Abstract
T follicular helper (Tfh) cells are crucial for B cell-mediated humoral immunity1. Although transcription factors, such as Bcl6, drive Tfh cell differentiation2,3, whether and how posttranscriptional and metabolic programs enforce Tfh cell programming are unclear. Here, we show that the cytidine diphosphate (CDP)-ethanolamine pathway coordinates the expression and localization of CXCR5 with Tfh responses and humoral immunity. Using in vivo CRISPR-Cas9 screening and functional validation, we uncover Etnk1, Pcyt2 and Selenoi – enzymes in the CDP-ethanolamine pathway for de novo phosphatidylethanolamine (PE) synthesis – as selective posttranscriptional regulators of Tfh cell differentiation, by promoting CXCR5 surface expression and functional effects. Tfh cells show unique lipid metabolic programs and PE distribution to the plasma membrane outer layer where it co-localizes with CXCR5. De novo PE synthesis via the CDP-ethanolamine pathway coordinates these events to prevent CXCR5 internalization and degradation. Genetic deletion of Pcyt2, but not the CDP-choline pathway enzyme Pcyt1a, in activated T cells impairs Tfh cell differentiation, associated with reduced humoral immune responses. Surface PE levels and CXCR5 expression on B cells also depend upon Pcyt2. Our results reveal that phospholipid metabolism coordinates posttranscriptional mechanisms for Tfh cell differentiation and humoral immunity, highlighting metabolic control of context-dependent immune signaling and effector programs.
Central carbon metabolism, such as glycolysis, supports the differentiation of multiple T cell subsets4,5, but Tfh-specific metabolic programming remains unclear. To systematically identify metabolic factors mediating Tfh development, we performed in vivo CRISPR-Cas9 screening using a pooled guide RNA (gRNA) library that targeted metabolism-associated genes6,7 (Extended Data Fig. 1a). SMARTA-transgenic T cells expressing Cas9 were transduced with gRNA library and transferred into C57BL/6 hosts, followed by LCMV Armstrong infection (Extended Data Fig. 1a). At day 7 post-infection, the representation of downregulated (i.e. Tfh positive regulators) or upregulated gRNAs in Tfh (CXCR5+SLAM−) versus Th1 (CXCR5−SLAM+) cells8,9 were examined (|log2 (Tfh/Th1 ratio)| > 0.5; adjusted P < 0.05). We identified 229 significantly altered genes, including known Tfh positive (Ric8a10 and Pik3cd11) or negative regulators (Tet212, Stat5a and Stat5b13) (Fig. 1a and Supplementary Table 1).
Figure 1. In vivo CRISPR-Cas9 screening reveals that CDP-ethanolamine pathway is critical for Tfh differentiation.
a, Scatterplot of gene enrichment (n = 6 sgRNAs per gene). b, c, mCherry+ and Ametrine+ sgRNA-transduced SMARTA cells were mixed at 1:2 and transferred into C57BL/6 recipients followed by LCMV infection. Analysis of donor-derived splenic Tfh (CXCR5+SLAM− or PSGL-1−Ly6C−) and Th1 (CXCR5−SLAM+ or PSGL-1+Ly6C+) cells at day 7 post-infection (n = 4 mice). d, Summary of CDP-ethanolamine pathway genes in Tfh generation. e, Heatmap of the enrichment of indicated genes ([log2 ratio (input/Tfh)]). f, g, Principal component analysis of lipidome (f) and quantification of PE content (g) in the indicated cells (n = 3 samples, each pooled from multiple mice). h, Distribution and the quantification of SMATRA cells in the splenic follicle at day 3 post-infection (sgNTC, n = 50 sections; sgPcyt2, n = 47 sections). Scale bar, 50 μm. i, Analysis of splenic GC B cells (B220+CD19+Fas+GL7+) and plasma cells (B220−CD138+) in LCMV-infected CD45.2+ mice receiving CD45.1+ wild-type or Pcyt2-deficient CXCR5+SLAM− Tfh cells (n = 4 mice). Data are representative of one (a, e), two (f–i), or at least three (b–d) independent experiments. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, and ***P < 0.001. Two-tailed paired Student’s t-test followed by Bonferroni correction (a), one-way ANOVA (c, g, i) or two-tailed unpaired Student’s t-test (h).
CDP-ethanolamine pathway in Tfh response
Etnk1 (encodes for ethanolamine kinase 1) and Pcyt2 (phosphate cytidylyltransferase 2) – two key components of the CDP-ethanolamine pathway that promotes de novo phosphatidylethanolamine (PE) biosynthesis14 – were the first and third top candidates as Tfh positive regulators, respectively (Fig. 1a). To establish cell-intrinsic effects, we used a dual transfer system6,7, where SMARTA cells expressing non-targeting control single gRNAs (sgNTC; mCherry+) and gene-specific gRNAs (sgRNAs; Ametrine+) were transferred into the same host, followed by LCMV infection (Extended Data Fig. 1b). As expected, targeting Prdm1 (encodes Blimp1)2 or Bcl62,3 increased and decreased Tfh accumulation, respectively (Extended Data Fig. 1c). Targeted sequencing and immunoblots revealed efficient disruption of Pcyt2 and Etnk1 (Extended Data Fig. 1d, e). Importantly, depletion of Etnk1 or Pcyt2 led to a marked reduction of CXCR5+SLAM− Tfh cells at day 7 post-infection (Fig. 1b, c), as well as PSGL-1−Ly6C− and CXCR5+PD-1+ Tfh cells15 (Fig. 1b and Extended Data Fig. 2a). In contrast, Th1 cells (CXCR5–SLAM+ or PSGL-1+Ly6C+) were increased (Fig. 1b and Extended Data Fig. 2b), revealing Etnk1 and Pcyt2 as selective Tfh positive regulators.
We next depleted additional enzymes involved in PE generation: Etnk2 (ethanolamine kinase 2), Selenoi (Selenoprotein I) and Cept1 (choline/ethanolaminephosphotransferase 1) (Fig. 1d and Extended Data Fig. 2c, d). Targeting Selenoi reduced Tfh cells (Extended Data Fig. 2e), whereas loss of Etnk2 or Cept1 had no effect (Fig. 1e and Extended Data Fig. 2d, e), even in Etnk1 or Selenoi-deficient backgrounds. Phosphatidylserine (PS) decarboxylation by PS decarboxylase (Pisd) also generates PE14, but targeting Pisd did not impact Tfh responses (Extended Data Fig. 2c–e). The inhibitory effects of Etnk1, Pcyt2 or Selenoi deletion on Tfh generation were verified in a protein-immunization model (Extended Data Fig. 2f). Collectively, Tfh generation depends upon Etnk1, Pcyt2 and Selenoi, indicating the critical requirement of de novo PE biosynthesis in Tfh responses.
Lipidomic analysis revealed that dynamic phospholipid profiles accompanied Tfh cell differentiation from naïve T cells (Fig. 1f and Supplementary Table 2), with increased PE, phosphatidylcholine (PC), and PS content (Fig. 1g and Extended Data Fig. 3a). Pcyt2 is a rate-limiting step of the CDP-ethanolamine pathway14, and accordingly, Pcyt2-deficient Tfh cells had decreased total PE, increased PC and unaltered PS content (Fig. 1g, Extended Data Fig. 3a and Supplementary Table 2), with PE being the most downregulated lipid species (Extended Data Fig. 3b). Moreover, 32 of the 44 significantly upregulated PE molecules in Tfh cells (versus naïve cells) were dampened by Pcyt2 deficiency (Extended Data Fig. 3c). Lipidomics also revealed the reduction of PE but not PC content upon Selenoi depletion (Extended Data Fig. 3d), including 47 ether-type and 32 diacyl-type PE (Supplementary Table 3a), thereby revealing Pcyt2 and Selenoi-dependent PE synthesis in Tfh cells.
We next explored the dynamics of PE biosynthesis and turnover via pulse-chase with ethanolamine (see methods). Tfh cells incorporated more [3H]-radiolabeled ethanolamine ([3H]-Etn) into lipids than Th1 cells (Extended Data Fig. 3e), indicative of increased PE synthesis. Further, after chasing with unlabeled ethanolamine, [3H] level was reduced in Tfh cells, accompanied by an increased release into the medium (Extended Data Fig. 3e). Thus, Tfh cells show greater PE synthesis and turnover than Th1 cells.
The Kennedy pathway encompasses CDP-ethanolamine and CDP-choline branches for PE and PC synthesis, respectively (Extended Data Fig. 4a)14. Depletion of genes in the CDP-choline pathway (Chka, Chkb, Pcyt1a, Pcyt1b, Chpt1, Chka and Chkb, Pcyt1a and Pcyt1b, or Cept1 and Chpt1) did not alter Tfh responses (Extended Data Fig. 4b, c), despite the reduction of total PC but not PE amounts in Tfh cells lacking Pcyt1a and Pcyt1b or the rate-limiting enzyme Pcyt1a16 (Extended Data Fig. 4d). Therefore, the CDP-choline pathway is dispensable for Tfh generation.
We next analyzed the cellular processes regulated by the CDP-ethanolamine pathway. Pcyt2-deficient cells displayed intact proliferation and survival (Extended Data Fig. 4e, f). However, depletion of Pcyt2 led to a marked reduction of T cells in the B cell follicle15 after LCMV infection (Fig. 1h and Extended Data Fig. 4g), indicating impaired T cell follicular recruitment. We then sorted Tfh cells at day 7 post-infection and transferred them into LCMV-infected mice and analyzed GC B and plasma cells 5 days later8 (Extended Data Fig. 4h). Mice that received Pcyt2-deficient Tfh cells exhibited significantly fewer GC B and plasma cells (Fig. 1i). Therefore, the CDP-ethanolamine pathway is critical for follicular recruitment and Tfh function.
Regulation of CXCR5 surface expression
To establish the underlying mechanisms, we profiled the transcriptome of wild-type and Pcyt2-deficient cells at day 3 post-infection. Gene-set enrichment analysis showed that the transcriptional signature of Tfh cells9 was not significantly altered by Pcyt2 deficiency (Extended Data Fig. 5a). At day 3 post-infection, Etnk1, Pcyt2 or Selenoi deficiency did not influence the respective Tfh or Th1-specific Bcl6 or T-bet2,3 (Fig. 2a and Extended Data Fig. 5b), despite the altered expression at later time points (Extended Data Fig. 5c). Also, Pcyt2 deficiency did not affect Bcl6 target genes17 or those directly downregulated by Bcl617 (Extended Data Fig. 5d, e), or the proportion of PSGL-1–Ly6C– cells (Extended Data Fig. 5f), suggesting intact initial Tfh programming. Additionally, Bcl6 overexpression in Pcyt2-deficient cells did not restore Tfh generation, despite an overall increase in Tfh cells (Fig. 2b). Moreover, deletion of Bcl6 or Pcyt2 reduced Tfh responses to a comparable degree (Extended Data Fig. 5g), whereas their co-deletion further impaired Tfh generation (Extended Data Fig. 5g). Thus, the CDP-ethanolamine pathway acts largely independently of Bcl6.
Figure 2. CDP-ethanolamine pathway regulates CXCR5 expression independently of Bcl6.
a, sgRNA-transduced SMARTA cells were transferred into C57BL/6 recipients followed by LCMV infection. Analysis of donor-derived splenic CXCR5+Bcl6+ cells at day 3 post-infection (left). Right, mean fluorescence intensity (MFI) of Bcl6 and CXCR5 (n = 4 mice). b, SMARTA cells were transduced with indicated retrovirus, followed by adoptive transfer and LCMV infection. Analysis of CXCR5+PD-1+ Tfh cells in donor-derived cells at day 3 post-infection (n = 3 mice). Numbers above the graph indicate the fold changes. c, Analysis of CXCR5 and PD-1 MFI on donor-derived cells at day 2 post-infection (n = 4 mice). d, Proportion of migrated CXCR5+SLAM− Tfh cells after 3 h of CXCL13 (n = 4 mice) or CCL21 treatment (n = 6 mice). e, Analysis of frequency of phosphorylated AKT (pAKT-S473)+ in donor-derived cells at day 3 post-infection (n = 4 mice). Data are representative of two (d) or at least three (a–c, e) independent experiments. Data are mean ± s.e.m. NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. One-way ANOVA (a) or two-tailed unpaired Student’s t-test (b–e).
Despite these observations, CXCR5+PD-1+ Tfh accumulation was markedly decreased at day 3 post-infection by CDP-ethanolamine pathway depletion (Extended Data Fig. 5h). To test whether these defects are associated with altered Tfh signature molecules15, we first analyzed expression of CXCR5 on Tfh cells (defined by Bcl6+ or PSGL-1–Ly6C–) at day 3 post-infection. While nearly all wild-type Bcl6+ cells co-expressed CXCR5, depletion of the CDP-ethanolamine pathway markedly reduced the percentage of CXCR5+Bcl6+ Tfh cells, and, more notably, CXCR5 expression on Bcl6+ cells (Fig. 2a), or among PSGL-1−Ly6C− Tfh cells (Extended Data Fig. 5f). At day 2 post-infection, Pcyt2 deficiency also reduced CXCR5+PD-1+ Tfh cells, associated with impaired CXCR5 but not PD-1 expression (Fig. 2c and Extended Data Fig. 5i, j). Importantly, CXCR5 overexpression significantly rectified the defective Tfh generation in the absence of Pcyt2 (Extended Data Fig. 5k). Therefore, the CDP-ethanolamine pathway promotes CXCR5 expression on early Tfh cells.
