Abstract
In insects the reserve proteins are stored in the oocytes into endocytic-originated vesicles named yolk organelles. VPS38/UVRAG and ATG14 are the variant regulatory subunits of two class-III ATG6/Beclin1 PI3K complexes that regulate the recruitment of the endocytic (complex II) and autophagic (complex I) machineries. In a previous work from our group, we found that the silencing of ATG6/Beclin1 resulted in the formation of yolk-deficient oocytes due to defects in the endocytosis of the yolk proteins. Because ATG6/Beclin1 is present in the two above-described PI3K complexes, we could not identify the contributions of each complex to the yolk defective phenotypes. To address this, here we investigated the role of the variant subunits VPS38/UVRAG (complex II, endocytosis) and ATG14 (complex I, autophagy) in the biogenesis of the yolk organelles in the insect vector of Chagas Disease Rhodnius prolixus. Interestingly, the silencing of both genes phenocopied the silencing of ATG6/Beclin1, generating 1) accumulation of yolk proteins in the hemolymph; 2) white, smaller, and yolk-deficient oocytes; 3) abnormal yolk organelles in the oocyte cortex; and 4) unviable F1 embryos. However, we found that the similar phenotypes were the result of a specific cross-silencing effect among the PI3K subunits where the silencing of VPS38/UVRAG and ATG6/Beclin1 resulted in the specific silencing of each other, whereas the silencing of ATG14 triggered the silencing of all three PI3K components. Because the silencing of VPS38/UVRAG and ATG6/Beclin1 reproduced the yolk-deficiency phenotypes without the cross silencing of ATG14, we concluded that the VPS38/UVRAG PI3K complex II was the major contributor to the previously observed phenotypes in silenced insects.
Altogether, we found that class-III ATG6/Beclin1 PI3K complex II (VPS38/UVRAG) is essential for the yolk endocytosis and that the subunits of both complexes are under an unknown transcriptional regulatory system.
Author summary
The oocytes of oviparous animals are highly specialized cells committed to the storage of nutrients (yolk) required for the maternally detached embryo development. Most oocytes undergo a massive growth during oogenesis due to their high endocytic activity of yolk macromolecules. In this work, we found that the components of two PI3K complexes are co-transcriptionally regulated, and that one specific PI3K complex (class-III PI3K complex II) is essential for the correct endocytosis and biogenesis of the yolk endocytic organelles. The molecular machinery and regulations that govern the yolk generation are mostly unknown. Thus, these findings are important in the context of general vector’s reproductive biology and for the elaboration of novel strategies for vector’s population control.
Introduction
The oocytes of oviparous animals are highly specialized cells dedicated to the storage of nutrients required for embryo development. Most oocytes undergo a massive growth in volume during oogenesis due to the endocytosis of yolk macromolecules which will be targeted for degradation in a regulated manner throughout embryogenesis [1–5].
Phosphoinositide 3-kinase (PI3K) family members are able to generate phosphatidylinositol-3P (PI3P), an important signal molecule involved in a variety of vesicle trafficking events, in a spatially- and timely-regulated manner. The controlled generation of this important signal inducer regulates diverse cellular processes including cell growth, proliferation, and survival [6–9]. Three classes of PI3K have been described, and each class includes several different PI3K complexes. Typically, PI3P produced by class-III PI3K complexes have been found in autophagosomes, endosomes and multivesicular bodies [6, 10–14]. Here, we focused on the role of the two autophagy related gene 6 (ATG6/Beclin1) class-III PI3K complexes, which are involved in the regulation of autophagy and the membrane dynamics of endocytosis [15].
ATG6/Beclin1 is an autophagy related gene component of two different class-III PI3K complexes. The invariant subunits of both complexes, in addition to ATG6/Beclin1, are the catalytic subunit vacuolar protein sorting 34 (VPS34) and the putative protein kinase vacuolar protein sorting 15 (VPS15), which were originally described in Saccharomyces cerevisiae but have been found in other species including plants and higher eucaryotes [16]. The distinct cellular functions of each ATG6/Beclin1 PI3K complex is related to the association of these complexes to different regulatory subunits vacuolar protein sorting 38/ UV irradiation resistance-associated gene (VPS38/UVRAG) or autophagy related gene 14 (ATG14). The complex I is composed by ATG6/Beclin1, VPS34, VPS15 and ATG14. It is formed on the pre-autophagosome structure and is involved in the recruitment of ATGs and regulation of autophagy initiation. The complex II is composed by ATG6/Beclin1, VPS34, VPS15 and VPS38/UVRAG. This complex is localized in the membrane of endosomes, being important to general vacuolar protein sorting [12, 17]. In brief, ATG14 is a key subunit in determining the role of the PI3K complex during autophagy, while VPS38 is essential to mobilize the vesicles along the endocytic route [15, 18, 19]. However, recent studies have demonstrated that not only the ATGs are involved in the regulation of other cellular functions, such as programmed cell death, aging, immune system, viral replication, tumorigenesis and others [20–33], but also that VPS38/UVRAG plays alternative functions that became more evident in the past few years. For example, VPS38/UVRAG participates in the regulation of the translocation of autophagy related gene 9 (ATG9) (a lipid scramblase that mediates autophagosomal membrane expansion [34]), to the autophagosome and its association with important effectors of retrograde vesicular transport [35, 36].
The participation of receptor mediated endocytosis in the yolk uptake and biogenesis of the yolk organelles have been investigated in oocytes of many species. In insects, the internalization of yolk proteins through the presence of a specialized endocytic cortex in the oocytes, which includes prominent microvilli, coated pits, coated vesicles, and endosomes have been shown in several species [37–44]. Immunodetection of the internalization of yolk proteins and markers of endocytosis (such as clathrin, α-adaptin, and yolk protein receptors) have been performed in Drosophila [44] and mosquito [45]. However, the regulations encompassing the recruitment of the endocytic machinery to specific sites of the oocyte cortex and the signals and regulations that govern the oocyte endocytic pathways and endosomal maturation have never been addressed.
In a previous work our group demonstrated that ATG6/Beclin1 is important for the uptake of yolk proteins, yolk organelles biogenesis and embryo viability in the insect vector R. prolixus [46]. Here, we investigate the role of the ATG6/Beclin1-PI3K complexes variant regulatory subunits: ATG14 and VPS38/UVRAG. We found that for both VPS38/UVRAG- and ATG14-silenced females, the yolk proteins are accumulated in the hemolymph as the result of poor uptake by the oocytes. As a consequence, the silenced mature oocytes are small, white (as opposed to control oocytes which are light red), present abnormal yolk organelles, as seen by light microscopy and flow cytometry, and do not support F1 embryo development. Because we learned that VPS38/UVRAG and ATG14 remarkably phenocopied each other and the effects of silencing ATG6/Beclin1 [46], we asked if there was a transcriptional regulation among these genes. Indeed, we found a specific cross-silencing among VPS38/UVRAG, ATG14 and ATG6/Beclin1 in all RNAi knockdown ovary samples, which explains the phenocopies and reveals the existence of transcriptional regulations among the subunits of PI3K complexes.
Material and methods
Ethics statement
All animal care and experimental protocols were approved by guidelines of the institutional care and use committee (Committee for Evaluation of Animal Use for Research from the Federal University of Rio de Janeiro, CEUA-UFRJ #01200.001568/2013-87, order number 155/13), under the regulation of the national council of animal experimentation control (CONCEA). Technicians dedicated to the animal facility conducted all aspects related to animal care under strict guidelines to ensure careful and consistent animal handling.
Bioinformatics
The sequence of Rhodnius prolixus VPS38/UVRAG (RPRC001388), ATG14 (RPRC001958) and ATG6/Beclin1 [46] were obtained from the R. prolixus genome and transcriptome databases (Rpro C3.2) from Vector Base (www.vectorbase.org) [47–49]. The Drosophila VPS38/UVRAG (Gene ID: 34735) and ATG14 (Gene ID: 43438) orthologues were used as the first template for identification. Values of similarity and identity were predicted using SIAS software (www.imed.med.ucm.es/Tools/sias.html) and compared to the sequences of Homo Sapiens UVRAG/VPS38 (Gene ID: 7405) and ATG14 (Gene ID: 2286) and Saccharomyces cerevisiae UVRAG/VPS38 (Gene ID: 851074) and ATG14 (Gene ID: 852425). Conserved domains were predicted using PFAM (http://pfam.xfam.org/) [50].
