the supernatant, CM obtained from strain 48 was additionally filtered through a 0.02-μm filter. Filtration did not affect the ESBL activity of the supernatant, and susceptible strain 33 still grew in the presence of cefotaxime when grown in conditioned medium demonstrating that OMVs are not the main mechanism for CTX-M-15 secretion (Fig. S3). These data demonstrated that CTX-M-15 is fully functional in the extracellular medium.
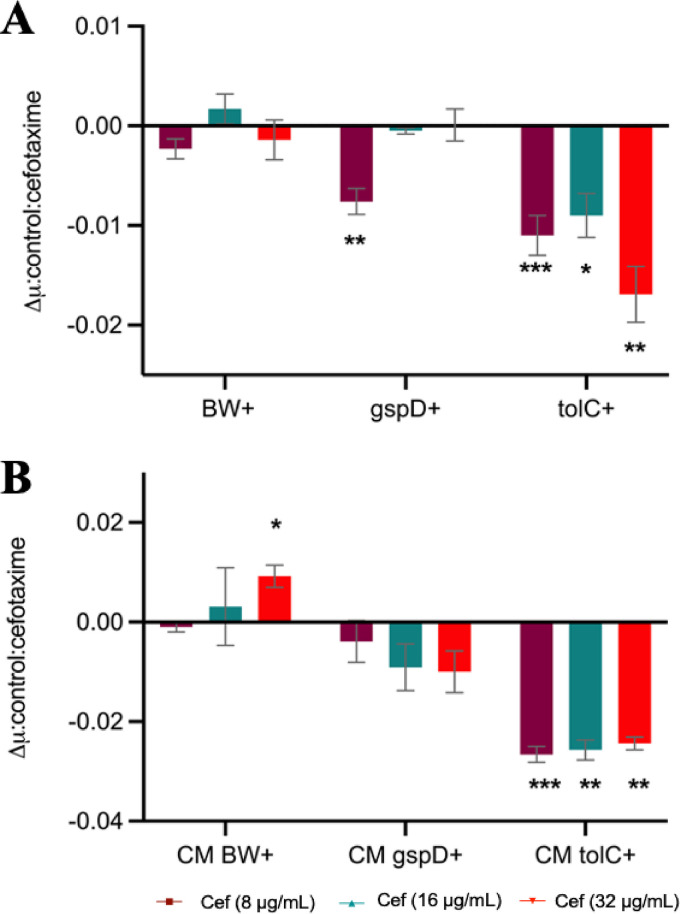
In agreement with their abundance in the XP of strain 48, in silico prediction revealed both CTX-M-15 and TEM contained the SP1 leader sequence required for translocation across the cytoplasmic membrane by the Sec pathway (Fig. S4 and Table S5). Strain 48 was predicted to contain four known secretion systems, T1SS, T2SS, T4SS, and T5SS, that therefore were candidates for CTX-M-15 secretion. T1SS and T2SS have potential to be involved in the secretion of hydrolytic enzymes, such as ESBLs. Mutant E. coli strains defective for genes required for either T1 secretion (tolC) or T2 secretion (gspD) were obtained from the Keio database, as was the parental wild type, E. coli BW 25113. pGEM-T-CTX-M-15, conferring cefotaxime resistance, was mobilized into all three strains (E. coli BW+, E. coli ΔtolC+, and E. coli ΔgspD+ strains), which again were subjected to growth in the presence of cefotaxime. In addition, all three strains, E. coli BW, E. coli ΔtolC, and E. coli ΔgspD strains, were transformed with an empty pGEM-T vector as controls. As expected, these three control strains failed to grow in the presence of cefotaxime (Fig. S5A). E. coli BW+, ΔtolC+, and ΔgspD+ strains all grew on various concentrations of cefotaxime, but mutation of either T1SS or T2SS significantly inhibited their growth rates (Fig. 4A and Fig. S5B). For the ΔgspD mutant, one cefotaxime concentration (8 μg/ml) showed a significant (P < 0.01) reduction in growth rate, while the ΔtolC+ strain displayed a significantly (P < 0.05) lower growth rate when challenged with all three concentrations of cefotaxime (Fig. 4A). To determine if this partial growth inhibition in either mutant was due to an inhibition by CTX-M-15 extracellular secretion, the growth of strain 33 on CM obtained from E. coli BW+, E. coli ΔtolC+, and ΔgspD+ cultures was monitored in the presence or absence of cefotaxime. Similar to the growth of both secretion mutants, the growth rate of strain 33 was significantly reduced by the presence of cefotaxime when cultured in CM obtained from either the ΔtolC+ or ΔgspD+ strain relative to the BW wild-type CM (Fig. 4B and Fig. S5C). Mutation of ΔtolC (T1SS) resulted in a greater sensitivity to cefotaxime for either the producer or the susceptible strain, indicating this secretion system is involved in, but not essential for, CTX-M-15 secretion. Given we also observed a smaller, albeit nonsignificant, effect when mutating gspD (T2SS), it is likely that both systems can facilitate CTX-M-15 secretion, with T1SS being the most important. Together, these data suggest secretion of CTX-M-15 is not a passive process and is facilitated by common secretion systems present in widespread bacteria.