After targeting the CDP-ethanolamine pathway enzymes, PD-1 expression was unaltered at day 2 (Fig. 2c) and modestly reduced at day 3 post-infection (Extended Data Fig. 5l). However, PD-1 antagonizes Tfh follicular recruitment18, so reduced PD-1 expression is likely a consequence of dampened CXCR5 expression15. ICOS, but not CD62L or CD44, expression was increased in the mutant cells (Extended Data Fig. 5l). Th1-associated CXCR3 showed a trending increase, while CCR7 expression was intact upon Pcyt2 depletion (Extended Data Fig. 5m, n). These results reveal a selective reduction of CXCR5 expression upon CDP-ethanolamine pathway depletion.
Accordingly, Pcyt2-deficient cells showed defective migration toward CXCL13, but not to the CCR7 ligand CCL21 (Fig. 2d). CXCR5 activates PI3K-AKT to promote Tfh motility18, so we measured phosphorylation of AKT (pAKT-S473)4,18. Pcyt2 deficiency markedly reduced pAKT-S473+ cells ex vivo (Fig. 2e) or after CXCL13 stimulation in vitro (Extended Data Fig. 5o). Collectively, CDP-ethanolamine pathway promotes CXCR5 expression and CXCR5-dependent cellular and molecular events.
PE surface distribution on Tfh membrane
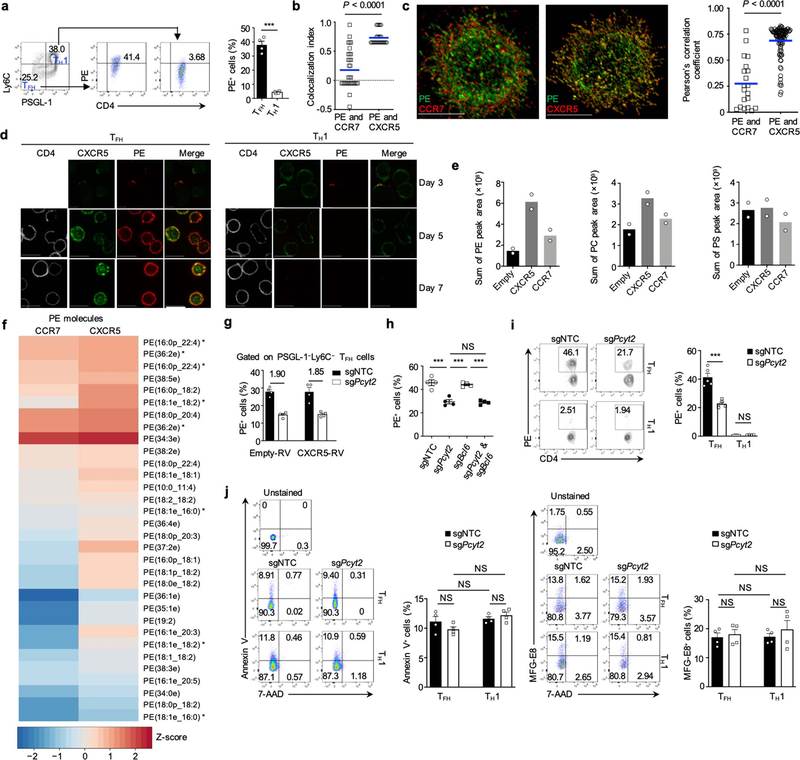
PE primarily resides in the inner leaflet19, except for during cell division or fusion20. However, context-dependent regulation and function of PE localization remain largely unexplored. Flow cytometry (Extended Data Fig. 6a) and confocal imaging (see methods; Fig. 3a) of staining with duramycin, a polypeptide that binds PE with high specificity on the outer leaflet of the plasma membrane in live cells19, revealed increased surface level of PE on the Tfh compared with Th1 plasma membrane. Imaging analysis also revealed the co-localization of surface PE and CXCR5 on Tfh cells (Fig. 3a). This observation was validated by high-resolution total internal reflection (TIRF)–stochastic optical reconstruction microscopy (STORM) imaging using independent algorithms to quantify co-localization (Fig. 3b and Extended Data Fig. 6b), and 3D confocal microscopy in combination with optical reassignment and deconvolution (Extended Data Fig. 6c), both of which revealed significantly stronger co-localization signals when comparing PE and CXCR5 with PE and CCR7. Moreover, we performed extracellular immunogold labeling combined with scanning electron microscopy and found the existence of discrete islands, where PE and CXCR5 were co-localized on the outer leaflet of the plasma membrane (Fig. 3c). We next assessed the temporal regulation of PE abundance during LCMV infection and observed a progressive accumulation of surface PE level during Tfh but not Th1 cell differentiation (Extended Data Fig. 6d). Thus, PE and CXCR5 show spatially and temporally coordinated expression in Tfh cell differentiation.
Figure 3. PE selectively distributes on the outer layer of Tfh membrane and stabilizes CXCR5.
a, Confocal microscopy and analyses of PE and CXCR5 co-localization and PE mean fluorescence intensity (MFI) on PSGL-1−Ly6C− Tfh cells and PSGL-1+Ly6C+ Th1 cells (Tfh, n = 99 cells; Th1 n = 85 cells). Scale bar, 10 μm. b, TIRF (total internal reflection fluorescence)–STORM (stochastic optical reconstruction microscopy) analysis and quantification (via NSInC) of PE and CCR7 or CXCR5 co-localization on PSGL-1−Ly6C− Tfh cells. Scale bar, 1 μm (PE : CCR7, n = 33 cells; PE : CXCR5, n = 25 cells). c, Scanning electron microscopy of PE and CXCR5 co-localization on the outer layer membrane of Tfh cells. Scale bar, 5 μm (n = 12 cells). Yellow rings show discrete islands. d, Image analysis of PE and CXCR5 levels on Tfh and Th1 cells (sgNTC Tfh, n = 120 cells; sgPcyt2 Tfh, n = 157 cells; sgNTC Th1, n = 72 cells; sgPcyt2 Th1, n = 45 cells). Scale bar, 10 μm. e, sgNTC and sgPcyt2-transduced SMARTA cells were supplemented with ether-type PE or diacyl-type PE, followed by adoptive transfer and LCMV infection. Quantification of CXCR5+PD-1+ Tfh cells and CXCR5 MFI on donor-derived cells (n = 5 mice). f, Images of Tfh cell CXCR5 internalization. Scale bar, 5 μm. g, CXCR5 internalization (see Methods; n = 4 mice). h, Immunoblot of CXCR5 in cells treated with cycloheximide (CHX) (n = 2 samples, each pooled from multiple mice). Data are representative of two (c, h) or at least three (a, b, d–g) independent experiments. Data are mean ± s.e.m. NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. One-way ANOVA (e) or two-tailed unpaired Student’s t-test (a–d, g, h).
To further examine PE and CXCR5 interaction, we quantified membrane-associated lipids bound to FLAG-tagged CXCR5 or CCR7 proteins, and found a higher abundance of PE bound to FLAG-CXCR5 (Extended Data Fig. 6e), including 32 PE molecules selectively bound to CXCR5 (Extended Data Fig. 6f). In contrast, PC was pulled down by CXCR5 with a modest increase over CCR7, while PS amounts did not show differences (Extended Data Fig. 6e). We next examined the signaling relationship of CXCR5 and PE, and found that the selective surface level of PE on Tfh cells was unaffected by CXCR5 expression or Bcl6 deficiency (Extended Data Fig. 6g, h). However, Pcyt2 deficiency decreased PE surface level on Tfh cells but not that of Th1 cells that contained little surface PE (Fig. 3d and Extended Data Fig. 6i). In contrast, wild-type or Pcyt2-deficient Tfh and Th1 cells showed comparable levels of surface PS (Extended Data Fig. 6j). Thus, Pcyt2 is required for the outer layer distribution of PE on Tfh cell membrane.
Ethanolamine serves as the initial substrate for the CDP-ethanolamine pathway14, so we next measured the incorporation of [3H]-Etn into cellular lipids. The incorporation of [3H]-Etn into Tfh cells was substantially reduced by Pcyt2 depletion (Extended Data Fig. 7a). To directly assess the functional contribution of PE to Tfh responses, we supplemented SMARTA cells with ether-type or diacyl-type PE before adoptive transfer (Extended Data Fig. 7b), and analyzed these mice at day 3 post-infection. PE supplementation enhanced cellular PE amounts in wild-type and Pcyt2-deficient cells (Extended Data Fig. 7c), and considerably rectified the downregulated PE molecules caused by Pcyt2 depletion (Extended Data Fig. 7c and Supplementary Table 3b), as well as the defective surface level of PE (Extended Data Fig. 7d). Importantly, PE supplementation reversed the defective CXCR5+ Tfh generation in Pcyt2-deficient cells (Fig. 3e and Extended Data Fig. 7e), indicating the role of PE for supporting these processes.
We explored additional processes that may underlie preferential surface presentation of PE on Tfh cells. The KEGG signature of ABC transporters was significantly (FDR < 0.05) upregulated in Tfh compared to Th1 cells (Extended Data Fig. 7f, g). To identify the possible underlying driver genes, we targeted 48 individual lipid translocases20 (Supplementary Table 4a) and found that 19 of the 48 lipid translocases reduced the total cellularity but not the frequency of Tfh cells (Supplementary Table 4b), suggesting that these lipid translocases are important for cell expansion but not Tfh cell differentiation.
To establish mechanisms of CXCR5 regulation, we performed internalization assays21 and observed internalization of surface CXCR5 in wild-type Tfh cells (Fig. 3f and Extended Data Fig. 8a). The internalization rate of CXCR5 but not PD-1 was accelerated upon Pcyt2 depletion (Fig. 3g and Extended Data Fig. 8b). Moreover, the majority of internalized CXCR5 in wild-type cells recycled back to the cell surface, but CXCR5 recycling was impaired by Pcyt2 deficiency (Extended Data Fig. 8c, d).
To test whether CXCR5 is rerouted for degradation in Pcyt2-deficient cells, we measured the half-life of CXCR5 after treatment with translational inhibitor cycloheximide. Although CXCR5 expression (Extended Data Fig. 8e) was relatively stable in wild-type Tfh cells, Pcyt2 deficiency reduced its half-life (Fig. 3h), indicating that the CDP-ethanolamine pathway promotes CXCR5 stability. Inhibition of lysosomal acidification with bafilomycin A1 (BafA1), but not of proteasome activity, stabilized the total amount of CXCR5 protein in Pcyt2-deficient cells (Extended Data Fig. 8f, g). However, CXCR5 surface expression was unaltered by BafA1 treatment (Extended Data Fig. 8h), consistent with an upstream effect of PE synthesis at shaping CXCR5 surface expression.
We further extended our analysis to human cells. In peripheral blood, surface level of PE was significantly higher on CXCR5+ memory Tfh than CXCR5+ central memory or CD45RA+ naïve-like T cells (Extended Data Fig. 8i). Additionally, in the tonsil, PE surface level on CXCR5+PD-1hi Tfh or transitional CXCR5+PD-1+ Tfh cells was significantly higher than that on CXCR5− non-Tfh or naïve-like T cells (Extended Data Fig. 8j). Therefore, PE surface distribution represents an evolutionarily conserved process.
Loss of Pcyt2 impairs humoral immunity
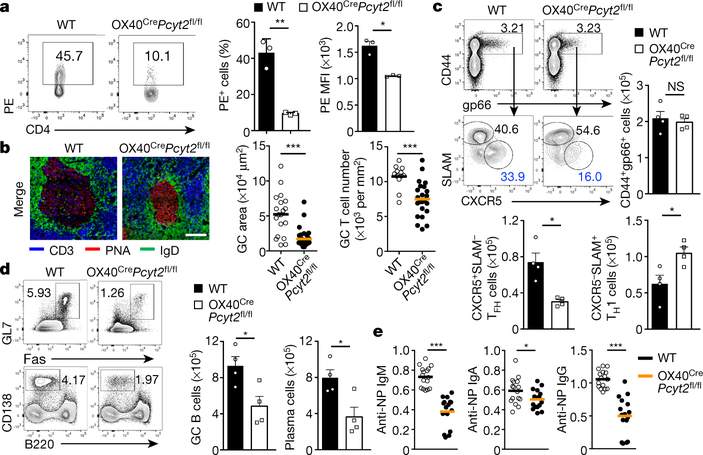
We generated OX40CrePcyt2fl/fl mice to delete Pcyt2 in activated CD4+ T cells4 (Extended Data Fig. 9a). OX40CrePcyt2fl/fl mice showed intact T cell development, in vitro proliferation and Th1, Th2, Th17 and Treg cell differentiation (data not shown). In contrast, homeostatic CXCR5+ Tfh cells and GC B cells in Peyer’s patches (PPs) and mesenteric lymph nodes (mLNs)4 were decreased in OX40CrePcyt2fl/fl mice (Extended Data Fig. 9b, c). Also, the frequency of PE+ Tfh cells was significantly decreased in OX40CrePcyt2fl/fl mice (Fig. 4a), while that of PD-1+ or Bcl6+ cells remained unchanged (Extended Data Fig. 9b). Additionally, OX40CrePcyt2fl/fl mice displayed smaller GC size and fewer T cells in GCs (Fig. 4b and Extended Data Fig. 9d). In contrast, mice with OX40Cre-mediated deletion of Pcyt1a16 showed intact Tfh responses (Extended Data Fig. 9e–g). We next generated mixed bone marrow (BM) chimeras, and found that Pcyt2-deficient donors had severely defective Tfh cells in this competitive environment (Extended Data Fig. 9h). Hence, impaired Tfh cell differentiation upon Pcyt2 deletion is a cell-intrinsic defect.
