Abstract
Acute myeloid leukemia (AML) is associated with a poor prognosis in elderly adults and currently lacks optimal treatment strategies. MicroRNAs (miRNAs or miRs) have increasingly been reported to be associated with AML progression; however, the mechanisms of action of miR-93 in AML with the involvement of disabled 2 (DAB2) are currently unknown. In the present study, miR-93 expression was assessed in patients with AML and in AML cell lines. The association between miR-93 expression and the pathological characteristics of patients with AML was analyzed. AML cells were then transfected to knockdown or overexpress miR-93 in order to elucidate its function in AML progression. The target gene of miR-93 was assessed using a dual-luciferase reporter gene assay. The expression levels of miR-93, DAB2 and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway-related proteins were measured and in vivo experiments were conducted to confirm the results. It was observed that miR-93 was highly expressed in patients with AML and in AML cells. The knockdown of miR-93 in HL-60 cells inhibited AML cell proliferation and resistance to apoptosis, while the overexpression of miR-93 in THP-1 cells led to contrasting results. Moreover, miR-93 targeted DAB2 to inactivate the PI3K/AKT pathway, and the overexpression of DAB2 reversed the effects of miR-93 on THP-1 cell growth. Tumor volume, tumor weight, and the positive expression of Ki67, survivin and p53 were increased in THP-1 cells overexpressing miR-93. On the whole, the present study demonstrates that miR-93 is highly expressed in AML cells, and that the suppression of miR-93 inhibits AML cell growth by targeting DAB2 and inhibiting the PI3K/AKT pathway.
Keywords: acute myeloid leukemia, microRNA-93, disabled 2, PI3K/AKT pathway, proliferation, apoptosis
Introduction
Leukemia is a fatal malignancy of hematopoietic stem cells and immature progenitors, presenting with increased leucocytes in the blood and bone marrow (1,2). Acute myeloid leukemia (AML), the most frequently diagnosed leukemia in adults (3), includes a group of malignant clonal disorders in the myeloid system with significant functional somatic cell mutation and chromosome translocation (4). Immature AML cells generally accumulate rapidly in the blood and sometimes metastasize to lymph nodes, spleen, liver and the central nervous system (5). Genetic diseases, ionizing radiation, cigarette smoking, benzene exposure, chemotherapy, viruses and a high gestational age at birth are positively associated with the occurrence of AML (6,7). It is known that >50% of patients with AML are >65 years of age and almost 30% are >75 years of age. Older-aged patients with AML generally have poor outcomes due to age-related factors, unfavorable cytogenetics, relapse following complete remission and drug resistance (8). Ample research has been performed with an aim to improve the prognosis of patients with AML; however, the standard treatment for most newly diagnosed AML subtypes has not changed over the past 40 years (9). Thus, there remains an urgent and clear need for the identification of novel AML biomarkers and therapies, as well as personalized treatment methods for patients with AML.
MicroRNAs (miRNAs or miRs), a class of small non-coding RNAs, which serve as critical post-transcriptional mediators of genes implicated in multiple fundamental processes, such as differentiation, proliferation, apoptosis and cancer drug resistance (10). miRNAs participate in hematopoietic cell differentiation, and an aberrant miRNA expression contributes to tumor progression, including leukemia (11). miR-93 belongs to the miR-106b-25 cluster, a subgroup of the miR-17-92 cluster (12). miR-93 expression has been demonstrated to be upregulated in patients with adult-T cell leukemia (13). The dysregulated expression of miR-93 has been shown to enhance the growth of leukemia stem cells, prevent differentiation and reduce apoptosis, thereby facilitating the progression of leukemia (14). A previous study reported the elevated expression of miR-93 in the serum of patients with AML (15). The detection of clinical samples from patients with AML has revealed that miR-93 expression positively correlates with vascular endothelial growth factor (VEGF) expression, while VEGF promotes AML progression (16). Moreover, accumulating studies have unveiled the promoting effects of miR-63 on tumor progression (15,17-19). miR-93 has been reported to function as an effective inhibitor of disabled 2 (DAB2) in lung cancer (20). DAB2 is a multifunctional scaffold protein that participates in cell growth, differentiation, homeostasis, cell-cell interaction, and platelet aggregation (21). DAB2 is a binding protein that regulates adhesion and mitogen-activated protein kinase activity in human chronic myeloid leukemia cells (22). A recent study revealed that the overexpression of DAB2 interactive protein inhibited the activation of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway (23). Martelli et al noted that targeting the PI3K/AKT/mTOR pathway with small molecule inhibitors may lead to decreased toxicity and improved the therapeutic effects in patients with AML (24). In addition, miR-93 has been shown to play a vital role in glioma progression via the PI3K/AKT pathway (25).
In view of the above-mentioned information, it was thus hypothesized speculated that miR-93, DAB2 and the PI3K/AKT pathway are involved in the biological process of AML progression. Therefore, the aim of the present study was to determine the roles of miR-93, DAB2 and the PI3K/AKT pathway in AML and their involvement in the progression of this disease.
Materials and methods
Ethics statement
The present study was approved and supervised by the Ethics Committee of the Third Affiliated Hospital of Sun Yat Sen University. All subjects signed an informed consent form. All animal experiments involving nude mice were approved by the Laboratory Animal Ethics Committee of the Third Affiliated Hospital of Sun Yat Sen University. Significant efforts were made to minimize both the number of animals and their suffering.
