Abstract
TRIM59 is a protein that is highly expressed in a variety of tumors and promotes tumor development. However, the use of TRIM59 as tumor diagnosis and prognosis biomarker has not been fully explored. We collected datasets from the cancer genome atlas (TCGA) and gene expression omnibus (GEO) to investigate its potential as a biomarker for diagnosis and prognosis. A total of 46 studies, including 11,558 patients were included in this study. Here, we showed that TRIM59 was significantly upregulated in 15 type of human solid tumors in comparison to their adjacent tissues. Receiver operating characteristic curve (ROC) results provided further evidence for the use of TRIM59 as a potential tumor diagnosis biomarker. Overall survival (OS) was compared between TRIM59 high expression and low expression groups. High expression of TRIM59 indicated a poor prognosis in multiple solid tumors. Taken together, these analyses showed that TRIM59 was upregulated in various types of tumors and had the potential to be used as a diagnostic and prognostic biomarker in human solid tumors.
Introduction
Cancer has always been a major global health concern and creates a heavy financial burden for patients and the health care system [1]. It is estimated that there might be 18.1 million new cancer cases and 9.6 million cancer deaths in 2018 [2]. Cancers are always characterized by a group of abnormal expressed genes, and these genes have the potential to be used as diagnosis markers or prognosis predictors [3, 4].
TRIM59 is a member of TRIM protein family, which is comprised of a RING domain, b-box domain, coiled helix domain, and c-terminal non-specific domain [5]. Recent studies have shown that TRIM59 played a role in epigenetic modification [6], embryonic development [7] and autophagy [8], and was upregulated in multiple tumors [9]. It has been found that down-regulation of TRIM59 significantly inhibited tumor growth in vitro experiments and animal models [10, 11]. However, no large-scale clinical studies have been performed to demonstrate the relationship between TRIM59 and tumor diagnosis and prognosis.
In this study, the cancer genome atlas (TCGA) and gene expression omnibus (GEO) datasets were used to analyze the expression of TRIM59 in tumors and evaluate the value of TRIM59 as a biomarker for tumor diagnosis and prognosis.
Materials and methods
Data collection and extraction
Datasets containing tumor samples and corresponding normal samples were downloaded from TCGA. Microarrays containing overall survival (OS) information were searched in the GEO database. Inclusion criteria were as follows: (1) should be human tumor samples and each dataset should contain at least 30 independent samples; (2) for the convenience of data processing, only microarray data performed on GPL570 platform was retained; (3) characteristics of studies and survival information were reported or could be determined. Exclusion criteria include: repetitive studies containing same datasets or patient cohorts. A total of 26 GEO datasets were included in the study, including 6 breast cancer datasets (GSE20685, GSE20711, GSE16446, GSE42568, GSE48390, GSE58812), 8 lung cancer datasets (GSE102287, GSE29013, GSE30219, GSE31210, GSE3141, GSE19188, GSE37745, GSE50081), 4 ovarian cancer datasets (GSE26193, GSE32062, GSE63885, GSE18520), 3 gastric cancer datasets (GSE15459, GSE57303, GSE62254), 1 pancreatic cancer dataset (GSE17891), 1 Ewing sarcoma dataset (GSE17679), 1 adrenocortical carcinoma dataset (GSE19750), 2 brain cancer dataset (GSE108474, GSE7696).
Data processing
Level 3 RNAseq and associated clinical information of each TCGA solid tumor project (LUAD-lung adenocarcinoma, BRCA-breast carcinoma, UCEC-uterine corpus endometrial carcinoma, LUSC-lung squamous cell carcinoma, HNSC-head and neck squamous cell carcinoma, KIRC-kidney renal clear cell carcinoma, PRAD-prostate adenocarcinoma, BLCA-bladder carcinoma, THCA-thyroid carcinoma, KIRP-kidney renal papillary cell carcinoma, LIHC-liver hepatocellular carcinoma, STAD-stomach adenocarcinoma, COAD-colon adenocarcinoma, READ-rectum adenocarcinoma, CHOL-cholangiocarcinoma, CESC-cervical squamous cell carcinoma and endocervical adenocarcinoma, ESCA-esophageal carcinoma, GBM-glioblastoma multiforme, LGG-lower grade glioma, OV-ovarian serous cystadenocarcinoma, PAAD-pancreatic adenocarcinoma, SARC-sarcoma, SKCM-skin cutaneous melanoma) was downloaded from UCSC Xena (https://xenabrowser.net/). Downloaded FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) gene expression data was log2 transformed for the convenience of comparison. Samples’ expression and clinical information was matched, samples without complete survival or expression information were excluded. TRIM59 expression in each project was presented with boxplot. As for GEO datasets, RMA (Robust Multichip Average) normalized gene expression data of GPL570 platform was downloaded and matched with associated clinical information.
