ABSTRACT
The global spread of antimicrobial-resistant bacteria has been one of the most severe threats to public health. The emergence of the mcr-1 gene has posed a considerable threat to antimicrobial medication since it deactivates one last-resort antibiotic, colistin. There have been reports regarding the mobilization of the mcr-1 gene facilitated by ISApl1-formed transposon Tn6330 and mediated rapid dispersion among Enterobacteriaceae species. Here, we developed a CRISPR/Cas9 system flanked by ISApl1 in a suicide plasmid capable of exerting sequence-specific curing against the mcr-1-bearing plasmid and killing the strain with chromosome-borne mcr-1. The constructed ISApl1-carried CRISPR/Cas9 system either restored sensitivity to colistin in strains with plasmid-borne mcr-1 or directly eradicated the bacteria harboring chromosome-borne mcr-1 by introducing an exogenous CRISPR/Cas9 targeting the mcr-1 gene. This method is highly efficient in removing the mcr-1 gene from Escherichia coli, thereby resensitizing these strains to colistin. The further results demonstrated that it conferred the recipient bacteria with immunity against the acquisition of the exogenous mcr-1 containing the plasmid. The data from the current study highlighted the potential of the transposon-associated CRISPR/Cas9 system to serve as a therapeutic approach to control the dissemination of mcr-1 resistance among clinical pathogens.
KEYWORDS: ISApl1, mcr-1, CRISPR/Cas9, transposon, antibiotic resistance
INTRODUCTION
Mobile genetic elements (MGEs) such as plasmids, transposons, integrons, and insertion sequences (ISs) play essential roles in disseminating antibiotic resistance genes (ARGs) among Gram-negative bacteria. The colistin resistance gene mcr-1 was first identified from a conjugative IncI2 plasmid in China and has been intensively reported globally from other plasmid types (1). There have been highly diverse plasmid types involved in the bearing of mcr-1, including the IncX4, IncI2, IncP, IncFII, and IncHI2 types (1–4). The IncX4 plasmid has been deemed one of the most prevalent types in Escherichia coli at a prevalence level ranging from 7.6% to 34% (5, 6). Except for the plasmid-borne mcr-1, chromosomal copies of mcr-1 have also been identified in E. coli strains isolated from retail meats and humans (7–9). Notably, a recent study reported an isolate found with three tandem copies of the mcr-1 element carried by ISApl1 (10). The ISApl1 belongs to the IS30 family and was first characterized in Actinobacillus pleuropneumoniae (11). There have been studies demonstrating that ISApl1 facilitated the mobilization of mcr-1 by forming a composite transposon Tn6330 and preferentially targeting AT-rich sequences (12–14). These results implied the active role of ISApl1 in the transposition of mcr-1 between plasmids and the bacterial chromosome and vice versa.
The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system was initially discovered as a bacterial immunity in archaea and bacteria against MGEs (15), yet it has now become the predominant technique for gene editing (16). This technique allows an easy-handling approach to knock out, insert, and mutate genes in more than 40 species with high-efficiency and low-cost procedures (17, 18). As to the broad applicability, the CRISPR/Cas9 system showed its potency to eradicate antibiotic resistance genes. Previous studies reported the employment of engineered plasmids to deliver the CRISPR/Cas9 system targeting mcr-1 in the recipient bacteria (19–21). However, notable demerits for the plasmid-based methods, commonly used in previous reports, are genetic instability and weak control toward the copy numbers of the established systems (22, 23). Alternatively, there have been studies to deliver the CRISPR/Cas9 system with a bacteriophage (24, 25). This strategy could specifically target bacterial genotypes from mixed bacterial populations in both in vitro and in vivo conditions (25–27), while the limited host ranges and emerging phage resistance might restrain the application of this phage-based strategy. Therefore, developing a novel delivery element for the CRISPR/Cas9 system has been of great scientific significance.
