In neurons, local protein synthesis is widespread, contributing to both the maintenance and plasticity of synapses.
Abstract
To supply proteins to their vast volume, neurons localize mRNAs and ribosomes in dendrites and axons. While local protein synthesis is required for synaptic plasticity, the abundance and distribution of ribosomes and nascent proteins near synapses remain elusive. Here, we quantified the occurrence of local translation and visualized the range of synapses supplied by nascent proteins during basal and plastic conditions. We detected dendritic ribosomes and nascent proteins at single-molecule resolution using DNA-PAINT and metabolic labeling. Both ribosomes and nascent proteins positively correlated with synapse density. Ribosomes were detected at ~85% of synapses with ~2 translational sites per synapse; ~50% of the nascent protein was detected near synapses. The amount of locally synthesized protein detected at a synapse correlated with its spontaneous Ca2+ activity. A multifold increase in synaptic nascent protein was evident following both local and global plasticity at respective scales, albeit with substantial heterogeneity between neighboring synapses.
INTRODUCTION
Biological compartments can function and adapt with autonomy by localizing cell biological organelles and machinery. Perhaps the best example of this is the neuronal synapse, where local control of protein production and distribution can allow, in principle, for the compartmentalization of synaptic function and plasticity (1–4). However, the prevalence, utility, and specificity of locally synthesized proteins are still not well understood. For example, is local protein synthesis constitutive during basal synaptic activity? And how are nascent proteins distributed among synapses that vary both in density and in activity level (5, 6)? In addition, the relationship between the machines (ribosomes), locally synthesized proteins, and synaptic features are not well understood. All of these questions are particularly important during plasticity, where local or global activity manipulations can result in a local or global change in synaptic strengths (3, 7). During plasticity, the underlying proteome remodeling concerns not only a few receptor complexes but also thousands of protein species (8). If plasticity is induced globally but proteins are supplied locally, what are the rules to allot nascent proteins to the synapse population? When plasticity is induced locally, how local is the change in protein distribution? Does it occur at the level of individual synapses, short segments of dendritic branches containing synaptic clusters, whole dendritic branches, etc.?
To address these questions, we first visualized and quantified the dendritic protein synthesis sites by labeling and measuring both assembled ribosomes and nascent proteins in mature, cultured rat hippocampal neurons using quantitative, multiplexed single-molecule localization imaging (DNA-PAINT) (9, 10). We monitored a cohort of newly synthesized proteins and correlated their spatial distribution with that of assembled ribosomes over time, following locally and globally induced plasticity. We observed widespread protein synthesis near synapses under both basal and stimulated conditions. During normal synaptic transmission, the amount of locally synthesized proteins detected at a synapse was correlated with its level of ongoing spontaneous activity. Plasticity induced by single-spine stimulations or by a global activity manipulation both resulted in a significant increase in local protein synthesis at respective spatial scales. However, the elevated nascent protein is not specific to stimulated spines but rather spread to neighboring synapses.
RESULTS
The ribosomal large and small subunits assemble during mRNA translation (11). To localize the sites of protein synthesis in synapses and dendrites of cultured hippocampal neurons, we colocalized ribosomal large and small subunits using quantitative, multiplexed, single-molecule localization microscopy (DNA-PAINT; Fig. 1A; see also fig. S1A and Materials and Methods) (12–14). To tag each subunit for single-molecule localization, we immunolabeled the endogenous ribosomal proteins RPS11 (epitope for 40S small subunit) and RPL36a (epitope for 60S large subunit) using primary antibodies and secondary antibodies, each labeled with a distinct single-strand DNA “docking” oligo for sequential multiplexed imaging [see fig. S1 (B to D) and Materials and Methods]. Tagged ribosomal subunits were detected by repeated, brief fluorescence detection events representing the transient hybridization between the docking and imager (complementary and fluorescent) oligos to form localization clusters (Fig. 1A), as previously described (12). Coincident detection of localization clusters from both subunits further identified assembled ribosomes (Fig. 1A, bottom right; see Materials and Methods). Using sucrose gradients that enrich ribosomes of different compositions (fig. S2A; see also Materials and Methods) (15), our method reliably distinguished between the enriched small-subunit fraction [isolated 40S clusters; Fig. 1B, left (green)], the enriched monosome [colocalizations that appeared isolated and uniform in size; Fig. 1B, middle; see also fig. S2 (B and C)], and the enriched polyribosome fraction (colocalizations that appeared in clusters, with an average magenta/green localization ratio of 0.95; Fig. 1B, right; see Materials and Methods). A significant difference in cluster size was also detected between the 40S fraction, 80S fraction, and polyribosome fraction (Fig. 1C and fig. S2, D and E). To validate the in situ sensitivity of ribosome detection, we induced the disassembly of polysomes in neurons with two different protein synthesis inhibitors (Fig. 1D and fig. S3A) (15, 16) and observed the predicted loss of the larger ribosome clusters (Fig. 1D). Furthermore, the degree of large and small subunit colocalization that we detected was significantly different from the colocalization obtained by chance (fig. S3, B to D). Together, these data indicate that our method can be used to reliably visualize translation sites in dendrites.
Fig. 1. Quantitative, multiplexed, single-molecule localization of assembled ribosomes in neuronal dendrites and synapses.
(A) A eukaryotic ribosome with RPL36a (magenta) and RPS11 (green; Protein Data Bank: 4V88). Bottom right: The coincidence detection of RPL36a and RPS11 by DNA-PAINT. (B) Representative images showing localizations of both subunits detected in a plated small-subunit fraction (left) and colocalizations detected in 80S (middle) and polyribosome fractions (right). Scale bar, 500 nm (C) Violin plots showing the size distributions of 40S localization clusters from the 40S fraction (0.014 ± 0.010 μm2), colocalized clusters from the 80S (0.026 ± 0.015 μm2), and the polyribosome fraction (0.062 ± 0.057 μm2). Analysis of variance (ANOVA) with post hoc Bonferroni test indicated significant differences (in μm2) among the three (***P< 0.001). (D) Ribosome size distribution (number of localizations per cluster) following puromycin (dark gray) or homoharringtonine (light gray) treatment. (E) Representative image of ribosome occupancy in dendrites (4300 μm2) with anti-Bassoon (synapse, blue) and ribosomal small (green) and large subunits (magenta). Scale bar, 2 μm. Dashed box indicates a branch at higher magnification (right inset; transecting dashed lines indicate segment boundaries). Scale bar, 1 μm. (F) A cumulative plot showing synapse fractions (y axis) in (E) that had ribosomes within a certain radius (x axis). (G) Representative dendritic segments (and branch point) illustrated as an overlay of synapse density (heatmap) and ribosome localizations (contour map). Scale bar, 2 μm. (H) Scatter plot showing the Pearson’s correlation between ribosome localization density and synapse density among 31 dendritic segments in (E) (r = 0.67; one-tailed test, ***P < 0.001).
We quantified the extent to which the detected ribosomes populated the dendrites and localized near synapses by coimmunolabeling with a synaptic marker (anti-Bassoon; Fig. 1E). The dendritic arbor shown in Fig. 1E was populated with synapses at an average density of ~1 per micrometer of dendrite (Fig. 1E; see also fig. S3E), as previously reported (17). Ribosomes populated the entire dendritic arbor; we detected ~2 ribosome clusters per micrometer of dendrite, corresponding to ~8 translating ribosomes/μm (the average ribosome cluster containing ~4 ribosomes; see Materials and Methods). This density is greater than that previously reported for polyribosomes using electron microscopy (~1 per μm with the requirement of ≥3 ribosomes detected in a cluster) (5). The detection and inclusion of monosomes (15) here [excluded in electron microscopy (EM) studies] likely contributed to the higher ribosome occupancy observed. We found that ~85% of synapses had one or more ribosome clusters associated with it (within a 1-μm radius from the local maxima of a Bassoon puncta; Fig. 1F; see Materials and Methods). Using heat and contour maps to depict the synapse and ribosome density distributions, respectively, we found that synapses and ribosomes tended to covary in their abundance (Fig. 1G), suggesting that local hotspots of translation form around synapse clusters (Fig. 1E, inset). We found a positive correlation (r = 0.67; Fig. 1H) between ribosome density and synapse density within individual dendritic segments that constitute a dendritic branch [see Fig. 3E (inset) for examples; see fig. S3F and Materials and Methods for dendrite segmentation]. Hence, local sites of protein synthesis congregate near synapses along dendrites, giving rise to ~2 translational sites per synapse.