ROC, AUC and SROC analyses
R package “OptimalCutpoints” was first used to determine the optimal cutpoint in each TCGA project, at which the sample types (tumor or normal) could be best distinguished. Then the sample types were predicted by the TRIM59 expression level according to the cut point of each project using the “Youden” prediction model [12]. ROC and associated 95% confidence intervals (CIs) of each project was also calculated using the “optimal.cutpoints” function of package “OptimalCutpoints”. Module “midas” of Stata 15.0 (StataCorp LLC, USA) was used for the meta-analysis of ROC curves of all the projects. Sensitivity and specificity of the meta-analysis were evaluated, and the summary of ROCs (SROC) was calculated. Deeks’ funnel plot asymmetry test was used to investigate potential publication bias in SROC analysis.
Overall survival analysis
Datasets regarding TRIM59 expression and clinical characteristics of human cancers were downloaded from TCGA and GEO databases. According to the TRIM59 expression level in each data set, the samples were divided into high expression and low expression groups in comparison to the median expression level. The "survival" package was used to calculate the hazard ratio (HR) and 95% CIs for the high-expression TRIM59 group versus the low-expression TRIM59 group.
Statistical analysis
All the analyses were performed on R (Version 3.6.1) and Stata 15.0 (StataCorp LLC, USA). Normalized expression data were downloaded from TCGA or GEO datasets. Unpaired t test was used for comparison between groups. As for ROC analysis, R package “OptimalCutpoints” was first used to calculate the optimal cutpoint for tumor diagnosis, then ROC and 95% CIs were calculated for each project. Stata module “medias” was used for ROC meta-analysis. R package “survival” was used for survival analysis and package “meta” was used for survival meta-analysis.
Results
TRIM59 was highly expressed in human tumors
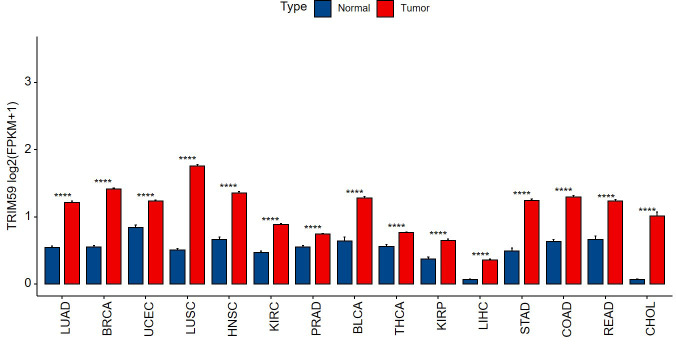
Previous studies have shown that TRIM59 was highly expressed in a variety of tumors and closely related to the occurrence and development of tumors. We analyzed 15 types of tumor datasets that included sufficient numbers of tumors and adjacent normal tissues in TCGA, confirming that TRIM59 was highly expressed in tumor samples in comparison to their adjacent tissues (Fig 1).
Fig 1. Relative expression of TRIM59 in human tumors based on TCGA database, comparisons were conducted using unpaired t test.