Figure 4. Genetic ablation of CDP-ethanolamine pathway disrupts humoral immunity.
a, Analysis of percentage of PE+ cells and PE mean fluorescence intensity (MFI, in PE+ cells) on Tfh cells from Peyer’s patches of WT and OX40CrePcyt2fl/fl mice (n = 3 mice). b, Immunohistochemistry of GCs in the mesenteric lymph nodes of WT and OX40CrePcyt2fl/fl mice. Right, GC area size (WT, n = 19 sections; OX40CrePcyt2fl/fl, n = 33 sections) and T cell number in GCs (WT, n = 12 sections; OX40CrePcyt2fl/fl, n = 23 sections). Scale bar, 50 μm. c, Analysis of gp66 tetramer-positive splenic CXCR5+SLAM− Tfh and CXCR5−SLAM+ Th1 cells in WT and OX40CrePcyt2fl/fl mice at day 7 post-infection (n = 4 mice). d, Analysis of GC B cells (B220+CD19+Fas+GL7+) and plasma cells (B220−CD138+) in WT and OX40CrePcyt2fl/fl mice at day 7 post-infection (n = 4 mice). e, Measurements of serum anti-NP immunoglobulins from WT and OX40CrePcyt2fl/fl mice at day 7 after NP-OVA + LPS immunization (n = 16, collected from 8 mice). Data are representative of two (c–e) or at least three (a, b) independent experiments. Data are mean ± s.e.m. NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. Two-tailed unpaired Student’s t-test (a–e).
Following LCMV challenge, antigen-specific CD4+ T cells were unchanged in OX40CrePcyt2fl/fl mice (Fig. 4c). However, antigen-specific Tfh cells were reduced, with a corresponding increase of Th1 cells (Fig. 4c and Extended Data Fig. 9i). Also, OX40CrePcyt2fl/fl mice had a much lower frequency of GC B and plasma cells (Fig. 4d). Furthermore, naïve T cells deficient for Etnk1 or Selenoi, generated via the retrogenic system, showed defective Tfh cell differentiation (Extended Data Fig. 9j), further supporting the importance of the CDP-ethanolamine pathway for Tfh generation.
Tfh responses promote antigen-specific immunoglobulin production1. OX40CrePcyt2fl/fl mice had reduced Tfh and GC B cells after NP-OVA + LPS immunization (Extended Data Fig. 9k). Moreover, there was a marked reduction of anti-NP-specific immunoglobulin secretion (Fig. 4e and Extended Data Fig. 9l). In contrast, no defect was observed in a model of T cell-independent immunoglobulin production (data not shown), indicating the selective requirement of Pcyt2 for T cell-dependent antibody responses.
Moreover, a large proportion of mature CXCR5+ naïve B cells was stained positive for surface PE, in contrast to immature B cells22 and naïve T cells that did not express CXCR5 or surface PE (Extended Data Fig. 10a). Peripheral B220+IgM+ cells, which contain transitional B cells, showed limited CXCR5 and PE co-expression (Extended Data Fig. 10a). We therefore generated Cd19CrePcyt2fl/fl mice and found that Pcyt2 deficiency in B cells decreased surface levels of PE and CXCR5 on B cells, associated with reduced chemotaxis toward CXCL13 (Extended Data Fig. 10b, c). Therefore, CDP-ethanolamine pathway also supports CXCR5 expression on B cells.
Here, we identify the CDP-ethanolamine pathway as a selective metabolic and posttranscriptional checkpoint to program Tfh cell differentiation, and establish the signaling function of de novo PE synthesis for underpinning effector T cell fate decisions (Extended Data Fig. 10d, e). The selectivity is further highlighted by the crucial requirement of all of the three enzymatic node genes in the CDP-ethanolamine pathway, but not the related CDP-choline pathway or PS decarboxylation, in Tfh responses. We demonstrate that plasma membrane-associated PE, which normally resides in the inner leaflet19, distributes to the outer layer of Tfh but not Th1 cells in a highly-regulated, temporal and spatial manner. Our findings address a long-standing question in Tfh biology of how the dynamic expression of CXCR5 is regulated, by establishing mechanisms shaping CXCR5 membrane localization and stability. Thus, the CDP-ethanolamine pathway likely serves as a new therapeutic target to tune Tfh responses and humoral immunity for vaccination or autoimmunity.
Methods
Mice, LCMV infection and NP-OVA immunization
Mice were housed and bred at the St. Jude Children’s Research Hospital Animal Resource Center in specific pathogen-free conditions. C57BL/6, CD45.1+, OX40Cre, Cd19Cre, Rag1–/–, Rosa26-Cas923 and SMARTA TCR transgenic24 mice were purchased from the Jackson Laboratory. Description of the construct design and generation of Pcyt2fl and Pcyt1afl mice has been detailed elsewhere25,26. Female and male mice were used at 6–10 weeks of age. Age and sex-matched mice were assigned randomly to experimental and control groups. BM chimeras were generated by transferring 1 × 107 T cell-depleted BM cells into sub-lethally irradiated (5.5 Gy) Rag1−/− mice. Mice remained on antibiotic (Baytril) water for 2–3 weeks, and after 6 weeks, reconstitution was determined by flow cytometry analysis of blood samples. For the generation of retrogenic mice27, lineage-negative BM cells were cultured in DMEM medium with 20% FBS, 20 ng/ml mouse IL-3 (Thermo Fisher Scientific), 50 ng/ml human IL-6 (BD Biosciences) and 50 ng/ml murine SCF (PeproTech). After 2 days, the BM cells were transduced with sgNTC, sgEtnk1 or sgSelenoi retrovirus, followed by culture in the DMEM medium supplemented with cytokines described above for another day. Then, a total of 0.5 × 106 cells were adoptively transferred into sub-lethally irradiated (5.5 Gy) CD45.1+ mice. Mice remained on Baytril water for 2–3 weeks, and after 6 weeks, reconstitution was determined by flow cytometry analysis of blood samples. For adoptive transfer, a total of 1 × 106 (for analysis at day 2 or 3) or 5 × 105 (for analysis at day 7) naïve CD45.1+ SMARTA cells (or retrovirus-transduced SMARTA cells) were adoptively transferred into naïve wild-type (CD45.2+) mice. For evaluation of Tfh function, 1 × 105 sorted wild-type or Pcyt2-deficient CXCR5+SLAM− Tfh cells were adoptively transferred into wild-type recipient mice 1 day after infection of the host mice with LCMV. For LCMV infection, 2 × 105 plaque-forming units (PFU) of LCMV Armstrong strain virus were in injected intraperitoneally for analysis of virus-induced Tfh responses as indicated in figure legends. For NP-OVA immunization, 100 μg NP-OVA, 10 μg LPS and alum adjuvant (10%) were mixed and injected intraperitoneally for analysis of NP-specific antibody response and Tfh cell differentiation4. For NP-AECM-FICOLL immunization, 40 μg NP-AECM-FICOLL in sterile PBS was injected intraperitoneally for analysis of T cell-independent NP-specific antibody response. Mice were on 12-hour light/dark cycles that coincide with daylight in Memphis, TN, USA. The St. Jude Children’s Research Hospital Animal Resource Center housing facility was maintained at 20–25 oC and 30–70 % humidity. All experiments with mice were conducted in accordance with the St. Jude Children’s Research Hospital institutional policies, and animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Human PBMC and tonsil mononuclear cell isolation and flow cytometry staining
Peripheral blood mononuclear cells (PBMCs) were isolated by using lymphocyte separation medium (Sigma-Aldrich). PBMCs (5 × 106) were stained on ice for 30 min with 2.5 μM duramycin-LC-biotin (D-1003, Molecular Targeting Technologies) followed by washing and staining with BV605 streptavidin (BD Biosciences), FITC anti-human CD45RO (Biolegend), APC anti-human CD45RA (Biolegend), PE-Cy7 anti-human CD4 (Biolegend), PE anti-PD-1 (Thermo Fisher Scientific), and BV421 anti-human CXCR5 (BD Biosciences) for 30 min. To analyze human tonsil Tfh cells28, tonsils were processed by mechanical disruption on 40 μm sterile cell culture strainer (Fisherbrand) and tonsil mononuclear lymphocytes were isolated using lymphocyte separation medium. Tonsil mononuclear cells (1 × 106) were stained at room temperature for 20 min with 0.5 μM duramycin-LC-biotin. Samples were acquired using a LSR Fortessa flow cytometer (BD Biosciences). All human studies were in compliance with the Declaration of Helsinki. Blood donors were recruited by the Blood Donor Center at St. Jude Children’s Research Hospital. Blood donors provided written consent for research use of their blood products not used in transfusions, which has been reviewed and approved by the Institutional Review Board at St. Jude Children’s Research Hospital. Tonsil donors were recruited by the Department of Otolaryngology at College of Medicine in the University of Tennessee Health Science Center (UTHSC). Tonsil donors provided written consent for their tonsil tissue to be used for translational immunology research, which has been approved and reviewed by the Institutional Review Board at UTHSC.
Cell culture and viral transduction
Naïve Cas9-expressing SMARTA cells were isolated from the spleen and pLN of Cas9-SMARTA mice using the naïve CD4+ T cell isolation kit (Miltenyi Biotec 130–104-453) according to the manufacturer’s instructions. Purified naïve SMARTA cells were activated in vitro for 18 h with 5 μg/ml anti-CD3 (2C11; Bio X Cell) and 5 μg/ml anti-CD28 (37.51; Bio X Cell) before viral transduction. Viral transduction was performed by spin-infection at 800 g at 25 oC for 3 h with 10 μg/ml polybrene (Sigma). Cells then cultured with human IL-2 (100 UI/ml; PeproTech) or mouse IL-7 (2 ng/ml; PeproTech) for 4 days. Transduced cells were sorted using a Reflection (i-Cyt) before adoptive transfer into recipients. sgRNAs were designed by using an online tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). sgRNAs used in this study were as follows or provided in Supplementary Table 4b: non-targeting control sgRNA: ATGACACTTACGGTACTCGT; sgEtnk1: GCCGCACTGGGACCCCCGGG; sgEtnk1 #2: GGTGCCGAAGCTCGACGTGA; sgEtnk2: GATGCATCGTTATTTCACGC; sgEtnk2 #2: TAGGTTGATCGCCTTAGAAA (which gave similar results as sgEtnk2; data not shown); sgPcyt2: GTGTAGGCCGGCGATGACGT; sgPcyt2 #2: GTGTCCACCACAGACCTCGT; sgSelenoi: GTCAAACAGCTCCCCTAACG; sgSelenoi #2: GTGTACTCCATCTTTGGACG (which gave similar results as sgSelenoi; data not shown); sgCept1: GAGATTTGGGGCAATCCATG; sgCept1 #2: TAAAACGGCTAGAAGAACAC (which gave similar results as sgCept1; data not shown); sgChka: AGTGCTCTTGCGGCTCTATG; sgChka #2: CTTGGACTCCAGCTCGCCTG (which gave similar results as sgChka; data not shown); sgChkb: ATCTCTACCAGGTTCATCTG; sgChkb #2: CAGAGCCAATCCCCAATCGG (which gave similar results as sgChkb; data not shown); sgPcyt1a: AATACGTATCTAATTGTGGG; sgPcyt1a #2: CTTTAGTAAGCCCTATGTCA (which gave similar results as sgCept1; data not shown); sgPcyt1b: GTGCCCTTGCGTGACCTGAG; sgPcyt1b #2: GCCTCCCTCAGAAACCATGG (which gave similar results as sgPcyt1b; data not shown); sgChpt1: GGAGGAGCACCGCTACACCG; sgChpt1 #2: GTAGAAGATGAGCACTAGTG (which gave similar results as sgChpt1; data not shown); sgPisd: GGGCCCTCGAGCCAATACAG; sgPisd #2: ACCAGCTGGTCACCCGGGAA (which gave similar results as sgPisd; data not shown). The coding sequence of Cxcr5 was obtained from Harvard PlasmID Database (Harvard PlasmID # MmCD00310701), and coding sequences of Abcf2, Abcg1, Abcd3, Abcd1, Abcc4, Abcg5, Abcb5, Abcb9, and Ano4 were purchased from GenScript. These sequences were subcloned into pMIG-II retroviral vector (Addgene # 52107), which were co-transfected into Plat-E cells with the helper plasmid pCL-Eco (Addgene # 12371) for the production of retrovirus.
Lentiviral sgRNA metabolic library CRISPR-Cas9 mutagenesis screening
Lentiviral and retroviral sgRNA vector design
The lentiviral sgRNA vector was generated from lentiGuide-puro vector by replacing the “EF-1α PuroR” fragment with a mouse PGK promoter-driven Ametrine (or GFP or mCherry) fluorescent protein6. The retroviral sgRNA vector was generated from pLMPd-Amt vector29 by replacing the miR30 shRNA cassette with the U6 promoter driven gRNA cassette from the lentiGuide-puro vector.