Study subjects
From October, 2016 to October, 2018, 28 patients with AML (16 males and 12 females, aged 19-71 years, with a median age of 46.2 years) diagnosed and treated at the Hematology Department of Third Affiliated Hospital of Sun Yat Sen University were enrolled in the present study. A total of 30 patients without malignant hematological diseases excluded by bone marrow examination during the same time period were included as the control group. All patients with AML were diagnosed according to the examinations of Morphology, Immunology, Cytogenetics and Molecular Biology (MICM), and were in accordance with the Criteria for Diagnosis and Therapeutic Effect of Hematological Diseases (3rd edition) (26). Mononuclear cells from the bone marrow of enrolled subjects were isolated and frozen in liquid nitrogen for future use. Mononuclear cells were isolated from the bone marrow using the hydroxyethyl starch precipitation method. Briefly, the bone marrow fluid was centrifuged at 450 × g for 10 min at room temperature to remove plasma and supplemented with the same volume of normal saline with plasma and mixed well. The mixture was then mixed with 6% hydroxyethyl starch (BA740193, Precedex; Hospira; Pfizer, Inc.) in a ratio of 1:4 and kept at room temperature for 40 min. The supernatant was centrifuged at 450 × g for 5 min at room temperature to obtain the cell precipitate, washed normal saline twice, and prepared into a cell suspension.
Cells and cell culture
AML cell lines (Kasumi-1, THP-1, and HL60, cat. nos. 3142C0001000000322, 3111C0001CCC 000057 and 3111C0001CCC000037 respectively) purchased from the Cell Bank of Experimental Medical Center of China Union Medical University (Beijing, China) and the normal bone marrow cell line (HS-5, cat. no. CRL-11882) purchased from the American Type Culture Collection (ATCC) were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% inactivated fetal bovine serum (FBS) in an incubator (37°C, 5% CO2). After cultured in a conventional manner, the cells in the exponential growth phase were obtained for use in subsequent experiments. 293T cells (ATCC, cat. no. CRL-1573) were cultured in Dulbecco's modified Eagle's medium (DMEM) under the same conditions.
Transfection and grouping
The sequences of the recombinant overexpression vector pZsGreen1-miR-93 (oe-miR-93), interference vector pLL3.7-anti-miR-93, and their negative control (NC) were designed by Shanghai GenePharma Co., Ltd. and synthesized by Invitrogen; Thermo Fisher Scientific, Inc. Following enzyme digestion, the vectors were sequenced by Shanghai Genechem Co., Ltd. Following amplification and extraction, the recombinant plasmid pZsGreen1-miR-93 was digested with restriction enzymes Kpn and BamHI overnight. Following amplification and extraction, the recombinant plasmid pLL3.7-anti-miR-93 was digested with restriction enzymes XbaI and NotI overnight. The digested products were electrophoresed respectively. The results of electrophoresis following digestion were all recombinant vectors as the digested sites of recombinant plasmids no longer existed. The results of DNA sequencing confirmed that the target sequence was inserted into the skeleton plasmid correctly, and the sequence was identical with the expected sequence.
One day prior to lentivirus infection, the THP-1, HL60 and HS-5 cells of the 3rd passage were seeded into 6-well plates at 3×105 cells/well. When the cell confluence reached 70%, the cells were transfected with 2.5 μg corresponding lentivirus vectors using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, Inc.). The cells were randomly assigned to the control group (cells were cultured routinely), oe-miR-93 or oe-NC group (THP-1 cells of the 3rd passage were cultured with the overexpression vector, pZsGreen1-miR-93 or its NC), and anti-miR-93 or anti-NC group (HL60 cells of the 3rd passage were cultured with interference vector pLL3.7-anti-miR-93 or its NC). The medium was refreshed at 24 h following transfection. Cell fluorescence was observed under a fluorescence microscope (FSX100, Olympus Corporation) after 72 h.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells were seeded in 96-well plates at a density of 5,000 cells/well in an incubator (37°C, 5% CO2). Following culture for 0, 24, 48 and 72 h, 20 μl of 5 mg/ml MTT solution (Sigma-Aldrich; Merck KGaA) were added to each well with subsequent incubation at 37°C for 4 h. Subsequently, 200 μl of dimethyl sulfoxide were added and the cells were placed in a vibrating apparatus for 10 min. The absorbance at 490 nm was measured using a microplate reader (Bio-Rad 680, Bio-Rad Laboratories, Inc.).
Ethynyl-2′-deoxyuridine (EdU) assay
A Cell-Light EdU fluorescence detection kit (Guangzhou RiboBio Co., Ltd.) was used to detect EdU-positive cells according to the manufacturer's instructions. The cells were seeded into 96-well plates (5,000 cells/well), added with 100 μl medium containing 50 μmol/l EdU, and cultured at 37°C with 5% CO2 for 2 h. The cells were then washed with PBS 3 times and each well was supplemented with 100 μl PBS solution containing 4% paraformaldehyde to fix the cells at room temperature for 30 min. Following PBS washing, each well was supplemented with 100 μl PBS solution containing 0.5% TritonX-100 to increase membrane permeability. Following PBS washing again, each well was added with 100 μl Apollo® staining reaction solution (provided with the kit) and cultured at room temperature in the dark for 30 min. The staining reaction solution was discarded and each well was supplemented with 100 μl DAPI (C1002, Beyotime Institute of Biotechnology, Inc.) at room temperature in the dark for 30 min. Following PBS washing, the staining effect was observed under fluorescence microscope (FSX100, Olympus Corporation) and images were collected. The images were analyzed using Olympus stream system. The total number of nuclei (blue) and proliferating cells (red) were counted respectively. EdU-positive cell rate=proliferating cells/total cells ×100%. The experiment was repeated three times.
Flow cytometry
Cells were centrifuged at room temperature and 1,000 × g for 1 min and collected following trypsin detachment, and 2 μl of 1 mg/ml RNase A (prepared by deionized water) were added at 37°C for 40 min to remove the RNA. Cells were treated with propidium iodide (PI) staining solution and incubated at room temperature without exposure to light for 20 min. Finally, the cell cycle distribution was detected using a flow cytometer (MoFloAstrios EQ, Beckman Coulter, Inc.).