Fig. 3. During basal activity, levels of nascent synaptic protein correlated with synaptic Ca2+ activity.
(A) Wide-field micrograph of a neuronal dendrite after 24 hours of upscaling immunolabeled with dendritic and synaptic reference markers (MAP2 in green and PSD95 in magenta). Scale bar, 5 μm. (B) Example of a synaptic mask generated by a custom-written local thresholding algorithm (see Materials and Methods) for the dendritic arbor shown in (A). (C) Synaptic nascent protein localizations (magenta) and dendritic and synaptic reference fluorescence (gray). Scale bar, 4 μm. (D) Cultured rat hippocampal neurons were transfected with GCaMP6s and PSD95-mCherry (see Materials and Methods) before basal Ca2+ activity imaging (followed by 15-min AHA labeling; top row). Subsequent fixation, labeling, and single-molecule localization were as described in Fig. 2. (E) Wide-field micrograph showing a representative dendrite with the overlay of GCaMP6s (green), PSD95-mCherry (magenta), and nascent proteins (black). Scale bar, 4 μm. (F) Higher-magnification image of a dendritic segment [green; indicated by box in (E)] and associated synapses (magenta). (G) Sum Ca2+ activity heatmap of the dendritic segment shown in (F) [also indicated by box in (E)]. (H) Overlay of PSD95-mCherry (magenta) and nascent proteins (black) for the dendritic segment shown in (F) and (G) [also indicated by box in (E)]. Scale bar, 0.5 μm. (I) Scatter plot showing significant correlations between synaptic protein localizations (Y) and synaptic Ca2+ events during basal Ca2+ activity imaging [see (A) and Materials and Methods] for 110 synapses with corresponding adjusted Pearson’s correlation coefficient r and P value ranges for one-tailed tests (***P < 0.001).
The proximity of ribosomes to synapses suggests that locally translated proteins may support the maintenance of the existing synaptic protein population. To address this question, proteins made from these local sites of translation must be labeled and visualized. We thus combined nascent protein metabolic labeling with DNA-PAINT in addition to ribosome localization. To tag nascent proteins for quantitative single-molecule localization, we used a brief (15 min) pulse of the noncanonical amino acid azidohomoalanine (AHA) [BioOrthogonal Non-Canonical Amino Acid Tagging (BONCAT); Fig. 2A; see Materials and Methods] (10, 18). At the cost of labeling all nascent proteins, a brief labeling period was used since prolonged labeling causes signal crowding that confounds individual clustering of nascent protein localizations (fig. S4A). After AHA labeling, neurons were fixed and AHA-tagged nascent proteins were labeled with a single-stranded DNA docking oligo using copper-free click chemistry (see Materials and Methods) (19). The samples were then immunolabeled for ribosome subunits (Fig. 1A) and dendritic and synaptic reference markers [using anti-microtubule-associated protein (MAP2) and anti-Bassoon antibodies]. Tagged nascent proteins and ribosomes in dendrites and synapses were detected sequentially using respective imager oligos (complementary and fluorescent) (Fig. 2A). As previously reported (18), without AHA treatment, we observed only a low level of background signal in the dendrites [Fig. 2 (B and C) and fig. S4 (B and C) indicate the corresponding dendrites]. In neurons treated with AHA, nascent proteins appeared as clusters of fluorescent localizations in dendrites (Fig. 2D). Our analyses indicated that a single copy of nascent protein was equivalent to ~7 ± 4 localizations (fig. S5; see also Materials and Methods) (20).
Fig. 2. Quantitative, multiplexed, single-molecule localization of locally synthesized nascent proteins and assembled ribosomes in neuronal dendrites and synapses.
(A) Nascent proteins were metabolically labeled with AHA and subsequently conjugated with a single-strand DNA barcode for visualization. Right scheme shows that recurring, transient, fluorescent DNA hybridization enables single-molecule localization of nascent proteins. TTX, tetrodotoxin. (B) Representative image showing nascent proteins detected following AHA labeling (top image). Box indicates the region enlarged in (D) (see fig. S4B for MAP2 immunostaining). Scale bar, 6 μm. (C) Representative image showing no nascent proteins detected following control treatment with methionine (see fig. S4C). Scale bar, 6 μm. (D) A dendritic segment (outlined by black dashed lines) containing clusters of nascent protein localizations. Scale bar, 0.5 μm. (E) Representative distributions of nascent protein (contour map) and ribosome localizations (heatmap) in a dendrite (4300 μm2). Scale bar, 2.5 μm. (F) Representative three-color colocalizations of nascent protein (blue) and ribosomal large- (magenta) and small-subunit (green) clusters. Black indicates overlap. Scale bar, 300 nm. (G) Scatter plot showing the Pearson’s correlation between nascent proteins and ribosomes (r = 0.73; 143 dendritic segments and more than six cell replicates; one-tailed test, ***P < 0.001). (H) Violin plots of nearest-ribosome distances (y) for 999 nascent protein clusters detected with 30-min chase (lavender) after metabolic labeling and 985 clusters detected without (gray) chase . Two-sample t test indicated a significant increase in distance following chase (**P < 0.01). (I) Representative dendritic segments showing the coincidence of ribosome hotspots (dark green) and nascent protein local maxima (magenta triangles) without and with chase following metabolic labeling. Scale bar, 2 μm.
We observed that nearly 30% of the nascent proteins were within 1 μm of a ribosome(s) and over 90% of the nascent proteins were detected within 5-μm radius of a nearby ribosome(s) (Fig. 2E and fig. S6A). In addition, local ribosome hotspots appeared to frequently overlap with high-density nascent protein domains (Fig. 2, E and F; see also Fig. 2I), while dendritic segments with low-ribosome occupancy often showed low levels of nascent proteins (Fig. 2E). We observed a significant, positive correlation (r = 0.73) between the position and level of nascent proteins and ribosomes among the dendritic segments (Fig. 2G). Overall, local differences in ribosome density appeared to account for local differences in nascent protein levels, suggesting that most of the proteins detected could arise from a local ribosome pool.
If the above-described nascent proteins were synthesized by nearby ribosomes, then imposing a chase (30 min of no label) following the brief metabolic label should diminish the spatial correlation because nascent proteins will have diffused. To test this, we compared the nearest-ribosome distances of individual nascent proteins in dendrites with and without a 30-min chase immediately following metabolic labeling. A significant increase in the nearest-ribosome distances was observed following the 30-min chase (Fig. 2H). We also noted that the local maxima of nascent protein contours were displaced from nearby ribosome hotspots [Fig. 2I, 30-min chase; see also fig. S6 (B to D)] following the chase. These data support the idea that on brief time scales, there is a tight spatial relationship between dendritic ribosomes and their nascent protein products.
