Inhibition of LPA1/ROCK enhances IFN-I/III production in multiple cell types against virus infection.
Abstract
Growing evidence indicates the vital role of lipid metabolites in innate immunity. The lipid lysophosphatidic acid (LPA) concentrations are enhanced in patients upon HCV or SARS-CoV-2 infection, but the function of LPA and its receptors in innate immunity is largely unknown. Here, we found that viral infection promoted the G protein–coupled receptor LPA1 expression, and LPA restrained type I/III interferon production through LPA1. Mechanistically, LPA1 signaling activated ROCK1/2, which phosphorylated IRF3 Ser97 to suppress IRF3 activation. Targeting LPA1 or ROCK in macrophages, fibroblasts, epithelial cells, and LPA1 conditional KO mice promoted interferon-induced clearance of multiple viruses. LPA1 was colocalized with the receptor ACE2 in lung and intestine. Together with previous findings that LPA1 and ROCK1/2 promoted vascular leaking or lung fibrosis, we propose that the current available preclinical drugs targeting the LPA1-ROCK module might protect from SARS-CoV-2 or various virus infections in the intestine or lung.
INTRODUCTION
Virus invasion triggers highly active lipid metabolism in the host cells, and growing evidence indicates that lipid metabolites might regulate antiviral responses (1). Currently, the emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes a pandemic worldwide, and several reports have identified that plasma concentrations of the simplest lysophospholipid lysophosphatidic acid (LPA) and its precursors are enhanced in coronavirus disease 2019 (COVID-19) patients compared to healthy population, especially in fatal cases (2, 3). Furthermore, plasma LPA levels are also enhanced in chronic hepatitis C virus (HCV)–infected patients, which are decreased after antiviral therapy, indicating a positive correlation between plasma LPA levels and HCV virus loads (4). Except for being a possible biomarker, it is unclear whether LPA signaling manipulates the innate antiviral responses to control virus infection.
LPA has numerous biological activities through binding with its G protein–coupled receptors (GPCRs), including LPA1 to LPA6, and the receptor LPA1 shows the highest affinity with LPA. The ligand receptor LPA-LPA1 signaling in lung induces fibroblast chemotaxis and enhances vascular leaking, which leads to macromolecule infiltration to bronchium and accelerates pulmonary fibrosis (5). Therefore, much effort has been made to inhibit LPA1 to treat idiopathic pulmonary fibrosis, including the phase 2 preclinical drug BMS-986020 (6). Nevertheless, data of coronavirus infection, including SARS, Middle East respiratory syndrome, and the COVID-19 pandemic, have suggested that patients might suffer substantial fibrotic consequences following infection (7). It would be important to elucidate whether targeting the LPA-LPA1 signaling pathway could regulate the innate antiviral responses.
After recognizing DNA or RNA viruses, the pattern recognition receptors in innate immune cells, including cGAS and RIG-I–like receptors (RLRs), can recruit their adaptor proteins STING and MAVS for TBK1 activation and phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3), resulting in type I interferon (IFN-I) transcription (8). In addition, IRF3 can also mediate IFN-III (i.e., IFN-λ2 and IFN-λ3 in mice) production in gastrointestinal and respiratory epithelial cells (ECs) to eliminate viruses (9). Studies have suggested that SARS-CoV-2 infects the ECs of lung and small intestine (10), which triggers IFN-III production while inducing very low levels of IFN-I in human intestinal organoids (11). Compared to IFN-I, IFN-III is more potent in preventing viral dissemination without causing inflammation during viral infections in epithelium (12). Therefore, how to evoke secretion of IFN-III and IFN-I is critical for the host against viral infections.
Here, we have identified that the expression of LPA1 was up-regulated in response to multiple virus infections. LPA binding to LPA1 inhibited IRF3-mediated type I and III IFN production and impaired host defense to eliminate viral infection. Targeting LPA1 with the specific inhibitor Ki16425 or the preclinical drug BMS-986020, as well as conditionally knockout (cKO) Lpa1 in intestinal ECs (IECs), could enhance IFN-I/III levels and antiviral responses. Critically, LPA1 colocalized with the SARS-CoV-2 receptor ACE2 in lung alveolar cells, bronchium, and small intestine, which might enable LPA1 as an ideal drug target specifically in SARS-CoV-2–infected organs. Mechanistically, LPA1 signaling activated ROCK1/2, and ROCK1/2 directly phosphorylated IRF3 Ser97 to reduce IRF3 activation and IFN production. Furthermore, targeting ROCK promoted IFN-mediated antiviral responses. Together, this study has elucidated the LPA1-ROCK module as a potential drug target against multiple virus infections.
RESULTS
LPA-induced signaling represses IFN-I/III production and impairs virus clearance
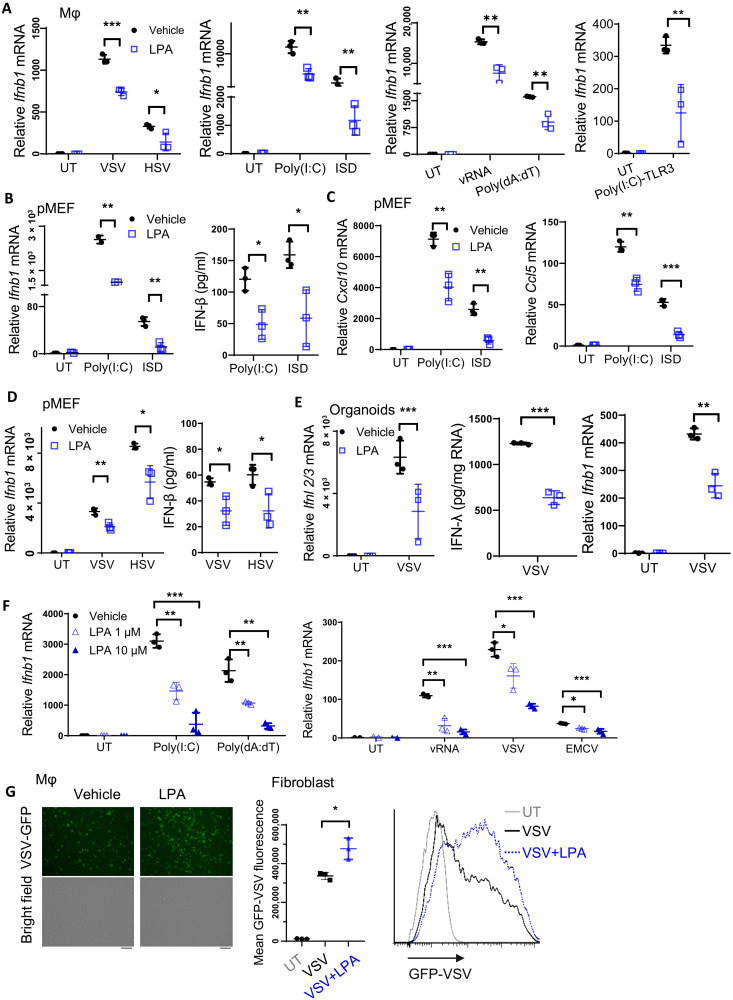
LPA displays dynamic changes in plasma of HCV-infected patients before and after receiving antiviral therapy, suggesting LPA as a biomarker to monitor HCV loads. In addition, plasma LPA concentrations are enhanced in COVID-19 patients (2, 3). We therefore asked whether LPA had any effect on antiviral responses. Various types of cells were pretreated with exogenous LPA, followed by infection with multiple viruses. In peritoneal macrophages (PEMs), LPA pretreatment inhibited Ifnb1 expression upon infection with the RNA virus vesicular stomatitis virus (VSV) and the DNA virus herpes simplex virus type 1 (HSV-1), as well as upon stimulation by transfection of polyinosine-polycytidylic acid [poly(I:C)] to trigger RIG-I/MDA5 activation or by ISD to trigger the cGAS-STING pathway (Fig. 1A). PEMs were transfected with the VSV genomic RNA (vRNA) for RIG-I activation or poly(dA:dT) for DNA sensor; alternatively, poly(I:C) was added directly into the culture medium, which could be internalized to the endosome to activate the Toll-like receptor 3 (TLR3) pathway. The expression levels of Ifnb1 (Fig. 1A) and the IFN-stimulated genes (ISGs), including Cxcl10 and Ccl5 (fig. S1A), were repressed, but the mRNA levels of Mx1 and Ifit1 were not affected (fig. S1B).
Fig. 1. LPA-induced signaling represses IFN-I/III production and impairs virus clearance.
(A) PEMs were pretreated with 10 μM LPA for 1 hour, followed by stimulation with VSV and HSV, or by transfection of poly(I:C), ISD, VSV genomic RNA (vRNA), and poly(dA:dT); alternatively, poly(I:C) was directly added into the culture medium to activate the TLR3 pathway for 4 or 6 hours. Ifnb1 mRNA level was determined by RT-qPCR. (B to D) pMEFs were pretreated with LPA and stimulated with poly(I:C) or ISD for 4 hours and VSV or HSV for 6 hours to check expression of IFN-β (B and D), Cxcl10, and Ccl5 (C). (E) LPA-pretreated intestinal organoids were stimulated with VSV for 5 hours to measure IFN-λ and Ifnb1 expression. IFN-λ secretion was normalized to the extracted total amount of RNA. (F) NIH-3T3 cells were pretreated with LPA for 1 hour and then stimulated with poly(I:C), poly(dA:dT), and vRNA for 4 hours, or VSV and EMCV for 6 hours to check Ifnb1 mRNA levels. (G) The amount of VSV-GFP was measured 12 hours after infection by microscopy in PEMs or by FACS analysis in NIH-3T3 cells that were pretreated with or without LPA. Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001 [unpaired Student’s t test in (A) to (E) and (G), one-way ANOVA followed by Tukey’s multiple comparisons test in (F)]. Data are from three (A to G) independent experiments (means ± SD) or are representative data (G).