FIG 4.
Potential role of T1SS in the secretion of CTX-M-15. (A) Growth rate (μ) of E. coli BW+, E. coli ΔgspD+, and E. coli ΔtolC+ strains in the presence of 8 μg/ml (in magenta), 16 μg/ml (in green), and 32 μg/ml (in red) cefotaxime. (B) Growth rate of strain 33 cultivated in CM obtained from E. coli BW+ (CM BW+), E. coli ΔgspD+ (CM ΔgspD+), and E. coli ΔtolC+ (CM ΔtolC+) in the presence of 8 μg/ml (in magenta), 16 μg/ml (in green), and 32 μg/ml (in red) cefotaxime. Graphs show the difference between the growth rate in the presence and the absence of antibiotic. The value indicates the means ± standard deviations from three biological replicates. t test was used to determine significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Secretion of ESBLs into the surrounding environment may reduce any damage to the cell wall by preventing entry of β-lactam into the periplasm, where they would be degraded by enzymes such as OXA or TEM. It is also likely to be an important ecological trait, enabling otherwise susceptible bacteria to survive long enough to allow mobilization of plasmid-encoded bla genes through conjugation (34, 35). While strain 48 possessed three annotated ESBLs, the data presented here prove that CTX-M-15 was the major secreted ESBL conferring resistance and providing a protective effect. This was further confirmed by exoproteomics, which revealed that CTX-M-15 was the third most abundant protein in the XP. The significant secretion of this resistance enzyme may provide improved protection against the antibiotic and explain how it became selected and transferred onto plasmids that were rapidly disseminated (1, 35–37).
After the gentle filtration of the supernatant, our data demonstrated CTX-M-15 remains fully functional and confers full protection to a susceptible strain. While the presence of ESBLs in OMVs has been linked to extracellular degradation of β-lactam antibiotics (15, 16, 38), to the best of our knowledge, this is the first-time secretion of an individual ESBL (i.e., CTX-M-15) has been linked to T1SS and proven to confer protection to susceptible bacteria. Our data clearly demonstrated CTX-M-15 was actively secreted into the extracellular milieu with no evidence that OMVs played a role. The metallo-β-lactamase NDM-1, which is the most widespread carbapenemase worldwide, has a lipobox proximal to its SP1 leader sequence that enables anchoring to the outer membrane and subsequent export in OMVs (38), a process known as lipidation. Removal of this lipobox inhibited NDM-1 export via OMVs, and the enzyme accumulated in the periplasm. Lipidation only occurs in a few ESBLs, such as BRO-1 (from the human pathogen Moraxella catarrhalis) and PenA (from Burkholderia pseudomallei) (39, 40), and does not include CTX-M-15, which may explain the lack of OMV involvement. While OMVs were implicated in the secretion and extracellular functioning of a nonlipidated serine β-lactamase CTX-M-1 (16), in our study, removal of vesicles by membrane filtration (0.02 μm) from the culture supernatant did not reduce the efficacy of CTX-M-15 to confer a protective effect, suggesting secretion occurred through an alternative mechanism. OMVs may play a large role in protecting ESBL integrity when inside a host, where biological fluids are likely to provide harsher conditions for enzyme activity. It should be noted that Gram-positive ESBLs are secreted and are stable to external attack; therefore, CTX-M-15 may also be resistant to harsher environmental conditions (41–43).