****p < 0.0001. FPKM (Fragments Per Kilobase of exon model per Million mapped fragments).
High expression of TRIM59 showed high efficacy in the diagnosis of human tumors
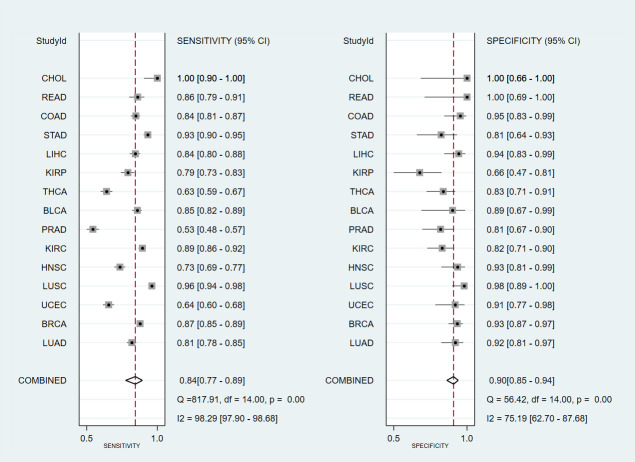
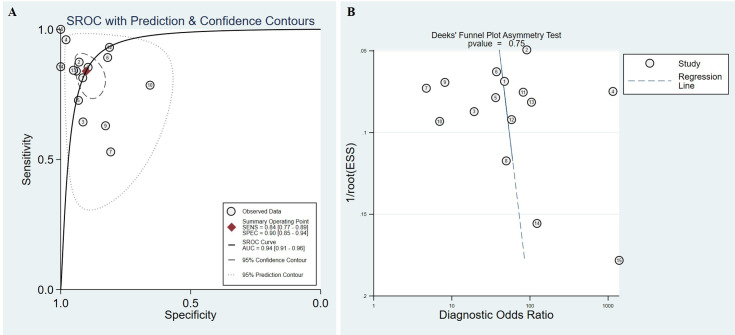
To explore the value of high expression of TRIM59 in tumors, we tested if TRIM59 could be used to identify healthy and tumor samples. ROC analysis was performed based on the data obtained from TCGA. Results presented in Table 1 demonstrated that TRIM59 showed high diagnosis efficacy in multiple cancers, especially in CHOL, with a prediction accuracy up to 100%. The pooled sensitivity and specificity were 0.84 (95% CI:0.77–0.89) and 0.90 (95% CI:0.85–0.94) respectively (Fig 2). Additionally, the area under ROC (AUC) of the SROC was 0.94 (95% CI:0.91–0.96) (Fig 3A). In order to test the potential publication bias, Deeks’ funnel plot asymmetry test was performed and revealed that no significant substantial publication bias was found in SROC analysis (p = 0.75) (Fig 3B). These results suggested that the high expression of TRIM59 could be used as a diagnostic marker in different types of human cancers.
Table 1. Characteristics of studies and AUC analyses.
| ID | Projects | AUC (95% CI) | Normal Samples | Tumor Samples | Cut-off value | FP | FN | TP | TN |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TCGA-LUAD | 0.928 (0.903, 0.952) | 59 | 526 | 0.768746 | 5 | 98 | 428 | 54 |
| 2 | TCGA-BRCA | 0.955 (0.94, 0.969) | 113 | 1104 | 0.882547 | 8 | 139 | 965 | 105 |
| 3 | TCGA-UCEC | 0.787 (0.737, 0.838) | 35 | 548 | 1.070402 | 3 | 195 | 353 | 32 |
| 4 | TCGA-LUSC | 0.985 (0.972, 0.998) | 49 | 501 | 0.846782 | 1 | 20 | 481 | 48 |
| 5 | TCGA-HNSC | 0.876 (0.837, 0.915) | 44 | 502 | 0.971722 | 3 | 137 | 365 | 41 |
| 6 | TCGA-KIRC | 0.912 (0.878, 0.946) | 72 | 535 | 0.573303 | 13 | 58 | 477 | 59 |
| 7 | TCGA-PRAD | 0.712 (0.644, 0.779) | 52 | 499 | 0.689123 | 10 | 235 | 264 | 42 |
| 8 | TCGA-BLCA | 0.905 (0.845, 0.965) | 19 | 411 | 0.828988 | 2 | 60 | 351 | 17 |
| 9 | TCGA-THCA | 0.742 (0.676, 0.809) | 58 | 510 | 0.690223 | 10 | 189 | 321 | 48 |
| 10 | TCGA-KIRP | 0.783 (0.706, 0.859) | 32 | 289 | 0.390211 | 11 | 62 | 227 | 21 |
| 11 | TCGA-LIHC | 0.921 (0.89, 0.952) | 50 | 374 | 0.104429 | 3 | 60 | 314 | 47 |
| 12 | TCGA-STAD | 0.951 (0.918, 0.984) | 32 | 375 | 0.691021 | 6 | 26 | 349 | 26 |
| 13 | TCGA-COAD | 0.941 (0.919, 0.963) | 41 | 471 | 0.943438 | 2 | 74 | 397 | 39 |
| 14 | TCGA-READ | 0.93 (0.889, 0.971) | 10 | 167 | 0.91301 | 0 | 24 | 143 | 10 |
| 15 | TCGA-CHOL | 1 (1, 1) | 9 | 36 | 0.287042 | 0 | 0 | 36 | 9 |
LUAD, lung adenocarcinoma; BRCA, breast carcinoma; UCEC, uterine corpus endometrial carcinoma; LUSC, lung squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; PRAD, prostate adenocarcinoma; BLCA, bladder carcinoma; THCA, thyroid carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; STAD, stomach adenocarcinoma; COAD, colon adenocarcinoma; READ, rectum adenocarcinoma; CHOL, cholangiocarcinoma.