To address how the above locally translated proteins distribute to synapses, we quantified the extent to which individual locally translated proteins occupied excitatory synapses during normal, ongoing network activity. Using synapse templates created by an excitatory synaptic marker [anti–postsynaptic density 95 (PSD95); Fig. 3, A and B; see also fig. S7], we extracted nascent synaptic proteins over a 6400-μm2 field of view and detected and quantified the nascent protein at each synapse on a dendrite (Fig. 3C). Although our 15-min labeling protocol inevitably resulted in an underdetection of nascent protein copies (see Materials and Methods and Discussion), we still detected ~8 copies of tagged nascent proteins per micrometer of dendrite (on average, 55 ± 11 localizations/μm with ~7 localization per copy; Fig. 3A), with 51 ± 6% detected within a 500-nm radius of a synapse (the PSD95 maxima) and 10 ± 5% overlapping with the PSD95. The number of nascent proteins present at individual synapses ranged from 0 to 10 (0 to 70 localizations) for the 1289 synapses measured from six cell replicates under control conditions. The number of nascent proteins detected within dendritic branches did not exhibit a gradient reflecting proximity to the cell soma (fig. S5C; see also Fig. 2B), consistent with the idea that they are synthesized locally. To examine the local protein allocation to synapses during basal activity, we recorded basal, spontaneous synaptic Ca2+ transients (500 frames at 1 Hz) before metabolic labeling and DNA-PAINT (Fig. 3D; see also Materials and Methods). Neurons were transfected with GCaMP6s (Ca2+-transient indicator; Fig. 3E, green) and PSD95-mCherry (synaptic reference marker; Fig. 3E, magenta) before Ca2+ imaging. We detected 4562 Ca2+ events distributed across 110 synapses in ~8 min of imaging. The subsequent detection of nascent protein (Fig. 3E) indicated that synapses that exhibited higher activity levels (Fig. 3, F and G) also had higher levels of locally synthesized protein (Fig. 3H). Overall, we found a significant correlation between the nascent synaptic proteins and basal synaptic Ca2+ events (Fig. 3I). Synaptic activity is thus a useful predictor of the locally synthesized protein levels observed under basal conditions. The above data indicate that local protein synthesis is prevalent during the ongoing maintenance of synapses, even in the absence of overt stimulations.
The above data suggest that nascent protein levels at synapses might sense activity levels. We next queried whether the local protein supply is altered by activity-dependent plasticity. We first examined a form of plasticity that is induced and expressed across the entire neuron, synaptic upscaling (7). Cultured hippocampal neurons were treated with tetrodotoxin (TTX; 2 μM) for 1 or 24 hours, representing an early “induction” time point (1 hour) and a time point when synaptic scaling had been established (24 hours) (2). At the end of the (1- or 24-hour) upscaling induction, neurons were metabolically labeled (4 mM AHA) for the last 15 min before fixation and processing to identify dendrites and synapses, as described above (Fig. 3, A and B). As observed previously (21), synapse density increased slightly after 1 and 24 hours of activity blockade (fig. S8) with a small but significant increase in mean synapse size (fig. S7) (22). We quantified the amount of nascent protein present and detected a significant, multifold increase in both dendrites (Fig. 4A) and synapses (Fig. 4B; see also fig. S9) after both 1 and 24 hours of upscaling. This indicates that local protein synthesis is globally elevated during synaptic scaling.
Fig. 4. A global increase in local protein supply was detected heterogeneously among synapses but homogeneously among dendritic segments of synapses during synaptic upscaling.
(A) Scatter plots indicating the tagged protein (locally synthesized, nascent; same below) density (localization per micrometer) for 23 dendritic branches from six untreated cells (gray), 34 branches from six cells with 1 hour of upscaling (lavender), and 29 branches from six cells with 24 hours of upscaling (purple). ANOVA with post hoc Bonferroni analysis indicated significant increases after both 1 and 24 hours of upscaling [***P < 0.001; same in (B)]. (B) Scatter plots indicating the mean synaptic protein, with significant increases after upscaling. n.s., not significant. (C to E) Example traces depicting the proteins at each synapse distributed along dendritic branches (y axis: nascent protein level; x axis: the series of synapses compressed and lined along the dendritic branches) from untreated and 1- and 24-hour-upscaled neurons. Purple arrows indicate example synapses with high nascent protein levels themselves but neighbored by synapses with low nascent protein levels. (F) Violin plots showing the wide distributions in nascent protein levels between synapses and the much tighter distributions (mean protein per synapse) between dendritic segments of synapses. White lines indicate mean values. Right inset depicts that local protein supply is similar among dendritic segments (pie chart indicates that segments containing more synapses obtain more proteins) but heterogeneous between neighboring synapses. (G to I) 3D scatter plots of (G) 1289 synapses from six untreated neurons, (H) 2955 synapses from six 1-hour upscaling neurons, and (I) 2625 synapses from six 24-hour upscaling neurons. Gray arrows in (H) indicate an apparent bifurcation of the synapse population.
While, at the population level, there was a scaling-induced increase in protein level at synapses and along dendrites, we observed significant diversity in synaptic protein levels within the dendrite (Fig. 4, C to E, and fig. S9, D to F). High-protein synapses did not appear clustered (Fig. 4, C to E; examples indicated by purple arrows) but were frequently neighbored by synapses with little to no detectable nascent protein [also schematized in Fig. 4F (right inset)]. Consequently, the average protein per synapse within a dendritic segment was very similar across all the segments (Fig. 4F) despite the high variability between individual synapses within a segment. Dendritic segments with highly clustered synapses [purple box in Fig. 4F (right inset)] became the local translational “hotspots” (fig. S10, lighter-colored segments). Hence, local translational hotspots are likely driven by the local clustering of synapses in number rather than the clustering of high-protein synapses [schematized in Fig. 4F (right inset)].
To better visualize the contribution of local protein supply at the synapse population level, for each synapse, we plotted its nascent protein level (x), the local synapse density (y), and the average nascent synaptic protein level present at its neighboring synapses (z) in a three-dimensional (3D) parameter space (Fig. 4, G to I, and fig. S11; see Materials and Methods). Overall, the greater than 1000 synapses that we measured did not segregate into discreet clusters in the 3D space. After 1 and 24 hours of upscaling (Fig. 4, H and I, respectively), the synapse population dilated significantly along all three axes in comparison to control, consistent with Fig. 4B and fig. S8 (synapse density increase), with the largest increase seen in the z axis, representing the increase in the neighboring synaptic protein level. Driven by a small number of extremely high-protein synapses, the population after 1 hour of upscaling appeared slightly bifurcated (indicated by two gray arrows in Fig. 4H): Extremely high-protein synapses occupied the high-protein-per-synapse region of the parameter space (bottom right corner), while their neighboring synapses extended into the high-local-protein-density region (top front corner). This distribution showed that protein was far from evenly distributed among neighboring synapses (as shown in Fig. 4C), reminiscent of competition between nearby synapses (see fig. S12 for illustrations of proposed modes of protein distribution in 3D) (23, 24). Compared to 1 hour of upscaling (Fig. 4E), the population contracted in the parameter space after 24 hours of upscaling, driven by a loss of extremely high-protein synapses (Fig. 4I). These observations were robust across dendrites and neurons within the same treatment group (fig. S12, A to C). Overall, these data indicate that synaptic upscaling elicited a global increase in local protein production, with significant heterogeneity in nascent protein levels at individual synapses.
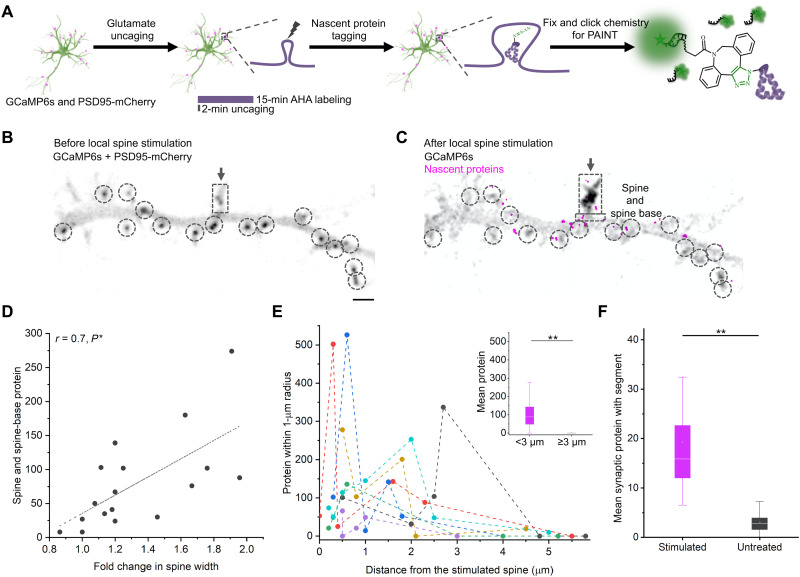
How are locally synthesized proteins allocated during locally induced forms of plasticity? To explore this, we elicited morphological plasticity at single spines using two-photon glutamate uncaging (Fig. 5A) combined with stimulation of the protein kinase A pathway, as previously described (23, 25). As before, neurons were transfected to express GCaMP6s and PSD95-mCherry before local spine stimulation, metabolic labeling, and DNA-PAINT (Fig. 5A; see Materials and Methods). As expected, local stimulation of an individual spine resulted in a Ca2+ increase in the stimulated spine, but not adjacent spines (fig. S13). As observed previously (25), this plasticity protocol resulted in an increase in spine size (Fig. 5, C and D). While spine size increased less than twofold (Fig. 5D, x axis), local protein supply to synapses increased by several-fold (Fig. 5D, y axis). We nevertheless detected a significant correlation between the spine and spine-base nascent protein level (indicated by dashed boxes in Fig. 5C) and the induced spine size increase (Fig. 5D).