Insects
Insects were maintained at a 28 ± 2°C controlled temperature, relative humidity of 70–80% and 12/12 h light and dark cycles. All females used in this work were obtained from our own insectarium where mated females are fed for the first time (as adult insects) in live-rabbit blood 14 to 21 days after the 5th instar nymph to adult ecdysis. After the first blood feeding, all adult insects in our insectarium are fed every 21 days. For all experiments, mated females of the second or third blood feeding were used, and dissections were carried out at different days after the blood meal depending on the experiment.
Dissection of the ovary parts, follicles, and chorionated oocytes
The different parts of the ovariole were carefully dissected in phosphate buffered saline (PBS) 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4 using fine tweezers and dissecting scissors under the stereo microscope according to [51], and the structures (tropharium, previtellogenic and vitellogenic follicles, and chorionated oocytes) were classified by length and morphology according to [52, 53]. The chorionated oocytes were dissected from the ovarioles (not from the oviducts). Because it is not known when oocyte maturation or activation occurs in R. prolixus, there is no way of knowing if the chorionated oocyte is still an immature oocyte or a mature egg. The terminology chorionated oocyte (and not chorionated egg) was chosen to refer to the oocytes right after the completion of choriogenesis because it matches to the terminology used in Drosophila [54, 55]. When chorionated oocytes were used, dissection was performed 7–12 days after the blood meal. All other organs and ovary parts were dissected 7 days after blood meal.
Extraction of RNA and cDNA synthesis
All samples were homogenized in Trizol reagent (Invitrogen) for total RNA extraction. Reverse transcription reaction was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using 1 μg of total RNA after RNase-free DNase I (Invitrogen) treatment, Multiscribe Reverse Transcriptase enzyme (2.5 U/μL) and random primers for 10 min at 25°C followed by 2 hours of incubation at 37°C. As a control for the DNAse treatment efficiency, we performed control reactions without the enzyme followed by testing the capacity of amplification by PCR.
PCR/RT-qPCR
Specific primers for the RpVPS38 and RpATG14 sequences were designed to amplify 323bp and 219bp fragments, respectively, in a PCR using the following cycling parameters: 10 min at 95°C, followed by 35 cycles of 15 s at 95°C, 45 s at 52°C and 30 s at 72°C and a final extension of 15 min at 72°C. Amplifications were observed in 2% agarose gels. Quantitative PCR (qPCR) was performed in a StepOne Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems) under the following conditions: 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 45 s at 60°C. qPCR amplification was performed using the specific primers described in S1 Table. All primers were used at a final concentration of 0.2 μM. The cDNAs were diluted 10X and used in the reactions. To exclude nonspecific amplification, blank reactions replacing de template (cDNA) for water were performed in all experiments. The relative expressions were calculated using the delta Ct (cycle threshold) obtained using the reference gene 18S (RPRC017412) and calculated 2-dCt. According to the minimum information for publication of quantitative RT-qPCR experiments (MIQE) Guidelines, normalization against a single reference gene is acceptable when the investigators present clear evidence that confirms its invariant expression under the experimental conditions [56]. S1 Fig shows the invariant expression of 18S in our experimental conditions. The primers for VPS38/UVRAG presented 84.52% efficiency with a slope of -3.759 and 25.01 Y-interception. The primers for ATG14 presented 100% efficiency with a slope of -3.28 and 25.872 Y-interception. For all RT-qPCRs, each biological replicate (n = 6–8) was prepared using a pool of samples dissected from 3 insects.
RNAi silencing
dsRNA was synthesized by MEGAScript RNAi Kit (Ambion Inc) using primers for RpVPS38/UVRAG and RpATG14 specific gene amplification with the T7 promoter sequence (S1 Table). dsRNA for the silencing of ATG6/Beclin1 was synthesized as previously described [46]. Unfed adult females were injected between the second and third thoracic segments using a 10 μl Hamilton syringe with 1 μg dsRNA (diluted in 1μl of water) and fed 2 days later. Knockdown efficiency was confirmed by qPCR at 7 days after blood meal in the ovary and fat body. The bacterial MalE gene was used as a control dsRNA [20]. Adult females injected with dsRNA were fed and transferred to individual vials. The mortality rates and the number of eggs laid by each individual were recorded daily and weekly, respectively. Additional measurements are described below.
PI3P detection by thin-layer chromatography (TLC) and quantification
Silenced and control females were fed with blood enriched with 32Pi [57] using a special feeder [58]. On the tenth day after a blood meal, the chorionated oocytes were collected and subjected to lipid extraction [59]. The thin-layer silica plates (G-60; 0±25 mm thickness) were pre-coated with trans-1,2-diaminocyclohexane-N,N,N’,N’- tetraacetic acid solution in water: methanol: 10M NaOH (1:2:0,1 v/v) and allowed to dry overnight [60]. The TLC developing solution was prepared by stirring together methanol (75 ml), CHC13 (60 ml), pyridine (45 ml), and boric acid (12 g) until the boric acid was dissolved. The plates were developed in a saturated tank and were run to the top [60]. The radioactivity was analyzed in a laser scanner Cyclone1Plus Storage Phosphor System (Perkin Elmer). Additionally, the plates were autoradiographed using X-ray film (Kodak T-Mat) and an intensifier screen. The cassette was stored at −70°C for 30 days. The autoradiography was then subjected to densitometric analysis. The phospholipids spots were identified by comparison with standards run in parallel according to [61]. Densitometry was performed using ImageJ software. Each biological replicate (n = 5) was prepared using a pool of 2 chorionated oocytes.
Egg homogenates and SDS-PAGE
Control and silenced eggs were collected at 24h of embryogenesis. Pools of 4 eggs were homogenized using a plastic pestle in 100 μl of PBS containing a cocktail of protease inhibitors (Aprotinin 0.3 μM, leupeptin 1 μg/μl, pepstatin 1 μg/μl, PMSF 100 μM and EDTA 1 mM). The samples were directly used, and 30 μg of total protein were loaded in each lane of a 13% SDS-PAGE. Gels were stained with silver nitrate [62]. For each biological replicate (n = 3), the samples were prepared using a pool of 4 eggs laid by different insects.
Hemolymph extraction and SDS-PAGE
The hemolymph was extracted from control and silenced females, 7 days after the blood meal, as originally described by Masuda e Oliveira, 1985 [63]. Approximately 10 μl of hemolymph per female was obtained by cutting one of the insects’ legs and applying gentle pressure to the abdomen. The hemolymph was collected using a 10μl pipette plastic tip. Once collected, the hemolymph was diluted 2x in PBS containing protease inhibitors (aprotinin 0.3 μM, leupeptin 1 μg/μl, pepstatin 1 μg/μl, PMSF 100 μM and EDTA 1 mM), and approximately 8 mg of phenylthiourea. The equivalent of 1 μl of hemolymph was loaded in each lane of a 15% SDS-PAGE. For each biological replicate (n = 4), the samples were prepared using a pool of 3 insects.
Determination of protein content
The total amount of protein in the silenced and control eggs and hemolymph samples was measured by the Lowry (Folin) method, using as standard control 1–5 μg of BSA [64] in a E-MAX PLUS microplate reader (Molecular devices) using SoftMax Pro 7.0 as software. For each biological replicate of the hemolymph samples (n = 8), the samples were prepared using a pool of 3 insects. Each biological replicate of the egg homogenates (n = 6) were prepared using a pool of 4 eggs laid by different insects.