As the principal function of CRISPR/Cas systems in archaea and bacteria is a defense against invasive DNA integration, including viruses, plasmids, and transposons (15). The Tn7-like transposons are widely found in the genomes of bacterial and archaeal species and have been proven to recruit CRISPR/Cas systems independently (28). An intriguing example is that the CRISPR/Cas systems work well with Tn7-like transposons in targeted transposition, facilitating the spread of genetic elements via phages and plasmids (28, 29). This nature highlighted the potential of a transposon as a tool to deliver the designed CRISPR/Cas systems (29, 30). Thus, we advanced the notion that the transposon-associated CRISPR/Cas system could contribute to the removal of antibiotic resistance. Until now, there has been no report on the development of transposon-associated CRISPR/Cas systems against antibiotic resistance.
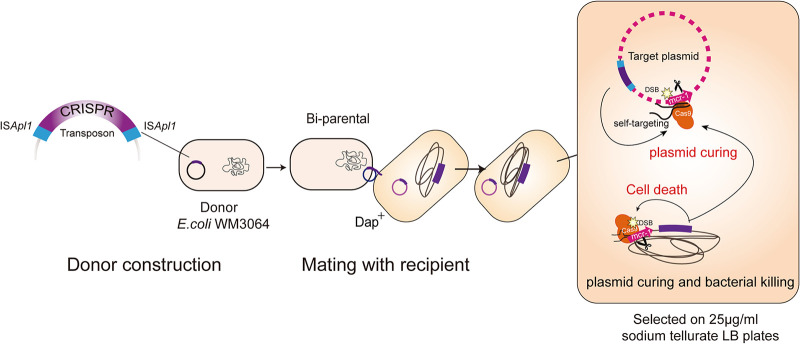
ISs and their associated transposons have been greatly reported to mediate the transposition of the resistance genes (31). However, their potential as a carrier to facilitate the CRISPR/Cas system for resistance gene editing and curing in bacteria has been rarely noticed and estimated. In the current study, we employed ISApl1 to construct a transposon-associated CRISPR/Cas system as a proof of concept in curing mcr-1 (Fig. 1). On the one hand, the system successfully eliminates the mcr-1-bearing plasmid, even resulting in strains immune from further acquisition of exogenous plasmids carrying mcr-1. On the other hand, it could specifically kill the bacteria in which the mcr-1 is located on the chromosome. This study provides a potent prototype of the transposon-associated CRISPR/Cas system in the fight against colistin resistance. And it might be further employed for the approaches against the antibiotic resistance of other types.
FIG 1.
The pISApl1-CRISPR/Cas9 system for plasmid curing and bacterial killing. A suicide plasmid containing the ISApl1-formed transposon carrying CRISPR/Cas9 was transferred to recipient bacteria by biparental mating. Donor cell E. coli WM3064 contains a chromosomal copy of the RP4 transfer machinery used to mobilize the suicide plasmids. Once inside the recipient cell, The Tn::ISApl1-CRISPR/Cas9 was integrated into the plasmid or chromosome in the recipient strain. Selection on antibiotic plates lacking DAP eliminates the E. coli WM3064 donors and retains recipients with an integrated ISApl1-CRISPR/Cas9 library. The coexpression of sgRNA and Cas9 is capable of plasmid curing and bacterial killing.
RESULTS
Construction of pISApl1-CRISPR/Cas9 plasmid.
The newly constructed pISApl1-CRISPR/Cas9 vector contained an R6K replication origin, which relies on the π protein. This suicide plasmid will survive only in bacterial hosts that express the π protein, such as E. coli WM 3064. The Cas9 expression was driven by the pLteto-1 promoter (32), and the single guide RNA (sgRNA) was driven by the constitutive promoter pJ23119. The introduction of the tpm gene allowed the selection of clinical multidrug-resistant (MDR) isolates. The recombinant plasmid pISApl1-CRISPR/Cas9 and the transposon ISApl1-CRISPR/Cas9 are illustrated in Fig. 2.
FIG 2.
Plasmid map of pISApl1-CRISPR/Cas9 and the transposon Tn::ISApl1/CRISPR/Cas9. (a) Plasmid pISApl1-CRISPR/Cas9 containing the cas9 gene with the promoter pLteto-1 (39) and the sgRNA with the synthetic J23119 promoter; F and R denote the locations of primers used for the sgRNA construct. (b) The Tn::ISApl1-CRISPR/Cas9, the cas9, and the tpm gene, along with sgRNA was flanked by two ISApl1 in the same orientation.