Lentiviral sgRNA metabolic library construction
The gene list of mouse metabolic library was based on the reported human metabolic genes30. A total of 6 gRNAs were designed for each mouse metabolic gene according to previously published selection criteria6,31 and were split into two sub-libraries (AAAQ05 and AAAR07, Supplementary Table 1), each containing 500 non-targeting controls. Oligonucleotides containing the guide sequence were synthesized (Custom Array), PCR amplified and cloned into the recipient vector via a Golden Gate cloning procedure, including 5 μl Tango Buffer (Thermo Fisher Scientific), 5 μl DTT (10 mM stock), 5 μl ATP (10 mM stock), 500 ng vector (pre-digested with Esp3I, gel-extracted, and isopropanol-precipitation purified), 100 ng insert PCR product, 1 μl Esp3I (Thermo Fisher Scientific ER0452), 1 μl T7 ligase (Enzymatics, 3,000 U/μl, L6020L), and water, up to 50 μl, and incubated in cycle (5 min at 37 °C and 5 min at 20 °C) for 100 times. The product was then purified by isopropanol precipitation and electroporated into STBL4 cells (Life Technologies 11635018). The distribution of the library was determined by Illumina sequencing.
In vivo screening
Lentivirus was produced by co-transfecting HEK293T cells with the lentiviral metabolic library plasmids, psPAX2 (Addgene plasmid # 12260) and pCAG4-Eco. At 48 h after transfection, virus was harvested and frozen at −80 °C. Naïve Cas9-expressing SMARTA cells were isolated from 6 Cas9-SMARTA mice and transduced at a MOI of 0.3 to achieve ~20% transduction efficiency. After viral transduction, cells were cultured with human IL-2 (140 IU/ml; PeproTech) for 3 days, then transferred to medium containing 2 ng/ml mouse IL-7 (PeproTech) for 1 day. Transduced cells expressing Ametrine were sorted using a Reflection sorter (i-Cyt), and an aliquot of 5 × 106 transduced SMARTA cells was saved as “input” (~500× cell coverage per sgRNA). Transduced SMARTA cells (1 × 106 cells per recipient) were i.v. transferred into recipient mice followed by LCMV infection 24 h later. Forty-five recipients were randomly divided into 3 groups as biological replicates in each sub-library screening. At 7 days after infection, donor-derived Tfh (CXCR5+SLAM−) and Th1 (CXCR5−SLAM+) cells were sorted using a Reflection sorter. On average, 5 × 106 SMARTA cells per sample (~500× cell coverage per sgRNA) were recovered for further analysis.
Sequencing library preparation
Genomic DNA was extracted by using the DNeasy Blood & Tissue Kits (Qiagen 69506). Primary PCR was performed by using the KOD Hot Start DNA Polymerase (Millipore 71086) and the following pair of Nextera NGS primers:
Nextera NGS-F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTTGTGGAAAGGACGAAACACCG; Nextera NGS-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCACTTTTTCAAGTTGATAACGG. Primary PCR products were purified using the AMPure XP beads (Beckman A63881). A second PCR was performed to add adaptors and indexes to each sample. Hi-Seq 50-bp single-end sequencing (Illumina) was performed.
Data processing
For data analysis, FastQ files obtained after sequencing were demultiplexed using the Hi-Seq Analysis software (Illumina). Single-end reads were trimmed and quality-filtered using the CLC Genomics Workbench v11 (Qiagen) and matched against sgRNA sequences from the sgRNA metabolic library. Read counts for sgRNAs were normalized against total read counts across all samples. For each sgRNA, the fold change (log2 ratio) for enrichment was calculated between each of the biological replicates and the input experiment. Gene ranking was based on the average enrichment among replicates in representation of 6 individual corresponding sgRNAs (combining two sub-libraries) in sgRNA metabolic sub-libraries, respectively. The gene level false discovery rate (FDR) adjusted P value was calculated among multiple sgRNAs of each gene, using a paired two-tailed t-test between log2 transformed average normalized read counts of Tfh cell (CXCR5+SLAM−), Th1 cell (CXCR5−SLAM+) or input cell samples, and a value of less than 0.05 was considered to be statistically significant.
Flow cytometry
For analysis of surface markers, cells were stained in PBS (Gibco) containing 2% (w/v) BSA (Sigma). Surface proteins were stained for 30 min on ice. Intracellular staining was performed with Foxp3/Transcription Factor Staining Buffer Set according to manufacturer’s instructions (eBioscience). Intracellular staining for cytokines was performed with the fixation/permeabilization kit (BD Biosciences). Caspase-3 staining was performed using instructions and reagents from the “Active Caspase-3 Apoptosis Kit” (BD Biosciences). 7AAD (Sigma) or fixable viability dye (eBioscience) was used for dead cell exclusion. MFG-E8-FITC (BLAC-FITC) staining32 was performed according to the manufacturer’s instruction (Haematologic Technologies). PE was stained with 0.5 μM duramycin-LC-biotin (Cat # D-1003, Molecular Targeting Technologies), followed by Alexa Fluor 568- or Alexa Fluor 647-conjugated streptavidin (Thermo Fisher Scientific). Peripheral transitional B cells33 were analyzed by surface staining of B220 and IgM (B220+IgM+). gp66 tetramer was provided by the National Institutes of Health Tetramer Facility. The following antibodies were used: anti-TCRβ (1:200, H57–597, eBioscience, 1105961–82), anti-CD45.2 (1:200, 104, eBioscience, 17–0454-82), anti-IFNγ (1:100, XMG1.2, eBioscience, 25–7311-41), anti-CXCR3 (1:100, CXCR3–173, eBioscience, 17–1831-82), anti-ICOS (1:200, 7E.17G9, eBioscience, 17–1942-82), anti-GL7 (1:100, GL-7, eBioscience, 48–5902-82), anti-CCR7 (1:75, 4B12, eBioscience, 12–1971-82), anti-IgM (1:200, ll/41, eBioscience, 17–5790-82), anti-IgD (1:200, 11–26, eBioscience, 11–5993-82), and PE-Cy7 streptavidin (1:300, eBioscience, 25–4317-82); anti-CD4 (1:200, RM4–5, Biolegend, 100557), anti-CD45.1 (1:100, A20, Biolegend, 110736), anti-CD44 (1:200, IM7, Biolegend, 103047), anti-SLAM (1:150, TC15–12F12.2, Biolegend, 115943), anti-CD138 (1:150, 281–2, Biolegend, 142513), and anti-PD-1 (1:150, RMP1–30, Biolegend, 109109); anti-CD8α (1:200, 53–6.7, SONY, 1103605); anti-B220 (1: 200, RA3–6B2, TONBO Bioscience, 65–0452-U025) and anti-CD62L (1:400, MEL-14, TONBO Bioscience, 25–0621-U025); biotin anti-CXCR5 (1:200, 2G8, BD Biosciences, 551960), anti-PSGL-1 (1:300, 2PH1, BD Biosciences, 562806), anti-Ly6C (1:200, AL-21, BD Biosciences, 553104), anti-Fas (1:200, Jo2, BD Biosciences, 561985), anti-Bcl6 (1:75, K112–91, BD Biosciences, 561522), anti-T-bet (1:75, O4–46, BD Biosciences, 561267), anti-active caspase-3 (1:75, C92–605, BD Biosciences, 560626), Annexin V (1:50, BD Biosciences, 556421), and PE streptavidin (1:400, BD Biosciences, 554061). For detection of phosphorylated signaling proteins, lymphocytes were rested in complete medium for 1 h, and then fixed with 1× Phosflow Lyse/Fix buffer, followed by permeabilization with Phosflow Perm buffer III (BD Biosciences) and staining with antibody recognizing phosphorylated AKT Ser 473 (1:50, D9E, Cell Signaling Technology, 5315S). For intracellular cytokine staining, T cells were stimulated for 4 h with PMA plus ionomycin in the presence of monensin (BD Biosciences) prior to staining. To monitor cell division, lymphocytes were labeled with CellTrace Violet (CTV; 1:1000, Life Technologies, C34557). Flow cytometry data were acquired on BD FACSDiva software (LSR II or LSR Fortessa) and analyzed using Flowjo software (Tree Star).
RNA isolation and real-time PCR
RNA was isolated using the RNeasy Micro Kit (Qiagen 70004) following the manufacturer’s instructions. RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific 4368813) according to manufacturer’s instructions. Real-time PCR analysis was performed on the QuantStudio 7 Flex System (Applied Biosystems) using the PowerSYBR Greeen PCR Master Mix (Thermo Fisher Scientific 4367659) and the following primers: Cxcr5-F: ATGAACTACCCACTAACCCTGG, Cxcr5-R: TGTAGGGGAATCTCCGTGCT; Bcl6-F: CCGGCACGCTAGTGATGTT, Bcl6-R: TGTCTTATGGGCTCTAAACTGCT; Pdcd1-F: ACCCTGGTCATTCACTTGGG, Pdcd1-R: CATTTGCTCCCTCTGACACTG; Etnk1-F: CTGTTCACAGATGGGATCACAA, Etnk1-R: GCCGTAAATCCTCACCAGAACTA; Pcyt2-F: TGGTGCGATGGCTGCTATG, Pcyt2-R: CCCTTATGCTTGGCAATCTCC; Selenoi-F: CTACTCCTGACATACTTCGACCC, Selenoi-R: CCACGACAATCCAAACCCAG; Pcyt1a-F: GATGCACAGAGTTCAGCTAAAGT, Pcyt1a-R: TGGCTGCCGTAAACCAACTG. Results were normalized to Actb expression.
Gene expression profiling
RNA samples were isolated from freshly-isolated CD45.1+ wild-type and Pcyt2-deficient SMARTA cells sorted from the spleens of CD45.2+ recipient mice (n = 3 biological replicates each group) and analyzed with the Clariom S mouse array. The expression signals were summarized using the robust multi-array average algorithm Affymetrix Expression Console v1.1, followed by differential expression analysis performed using R package limma v.3.34.9. All the plots were generated using R package ggplot2 v.2.2.1. Differentially expressed (DE) transcripts were identified by ANOVA (Partek Genomics Suite version 6.5), and the Benjamini-Hochberg method was used to estimate the FDR as described34. DE genes were defined by |log2 FC| ≥ 0.5. Gene set enrichment analysis (GSEA) was performed as described previously35 using the “Hallmark” database, or the KEGG signature of ABC transporters. For GSEA using manually curated gene signatures from public datasets, the dataset GSE213809 was used for generating ‘Tfh gene signatures’ (< 5% FDR). Because the total number of upregulated and downregulated genes was more than 200, we ranked genes by their FC (log2-transformation of their expression in Tfh versus non-Tfh cells) and used the top 200 upregulated genes as ‘Tfh gene signatures’. GSE72188 was used to curate Bcl6 target genes17, as well as 48 Tfh-specific genes that are directly downregulated by Bcl6 (Bcl6 mainly functions as a transcriptional repressor for the commitment of Tfh cell lineage36) by combining ChIP-Seq, RNA-Seq and microarray datasets17.
Immunoblot analysis
Cells were lysed in RIPA buffer (Thermo Fisher Scientific 89900), resolved in 4–12% Criterion XT Bis-Tris Protein Gel (Bio-Rad 3450124) and transferred to PVDF membrane (Bio-Rad 1620177). Membranes were blocked using 5% BSA for 1 h and then incubated for overnight with the following antibodies: anti-Pcyt2 (14827–1-AP) (Proteintech), anti-Etnk1 (ab236615) (Abcam), anti-Etnk2 (H00055224-B01P) (Novus), anti-Selenoi (H00085465-A01) (Abnova), anti-Cept1 (PA5–23876) (Thermo Fisher Scientific), anti-Chka (13520–1-AP) (Thermo Fisher Scientific), anti-Chkb (sc-398957) (Santa Cruz), anti-Pcyt1a (HPA035428–100UL) (sigma-aldrich), anti-Pcyt1b (13765–1-AP) (Thermo Fisher Scientific), anti-Chpt1 (sc-515577) (Santa Cruz), anti-Pisd (sc-390070) (Santa Cruz), anti-Tubulin (#2146) (Cell Signaling Technology), anti-GAPDH (D16H11) (Cell Signaling Technology), anti-CXCR5 (EPR23463–30, catalog 254415) (Abcam), or anti-β-actin (8H10D10) (Cell Signaling Technology). Membranes were washed 3 times with TBST and then incubated with 1:5,000 diluted HRP conjugated anti-mouse IgG (W402B) or anti-rabbit IgG (W401B) (from Promega) for 1 h. After three additional washes with TBST, the membranes were imaged using the ODYSSEY Fc Analyzer (LI-COR). To measure the changes of CXCR5 expression after inhibition of proteasome or lysosome activity, SMARTA cells were incubated with the MG-132 proteasome inhibitor (4 μM; Sigma-Aldrich) or a lysosome inhibitor bafilomycin A1 (BafA1; 0.1 and 0.2 μM; Sigma-Aldrich) for 10 h, followed by lysis and subsequent immunoblot analysis with the indicated antibodies.
CXCR5 internalization assay
The internalization of surface CXCR5 molecules was measured as described21. Briefly, cells were collected with Click’s medium (Irvine Scientific), placed on ice and washed once in HBSS (Gibco) containing 2% (w/v) FBS (Gibco). Cell surface CXCR5 was first labeled using a non-conjugated antibody (clone 2G8) at room temperature for 15 min and unbound antibody was removed via washing twice with cold HBSS. Cells were resuspended in HBSS on ice and a baseline sample removed and kept on ice. For confocal imaging, surface CXCR5 antibody was then stained with Alexa Fluor 647-conjugated anti-rat secondary antibody for 30 min on ice. Samples were washed twice and transferred to 37 °C for CXCR5 internalization. For flow cytometry-based detection, cells were then incubated at 37 °C with 5% CO2, and samples were removed at the indicated time points and immediately diluted in ice-cold HBSS to stop further endocytosis. Cells were washed twice and the remaining surface CXCR5 antibody was stained with Alexa Fluor 647-conjugated anti-rat secondary antibody for 30 min on ice. Samples were washed twice and analyzed immediately.