As ECs play a crucial role against viruses in the mucosal sites, we next determined how LPA affected antiviral function in primary mouse embryo fibroblasts (pMEFs). Similar to the phenotype in macrophages, in poly(I:C) and ISD-treated pMEFs (Fig. 1, B and C) or in VSV- and HSV-infected pMEFs (Fig. 1D and fig. S1C), LPA suppressed IFN-β and ISG expression at mRNA or protein levels. Consistently, LPA exerted the inhibitory effects on IFN-β expression in the human cell lines HCT116 and human embryonic kidney (HEK) 293A, which are derived from intestine and kidney, respectively (fig. S1D).
IFN-III displays a predominant role to clear the virus in the mucosal surface, including small intestine (13). We prepared the organoids derived from small intestine and found that IFN-III was induced at high levels upon VSV infection (Fig. 1E). LPA treatment repressed Ifnl2/3 mRNA expression and secretion as well as Ifnb1 production in the intestinal organoids (Fig. 1E). Moreover, LPA exerted the dose-dependent suppressive effect on IFN expression (Fig. 1F) and ISG expression (fig. S1E) in response to various stimuli or infection with VSV and encephalomyocarditis virus (EMCV). In agreement with the suppressed IFN production, LPA treatment enhanced the amount of VSV–green fluorescent protein (GFP) in macrophages as measured under fluorescence microscopy or in fibroblasts by fluorescence-activated cell sorting (FACS) (Fig. 1G). Together, we have elucidated that the bioactive lipid LPA substantially represses both IFN-I and IFN-III production in different cell types to facilitate multiple virus infection.
Targeting the receptor LPA1 promotes production of IFNs and ISGs against various virus infections
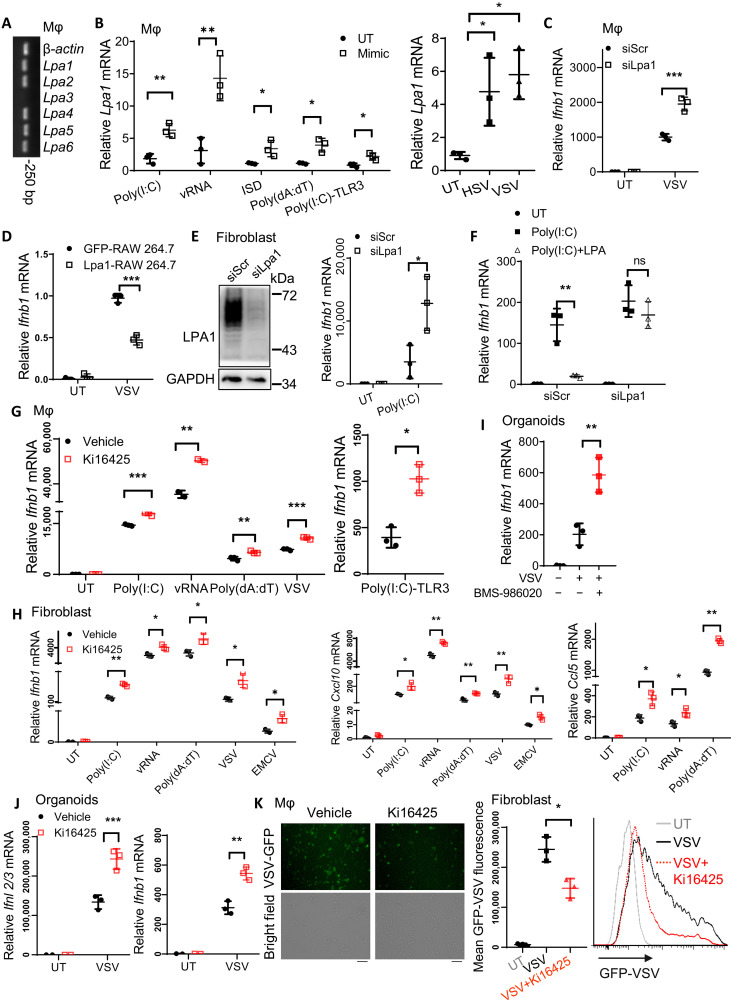
LPA signaling can be transduced through the different GPCRs including LPA1 to LPA6 in different cell types. We asked which LPA receptor was dominantly involved in the innate immunity against infections. Except for Lpa3 (Fig. 2A) and Lpa4 (fig. S2A), the other LPA receptors were expressed in PEMs and IECs. Only Lpa1 expression was consistently enhanced upon treatment with various stimuli, including virus mimics or different types of viruses (Fig. 2B). In contrast, expression of the other receptors remained unchanged or decreased (fig. S2B). Considering LPA1 having the highest affinity with LPA, we proposed a potential role of LPA1 in response to virus infection. We therefore silenced Lpa1 in macrophages (fig. S2C), which enhanced mRNA expression of Ifnb1 (Fig. 2C) and ISG secretion (fig. S2D) upon VSV infection or poly(I:C) treatment (fig. S2E). In RAW264.7 cells, which were generated to stably overexpress LPA1 (fig. S2F), Ifnb1 mRNA levels were reduced in response to VSV infection (Fig. 2D). In fibroblast cells, which silence Lpa1 expression with small interfering RNA (siRNA), Ifnb1 production was also enhanced upon poly(I:C) stimulation (Fig. 2E). LPA1 knockdown could abolish the inhibitory effect of LPA on Ifnb1 expression (Fig. 2F). This confirms that LPA is dependent on the receptor LPA1 to inhibit IFN production upon treatment with various infections or viral mimics.
Fig. 2. Targeting the receptor LPA1 promotes production of IFNs and ISGs against various virus infections.
(A and B) Lpa1-6 mRNA levels in resting PEMs (A) or in PEMs after stimulation with the indicated pathogen-associated molecular pattern mimics for 4 hours or viruses for 6 hours (B). (C and D) Ifnb1 mRNA levels in VSV-infected PEMs that were transfected with siRNA to knock down LPA1 (C) or in RAW264.7 cells overexpressing LPA1 (D). (E and F) LPA1 was knocked down in NIH-3T3 cells followed by transfection of poly(I:C) for 4 hours (E) or pretreated with LPA (5 μM in DMEM without FBS) for 1 hour followed by transfection of poly(I:C) to check Ifnb1 mRNA levels (F). (G to J) Macrophages (G), NIH-3T3 cells (H), or the intestinal organoids (I and J) were pretreated with 10 μM Ki16425 or BMS-986020 followed by the indicated stimulation for 4 hours or VSV and EMCV infection for 6 hours to check Ifnb1, Ifnl2/3, Cxcl10, and Ccl5 mRNA levels. (K) The amount of VSV-GFP was measured 12 hours after infection in Ki16425-pretreated PEMs by microscopy or in NIH-3T3 cells by FACS analysis. Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001 [unpaired Student’s t test in (B) (left), (C) to (E), and (G) to (K); one-way ANOVA followed by Tukey’s multiple comparisons test in (B) (right); two-way ANOVA followed by Sidak’s multiple comparisons test in (F)]. Data are from three (B to K) independent experiments (means ± SD) or are representative data (A, E, and K).
Because GPCR is one of the largest and promising candidates for drug development, we next tested the potential application of the LPA1/3 inhibitor Ki16425 against virus infection. Since LPA3 expression was undetectable in macrophages and ECs, Ki16425 could selectively act on LPA1 in these two cell types. In macrophages, Ki16425 treatment enhanced expression of Ifnb1 (Fig. 2G) and the ISGs including Cxcl10 or Ccl5 (fig. S2G) after treatment with various stimuli to activate cytosolic DNA and RNA sensors as well as the TLR3 pathway. Ki16425 treatment also promoted Ifnb1, Cxcl10, or Ccl5 expression in fibroblast cells (Fig. 2H) or in the human cell lines, including HCT116 and HEK293A cells (fig. S2H). In intestinal organoids, inhibition of LPA1 by the preclinical drug BMS-986020 (Fig. 2I) or by Ki16425 (Fig. 2J) could substantially elevate IFN-III or IFN-I expression. In agreement with this, Ki16425 treatment enhanced virus clearance as shown by the reduced mean fluorescence of VSV-GFP (Fig. 2K). Together, we have identified that targeting LPA1 by the inhibitor Ki16425 or the preclinical drug BMS-986020 can enhance IFN-I and IFN-III expression in multiple cell types as well as in intestinal organoids against virus infection.