Elimination of the mcr-1 bearing plasmid using pISApl1-CRISPR/Cas9.
To verify the potency of the ISApl1-CRISPR/Cas system in eliminating the plasmid-borne mcr-1, the strains artificially or naturally bearing the mcr-1 plasmid were used as the model strains. The results showed that the ISApl1-CRISPR/Cas9 was efficiently transferred into the recipient to cut the mcr-1 harboring plasmids (Table 1), presumably by an unrepairable double-strand break (DSB) caused by highly efficient ISApl1-CRISPR/Cas9 cleavage. The successful elimination of the mcr-1 was genotypically and phenotypically approved (see Fig. S4 in the supplemental material).
TABLE 1.
The transposition efficiency of pISApl1-CRISPR/Cas9 into recipients and the mcr-1 gene and plasmid curing efficiencya
| Strain | Target gene | Target sequence (N20) | Transposition efficiency (transconjugants/recipient) | Curing efficiency (%)b |
|---|---|---|---|---|
| E. coli C600 | (5.8 ± 0.6) × 10−4 | |||
| E. coli C600(pUC19-mcr-1) | mcr-1 | GCGGCATTCGTTATAAGGAT | (7.4 ± 0.5) × 10−4 | 100 ± 0 |
| E. coli CSZ4 | mcr-1 | GCGGCATTCGTTATAAGGAT | (8.9 ± 0.3) × 10−4 | 99.3 ± 1.5 |
| E. coli CSZ4 | IncX4 replication gene | AGACTCAAATTCATTGAATC | (6.7 ± 0.8) × 10−4 | 98.5 ± 1.2 |
The donor strain used was E. coli WM3064. Selective plating was done on ST25 plates selected for the transconjugants. Transposition efficiency was calculated as the number of transconjugants per recipient.
The values shown are the mean ± SD from three independent experiments.
Bactericidal effect of pISApl1-CRISPR/Cas9 on chromosome-borne mcr-1 strains.
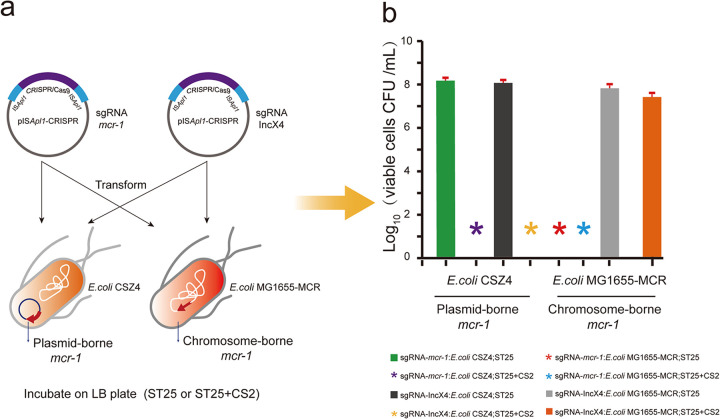
In this section, we further applied the ISApl1-CRISPR/Cas9 system to the strain that harbored the chromosome-borne mcr-1. Impressively, a very remarkable reduction in the viability of mcr-1-positive cells was observed as no detectable colonies on the selective agar. However, the ISApl1-CRISPR/Cas9(sgRNA::IncX4) exerted no impact on cell viability (Fig. 3). The system was supposed to cut the chromosome at the site of mcr-1 and then lead to cell death of mcr-1-positive bacteria. The results suggested the specificity of the system with high efficiency and accuracy.
FIG 3.
(a) Schematic of mobilizable ISApl1-CRISPR/Cas9-mediated plasmid curing and cell killing. (b) E. coli WM3064 donor cells possessing pISApl1-CRISPR/Cas9-mcr-1 or pISApl1-CRISPR/Cas9-IncX4 were mated at a donor/recipient ratio of 1:1 with strain E. coli CSZ4 harboring a plasmid-borne mcr-1 or strain E. coli MG1655-MCR in which mcr-1 is located on the chromosome. Cultures were plated on selective LB agar sodium tellurite (25 μg/ml) in the presence and absence of colistin (2 μg/ml). The mobilization of the pISApl1-CRISPR/Cas9 construct into cells containing the mcr-1 gene resulted in plasmid curing and cell death. Mean ± standard error (SE). Asterisks denote the absence of detectable transconjugant colonies.