CXCR5 recycling assay
The recycling of surface CXCR5 molecules was measured as described21,37. Briefly, cell surface CXCR5 was labeled with biotin-CXCR5 (clone 2G8), and unbound antibody was removed via washing with cold HBSS (Gibco) containing 2% (w/v) FBS (Gibco). The cells were then incubated in 30 oC to allow endocytosis of antibody-labeled CXCR5 for 30 min. After washing, T cells were treated with proteinase K (1 mg/ml) for 3 min in 37 oC, which efficiently removed the noninternalized CXCR5 on the cell surface. A baseline sample was kept on ice and cells were returned to 30 °C to allow CXCR5 recycling. The recovery of surface CXCR5 expression was measured by removing the samples at the indicated time points and immediately diluting in ice-cold HBSS to stop further recycling. Cells were washed twice and the recycled surface CXCR5 antibody was stained with PE-conjugated streptavidin (554061, BD Biosciences) for 30 min on ice. Samples were washed twice and analyzed by flow cytometry immediately.
Transwell migration assay
T cells were rested in the RPMI 1640 medium (Gibco) containing 0.1% fatty acid-free BSA (Sigma-Aldrich) at 37 °C for 30 min before being loaded as a 100 μl suspension of 106 cells into the upper chamber of a 24-well transwell plate (3421, Corning). Recombinant CXCL13 or CCL21 (Peprotech) of indicated concentrations was added to the bottom wells before the cells were incubated at 37 °C for 3 h. Cells that had migrated to the bottom wells were enumerated by flow cytometry with CountBright Absolute Counting Beads (C36950, Thermo Fisher Scientific).
Immunofluorescence
Tissues were fixed in 2% paraformaldehyde, 0.1% Triton-100 and 1% DMSO for 24 h prior to cryoprotection. Tissues were sectioned at 10 μm thickness and blocked in buffer comprised of PBS containing 2% BSA and 5% goat serum, and stained overnight at 4 °C with blocking buffer containing the following antibodies or lectins where indicated: rabbit anti-GFP/Ametrine (Rockland; catalog 600–401-215), anti-IgD (Biolegend; clone 11–26c.2a), anti-CD3 (Biolegend; clone 17A2), or biotin-peanut agglutinin (VectorLabs; catalog B-1075). The cells were stained with the appropriate secondary reagents for detection: Alexa Fluor 488-conjuagted goat anti-rabbit or Alexa Fluor 488-conjugated streptavidin (Thermo Fisher Scientific). Widefield fluorescence microscopy was performed using a motorized Nikon TiE inverted microscope equipped with a 40× 1.3NA Plan Fluor objective, standard FITC, TRITC and Cy5 filter sets, and an EMCCD camera (Andor). Analysis was performed using NIS Elements software, wherein images were segmented based on channel intensity, and further refined using a spot identification algorithm to identify the number of cells within the manually-defined area of the germinal center.
Sorted cells were incubated on ice in phenol red-free RPMI containing rabbit anti-CXCR5 (Abcam; catalog 254415), 0.5 μM duramycin-LC-biotin (D-1003, Molecular Targeting Technologies), or Alexa Fluor 488-conjugated cholera toxin subunit β (ThermoFisher; catalog C34775), and in select experiments either Alexa Fluor 488-conjugated anti-CD4 (Biolegend; clone RM4–5) or Alexa Fluor 488-conjugated anti-CD45 (Biolegend; clone 30-F11). Cells were washed, followed by incubation with Alexa Fluor 647-labeled goat anti-rabbit and Alexa Fluor 568-conjugated streptavidin (Thermo Fisher Scientific). Cells were washed and immediately imaged using a Marianis spinning disk confocal microscope (Intelligent Imaging Innovations) equipped with a 100× 1.45NA objective, 488 nm, 561 nm and 647 nm laser lines and Prime 95B CMOS camera (Photometrics), and analyzed using Slidebook software (Intelligent Imaging Innovations). In selected experiments, cells were fixed with 2% paraformaldehyde for 15 min prior to permeabilization with 0.1% Triton-100 for 3 min. Cells were subsequently blocked in PBS containing 2% BSA followed by staining with CXCR5 or duramycin-LC-biotin. Detection of the signals was achieved using Alexa Fluor 647-conjugated goat anti-rabbit secondary reagents or Alexa Flour 568-conjugated streptavidin and Alexa Fluor 488-labeled phalloidin (Thermo Fisher Scientific).
TIRF–STORM imaging
Tfh cells were sorted and incubated with 0.5 μM duramycin-LC-biotin (D-1003, Molecular Targeting Technologies), rabbit anti-CXCR5 (Abcam; catalog 254415, 1:500 dilution) and rabbit anti-CCR7 (Abcam; catalog 32527, 1:200 dilution) in RPMI containing fetal bovine serum for 30 min on ice. Cells were washed twice and incubated for 30 min on ice with Alexa Fluor 405-Alexa Fluor 647-conjugated streptavidin and Cy3-Alexa Fluor 647-conjugated donkey anti-rabbit IgG, prepared as previously described38. TCR-stimulatory planar lipid bilayers containing anti-CD3 antibody and the recombinant integrin ligand ICAM-1 were used for TIRF microscopy as we previously described39. STORM acquisition of at least 10 fields per experiment was performed using a Nikon N-STORM system comprised of an inverted TiE stand, motorized TIRF slider, 100× 1.45 NA objective, DU-897 EMCCD camera, Agilent laser launch and appropriate optics, as previously reported40. To facilitate robust statistical analysis of STORM or other co-ordinate-based data, we developed a new algorithm denoted as Normalized Spatial Intensity Correlation (NSInC; Liu X et al., St. Jude Children’s Research Hospital, unpublished), which has been applied successfully41. When the co-localization analysis using NSInC was directly compared with Coordinate-Based Colocalization (CBC) method42, the default radius value, r = 250 nm, was used. For either NSInC or CBC analysis, a value of 1 indicates complete co-localization, a value of 0 represents spatial randomness, and value of −1 indicates inter-molecular exclusion.
3-dimensional (3D) super resolution confocal imaging
Sorted Tfh cells were allowed to settle onto poly-L-lysine coated 12 mm glass coverslips for 15 min at 37 oC. Cells were fixed with 4% PFA and 0.1% glutaraldehyde in PBS for 20 min at room temperature, and were subsequently washed and blocked with PBS containing 1% BSA prior to incubation with duramycin-LC-biotin, rabbit anti-CCR7 and rabbit anti-CXCR5 as noted above, followed by detection with Alexa Fluor 568-conjugated streptavidin and Alexa Fluor 647-conjugated donkey anti-rabbit antibody. Cells were mounted with hardset mounting media (ThermoFisher) and were subsequently imaged using a Marianas confocal microscope (Intelligent Imaging Innovations) equipped with a CSU-W spinning disk (Yokogawa) with Super Resolution via Optical Reassignment43 (SoRA), a 100× 1.45 NA oil objective and Prime95B camera (Photometrics). Images were deconvolved with Microvolution software, and analyzed using Slidebook (Intelligent Imaging Innovations) and Imaris (Bitplane) software.
Immunogold scanning electron microscopy (SEM)
Sorted Tfh cells were plated on poly-lysine-coated 18 mm glass coverslips and allowed to settle for 15 min at 37 oC prior to fixation with 4% PFA and 0.1% glutaraldehyde in PBS for 20 min. Following blocking with PBS containing 1% BSA, cells were surface labeled with duramycin-LC-biotin and rabbit anti-CXCR5 as noted above. Biotinylated duramycin was subsequently identified using unlabeled streptavidin followed by goat anti-streptavidin (Vector Labs; catalog BA-0500). Surface moieties were detected with 10 nm-colloidal gold labeled donkey anti-goat (Abcam; catalog 41496) and 25 nm-colloidal gold labeled donkey anti-rabbit (Abcam; catalog 41514). Fixed, gold-labeled samples were buffer washed, then post-fixed in 0.1% aqueous osmium tetroxide for 20 min, washed with ddH20, dehydrated with an ascending ethanol series, and critical point dried (autosamdri 931, Tousimis, Rockville, MD). Dried samples were carbon coated and imaged in a ThermoFisher Scientific Helios NanoLab 660 operating in immersion mode. Samples were imaged with both the through lens detector (TLD; 2 kV, 25 pA) and the concentric backscatter detector (CBS; 10 kV, 0.20 nA) at a working distance of 7 mm. Detection on the CBS was limited to the inner most segment of the detector. Magnification matched images were acquired from the TLD and CBS detectors; higher magnification CBS images were acquired to differentiate the two gold labels. SEM images44 were processed using NIS Elements software (Nikon Instruments), specifically employing an artificial-intelligence powered denoising function to clearly delineate immunogold particles identified using the backscatter detector. Images were subsequently analyzed using Imaris (Bitplane, Inc.) software and a spot-fitting function to identify 10 and 25 nm gold particles, respectively. Co-ordinate-based analysis of spatial arrangement and clustering was performed using the NSInC algorithm as described for STORM imaging above.
Lipidomic analysis
Lipid extraction
Equal numbers (3 × 106) of naïve CD4+, Th1 (CXCR5−SLAM+) and Tfh (CXCR5+SLAM−) cells, with the indicated genotypes, were washed with ice-cold PBS, flash-frozen in liquid nitrogen and then stored at −80 °C until they were processed for extraction of total lipids. A modified Folch extraction procedure45 was used for the extraction of total lipids from purified naïve CD4+, Th1 or Tfh cells or anti-FLAG M2 beads. Briefly, 1 ml of chloroform-methanol (2:1, v/v) was added to the cells or beads and mixed by vortexing. Next, 200 μl of saline was added, and the tubes were mixed for 30 sec in a Bead Ruptor Elite (OMNI International) for 30 sec at 8 m/s. The homogenate incubated at room temperature for 30 sec and then centrifuged for 10 min at 21,000 g at 4 °C. After centrifugation, the lower organic-phase layer was transferred to a new tube and evaporated to dryness under a stream of liquid nitrogen. The dried lipid extracts were dissolved with 35 μl of chloroform-methanol (2:1, v/v), transferred to autosampler vials and analyzed by LC-MS (10 μl per injection).
LC-MS lipid profiling
LC separations were performed with a Vanquish Horizon UHPLC (Thermo Fisher Scientific) using stepped-gradient conditions as follows: 0–4.5 min, 45 to 60%, B; 4.5–5 min, 60 to 70%, B; 5–8 min, 70%, B; 8–19 min, 70 to 75%, B; 19–20 min, 75 to 90%, B; 20–33 min, 90 to 95%, B; 33–34 min, 90 to 100%, B; 34–39 min, 100%, B; 39–40 min, 100 to 45%, B; 40–45 min, 45%, B. Mobile phase A was water/acetonitrile (60:40, v/v) and mobile phase B was IPA/acetonitrile (90:10, v/v); both A and B contained 10 mM ammonium acetate. The column used was a Thermo Fisher Scientific Accucore C30 (2.1 × 250 mm, 2.6 μm) operated at 50 °C. The flow rate was 250 μl/min and the injection volume was 10 μl. A Thermo Fisher Scientific Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (QE-MS) equipped with a HESI-II probe was employed as detector, and data were collected using Xcalibur 4.0 (Thermo Fisher Scientific). For each sample, two chromatographic runs were carried out subsequently, and separate data were acquired for negative and positive ions. The QE-MS was operated using a data-dependent LC-MS/MS method (Top-15 dd-MS2) for both positive and negative ion modes. The mass spectrometer was operated at a resolution of 140,000 (FWHM, at m/z 200), AGC targeted of 1 × 106, and max injection time 80 msec. The instrument’s operating conditions were: scan range 100–1,500 m/z; sheath gas flow 45; aux gas flow 8; sweep gas 2; spray voltage 3.6 kV for positive mode and 2.5 kV for negative mode; capillary temperature equal to 320 °C; S-lenses RF level 50; aux gas heater equal to 320 °C. For the Top-15 dd-MS2 conditions a resolution of 35,000 was used, AGC targeted of 1 × 105, max injection time 50 msec, MS2 isolation width 1.0 m/z, NCE 35.
Data processing
The Thermo Fisher Scientific LipidSearch software (version 4.1) was used for identification and relative quantification of lipids with the following parameters: precursor and product ion mass tolerance of ± 5 ppm; main adducts search (M+H, M-H, M+NH4, M+CH3COO, M+2H, M-2H, M+Na) for mass. All lipid sub-classes were searched within major lipid classes (phospholipids, sphingolipids, glycerolipids and neutral lipids). All individual data files were searched for product ion MS/MS spectra of lipid precursor ions. The MS/MS predicted fragmented ions for all precursor adducts as measured within 5 ppm of mass tolerance. The product ions that matched the predicted fragment ions within 5 ppm of mass tolerance were used to calculate a match-score, and those candidates providing the highest quality match were determined. Next, the search results from the individual positive or negative ions from each sample group were aligned within a defined retention time window (± 0.25 min), and the data were merged for each annotated lipid.