Light microscopy
Opercula of recently dissected chorionated oocytes were carefully detached using a sharp razor blade under the stereomicroscope. The opercula-free oocytes were fixed by immersion in 4% freshly prepared formaldehyde in 0.1 M cacodylate buffer, pH 7.2 for 12 h at room temperature. Samples were washed 3 times for 10 minutes in the same buffer and embedded in increasing concentrations (25%, 50%, 75% and 100%) of OCT compound medium (Tissue-TEK) plus 20% glucose as a cryoprotectant, for 12 h for each of the concentrations. Once infiltrated in pure OCT, 15–30 μm transversal sections of the chorionated oocytes were obtained in a cryostat. The slides were mounted in glycerol 50% followed by observation in a Zeiss Axio Imager D2 equipped a Zeiss Axio Cam MrM operated in a differential interferential contrast (DIC) mode. For each biological replicate (n = 3), one chorionated oocyte was dissected from a different insect and processed as described above.
Yolk organelles suspension and flow cytometry
Suspensions of yolk organelles were obtained by gently disrupting of recently dissected chorionated oocytes in PBS (2 oocytes in 250μl of buffer). The population profiles of the yolk organelles were acquired on a FACS Calibur instrument (BD Bioscience, USA) powered by CellQuest Pro software v5.1 and analyzed using Flowing Software 2.5.1. For each biological replicate (n = 5), the samples were prepared using a pool of 2 chorionated oocytes dissected from different insects.
Statistics
The relative expression and ΔCt values were calculated from obtained Ct (cycle threshold) values. The Ct mean values obtained from the experiments were compared using One-way ANOVA followed by Tukey’s multiple comparison test. Differences were considered significant at P< 0.05. The relative expression values (2-ΔCt) were used only for graph construction. Other results were analyzed by Student’s t-Test for the comparison of two different conditions and One-way ANOVA followed by Tukey’s test for the comparison among more than two conditions. Differences were considered significant at p<0.05. All statistical analyses were performed using the Prism 7.0 software (GraphPad Software).
Results
VPS38/UVRAG and ATG14 maternal RNAs are accumulated in the oocytes
In a previous work, our group described that silencing of ATG6/Beclin1 in vitellogenic females resulted in the production of white/small oocytes and eggs due to the abnormal uptake of yolk components from the hemolymph. ATG6/Beclin1 is part of two class-III PI3K complexes, that signal to different intracellular routes: endocytosis and autophagy. To further investigate each of the ATG6/Beclin1 PI3K complexes were contributing to the ATG6/Beclin1 phenotypes, we decided to investigate the silencing effects of VPS38/UVRAG and ATG14, the variant subunits present in each of the ATG6/Beclin1 PI3K complexes. We found one isoform of the genes VPS38/UVRAG (RPRC001388) and ATG14 (RPRC001958) in the Rhodnius genome assembly (RproC3), with a total of 8 and 3 exons, respectively (S2 Fig). VPS38/UVRAG and ATG14 predicted proteins are 70 and 80% similar to their human orthologs (S3 Fig), presenting their expected conserved domains (CC and C2, Pfam: PF04111 and PF00168) (Fig 1A and 1B). RT-qPCR showed that the ovary of R. prolixus expresses approximately 6x more VPS38/UVARG than the midgut, fat body and flight muscle (Fig 1C), whereas ATG14 is 5x more expressed in the ovary than in the flight muscle and midgut (Fig 1D). Within the ovary, the highest levels of VPS38/UVRAG and ATG14 mRNAs were detected in the tropharium, which expresses an average of 2x the mRNA levels of both genes detected in the developing follicles (pre-vitellogenic and vitellogenic follicles) and chorionated oocytes (Fig 1C and 1D).
Fig 1. VPS38/UVRAG and ATG14 are conserved and highly expressed in the ovaries.
A-B. Diagram of the predicted conserved functional domains of human, yeast and Rhodnius prolixus VPS38/UVRAG and ATG4. C2, a protein structural domain involved in targeting PI3K to the cell membrane, CC, coiled coil domain. C-D. RT-qPCR showing the relative expression of VPS38 and ATG4 in different organs and during oogenesis of vitellogenic females. MG, Midgut; FB, Fat body; FM, flight muscle; Ov, Ovary. Troph, tropharium; PreVit, pre-vitellogenic follicles; Vit, vitellogenic follicles; Chor, chorionated oocytes. The relative expression was quantified using the ΔCT method with Rp18S as endogenous control. Graphs show mean ± SEM (n = 6). * p<0.05, **p<0.01, One Way ANOVA.
Silencing of VPS38/UVRAG and ATG14 results in the accumulation of yolk proteins in the hemolymph and formation of small yolk-deficient oocytes
Double stranded RNAs designed to target specific regions of the VPS38/UVRAG and ATG14 mRNAs were injected to the insects hemocoel 2 days before the blood meal. Nine days later (7 days after the blood meal), the ovaries and fat bodies of control and silenced females were dissected, and their mRNA contents were analyzed by RT-qPCR. The VPS38/UVRAG expression levels were reduced by 70% in the ovary and by 50% in the fat body (Fig 2A), whereas the ATG14 expression levels were decreased by 80% in the ovary and by 90% in the fat body (Fig 2B). Because PI3P is the product of PI3K complexes, we also tested the levels of PI3P in the ovaries of silenced females. We found that silenced ovaries presented a reduction of 60%, approximately, in their PI3P levels suggesting that their general PI3K activities were compromised (Figs 2C and S4). Despite the systemic knockdown efficiency, no alterations in the insect’s main physiological characteristics such as digestion and longevity were observed in silenced animals when compared to control ones (S5 Fig).
Fig 2. RNAi knockdown of VPS38/UVRAG and ATG14 results in reduced levels of PI3P in the ovary.
A-B. RT-qPCR showing the relative expression levels of VPS38/UVRAG and ATG14 in the ovary and fat body of control (dsMal) and silenced females. The organs were dissected 7 days after the blood meal (n = 8). The expression levels of VPS38/UVRAG were reduced by 70% in the ovary and by 50% in the fat body. The expression levels of ATG14 were reduced by 80% in the ovary and by 90% in the fat body. C. Densitometric measurements of PI3P detection by TLC in the chorionated oocytes of control and silenced insects (n = 5). * p<0.05, **p<0.01, One Way ANOVA.
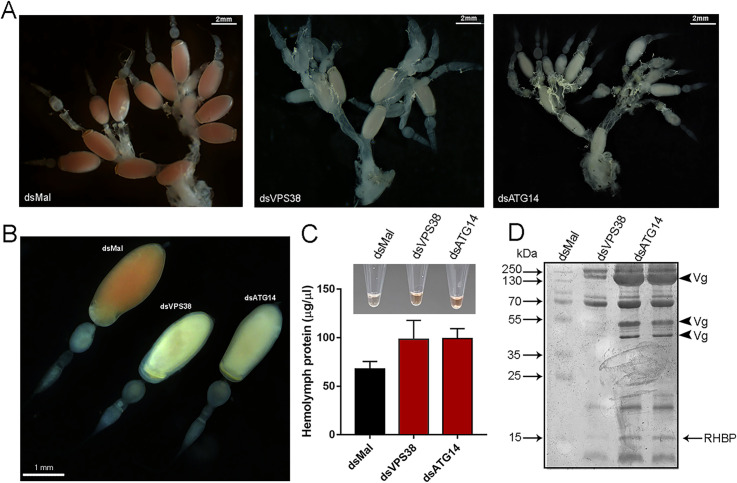
When the silenced vitellogenic females were dissected, however, we observed that knockdown of both VPS38/UVRAG and ATG14 resulted in the production of small and white oocytes in the ovaries, remarkably similar to the pattern that we previously observed for the silencing of ATG6/Beclin1 [46] (Fig 3A and 3B). To test if the formation of the white/small oocytes was due to a deficiency in taking the yolk proteins from the hemolymph, as it happens in ATG6/Beclin1 insects [46], we quantified the hemolymph yolk protein content in control and silenced females. Higher concentrations of Rhodnius heme binding protein (RHBP) were effortlessly noticeable in the reddish hemolymph extracted from silenced females, when compared to the control female’s transparent hemolymph (Fig 3C, detail image). RHBP is known for being the red molecule that provides the roseate color of R. prolixus oocytes and eggs [5, 65, 66]. For both VPS38/UVRAG and ATG14, the overall levels of protein in the hemolymph were increased by 30% (Fig 3C), and the fragments compatible with the major yolk proteins Vitellogenin [63] (Vg) (arrowheads) and RHBP [5, 65] were accumulated in the hemolymph (Fig 3D).