The cells were centrifuged and collected following trypsin detachment. To each tube, 100 μl 1X Annexin binding buffer was added to resuspend the cell precipitates. Alexa Fluor 488 Annexin V-fluorescein isothiocyanate (FITC) and 1 μl of 100 mg/ml PI staining solution were mixed and added to the cells, followed by incubation at room temperature for 15 min without light exposure. Apoptotic cells were measured with a flow cytometer (MoFloAstrios EQ, Beckman Coulter, Inc.).
Hoechst 33258 staining
THP-1 and HL60 cells in exponential phase were seeded at 1×105 cells/ml into 6-well plates on a coverslip. Following culture at 37°C with 5% CO2 for 24 h, the cells were fixed for 10 min, and stained with Hoechst 33258 dye (Shanghai, Beyotime Biotechnology Co., Ltd.) for 5 min in the dark. The cells were then placed on a slide covered in an anti-fluorescence quenching agent (E675011-0005, BBI Life Sciences Corporation). The slides were observed and photographed using a fluorescence microscope (FSX100, Olympus Corporation).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from bone marrow monocytes, leukemic cell lines and normal bone marrow cells, and the extracted high-quality RNA was examined using ultraviolet analysis and formaldehyde denaturation electrophoresis. A total of 1 μg RNA was reverse transcribed into cDNA using avian myeloblastosis virus reverse transcriptase (M5101, Promega, Corporation). qPCR was conducted using the SYBR-Green system (Beijing Labest Biotechnology Co., Ltd.) with U6 as an internal reference gene for miR-93 and β-actin as an internal reference gene for DAB2 and other genes. PCR primers were designed and synthesized by Shanghai Sangon Biotechnology Co., Ltd. and are presented in Table I. The PCR system included 1.0 μl cDNA, 10 μl 2X SYBR-Green mix, 0.5 μl of forward primer (10 μM), 0.5 μl reverse primer (10 μM), and was supplemented into 20 μl with RNase-free water. The PCR conditions were as follows: Pre-denaturation at 94°C for 5 min, 40 cycles of denaturing at 94°C for 40 sec, annealing at 60°C for 40 sec, and extension at 72°C for 60 sec, and a final extension at 72°C for 10 min. The products were verified by agarose gel electrophoresis. The data were analyzed using the 2−ΔΔCq method (27), indicating multiple associations between the experimental group and the control group. ΔΔCt=[Ct (target gene)-Ct (internal reference gene)] experimental group-[Ct (target gene)-Ct (internal reference gene)] control group.
Table I.
Primer sequences used for RT-qPCR.
| Genes | Sequences |
|---|---|
| miR-93 | F: 5′-GAAGATCTACCTTCACTGAGAGGGTGGT-3′ |
| R: 5′-CGGAATTCACCAGACCCTTTTGAACGCC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| DAB2 | F: 5′-TGGACGATGTGCTCTATGCC-3′ |
| R: 5′-GGATGGTGATGGTTTGGTAG-3′ | |
| TGFBR2 | F: 5′-CCTGAGAGGAGAT GAAGTAG-3′ |
| R: 5′-CTTAAAGTATAAGAGCCTGCA-3′ | |
| ITGB8 | F: 5′-CGTGACTTTCGTCTTGGATTTGG-3′ |
| R: 5′-TCCTTTCGGGGTGGATGCTAA-3′ | |
| CDKN1A | F: 5′-CGATGGAACTTCGACTTTGTCA-3′ |
| R: 5′-GCACAAGGGTACAAGACAGTG-3′ | |
| BTG3 | F: 5-ATGAAGAATGAAATTGCTGCCGTTG-3′ |
| R: 5′-GTGAGGTGCTAACATGTGAGGATT-3′ | |
| β-actin | F: 5-TGAGCGCGGCTACAGCTT-3′ |
| R: 5′-TCCTTAATGTCACGCACGATTT-3′ |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR-93, microRNA-93; DAB2, disabled 2; TGFBR2, transforming growth factor beta receptor II; ITGB8, Integrin beta8; CDKN1A, cyclin-dependent kinase (CDK) inhibitor p21; BTG3, B-cell translocation gene 3; F, forward; R, reverse.
Western blot analysis
Protein was extracted from the cells and the concentration was determined using a bicinchoninic acid kit (Boster Biological Technology Co., Ltd.), following the manufacturer's instructions. The extracted proteins were mixed with loading buffer, boiled at 95°C for 10 min, loaded (each for 30 μg) and separated via 10% polyacrylamide gel electrophoresis (Boster Biological Technology Co., Ltd.). Subsequently, the proteins were transferred onto polyvinylidene fluoride membranes and the membranes were incubated in 5% bovine serum albumin (BSA) at room temperature for 1 h. Membranes were incubated with the following primary antibodies at 4°C overnight: p21 (1:500, ab218311), cyclin D1 (1/200, ab16663), caspase3 (1:500, ab13847), Bax (1:1,000, ab32503), Bcl-2 (1:500, ab185002), DAB2 (2 μg/ml, ab33441), transforming growth factor beta receptor II (TGFBR2, 1:2,000, ab186838), integrin beta8 (ITGB8, 1 μg/ml, ab80673), cyclin-dependent kinase (CDK) inhibitor p21 (CDKN1A, 1:500, ab102013), B-cell translocation gene 3 (BTG3, 1:1,000, ab197399), AKT (1:10,000, ab179463), p-AKT (1:500, ab38449), PI3K (1:1,000, ab32089) and p-PI3K (1:500, ab182651) (all from Abcam). The membranes were rinsed using Tris-buffered saline containing 0.05% Tween-20 (TBST) and then incubated for 1 h with the secondary antibody (1:5,000, ZB-2301, ZSGB-Bio Co., Ltd.) at room temperature. The membranes were then washed with TBST and developed using a chemiluminescence reagent. Protein bands were visualized using the Gel Doc EZ imager (Bio-Rad Laboratories, Inc.). The target band was examined using ImageJ software (National Institutes of Health) for gray value analysis.