LPA1 deficiency in ECs protects the host against various virus infections in vivo
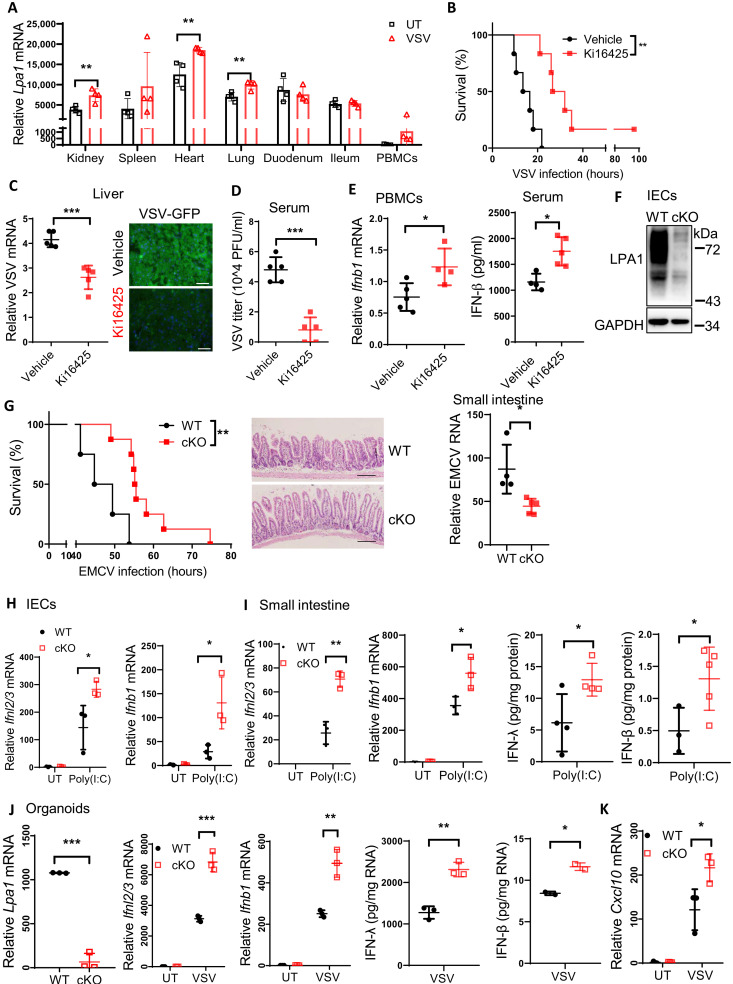
Because half amount of plasma LPA is generated from lysophospholipids by the lysophospholipase autotaxin (ATX) (14), we pretreated mice with the ATX inhibitor PF-8380 to reduce LPA concentrations. PF-8380 treatment promoted IFN-β1 secretion in serum of VSV-infected mice (fig. S3A). We also noted that Lpa1 expression was induced in multiple organs, tissues, and peripheral blood mononuclear cells (PBMCs) upon VSV infection in vivo (Fig. 3A); therefore, the LPA1 inhibitor Ki16425 was applied to test the in vivo protection. Ki16425 treatment increased the survival rates (Fig. 3B) and attenuated the amount of VSV mRNA and the fluorescence intensity of GFP-VSV in liver (Fig. 3C) as well as VSV titers in serum (Fig. 3D). Consistently, Ki16425 treatment enhanced Ifnb1 mRNA expression in PBMCs and serum IFN-β concentrations (Fig. 3E). These data suggest that the LPA1 inhibitor has the systematic benefit against virus infection.
Fig. 3. LPA1 deficiency in ECs protects the host against various virus infections in vivo.
(A) Lpa1 mRNA levels in kidney, spleen, heart, lung, duodenum, ileum, and PBMCs from mice infected with or without VSV for 18 hours (n = 4). (B to E) WT mice were pretreated with or without Ki16425 followed by VSV-GFP infection at lethal doses to measure the survival rates (B) (n = 6), the amount of VSV in liver (C) (scale bars, 50 μm) or in serum (D), and IFN-β levels in PBMCs or in serum (E). (F and G) LPA1 expression in IECs from WT and cKO mice (F). WT and cKO mice were intraperitoneally infected with EMCV (WT, n = 4; cKO, n = 8) to measure the survival rates, H&E staining, and EMCV mRNA levels of small intestines. Scale bars, 100 μm (G). (H and I) WT and cKO mice were injected with poly(I:C) for 2 hours, and IECs (H) or small intestine (I) were isolated to check IFN-β and IFN-λ expression. (J and K) The intestine organoids from WT and cKO mice were infected with VSV for 4 hours to check IFN-β, IFN-λ, and Cxcl10 expression. *P < 0.05, **P < 0.01, ***P < 0.001 [unpaired Student’s t test in (A), (C) to (E), and (G) to (K); log-rank test in (B) and (G)]. Each dot (A to E and G to I) represents an individual mouse. Data are from three (J and K) independent experiments (means ± SD) or are representative of three (A to I) independent experiments.
To determine LPA1 function in vivo, we generated Lpa1fl/fl mice with the two loxP sites flanking exon 3 (fig. S3B). Although LPA1 in macrophages could regulate type I IFN production, we noted that Lpa1 expression levels were much higher in IECs and fibroblast cells than those in PEMs (fig. S3C), and Lpa1 mRNA levels in IECs were enhanced by VSV infection (fig. S3D). To examine the potential role of LPA1 in ECs, Lpa1fl/fl mice were crossbred with the Villin-cre mice to specifically deplete Lpa1 in IECs (Fig. 3F). Although LPA1 null mice are reported with the impaired intestine integrity (15), Lpa1 cKO IECs expressed normal mRNA levels of tight junction proteins including E-cadherin, Claudin-2, and Claudin-4 (fig. S3E). The structure of small intestine remained intact in Lpa1 cKO mice, and the cKO organoids derived from crypts also developed normally as those from wild-type (WT) mice (fig. S3F).
We applied EMCV to generate a mouse model with intestinal infection. Lpa1 cKO mice improved the survival rates with longer villi, and the intestine structure was more intact with the decreased amount of EMCV (Fig. 3G) compared to that of WT mice. After challenge with VSV, Lpa1 cKO mice also prolonged the survival rates (fig. S3G). This suggests that LPA1 deficiency could protect the host from different viral infections in vivo.
Next, WT or Lpa1 cKO mice were injected with poly(I:C), and IECs were isolated to determine how LPA1 affected IFN expression in vivo. Lpa1 deficiency enhanced the mRNA levels of IFN-I, IFN-III (Fig. 3H), and ISGs (fig. S3H) in IECs. In addition, the intestine tissue isolated from Lpa1 cKO mice expressed higher levels of IFN-I, IFN-III (Fig. 3I), and Cxcl10 (fig. S3I). Furthermore, the organoids from WT or Lpa1 cKO mice were exposed to VSV infection. cKO organoids expressed higher levels of IFN-I, IFN-III (Fig. 3J), and Cxcl10 (Fig. 3K). Together, targeting the LPA-LPA1 axis genetically in IECs or by the LPA1 inhibitor could boost the production of IFN-I/III and ISG to attenuate virus infection.
Targeting LPA1 facilitates clearance of SARS-CoV-2 and ZIKV
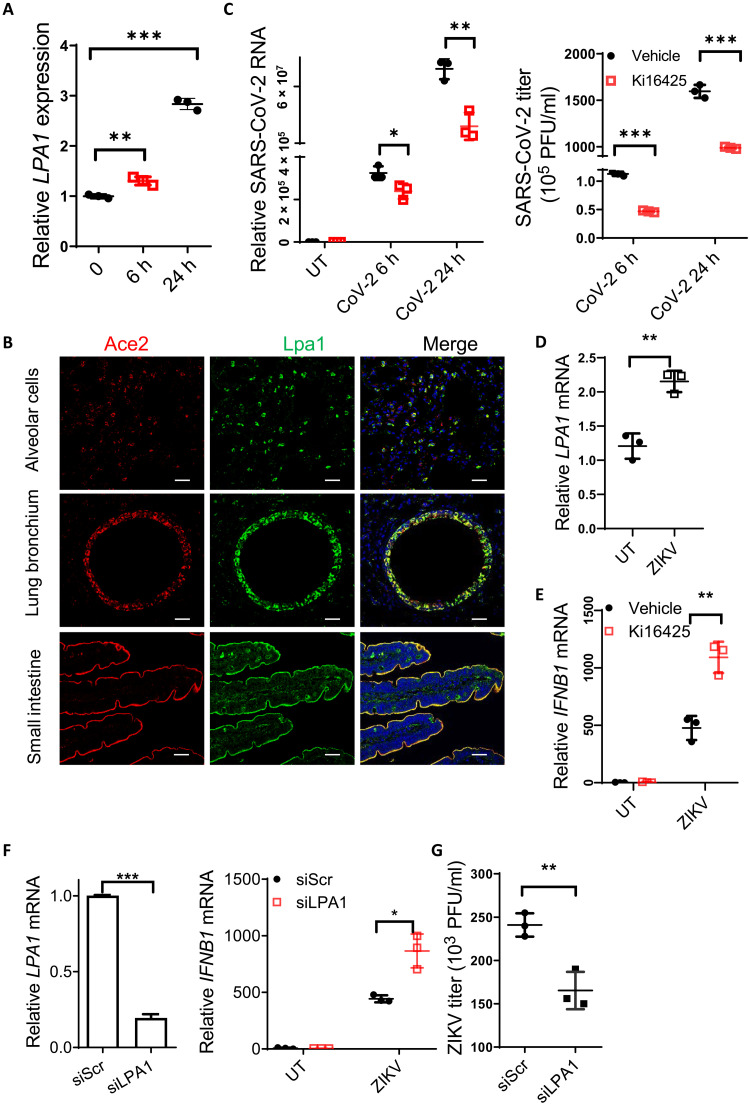
Clinical reports have suggested that SARS-CoV-2 mainly infects the epithelial system via the receptor Ace2, which is highly expressed in lung, intestine, and kidney (16), resulting in function failure of multiple organs. Recent studies have reported that plasma LPA levels were enhanced in COVID-19 patients (2, 3). We noted that expression of the receptor LPA1 was induced by SARS-CoV-2 infection in vitro (Fig. 4A). Lpa1 displayed similar expression pattern with Ace2, i.e., expressed at high levels in small intestine, kidney, heart, and lung (fig. S4A). We further provided the first evidence by immunofluorescence staining showing the colocalization of LPA1 with ACE2 in the mucosal epithelium, including lung alveolar cells, bronchium, and small intestine in mice (Fig. 4B) but not in kidney proximal tubular cells (fig. S4B). These data indicate that the LPA1 inhibitor could exhibit organ specificity in some major organs infected by SARS-CoV-2. Because we observed that targeting LPA1 could induce much higher levels of IFN-III production especially in ECs, we next determined whether the LPA1 inhibitors could block SARS-CoV-2 replication. Ki16425-treated Calu-3 cells decreased the amount of SARS-CoV-2 inside the cells (left) as well as SARS-CoV-2 virions released to the culture medium (right) (Fig. 4C). A549 cells overexpressing hACE2 were generated and stimulated with SARS-CoV-2 pseudovirus to examine IFN-III expression. Addition of LPA could inhibit IFNL1 mRNA levels (fig. S4C), while treatment with the LPA1 inhibitor Ki16425 or BMS-986020 enhanced IFN-III expression (fig. S4D). Ki16425 also promoted IFN expression in lungs of the K18-hACE2 transgenic mice upon SARS-CoV-2 pseudovirus infection (fig. S4E). This suggests that targeting LPA1 protects against SARS-CoV-2 infection by enhancing IFN-mediated antiviral responses.