In contrast, we discovered that mutation of key genes required for either type I or type II secretion inhibited the efficacy of CTX-M-15-induced protection, both to the producer and the susceptible strains. Interestingly, our data proved that T1SS played a role in the direct secretion of CTX-M-15 despite the fact that this ESBL contains a leader sequence for localization in the periplasm. The differential effect of mutants on CTX-M-15-induced protection suggests a more direct role for the T1SS. The T2SS may also play a role in secretion of CTX-M-15, although such a small amount of proteins in the XP could be explained by spontaneous release of the periplasmic proteins. Indeed, out of the top five most abundant proteins in the XP, OsmY and two ligand binding proteins were also predicted to be periplasmic (44, 45) but were still found in the culture supernatant. Exoproteomics often captures a range of periplasmic proteins, especially ligand binding proteins (46–48), which suggests that the outer membrane is leaky. The mechanism of secretion for OsmY has not been studied, but it was used in biotechnology to deliver proteins into the medium via C-terminal fusion (49, 50). Reports concerning secretion in E. coli remain elusive mainly because nonpathogenic laboratory strains generally express a small amount of proteins in the culture medium (51, 52). Identifying the causal mechanism for CTX-M-15 secretion could help develop novel therapeutic drugs to block secretion, as we have proven it is essential for resistance under exposure to cefotaxime.
We have strong evidence based on the presence of enzyme, bioinformatics, and mutant studies that T1SS and not T2SS is responsible for the secretion of all CTX-M-15 in the exoproteome, but further work is required to consolidate this observation and fully establish the secretion pathway for CTX-M-15. It is feasible that secretion of CTX-M-15 represents an evolutionary advantage, as no damage would occur to the cell wall if the antibiotic is disabled outside the cell, in opposition to hydrolysis in the periplasm. The role of environmental contamination in the transmission of Enterobacteriaceae and, in particular, E. coli ST131 is increasingly recognized. However, factors influencing duration of survival in the environment have not yet been extensively studied.
MATERIALS AND METHODS
Bacterial strains and growth medium.
Bacterial strains used in this study are listed in the supplemental information (see Table S1). Environmental E. coli strains were both isolated from the River Sowe, Coventry, United Kingdom, namely, E. coli ST3576 O8:H7 (strain 33) and E. coli ST131 O25:H4 (strain 48). Commercial laboratory strains of E. coli JM109, E. coli BW2511, E. coli JW5503, and E. coli JW5707 were also used. Cells were routinely grown in lysogeny broth (LB) liquid (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter sodium chloride) or LB agar medium (addition of 15 g/liter agar). The following antibiotics were supplemented when required: 8 μg/ml cefotaxime, 100 μg/ml ampicillin, and 5 μg/ml kanamycin. Additionally, culture medium for JM109 was supplemented with isopropyl β-d-thiogalactosidase (IPTG) (0.1 M) and X-galactosidase (20 mg/ml) to induce expression of recombinant CTX-M-15. Cells were incubated at 37°C with either shaking (200 rpm) or static conditions.
Antimicrobial phenotypic screening.
Oxid antibiotic discs were used to determine phenotypic resistance profiles. Strains 33 and 48 were streaked on LB agar plates, and discs containing either 25 μg ampicillin, 5 μg cefotaxime, 10 μg imipenem, 30 μg chloramphenicol, or 8 μg erythromycin were added on top of the plates. All of the plates were incubated overnight at 37°C.
ESBL genotypic screening.
Single colonies of strains 33 and 48 were picked and individually inoculated into 10 ml LB and incubated overnight at 37°C with shaking at 150 rpm. Cultures were then centrifuged at (1,500 rpm for 10 min) and supernatant discarded. Pellets were resuspended in 500 μl phosphate-buffered saline and used for DNA extraction using the MPBio FastDNA spin kit by following the manufacturer’s guidelines. Specific primers for amplification of ESBL genes, blaCTX-M-15, blaTEM, and blaOXA, were designed from the Illumina sequencing done previously (53) (Table S2). PCRs were done using 12.5 μl Master Mix 2× (Promega), 1.25 μl dimethyl sulfoxide, 0.8 μM forward primer, 0.8 μM reverse primer, 2 μl DNA template, and distilled H2O for a final PCR volume of 25 μl. PCR was performed at an initial denaturation temperature of 95°C for 5 min, followed by 34 cycles of denaturation at 95°C for 30 s, annealing temperature at 55 to 66°C (depending on the primer set) for 30 s, and extension for 1 min 50 s. A final extension was performed at 72°C for 5 min.