Bioinformatics analysis
The target genes of miR-93 were predicted through TargetScan (http://www.targetscan.org/vert_71/) (28), miRBase (http://www.mirbase.org/) (29), MicroRNA.org (http://www.microrna.org) (30), and PicTar (https://pictar.mdc-berlin.de/) (31).
Dual-luciferase reporter gene assay
The DAB2 3′-untranslated region (3′UTR) sequence containing the binding site of miR-93 was synthesized and a DAB2 3′UTR wild-type plasmid (DAB2-WT) was constructed. Based on DAB2-WT, the DAB2 3′UTR mutant-type (MUT) plasmid (DAB2-MUT) was constructed. The plasmids were purchased from Promega Corporation. DAB2-WT and DAB2-MUT plasmids were mixed with NC and miR-93 mimics (GenePharma Co., Ltd.) respectively, and then transfected into 293T cells using Lipofectamine® 3000. Each group was co-transfected with 800 ng recombinant plasmid, and the final concentration of mimics was 20 nmol/l. The cells were collected and lysed following transfection for 48 h. Luciferase activity was measured using a luciferase detection kit (BioVision, Inc.) using a Glomax 20/20 luminometer (Promega Corporation). With Renilla luciferase as the internal reference, the relative light unit (RLU) measured by Firefly luciferase was divided by the RLU measured by Renilla luciferase to normalize the data of each group.
Xenograft tumors in nude mice
BALB/c male nude mice (aged, 4-6 weeks; weighing, 16-18 g; Beijing Vital River Laboratory Animal Technology Co., Ltd.) were raised in a specific pathogen-free animal center. A volume of 1×107 THP-1 cells/100 μl was subcutaneously injected into the armpit of the right forelimb of each nude mouse. Tumor volume was measured every 7 days according to the following formula: V=L × W2 × 0.5, where L represents the long diameter of the tumor and W represents the short diameter. Tumor growth curves were drawn. Mice were euthanized by cervical dislocation 35 days later and tumors were peeled off along the subcutaneous side and weighed for immunohistochemistry.
Immunostaining
For immunofluorescence, the cells were fixed with 4% paraformaldehyde solution and treated with Triton X-100 (Guangzhou Eskin Biotechnology Co., Ltd.). The cells were then incubated with p-AKT (1:100, ab38513, Abcam) antibody at 4°C overnight followed by incubation with FITC-labeled goat anti-rabbit immunoglobulin G (IgG, 1:200, ab150077, Abcam). The cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI). Briefly, the slide was added with DAPI (1 μg/ml, Beyotime Institute of Biotechnology), incubated at room temperature in the dark, and washed with PBS. The cell fluorescence and localization were observed under an inverted fluorescence microscope (FSX100, Olympus Corporation). The fluorescence intensity of p-AKT was quantified using the Olympus Stream system.
For immunohistochemistry, after weighing, the tumors were fixed in 4% paraformaldehyde for 24 h, dehydrated in an alcohol gradient, embedded in paraffin, sectioned at 4 μm, dewaxed with xylene and ethanol, and placed in 0.01 mol/l citric acid buffer. Subsequently, the sections were incubated with 3% H2O2 for 15 min at 37°C, rinsed with distilled water for 5 min, immersed in PBS for 5 min, and incubated with primary antibodies (Ki67: 1:500, ab92742, Abcam; survivin: 1:1,000, ab76424, Abcam; p53: 1:100, ab32389, Abcam) at 4°C overnight. The following day, the sections were incubated with the corresponding secondary antibodies (goat anti-rabbit IgG H&L (HRP): 1:1,000, ab6721, Abcam) and placed in a constant temperature box at 37°C for 40 min. The sections were developed with 2,4-diaminobutyric acid, counterstained with hematoxylin at room temperature for 1 min, differentiated, washed with cold water, dehydrated and sealed. Positively stained sections were observed and photographed under an optical microscope (Olympus Corporation).
Statistical analysis
SPSS 21.0 (IBM Corp.) was used for data analysis. The Kolmogorov-Smirnov test revealed that the data were normally distributed. The data are presented as the means ± standard deviation, or count and percentage (categorical variables). The diagnostic efficacy of miR-93 in patients with AML was analyzed using the receiver operating characteristic (ROC) curve. Correlation analysis was performed using the Pearson's correlation coefficient. Comparisons between 2 groups were performed using the Student's t-test or Chi-squared test, and comparisons among multiple groups were performed using one-way analysis of variance (ANOVA), followed by Tukey's multiple comparisons test. P-values were obtained using a two-tailed test, and P<0.05 was considered to indicate a statistically significant difference.