Fig 3. Silencing of VPS38/UVRAG and ATG14 results in small and white oocytes.
A. Representative images of the ovaries of vitellogenic females injected with dsMal, dsVPS38/UVRAG and dsATG14. B. Representative image of ovarioles dissected from females previously injected with dsMal, dsVPS38/UVRAG and dsATG14. C. Image: representative image of the hemolymph samples extracted from females previously injected with dsMal, dsVPS38/UVRAG and dsATG14. Graph: total amount of protein in the hemolymph from silenced and control females. Graph shows mean ± SEM (n = 8). D. 13% SDS-PAGE showing the protein profile of the hemolymph from silenced and control females. All experiments were performed 7 days after the blood meal. Arrowheads, Vg, vitellogenin subunits; RHPB, Rhodnius heme binding protein (n = 4).
VPS38/UVRAG and ATG14 silenced eggs are yolk-deficient and do not support embryo development
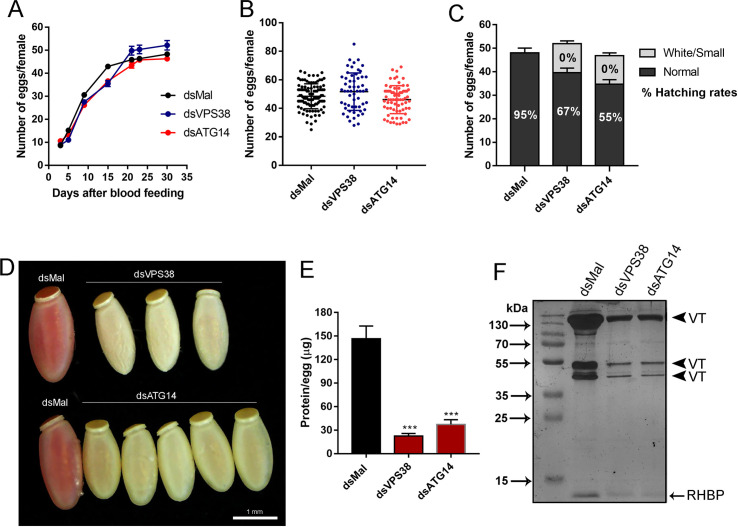
Despite the reduced size and abnormal color of the VPS38/UVRAG and ATG14 silenced oocytes (Fig 3B), the silenced females laid similar number of eggs as control females (Fig 4A and 4B). Fig 4C shows the F1 phenotypic distribution for control and silenced females. For VPS38/UVRAG an average of 11 eggs/ female (22% of the total number of eggs) were white/small, and for ATG14 an average of 10 eggs/female (27% of the total number of eggs) presented the white/small phenotype (Fig 4C). All eggs presenting the white/small phenotype (Fig 4D) were not viable (0% hatching rate), whereas the apparently morphologically normal eggs laid by silenced females decreased their viability to 67% and 55% for VPS38/UVRAG and ATG4, respectively (Fig 4C, numbers inside the bars) (S6 Fig).
Fig 4. Silencing of VPS38/UVRAG and ATG14 does not alter oviposition but decreases embryo viability.
A. Number of eggs laid per female over 4 weeks after the blood meal. Graph shows mean ± SEM (n = 52). B. Total of eggs laid by silenced and control females. Graph shows mean ± SEM (n = 52). C. Phenotypic distribution of the F1 eggs observed after knockdown of VPS38/UVRAG and ATG14. Percentage (%) of hatching per phenotype is also showed inside the bars. D. Representative image of the phenotypes observed in the F1 eggs laid by control and silenced females. E. Total amount of protein from control and VPS38/UVRAG and ATG14 silenced eggs, measured by the Lowry method. Graph shows mean ± SEM (n = 6). ***p<0.001, One Way ANOVA. F. 13% SDS-PAGE showing the protein profile of freshly laid eggs from control and silenced females. Arrowheads, VT, Vitellin subunits; RHPB–Rhodnius Heme Binding Protein (n = 3).
VPS38/UVRAG and ATG14 white/small eggs (Fig 4D) accumulated only 15% and 20%, respectively, of the total protein found in the eggs produced by control females (Fig 4E), and the accumulation of the main yolk proteins, vitellin (arrowheads) and RHBP (arrow), was severely compromised as seen by SDS-PAGE (Fig 4F). The approximated predicted volumes of the silenced white eggs are 50–60% smaller than control eggs, with an average volume of 0.82 ± 0.01mm3 for ATG14 and of 0.67 ± 0.03mm3 for VPS38/UVRAG versus 1.58 ± 0.04 mm3 of the eggs laid by control females.
VPS38/UVRAG and ATG14 are important for the biogenesis of the yolk organelles in the oocyte cortex
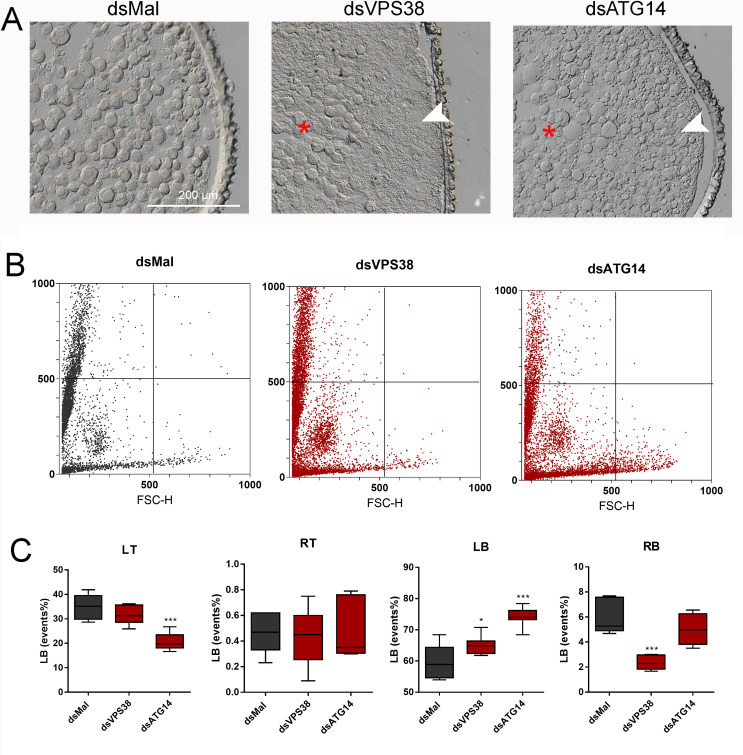
Transversal cryosections of chorionated oocytes showed an accumulation of larger yolk organelles in the core cytoplasm (red asterisk) and an irregular distribution of smaller and abnormal organelles in the periphery of the oocytes (white arrowhead) (Fig 5A). This morphology is similar to what we previously observed in ATG6/Beclin1-silenced oocytes and eggs [46] and suggests problems in the biogenesis/ sorting of the yolk organelles, compatible with the hypothesis that silenced oocytes were not able to properly recruit the endocytosis machinery during oogenesis. To further investigate the characteristics of the yolk organelles formed in the oocytes of silenced females, we used flow cytometry to get further insight into the changes in the population of organelles in terms of size (FSC) and internal complexity (SCC), which is an unprecedent attempt. Isolation of the yolk organelles in R. prolixus have been performed before and the vesicles can vary in size from 500 nm to 40 μm [67, 68]. Fig 5B shows representative dot-plots of the yolk organelles present in chorionated oocytes from control and silenced females. The representative plots allowed the identification of a broad range of events and showed that although the organelles are dispersed over a large range of sizes and internal complexities, the organelles from control and silenced samples presented different profile as quantified in Fig 5C. For both VPS38/UVRAG and ATG14, there is an increase in frequency of the organelles in the left bottom quadrant (LB), indicating reduced complexity and size in the general population of the yolk organelles obtained from silenced samples. Interestingly, for ATG14 the high complex and smaller organelles are decreased in frequency (left top quadrant, LT), whereas for VPS38/UVRAG the low complex and larger organelles are less frequent (right bottom quadrant, RB).