The integrated ISApl1-CRISPR/Cas9 system blocks plasmid acquisition.
The CRISPR/Cas system conferred adaptive immunity against the incorporation of exogenous genetic elements in many bacteria and most archaea. We, therefore, examined whether an integrated CRISPR/Cas9 system was capable of precluding the acquisition mcr-1. The plasmids bearing mcr-1 from E. coli CSZ4, Salmonella enterica 19E0341, and Klebsiella pneumoniae strain YZ01 were conjugated into the pUC19-mcr-1-cured strain, E. coli C600, which contained the integrated transposon ISApl1-CRISPR/Cas9(sgRNA::mcr-1). As shown in Fig. 4, genetic incorporation of ISApl1-CRISPR/Cas9 curtailed the ARG transfer to the strains yet did not acquire any of the donor strain plasmids, whereas the control E. coli C600 demonstrated particularly high frequencies of mcr-1 acquisition.
FIG 4.
(a) Schematic diagram of the immune system against foreign plasmids of E. coli C600 with ISApl1-CRISPR/Cas9 integrated into the chromosome. (b) The strain E. coli C600 with the chromosome bearing ISApl1-CRISPR/Cas9-ISApl1 and the parental strain E. coli C600 were used as recipients conjugated with donor strains E. coli CSZ4, S. enterica 19E0341, and K. pneumoniae YZ01, respectively. The bar chart represented the results when E. coli C600 was used as a recipient (positive control). The asterisks denote the chromosome-borne ISApl1-CRISPR/Cas9-ISApl1 strain used as recipient. Each experiment was performed in triplicate. Data points represent the mean values of three biological replicates with error bars showing standard deviation (SD).
Escape mutant analysis.
A handful of cells of E. coli CSZ4 revealed colistin resistance after receiving the ISApl1-CRISPR/Cas9(sgRNA::mcr-1) system. We, therefore, analyzed their cas9 along with the sgRNA region by PCR. The Sanger sequencing results showed 5 of them with a 510-bp deletion, while 3 demonstrated another 511-bp deletion that includes the sgRNA (Fig. 5).
FIG 5.
Characterization of escape mutants that tolerated transformation of the pISApl1-CRISPR/Cas9 construct. (a) The CRISPR/Cas9 region of the escape mutants was amplified by a high-fidelity enzyme and followed by sanger sequencing. (b, c) The results show that 510 or 511 deletions at the same sgRNA area led to pISApl1-CRISPR/Cas9 inactivation in the successful transformants.
DISCUSSION
The occurrence and dissemination of colistin resistance gene mcr-1 in bacteria severly challenged the current treatment of clinical infections. Therefore, it is essential to develop new methods and strategies to tackle the distribution of mcr-1, and the restoration of bacterial sensitivity to colistin would be of great scientific significance. Transposons (Tn) and insertion sequences (IS) are often associated with the transmission of ARGs. They are discrete DNA segments that are able to move their flanked sequences and themselves per se randomly to new locations in identical or different DNA (31). In our previous study, we showed that an ISApl1-formed transposon Tn6330 mediated the mcr-1 gene transposition at an exceptionally high level (12). This also corresponded to the investigation of Laurent Poirel and colleagues (14), indicating that the ISs and associated Tns might be powerful tools to expedite the CRISPR/Cas editing in bacterial species. Based on our research foundation of ISApl1, we developed an ISApl1-based transposon carrying the CRISPR/Cas9 system to combat the dispersion of mcr-1. Once introduced into the targeted strains, the ISApl1-CRISPR/Cas9 cassette randomly integrated into either the chromosome or plasmid or even both in some cases. Current antibiotics tend to be broad-spectrum, leading to indiscriminate killing of commensal bacteria and stimulating drug resistance evolution (24). In our study, the sgRNA was designed to guide the Cas9 protein specifically to the site of mcr-1 and induce DSBs. The RNA-guided nuclease (RGN) targeting this resistance gene is delivered efficiently using ISApl1-CRISPR/Cas9 carrying suicide plasmids by conjugation. It introduced the DBSs to the DNA segment involving the mcr-1 gene specifically, making it barely possible to result in indiscriminate killing. Considering that some strains carry mcr-1 on their chromosomes, we sought to determine the impact of the constructed system on strains with chromosome-harbored mcr-1. The results indicated that the system mediated direct killing in the strains of the type as mentioned above (Fig. 3b). This implied that RNA-guided nucleases (RGNs) target specific DNA sequences regardless of whether they are on plasmid or chromosome.