Fig. 5. Single-spine stimulation induced a local increase in nascent protein among nearby synapses.
(A) Cultured rat hippocampal neurons [12 days in vitro (DIV)] were transfected with GCaMP6s and PSD95-mCherry (see Materials and Methods) before single-spine stimulation (two-photon glutamate uncaging in AHA-containing buffer). Subsequent fixation, labeling, and single-molecule localization were as described in Fig. 2 (see also Materials and Methods). (B) Spinning disk micrograph with overlay of GCaMP6s (maximum projection in Z) and PSD95-mCherry showing the spine of interest before local stimulation (dashed box) and its neighboring synapses (encircled dark puncta). Scale bar, 2 μm. (C) Overlay of spinning disk GCaMP6s micrograph (maximum projection in time; gray) and single-molecule localization micrograph of locally translated nascent proteins (magenta). Dashed boxes indicate the spine and spine base of the stimulated spine. Dashed circles indicate its neighboring synapses. (D) Scatter plot showing significant correlations between nascent protein (localizations of locally synthesized proteins) in stimulated spine and spine base (Y) and δF/F GCaMP6s fluorescence during the stimulation of 17 spines from six preparations (X; see Materials and Methods) with corresponding adjusted Pearson’s correlation coefficient r and P value ranges for one-tailed tests (*P < 0.05). (E) Scatter plots showing the nascent protein (localizations of locally synthesized proteins; Y) in the neighboring synapses of six stimulated spines (one color for each set of neighboring synapses) distributed along the dendrite (X). Inset box plot: two-sample t tests indicated a significant increase of local protein supply among neighboring synapses within 3 μm of the stimulated spine compared to those greater than 3 μm away from the stimulated spine (**P < 0.01). (F) Box plot showing the mean synaptic protein (per synapse; normalized per cell) within each segment for 9 segments containing stimulated spines and 119 untreated segments. Two-sample t tests indicated a significant increase of local protein supply for segments containing stimulated spines (**P < 0.01).
To investigate whether the increase in nascent protein at the stimulated synapses was specific, all the identified synapses within 15-μm distance from the stimulated spine were analyzed (Fig. 5, B and C). Figure 5C shows the distribution of nascent proteins (magenta) within this dendritic segment after local stimulation (the same set of synapses encircled as in Fig. 5B). To reveal the protein distribution among these neighboring synapses, their nascent protein allocation (Fig. 5E; see Materials and Methods) was plotted relative to their distances from the corresponding stimulated spines (each color series represents a set of neighbors near a stimulated spine replicate; seven stimulated spine replicates with at least five neighbors within 5 μm are shown; see Materials and Methods). Neighboring synapses nearer to the stimulated spine (e.g., <3 μm) showed heterogeneous but significantly higher protein allocation on average compared to those farther away (Fig. 5E, inset). Consistently, at a larger scale, dendritic segments containing stimulated spines exhibited significantly higher mean protein (per synapse) compared to control untreated segments (Fig. 5F; see also fig. S14) across the dendrite. At the same time, neighboring segments did not significantly rise above or sink below the remaining untreated segments (fig. S14), implying locally elevated protein synthesis rather than a depletion of proteins from surrounding segments. Together, these data indicate that although the spine size changes were specific to the stimulated spine, the nascent proteins stimulated by plasticity induction were not specific.
DISCUSSION
Decades of research have detected thousands of mRNAs, the essential elements of the protein synthesis machinery, and evidence for protein synthesis in neuronal processes (1, 5, 15, 26, 27). Locally synthesized nascent proteins have been visualized (28, 29). However, a persistent open question has been the prevalence of local protein synthesis and whether locally synthesized proteins contribute to the proteome to offset ongoing protein turnover. In addition, the specificity of locally synthesized proteins during plasticity has not been addressed. Here, we used metabolic labeling and DNA-PAINT to spatially dissect and quantify the contribution of local protein synthesis at neuronal synapses during basal activity and following plasticity. Although our method required subsampling (low concentration of AHA for just 15 min), we detected nascent proteins throughout the dendrites in the absence of any exogenous stimulation and ribosomes at 85% of all synapses. A multifold increase in nascent proteins was observed at synapses following both global synaptic upscaling and local single-spine plasticity. Rather than being synapse specific, the local protein supply appeared to be produced and redistributed among neighboring synapses. Local protein synthesis thus occurs during maintenance while harboring a multifold capacity to supply proteins for synapses during plasticity.
How abundant is local translation in neurons? Here, we detected ~8 ribosome copies per micrometer of dendrite and ~8 tagged copies of nascent proteins per micrometer of dendrite within 15 min of metabolic labeling. Note that since ribosomes cluster around synapses, these numbers could increase by nearly 10-fold at synapses. Taking the necessary subsampling into consideration, these estimates give a lower bound of the local translation that occurs in dendrites. Furthermore, each neuron establishes to 10,000 synapses with other neurons, giving rise to the active local translation of 103 to 104 transcripts within 15 min. Last, we tagged ~1 copy of nascent protein per ribosome copy within a 15-min labeling period. Considering that a translating ribosome makes, on average, one copy of protein per minute (~5 amino acids per second; ~300 amino acids for an average protein) (30), we estimated a sample rate of ~10% for locally synthesized proteins (also see Materials and Methods). Taking this into account, we further estimate that roughly 80 protein copies are locally synthesized per micrometer of dendrite within just 15 min.
Where do locally synthesized proteins go after being made by local ribosomes? While biophysical measurements and simulations have provided important information on how proteins move in the cytosol (31, 32), endogenous proteins made from local ribosomes may exhibit different movements because of their spatial confinement and molecular crowding (e.g., in spines versus in dendritic shafts) (33), as well as local protein-protein interaction mechanisms engaged to capture them (34). Thus far, it has been difficult to visualize and resolve the distribution (at a proteome-wide scale) of individual, locally synthesized, endogenous proteins. To directly probe this question, we localized ribosomes together with the nascent proteins tagged in distal dendrites using brief metabolic labeling. Hence, the locally synthesized protein cohort was spatially and temporally “dissected” from the somatic influx of nascent proteins. While some fast-diffusing proteins from the soma may have reached our fields of view, we did not observe a gradient of nascent proteins as a function of distance from the cell body; rather, we detected a tight spatial relationship between dendritic ribosomes and nascent proteins. These suggest that, although individual protein species may differ markedly in their addressing from local sources, the locally synthesized proteome, as a whole, tends to distribute locally.