Fig 5. Knockdown of VPS38/UVRAG and ATG14 results in abnormal distribution of the yolk organelles.
A. Representative images of cross-sections from chorionated oocytes were observed in the light microscope. The images show an accumulation of larger yolk organelles in the core cytoplasm (red asterisk) and an irregular distribution of smaller/abnormal organelles in the periphery of silenced oocytes (white arrowheads). Bars: 200 μm. (n = 3). B. Flow cytometry FSC x SSC dot-plots of the yolk organelles extracted from chorionated oocytes obtained from control and silenced females. The plots are representative of five experiments (n = 5). C. Quantification of the organelles (events) frequency in each quadrant of the plots shown in B (RT, Right top; LT, left top; RB, right bottom; LB, left bottom). (n = 5) *p<0.05,***p<0.001, One Way ANOVA.
VPS38/UVRAG, ATG14 and ATG6/Beclin1 are transcriptionally regulated
Because the phenotypes of both VPS38/UVRAG and ATG14 were remarkably similar to each other and to the phenotypes of silencing ATG6/Beclin1 [46], we decided to test if the RNAi silencing of one of those genes was, somehow, interfering in the expression of the other genes. Interestingly, we found that the silencing of VPS38/UVRAG and ATG6/Beclin1, resulted in the silencing of both VPS38 and ATG6, and did not trigger the silencing of ATG14, whereas the silencing of ATG14 resulted in the silencing ATG14, VPS38/UVRAG and ATG6/beclin1 (Fig 6). As a control we tested the expression levels of other genes, non-related to the class III PI3K complexes, such as ATG8/LC3, ATG1/ULK1, the elongation factor 1-alpha (EF1) and the unfolded protein response sensor PKR-like ER kinase (PERK). We found that, with the exception of ULK1/ATG1 in VPS38/UVRAG-silenced samples, which was also moderately silenced, PI3K non-related genes showed no significant changes in expression levels in the silenced samples for VPS38/UVRAG, ATG14 and ATG6/Beclin1 (Fig 6).
Fig 6. VPS38/UVRAG, ATG14 and ATG6/Beclin1 are transcriptionally regulated in the oocytes of Rhodnius prolixus.
Oocytes dissected from silenced females were tested for the expression levels of the other class III PI3K components, and for the control genes ATG8/LC3, ATG1/ULK1, EF1 and PERK (non-related to the class III PI3K complexes). A. Expression levels of different genes in the ovaries of females silenced for VPS38/UVRAG (dsVPS38). B. Expression levels of different genes in the ovaries of females silenced for ATG14 (dsATG14). C. Expression levels of different genes in the ovaries of females silenced for ATG6/Beclin1 (dsATG6/Beclin1). Graphs show mean ± SEM. (n = 8) **p<0.01,***p<0.001. One Way ANOVA. D. Model for the transcriptional regulations between ATG6/Beclin1, VPS38/UVRAG and ATG14.
Discussion
In 2005, the WHO created the department of Neglected Tropical Diseases (NTDs), recognizing their importance and aiming to manage their incidence mostly in Africa and Latin America (http://www.who.int/neglected_diseases/en/). Rhodnius prolixus is one of the main vectors of Chagas disease, which is one of the several NTDs that are important in Latin America. Currently 6 to million people are estimated to be infected by Chagas Disease, and vector control is still the most useful method to prevent this illness. Because embryo development relies entirely on the buildup of the yolk nutritional reserves, the proper biogenesis of the yolk organelles is indispensable for vector reproduction representing a promising target for interference in vector’s reproduction.
The critical roles of the ATG6/Beclin1-associated complexes in the endocytic trafficking and autophagy pathways have been described before in models such as Drosophila and C. elegans [69–72]. Interestingly, in the hard tick Haemaphysalis longicornis depletion of ATG6/Beclin1 also results in a repression of Vg uptake by the oocytes [73] and in Caenorhabditis elegans, disruption of ATG6/Beclin1 results in defects in the secretion and localization of the lipoprotein receptor-related protein 1 (LRP1) [74]. Both LRP1 and Vg are important for oogenesis and their disruption might also be the result impaired endocytic activities in those models.
Similar to the results previously obtained by our group with ATG6/Beclin1 [46], we found that the systemic silencing of both VPS38/UVRAG and ATG14 resulted in specific phenotypes of impaired yolk uptake in vitellogenic females. This is compatible with the fact those genes are highly expressed in the ovaries and accumulated in the oocytes. Also, it is interesting to note that the elevated expressions of VPS38/UVRAG and ATG14 in the tropharium suggest that the nurse cells (located in the tropharium and connected to the oocytes by cytoplasmic bridges) are indeed an important site of maternal mRNAs transcription and delivery to the transcriptionally inactive developing oocytes [75], and this pattern has been previously observed for other ATGs in R. prolixus [76–78].
The silencing of VPS38/UVRAG and ATG14 resulted in altered biogenesis of the yolk organelles as observed in histology sections, and a shift to the formation of smaller and less granular organelles in the oocytes of silenced females could be observed using flow cytometry. It is possible that yolk organelles differences in contents may be detectable as changes in internal complexity. For example, the yolk proteins in Xenopus are packed in some yolk organelles in a crystalline structure [79], and internal vesicles and membranes have been observed inside some specific organelles in the oocytes of chicken and echinoderms [80, 81]. Because we do not know much about the nature of the distinct yolk organelles in R. prolixus, observing their overall pattern of relative size and internal complexity showed that indeed those organelles are diverse, and most importantly for this work, this pattern was altered in the oocytes produced by silenced females. There is no previous reference for this experiment or specific fluorescence labeling available for yolk organelles. Thus, it is not possible to rule out the detection of aggregates as typically performed in cell culture experiments using flow cytometry. Our lab intends to further explore this methodology and develop labels to detect specific yolk organelles under different conditions.
The phospholipid PI3P is the key signal generated by the PI3K complexes to recruit the specific effector proteins that promote endocytosis or autophagy [16, 82]. Because both VPS38/UVRAG and ATG14 are components of PI3K complexes, it was expected that their silencing would result in some decrease in the levels of PI3P, suggesting that the RNAi silencing of those genes resulted in at least some reduction of their protein activity. Plus, as previously observed for the silencing of ATG6/Beclin1, the lack of that signal molecule is likely the reason why silenced oocytes were not able to properly uptake the yolk proteins. The participation of endocytosis in the yolk uptake is not disputed and have been previously described in the oocytes of other insects [37–45]. Because PI3P is typically highly concentrated in early endosomes [83], and the mature yolk organelles are notably not formed in the oocytes of silenced females (Fig 5), it is likely that the silencing of the ATG6-PI3K complexes impair the endocytic flux at early steps of endosomal sorting. However, the identification of the different endocytic stages that could be compromised in the oocytes of silenced insects remains to be investigated.