One nature of the translocation of the transposon is its random integration into the bacterial genome. This feature leads to nonmarkerless deletion, which means it is impossible to remove the CRISPR/Cas9 elements from the strains that receive editing. However, given the fact that the CRISPR/Cas system is an adaptive immune system of bacteria and archaea against foreign DNA, the genomically incorporated ISApl1-CRISPR/Cas9 system may confer bacterial immunity from further acquisition of eliminated DNAs. Here, we present an interesting example that the employment of transposon-associated CRISPR/Cas9 offered the recipients stable resistance to foreign integration of mcr-1 from other bacteria. This “vaccine-like” effect may provide a robust scientific approach for controlling the transmission of the mcr-1 gene. In CRISPR/Cas editing, the off-target property of the system has been a bottleneck for its successful application (33). In our study, we demonstrated the sequence-specific removal of mcr-1-harboring strains using a transposon-associated CRISPR/Cas9 system. We observed highly efficient removal of the mcr-1 gene (>98% as shown above) and promising specificity by the system. However, a small number of the transformed cells of E. coli CSZ4 remained colistin resistant after CRISPR/Cas9 editing. Sequencing of these survivors revealed consistent loss of sgRNA targeting in the transformed transposon ISApl1-CRISPR/Cas9. This insight is consistent with the findings of a previous study showing loss or inactivation of CRISPR elements under evolutionary pressure (34).
Compared to the several previous studies that require specific reagents (e.g., arabinose, anhydrous tetracycline, rhamnose) to induce the CRISPR/Cas9 system to cure the antibiotic resistance genes (19, 20, 35, 36), the system established in the current study enjoys the benefit of autonomous transposition and the self-driven function of CRISPR/Cas editing, which is particularly meaningful for its in vivo applications. Since the concentrations of the inducing reagents are hard to control in the in vivo conditions, the self-driven expression of Cas9 will be ideal in practical applications. There is an argument that the continuously expressed Cas9 protein will limit the transformation efficiency of the CRISPR/Cas-containing plasmids. Fortunately, the ISApl1-CRISPR/Cas9 system in this study maintained a high level of transposition efficiency. This is probably because IS-formed transposon partially compensates for the efficiency reduction. To conclude, the CRISPR/Cas9 system mobilized by transposon to exclude antibiotic resistance genes was first established in the present study. This novel strategy demonstrated substantially high efficiency in removing mcr-1 genes from clinical or lab mcr-1-positive strains and kill the strains with chromosomal mcr-1 genes. Moreover, the integration of the CRISPR/Cas9 system conferred immunity to the cured bacteria from the further acquisition of mcr-1 genes. With the efficiency and safety observed, the reported system showed great potential in MDR bacteria treatment and expanded the arsenal of tools against a possible superbug crisis.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 2, and primers can be found in Table S1 in the supplemental material. E. coli strain WM3064 was used as the donor strain in the study. All strains were cultured at 37°C in Luria Bertani (LB) medium, and diaminopimelic acid (DAP) was supplemented at 0.3 mM for the conjugation assays. When necessary for the selection of transformants, the antibiotics were added at the following concentrations: colistin at 2 μg/ml (abbreviated to CS2), chloramphenicol at 25 μg/ml (abbreviated to C25), and sodium tellurite at 25 μg/ml (abbreviated to ST25).
TABLE 2.