Antibiotic resistance screening.
Growth curves were implemented in 96-well plates with 200 μl culture per well and incubated at 37°C with shaking (200 rpm) in a microplate reader (POLARstar Omega; BMG Labtech). As inoculum, overnight starter cultures of each bacterial strain (5 ml) were diluted to an initial concentration of 3 × 107 cells/ml. Culture media were supplemented with 0, 8, 16, 32, or 64 μg/ml cefotaxime (VWR International Ltd.). Cell proliferation was determined by measuring the optical density at 600 nm for 8 or 12 h every 15 min. Each condition was set up in triplicate. Exponential growth rates were calculated for the growth of E. coli BW+, E. coli ΔtolC+, and E. coli ΔgspD+ strains and for the protective effect on strain 33 by using the formula P(t) = P0ert, where P(t) is the amount of cell number at time t, P0 the initial cell number, r the growth rate, and t the number of periods (54). Two-sample t test was performed to compare the significance of the growth rate differences.
Generation of conditioned medium.
Strain 48, or the engineered laboratory E. coli strains harboring CTX-M-15, were grown in the presence of cefotaxime (8 μg/ml). After overnight growth, cells were removed by pelleting (3,228 × g for 15 min). Supernatant was carefully filtered through a 0.22-μm membrane (Fisher Scientific) to avoid cell lysis. Conditioned medium (CM) was diluted 1:1 (vol/vol) with fresh LB medium and supplemented with various concentrations of cefotaxime. Overnight cultures of susceptible strain 33 were inoculated (1%, vol/vol) in the conditioned medium and grown as described above.
Cloning of the ESBLs, blaCTX-M-15, blaTEM, and blaOXA.
A full list of primers used in this study is presented in Table S2, and the genes blaCTX-M-15, blaTEM-1, and blaOXA-1 were cloned from strain 48 into the cloning vector pGEM-T easy (Promega, UK) using the HiFi assembly kit (New England, Biolabs). The newly constructed plasmids pGEM-CTX-M-15, pGEM-TEM, and pGEM-OXA, and control empty-vector pGEM-T, were transformed into E. coli JM109.
Detection of β-lactamase activity by the nitrocefin assay.
β-Lactamase activity was assessed by colorimetric assay using the chromogenic cephalosporin compound nitrocefin (Thermo Scientific). Strain 33 and strain 48 were inoculated in LB or M9 minimal medium (33.9 g/liter Na2HPO4, 15 g/liter KH2PO4, 5 g/liter NH4Cl, 2.5 g/liter NaCl, 20% glucose, 1 M MgSO4, 1 M CaCl2) supplemented with 0, 2, 4, or 8 μg/ml cefotaxime. Strains were grown at 37°C until mid-exponential phase, and supernatant was collected by first removing cells (4,000 rpm for 15 min) and then gentle filtration through a 0.22-μm membrane (Fisher Scientific) to prevent cell lysis and remove intact cells. Supernatants were incubated with 15 μg/ml nitrocefin (stock concentration, 500 μg/ml) at room temperature (∼22°C) for 30 min.
Determination of blaCTX-M-15 transcription in strain 48.
Strain 48 was grown at 37°C in LB supplemented with 0 or 8 μg/ml cefotaxime. Diluted cultures were grown at 37°C with shaking (200 rpm) to exponential phase before RNA extraction. A detailed protocol for extraction, reverse transcription, and quantitative PCR can be found in the supplemental material.
Preparation of exoproteome, total proteome samples, and liquid chromatography tandem mass spectrometry analysis.
Exoproteomes and total proteomes of strain 48 were prepared by adapting the protocol described in Christie-Oleza and Armengaud (47). Briefly, Strataclean beads (Agilent) were used to isolate proteins instead of trichloroacetic acid (TCA) precipitation. A detailed procedure is provided in the supplemental material.