Fig 2. Forest plot showing mean sensitivity and specificity with corresponding heterogeneity statistics for the prediction of sample types with the expression level of TRIM59.
Fig 3. Summary ROC analysis.
(A) Summary ROC for the evaluation of prediction efficacy of TRIM59, with confidence and prediction regions around mean operating sensitivity and specific point. The ID in the figure corresponds to the projects in Table 1. (B) Deeks’ funnel plot asymmetry test showing the potential publication bias. The ID in the figure corresponds to the projects in Table 1.
Expression of TRIM59 indicated prognosis in human tumors
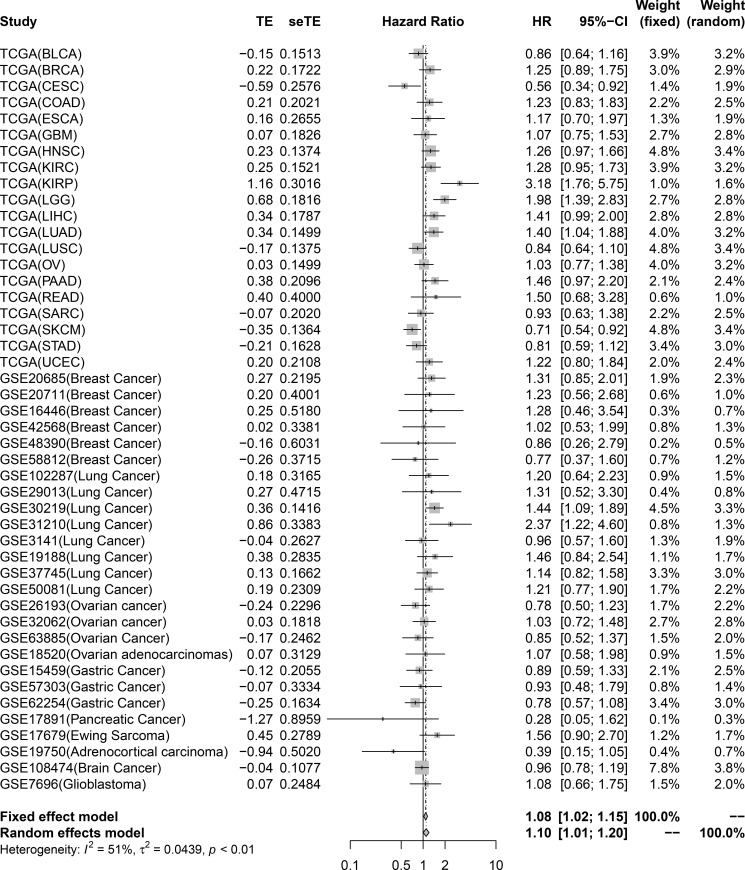
To investigate the relationship between TRIM59 and tumor prognosis, OS meta-analysis was conducted. A total of 21 TCGA and 26 GEO datasets containing 11,558 patients were included (Table 2). High expression of TRIM59 indicated poor prognosis in KIRP (HR 3.18; 95%CI 1.76–5.75), LGG (HR 1.98; 95%CI 1.39–2.83), LUAD (HR 1.40; 95%CI 1.04–1.88), lung cancer (GSE30219: HR 1.44; 95%CI 1.09–1.89; GSE31210: HR 2.37; 95%CI 1.22–4.60), and indicated good prognosis in CESC (HR 0.56; 95%CI 0.34–0.92) and SKCM (HR 0.71; 95%CI 0.54–0.92) (Fig 4).
Table 2. Characteristics of studies in OS analysis.