Fig. 4. Targeting LPA1 facilitates clearance of SARS-CoV-2 and ZIKV.
(A) Calu-3 cells were infected with SARS-CoV-2 to assess LPA1 mRNA levels. (B) Immunostaining of Ace2 (red), Lpa1 (green), and 4′,6-diamidino-2-phenylindole (DAPI) (blue) in mouse lung alveolar cells, bronchium, and small intestine. Scale bars, 20 μm. (C) Calu-3 cells were pretreated with Ki16425 followed by SARS-CoV-2 infection to check the amount of SARS-CoV-2 in cells (left) or in the culture medium (right). (D) LPA1 mRNA levels in ZIKV-infected A549 cells. (E to G) A549 cells were pretreated with Ki16425 (E) or knocked down LPA1 (F), followed by ZIKV infection for 24 hours to check IFNB1 mRNA levels (E and F) or ZIKV titers in the culture medium (G). *P < 0.05, **P < 0.01, ***P < 0.001 [unpaired Student’s t test in (C) and (D) to (G); one-way ANOVA followed by Tukey’s multiple comparisons test in (A)]. Data are from three (A and C to G) independent experiments (means ± SD) or are representative of three independent experiments (B).
Considering that IFN-III is critical to prevent Zika virus (ZIKV) infection in human placental trophoblasts (17), and IFN-I can prevent ZIKV infection in adult mice (18), we next tested the function of LPA1 signaling in response to ZIKV infection. Upon ZIKV infection, LPA1 expression was increased in A549 cells (Fig. 4D). Targeting LPA1 in A549 cells via the inhibitor Ki16425 (Fig. 4E) or silencing LPA1 expression (Fig. 4F) could enhance IFNB1 expression in response to ZIKV infection. Consistently, ZIKV titers were reduced as measured by the plaque assay after silencing LPA1 (Fig. 4G and fig. S4F). Together, inhibition of LPA-LPA1 signaling can boost IFN-I/III production against infections by SARS-CoV-2 and ZIKV.
LPA-LPA1 signaling disrupts IRF3 activation
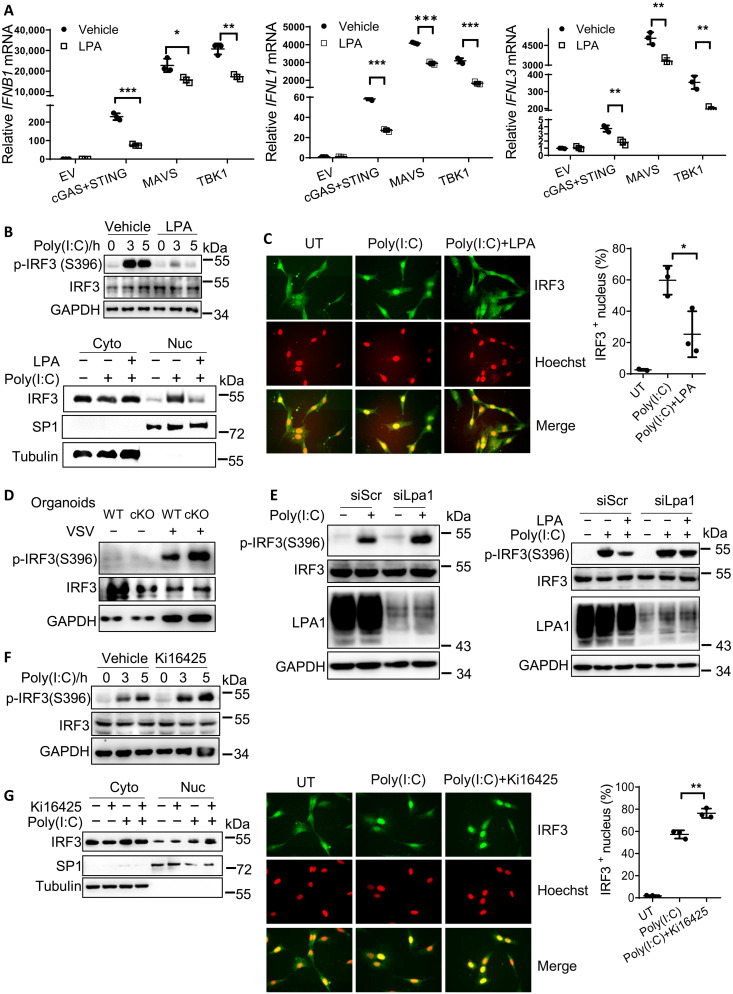
To clarify how LPA regulated IFN expression, HEK293A cells were transfected with the plasmids expressing cGAS and STING, MAVS, and TBK1, which could turn on the transcription of IFN-I and IFN-III. Addition of LPA suppressed the signaling molecule–induced IFN-I (i.e., IFNB1) and IFN-III (i.e., IFNL1/3) expression (Fig. 5A). This indicates that LPA1 signaling should affect the signaling molecule downstream of TBK1 in both cGAS-STING and RIG-I/MAVS pathways. Although both IRF1 and IRF3 are responsible for IFN-III transcription upon infection, IRF1 is dispensable for IFN-I expression at early phase of infection (19). We therefore mainly asked whether LPA reduced IRF3 activation to block both IFN-I/III productions. IRF3 activation can be measured by IRF3 phosphorylation at Ser396 (p-IRF3 S396), which then forms dimerization to enter the nucleus. LPA treatment repressed p-IRF3 S396 levels as well as IRF3 translocation to the nucleus by Western blotting (Fig. 5B) or by immunofluorescence staining (Fig. 5C) in response to poly(I:C)-induced RIG-I/MDA5 activation. Consistently, LPA1 deficiency in organoids, fibroblast cells, and PEM enhanced p-IRF3 S396 levels upon various stimulations (Fig. 5, D and E, left, and fig. S5A). Furthermore, LPA-induced suppression of p-IRF3 S396 was abrogated once Lpa1 was silenced (Fig. 5E, right), proving that LPA inhibited IRF3 activation through the receptor LPA1. Consistently, when LPA1 was targeted by the inhibitor Ki16425, p-IRF3 S396 levels and the amount of IRF3 into the nucleus were increased (Fig. 5, F and G) upon poly(I:C) transfection. Together, LPA-LPA1 signaling suppresses IRF3 phosphorylation and activation to block IFN-I and IFN-III production.
Fig. 5. LPA-LPA1 signaling disrupts IRF3 activation.
(A) HEK293A cells were transfected with plasmids expressing empty vector, cGAS + STING, MAVS, and TBK1 for 24 hours, followed by 10 μM LPA treatment for 4 hours, to check IFNB1, IFNL1, and IFNL3 mRNA levels. (B and C) NIH-3T3 cells were pretreated with LPA and stimulated with poly(I:C) to check p-IRF3 and total IRF3 levels, or the amount of IRF3 in the nucleus by Western blotting. (B) Specificity protein 1 (SP1) and α-tubulin were used to indicate the nucleus and cytoplasm. Alternatively, the amount of IRF3 in the nucleus was analyzed by immunofluorescence microscopy. (C) IRF3 in green and Hoechst in red; >100 cells were measured. (D and E) WT and cKO organoids were infected with VSV (D) or LPA1 was knocked down in NIH-3T3 cells, pretreated without (E) (left) or with (E) (right) LPA followed by transfection of poly(I:C) to measure p-IRF3 and total IRF3 levels. (F and G) NIH-3T3 cells were treated with or without 10 μM Ki16425 for 1 hour, followed by transfection of poly(I:C) to measure p-IRF3 levels (F) or the amount of IRF3 in the nucleus (G) by Western blotting or immunofluorescence staining. *P < 0.05, **P < 0.01, ***P < 0.001 [unpaired Student’s t test in (A), (C), and (G)]. Data are from three (A, C, and G) independent experiments (means ± SD) or are representative of three independent experiments (B to G).