How “local” is local protein synthesis? While early thinking posited the idea of a dedicated toolkit of mRNAs and ribosomes for an individual synapse (35), emerging data rather suggest a local sharing of resources: a local “neighborhood” of synapses populated by translating ribosomes, mRNAs (36, 37), and potentially other essential organelles such as mitochondria (25) and endoplasmic reticulum (4). In our experiments, despite the observation that most nascent proteins were initially detected close to ribosomes, chase experiments revealed that a significant fraction of the locally synthesized proteome appeared somewhat mobile and spread over time. Previous studies have found that even scaffold proteins such as PSD95 are constantly exchanged between neighboring synapses (38). The neighborhood specificity of local protein synthesis is further evident during plasticity: Local clusters of synapses obtained similar level of proteins on average during global upscaling, while single-spine stimulation elicited an increase of local protein supply that “spilled over” to neighboring spines. This local sharing of protein supply may prime adjacent synapses for subsequent plasticity (8). Many studies have suggested that mRNAs are not present in sufficient copy numbers to populate all synapses (e.g., one calcium/calmodulin-dependent protein kinase II mRNA per 5 μm, rather than per synapse) (36). The local sharing of nascent protein is therefore implied by both the densities of mRNAs and ribosomes and the nascent proteins detected in our single-spine plasticity experiments. These provide a potential molecular basis for clustered and heterosynaptic plasticity observed to underlie various forms of plasticity and memory (6, 39, 40).
MATERIALS AND METHODS
Cell culture
Dissociated rat hippocampal neuron cultures were prepared and maintained as previously described (28). Briefly, rat hippocampi were dissected from postnatal day 0 to 1 pups of either sex (Sprague-Dawley strain; Charles River Laboratories), dissociated with papain (Sigma-Aldrich), and then plated at a density of 30 × 103 cells/cm2 on poly-d-lysine–coated glass-bottom petri dishes (MatTek). Neurons were maintained (until day of experiment) in a humidified atmosphere at 37°C and 5% CO2 in growth medium (Neurobasal-A supplemented with B27 and GlutaMAX-I; Life Technologies) for 13 to 21 days in vitro (DIV) to ensure synapse maturation. For polysome profiling, because of the larger quantity of cells needed, dissociated rat cortical neuron cultures were prepared and maintained similar to above. All experiments complied with National Animal Care Guidelines and the guidelines issued by the Max Planck Society and were approved by local authorities.
Ribosome profiling
Harringtonine (LKT Laboratories) was prepared at a final concentration of 2 μM (2 mM stock, 100% ethanol) and incubated with cells for 30 min at 37°C. Puromycin (Thermo Fisher Scientific) was prepared at a final concentration of 500 μM and incubated with cells for 10 min at 37°C. After treatments, cells were immediately placed on ice, washed with ice-cold phosphate-buffered saline (PBS) containing cycloheximide (100 μg/ml), lysed, and scraped in polysome lysis buffer [20 mM tris (pH 7.4), 150 mM NaCl, 5 mM MgCl2, TURBO DNase (24 U/ml), cycloheximide (100 μg/ml), 1% Triton X-100, 1 mM dithiothreitol (DTT), RNasin Plus RNase Inhibitor (200 U/ml), and 8% glycerol]. After scraping, the lysates were pipetted up and down until homogenization was clear with a 0.4 × 20 mm syringe needle (HSW FINE-JECT) on ice. The lysates were then centrifuged at 10,000g for 10 min at 4°C. The supernatant was used for ribosome fractionation.
For sucrose gradients, all solutions were prepared in gradient buffer [20 mM tris (pH 7.5), 8% glycerol, 150 mM NaCl, 5 mM MgCl2, cycloheximide (100 μg/ml), and 1 mM DTT]. Gradients were prepared by sequentially adding different sucrose concentrations (in order from first added to last: 8 ml of 55%, 0.5 ml of 50%, 0.5 ml of 40%, 0.5 ml of 30%, 0.5 ml of 20%, and 0.5 ml of 10%) into the same thin-wall polypropylene tube (Beckman, catalog no. 331372). After the addition of each sucrose solution, tubes were placed at −80°C to freeze the content before the next layer. The gradients were stored at −80°C and left to equilibrate at 4°C overnight. Then, 0.5 to 1.5 optical density (measured with NanoDrop at 260 nm) of the lysates were loaded on top of the gradients and spun at 36,000 rpm at 4°C for 2 hours with a SW 41 Ti rotor (Beckman). Fractions from each sample were collected every 7 s using a density gradient fractionation system (Teledyne ISCO; intensity used, 1), chased by 60% sucrose and 10% glycerol in water at 850 μl/min, and continuous monitoring at 254 nm using a UA-6 detector.
Fractions of 125 μl corresponding to the 40S, 80S, or the polysome peaks were collected and pooled as the enriched 40S, 80S, and polysome fractions, respectively. For plating ribosome fractions, diluted fractions were incubated in (1:100 dilution in the above lysis buffer without TURBO DNase, glycerol, and protease inhibitor cocktails) poly-d-lysine–coated MatTek dishes for 2 hours before the liquid was removed, and 4% paraformaldehyde in PBS containing cycloheximide (100 μg/ml) was added to fix the samples. After fixation, the dishes were washed three times using PBS buffer (pH 7.4).
Metabolic labeling with AHA and synaptic scaling
Neurons (18 to 21 DIV) on MatTek dishes were incubated in the growth medium described above (for upscaling, 2 μM TTX was used for 1 or 24 hours) at 37°C and 5% CO2. Neurons were incubated in methionine-free Neurobasal-A (custom-made by Life Technologies) supplemented with 4 mM AHA for 15 min (10). For upscaling, this step started 15 min before each treatment ended, and 2 μM TTX was added. In methionine control experiments, AHA was replaced by 4 mM methionine (Sigma-Aldrich). Subsequently, cells were washed twice with Neurobasal-A, fixed in paraformaldehyde-sucrose (4% paraformaldehyde; Alfa Aesar) and 4% sucrose in PBS supplemented with Mg2+ and Ca2+ at room temperature for 20 min, washed again, permeabilized with 0.5% Triton X-100 in 1 × PBS (pH 7.4) for 15 min, and blocked with blocking buffer (4% goat serum in 1× PBS) for 1 hour. To optimize conditions for the subsequent click reaction, neurons were equilibrated by washes with 1× PBS (pH 7.8).
The BONCAT part of the assay was performed as described previously (28) with the following modification: We used a previously reported single-stranded DNA oligo sequence (P1 docking oligo) (12) modified to carry a more reactive alkyne, dibenzocyclooctyne (DBCO; GeneLink), as a tag in the copper-free azide-alkyne cycloaddition click reaction (19). For the Cu-free click reaction, 0.4 μM P1-DBCO tag was prepared in 1× PBS (pH 7.8) before application to the cells and the click chemistry was performed overnight at room temperature. After the click reaction, cells were washed two times with PBS (pH 7.8) and 0.5% Triton in PBS and three times with PBS (pH 7.4) before immunofluorescence labeling with various markers (see below). We estimated the undersampling rate of our metabolic labeling on the basis of the measured synaptic nascent protein localizations (see below the “Data analyses” section).
Immunofluorescence labeling
Neurons were washed three times in PBS before blocking in PBS containing 4% goat serum (Gibco) for 1 hour. Neurons were incubated overnight with guinea pig antibodies, anti-MAP2 (1:2000; 188004, Synaptic Systems), or chicken antibodies, anti-MAP2 (1:2000; ab5392, Abcam), and mouse antibodies, anti-PSD95 (1:1000; MA1-046, Thermo Fisher Scientific), or guinea pig antibodies, anti-Bassoon (1:1000; 141004, Synaptic Systems), in PBS containing 4% goat serum (Gibco) at 4°C to stain the dendrites for morphology and synapses, respectively. To stain ribosomal subunits, mouse antibodies, RPL36a (1:500 overnight, or 1:1000, 3 hours for subsampling; sc-100831, Santa Cruz Biotechnology), and rabbit antibodies, RPS11 (1:500 overnight, or 1:1000, 3 hours for subsampling; A303-936A, Bethyl Laboratories), were used. The samples were then washed three times in PBS (5 min each) before incubation for 1 hour with anti–guinea pig antibody conjugated with Alexa Fluor 488 (1:1000; Nanoprobes) or anti-chicken antibody conjugated with Alex Fluor 405 (1:1000; Nanoprobes) and anti-mouse antibody with Alexa Fluor 546 (1:1000; Nanoprobes) or anti–guinea pig antibody with Alexa Fluor 488 (1:1000; Nanoprobes). For secondary antibody staining of ribosome for DNA-PAINT, anti-mouse antibodies conjugated with P1 imager oligo and anti-rabbit antibodies conjugated with P5 imager oligo were used (1:1000; custom-made, as previously reported) (9). For ribosome-nascent protein colocalization, anti-mouse antibodies conjugated with P3 imager oligo and anti-rabbit antibodies conjugated with P5 imager oligo were used for ribosomal subunits. Neurons were washed three times in PBS (5 min each). All steps were performed at room temperature. Neurons were then stored in PBS at 4°C for up to 3 weeks until DNA-PAINT imaging.