| Study | Country | Year | Patients | HR | Lower | Upper |
|---|---|---|---|---|---|---|
| TCGA (BLCA) | USA | 2014 | 402 | 0.86 | 0.64 | 1.16 |
| TCGA (BRCA) | USA | 2014 | 1006 | 1.25 | 0.89 | 1.75 |
| TCGA (CESC) | USA | 2014 | 264 | 0.56 | 0.34 | 0.92 |
| TCGA (COAD) | USA | 2014 | 440 | 1.23 | 0.83 | 1.83 |
| TCGA (ESCA) | USA | 2014 | 144 | 1.17 | 0.70 | 1.97 |
| TCGA (GBM) | USA | 2014 | 152 | 1.07 | 0.75 | 1.53 |
| TCGA (HNSC) | USA | 2014 | 496 | 1.26 | 0.97 | 1.66 |
| TCGA (KIRC) | USA | 2014 | 522 | 1.28 | 0.95 | 1.73 |
| TCGA (KIRP) | USA | 2014 | 284 | 3.18 | 1.76 | 5.75 |
| TCGA (LGG) | USA | 2014 | 510 | 1.98 | 1.39 | 2.83 |
| TCGA (LIHC) | USA | 2014 | 360 | 1.41 | 0.99 | 2.00 |
| TCGA (LUAD) | USA | 2014 | 492 | 1.40 | 1.04 | 1.88 |
| TCGA (LUSC) | USA | 2014 | 488 | 0.84 | 0.64 | 1.10 |
| TCGA (OV) | USA | 2014 | 294 | 1.03 | 0.77 | 1.38 |
| TCGA (PAAD) | USA | 2014 | 174 | 1.46 | 0.97 | 2.20 |
| TCGA (READ) | USA | 2014 | 158 | 1.50 | 0.68 | 3.28 |
| TCGA (SARC) | USA | 2014 | 258 | 0.93 | 0.63 | 1.38 |
| TCGA (SKCM) | USA | 2014 | 458 | 0.71 | 0.54 | 0.92 |
| TCGA (STAD) | USA | 2014 | 378 | 0.81 | 0.59 | 1.12 |
| TCGA (UCEC) | USA | 2014 | 540 | 1.22 | 0.80 | 1.84 |
| GSE20685 (Breast Cancer) | China | 2011 | 327 | 1.31 | 0.85 | 2.01 |
| GSE20711 (Breast Cancer) | Canada | 2011 | 88 | 1.23 | 0.56 | 2.68 |
| GSE16446 (Breast Cancer) | Canada | 2010 | 107 | 1.28 | 0.46 | 3.54 |
| GSE42568 (Breast Cancer) | Ireland | 2013 | 104 | 1.02 | 0.53 | 1.99 |
| GSE48390 (Breast Cancer) | China | 2014 | 81 | 0.86 | 0.26 | 2.79 |
| GSE58812 (Breast Cancer) | France | 2015 | 107 | 0.77 | 0.37 | 1.60 |
| GSE102287 (Lung Cancer) | USA | 2017 | 66 | 1.20 | 0.64 | 2.23 |
| GSE29013 (Lung Cancer) | USA | 2011 | 55 | 1.31 | 0.52 | 3.30 |
| GSE30219 (Lung Cancer) | France | 2013 | 293 | 1.44 | 1.09 | 1.89 |
| GSE31210 (Lung Cancer) | Japan | 2011 | 226 | 2.37 | 1.22 | 4.60 |
| GSE3141 (Lung Cancer) | USA | 2005 | 111 | 0.96 | 0.57 | 1.60 |
| GSE19188 (Lung Cancer) | Netherlands | 2010 | 82 | 1.46 | 0.84 | 2.54 |
| GSE37745 (Lung Cancer) | Sweden | 2012 | 196 | 1.14 | 0.82 | 1.58 |
| GSE50081 (Lung Cancer) | Canada | 2013 | 181 | 1.21 | 0.77 | 1.90 |
| GSE26193 (Ovarian cancer) | France | 2011 | 107 | 0.78 | 0.50 | 1.23 |
| GSE32062 (Ovarian cancer) | Japan | 2012 | 260 | 1.03 | 0.72 | 1.48 |
| GSE63885 (Ovarian Cancer) | Poland | 2014 | 75 | 0.85 | 0.52 | 1.37 |
| GSE18520 (Ovarian adenocarcinomas) | USA | 2009 | 53 | 1.07 | 0.58 | 1.98 |
| GSE15459 (Gastric Cancer) | Switzerland | 2009 | 192 | 0.89 | 0.59 | 1.33 |
| GSE57303 (Gastric Cancer) | China | 2014 | 34 | 0.93 | 0.48 | 1.79 |
| GSE62254 (Gastric Cancer) | USA | 2015 | 295 | 0.78 | 0.57 | 1.08 |
| GSE17891 (Pancreatic Cancer) | United Kingdom | 2011 | 21 | 0.28 | 0.05 | 1.62 |
| GSE17679 (Ewing Sarcoma) | Finland | 2011 | 88 | 1.56 | 0.90 | 2.70 |
| GSE19750 (Adrenocortical carcinoma) | USA | 2013 | 22 | 0.39 | 0.15 | 1.05 |
| GSE108474 (Brain Cancer) | USA | 2018 | 487 | 0.96 | 0.78 | 1.19 |
| GSE7696 (Glioblastoma) | Switzerland | 2008 | 80 | 1.08 | 0.66 | 1.75 |
BLCA, bladder carcinoma; BRCA, breast carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma.