Peptide identification and comparative proteomic analysis.
A custom database was made with the genome of strain 48 by using Prokka v1.14.5 for annotation (59), and MASCOT was used to assign peptide to protein by using the custom database. Identified proteins were further analyzed using Scaffold (55) (protein threshold, 99.9%; minimum peptide 2, peptide threshold, 80%). The normalized spectral abundance factor (NSAF) (56) was calculated for each protein to compare the abundance for all proteins. Two-sample t test was used to determine if the presence of antibiotic significantly impacted protein abundance.
In silico prediction of protein localization and secretion pathways.
Analysis was done on the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP-5.0/) and enabled the prediction of the presence and the location of cleavage sites in the three β-lactamase proteins CTX-M-15, TEM, and OXA using the Fasta sequences generated in-house (see the supplemental material) (57). The TXSScan web tool (https://galaxy.pasteur.fr/) (58) was used for prediction of the presence of secretion systems in strain 48 using the genome of E. coli 48.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support from the Natural Environment Research Council (grant NE/N019857/1). S.R. was supported by a CENTA NERC studentship.
We declare that we have no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Severine Rangama, Email: m.rangama@warwick.ac.uk.
Elizabeth M. H. Wellington, Email: e.m.h.wellington@warwick.ac.uk.
REFERENCES
- 1.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 2.Canton R, Coque TM. 2006. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol 9:466–475. 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect 14(Suppl 1):33–41. 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 4.Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 13:19–44. [PubMed] [Google Scholar]
- 5.Pitout JD, DeVinney R. 2017. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res 6:F1000 Faculty Rev-195. [PMC] 10.12688/f1000research.10609.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkey PM, Jones AM. 2009. The changing epidemiology of resistance. J Antimicrob Chemother 64(Suppl 1):i3–i10. 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 8.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321. 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53:4472–4482. 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK. 2019. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci Rep 9:4224. 10.1038/s41598-019-39730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brook I. 2004. Beta-lactamase-producing bacteria in mixed infections. Clin Microbiol Infect 10:777–784. 10.1111/j.1198-743X.2004.00962.x. [DOI] [PubMed] [Google Scholar]
- 13.Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 61:839–846. 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 14.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciofu O, Beveridge T, Walther-Rasmussen J, Hoiby N, Kadurugamuwa J. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45:9–13. 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 16.Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, Lazarte JMS, Kim J, Seo JS, Kim JH, Song JW, Jung HS, Kim GJ, Lee YJ, Lim SK, Jung TS. 2018. Outer membrane vesicles from beta-lactam-resistant Escherichia coli enable the survival of beta-lactam-susceptible E. coli in the presence of beta-lactam antibiotics. Sci Rep 8:5402. 10.1038/s41598-018-23656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson C, Matos CF, Beck D, Ren C, Lawrence J, Vasisht N, Mendel S. 2011. Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim Biophys Acta 1808:876–884. 10.1016/j.bbamem.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4:10.1128/microbiolspec.VMBF-0012-2015. [PMC] 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsirigotaki A, De Geyter J, Sostaric N, Economou A, Karamanou S. 2017. Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15:21–36. 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 20.Piddock LJV. 2006. Clinically relevant bacterial chromosomally encoded multi-drug resistance efflux pumps. Clin Microbiol Rev 19:382–402. 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni R, Dhakal BK, Slechta ES, Kurtz Z, Mulvey MA, Thanassi DG. 2009. Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli. PLoS One 4:e4752. 10.1371/journal.pone.0004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc Natl Acad Sci USA 99:7066–7071. 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Smith P, Schuster M. 2019. Public goods and cheating in microbes. Curr Biol 29:R442–R447. 10.1016/j.cub.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Lidbury I, Murphy ARJ, Fraser TD, Bending GD, Jones AME, Moore JD, Goodall A, Tibbett M, Hammond JP, Scanlan DJ, Wellington EMH. 2017. Identification of extracellular glycerophosphodiesterases in Pseudomonas and their role in soil organic phosphorus remineralisation. Sci Rep 7:2179. 10.1038/s41598-017-02327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reintjes G, Arnosti C, Fuchs BM, Amann R. 2017. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J 11:1640–1650. 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, Pascual-Garcia A, Cordero OX. 2019. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol 29:1528–1535. 10.1016/j.cub.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 28.Whitmer GR, Moorthy G, Arshad M. 2019. The pandemic Escherichia coli sequence type 131 strain is acquired even in the absence of antibiotic exposure. PLoS Pathog 15:e1008162. 10.1371/journal.ppat.1008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 31.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:9. 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 85:11–19. 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, Choroszy-Krol I. 2019. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog 11:10. 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De La Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol 8:128–133. 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 35.Amos GC, Hawkey PM, Gaze WH, Wellington EM. 2014. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J Antimicrob Chemother 69:1785–1791. 10.1093/jac/dku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amos GCA, Ploumakis S, Zhang L, Hawkey PM, Gaze WH, Wellington EMH. 2018. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J 12:681–691. 10.1038/s41396-017-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol 12:516–522. 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bootsma HJ, Van Dijk H, Verhoef J, Fleer A, Mooi FR. 1996. Molecular characterization of the BRO beta-lactamase of Moraxella (Branhamella) catarrhalis. Antimicrob Agents Chemother 40:966–972. 10.1128/AAC.40.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randall LB, Dobos K, Papp-Wallace KM, Bonomo RA, Schweizer HP. 2015. Membrane-bound PenA beta-lactamase of Burkholderia pseudomallei. Antimicrob Agents Chemother 60:1509–1514. 10.1128/AAC.02444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anne J, Economou A, Bernaerts K. 2017. Protein secretion in Gram-positive bacteria: from multiple pathways to biotechnology. Curr Top Microbiol Immunol 404:267–308. 10.1007/82_2016_49. [DOI] [PubMed] [Google Scholar]
- 42.Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, Kim YK, Roh TY, Gho YS. 2013. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrob Agents Chemother 57:2589–2595. 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Defourny KAY, Smid EJ, Abee T. 2018. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1502. 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yim HH, Villarejo M. 1992. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol 174:3637–3644. 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh BH, Kang CH, De Bondt H, Kim SH, Nikaido K, Joshi AK, Ferro-Luzzi Ames G. 1995. The bacterial periplasmic histidine-binding protein. J Biol Chem 270:16097–14143. 10.1016/S0021-9258(17)41754-6. [DOI] [PubMed] [Google Scholar]
- 46.Lidbury I, Krober E, Zhang Z, Zhu Y, Murrell JC, Chen Y, Schafer H. 2016. A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ Microbiol 18:2754–2766. 10.1111/1462-2920.13354. [DOI] [PubMed] [Google Scholar]
- 47.Christie-Oleza JA, Armengaud J. 2010. In-depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography-tandem mass spectrometry: the Ruegeria pomeroyi DSS-3 case-study. Mar Drugs 8:2223–2239. 10.3390/md8082223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christie-Oleza JA, Armengaud J. 2015. Proteomics of the Roseobacter clade, a window to the marine microbiology landscape. Proteomics 15:3928–3942. 10.1002/pmic.201500222. [DOI] [PubMed] [Google Scholar]
- 49.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD. 2011. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci USA 108:19949–19954. 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian ZG, Xia XX, Choi JH, Lee SY. 2008. Proteome-based identification of fusion partner for high-level extracellular production of recombinant proteins in Escherichia coli. Biotechnol Bioeng 101:587–601. 10.1002/bit.21898. [DOI] [PubMed] [Google Scholar]
- 51.Papanikou E, Karamanou S, Economou A. 2007. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol 5:839–851. 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 52.Kotzsch A, Vernet E, Hammarström M, Berthelsen J, Weigelt J, Gräslund S, Sundström M. 2011. A secretory system for bacterial production of high-profile protein targets. Protein Sci 20:597–609. 10.1002/pro.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill G. 2016. Investigating WWTP impact on antibiotic resistance within UK river systems. Doctoral Dissertation University of Warwick, Coventry, UK. [Google Scholar]
- 54.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 55.Searle BC. 2010. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10:1265–1269. 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 56.Zhu W, Smith JW, Huang CM. 2010. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol 2010:840518. 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 58.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.00663-21-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)