Results
High miR-93 expression in patients with AML and AML cells
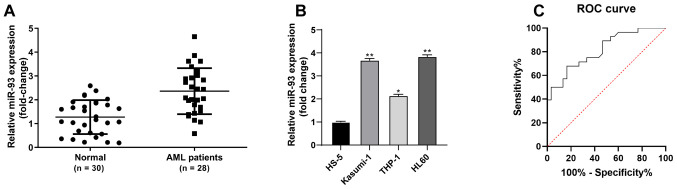
The present study sought to investigate the association between miR-93 expression and AML. The results of RT-qPCR revealed that miR-93 expression was notably higher in bone marrow-derived monocytes from patients with AML than in non-malignant hematological patients (P<0.05, Fig. 1A). miR-93 expression was also upregulated in AML cells (Kasumi-1, THP-1 and HL-60) compared to normal bone marrow cells (HS-5) (all P<0.05). miR-93 exhibited the highest expression in HL60 cells and the lowest in the THP-1 cells (Fig. 1B). Of the patients with AML, 14 were allocated to the low expression group and 14 to the high expression group, with a median value of miR-93 expression as the critical threshold, as previously described (32). The distribution of sex, age French-American-British (FAB) classification and white blood cells (WBC) were analyzed. The WBC levels of patients with a high miR-93 expression were significantly higher than those with a low miR-93 expression (P<0.05); however, there were no significant differences in sex, age and FAB classification between the 2 groups (P>0.05; Table II). The diagnostic efficacy of miR-93 in patients with AML was analyzed using the ROC curve, and the area under the curve was 0.8143 (sensitivity, 0.6786; specificity, 0.8333), indicating that miR-93 expression was clinically significant for the diagnosis of AML (Fig. 1C).
Figure 1.
miR-93 is highly expressed in patients with AML and AML cells. (A) miR-93 expression in AML and non-malignant hematological patients was examined by RT-qPCR. (B) miR-93 expression in AML cells (Kasumi-1, THP-1 and HL-60) and normal bone marrow cells (HS-5) was examined by RT-qPCR. (C) Clinical significance of miR-93 expression in the diagnosis of AML was analyzed by ROC curve. Each experiment was performed 3 times, independently. Data are presented as the means ± standard deviation. Data in (A) were analyzed using a t-test, and data in (B) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05 and **P<0.01, compared with HS-5 cells. miR-93, microRNA-93; AML, acute myeloid leukemia; ROC, receiver operating characteristic.
Table II.
Association between miR-93 expression and the clinical characteristics of patients with AML.
| Clinical data | miR-93 expression in AML tissues
|
χ2/F-value | P-value | |
|---|---|---|---|---|
| Low expression (n=14) | High expression (n=14) | |||
| Sex | ||||
| Male | 9 (64.29%) | 7 (50.00%) | 0.583 | 0.445 |
| Female | 5 (35.71%) | 7 (50.00%) | ||
| Age, years; median (range) | 45.5 (19-63) | 45.0 (34-71) | 1.341 | 0.897 |
| FAB classification | ||||
| M0 | 1 (7.14%) | 2 (14.29%) | 5.082 | 0.406 |
| M1 | 8 (57.14%) | 3 (21.43%) | ||
| M2 | 3 (21.43%) | 4 (28.57%) | ||
| M3 | 0 (0.00%) | 2 (14.29%) | ||
| M4 | 1 (7.14%) | 2 (14.29%) | ||
| M5 | 1 (7.14%) | 1 (7.14%) | ||
| M6 | 0 (0.00%) | 0 (0.00%) | - | - |
| WBC (×109/l); median (range) | 48.13 (30.98-122.85) | 85.69 (21.59-166.16) | 1.272 | <0.001 |
AML, acute myeloid leukemia; miR-93, microRNA-93; FAB, French-American-British; WBC, white blood cells. Sex and FAB classification were examined using a Chi-squared test; Age and WBC were examined using a t-test.
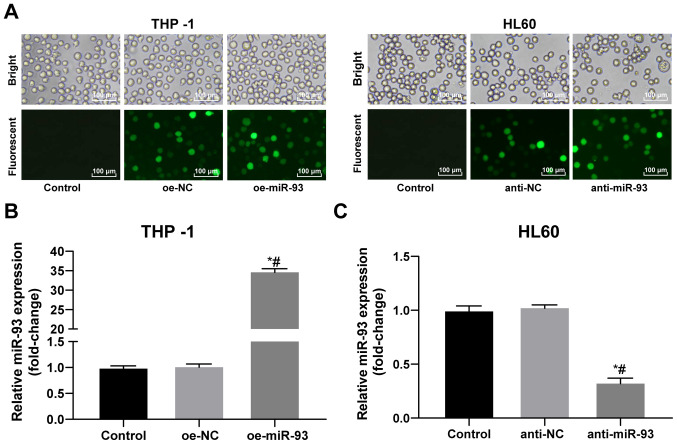
Construction of miR-93 lentiviral expression vector and lentiviral interference vector
Following 72 h of transfection with a recombinant lentivirus, the THP-1 and HL60 cells grew well. Compared with the control group, the transfected cells exhibited extensive green fluorescence (Fig. 2A). miR-93 expression in the THP-1 and HL60 cells following transfection was detected by RT-qPCR. miR-93 expression was increased in the THP-1 cells and decreased in the HL60 cells, indicating successful transfection (all P<0.05; Fig. 2B and C).
Figure 2.
Successful constructions of miR-93 lentiviral expression vector and lentiviral interference vector. (A) Transfection efficiency of lentivirus was identified under a fluorescent microscope. (B and C) Transfection efficiency of miR-93 in THP-1 and HL60 cells was determined by RT-qPCR. Each experiment was performed 3 times, independently. Data are presented as the means ± standard deviation. Data in (B) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC or anti-NC group. miR-93, microRNA-93; NC, negative control.