Fig 4. Forest plot showing the overall survival significance of TRIM59.
Patients were divided into high expression and low expression groups based on the median expression levels of TRIM59, then overall survival was compared between the high expression and low expression group.
Discussion
In this study, we used TCGA datasets to demonstrate that TRIM59 was upregulated in multiple cancers in comparison to the adjacent normal tissues. To explore the diagnosis efficacy of TRIM59, we performed ROC and SROC analyses to identify that high expression of TRIM59 showed a high diagnosis efficacy in each tumor type (AUC > 0.5), and with an AUC of summary ROC of 0.94 (0.91–0.96). We also combined GEO datasets and performed a meta-analysis to reveal that high expression of TRIM59 showed significant (p<0.05) poor prognosis in KIRP, LGG, LUAD, lung cancer and showed better prognosis in CESC and SKCM. Especially in LUAD, the expression of TRIM59 in tumors with better prognosis was still higher than that in adjacent tissues (S1 Fig).
TRIM59 is closely related to cancers. A previous study used Immunohistochemistry (IHC) to determine the expression of TRIM59 in 291 cases of 37 tumor types, and found that TRIM59 expression was upregulated in tumor samples, particularly in lung, breast, liver, skin, tongue and mouth (squamous cell cancer) and endometrial cancers [9]. In subsequent studies, it was shown that upregulation of TRIM59 can promote tumor growth in tumor cell lines and animal models, while downregulation had the opposite effect. These cancers include pancreatic cancer [13], cholangiocarcinoma [14], ovarian cancer [15, 16], lung cancer [17–19], breast cancer [10, 20, 21], euroblastoma [22], medulloblastoma [23], hepatocellular carcinoma [24], glioblastoma [11], colorectal cancer [25, 26], bladder cancer [21], prostate cancer [27], cervical cancer [28], osteosarcoma [29], gastric cancer [30]. However, results from these studies were limited by a small number of tumor samples, or the use of in vitro experimentation or animal models could not fully address the relationship between TRIM59 and human cancers. Thanks to the gene expression data and associated prognosis information provided in public database, we identified that TRIM59 was highly expressed in most solid tumors and could indicate the prognosis in several cancers.
This study is a meta-analysis of multiple solid tumors, indicated that TRIM59 has the potential to be used as a diagnostic molecule for a variety of tumors, and special attention should be paid to the abnormal high expression of TRIM59 in specific tissues. Moreover, it plays a prognostic role in specific tumors, especially in KIRP/LGG/LUAD/Lung cancer/CESC/SKCM. Detection of TRIM59 expression in these tumor tissues is helpful for us to evaluate the prognosis of patients.
The limitation of this study lies in the fact that it is only the data analysis of biological database, and further verification of TRIM59 expression level and follow-up information are needed in clinical samples for each specific tumor type. Basic research on TRIM59 is also needed to improve our understanding of tumors.
Conclusions
In this study, based on TCGA datasets, we revealed that TRIM59 was upregulated in 15 types of solid tumors. Additionally, TRIM59 have a high efficacy in diagnosis and prognosis prediction in various tumor types. TRIM59 have the potential to be used as diagnosis marker or prognosis predictor in tumors.
Supporting information
(TIF)
Data Availability
The data that support the findings of this study were derived from the following resources available in the public domain: https://www.ncbi.nlm.nih.gov/geo (Accession numbers: GSE20685, GSE20711, GSE16446, GSE42568, GSE48390, GSE58812, GSE102287, GSE29013, GSE30219, GSE31210, GSE3141, GSE19188, GSE37745, GSE50081, GSE26193, GSE32062, GSE63885, GSE18520, GSE15459, GSE57303, GSE62254, GSE17891, GSE17679, GSE19750, GSE108474, GSE7696); https://xenabrowser.net (Projects: LUAD, BRCA, UCEC, LUSC, HNSC, KIRC, PRAD, BLCA, THCA, KIRP, LIHC, STAD, COAD, READ, CHOL, CESC, ESCA, GBM, LGG, OV, PAAD, SARC, SKCM)
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 81871245) and Department of Education of Jilin Province (JJKH20190095KJ).