G12/13-ROCK1/2 is downstream of LPA1 to inhibit IRF3 activation
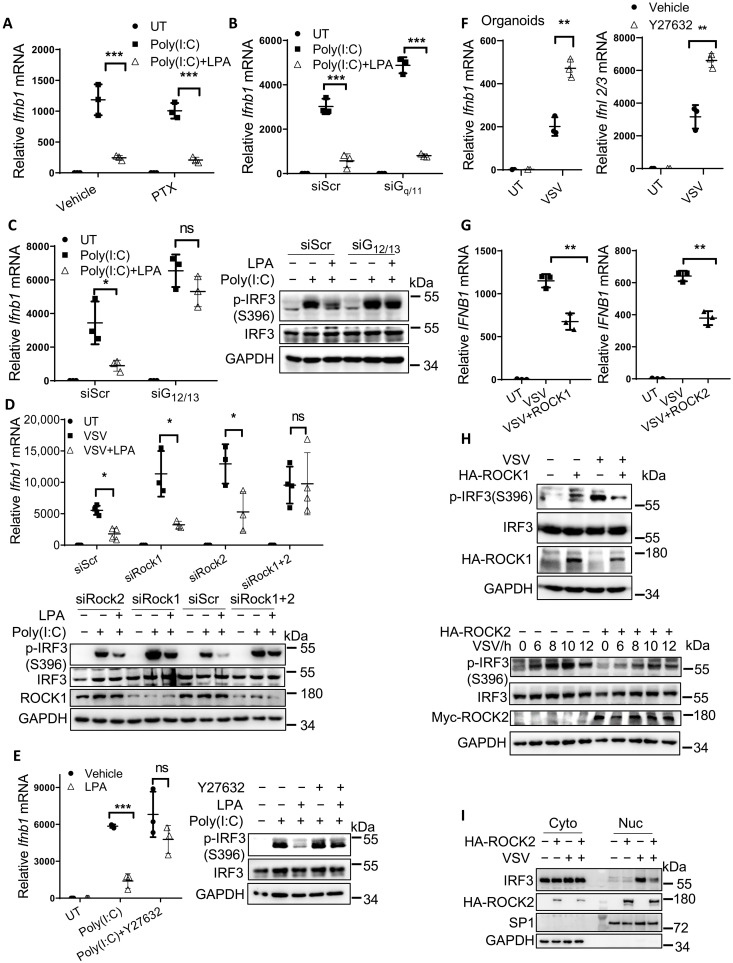
We next dissected LPA1-IRF3 signaling in detail. Once activated, GPCRs activate several heterotrimeric G proteins to transduce intracellular signaling. To determine whether Gα proteins were involved in IFN regulation by LPA, we used pertussis toxin (PTX) to block Gi/o activation (Fig. 6A and fig. S6A) or used siRNA-transfected NIH-3T3 cells to silence Gq/11 expression (Fig. 6B and fig. S6B), which unexpectedly failed to influence LPA-mediated inhibition on Ifnb1 production upon poly(I:C) transfection. In contrast, knockdown of G12/13 (fig. S6C) could abrogate LPA-mediated inhibition on Ifnb1 expression and p-IRF3 S396 levels (Fig. 6C), indicating that G12/13 is downstream of LPA1 to repress IRF3 activation.
Fig. 6. G12/13-ROCK1/2 is downstream of LPA1 to inhibit IRF3 activation.
(A to E) NIH-3T3 cells were transfected with siRNAs to knock down Gq/11 (B), G12/13 (C), and Rock1 and Rock2 (D) or pretreated with the inhibitor PTX (100 ng/ml) (A) and Y27632 (10 μM) (E), with or without 10 μM LPA treatment for 1 hour, followed by stimulation with poly(I:C) or VSV. The expression levels of Ifnb1, p-IRF3, and total IRF3 were examined. (F) The intestine organoids were pretreated with Y27632 and then infected with VSV for 5 hours to check the mRNA levels of Ifnb1 and Ifnl2/3. (G to I) HEK293T cells were transfected with plasmids expressing ROCK1 and ROCK2 stimulated with VSV to check the levels of IFNB1 mRNA (G), p-IRF3, total IRF3 (H), and the amount of IRF3 in the nucleus (I). *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant (P > 0.05) [unpaired Student’s t test in (F) and (G); two-way ANOVA followed by Sidak’s multiple comparisons test in (A) to (E)]. Data are from three (A to G) independent experiments (means ± SD) or are representative of three independent experiments (H and I).
Previous research suggested that sphingosine-1-phosphate (S1P) and LPA could activate yes-associated protein (YAP) through the G12/13-coupled GPCRs in HEK293A to facilitate cell migration. Because YAP was also reported to suppress IFN-I production (20), we tested whether YAP was the downstream effector upon LPA treatment. As noted, after silencing YAP in NIH-3T3 cells, LPA still repressed Ifnb1 production and p-IRF3 S396 levels (fig. S6D), indicating that other molecules might mediate the inhibition of LPA1-G12/13 signaling on IFN production.
Rho guanosine triphosphate (GTPase) was reported downstream of G12/13, and the autoinhibition of ROCK1 or ROCK2 is then released by binding to Rho GTPase (21). After we silenced both ROCK1 and ROCK2 (fig. S6E), or used the ROCK inhibitor Y27632, LPA treatment no longer repressed Ifnb1 production and p-IRF3 S396 levels (Fig. 6, D and E). In contrast, by knocking down ROCK1 alone or ROCK2 alone, LPA treatment still showed its inhibitory effect (Fig. 6D), suggesting that ROCK1 and ROCK2 are both needed in LPA1 signaling.
Current understanding has revealed that ROCK1/2 are mainly involved in cell cytoskeleton change, including cell adhesion, migration, and cytokinesis (21). However, the function of ROCK1/2 in antiviral responses remains unknown. We observed that silencing ROCK1 or ROCK2 alone could enhance Ifnb1 expression (Fig. 6D). Furthermore, the ROCK inhibitor Y27632 increased both IFN-I and IFN-III production in VSV-infected intestine organoids (Fig. 6F). Overexpression of ROCK1 and ROCK2 in HEK293T cells could suppress IFNB1 expression upon VSV infection (Fig. 6G). Mechanistically, p-IRF3 S396 (Fig. 6H) and IRF3 translocation into the nucleus (Fig. 6I) were repressed by overexpression of ROCK1 and ROCK2. Consistent with the phenotype from different cell types, mice pretreated with the ROCK inhibitor Y27632 displayed better survival rates after VSV infection (fig. S6F). Hence, we have elucidated that ROCK1/2 are downstream of LPA1 signaling to suppress IFN-I/III production and antiviral responses.
ROCK directly binds and phosphorylates IRF3 at Ser97 to inhibit IFN production
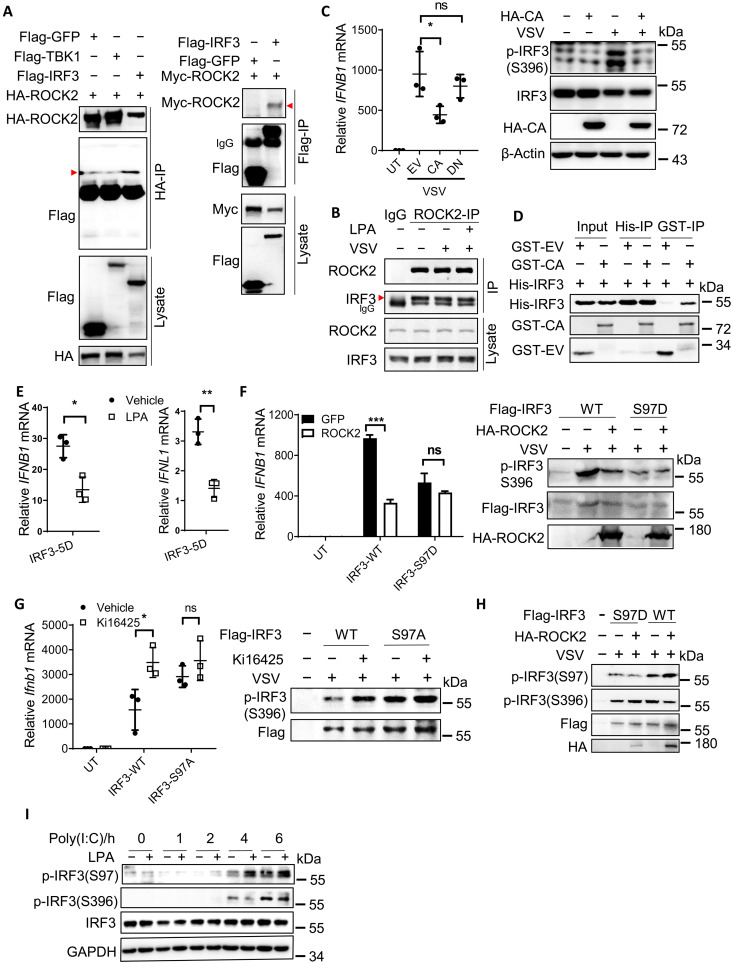
To dissect how ROCK kinases inhibit IRF3 activation, we overexpressed Flag-tagged TBK1 or IRF3 together with ROCK2 in HEK293T cells. Hemagglutinin (HA)–tagged ROCK2 coimmunoprecipitated with Flag-tagged IRF3 but not TBK1, and Flag-tagged IRF3 could also coimmunoprecipitate with Myc-tagged ROCK2 (Fig. 7A). Furthermore, anti-ROCK2 antibody precipitated endogenous IRF3 in NIH-3T3 cells, and the IRF3-ROCK2 binding was not affected by LPA treatment or VSV infection (Fig. 7B). In the resting state, the ROCK kinase domain (i.e., named CA) can be inhibited by the Rho-binding domain and PH domain (i.e., named DN) (fig. S7A) (21). Because the kinase activity of ROCKs was required for LPA1 signaling (Fig. 6E), we therefore overexpressed the truncated versions of ROCK2. The ROCK2 CA domain, but not the DN fragment, suppressed IFNB1 expression and p-IRF3 S396 levels in VSV-infected HEK293T cells (Fig. 7C). Next, His-tagged IRF3 and glutathione S-transferase (GST)–tagged ROCK2 CA domain were purified and incubated together in vitro. His-tagged IRF3 could pull down the GST-tagged ROCK2 CA domain and vice versa (Fig. 7D), suggesting a direct interaction between the ROCK kinase domain and IRF3.
Fig. 7. ROCK directly binds and phosphorylates IRF3 at Ser97 to inhibit IFN production.