The previous work from our group has shown that silencing of ATG6/Beclin1 is important for the yolk protein uptake, yolk organelles biogenesis and embryo viability, resulting in the accumulation of yolk proteins in the hemolymph and the production of small/white oocytes, which accumulated only 20% of the amount of yolk proteins detected in control oocytes [46]. Even though the ATG6/Beclin1 data indicated that the yolk endocytosis was compromised, we could not exclude the contribution of both complexes to the observed phenotypes, especially because the regulations encompassing the formation of each of the complexes are still unclear [15, 84]. To address this, in this work, we tested the silencing of the ATG6/Beclin1 PI3K complexes variant regulatory subunits: ATG14 (complex I, autophagy) and VPS38/UVRAG (complex II, endocytosis). Surprisingly, the silencing of both genes phenocopied the silencing of ATG6/Beclin1, impairing any additional phenotype-based interpretations on the contribution of each of the PI3K complexes to the previously observed compromised yolk accumulation. However, we found that the similar phenotypes were the result of a specific cross-silencing effect among the PI3K subunits VPS38/UVRAG, ATG14 and ATG6/Beclin1. Fig 6 shows that the silencing of VPS38/UVRAG and ATG6/Beclin1 resulted in the specific silencing of each other, whereas the silencing of ATG14 triggered the silencing of all three PI3K components. This data is interesting in the sense that it allows the speculation that the subunits of both PI3K complexes are under a circuit of gene regulatory network that shape the effects of PI3P production and signals during oogenesis. Gene regulation networks consist of transcription factors and regulatory elements that altogether control spatiotemporal patterns of gene expression [85]. Furthermore, the fact that the expression of ATG14 is not altered by downregulation of VPS38/UVRAG and ATG6/Beclin1, allowed us to infer that the impaired yolk uptake phenotype (accumulated yolk protein in the hemolymph and the production of small/white oocytes and eggs), is, indeed, mostly the effect of silencing the VPS38 PI3K complex II, as in VPS38/UVRAG- and ATG6-silenced insects the very same phenotypes were reproduced without the cross silencing of ATG14. It is possible that the ATG14 PI3K complex is maternally loaded during oogenesis but activated only at later processes of oogenesis or early development such as egg activation, fertilization, and early yolk degradation. Activation of the autophagic flux after fertilization has been shown in mice, where it was associated with mRNA degradation for the maternal-zygotic transition [86]. In C. elegans, paternal mitochondria are degraded via autophagy after fertilization [87], and, in Drosophila, ATG mutants have varied phenotypes presenting compromised embryogenesis [88]. Yolk degradation, specifically, was associated with autophagic mechanisms in 2015 in Drosophila. In this work, the authors showed that target of rapamycin (TOR) and ATG1 are important for the yolk catabolism and for the formation of autophagosomes [89].
Considering that the three genes in question, and their predicted proteins, are quite divergent both in terms of sequence (the highest similarity value of 13,65% is between VPS38/UVRAG and ATG14) and function, and that all primers and dsRNA fragments were tested in silico for potential cross pairing with similar sequences against the Rhodnius transcriptome database, the possibility of primers and dsRNAs cross targeting to produce the cross-silencing effects is not supported. However, general off targets effects of RNAi have been described and cannot be ruled out in any RNAi silencing experiment [90–92]. For this reason, as a way to access if the cross-silencing regulation was specific for the subunits of the PI3K complexes, non-related genes were also tested, and, in general, showed no silencing tendency. Interestingly, however, the silencing of VPS38/UVRAG triggered the downregulation of ULK1/ATG1, and this did not occur in ATG14 or ATG6/Beclin1 silenced females, where VPS38/UVRAG was also cross-silenced. Because the crosstalk between endocytosis and autophagy is known and described in different models, it is possible that the signals for endocytosis also co-regulate the biogenesis of autophagosomes through the transcriptional regulation of ULK1/ATG1. The absence of the same effect in the samples where VPS38 was cross silenced (dsATG6/Beclin1 and dsATG14) might be due to the fact that gene regulatory networks are wired in a temporal regulated manner, so that the timing of the transcriptional downregulation was not enough to trigger the effect on ULK1/ATG1 observed when VPS38 itself was silenced. Experiments to test the levels of all three transcripts and the lipidation of ATG8/LC3 (as a readout of autophagosome formation) throughout the vitellogenesis period could help illuminate this hypothesis.
Altogether, we found the existence of a transcriptional regulatory system between the subunits of class-III PI3K complexes I and II, and that PI3P signals emerging from the class-III PI3K complexes are critical regulators of the yolk organelles biogenesis in the oocytes of the vector Rhodnius prolixus.
Supporting information
18s Cts obtained from different samples and conditions. (A) RNA extracted from the ovary, midgut, fat body and flight muscle. (B) RNA extracted from the different parts of the ovariole (troph, tropharium; PV, previtellogenic follicle; Vit, Vitellogenic follicle; Chor, chorionated oocyte). (C) RNA extracted from the ovary and fat body of control (dsMal) and silenced insects. All samples were dissected 7 days after the blood meal.
(TIF)
Sequence information was obtained from Vector Base (https://www.vectorbase.org/).
(TIF)
Reference sequence Rp–Rhodnius prolixus. Dm–Drosophila melanogaster; Hs–Homo sapiens; Sc—Saccharomyces cerevisiae.
(TIF)
Control and silenced females were fed with a 32P-enriched blood meal. The chorionated oocytes were collected and subjected to lipid extraction followed by TLC and autoradiography. Arrows indicate the spots for PI3P and PI4P as previously described by [61].
(TIF)
A. Effect of VPS38/UVRAG and ATG14 knockdown in the blood protein amount in the midgut of the females during digestion (n = 3). B. Survival curves of control and silenced females (n = 3). Graph shows mean ± SEM. p = 0.57 (VPS38) and p = 0.43 (ATG14), Log-rank (Mantel-Cox) test.
(TIF)
The major hatching phenotypes were observed in eggs produced during the second and third cycles of oviposition.
(TIF)
All sequences were obtained from Vector Base (https://www.vectorbase.org/) and primers were synthesized by Macrogen or IDT technologies.
(DOCX)
Acknowledgments
The authors thank Yasmin Gutierrez, Mariana Martins and Yan Mendonça for the careful care of our lab insectarium and CENABIO-UFRJ and PIA-IBCCF/UFRJ for providing equipment and facilities.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by the following grants. Fundação Carlos Chagas Filho De Amparo À Pesquisa Do Estado Do Rio De Janeiro (FAPERJ) (JCNE E-26/2031802017; http://www.faperj.br/) to I.R.; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (INCT-EM 16/2014; http://cnpq.br/) to I.R.; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (https://www.gov.br/capes/pt-br) to I.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Santos R, Mariano AC, Rosas-Oliveira R, Pascarelli B, Machado EA, Meyer-Fernandes JR, et al. Carbohydrate accumulation and utilization by oocytes of Rhodnius prolixus. Arch Insect Biochem Physiol. 2008. doi: 10.1002/arch.20217 [DOI] [PubMed] [Google Scholar]
- 2.Atella GC, Arruda MABCF, Masuda H, Gondim KC. Fatty Acid Incorporation by Rhodnius prolixus Midgut. Arch Insect Biochem Physiol. 2000;107: 99–107. doi: [DOI] [PubMed] [Google Scholar]
- 3.Braz GRC, Abreu L, Masuda H, Oliveira PL. Heme biosynthesis and oogenesis in the blood-sucking bug, Rhodnius prolixus. Insect Biochem Mol Biol. 2001. doi: 10.1016/s0965-1748(00)00129-6 [DOI] [PubMed] [Google Scholar]
- 4.Gondim KC, Oliveira PL, Masuda H. Lipophorin and oögenesis in Rhodnius prolixus: Transfer of phospholipids. J Insect Physiol. 1989. doi: 10.1016/0022-1910(89)90032-2 [DOI] [Google Scholar]
- 5.Machado EA, Oliveira PL, Moreira MF, De Souza W, Masuda H. Uptake of Rhodnius heme-binding protein (RHBP) by the ovary of Rhodnius prolixus. Arch Insect Biochem Physiol. 1998. doi: [DOI] [PubMed] [Google Scholar]