References
- 1.Gelband H, Sankaranarayanan R, Gauvreau CL, Horton S, Anderson BO, Bray F, et al. Costs, affordability, and feasibility of an essential package of cancer control interventions in low-income and middle-income countries: key messages from Disease Control Priorities, 3rd edition. Lancet. 2016;387(10033):2133–44. Epub 2015/11/19. doi: 10.1016/S0140-6736(15)00755-2 . [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. Epub 2018/09/13. doi: 10.3322/caac.21492 . [DOI] [PubMed] [Google Scholar]
- 3.Chistiakov DA, Myasoedova VA, Grechko AV, Melnichenko AA, Orekhov AN. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin Cancer Biol. 2018;52(Pt 1):9–16. Epub 2018/01/24. doi: 10.1016/j.semcancer.2018.01.012 . [DOI] [PubMed] [Google Scholar]
- 4.Ding W, Chen G, Shi T. Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis. Epigenetics. 2019;14(1):67–80. Epub 2019/01/31. doi: 10.1080/15592294.2019.1568178 ; PubMed Central PMCID: PMC6380428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64(1):64–71. Epub 2011/12/02. doi: 10.1002/iub.580 . [DOI] [PubMed] [Google Scholar]
- 6.Spolnicka M, Pospiech E, Peplonska B, Zbiec-Piekarska R, Makowska Z, Pieta A, et al. DNA methylation in ELOVL2 and C1orf132 correctly predicted chronological age of doi: 10.1007/s00414-017-1636-0from three disease groups. International journal of legal medicine. 2018;132(1):1–11. PubMed Central PMCID: PMC5748441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su X, Wu C, Ye X, Zeng M, Zhang Z, Che Y, et al. Embryonic lethality in mice lacking Trim59 due to impaired gastrulation development. Cell death & disease. 2018;9(3):302. doi: 10.1038/s41419-018-0370-y; PubMed Central PMCID: PMC5833458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han T, Guo M, Gan M, Yu B, Tian X, Wang JB. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy. 2018;14(12):2035–48. Epub 2018/09/21. doi: 10.1080/15548627.2018.1491493 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khatamianfar V, Valiyeva F, Rennie PS, Lu WY, Yang BB, Bauman GS, et al. TRIM59, a novel multiple cancer biomarker for immunohistochemical detection of tumorigenesis. BMJ Open. 2012;2(5). Epub 2012/10/11. doi: 10.1136/bmjopen-2012-001410; PubMed Central PMCID: PMC3488719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan P, Ye Y, He L, Xie J, Jing J, Ma G, et al. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018;16(11):e3000051. Epub 2018/11/09. doi: 10.1371/journal.pbio.3000051; PubMed Central PMCID: PMC6245796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang Y, Li Y, Song L, Alvarez AA, Zhang W, Lv D, et al. TRIM59 Promotes Gliomagenesis by Inhibiting TC45 Dephosphorylation of STAT3. Cancer research. 2018;78(7):1792–804. doi: 10.1158/0008-5472.CAN-17-2774 ; PubMed Central PMCID: PMC5882560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youden WJ. Index for rating diagnostic tests—Youden—2006—Cancer—Wiley Online Library. Cancer. 1950;3(1):32–5. doi: [DOI] [PubMed] [Google Scholar]
- 13.Li R, Weng L, Liu B, Zhu L, Zhang X, Tian G, et al. TRIM59 predicts poor prognosis and promotes pancreatic cancer progression via the PI3K/AKT/mTOR-glycolysis signaling axis. J Cell Biochem. 2019. Epub 2019/11/07. doi: 10.1002/jcb.29433. [DOI] [PubMed] [Google Scholar]
- 14.Shen H, Zhang J, Zhang Y, Feng Q, Wang H, Li G, et al. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene. 2019;698:50–60. Epub 2019/03/02. doi: 10.1016/j.gene.2019.02.044 . [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Zhou Z, Wang X, Zhang X, Chen Y, Bai J, et al. TRIM59 Is a Novel Marker of Poor Prognosis and Promotes Malignant Progression of Ovarian Cancer by Inducing Annexin A2 Expression. Int J Biol Sci. 2018;14(14):2073–82. Epub 2018/12/27. doi: 10.7150/ijbs.28757 ; PubMed Central PMCID: PMC6299375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Zhang H, Wang Y, Zhang P, Qi Y. Tripartite Motif-Containing Protein 59 (TRIM59) Promotes Epithelial Ovarian Cancer Progression via the Focal Adhesion Kinase(FAK)/AKT/Matrix Metalloproteinase (MMP) Pathway. Med Sci Monit. 