(A) HEK293T cells were transfected with HA-ROCK2 (left) and Myc-ROCK2 (right), along with Flag-tagged GFP or TBK1 or IRF3, followed by immunoprecipitation (IP) and immunoblotting using anti-HA, anti-Flag, or anti-Myc antibodies. (B) NIH-3T3 cells were pretreated with or without LPA and infected with VSV followed by immunoprecipitation using anti-ROCK2 antibody to assess its endogenous protein binding with IRF3. (C) HEK293T cells were transfected with HA-ROCK2-CA or DN fragments followed by VSV infection to check the expression levels of IFNB1, p-IRF3, and total IRF3. (D) The GST-ROCK2 CA kinase domain and His-IRF3 proteins were purified and incubated together, followed by a pull-down assay using glutathione Sepharose or Ni-NTA agarose. (E) HEK293A cells were transfected with the plasmid expressing IRF3-5D and treated with or without LPA to check IFNB1 and IFNL1 mRNA levels. (F and H) HEK293T cells were transfected with HA-ROCK2, together with Flag-tagged IRF3 or the S97D mutant, followed by VSV stimulation to assess the expression levels of IFNB1, p-IRF3, and total IRF3 (F), as well as p-IRF3 S97 versus S396 (H). (G) NIH-3T3 cells stably expressing Flag-tagged IRF3 or the S97A mutant were generated and treated with or without Ki16425, followed by VSV infection to check the expression levels of IFNB1 and p-IRF3. (I) IRF3-knockout HCT116 cells were transfected with IRF3 and treated with LPA, followed by stimulation with poly(I:C) to check the expression of p-IRF3 S97. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant (P > 0.05) [unpaired Student’s t test in (E); one-way ANOVA followed by Tukey’s multiple comparisons test in (C); two-way ANOVA followed by Sidak’s multiple comparisons test in (F) and (G)]. Data are from three (C, E, and G) independent experiments (means ± SD) or are representative of three independent experiments (A to D, F, H, and I).
As a serine/threonine kinase, could ROCK2 phosphorylate IRF3 to perturb IRF3 activation? As noted, LPA treatment even inhibited the constitutively active IRF3-5D–induced IFN-I/III production in HEK293A cells (Fig. 7E); we therefore hypothesized that ROCK1/2 might phosphorylate some serine/threonine sites that inhibit IRF3 activation. On the basis of others’ findings, phosphorylation of S82, T253, S336/S339, and S97 could negatively regulate IRF3 activation (22). HEK293T cells were then transfected with the plasmids expressing these IRF3 mutants followed by VSV infection. Only the mutants expressing S97A (mimicking the dephosphorylation state) and S97D (mimicking the phosphorylation state) were involved in ROCK2-induced inhibition on IFNB1 expression (Fig. 7F and fig. S7B). For example, overexpression of ROCK2 inhibited p-IRF3 S396 levels in WT cells but failed to further reduce p-IRF3 S396 levels in IRF3-S97D–expressing cells (Fig. 7F) upon VSV stimulation. Moreover, S97A could enhance Ifnb1 production and p-IRF3 S396 levels compared to the vector control cells (Fig. 7G), while Ki16425 treatment failed to further promote Ifnb1 production and p-IRF3 S396 levels in S97A-expressing cells (Fig. 7G). Together, LPA1 and the downstream ROCK1/2 suppress IRF3 activation via phosphorylation of IRF3 Ser97.
We next examined whether IRF3 Ser97 could be phosphorylated by ROCK1/2 activated by LPA1 signaling. WT and S97D IRF3 were overexpressed together with or without ROCK2 followed by VSV infection, and cells were prepared to detect the phosphorylation levels of IRF3 Ser97 as in a previous report (22). ROCK2 overexpression could enhance p-IRF3 S97 levels and reduce IRF3 S396 levels in cells expressing WT IRF3 (Fig. 7H, lane 5 versus lane 4), but not in IRF3 S97D–expressing cells. Furthermore, LPA treatment elevated p-IRF3 S97 levels in HCT116 cells upon poly(I:C) stimulation (Fig. 7I). Together, upon LPA treatment, ROCK1/2 phosphorylate the inhibitory site of IRF3 at Ser97 to suppress IRF3 activation (fig. S7C).
DISCUSSION
As the simplest glycerophospholipid, LPA is distributed in many organs, including lung and intestine (23). LPA is suggested to act as an inflammation mediator in airway epithelium (24). As the LPA receptor, LPA1 is highly expressed in vascular, lung, intestine, heart, and brain. Coevolved with the sophisticated development of vascular, nervous, and immune system from invertebrates to vertebrates (25), LPA1 can promote neuroblast differentiation and vascular endothelial permeability. However, LPA1 function remains unclear in the immune system. The intronic polymorphisms of LPA1 (rs10980684) alter IFN-α activity in systemic lupus erythematosus (SLE) patients who display high concentrations of autoantibody to RNA binding proteins (26), but no data are available about how LPA1 regulates IFN production. Here, our study has demonstrated that LPA-LPA1 signaling represses IFN-I and IFN-III production and reduces antiviral defense in various cell types. Upon binding LPA, LPA1 activates G12/13 and is dependent on ROCK1/2 to phosphorylate IRF3 at Ser97, resulting in the reduced IRF3 activation in macrophages and ECs (fig. S7C model). Targeting LPA1 via its specific inhibitors therefore enhances IFN production.
LPA is synthesized by the secreted enzyme ATX, and we found that inhibition of ATX could also promote IFN production in vivo (fig. S3A). In various inflammatory autoimmune diseases, including idiopathic pulmonary fibrosis, rheumatoid arthritis, and multiple sclerosis, the expression levels of LPA and ATX in plasma are increased (27). We hypothesize that the elevated LPA and LPA1 could prevent excessive production of IFN in several types of cells, which help to restrain the development of autoimmune diseases; on the other hand, LPA can act as a growth factor to contribute to tissue repair after viral infection. These hypotheses need further investigation.
Regarding the SARS or SARS-CoV-2 pandemic and devastating damages to severe patients, researchers are searching for effective drugs or vaccines (28). The bioactive lipids, including prostaglandin E2 and S1P, recently attract great attention on treating SARS-CoV-2 infection because targeting their receptors or biosynthetic enzymes could regulate antiviral or inflammatory responses (29). In line with this, we recommend more attention to the bioactive lipid LPA because the LPA1-ROCK module acts as the ideal drug targets based on the following reasons: (i) LPA is reported to be elevated in COVID-19 patients, especially in the fatal cases (2, 3). We found that LPA1 colocalizes with ACE2 in the lung and intestine epithelium. As the downstream effector of LPA1, ROCK1/2 expression is also elevated in lungs of COVID-19 patients (30). Furthermore, ROCK1/2 are suggested to promote influenza virus internalization (31, 32). (ii) Previous studies suggest that LPA treatment enhances endothelial permeability and vascular leakage via ROCK1/2 (33), and the enhanced expression of ROCK1/2 is related with the pathological intussusceptive angiogenesis due to the endothelial injury (30). LPA1 or ROCK1/2 deficiency could reduce vascular leakage and attenuate fibrosis in experimental mouse lung fibrosis (5, 34). Because clinical studies have identified lung fibrosis in some SARS-CoV– or SARS-CoV-2–infected patients (7), targeting LPA1 or ROCK1/2 might have the second protection by limiting vascular leakage or pulmonary fibrosis in severe COVID-19 patients. Together, we propose that the LPA1 or ROCK1/2 inhibitors including the preclinical drugs could provide multiple protection to treat severe viral infections.
Mechanistically, this study has revealed that ROCK1/2 directly bind and phosphorylate IRF3 Ser97 to repress IRF3 activation. As a critical inhibitory site, Ser97 phosphorylation is erased by the phosphatase PTEN in response to virus infection to facilitate IRF3 translocation to the nucleus (22). Here, we have identified ROCK1/2 as the potential writer for Ser97 phosphorylation, resulting in the indispensable role by ROCK1/2 to inhibit IFN-I/III expression. The classical role of ROCKs is to phosphorylate a series of substrates for manipulating actin filament dynamics contributing to cell migration (21). This study has therefore broadened our understanding of ROCK1/2 function in innate immunity against IFN-related infections or possible autoimmune diseases.
In conclusion, the bioactive lipid LPA binding the GPCR receptor LPA1 can activate ROCK1/2 kinases to phosphorylate the inhibitory site Ser97 in IRF3, resulting in the suppressed IFN-I/III production and viral clearance. This finding provides potential drug targets to enhance antivirus responses, especially for treating severe virus infections.
MATERIALS AND METHODS
Mice
WT male mice were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences (CAS). K18-hACE2 mice, which express the human ACE2 protein under the K18 promoter, were purchased from GemPharmatech Co. Ltd. The Villin-cre mice were provided by J. Qin. The third exon of Lpa1 was flanked with two loxP sites to generate Lpa1fl/fl mice. All mice were on the C57BL/6 background and bred under specific pathogen–free conditions, and animal studies (protocol no. IBCB0057) were approved by the Animal Care Facility of Shanghai Institute of Biochemistry and Cell Biology (SIBCB), CAS. The primers were designed for genotyping as follows: LoxP1-F, 5′-GGGTCATGCTAACCACAGGAA-3′; LoxP1-R, 5′-AAAAGGTCTCTGCTTGGTGG-3′; LoxP2-F, 5′- ATATGCTAAATATTGGTCTCTGTGC-3′; LoxP2-R, 5′-TGAAGCAAAGTCCTAAGAGTGAGA-3′.
Isolation of PEM and pMEFs
PEMs were isolated from peritoneal cavity of mice intraperitoneally injected with 3 ml of 3% Brewer thioglycollate medium. To prepare pMEFs, 13.5-day embryos were isolated from euthanized pregnant female mice that were previously soaked with 70% ethanol. The head above the eyes, limbs, and liver were removed and chopped up in phosphate-buffered saline (PBS). The pieces were digested by adding equivalent 0.25% trypsin-EDTA at 37°C for 3 min. After pipetting up and down for several times, digestion was stopped by adding equivalent fetal bovine serum (FBS)–containing medium and cell suspension was seeded for 12 hours in the incubator. Cells were digested for freezing or further experiment.