Compound Discoverer 3.1 (CD31; Thermo Fisher Scientific) software was used for multivariate statistical analysis and differential expression analysis of individual molecules using a pre-defined workflow from CD31: untargeted metabolomics with statistics; detect ID of unknowns using online databases; maps compounds to biological pathways using Metabolika. The spectral alignment node used the adaptative curve algorithm, with a maximum shift of 2 min. The compound detection option was set for a signal-to-noise ratio ≥ 3; min peak intensity 1 × 106; parent ion mass tolerance of ± 5 ppm; retention time tolerance of 0.25 min; and preferred fragments data selection M+H, M-H. The data were normalized by the constant mean algorithm. Metabolite identifications and pathway analysis features were based on the above indicated workflow. The mass-to-charge (m/z) data from positive or negative ion modes processed from CD31 was combined and then formatted to comma-separated value (CSV) file and imported into MetaboAnalyst (version 4.0). The peak areas were normalized using the parameters for sample normalization sum, data transformation log10 and range data scaling. After normalization, the statistical analysis using multiparametric ANOVA and principal component analysis (PCA) were evaluated and heatmaps were generated using default clustering algorithms.
Immunoprecipitation of CXCR5 or CCR7 for lipid binding
Naïve SMARTA cells were transduced with FLAG-tagged CXCR5-RV-GFP, CCR7-RV-GFP or empty vector retrovirus and transferred into B6 recipients followed by LCMV infection. At 7 days later, donor SMARTA cells were sorted, washed in ice-cold PBS, centrifuged for 2 min at 1,000 r.p.m., and resuspended in 1 ml of ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5; 200 mM NaCl; 1.5 mM MgCl2; 0.5 mM DTT; 1.0 mM, AEBSF; complete-mini EDTA-free protease inhibitor cocktail ± 0.5% DDM). Following immunoprecipitation with anti-FLAG M2 magnetic beads (Sigma-Aldrich), lipids were extracted and subjected to LC-MS/MS to quantitatively measure the lipids bound to CXCR5 or CCR7, as follows. Briefly, the cell suspensions were placed on ice and sonicated for 4−5 sec to obtain a homogeneous solution. Cell extracts were centrifuged for 5 min at 21,000 g at 4 °C to remove cellular debris, and protein concentrations in supernatants were measured using the Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific). Total proteins (2 mg; adjusted to a total volume of 500 μl of lysis buffer) were then incubated with 15 μl of Protein A/G Plus Agarose Beads (Santa Cruz Biotechnology) at 4 °C for 30 min with constant rotation to remove proteins and lipids that bound nonspecifically to the beads. Following centrifugation of the beads, 15 μl of anti-FLAG M2 magnetic beads (Sigma-Aldrich) were added and incubated at 4 °C with constant rotation for 3–4 h. The anti-FLAG M2 beads were centrifuged for 30 sec at 12,000 g at 4 °C, washed three times with ice-cold PBS and stored at −80 °C until processing for extraction of the lipids bound to the immunoprecipitated CXCR5 or CCR7.
[3H]-labeled ethanolamine incorporation and lipid turnover
PE biosynthesis and turnover were measured as described46. Equal numbers (2× 106) of Tfh or Th1 cells were suspended in RPMI 1640 medium (Gibco) containing 0.5% BSA (Sigma-Aldrich) and final concentration of 1 μCi/ml [1,2-3H]-ethanolamine hydrochloride ([3H]-Etn) (specific radioactivity, 18 Ci/mmol) (Moravek). Non-radiolabeled reference conditions were prepared using unlabeled ethanolamine (Sigma-Aldrich) at an equivalent concentration of 5.49 ng/ml. For [3H]-Etn incorporation assay, Tfh and Th1 cells were pulsed with [3H]-Etn for 3 h, followed by lipid extraction. For lipid turnover assay, cells were pulsed with [3H]-Etn for 3 h, extensively washed twice with ice-cold PBS, and then chased with unlabeled ethanolamine for another 3 h before lipid extraction. Cells and medium were separated by a brief centrifugation (2 min at 1,000 r.p.m.), cells were washed twice with ice-cold PBS, and cell pellets and medium were then flash-frozen in liquid nitrogen and stored at −80 °C until the extraction of lipids. Processing for liquid scintillation counting (LSC) to measure the amount of radioactivity remaining in the cells was performed, and the amount released into the cell culture medium was then analyzed.
After isolating lipids as indicated above, the dried extracts of total lipids were dissolved with 50 μl of chloroform-methanol (2:1, v/v), transferred into a 7 ml pico glass liquid scintillation vial (PerkinElmer) to measure the [3H]-Etn incorporated into lipid molecules by LSC. The collected cell culture medium was placed into an LSC glass vial and evaporated to dryness under a stream of nitrogen at RT. Next, the dried material was dissolved in 100 μl of 20% acetonitrile to measure the [3H] label released by the cells into the cell culture medium. The dissolved material in the scintillation vials was mixed with 7 mL of ProSafe FC+ High Efficiency LSC cocktail (Meridian Biotechnologies) and then the presence of radioactivity was quantitatively measured as counts per minute (CPM) in a Tri-Carb 4910 TR Liquid Scintillation Counter (PerkinElmer).
Lipid supplementation
All lipids were purchased from Avanti Polar Lipids: 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (diacyl-type PE, 850705) and 1-O-hexadecanyl-2-O-(9Z-octadecenyl)-sn-glycero-3-phosphoethanolamine (ether-type PE, 999974). The ether-type or diacyl-type PE47 used for lipid supplementation assays were dissolved in chloroform, evaporated under a gentle stream of nitrogen and then immediately dissolved in an ethanol vehicle (0.1%). The indicated sgRNA-transduced SMARTA cells were supplemented with the lipids in the indicated concentration, followed by transfer of these cells to C57BL/6 recipient mice and LCMV infection. The lipids in culture medium were replenished every 2 days (specifically, lipid was supplemented at days −5, −3 and −1 prior to adoptive transfer and LCMV infection).
Serum antibodies
Sera from immunized mice were collected, and antigen-specific IgM, IgG, IgG1, IgG2a, IgG2b, IgG2c, IgG3 and IgA were measured by using ELISA, as described4. Briefly, isotype-specific antibodies to NP were measured in plates coated with NP (8)-BSA (Biosearchtech) using the SBA Clonotyping System (Southern Biotech). Titers were presented as the maximum serum dilution exceeding 1.5-fold above the average background.
Statistical analysis
For biological experiment (non-omics) analyses, data were analyzed using Prism 8 software (Graphpad) by two-tailed unpaired Student’s t-test or one-way ANOVA with Newman-Keuls’s test. P < 0.05 was considered as significant. Data are presented as mean ± s.d. or mean ± s.e.m.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Microarray data of wild-type and Pcyt2-deficient cells at day 3 after LCMV infection have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number: GSE147190 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147190). For GSEA using manually curated gene signatures from public datasets, the dataset GSE213809 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21380) and GSE7218817 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72188) from NCBI GEO database were used to generate ‘Tfh gene signatures’ (< 5% FDR). Source data are provided with this paper.
Code availability
The Code used for colocalization index analysis of STORM or other co-ordinate-based data has been applied successfully41 and the accessible link of the code is https://rdrr.io/cran/colocalization/.
Extended Data
Extended Data Figure 1. Pooled in vivo CRISPR-Cas9 screening and an in vivo dual transfer system to identify and validate potential regulators of Tfh cells.
a, Diagram of the screening system. Naïve Cas9-expressing SMARTA cells were transduced with lentiviral-derived sgRNA metabolic library, expanded in vitro, and transferred into C57BL/6 hosts, which were then infected with LCMV 24 h later. At day 7 post-infection, splenic Tfh (CXCR5+SLAM−) and Th1 (CXCR5−SLAM+) cells were purified and those gRNAs that were upregulated (corresponding to negative regulators of Tfh responses) or downregulated (corresponding to positive regulators of Tfh responses) in Tfh versus Th1 cells [|log2 ratio (Tfh/Th1)| > 0.5; adjusted P < 0.05] were determined for metabolism-related genes that establish Tfh over Th1 cell differentiation. b, Diagram of in vivo dual transfer system. SMARTA cells transduced with sgRNA viral vectors expressing distinct fluorescent proteins were mixed and transferred into the same C57BL/6 hosts, followed by LCMV infection and experimental analyses. c, SMARTA cells transduced with the indicated sgRNA viral vectors (Ametrine+) were mixed at a 2:1 ratio with sgNTC (mCherry+)-transduced SMARTA cells and transferred into C57BL/6 hosts, followed by LCMV infection. Analyses of the proportion of donor-derived Tfh (CXCR5+SLAM− or PSGL-1−Ly6C−) and Th1 (CXCR5−SLAM+ or PSGL-1+Ly6C+) cells and quantification of relative Tfh cell percentage and number (right lower) in the spleen at day 7 (n = 4 mice). d, Insertion and deletion (indel) mutations after CRISPR-Cas9 targeted disruption in SMARTA cells transduced with sgNTC or sgPcyt2, via deep sequencing analysis of indels generated at the exonic target site of the Pcyt2 gene, including 74% of indel events in sgPcyt2-transduced cells compared to 0.6% in sgNTC-transduced cells. e, Immunoblot analyses of Etnk1 and Pcyt2 in splenic SMARTA cells at day 3 post-infection. Asterisk (*), non-specific band; arrow, the target band. Data are representative of two (d, e), or at least three (c) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (c). ***P < 0.001. Numbers in quadrants or gates indicate percentage of cells.
Extended Data Figure 2. Validation of the effects of CDP-ethanolamine pathway genes on Tfh generation during both virus infection and protein immunization.
a, b, Quantification of relative proportions and numbers of CXCR5+PD-1+ Tfh cells (a) and CXCR5−SLAM+ Th1 cells (b) in donor-derived cells from spleen of mice receiving the indicated sgRNA-transduced SMARTA cells at day 7 after LCMV infection (n = 4 mice). c, Insertion and deletion (indel) mutations after CRISPR-Cas9 targeted disruption in SMARTA cells transduced with the indicated sgRNAs via deep sequencing analysis of indels generated at the exonic target sites of the indicated genes. d, Immunoblot analyses of indicated proteins in the indicated sgRNA-transduced SMARTA cells isolated from the spleen at 3–5 days after adoptive transfer and LCMV infection. Non-specific bands are indicated by an asterisk (*); target bands are indicated by an arrow. e, SMARTA cells transduced with the indicated sgRNA-expressing vectors (Ametrine+) were mixed at a 2:1 ratio with sgNTC (mCherry+)-transduced SMARTA cells and transferred into C57BL/6 hosts, followed by LCMV infection. Quantification of donor-derived Tfh and Th1 cells from the host spleen at day 7 post-infection (n = 4 mice). f, OT-II cells transduced with the indicated sgRNAs were transferred into C57BL/6 hosts, followed by NP-OVA + LPS in alum immunization. Quantification of Tfh cell percentage and number in the spleen at day 7 (n = 4 mice). Data are representative of two (c–e), or at least three (a, b, f) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (a, b, e, f). NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001.
Extended Data Figure 3. Dynamic regulation of PE metabolic programs in Tfh cells.
a, Lipidomic analysis of the lipid content (phosphatidylcholine, PC; phosphatidylserine, PS) in naïve (CD4+) T cells, wild-type Tfh (transduced with sgNTC), and Pcyt2-deficient Tfh cells (n = 3 samples, each pooled from multiple mice). b, c The dependence of Pcyt2 for PE alterations during Tfh cell differentiation from naïve CD4+ T cells. Heatmap showing the 39 significantly downregulated PE molecules in Pcyt2-deficient Tfh cells as compared with wild-type Tfh cells (log2 ratio ≥ 0.5, P < 0.05) (b). Venn diagram showing the overlap of upregulated (left) or downregulated (right) PE molecules in wild-type Tfh versus naïve CD4+ T cells compared to wild-type versus Pcyt2-deficient Tfh cells (c) (n = 3 samples, each pooled from multiple mice). d, Lipidomic analysis of the lipid content (PE and PC) in wild-type and Selenoi-deficient Tfh cells (n = 3 samples, each pooled from multiple mice). e, Primary Tfh and Th1 cells were sorted from LCMV-infected mice and incubated with [3H]-Etn (2 μCi/ml) for 3 h. To assess PE synthesis, lipids were extracted and [3H]-Etn incorporation into Tfh cells was assessed with a scintillation counter (see synthesis column, left graph). To assess PE turnover, after 3 h of [3H]-Etn incubation, cells were washed and chased with unlabeled ethanolamine for another 3 h (see turnover column, left graph). The presence of [3H] radioactivity in cells and culture medium (right graph) was measured with a scintillation counter (n = 4 samples, each pooled from multiple mice). CPM, counts per million. Data are representative of at least two independent experiments (a–e). Data are mean ± s.e.m. P values are determined one-way ANOVA (a) or by two-tailed unpaired Student’s t-test (d, e). NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001.