- 6.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002. doi: 10.1126/science.296.5573.1655 [DOI] [PubMed] [Google Scholar]
- 7.Hawkins PT, Stephens LR. PI3K signalling in inflammation. Biochimica et Biophysica Acta—Molecular and Cell Biology of Lipids. 2015. doi: 10.1016/j.bbalip.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 8.Yu JSL, Cui W. Proliferation, survival and metabolism: The role of PI3K/AKT/ mTOR signalling in pluripotency and cell fate determination. Development (Cambridge). 2016. doi: 10.1242/dev.137075 [DOI] [PubMed] [Google Scholar]
- 9.Werner M, Hobeika E, Jumaa H. Role of PI3K in the generation and survival of B cells. Immunological Reviews. 2010. doi: 10.1111/j.1600-065X.2010.00934.x [DOI] [PubMed] [Google Scholar]
- 10.Petiot A, Ogier-Denis E, Blommaart EFC, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pa7thways that control macroautophagy in HT-29 cells. J Biol Chem. 2000. doi: 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- 11.Backer JM. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochemical Journal. 2008. doi: 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- 12.Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nature Reviews Molecular Cell Biology. 2019. doi: 10.1038/s41580-019-0129-z [DOI] [PubMed] [Google Scholar]
- 13.Thoresen SB, Pedersen NM, Liestøl K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010. doi: 10.1016/j.yexcr.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 14.Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase-Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003. doi: 10.1042/BJ20030826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itakura E, Kishi C, Kinji I, Mizushima N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19: 5360–5372. doi: 10.1091/mbc.e08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raiborg C, Schink KO, Stenmark H. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J. 2013;280: 2730–2742. doi: 10.1111/febs.12116 [DOI] [PubMed] [Google Scholar]
- 17.Itakura E, Mizushima N. Atg14 and UVRAG: Mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009. doi: 10.4161/auto.5.4.8062 [DOI] [PubMed] [Google Scholar]
- 18.Obara K, Ohsumi Y. Atg14: A key player in orchestrating autophagy. International Journal of Cell Biology. 2011. doi: 10.1155/2011/713435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015. doi: 10.1038/nature14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen IA, Attardo GM, Roy SG, Raikhel AS. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J Biol Chem. 2005;280: 20565–20572. doi: 10.1074/jbc.M500712200 [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008. doi: 10.1038/sj.cdd.4402233 [DOI] [PubMed] [Google Scholar]
- 22.Ren C, Finkel SE, Tower J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2009. doi: 10.1016/j.exger.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang GM, Tan Y, Wang H, Peng L, Chen HT, Meng XJ, et al. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Molecular Cancer. 2019. doi: 10.1186/s12943-019-0944-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munch D, Rodriguez E, Bressendorff S, Park OK, Hofius D, Petersen M. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy. 2014. doi: 10.4161/auto.29406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codogno P, Meijer AJ. Autophagy and signaling: Their role in cell survival and cell death. Cell Death and Differentiation. 2005. doi: 10.1038/sj.cdd.4401751 [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nature Cell Biology. 2010. doi: 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yordy B, Tal MC, Hayashi K, Arojo O, Iwasaki A. Autophagy and selective deployment of Atg proteins in antiviral defense. International Immunology. 2013. doi: 10.1093/intimm/dxs101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Münz C. Autophagy proteins in viral exocytosis and anti-viral immune responses. Viruses. 2017. doi: 10.3390/v9100288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajawat YS, Bossis I. Autophagy in aging and in neurodegenerative disorders. Hormones. 2008. doi: 10.14310/horm.2002.1111037 [DOI] [PubMed] [Google Scholar]
- 30.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012. doi: 10.1016/j.chom.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in arabidopsis. Plant Cell. 2009. doi: 10.1105/tpc.109.068635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011. doi: 10.1111/j.1365-313X.2011.04546.x [DOI] [PubMed] [Google Scholar]
- 33.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006. doi: 10.1038/ncb1482 [DOI] [PubMed] [Google Scholar]
- 34.Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020;27. doi: 10.1038/s41594-020-00518-w [DOI] [PubMed] [Google Scholar]
- 35.He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013. doi: 10.1038/ncb2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008. doi: 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raikhel AS, Dhadialla TS. Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol. 1992;37: 217–251. doi: 10.1146/annurev.en.37.010192.001245 [DOI] [PubMed] [Google Scholar]
- 38.Roy S, Saha TT, Zou Z, Raikhel AS. Regulatory Pathways Controlling Female Insect Reproduction. Annu Rev Entomol. 2018;63: 489–511. doi: 10.1146/annurev-ento-020117-043258 [DOI] [PubMed] [Google Scholar]
- 39.Anderson KL, Woodruff RI. A gap junctionally transmitted epithelial cell signal regulates endocytic yolk uptake in Oncopeltus fasciatus. Dev Biol. 2001;239: 68–78. doi: 10.1006/dbio.2001.0433 [DOI] [PubMed] [Google Scholar]
- 40.Postlethwait JH, Giorgi F. Vitellogenesis in insects. Dev Biol (N Y 1985). 1985;1: 85–126. doi: 10.1007/978-1-4615-6814-8_2 [DOI] [PubMed] [Google Scholar]
- 41.Raikhel AS, Dhadialla TS, Cho W, Hays AR, Koller CN. Aedes aegypti,. 1990. [Google Scholar]
- 42.Brooks RA, Woodruff RI. Calmodulin transmitted through gap junctions stimulates endocytic incorporation of yolk precursors in insect oocytes. Dev Biol. 2004;271: 339–349. doi: 10.1016/j.ydbio.2004.03.037 [DOI] [PubMed] [Google Scholar]
- 43.Ferenz H-J. Receptor-Mediated Endocytosis of Insect Yolk Proteins. In: Hagedorn HH, Hildebrand JG, Kidwell MG, Law JH, editors. Molecular Insect Science. Boston, MA: Springer US; 1990. pp. 131–138. doi: 10.1007/978-1-4899-3668-4_16 [DOI] [Google Scholar]
- 44.Richard DS, Gilbert M, Crum B, Hollinshead DM, Schelble S, Scheswohl D. Yolk protein endocytosis by oocytes in Drosophila melanogaster: Immunofluorescent localization of clathrin, adaptin and the yolk protein receptor. J Insect Physiol. 2001;47: 715–723. doi: 10.1016/s0022-1910(00)00165-7 [DOI] [PubMed] [Google Scholar]
- 45.Raikhel AS, Dhadialla TS, Cho W-L, Hays AR, Koller CN. Biosynthesis and Endocytosis of Yolk Proteins in the Mosquito. In: Hagedorn HH, Hildebrand JG, Kidwell MG, Law JH, editors. Molecular Insect Science. Boston, MA: Springer US; 1990. pp. 147–154. doi: 10.1007/978-1-4899-3668-4_18 [DOI] [Google Scholar]
- 46.Vieira PH, Bomfim L, Atella GC, Masuda H, Ramos I. Silencing of RpATG6 impaired the yolk accumulation and the biogenesis of the yolk organelles in the insect vector R. prolixus. PLoS Negl Trop Dis. 2018;12: 1–19. doi: 10.1371/journal.pntd.0006507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribeiro JMC, Genta FA, Sorgine MHF, Logullo R, Mesquita RD, Paiva-Silva GO, et al. An Insight into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius prolixus. PLoS Negl Trop Dis. 2014;8: 27. doi: 10.1371/journal.pntd.0002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43: D707–13. doi: 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc Natl Acad Sci. 2015;112: 14936–14941. doi: 10.1073/pnas.1506226112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49: D412–D419. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huebner E, Injeyan H. Patency of the follicular epithelium in Rhodnius prolixus: a reexamination of the hormone response and technique refinement. Can J Zool. 1980;58: 1617–1625. [Google Scholar]
- 52.Bjornsson CS, Huebner E. Extracellular H+ dynamics during oogenesis in Rhodnius prolixus ovarioles. J Exp Biol. 2004;207: 2835–2844. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15235012 doi: 10.1242/jeb.01089 [DOI] [PubMed] [Google Scholar]