Transfection, Ca2+ imaging, and local spine stimulation
Experiments were conducted similar to previously described (25). Briefly, transfections were carried out 12 DIV with CombiMag (OZ Biosciences) and Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Live cell imaging was carried out using 13 DIV hippocampal neurons. All live cell imaging was performed at 32°C, in E4 imaging buffer [120 mM NaCl, 3 mM KCl, 10 mM Hepes (pH 7.4), 3 mM CaCl2, 1 mM MgCl2, and 10 mM glucose]. Glutamate uncaging experiments used a modified E4 buffer lacking MgCl2 and containing 4 mM CaCl2.
All live cell imaging used an inverted spinning disk confocal microscope (3i imaging systems; model CSU-X1) using the SlideBook 5.5 software. Images were acquired with a Plan-Apochromat ×63/1.4 oil differential interference contrast objective at laser powers 1.1 mW (488 nm) and 0.8 mW (561 nm) for basal Ca2+ imaging and glutamate uncaging experiments with an Evolve 512 camera (Photometrics). We used 488-nm excitation and 525/30-nm emission filters for imaging GCaMP6s, and 561-nm excitation and 617/73-nm emission filters were used for imaging PSD95-mCherry. Images were analyzed using ImageJ (see the “Data analyses” section for details). OriginPro 2019 was used for data analysis, statistical testing, and plotting graphs.
Transfected neurons were identified by GCaMP6s fluorescence (i.e., calcium transients). For basal Ca2+ activity imaging, 500 frames were acquired at 1 Hz (five planes each spanning 5 μm in Z) in E4 buffer containing 1 μM TTX followed by 15-min metabolic labeling with AHA (in E4 buffer; 4 mM) before fixation. Subsequent steps are as described in the “Metabolic labeling with AHA and synaptic scaling” section. For glutamate uncaging experiments, spines were identified using PSD95-mCherry fluorescence. Immediately before glutamate uncaging, neurons were treated with 1 μM TTX (citrate salt, 2 mM stock made in water), 50 μM forskolin (100 mM stock made in dimethyl sulfoxide; Tocris Bioscience), 4 mM AHA, and 2 mM 4-methoxy-7-nitroindolinyl–caged l-glutamate (100 mM stock made in E4 buffer; Tocris Bioscience) in modified E4 buffer lacking Mg2+ (see above). Glutamate uncaging was carried out with a 720-nm multiphoton laser (Chameleon, Coherent) and a Pockels cell (Conoptics) for controlling the uncaging pulses. Spines at least 50 μm away from the cell body were chosen for uncaging experiments. To test a spine’s response to an uncaging pulse, an uncaging spot (∼2 μm2) close to a spine head was selected, and two to three uncaging pulses at 10-ms pulse duration per pixel and 2.5-mW power were delivered to confirm spine-specific calcium transients. During uncaging, 60 uncaging pulses at 0.5 Hz with 10-ms pulse duration per pixel at 2.5-mW power were used. After uncaging, neurons were left in the same AHA-containing buffer for ~13 min, adding up to a total of 15 min of AHA incubation before washing with modified E4 buffer and fixation. Following fixation, neurons were processed as described in the “Metabolic labeling with AHA and synaptic scaling” section.
Superresolution microscopy
For DNA-PAINT imaging, the imaging buffer contains 500 nM P1, P3, or P5 conjugated with Atto655 (Eurofins Genomics) in 500 mM NaCl in PBS (pH 7.4) (20). For ribosome-nascent protein colocalization, 2 nM P1 was used for nascent protein PAINT to shorten the duration of multiplexed imaging. Immunolabeled cultured neurons containing 90-nm gold fiducial markers (A1190, Nanoparz) were imaged on an N-STORM system (Nikon, Japan): an Eclipse Ti-E inverted microscope, equipped with a Perfect Focus System and a motorized x-y stage. Total internal reflection fluorescence (TIRF) and highly inclined and laminated optical sheet (HILO) (20) configurations were adjusted using a motorized TIRF illuminator in combination with a ×100 oil-immersion objective [CFL Apo TIRF; 1.49 numerical aperture (NA)] with a final pixel size of 158 nm. For imaging, 647-nm excitation wavelength was used and housed in a MLC400B (Agilent) laser combiner. An optical fiber guided the laser beam to the microscope body and via a dichroic mirror (T660LPXR, Chroma) to the sample plane. Fluorescence emission was separated from excitation light via a band-pass filter (ET705/72m, Chroma) and detected by an iXon Ultra electron multiplying charge-coupled device (EMCCD) camera (DU-897U-CS0-23 #BV, Andor). The software NIS-Elements Ar/C and μManager were used to control the setup and the camera. Alternatively, immunolabeled cultured neurons prepared similarly were imaged on a Leica DMi8 S system with Infinity TIRF HP, Infinity Scanner, and an iXon Ultra 888 EMCCD camera (Andor). Oil-immersion objective (×100; HC PL APO CORR TIRF; 1.47 NA) was used in combination with a motorized TIRF illuminator. For imaging, 638-nm excitation wavelength was used (150 mW), and LAS X software package was used for image acquisition.
Wide-field micrographs of the MAP2 and PSD95 (or Bassoon) reference markers (see the “Immunofluorescence labeling” section) were obtained for nonproximal dendrites (at least one branching point away from the soma) before DNA-PAINT as summed projections of 2-μm-thick Z-stacks to capture the entire dendritic volume. The chosen fields of view typically contained clearly separated dendrites that originated from the same neuron. HILO illumination was used for superresolution acquisition with a power of 30 to 40 mW, which was determined directly after the objective and under wide-field configuration. For the N-STORM system, the intensity density (45% intensity of 647-nm laser) was 0.9 kW/cm2. Time-lapse datasets with 50,000 frames and 16-bit depth were acquired for nascent protein localization at 5-Hz frame rate and 5-MHz camera readout bandwidth (preamplification, 3; electron multiplying gain, 4). For the DMi8 S system, time-lapse datasets with 50,000 frames and 16-bit depths were acquired for ribosomal subunit localization (25,000 frames for nascent protein PAINT in ribosome-nascent protein colocalization) at 5-Hz frame rate and 10-MHz camera readout bandwidth (gain list, 2; electron multiplying gain, 100).
Data analyses
DNA-PAINT acquisitions were reconstructed with Picasso:Localize, a module of the Picasso software (12), by applying a minimal net gradient of 2500 (for N-STORM) or 15,000 (for DMi8 S). With Picasso:Render, drift corrections were applied in two subsequent fashions: First, a drift correction based on the redundant cross-correlation, with a segmentation of 200, was applied. Second, fiducial markers (gold beads) were manually selected, localized, and used for drift correction. Drift-corrected data were filtered using Picasso:Filter. Raw localizations within the same location were further filtered temporally on the basis of their average frame numbers and SD of frame numbers to eliminate background signal due to the unspecific binding of the imager oligo to a random target. These background signals are often clustered temporally rather than distributed through the imaging course, resulting in lower or higher average frame numbers and lower frame number variance compared to real signal. Afterward, raw localizations within a maximal distance of 6× measured localization precision and showing a maximum number of transient dark frames of 20 were linked together, resulting in a single, linked localization event (referred to as “localization”). Localization precision (Nearest neighbor based analysis values) was determined to be 13.1 nm, as previously described using nearest neighbor–based analyses (41).