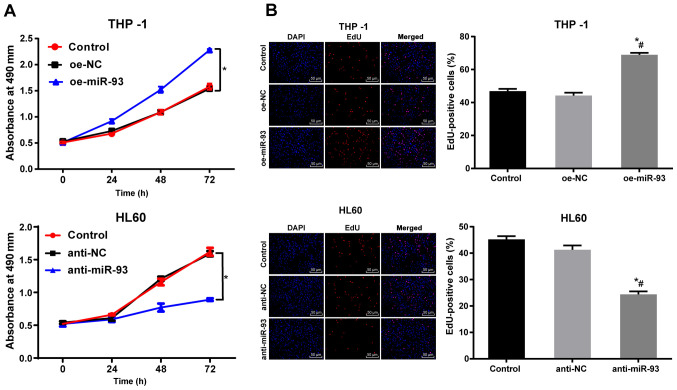
Anti-miR-93 vector inhibits AML cell proliferation in vitro
The proliferation of the THP-1 and HL60 cells following transfection was measured by MTT assay. There was no marked difference in THP-1 and HL60 cell proliferation between the NC and control groups (P>0.05). Compared with the NC cells, the proliferation of the THP-1 cells transfected with oe-miR-93 was markedly increased, while the proliferation of the HL60 cells transfected with anti-miR-93 was notably decreased (P<0.05, Fig. 3A). The DNA replication activity of the AML cells was measured using an EdU assay. There were no significant differences in DNA replication activity or in the percentage of EdU-positive cells between the NC and control groups (P>0.05). Compared with the NC group, DNA replication and the percentage of EdU-positive THP-1 cells transfected with oe-miR-93 were markedly increased (P<0.05). However, DNA replication and the percentage of EdU-positive HL60 cells transfected with anti-miR-93 was significantly decreased (P<0.05, Fig. 3B). These results suggest that the overexpression of miR-93 promotes AML cell proliferation in vitro.
Figure 3.
Anti-miR-93 inhibits AML cell proliferation in vitro. The expression of miR-93 was upregulated in THP-1 cells by oe-miR-93 and downregulated in HL60 cells by anti-miR-93. (A) Cell viability was measured by MTT assay. (B) DNA replication activity of cells and the rate of EdU-positive cells were measured by EdU assay. Each experiment was performed 3 times, independently. Data are presented as the means ± standard deviation. Data in (A) were analyzed using a t=test. Data in (B) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC or anti-NC group. miR-93, microRNA-93; NC, negative control; AML, acute myeloid leukemia.
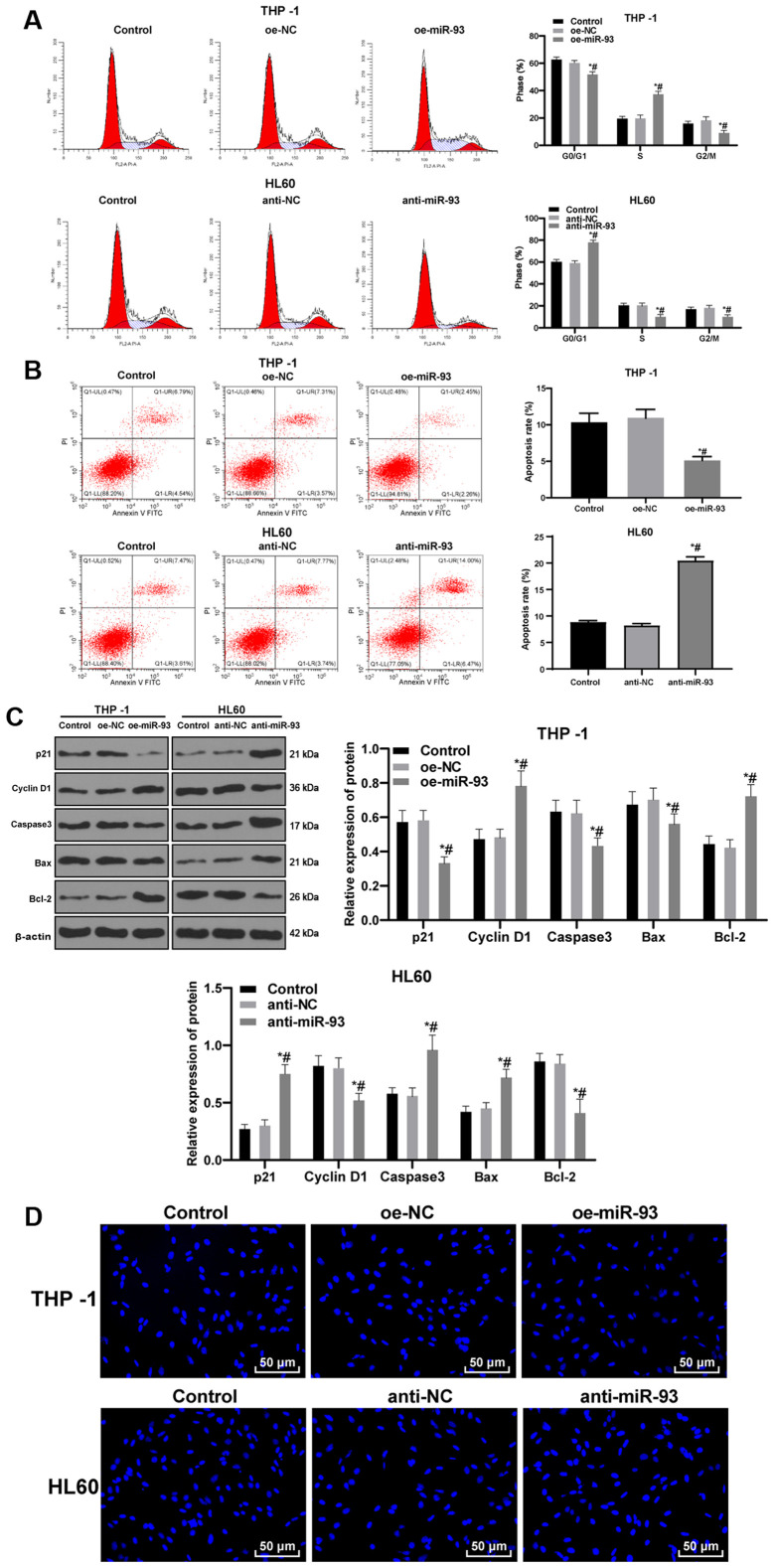
Anti-miR-93 promotes AML cell cycle arrest and apoptosis
Cell cycle distribution and apoptosis were measured by flow cytometry. No significant differences were found in cell cycle distribution and apoptosis between the control and NC groups (P>0.05). Compared with the NC cells, the proportion of THP-1 cells transfected with oe-miR-93 in the G0/G1 phase was decreased and the proportion in the S phase were increased. The proportion of HL60 cells transfected with anti-miR-93 in the G0/G1 phase were increased and those in the S phase were decreased (all P<0.05; Fig. 4A). Relative to that in the NC cells, the apoptotic rate of the THP-1 cells transfected with oe-miR-93 was markedly decreased, while the apoptotic rate of the HL60 cells transfected with anti-miR-93 was markedly increased (both P<0.05; Fig. 4B). In addition, the levels of the cycle-related proteins, p21 and cyclin D1, and the apoptosis-related proteins, caspase3, Bax and Bcl-2, were detected by western blot analysis. miR-93 overexpression promoted cell cycle progression and inhibited apoptosis, while miR-93 inhibition exerted the opposite effect (Fig. 4C).