Plasmids and strains used in this study
| Strain or plasmid | Description | Reference or study |
|---|---|---|
| Strain | ||
| E. coli C600 | Conjugation recipient, F− tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 λ− | 40 |
| E. coli C600(pUC19-mcr-1) | E. coli C600 harboring pUC19-mcr-1 | This study |
| E. coli CSZ4 | Clinical isolate harboring an IncX4 plasmid pCSZ4 bearing mcr-1 | 1 |
| E. coli MG1655-MCR | E. coli MG1655(ΔrecA::Kan), chromosome bearing mcr-1 | 12 |
| Salmonella enterica 19E0341 | Clinical isolate harboring an IncI2 plasmid bearing mcr-1 | 41 |
| Klebsiella pneumoniae YZ01 | Clinical isolate harboring an IncI2 plasmid bearing mcr-1 | This study |
| Plasmid | ||
| pISApl1-CRISPR/Cas9 | Suicide plasmid harboring Tn::ISApl1-CRISPR/Cas9, R6K ori, mobRP4 | This study |
| pUC19-mcr-1 | pUC19 derivative harboring mcr-1 | This study |
Plasmid construction.
We constructed a suicide plasmid consisting of the CRISPR/Cas9 cassette flanked by ISApl1, which possessed the RP4oriT fragment from pCVD442 (37) and the R6K replication origin from pSV03 (12). Traditional antibiotic selection markers were replaced by the Acinetobacter baylyi-derived tellurite resistance marker gene tpm, encoding a thiopurine S-methyltransferase, to allow selection for chromosomal plasmid integration with sodium tellurite (38). Plasmid construction is described in greater detail in Appendix S1 in the supplemental material.
sgRNA design and cloning.
A 20-nt base-pairing region (N20) of an sgRNA was designed using the Custom Dicer-Substrate siRNA (DsiRNA) tool (https://eu.idtdna.com/site/order/designtool/index/crispercustom). The sgRNA fragments with N20 were amplified with primers AATACTAGT-N20- GTTTTAGAGCTAGAAATAGC and GGACTGCAGGCAACGTTCAA (restriction sites are underlined) using pISApl1-CRISPR/Cas9 plasmid as the template. The PCR products were successively ligated to predigested pISApl1-CRISPR/Cas9 plasmid at the SpeI and PstI sites, generating pISApl1-CRISPR/Cas9 plasmids containing sgRNAs specific for mcr-1 and IncX4. The cloning procedure of the targeted sgRNA into the pISApl1-CRISPR/Cas9 was shown in Fig. S3 in the supplemental material.
Transposition assays.
The transposition was evaluated by the biparental mating assay with the pISApl1-CRISPR/Cas9-bearing E. coli WM3064 as the donor. In brief, donor strains were grown overnight at 37°C in LB supplemented with 300 μM DAP, and recipients were cultivated until the early stationary phase. Donors and recipients (100 μl of each) were combined in 700 μl LB and then centrifuged for 2 min at 7,000 × g. The cell pellets were washed with phosphate-buffered saline (PBS) three times and then resuspended in 50 μl LB. The mixture was, after that, transferred onto an MF HAWG01300 cellulose membrane (Millipore, Burlington, MA, USA) and placed on the prewarmed LB agar and incubated at 37°C for 12 h. The transconjugants were liberated from the membrane by vortexing in PBS and then serially diluted and plated on ST25 LB and antibiotic-free LB agar plates to calculate the transposition efficiency. To calculate the viable cell CFU of the plasmid-harboring mcr-1 and chromosome-harboring mcr-1 strains, the transconjugants were then serially diluted and plated on ST25 LB agar plates or CS2+ST25 LB agar plates.
In vitro plasmid curing efficiency evaluation.
The strains E. coli CSZ4 and E. coli C600(pUC19-mcr-1), which naturally or artificially harbored the mcr-1 gene and the E. coli C600 strain, were used in the conjugation assay to assess the plasmid curing and transposition efficiency of pISApl1-CRISPR/Cas9-mcr-1. To confirm the loss of the pCSZ4 and pUC19-mcr-1 in the successful transformants, fifty colonies grown on ST25 plates were randomly selected and subjected to PCR detection using primers IncX4-TF+IncX4-TR and mcr-TF+mcr-TR targeting the replication gene and mcr-1 gene of pCSZ4, respectively, and the primers UC19-TF+UC19-TR and mcr-TF+mcr-TR targeting the replication gene and mcr-1 gene of pUC19-mcr-1, respectively. The plasmid curing efficiency was calculated based on the PCR detection results. The experiments were technically repeated twice with three biological replicates.