Extended Data Figure 4. CDP-ethanolamine pathway but not CDP-choline pathway is required for Tfh cell differentiation.
a, Diagram summary of the effects of single or combined deficiency of genes in the CDP-ethanolamine and CDP-choline pathways on Tfh cell differentiation after LCMV infection. b, Immunoblot analyses of indicated proteins in the indicated sgRNA-transduced SMARTA cells isolated from the spleen at 3–5 days after adoptive transfer and LCMV infection. The non-specific band is indicated by an asterisk (*); the target band is indicated by an arrow. c, Summary of the relative proportions of Tfh (CXCR5+SLAM−) and Th1 (CXCR5−SLAM+) cells in donor-derived cells in spleen of mice receiving the indicated sgRNA-transduced SMARTA cells at day 7 after LCMV infection (n = 4 mice). d, Lipidomic analysis of the lipid content (PC and PE) in wild-type Tfh (transduced with sgNTC), Pcyt1a-deficient Tfh and Pcyt1a and Pcyt1b-doubly deficient Tfh cells (n = 3 samples, each pooled from multiple mice). e, Flow cytometry analysis (left) and summary of the proportion of wild-type and Pcyt2-deficient CXCR5+ SMARTA cells (middle) and their dilution of CellTrace Violet (CTV; right) at day 2 after LCMV infection (n = 3 mice). f, Summary of apoptotic wild-type and Pcyt2-deficient SMARTA cells as analyzed by Annexin V and 7AAD staining (or unstained) in freshly-isolated splenocytes at day 2 after LCMV infection (n = 3 mice). g, Distribution of SMARTA cells in the follicle in the spleen at day 3 post-infection. Scale bar, 50 μm (n = 50 sections). h, Diagram of Tfh cell effector functional assay. sgNTC (mCherry+) and sgPcyt2 (Ametrine+)-transduced SMARTA cells (CD45.1+) were transferred (1st transfer) into C57BL/6 hosts (CD45.2+), followed by LCMV infection. Seven days later, the fully differentiated wild-type or Pcyt2-deficient CXCR5+SLAM− Tfh cells (CD45.1+) were sorted and equal numbers of these cells were transferred (2nd transfer) into LCMV-infected mice (CD45.2+; this transfer occurred at day 1 after LCMV infection). GC B cell and plasma cell formation was analyzed at day 5 after the 2nd adoptive transfer. Data are representative of one (d) or at least two (b, c, e–g) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (c, d) or by two-tailed unpaired Student’s t-test (e, f). NS, not significant; **P < 0.01 and ***P < 0.001. Numbers in gates indicate percentage of cells.
Extended Data Figure 5. CDP-ethanolamine pathway regulates Tfh cell differentiation independently of Bcl6–T-bet axis.
a, sgNTC or sgPcyt2-transduced SMARTA cells (CD45.1+) were transferred into C57BL/6 recipients (CD45.2+) that were subsequently infected with LCMV. CD45.1+ cells were isolated at day 3 for transcriptional profiling by microarray. Gene set enrichment analysis (GSEA) of the Tfh signature in Pcyt2-deficient compared to wild-type T cells (n = 3 samples, each pooled from multiple mice). b, c, sgNTC, sgEtnk1, sgPcyt2, or sgSelenoi-transduced SMARTA cells were transferred into C57BL/6 recipients, followed by infection with LCMV. Quantification of the percentages of Bcl6+T-bet– and Bcl6–T-bet+ cells among donor-derived T cells in the spleen at day 3 (b) and day 7 (c) post-infection (n = 4 mice). d, GSEA of Bcl6 target genes (identified by ChIP-Seq17) in Pcyt2-deficient compared to wild-type T cells (samples are the same as Extended Data Fig. 5a; n = 3 samples, each pooled from multiple mice). e, GSEA of the Tfh-specific genes that are directly downregulated by Bcl6 (in total 48 genes, identified by combining ChIP-Seq, RNA-Seq and microarray datasets17) in Pcyt2-deficient compared to wild-type T cells (samples are the same as Extended Data Fig. 5a; n = 3 samples, each pooled from multiple mice). f, Analysis and quantification of PSGL-1−Ly6C− cells, CXCR5+ cells and CXCR5 MFI among PSGL-1−Ly6C− Tfh cells at day 3 post-infection (n = 4 mice). g, sgNTC, sgPcyt2, sgBcl6, or sgPcyt2 and sgBcl6-transduced SMARTA cells were transferred into C57BL/6 recipients that were subsequently infected with LCMV. Quantification of the proportion of Tfh cells (CXCR5+PD-1+ and CXCR5+SLAM−) in donor-derived cells from the spleen at day 7 post-infection (n = 4 mice). h, sgNTC, sgEtnk1, sgPcyt2, or sgSelenoi-transduced SMARTA cells (CD45.1+) were transferred into C57BL/6 recipients (CD45.2+) that were subsequently infected with LCMV. Quantification of the proportion of Tfh cells (CXCR5+PD-1+) in donor-derived T cells from the spleen at day 3 post-infection (n = 4 mice). i–o, SMARTA cells transduced with the indicated sgRNAs were transferred into C57BL/6 recipients, followed by infection with LCMV. i, Quantification of the frequency and mean fluorescence intensity (MFI) of CXCR5 on Tfh cells (PSGL-1−Ly6C−) in the spleen at day 2 post-infection (n = 3 mice). j, Quantification of the frequency and MFI of PD-1 on Tfh cells (PSGL-1−Ly6C−) in the spleen at day 2 post-infection (n = 3 mice). k, SMARTA cells were transduced with sgNTC or sgPcyt2 alone or in combination with a retrovirus overexpressing empty vector (Empty-RV) or CXCR5 (CXCR5-RV), followed by adoptive transfer into C57BL/6 mice that were then infected with LCMV. Analysis (upper) and quantification (lower) of the proportion of Tfh cells (CXCR5+PD-1+ and PSGL-1−Ly6C−) in donor-derived cells from the spleen at day 7 post-infection (n = 4 mice in CXCR5-RV group of CXCR5+PD-1+ Tfh data, n = 3 mice in other groups). l, MFI of PD-1, ICOS, Ly6C, PSGL-1, CD44, and CD62L on CD45.1+ SMARTA cells at day 3 post-infection (n = 4 mice). m, Quantification of the frequency and MFI of CXCR3 on CD45.1+ SMARTA cells at day 3 post-infection (n = 3 mice). n, MFI of CCR7 expression on donor-derived T cells from the host spleen at day 3 post-infection (n = 3 mice). o, Quantification of phosphorylated AKT (pAKT-S473) levels in purified wild-type or Pcyt2-deficient Tfh cells (CXCR5+SLAM−) that were stimulated with CXCL13 for 3 h (n = 4 mice). Data are representative of one (a, d, e), at least two (b, c, g, h), or at least three (f, i–o) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (b, c, f–h, k, l), or by two-tailed unpaired Student’s t-test (i, j, m–o). NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. Numbers in gates indicate percentage of cells.
Extended Data Figure 6. PE is selectively distributed on the outer layer of the Tfh but not Th1 cell plasma membrane.
a, Analysis and quantification of surface level of PE on Tfh (PSGL-1−Ly6C−) and Th1 (PSGL-1+Ly6C+) cell membrane at day 5 post-infection (n = 4 mice). b, Quantification of co-localization index of PE and CCR7 or CXCR5 on PSGL-1−Ly6C− Tfh cells in Fig. 3b using the Coordinate-Based Colocalization (CBC) algorithm (PE : CCR7, n = 33 cells; PE : CXCR5, n = 25 cells). A co-localization index value of 1 indicates complete co-localization, a value of 0 represents spatial randomness, and value of −1 indicates inter-molecular exclusion. c, 3D super resolution confocal image of the entire Tfh cell on poly-L-lysine-coated coverslips. Surface staining was reconstructed using Imaris software. Scale bar, 2 μm. Layers are translucent to readily visualize both colors. Right, quantification of co-localization index of PE and CCR7 or PE and CXCR5 on PSGL-1−Ly6C− Tfh cells (n = 4 mice and 20 cells were quantified). d, Confocal microscopy imaging of PE distribution on Tfh (PSGL-1−Ly6C−) and Th1 (PSGL-1+Ly6C+) cells at the indicated time points after LCMV infection. Scale bar, 10 μm (n = 3 mice). e, f, Lipids associated with FLAG-CXCR5 or FLAG-CCR7 expressed in SMARTA cells isolated from LCMV-infected mice were immunoprecipitated by anti-FLAG M2 magnetic beads and quantified using LC-MS/MS. Analysis of lipid (PE, PC and PS) content (e) and heatmap showing significantly changed (P < 0.05) PE molecules (f) in the indicated groups (n = 2 samples, each pooled from multiple mice). Asterisks indicate PE molecules with the exact mass. g, SMARTA cells were transduced with the indicated sgRNA and the retrovirus overexpressing empty vector (Empty-RV) or CXCR5 (CXCR5-RV), followed by adoptive transfer into C57BL/6 mice and LCMV infection. Quantification of the proportion of PE+ cells in PSGL-1−Ly6C− Tfh cells in donor-derived cells from the spleen at day 7 (n = 4 mice). h, SMARTA cells were transduced with the indicated sgRNAs, followed by adoptive transfer into C57BL/6 mice and LCMV infection. Quantification of the proportion of PE+ cells in donor-derived PSGL-1−Ly6C− Tfh cells from the spleen at day 7 post-infection (n = 4 mice). i, Analysis and quantification of PE outer layer membrane distribution on freshly-isolated wild-type and Pcyt2-deficient Tfh (PSGL-1−Ly6C−) and Th1 (PSGL-1+Ly6C+) cells (Tfh, n = 5 mice; Th1, n = 4 mice). j, sgNTC or sgPcyt2-transduced SMARTA cells were transferred into C57BL/6 recipients that were subsequently infected with LCMV. Phosphatidylserine (PS) exposure in intact Tfh (PSGL-1−Ly6C−) and Th1 (PSGL-1+Ly6C+) cell membrane was assessed by Annexin V and 7AAD (left) or MFG-E8 and 7AAD (right) staining analysis at day 5 post-infection (n = 4 mice). Data are representative of two (e–h, j), or at least three (a–d, i) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (h, j), or two-tailed unpaired Student’s t-test (a–c, g, i). NS, not significant; ***P < 0.001. Numbers in quadrants or gates indicate percentage of cells.
Extended Data Figure 7. PE regulates CDP-ethanolamine pathway-mediated Tfh responses.
a, Wild-type and Pcyt2-deficient Tfh cells were sorted from LCMV-infected mice and incubated with [3H]-Etn (2 μCi/ml) for 3 h. Lipids were extracted and [3H]-Etn incorporated into Tfh cells was assessed with a scintillation counter (n = 4 samples). b–e, Lipid add-back/rescue assay. The sgNTC and sgPcyt2-transduced SMARTA cells were supplemented with the indicated lipids, followed by transfer of these cells to C57BL/6 mice and LCMV infection. b, Diagram showing the time points of lipid treatment. PE was supplemented at days −5, −3 and −1 prior to adoptive transfer, and LCMV-infected mice were analyzed at day 3 post-infection (to capture the effects induced by in vitro PE supplementation). c, Lipidomic analysis showing the PE content in the indicated groups (left), and Venn diagram showing the rescued PE molecules by diacyl-type and ether-type PE supplementation (right) at day 3 post-infection. d, Analysis and quantification of the proportion of PE+ cells in donor-derived Tfh cells (PSGL-1−Ly6C−) from the spleen (n = 5 mice). e, Analysis and quantification of the proportion of donor-derived CXCR5+SLAM− Tfh cells from the spleen (n = 5 mice). f, Gene set enrichment analysis (GSEA) plot for KEGG signature of ABC transporters in Tfh versus Th1 cells (from a public dataset GSE7485448). g, Heatmap showing the top 22 leading-edge genes from the enrichment plot in (f). Data are representative of two (a, c) or at least three (d, e) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (d, e) or two-tailed unpaired Student’s t-test (a). *P < 0.05, **P < 0.01, and ***P < 0.001. Numbers in gates indicate percentage of cells.
Extended Data Figure 8. PE stabilizes surface CXCR5 and prevents it from being targeted for lysosome-mediated degradation.
a, The internalization of surface CXCR5 over time was traced by confocal imaging. b, Wild-type or Pcyt2-deficient SMARTA cells were isolated from LCMV-infected mice and labeled with unconjugated PD-1-specific antibody before incubation at 37 °C. The amount of surface PD-1 remaining over time was detected by fluorophore-conjugated secondary antibody staining (n = 3 mice). c, Flow cytometry analysis of CXCR5 surface expression showed proteinase K treatment (0.1 mg/ml) efficiently removed the non-internalized CXCR5 on the cell surface. d, CXCR5 recycling assay. Cell surface CXCR5 was labeled with unconjugated anti-CXCR5 antibody and incubated at 30 °C to allow internalization of antibody-labeled CXCR5. After washing, the remaining surface-bound antibody was stripped by resuspension in proteinase K, followed by washing and incubation at different time points to allow CXCR5 recycling. The amount of surface CXCR5 recycled over time was detected by fluorophore-conjugated secondary antibody staining and analyzed by flow cytometry (left). Right, quantification of percentage of recycled CXCR5 (relative ratio to untreated cells) (n = 4 mice). e, Validation of anti-CXCR5 antibody (clone number, EPR23463–30) by immunoblot analysis of CXCR5 expression in wild-type and CXCR5-deficient SMARTA cells that were isolated from LCMV-infected mice. f, Immunoblot analysis of CXCR5 and Pcyt2 expression in wild-type and Pcyt2-deficient SMARTA cells that were isolated from LCMV-infected mice and treated with the indicated concentrations of Bafilomycin A1 (BafA1) for 8 h. g, Immunoblot analysis of CXCR5 expression in wild-type and Pcyt2-deficient SMARTA cells isolated from LCMV-infected mice after treatment with the proteasome inhibitor MG-132. h, Flow cytometry analysis of CXCR5 surface expression on wild-type and Pcyt2-deficient SMARTA cells that were isolated from LCMV-infected mice and treated with the indicated concentrations of BafA1 for 8 h. i, Quantification of surface PE levels on human CXCR5+ memory Tfh, CXCR5+ central memory (Tcm) and CD45RA+ naïve-like T cells in peripheral blood (n = 4 donors). j, Analysis and quantification of surface PE levels on human tonsil CXCR5+PD-1hi Tfh cells, transitional CXCR5+PD-1+ Tfh cells, CXCR5− non-Tfh and CD45RA+ naïve-like T cells (n = 8 donors). Data are representative of two (b, e–h) or at least three (a, c, d, i, j) independent experiments. P values are determined by one-way ANOVA (i, j) or two-tailed unpaired Student’s t-test (b, d). NS, not significant; *P < 0.05 and ***P < 0.001. Numbers in gates indicate percentage of cells.