Cells and reagents
pMEFs, PEMs, HCT116, Calu-3, HEK293A, HEK293T, and NIH-3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, penicillin-streptomycin (100 U/ml), and 2 mM l-glutamine. For LPA treatment, cells were starved in Dulbecco’s modified Eagle’s medium (DMEM) containing no or 1% FBS for at least 2 hours.
Virus infection and poly(I:C) injection in vivo
For inhibitor treatment, Y27632 (5 mg/kg per day), Ki16425 (20 mg/kg per day), or PF-8380 (15 mg/kg per day) was intraperitoneally injected to 6- to 8-week-old mice for 4 days before virus infection. Mice were infected with VSV intraperitoneally [lethal dose at 2 × 105 plaque-forming units (PFU) per mouse] or intravenously (lethal dose at 1.2 × 105 PFU per mouse and sublethal dose at 7 × 104 PFU per mouse). Serum IFN-β was measured by enzyme-linked immunosorbent assay (ELISA). PBMCs were isolated from blood using lymphocyte separation medium; VSV titer in serum was determined by plaque-forming assay; EMCV (3 × 105 PFU per mouse) was intraperitoneally injected, and small intestine was collected to analyze EMCV mRNA 16 hours after infection; 100 μg of poly(I:C) was intraperitoneally injected to 3-week-old mice for 2 or 4 hours, and IECs and small intestine were collected for total RNA extraction or ELISA. SARS-CoV-2 pseudovirus [4 × 104 transduction units (TU)] was intranasally given to the K18-hACE2 mice, and lung tissue was collected for quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis 12 hours after infection. All work was performed in an animal biosafety level 2 (ABSL2) facility.
VSV/ZIKV/SARS-CoV-2 plaque assay
A549 cells were infected with ZIKV for 24 hours to measure IFNB1 mRNA levels by RT-qPCR. Forty-eight hours after infection, ZIKV titers in the culture medium were measured by the plaque assay in Vero E6 cells. Vero E6 cells at 20% confluency were incubated with ZIKV for 1.5 hours in DMEM without FBS. Four days later, the semisolid agarose was stained with crystal violet to count the plaques. To measure serum VSV-GFP titers from the infected mice, HEK293T cells were incubated with the diluted serum in DMEM without FBS, which was replaced by FBS-containing medium with 1.6% low–melting point agarose 3 hours later. Cells were cultured for 2 days after solidification, and the GFP colonies of HEK293T under immunofluorescence microscopy were counted to calculate VSV titers. Alternatively, 0.1% crystal violet in 10% formaldehyde was added to the semisolid agarose and incubated for 2 hours. The plaques were counted after removing the agarose. For SARS-CoV-2 infection, Calu-3 cells were infected for the indicated time, the titer in the culture medium was measured in Vero E6 cells, and all work was performed in a biosafety cabinet under BSL3 conditions.
IEC isolation
The small intestine was washed with cold PBS until the flushing fluid was clear; the intestine was then opened longitudinally, and cut into small pieces. The tissue was shaken slowly in PBS containing 5 mM EDTA at 4°C for 30 min. After shaking the tube, the villi were collected by passing the sample through a 70-μm cell strainer to filter out the crypts and further lysed for immunoblotting analysis or total RNA extraction.
Infection of intestinal organoids
Matrigel was washed once with cold PBS and then melted with cell recovery solution for 40 min on ice. Mouse intestinal organoids were collected at 150g, 4°C for 3 min. The buds were broken up by pipetting up and down for 15 times and spun at 50g for 3 min. The organoid pellet was resuspended with Matrigel and put to a 24-well plate. For VSV infection, the broken buds were stimulated with VSV in the complete DMEM/F12 medium. The supernatants were collected for ELISA, and the pellets were lysed for RNA extraction or immunoblotting analysis.
Tissue histology
The small intestine was embedded in paraffin after fixing in 4% paraformaldehyde. Tissue was cut into 3.5-μm sections and stained with hematoxylin and eosin (H&E) solution. H&E staining was observed under an Olympus BX51 microscope; for frozen sections of liver and small intestine, tissues were fixed in 4%paraformaldehyde followed by 30% sucrose overnight and then embedded in optimal cutting temperature compound at −80°C. Tissues were cut into 8-μm sections to observe GFP-VSV signal in liver or for immunofluorescence staining, and images were acquired under a Leica SP8 WLL confocal microscope.
Quantitative RT-PCR
Total RNA was isolated by TRIzol reagent and reverse-transcribed by reverse transcriptase M-MLV. Real-time PCR was performed in Bio-Rad CFX96 machine using SYBR Green PCR Master Mix. The relative expression of target genes was normalized to the housekeeping gene coding β-actin. For SARS-CoV-2, all experiments were performed under BSL3 conditions.
Pulldown assay
GST-ROCK2 CA kinase was incubated with the purified his-hIRF3 at an equal molar amount in NP-40 buffer [20 mM tris-Cl, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 0.1% NP-40 (pH 7.4)] at 4°C for 3 hours followed by incubation with glutathione Sepharose or Ni agarose for 2 hours. After washing three times, the beads were combined with the SDS loading buffer for immunoblotting analysis.
Subcellular fractionation
Cell pellets were frozen at −20°C for 5 min and lysed with buffer 1 [10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM NaF, 2 mM NaVO3, and 0.1% (v/v) NP-40 (pH 7.9)]. After centrifugation at 6000 rpm for 10 min at 4°C, the supernatants were collected as cytoplasmic samples. After washing three times with buffer 1, the nucleus pellet was lysed in buffer 2 [20 mM Hepes, 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 25% (v/v) glycerol, 0.5 mM DTT, 1 mM PMSF, 5 mM NaF, 2 mM NaVO3, and 0.1% (v/v) NP-40 (pH 7.9)]. After centrifugation at 12,000 rpm for 1 min, the supernatants were collected as nucleus samples.
Immunofluorescence
Cells were fixed for 10 min with 4% paraformaldehyde at room temperature and permeabilized for 15 min followed by blocking with 1% bovine serum albumin in PBS for 30 min. Samples were incubated with anti-IRF3 antibody for 1.5 hours followed by secondary antibody and Hoechst for 1 hour. IRF3+ nucleus was counted in different fields under an Olympus microscope.
Protein expression and purification
Escherichia coli strain BL21(DE3) was transduced with the plasmid pET-6*His-hIRF3 and grown in LB medium (OD600 = 0.6) followed by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside at 16°C for 16 hours. For purification, E. coli was collected and resuspended in the high-salt lysis buffer [25 mM tris (pH 7.4) and 500 mM NaCl]. After sonication and centrifugation, the supernatants were incubated with Ni agarose in the lysis buffer with 10 mM imidazole. Purified IRF3 was eluted by 300 mM imidazole. Desalting column was used to exclude imidazole from the eluted IRF3.
Detection of intestinal cytokine production
The intestine (1.5 cm) was homogenized in cold PBS containing protease inhibitors using a glass homogenizer for 20 times and then centrifuged at 13,000 rpm for 10 min. The supernatants were applied for ELISA measurement, and the final concentration was normalized to the total amount of protein. For samples from the culture medium of organoids, ELISA readings were normalized to extracted total RNA.
Coimmunoprecipitation and immunoblotting
The indicated plasmids were transfected in HEK293T cells for 24 hours, and cells were lysed for 15 min in cold lysis buffer [25 mM tris-Cl, 150 mM NaCl, 1% (v/v) NP-40, 5 mM EDTA, and protease inhibitor cocktail (pH 7.2)]. The cell lysate was centrifuged at 13,000 rpm for 15 min at 4°C. The supernatants were incubated with Flag-, HA–, or Myc-conjugated beads for 2 hours at 4°C. After washing, beads were combined with SDS loading buffer for immunoblotting analysis. To endogenously immunoprecipitate ROCK2 in NIH-3T3 cells, the cell lysate was incubated with anti-ROCK2 antibody or the rabbit monoclonal antibody (mAb) immunoglobulin G (IgG) isotype control at 4°C overnight, followed by incubation with protein A/G beads for 2 hours. For immunoblotting, proteins were separated in SDS–polyacrylamide gel electrophoresis and detected with specific primary antibody and secondary horseradish peroxidase–conjugated anti-mouse or rabbit antibody. Proteins were visualized using the enhanced chemiluminescence reagent. For anti-LPA1 immunoblotting, protein samples were not boiled.
RNA interference
For NIH-3T3 cells, 0.07 million cells were seeded in one well of a 24-well plate and transfected with synthetic siRNA or Dharmacon siRNA (40 pm) with Lipofectamine RNAiMAX Reagent for 48 hours according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed in GraphPad Prism. Student’s unpaired t test was used to compare two groups. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was applied to analyze multiple groups. Two-way ANOVA followed by Sidak’s multiple comparisons test was used for data with two independent variables. For the comparison of mice survival rates, log-rank test was used. P < 0.05 was considered significant. The value of n represented the number of mice in each group.