In vitro sequence-specific bacterial killing using pISApl1-CRISPR/Cas9.
To test the bacterial killing capacity of ISApl1-CRISPR/Cas9, the pISApl1-CRISPR/Cas9 targeting mcr-1 was introduced into E. coli MG1655-MCR by conjugation, and the viable cells after treatment were calculated from the ST25 or ST25+CS2 agar plates. The experiments were technically repeated twice with three biological replicates.
Sequencing of escape mutants.
For plasmid curing, the transformants spread on LB agar plates with ST25 were verified by genotypes and phenotypes. Through PCR, we noticed that a minimal number of colonies were positive and that these colonies could grow on the agar plate of CS2. Then genomic DNA was extracted from escape mutants using a TIANamp bacteria DNA kit (Tiangen, Beijing, China). The Cas9+sgRNA containing regions of each escape mutant were amplified using primers mut-Cas9F and mut-Cas9R and subjected to DNA sequencing.
The ISApl1-CRISPR/Cas9 system preventing plasmid acquisition.
In this section, we used the plasmid-cured strain E. coli C600(ISApl1-CRISPR/Cas9) as the recipient in conjugation assay to see whether the integration of ISApl1-CRISPR/Cas was able to impede the acquisition of mcr-1 from model strains, and the donors were E. coli CSZ4, Salmonella enterica 19E0341, and Klebsiella pneumoniae YZ01, respectively. The conjugation assay was performed as described above. The blockade of mcr-1 transfer was evaluated by conjugation efficiency. The experiments were twice technically repeated with at least three biological replicates.
Accession number(s).
The complete nucleotide sequence of pISApl1-CRISPR/Cas9 was deposited in GenBank under accession number MW811192.
ACKNOWLEDGMENTS
This work was supported by the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (grant 2019BT02N054). This work was supported by the Guangdong Major Project of Basic and Applied Basic Research, no. 2020B0301030007. This work was supported in part by the Program for Innovative Research Team in the University of Ministry of Education of China (IRT_17R39).
We declare no competing interests.
J.S. and X.-P.L. designed the study. Y.-Z.H., J.-R.Y., B.H., and X.K. performed the experiments. Y.-Z.H. and J.S. analyzed the data. Y.-Z.H. made the figures. Y.-Z.H. wrote this manuscript. J.S., H.R., and X.-P.L. edited and revised the manuscript. Y.-H.L. coordinated the whole project. All authors contributed to the article and approved the submitted version.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, Li SM, Liao XP, Feng Y, Liu YH. 2017. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep 7:424. doi: 10.1038/s41598-017-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Xie M, Lv J, Chan W-C, Chen S. 2017. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother 72:696–699. doi: 10.1093/jac/dkw509. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Dobiasova H, Dolejska M. 2016. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and beta-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J Antimicrob Chemother 71:2118–2124. doi: 10.1093/jac/dkw144. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Li XP, Fang LX, Sun RY, He YZ, Lin J, Liao XP, Feng Y, Liu YH. 2018. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents 51:842–847. doi: 10.1016/j.ijantimicag.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang YW, Kreiswirth BN, Du H, Chen L. 2016. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother 60:5033–5035. doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veldman K, van Essen-Zandbergen A, Rapallini M, Wit B, Heymans R, van Pelt W, Mevius D. 2016. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother 71:2340–2342. doi: 10.1093/jac/dkw181. [DOI] [PubMed] [Google Scholar]
- 10.Yu CY, Ang GY, Chong TM, Chin PS, Ngeow YF, Yin WF, Chan KG. 2017. Complete genome sequencing revealed novel genetic contexts of the mcr-1 gene in Escherichia coli strains. J Antimicrob Chemother 72:1253–1255. doi: 10.1093/jac/dkw541. [DOI] [PubMed] [Google Scholar]