Extended Data Figure 9. Pcyt2 deficiency in activated T cells reveals selective impairments of Tfh accumulation associated with reduced GC responses.
a, Pcyt2 mRNA expression in freshly-isolated (day 0) naïve CD4+ T cells from wild-type and OX40CrePcyt2fl/fl mice, or after in vitro anti-CD3/CD28 antibody stimulation for the indicated times (n = 4 samples). b, Analysis and quantification of Tfh cells (CXCR5+Bcl6+ or CXCR5+PD-1+) among B220−CD4+TCRβ+ cells in Peyer’s patches (PPs) and mesenteric lymph nodes (mLNs) from WT and OX40CrePcyt2fl/fl mice (8 weeks old; CXCR5+Bcl6+ Tfh cells, n = 4 mice; CXCR5+PD-1+ Tfh cells, n = 3 mice). c, Analysis and quantification of GC B cells (Fas+GL7+) among B220+CD19+ cells in PPs and mLNs from WT and OX40CrePcyt2fl/fl mice (n = 3 mice). d, Immunohistochemistry of GCs in the mLNs of WT and OX40CrePcyt2fl/fl mice. Scale bar, 50 μm (n = 3 mice). e, Pcyt1a mRNA expression in freshly-isolated (day 0) naïve CD4+ T cells from WT and OX40CrePcyt1afl/fl mice, or after in vitro stimulation with anti-CD3/CD28 antibodies for indicated times (n = 3 samples). f, Analysis and quantification of Tfh cells (CXCR5+Bcl6+ or CXCR5+PD-1+) among B220−CD4+TCRβ+ cells in PPs and mLNs from WT and OX40CrePcyt1afl/fl mice (8 weeks old; n = 3 mice). g, Analysis and quantification of GC B cells (Fas+GL7+) among B220+CD19+ cells in PPs and mLNs from WT and OX40CrePcyt1afl/fl mice (n = 3 mice). h, Mixed bone marrow (BM) chimeras were constructed by mixing BM cells from WT or OX40CrePcyt2fl/fl mice and CD45.1+ ‘spike’ mice followed by injection into sub-lethally irradiated Rag1–/– recipient mice. Quantification of Tfh cells (CXCR5+Bcl6+) among CD45.1+B220–CD4+TCRβ+ or CD45.2+B220–CD4+TCRβ+cells in the PPs and mLNs under steady state (n = 3 mice). i, Analysis and quantification of gp66 tetramer-positive CXCR5+PD-1+ Tfh cells in the spleen of WT and OX40CrePcyt2fl/fl mice at day 7 post-infection (n = 4 mice). j, Retrogenic mouse-derived naïve CD4+ T cells deficient for Etnk1 or Selenoi were transferred into C57BL/6 hosts, followed by LCMV infection. Analyses of the proportion of donor-derived Tfh (CXCR5+SLAM−, CXCR5+PD-1+ or PSGL-1−Ly6C−) cells (left) and quantification of Tfh cell percentage and number (right) in the spleen at day 7 post-infection (n = 4 mice). k, Quantification of numbers of Tfh cells (CXCR5+PD-1+, CXCR5+ICOS+ or CXCR5+Ly6C−) or GC B cells in the spleen from WT and OX40CrePcyt2fl/fl mice at day 7 after intraperitoneal immunization with NP-OVA + LPS in alum (n = 4 mice). l, Measurements of anti-NP immunoglobulins in the serum from WT and OX40CrePcyt2fl/fl mice at day 7 after NP-OVA + LPS immunization (n = 16, collected from 8 mice). Data are representative of two (a, e–l) or at least three (b–d) independent experiments. Data are mean ± s.e.m. P values are determined by one-way ANOVA (j) or by two-tailed unpaired Student’s t-test (b, c, f–i, k, l). NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. Numbers in gates indicate percentage of cells.
Extended Data Figure 10. Surface PE distribution and CXCR5 expression on B cells are dependent upon the CDP-ethanolamine pathway.
a, Analysis and quantification of the proportions of CXCR5+PE+ cells on mature conventional B cells (B220hiIgDhiIgMlo) or total B220+IgM+ B cells from the spleen, immature B cells (B220+IgM+ cells from the bone marrow (BM)), as well as splenic naïve CD4+ T cells (n = 3 mice). b, Quantification of CXCR5 and PE mean fluorescence intensities (MFIs) among B220+ cells in spleen, peripheral lymph nodes (pLNs), mesenteric lymph nodes (mLNs), and Peyer’s patches (PPs) of wild-type (WT) and Cd19CrePcyt2fl/fl mice (n = 3 mice). c, Proportion of migrated B cells from the spleen of WT and Cd19CrePcyt2fl/fl mice was assessed by flow cytometry after CXCL13 treatment for 3 h (n = 3 mice). d, Two-step model of Tfh cell differentiation. Expression of Bcl6 and other transcription factors in activated CD4+ T cells orchestrates the initiation and commitment to the Tfh program, leading to the induction of chemokine receptor CXCR5 (left cell). Right cell: In Bcl6/CXCR5-expressing Tfh cells, the CDP-ethanolamine pathway (composed of Etnk1, Pcyt2 and Selenoi), which mediates de novo phosphatidylethanolamine (PE) synthesis, acts as a critical posttranscriptional program for the functional maintenance of the Tfh program. Maintenance of the Tfh program is essential for germinal center (GC) responses, plasma cell formation, and antigen-specific immunoglobulin secretion. In contrast, the CDP-choline pathway that promotes phosphatidylcholine (PC) synthesis and the Pisd-dependent decarboxylation of phosphatidylserine (PS) are dispensable for Tfh accumulation, as indicated by gray shading. e, Mechanistically, the CDP-ethanolamine pathway controls Tfh responses by stabilizing CXCR5 surface expression, CXCL13–CXCR5-mediated signaling events and cellular trafficking to B cell follicles. De novo synthesis of PE downstream of the CDP-ethanolamine pathway enables PE to interact with CXCR5 and prevents its targeting for lysosome-mediated degradation. Depletion of the CDP-ethanolamine pathway reduces the stability, accelerates the internalization rate, and decreases the surface recycling of CXCR5. Data are representative of at least two (a–c) independent experiments. Data are mean ± s.e.m. P values are determined by two-tailed unpaired Student’s t-test (b, c). NS, not significant; *P < 0.05 and **P < 0.01. Numbers in gates indicate percentage of cells.
Supplementary Material
Acknowledgements
The authors acknowledge J. Saravia for human samples, Y. Wang and J. Raynor for editing of the manuscript, M. Hendren for animal colony management, the St. Jude Immunology FACS core facility for cell sorting, Center for Advanced Genome Engineering (CAGE) for testing sgRNA deletion efficiency, and the Tetramer Facility of the National Institutes of Health for gp66. Electron microscopy support was provided by the Cell & Tissue Imaging Center from SJCRH and NCI P30 CA021765. This work was supported by National Institutes of Health (NIH) AI105887, AI131703, AI140761, AI150241, AI150514 and CA221290, and Lupus Research Alliance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests
H. Chi is a consultant for Kumquat Biosciences, Inc.
Additional information
Supplementary information The online version contains supplementary material available.
References
- 1.Crotty ST Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston RJ et al. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science 325, 1006–1010, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurieva RI et al. Bcl6 Mediates the Development of T Follicular Helper Cells. Science 325, 1001–1005, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity 45, 540–554, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman NM, Boothby MR & Chi H Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20, 55–70, (2020). [DOI] [PubMed] [Google Scholar]
- 6.Wei J et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H et al. In vivo CRISPR screening reveals nutrient signaling processes underpinning CD8(+) T cell fate decisions. Cell 184, 1245–1261 e1221, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol 16, 991–999, (2015). [DOI] [PubMed] [Google Scholar]
- 9.Choi YS et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 16, 980–990, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boularan C et al. B Lymphocyte-Specific Loss of Ric-8A Results in a Galpha Protein Deficit and Severe Humoral Immunodeficiency. J Immunol 195, 2090–2102, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preite S et al. Hyperactivated PI3Kdelta promotes self and commensal reactivity at the expense of optimal humoral immunity. Nat Immunol 19, 986–1000, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemonnier F et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120, 1466–1469, (2012). [DOI] [PubMed] [Google Scholar]
- 13.Johnston RJ, Choi YS, Diamond JA, Yang JA & Crotty S STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med 209, 243–250, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calzada E, Onguka O & Claypool SM Phosphatidylethanolamine Metabolism in Health and Disease. Int Rev Cell Mol Biol 321, 29–88, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haynes NM et al. Role of CXCR5 and CCR7 in Follicular Th Cell Positioning and Appearance of a Programmed Cell Death Gene-1 High Germinal Center-Associated Subpopulation. J Immunol 179, 5099–5108, (2007). [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Magdaleno S, Tabas I & Jackowski S Early embryonic lethality in mice with targeted deletion of the CTP:phosphocholine cytidylyltransferase alpha gene (Pcyt1a). Mol Cell Biol 25, 3357–3363, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X et al. Genome-wide Analysis Identifies Bcl6-Controlled Regulatory Networks during T Follicular Helper Cell Differentiation. Cell Rep 14, 1735–1747, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J et al. PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity 49, 264–274, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao M Lantibiotics as probes for phosphatidylethanolamine. Amino Acids 41, 1071–1079, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irie A, Yamamoto K, Miki Y & Murakami M Phosphatidylethanolamine dynamics are required for osteoclast fusion. Sci Rep 7, 46715, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burr ML et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H et al. Discrete roles and bifurcation of PTEN signaling and mTORC1-mediated anabolic metabolism underlie IL-7–driven B lymphopoiesis. Sci Adv 4, 1–19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online References
- 23.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oxenius A, Bachmann MF & Zinkernagel RMH, H. Virus-specific MHC class II-restricted TCR- transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol 28, 390–400, (1998). [DOI] [PubMed] [Google Scholar]
- 25.Leonardi R, Frank MW, Jackson PD, Rock CO & Jackowski S Elimination of the CDP-ethanolamine Pathway Disrupts Hepatic Lipid Homeostasis. J Biol Chem 284, 27077–27089, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D et al. Macrophages deficient in CTP:Phosphocholine cytidylyltransferase-alpha are viable under normal culture conditions but are highly susceptible to free cholesterol-induced death. J Biol Chem 275, 35368–35376, (2000). [DOI] [PubMed] [Google Scholar]
- 27.Bettini ML, Bettini M, Nakayama M, Guy CS & Vignali DA Generation of T cell receptor-retrogenic mice: improved retroviral-mediated stem cell gene transfer. Nat Protoc 8, 1837–1840, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locci M et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39, 758–769, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanson KR et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9, 5416, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanayama R et al. Autoimmune Disease and Impaired Uptake of Apoptotic Cells in MFG-E8–Deficient Mice. Science 304, 1147–1150, (2004). [DOI] [PubMed] [Google Scholar]
- 33.Todo T et al. Anti-CD3e induces splenic B220lo B-cell expansion following anti-CD20 treatment in a mouse model of allosensitization. Int Immunol 24, 529–538, (2012). [DOI] [PubMed] [Google Scholar]
- 34.Zeng H et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrestha S et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol 16, 178–187, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D et al. The Transcriptional Repressor Bcl-6 Directs T Follicular Helper Cell Lineage Commitment. Immunity 31, 457–468, (2009). [DOI] [PubMed] [Google Scholar]
- 37.Li W et al. Chimeric Antigen Receptor Designed to Prevent Ubiquitination and Downregulation Showed Durable Antitumor Efficacy. Immunity 53, 456–470 e456, (2020). [DOI] [PubMed] [Google Scholar]
- 38.Bates M, Dempsey BHGT & Zhuang X Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes. Science 317, 1749–1753, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy CS et al. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol 14, 262–270, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liedmann S et al. Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat Commun 5, 5645, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samir P et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malkusch S et al. Coordinate-based colocalization analysis of single-molecule localization microscopy data. Histochem Cell Biol 137, 1–10, (2012). [DOI] [PubMed] [Google Scholar]
- 43.Azuma T & Kei T Super-resolution spinning-disk confocal microscopy using optical photon reassignment. Opt Express 23, 15003–15011, (2015). [DOI] [PubMed] [Google Scholar]
- 44.Guagliardi LE et al. Co-localization of molecules involved in antigen processing and presentation in an early endocytic compartment. Nature 343, 133–139, (1990). [DOI] [PubMed] [Google Scholar]
- 45.Folch J, Lees M & Stanley GHS A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226, 497–509, (1957). [PubMed] [Google Scholar]
- 46.PASTERNAK CA & BERGERON JJM Turnover of Mammalian Phospholipids. Biochem. J 119, 473–480, (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keckesova Z et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 543, 681–686, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Microarray data of wild-type and Pcyt2-deficient cells at day 3 after LCMV infection have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number: GSE147190 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147190). For GSEA using manually curated gene signatures from public datasets, the dataset GSE213809 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21380) and GSE7218817 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72188) from NCBI GEO database were used to generate ‘Tfh gene signatures’ (< 5% FDR). Source data are provided with this paper.