Figure 4.
Anti-miR-93 promotes cell cycle arrest and the apoptosis of AML cells. The expression of miR-93 was upregulated in THP-1 cells and downregulated in HL60 cells. (A) Cell cycle distribution was measured by flow cytometry. (B) Relative cell apoptosis was detected by flow cytometry. (C) Levels of the cycle-related proteins, p21 and cyclin D1, and the apoptosis-related proteins, caspase3, Bax and Bcl-2, were detected by western blot analysis. (D) Representative images of Hoechst 33258 staining. Each experiment was performed 3 times, independently. Data are presented as the means ± standard deviation. Data in (A-C) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC or anti-NC group. miR-93, microRNA-93; NC, negative control; AML, acute myeloid leukemia.
Following Hoechst 33258 staining, cells in the control and NC groups exhibited normal, full and clear nuclei, presenting a round or oval shape. Compared with the NC cells, the THP-1 cells transfected with oe-miR-93 exhibited slightly deformed nuclei. However, the HL60 cells transfected with anti-miR-93 exhibited evidently deformed nuclei, unclear nucleus margins, nucleus shrinkage or fragmentation, chromatin concentration and aggregation, and marginalization. Under the inverted fluorescence microscope, the nuclei exhibited bright blue spots, or were enveloped by the nucleus membrane as apoptotic bodies, showing typical morphological changes of apoptosis (Fig. 4D).
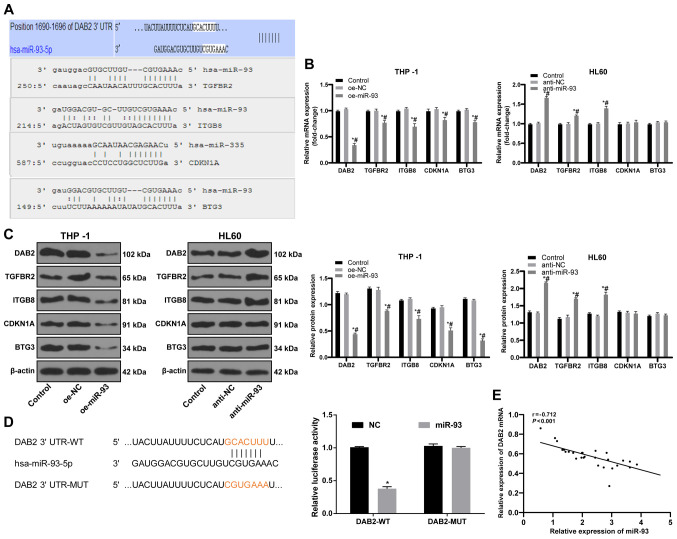
miR-93 targets DAB2
To elucidate the downstream mechanisms of miR-93 in AML, data from the bioinformatics databases, miRBase, microRNA.org, PicTar and TargetScan, were used to predict the target genes of miR-93. Targets related to tumor growth were DAB2, TGFBR2, ITGB8, CDKN1A and BTG3 (Fig. 5A); of these targets, DAB2 can affect the proliferation of leukemia cells (33). The results of RT-qPCR and western blot analysis were employed to measure the levels of target genes. DAB2 expression in the THP-1 cells overexpressing miR-93 was decreased, while DAB2 expression in the HL60 cells in which miR-93 was knocked down was significantly increased (both P<0.05; Fig. 5B and C). A dual-luciferase reporter gene assay further indicated that DAB2 was a target gene of miR-93 (P<0.05; Fig. 5D). DAB2 mRNA expression and miR-93 expression in the bone marrow mononuclear cells of 28 patients with AML were detected by RT-qPCR. Pearson's correlation analysis revealed a negative correlation between DAB2 mRNA and miR-93 expression (r=−0.712, P<0.01; Fig. 5E).
Figure 5.
DAB2 is a target gene of miR-93. (A) The major target genes of miR-93 related to tumor growth were predicted by bioinformatics databases. (B and C) mRNA and protein expression levels of target genes detected by RT-qPCR and western blot analysis. (D) Target relation of miR-93 and DAB2 was verified by dual-luciferase reporter gene assay. (E) The correlation between DAB2 mRNA and miR-93 expression in bone marrow mononuclear cells of 28 AML patients was determined by Pearson's correlation analysis. Each experiment was performed 3 times, independently. Data are presented as the means ± standard deviation. Data in (B and C) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. Data in (D) were analyzed using a t-test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC or anti-NC group. miR-93, microRNA-93; NC, negative control; DAB2, disabled 2.