- 53.Nunes-da-fonseca R, Berni M, Pane A, Araujo HM. Rhodnius prolixus: From classical physiology to modern developmental biology. 2017; 1–11. doi: 10.1002/dvg.22995 [DOI] [PubMed] [Google Scholar]
- 54.Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Dev Biol. 2001;234: 416–424. doi: 10.1006/dbio.2001.0246 [DOI] [PubMed] [Google Scholar]
- 55.Horner VL, Wolfner MF. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev Biol. 2008;316: 100–109. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18304524 doi: 10.1016/j.ydbio.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55: 611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 57.Gondim K, Oliveira P, Coelho H, Masuda H. Lipophorin from Rhodnius prolixus: purification and partial characterization. Insect Biochem. 1989;19: 153–161. [Google Scholar]
- 58.Garcia ES, Macarini JD, Garcia ML, Ubatuba FB. [Feeding of Rhodnius prolixus in the laboratory]. An Acad Bras Cienc. 1975;47: 537–545. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=829020 [PubMed] [Google Scholar]
- 59.Blight EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37: 911–917. doi: 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 60.Malaquias AT, Oliveira MM. Phospholipid signalling pathways in Trypanosoma cruzi growth control. Acta Trop. 1999;73: 93–108. doi: 10.1016/s0001-706x(99)00016-9 [DOI] [PubMed] [Google Scholar]
- 61.Walsh JP, Caldwell KK, Majerus PW. Formation of phosphatidylinositol 3-phosphate by isomerization from phosphatidylinositol 4-phosphate. Proc Natl Acad Sci U S A. 1991;88: 9184–9187. doi: 10.1073/pnas.88.20.9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merril CR, Dunau ML, Goldman D. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal Biochem. 1981;110: 201–207. doi: 10.1016/0003-2697(81)90136-6 [DOI] [PubMed] [Google Scholar]
- 63.Masuda H, Oliveira PL. Characterization of vitellin and vitellogenin of Rhodnius prolixus. identification of phosphorilated compounds in the molecule. Insect Bichemistry Mol Biol. 1985;15: 543–550. [Google Scholar]
- 64.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193: 265–275. [PubMed] [Google Scholar]
- 65.Oliveira PL, Kawooya JK, Ribeiro JM, Meyer T, Poorman R, Alves EW, et al. A heme-binding protein from hemolymph and oocytes of the blood-sucking insect, Rhodnius prolixus. Isolation and characterization. J Biol Chem. 1995;270: 10897–10901. doi: 10.1074/jbc.270.18.10897 [DOI] [PubMed] [Google Scholar]
- 66.Walter-Nuno AB, Oliveira MP, Oliveira MF, Gonçalves RL, Ramos IB, Koerich LB, et al. Silencing of maternal heme-binding protein causes embryonic mitochondrial dysfunction and impairs embryogenesis in the blood sucking insect rhodnius prolixus. J Biol Chem. 2013;288: 29323–29332. doi: 10.1074/jbc.M113.504985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramos IB, Miranda K, de Souza W, Oliveira DMP, Lima a PC a, Sorgine MHF, et al. Calcium-regulated fusion of yolk granules is important for yolk degradation during early embryogenesis of Rhodnius prolixus Stahl. J Exp Biol. 2007;210: 138–48. doi: 10.1242/jeb.02652 [DOI] [PubMed] [Google Scholar]
- 68.Ramos I, Gomes F, Koeller CM, Saito K, Heise N, Masuda H, et al. Acidocalcisomes as calcium- and polyphosphate-storage compartments during embryogenesis of the insect Rhodnius prolixus Stahl. PLoS One. 2011;6: e27276. doi: 10.1371/journal.pone.0027276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181: 655–666. doi: 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruck A, Attonito J, Garces KT, Núnez L, Palmisano NJ, Rubel Z, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7: 386–400. doi: 10.4161/auto.7.4.14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shravage B V., Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140: 1321–1329. doi: 10.1242/dev.089490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fischer JA, Eun SH, Doolan BT. Endocytosis, endosome trafficking, and the regulation of Drosophila development. Annu Rev Cell Dev Biol. 2006;22: 181–206. doi: 10.1146/annurev.cellbio.22.010605.093205 [DOI] [PubMed] [Google Scholar]
- 73.Kawano S, Umemiya-Shirafuji R, Boldbaatar D, Matsuoka K, Tanaka T, Fujisaki K. Cloning and characterization of the autophagy-related gene 6 from the hard tick, Haemaphysalis longicornis. Parasitol Res. 2011;109: 1341–1349. doi: 10.1007/s00436-011-2429-x [DOI] [PubMed] [Google Scholar]
- 74.Roudier N, Lefebvre C, Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. 2005;6: 695–705. doi: 10.1111/j.1600-0854.2005.00309.x [DOI] [PubMed] [Google Scholar]
- 75.Kunkel JG. Models of pattern formation in insect oocytes. In Vivo (Brooklyn). 1991;5: 443–456. [PubMed] [Google Scholar]
- 76.Bomfim L, Ramos I. Deficiency of ULK1/ATG1 in the follicle cells disturbs ER homeostasis and causes defective chorion deposition in the vector Rhodnius prolixus. FASEB J Off Publ Fed Am Soc Exp Biol. 2020;34: 13561–13572. doi: 10.1096/fj.202001396R [DOI] [PubMed] [Google Scholar]
- 77.Anna, Santos; Isabela R. ATG3 is important for the chorion ultrastructure during oogenesis in the insect vector Rhodnius prolixus. Forntiers Physiol. 2021;In press. doi: 10.3389/fphys.2021.638026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira J, Diogo C, Fonseca A, Bomfim L, Cardoso P, Santos A, et al. Silencing of RpATG8 impairs the biogenesis of maternal autophagosomes in vitellogenic oocytes, but does not interrupt follicular atresia in the insect vector Rhodnius prolixus. PLoS Negl Trop Dis. 2020;14: e0008012. doi: 10.1371/journal.pntd.0008012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karasaki S; Komoda T. Electron Micrographs of a crystalline lattice Structure in Yolk Platelets of the Amphibian Embryo. Nature. 1958;4506: 407–408. doi: 10.1038/181407a0 [DOI] [PubMed] [Google Scholar]
- 80.Whalley T, Grossmann JG, Wess TJ. An analysis of the lamellar structure of sea urchin egg cortical granules using X-ray scattering. FEBS Lett. 2000;482: 242–246. doi: 10.1016/s0014-5793(00)02068-8 [DOI] [PubMed] [Google Scholar]
- 81.Ramos IB, Miranda K, Ulrich P, Ingram P, LeFurgey A, Machado E a, et al. Calcium- and polyphosphate-containing acidocalcisomes in chicken egg yolk. Biol Cell. 2010;102: 421–34. doi: 10.1042/BC20100011 [DOI] [PubMed] [Google Scholar]
- 82.Daste F, Walrant A, Holst MR, Gadsby JR, Mason J, Lee J-E, et al. Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature. J Cell Biol. 2017;216: 3745–3765. doi: 10.1083/jcb.201704061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naslavsky N, Caplan S. The enigmatic endosome—Sorting the ins and outs of endocytic trafficking. J Cell Sci. 2018;131. doi: 10.1242/jcs.216499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKnight NC, Yue Z. Beclin 1, an Essential Component and Master Regulator of PI3K-III in Health and Disease. Curr Pathobiol Rep. 2013;1: 231–238. doi: 10.1007/s40139-013-0028-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halfon MS. Perspectives on Gene Regulatory Network Evolution. Trends Genet. 2017;33: 436–447. doi: 10.1016/j.tig.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamamoto A, Mizushima N, Tsukamoto S. Fertilization-Induced Autophagy in Mouse Embryos is Independent of mTORC11. Biol Reprod. 2014;91: 1–7. doi: 10.1095/biolreprod.113.115816 [DOI] [PubMed] [Google Scholar]
- 87.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science (80-). 2011;334: 1144–1147. doi: 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- 88.McPhee CK, Baehrecke EH. Autophagy in Drosophila melanogaster Christina. Biochim Biophys Acta. 2010;1793: 1452–1460. doi: 10.1016/j.bbamcr.2009.02.009.Autophagy [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhn H, Sopko R, Coughlin M, Perrimon N, Mitchison T. The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development. 2015;142: 3869–3878. doi: 10.1242/dev.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartoszewski R, Sikorski AF. Editorial focus: understanding off-target effects as the key to successful RNAi therapy. Cell Mol Biol Lett. 2019;24: 69. doi: 10.1186/s11658-019-0196-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu KY, Palli SR. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu Rev Entomol. 2020;65: 293–311. doi: 10.1146/annurev-ento-011019-025224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Senthil-Kumar M, Mysore KS. Caveat of RNAi in plants: the off-target effect. Methods Mol Biol. 2011;744: 13–25. doi: 10.1007/978-1-61779-123-9_2 [DOI] [PubMed] [Google Scholar]