2019;25:3366–73. Epub 2019/05/08. doi: 10.12659/MSM.916299 ; PubMed Central PMCID: PMC6519306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng B, Liang M, Qin L, Zhao W, Wang H, Wang L, et al. An TRIM59-CDK6 axis regulates growth and metastasis of lung cancer. J Cell Mol Med. 2019;23(2):1458–69. Epub 2018/12/06. doi: 10.1111/jcmm.14052 ; PubMed Central PMCID: PMC6349187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao L, Du B, Xi X. TRIM59 is a novel potential prognostic biomarker in patients with non-small cell lung cancer: A research based on bioinformatics analysis. Oncol Lett. 2017;14(2):2153–64. Epub 2017/08/10. doi: 10.3892/ol.2017.6467 ; PubMed Central PMCID: PMC5530082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhan W, Han T, Zhang C, Xie C, Gan M, Deng K, et al. TRIM59 Promotes the Proliferation and Migration of Non-Small Cell Lung Cancer Cells by Upregulating Cell Cycle Related Proteins. PLoS One. 2015;10(11):e0142596. Epub 2015/11/26. doi: 10.1371/journal.pone.0142596; PubMed Central PMCID: PMC4658198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Dong Y, Zhao L, Su L, Diao K, Mi X. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol Carcinog. 2018;57(12):1792–802. Epub 2018/09/04. doi: 10.1002/mc.22897 . [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Yang WB. Down-regulation of tripartite motif protein 59 inhibits proliferation, migration and invasion in breast cancer cells. Biomed Pharmacother. 2017;89:462–7. Epub 2017/03/02. doi: 10.1016/j.biopha.2017.02.039 . [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Chen W, Ye M, Tan W, Jia B. TRIM59 knockdown inhibits cell proliferation by down-regulating the Wnt/beta-catenin signaling pathway in neuroblastoma. Biosci Rep. 2019;39(1). Epub 2018/11/06. doi: 10.1042/BSR20181277; PubMed Central PMCID: PMC6340953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao R, Lv G, Zhang C, Wang X, Chen L. TRIM59 induces epithelial-to-mesenchymal transition and promotes migration and invasion by PI3K/AKT signaling pathway in medulloblastoma. Oncol Lett. 2018;15(6):8253–60. Epub 2018/05/29. doi: 10.3892/ol.2018.8432 ; PubMed Central PMCID: PMC5950029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun G, Sui X, Han D, Gao J, Liu Y, Zhou L. TRIM59 promotes cell proliferation, migration and invasion in human hepatocellular carcinoma cells. Pharmazie. 2017;72(11):674–9. Epub 2018/02/15. doi: 10.1691/ph.2017.7659 . [DOI] [PubMed] [Google Scholar]
- 25.Wu W, Chen J, Wu J, Lin J, Yang S, Yu H. Knockdown of tripartite motif-59 inhibits the malignant processes in human colorectal cancer cells. Oncol Rep. 2017;38(4):2480–8. Epub 2017/08/30. doi: 10.3892/or.2017.5896 . [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Ji B, Feng Y, Zhang Y, Ji D, Zhu C, et al. TRIM59 facilitates the proliferation of colorectal cancer and promotes metastasis via the PI3K/AKT pathway. Oncol Rep. 2017;38(1):43–52. Epub 2017/05/24. doi: 10.3892/or.2017.5654 ; PubMed Central PMCID: PMC5492839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin WY, Wang H, Song X, Zhang SX, Zhou PS, Sun JM, et al. Knockdown of tripartite motif 59 (TRIM59) inhibits tumor growth in prostate cancer. Eur Rev Med Pharmacol Sci. 2016;20(23):4864–73. Epub 2016/12/17. . [PubMed] [Google Scholar]
- 28.Aierken G, Seyiti A, Alifu M, Kuerban G. Knockdown of Tripartite-59 (TRIM59) Inhibits Cellular Proliferation and Migration in Human Cervical Cancer Cells. Oncol Res. 2017;25(3):381–8. Epub 2016/09/24. doi: 10.3727/096504016X14741511303522 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang J, Xing D, Li Z, Shen J, Zhao H, Li S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol Med Rep. 2016;13(6):5200–6. Epub 2016/04/29. doi: 10.3892/mmr.2016.5183 . [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH, et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014;147(5):1043–54. Epub 2014/07/22. doi: 10.1053/j.gastro.2014.07.021 . [DOI] [PubMed] [Google Scholar]