Coincidence detection thresholds the interdistances between small- and large-subunit clusters at 100 nm based on the estimates of oligo docking site interdistance (maximum, ~50 nm), localization precision (see above paragraph), and corrected image drift (based on fiducial marker separation after drift correction). Without primary or secondary antibodies, only a low level of background signal was observed in the dendrites (fig. S1G). Prolonged labeling created too much signal, making it impossible to isolate and analyze individual clusters of ribosomal localizations (fig. S1E). Subsampling was thus optimized in situ (fig. S1F). Using purified monosomes plated on a coverslip (fig. S2, B and C), subsampling is estimated to detect 31% RPS11, 33% RPL36a, 10% of monosomes (i.e., individual 80S ribosomes), and, by calculation, most polysomes (e.g., a calculated ~70% for a polysome with four 80S ribosomes, an average ribosome cluster).
Synaptic regions were determined using a custom-written algorithm and the diffraction-limited PSD95 or Bassoon immunolabeling signal. To identify the positions of excitatory synapses, local PSD95 (or Bassoon) puncta maxima and minima were identified and normalized to the same intensity range (0 for minima and 255 for maxima). Pixels of PSD95 puncta with over 12% intensity (30 in normalized intensity) of its associated, normalized local maximum (255 in normalized intensity) were selected to define the synaptic compartments. These local intensity thresholds resulted in puncta size estimates that were less affected by local intensity and background differences (e.g., the phenomena where brighter puncta appear larger by eye while less bright puncta appear smaller; fig. S3, right table) because of heterogeneity in staining or focusing. Puncta were eliminated if they are on the soma or more than 2 μm away from a dendritic shaft marked by MAP2 signal; Puncta with sizes smaller than 0.15 μm2 were excluded from size-based analyses because of significant inaccuracy of size measurements at smaller spatial scales. Dendrites with a high level of background signal outside MAP2-labeled regions, dendrites containing overlapping signals with AHA labeling from glia, out-of-focus dendritic branches, and branches shorter than 5 μm were excluded from analyses. All in all, this created a synapse mask with a measured, average synapse density and synapse size distribution consistent with published values (17). This mask effectively enriched protein signals allocated into the synaptic area by excluding adjacent regions such as nearby proteins in the shaft (e.g., spine base). To evaluate adjacent regions such as the spine neck and base, a mask was generated to include an area of 1 μm in diameter centered on the PSD intensity maxima; analyses showed qualitatively consistent results. All localizations <2.5 μm from a skeletonized line that traversed the center of the MAP2-labeled dendrites were included as total dendritic localizations.
Synaptic nascent protein localizations were further identified by a custom-written, noise-tolerant cluster identification algorithm (Density-based spatial clustering of applications with Noise based) (42), the spatial-distance parameter (r) of which was determined using the measured localization precisions for each dendrite. In particular, any cluster with <3 localizations within r was excluded from clustering, which is more selective than DBSCAN. It is therefore more robust against the inclusion of noise localizations. The PAINT signal of a single docking oligo (further approximated as a single protein copy due to the low incorporation efficiency of AHA) was determined as follows: We plotted the number of linked localization distributions (see fig. S2) for all the identified synaptic clusters and attributed the smallest population of clusters as the signal of a single protein copy; we further determined the average dark time for a single-protein copy from this population (containing ≤11 linked localizations) (fig. S2) and found it comparable with previously reported benchmark test of a single docking oligo based on DNA origami (with identical setup) (20); hence, we calculated that a single protein copy corresponds to roughly 7 ± 4 linked localizations. Because of these approximations and the known difficulties in obtaining accurate molecule counting (43), in the main text, we directly use the number of linked localizations as a proxy measure for protein copy numbers. In parallel, the widely used DBSCAN cluster identification was also implemented to double-check the robustness of our subsequent analyses. Afterward, Picasso:Render and ImageJ were used for dendritic-branch level analyses. A 1D dendritic tree was constructed on the basis of the MAP2-immunolabeling signal. Synapses and localization positions were then mapped onto the closest dendritic branch. The local synapse and synaptic nascent protein densities per unit length were obtained by applying smoothing with a Gaussian kernel with parameter σ = 2 μm. Note that for protein density, this contribution is proportional to its synaptic localization counts. To define dendritic segments, borders were drawn at the turning points of the smoothed synapse density, such that segments with a range of low and high synapse densities were identified (fig. S3F). The majority of dendritic segments were <10 μm in length, containing <10 synapses. We constrain segment size to be at least 0.5 μm and the slope of the synapse density at the turning points to be >0.1 synapse/μm2 for partitioning into a new segment. The synapse densities and synaptic nascent protein densities were measured using the corresponding total numbers divided by the dendritic lengths. To estimate the undersampling rates due to metabolic labeling and click chemistry, we note that in untreated groups, synaptic nascent protein localization was ~1 to 2 per synapse. A synapse contains approximately 104 to 105 protein copies with an average half-life of 5 DIV (44). This gives an estimate of 1 to 10 proteins being renewed every 15 min, which amounts to ~7 to 70 linked localizations per synapse (fig. S5; assuming ~1 localization per synapse in untreated). This yields an undersampling estimate of 86 to 98.6%, meaning that we likely acquire data from 1.4 to 14% of the synaptic nascent protein pool. Undersampling is crucial for avoiding high-density, overlapping localizations, which may introduce inaccuracies in quantification (see fig. S4).
To count the synaptic Ca2+ events during GCaMP6s imaging, a maximum intensity projection image was generated from the GCaMP6s time-lapse images. A region of interest (ROI) was chosen for each PSD95-mCherry–positive spine, and a temporal fluorescence profile over 500 frames of imaging was generated for each ROI. Using automatic peak identification in OriginLab 2019, the number of Ca2+ events was counted for each ROI.
To measure changes in spine size, a line crossing the center of the spine head was drawn, as described previously (25). The pixel intensity profile along the line was fit to a Gaussian to obtain the full width at half maximum, which is defined as the spine head width and used as a proxy for spine head size changes.
For glutamate uncaging experiments, the shape of the uncaged spine was traced using the GCaMP6s signal, including 500 nm into the dendritic shaft, ±1 μm upstream and downstream. δF was calculated by subtracting the average projection GCaMP6s signal before uncaging (F) from the maximum projection GCaMP6s signal during uncaging. δF/F was normalized by the stimulated spine area and used as a measure for Ca2+ transients due to uncaging. The number of nascent protein localizations (20,000 frames; 5 nM P1 imager) within the defined spine and base area was counted. As control, unstimulated PSD95-mCherry–positive spines from adjacent dendritic branches were measured similarly.
Constructs
PSD95-mCherry construct was obtained in-house (45); GCaMP6s was purchased from Addgene (plasmids 40753).
Reagents
Unless noted otherwise, all substances were molecular biology or cell culture grade and purchased from Sigma-Aldrich or Roth. TTX citrate was used from stock solutions (Tocris Bioscience; 2 mM in H2O and 2 μM final).
Statistical analysis
Two-sample t tests were used for comparisons between two groups. Comparisons between more than two groups were conducted using analysis of variance (ANOVA) with Bonferroni post hoc analysis. One-tailed tests were used for Pearson’s correlations. For all statistical tests, a P value of less than 0.05 was considered significant. For experiments that involved comparisons between drug treatments, at least six cell replicates from four preparations were measured for each treatment group. Otherwise, at least three cell replicates from three preparations were measured for each experiment.