Acknowledgments
We thank L. Zhang (School of Medicine, Tsinghua University) and B. Sun (Shanghai Institute of Biochemistry and Cell Biology) for providing SARS-CoV-2 pseudovirus, D. Guo (Zhongshan School of Medicine, Sun Yat-sen University) for providing polyclonal antibody to human p-IRF3 Ser97, A. Meng (Tsinghua University) for providing pCMV5-ROCK2 plasmids, and Y. Zhao (University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania) for providing pcDNA3.1-LPA1 plasmid. Funding: This work was supported by grants from the Ministry of Science and Technology of China (2018YFA0800702, 2016YFC0905902), National Natural Science Foundation of China (81930038, 81825011, 81630043, 180060957, 81801575, 81961160738). We thank the Genome Tagging Project (GTP) Center, Shanghai Institute of Biochemistry and Cell Biology, CAS for technical support. Hongyan Wang is supported by the National Science Foundation for Distinguished Young Scholars. We appreciate the Animal Core Facility, Core Facility of Molecular Biology (CFMB), and Core Facility of Cell Biology in SIBCB for technical support and help. Author contributions: C.Z. and W.L. performed most of the experiments and statistical analysis. X.L. and X.X. performed SARS-CoV-2 infection. Z.X. and J.X. generated the Lpa1fl/fl mice. L.Q. performed ZIKV infection. Hui Wang, Y.Z., C.D., X.Z., and S.C. helped with experiments. Hongyan Wang, J.W., B.W., C.Z., W.L., J.L., Q.Z., J.Q., B.S., and J.C. designed the study. C.Z. and Hongyan Wang wrote the paper. All authors approved the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The Lpa1fl/fl Villin-cre mice can be provided by Hongyan Wang pending scientific review and a completed material transfer agreement. Requests for the mice should be submitted to hongyanwang@sibcb.ac.cn. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.York A. G., Williams K. J., Argus J. P., Zhou Q. D., Brar G., Vergnes L., Gray E. E., Zhen A., Wu N. C., Yamada D. H., Cunningham C. R., Tarling E. J., Wilks M. Q., Casero D., Gray D. H., Yu A. K., Wang E. S., Brooks D. G., Sun R., Kitchen S. G., Wu T.-T., Reue K., Stetson D. B., Bensinger S. J., Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163, 1716–1729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu D., Shu T., Yang X., Song J.-X., Zhang M., Yao C., Liu W., Huang M., Yu Y., Yang Q., Zhu T., Xu J., Mu J., Wang Y., Wang H., Tang T., Ren Y., Wu Y., Lin S.-H., Qiu Y., Zhang D.-Y., Shang Y., Zhou X., Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 7, 1157–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song J.-W., Lam S. M., Fan X., Cao W.-J., Wang S.-Y., Tian H., Chua G. H., Zhang C., Meng F.-P., Xu Z., Fu J.-L., Huang L., Xia P., Yang T., Zhang S., Li B., Jiang T.-J., Wang R., Wang Z., Shi M., Zhang J.-Y., Wang F.-S., Shui G., Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 32, 188–202.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostadinova L., Shive C. L., Judge C., Zebrowski E., Compan A., Rife K., Hirsch A., Falck-Ytter Y., Schlatzer D. M., Li X., Chance M. R., Rodriguez B., Popkin D. L., Anthony D. D., During Hepatitis C Virus (HCV) infection and HCV-HIV coinfection, an elevated plasma level of autotaxin is associated with lysophosphatidic acid and markers of immune activation that normalize during interferon-free HCV therapy. J. Infect. Dis. 214, 1438–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tager A. M., LaCamera P., Shea B. S., Campanella G. S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B. A., Kim N. D., Hart W. K., Pardo A., Blackwell T. S., Xu Y., Chun J., Luster A. D., The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14, 45–54 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Kihara Y., Mizuno H., Chun J., Lysophospholipid receptors in drug discovery. Exp. Cell Res. 333, 171–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George P. M., Wells A. U., Jenkins R. G., Pulmonary fibrosis and COVID-19: The potential role for antifibrotic therapy. Lancet Respir. Med. 8, 807–815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onomoto K., Onoguchi K., Yoneyama M., Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell. Mol. Immunol. 18, 539–555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S., Baldridge M. T., Interferon-lambda: A potent regulator of intestinal viral infections. Front. Immunol. 8, 749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., Cao Y., Yousif A. S., Bals J., Hauser B. M., Feldman J., Muus C., Wadsworth M. H. II, Kazer S. W., Hughes T. K., Doran B., Gatter G. J., Vukovic M., Taliaferro F., Mead B. E., Guo Z., Wang J. P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J. M. S., Taylor C. J., Lin B., Waghray A., Mitsialis V., Dwyer D. F., Buchheit K. M., Boyce J. A., Barrett N. A., Laidlaw T. M., Carroll S. L., Colonna L., Tkachev V., Peterson C. W., Yu A., Zheng H. B., Gideon H. P., Winchell C. G., Lin P. L., Bingle C. D., Snapper S. B., Kropski J. A., Theis F. J., Schiller H. B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J. L., Fortune S. M., Berger B., Finberg R. W., Kean L. S., Garber M., Schmidt A. G., Lingwood D., Shalek A. K., Ordovas-Montanes J.; HCA Lung Biological Network , SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., Ravelli R. B. G., van Schayck J. P., Mykytyn A. Z., Duimel H. Q., van Donselaar E., Riesebosch S., Kuijpers H. J. H., Schippers D., van de Wetering W. J., de Graaf M., Koopmans M., Cuppen E., Peters P. J., Haagmans B. L., Clevers H., SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokunina-Olsson L., Alphonse N., Dickenson R. E., Durbin J. E., Glenn J. S., Hartmann R., Kotenko S. V., Lazear H. M., O’Brien T. R., Odendall C., Onabajo O. O., Piontkivska H., Santer D. M., Reich N. C., Wack A., Zanoni I., COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 217, e20200653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pott J., Mahlakõiv T., Mordstein M., Duerr C. U., Michiels T., Stockinger S., Staeheli P., Hornef M. W., IFN-λ determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U.S.A. 108, 7944–7949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M., Okudaira S., Kishi Y., Ohkawa R., Iseki S., Ota M., Noji S., Yatomi Y., Aoki J., Arai H., Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 281, 25822–25830 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Lin S., Han Y., Jenkin K., Lee S.-J., Sasaki M., Klapproth J.-M., He P., Yun C. C., Lysophosphatidic acid receptor 1 is important for intestinal epithelial barrier function and susceptibility to colitis. Am. J. Pathol. 188, 353–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamming I., Timens W., Bulthuis M. L. C., Lely A. T., Navis G. J., van Goor H., Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer A., Lennemann N. J., Ouyang Y., Bramley J. C., Morosky S., Torres De Azeved Marques E. Jr., Cherry S., Sadovsky Y., Coyne C. B., Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19, 705–712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazear H. M., Govero J., Smith A. M., Platt D. J., Fernandez E., Miner J. J., Diamond M. S., A mouse model of Zika virus pathogenesis. Cell Host Microbe 19, 720–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odendall C., Dixit E., Stavru F., Bierne H., Franz K. M., Durbin A. F., Boulant S., Gehrke L., Cossart P., Kagan J. C., Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q., Meng F., Chen S., Plouffe S. W., Wu S., Liu S., Li X., Zhou R., Wang J., Zhao B., Liu J., Qin J., Zou J., Feng X.-H., Guan K.-L., Xu P., Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 19, 362–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riento K., Ridley A. J., Rocks: Multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4, 446–456 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Li S., Zhu M., Pan R., Fang T., Cao Y.-Y., Chen S., Zhao X., Lei C.-Q., Guo L., Chen Y., Li C.-M., Jokitalo E., Yin Y., Shu H.-B., Guo D., The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 17, 241–249 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Yun C. C., Kumar A., Diverse roles of LPA signaling in the intestinal epithelium. Exp. Cell Res. 333, 201–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y., Natarajan V., Lysophosphatidic acid (LPA) and its receptors: Role in airway inflammation and remodeling. Biochim. Biophys. Acta 1831, 86–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hla T., Genomic insights into mediator lipidomics. Prostaglandins Other Lipid Mediat. 77, 197–209 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Kariuki S. N., Franek B. S., Kumar A. A., Arrington J., Mikolaitis R. A., Utset T. O., Jolly M., Crow M. K., Skol A. D., Niewold T. B., Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res. Ther. 12, R151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magkrioti C., Galaris A., Kanellopoulou P., Stylianaki E.-A., Kaffe E., Aidinis V., Autotaxin and chronic inflammatory diseases. J. Autoimmun. 104, 102327 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Yang P., Wang X., COVID-19: A new challenge for human beings. Cell. Mol. Immunol. 17, 555–557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theken K. N., FitzGerald G. A., Bioactive lipids in antiviral immunity. Science 371, 237–238 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Ackermann M., Verleden S. E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W. W., Li V. W., Mentzer S. J., Jonigk D., Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 383, 120–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujioka Y., Tsuda M., Nanbo A., Hattori T., Sasaki J., Sasaki T., Miyazaki T., Ohba Y., A Ca2+−dependent signalling circuit regulates influenza A virus internalization and infection. Nat. Commun. 4, 2763 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Zhang C., Wu Y., Xuan Z., Zhang S., Wang X., Hao Y., Wu J., Zhang S., p38MAPK, Rho/ROCK and PKC pathways are involved in influenza-induced cytoskeletal rearrangement and hyperpermeability in PMVEC via phosphorylating ERM. Virus Res. 192, 6–15 (2014). [DOI] [PubMed] [Google Scholar]
- 33.van Nieuw Amerongen G. P., Vermeer M. A., van Hinsbergh V. W., Role of RhoA and Rho kinase in lysophosphatidic acid-induced endothelial barrier dysfunction. Arterioscler. Thromb. Vasc. Biol. 20, E127–E133 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Knipe R. S., Probst C. K., Lagares D., Franklin A., Spinney J. J., Brazee P. L., Grasberger P., Zhang L., Black K. E., Sakai N., Shea B. S., Liao J. K., Medoff B. D., Tager A. M., The Rho kinase isoforms ROCK1 and ROCK2 each contribute to the development of experimental pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 58, 471–481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3