- 11.Tegetmeyer HE, Jones SC, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353. doi: 10.1016/j.vetmic.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 12.He Y-Z, Li X-P, Miao Y-Y, Lin J, Sun R-Y, Wang X-P, Guo Y-Y, Liao X-P, Liu Y-H, Feng Y, Sun J. 2019. The ISApl12 dimer circular intermediate participates in mcr-1 transposition. Front Microbiol 10:15. doi: 10.3389/fmicb.2019.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother 60:6973–6976. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Kieffer N, Nordmann P. 2017. In vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother 61:e00127-17. doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faure G, Shmakov SA, Yan WX, Cheng DR, Scott DA, Peters JE, Makarova KS, Koonin EV. 2019. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 17:513–525. doi: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, Dyall-Smith M, Marchfelder A. 2012. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem 287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hryhorowicz M, Lipiński D, Zeyland J, Słomski R. 2017. CRISPR/Cas9 immune system as a tool for genome engineering. Arch Immunol Ther Exp (Warsz) 65:233–240. doi: 10.1007/s00005-016-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kick L, Kirchner M, Schneider S. 2017. CRISPR-Cas9: from a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered 8:280–286. doi: 10.1080/21655979.2017.1299834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong H, Xiang H, Mu D, Wang D, Wang T. 2019. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int J Antimicrob Agents 53:1–8. doi: 10.1016/j.ijantimicag.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Wang P, He D, Li B, Guo Y, Wang W, Luo X, Zhao X, Wang X. 2019. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J Antimicrob Chemother 74:2559–2565. doi: 10.1093/jac/dkz246. [DOI] [PubMed] [Google Scholar]
- 21.Wan P, Cui S, Ma Z, Chen L, Li X, Zhao R, Xiong W, Zeng Z. 2020. Reversal of mcr-1-mediated colistin resistance in Escherichia coli by CRISPR-Cas9 system. Infect Drug Resist 13:1171–1178. doi: 10.2147/IDR.S244885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyo KE, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 23.Friehs K. 2004. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- 24.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA 112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji W, Lee D, Wong E, Dadlani P, Dinh D, Huang V, Kearns K, Teng S, Chen S, Haliburton J, Heimberg G, Heineike B, Ramasubramanian A, Stevens T, Helmke KJ, Zepeda V, Qi LS, Lim WA. 2014. Specific gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth Biol 3:929–931. doi: 10.1021/sb500036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters JE, Makarova KS, Shmakov S, Koonin EV. 2017. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci USA 114:E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petassi MT, Hsieh S-C, Peters JE. 2020. Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. bioRxiv doi: 10.1101/2020.07.02.184150. [DOI] [PMC free article] [PubMed]
- 30.Ma W, Xu YS, Sun XM, Huang H. 2021. Transposon-associated CRISPR-Cas system: a powerful DNA insertion tool. Trends Microbiol 29:565–568. doi: 10.1016/j.tim.2021.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Yu H, Xie M, Chen K, Dong N, Lin D, Chan EW-C, Chen S. 2018. Genetic basis of chromosomally-encoded mcr-1 gene. Int J Antimicrob Agents 51:578–585. doi: 10.1016/j.ijantimicag.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4:E264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet 9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao M, He Y, Zhang H, Liao XP, Liu YH, Sun J, Du H, Kreiswirth BN, Chen L. 2020. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 64:e00843-20. doi: 10.1128/AAC.00843-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauritsen I, Porse A, Sommer MOA, Norholm MHH. 2017. A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb Cell Fact 16:135. doi: 10.1186/s12934-017-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Trebosc V, Gartenmann S, Royet K, Manfredi P, Tötzl M, Schellhorn B, Pieren M, Tigges M, Lociuro S, Sennhenn PC, Gitzinger M, Bumann D, Kemmer C. 2016. A novel genome editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob Agents Chemother 60:7263–7271. doi: 10.1128/AAC.01275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 41.Li XP, Sun RY, Song JQ, Fang LX, Zhang RM, Lian XL, Liao XP, Liu YH, Lin J, Sun J. 2020. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int J Antimicrob Agents 55:105806. doi: 10.1016/j.ijantimicag.2019.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.01054-21-s0001.pdf, PDF file, 13.5 MB (13.5MB, pdf)