Acknowledgments
We thank G. Tushev, F. Kretschmer, and L. Anneser for assistance in coding; A. R. Dörrbaum and S. T. Dieck for instructions during metabolic labeling and synaptic upscaling; C. Böger and A. S. Hafner for advice on image acquisition and analysis of DNA-PAINT; I. Bartnik, N. Fuerst, and D. Vogel for the preparation of cultured neurons; A. Schwartz for EM imaging; the Max Planck Institute for Biophysics for sharing EM facility; A. Koegel and E. Conti for the gift of purified ribosomes; and G. Laurent for inputs during manuscript preparation. Funding: C.S. is supported by an EMBO long-term postdoctoral fellowship (EMBO ALTF 860-2018) and HFSP Cross-Disciplinary Fellowship (LT000737/2019-C). C.S. and A.N. are both supported by Joachim Herz Stiftung Add-on Fellowship for Interdisciplinary Life Science (project number 850027). T.T. is funded by the Max Planck Society, the DFG CRC1080: Molecular and Cellular Mechanisms of Neural Homeostasis, and the fellowship of the Behrens-Weise-Foundation. M.H. is funded by the German Science Foundation (grants SFB 807, SFB902, HE 6166/11-1, and EXC115) and the Bundesministerium für Bildung und Forschung (BMBF:eBio). E.M.S. is funded by the Max Planck Society, an Advanced Investigator award from the European Research Council (grant 743216), DFG CRC 1080: Molecular and Cellular Mechanisms of Neural Homeostasis, and DFG CRC 902: Molecular Principles of RNA-based Regulation. Author contributions: Conceptualization: C.S. and E.M.S. Experimentation and image analyses: C.S., V.R., and C.M.F. Data analyses: C.S. and A.N. Coding: A.N. Manuscript writing: C.S. and E.M.S. Manuscript editing: all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
REFERENCES AND NOTES
- 1.Hafner A.-S., Donlin-Asp P. G., Leitch B., Herzog E., Schuman E. M., Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364, eaau3644 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Sutton M. A., Ito H. T., Cressy P., Kempf C., Woo J. C., Schuman E. M., Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell 125, 785–799 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Rangaraju V., Dieck S. T., Schuman E. M., Local translation in neuronal compartments: How local is local? EMBO Rep. 18, 693–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirillo M. A., Waters M. S., Lindsey L. F., Bourne J. N., Harris K. M., Local resources of polyribosomes and SER promote synapse enlargement and spine clustering after long-term potentiation in adult rat hippocampus. Sci. Rep. 9, 3861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostroff L. E., Fiala J. C., Allwardt B., Harris K. M., Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron 35, 535–545 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Kerlin A., Boaz M., Flickinger D., MacLennan B. J., Dean M. B., Davis C., Spruston N., Svoboda K., Functional clustering of dendritic activity during decision-making. eLife 8, e46966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turrigiano G. G., The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell 135, 422–435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schanzenbächer C. T., Sambandan S., Langer J. D., Schuman E. M., Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnitzbauer J., Strauss M. T., Schlichthaerle T., Schueder F., Jungmann R., Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Dieterich D. C., Lee J. J., Link A. J., Graumann J., Tirrell D. A., Schuman E. M., Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2, 532–540 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishnan V., Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Jungmann R., Avendaño M. S., Woehrstein J. B., Dai M., Shih W. M., Yin P., Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M., The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 (2011). [DOI] [PubMed] [Google Scholar]
- 14.D. Sehnal, A. Rose, J. Koca, S. Burley, S. Velankar, Mol*: Towards a common library and tools for web molecular graphics, in Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data (MolVa ‘18) (Eurographics Association, 2018), pp. 23–33. [Google Scholar]
- 15.Biever A., Glock C., Tushev G., Ciirdaeva E., Dalmay T., Langer J. D., Schuman E. M., Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367, eaay4991 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Schneider T., Hung L.-H., Schreiner S., Starke S., Eckhof H., Rossbach O., Reich S., Medenbach J., Bindereif A., CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep. 6, 31313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris K., Stevens J., Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: Serial electron microscopy with reference to their biophysical characteristics. J. Neurosci. 9, 2982–2997 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dieterich D. C., Hodas J. J. L., Gouzer G., Shadrin I. Y., Ngo J. T., Triller A., Tirrell D. A., Schuman E. M., In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 13, 897–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jewett J. C., Sletten E. M., Bertozzi C. R., Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–3690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böger C., Hafner A.-S., Schlichthärle T., Strauss M. T., Malkusch S., Endesfelder U., Jungmann R., Schuman E. M., Heilemann M., Super-resolution imaging and estimation of protein copy numbers at single synapses with DNA-point accumulation for imaging in nanoscale topography. Neurophoton 6, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wierenga C. J., Walsh M. F., Turrigiano G. G., Temporal regulation of the expression locus of homeostatic plasticity. J. Neurophysiol. 96, 2127–2133 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Minerbi A., Kahana R., Goldfeld L., Kaufman M., Marom S., Ziv N. E., Long-term relationships between synaptic tenacity, synaptic remodeling, and network activity. PLOS Biol. 7, e1000136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govindarajan A., Israely I., Huang S.-Y., Tonegawa S., The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron 69, 132–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sajikumar S., Morris R. G. M., Korte M., Competition between recently potentiated synaptic inputs reveals a winner-take-all phase of synaptic tagging and capture. Proc. Natl. Acad. Sci. U.S.A. 111, 12217–12221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangaraju V., Lauterbach M., Schuman E. M., Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 176, 73–84.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Kang H., Schuman E. M., A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273, 1402–1406 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Holt C. E., Martin K. C., Schuman E. M., Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 26, 557–566 (2019). [DOI] [PubMed] [Google Scholar]
- 28.tom Dieck S., Kochen L., Hanus C., Heumüller M., Bartnik I., Nassim-Assir B., Merk K., Mosler T., Garg S., Bunse S., Tirrell D. A., Schuman E. M., Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aakalu G., Smith W. B., Nguyen N., Jiang C., Schuman E. M., Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30, 489–502 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Sharma A. K., Sormanni P., Ahmed N., Ciryam P., Friedrich U. A., Kramer G., O’Brien E. P., A chemical kinetic basis for measuring translation initiation and elongation rates from ribosome profiling data. PLOS Comput. Biol. 15, e1007070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reshetniak S., Ußling J.-E., Perego E., Rammner B., Schikorski T., Fornasiero E. F., Truckenbrodt S., Köster S., Rizzoli S. O., A comparative analysis of the mobility of 45 proteins in the synaptic bouton. EMBO J. 39, e104596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartori F., Hafner A.-S., Karimi A., Nold A., Fonkeu Y., Schuman E. M., Tchumatchenko T., Statistical laws of protein motion in neuronal dendritic trees. Cell Rep. 33, 108391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choquet D., Triller A., The role of receptor diffusion in the organization of the postsynaptic membrane. Nat. Rev. Neurosci. 4, 251–265 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Bloodgood B. L., Sabatini B. L., Neuronal activity regulates diffusion across the neck of dendritic spines. Science 310, 866–869 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Steward O., Schuman E. M., Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 24, 299–325 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Cajigas I. J., Tushev G., Will T. J., tom Dieck S., Fuerst N., Schuman E. M., The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tushev G., Glock C., Heumüller M., Biever A., Jovanovic M., Schuman E. M., Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98, 495–511.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Gray N. W., Weimer R. M., Bureau I., Svoboda K., Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLOS Biol. 4, e370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh W. C., Parajuli L. K., Zito K., Heterosynaptic structural plasticity on local dendritic segments of hippocampal CA1 neurons. Cell Rep. 10, 162–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Govindarajan A., Kelleher R. J., Tonegawa S., A clustered plasticity model of long-term memory engrams. Nat. Rev. Neurosci. 7, 575–583 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Endesfelder U., Malkusch S., Fricke F., Heilemann M., A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem. Cell Biol. 141, 629–638 (2014). [DOI] [PubMed] [Google Scholar]
- 42.A. Nold, C. Sun, M. Heilemann, E. Schuman, T. Tchumatchenko, Unbiased choice of global clustering parameters in single-molecule localization microscopy. bioRxiv 2021.02.22.432198 [Preprint]. 2021. 10.1101/2021.02.22.432198. [DOI] [PMC free article] [PubMed]
- 43.Dietz M. S., Heilemann M., Optical super-resolution microscopy unravels the molecular composition of functional protein complexes. Nanoscale 11, 17981–17991 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Dörrbaum A. R., Kochen L., Langer J. D., Schuman E. M., Local and global influences on protein turnover in neurons and glia. eLife 7, e34202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanus C., Geptin H., Tushev G., Garg S., Alvarez-Castelao B., Sambandan S., Kochen L., Hafner A.-S., Langer J. D., Schuman E. M., Unconventional secretory processing diversifies neuronal ion channel properties. eLife 5, e20609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14