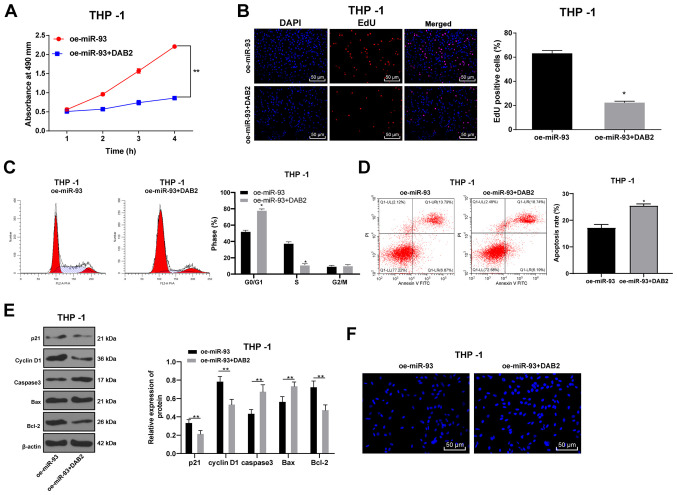
DAB2 reverses the promoting effects of miR-93 overexpression on AML cell growth
To confirm that miR-93 regulates the growth of AML cells by targeting DAB2, the DAB2 overexpression plasmid was transfected into THP-1 cells overexpressing miR-93. The THP-1 cells were transfected with DAB2, and the proliferation of the THP-1 cells was detected by MTT, EdU and colony formation assays. Compared with the oe-miR-93 group, the proliferative ability of the THP-1 cells in the oe-miR-93 + DAB2 group was decreased (all P<0.05), indicating that the overexpression of DAB2 inhibited AML cell proliferation in vitro (Fig. 6A and B).
Figure 6.
DAB2 reverses the promoting effects of the overexpression of miR-93 on AML cell growth. The expression of DAB2 was upregulated in THP-1 cells overexpressing miR-93. (A) Cell viability was measured by MTT assay. (B) DNA replication of AML cells was measured by EdU assay. (C) Cell cycle was detected by flow cytometry. (D) Cell apoptosis was measured by flow cytometry. (E) Levels of cell cycle-related proteins and apoptosis-related proteins were detected by western blot analysis. (F) Hoechst 33258 staining. Each experiment was performed 3 times, independently. The data are presented as the means ± standard deviation. Data in (A–E) were analyzed using a t-test. *P<0.05 and **P<0.01, compared with the oe-miR-93 group. miR-93, microRNA-93; NC, negative control; AML, acute myeloid leukemia; DAB2, disabled 2.
Flow cytometry was used to assess cell cycle distribution and the apoptotic rate. The proportion of cells in the G0/G1 phase markedly increased in the oe-miR-93 + DAB2 group, while the proportion of cells in the S phase substantially decreased as compared with those in the oe-miR-93 group (P<0.05; Fig. 6C). The apoptotic rate in the oe-miR-93 + DAB2 group was significantly higher than that in the oe-miR-93 group (Fig. 6D). In addition, the levels of the cycle-related proteins, p21 and cyclin D1, and the apoptosis-related proteins, caspase3, Bax, and Bcl-2, were detected by western blot analysis. It was found that oe-miR-93 + DAB2 treatment promoted cell cycle progression and inhibited apoptosis, whereas oe-miR-93 treatment alone had the opposite effect (all P<0.05; Fig. 6E).
Hoechst 33258 staining revealed that compared with the oe-miR-93 group, the oe-miR-93 + DAB2 group exhibited obvious deformed nuclei, nuclear nucleus margin, nucleus shrinkage or fragmentation, chromatin concentration and aggregation and marginalization. Under the inverted fluorescence microscope, the nuclei exhibited bright blue spots, or were wrapped by the nucleus membrane into apoptotic bodies, showing typical morphological changes of apoptosis (P<0.05; Fig. 6F).
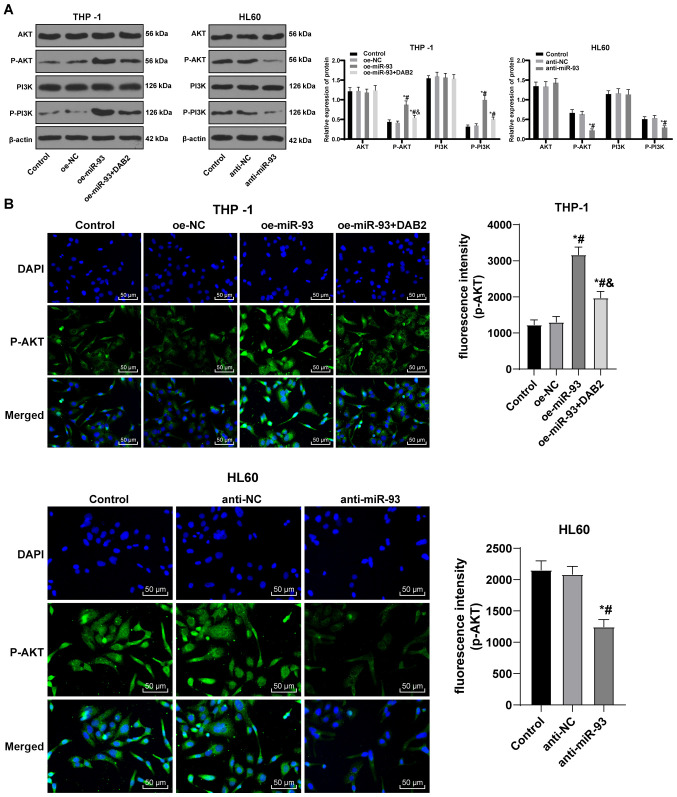
miR-93 promotes the activation of the PI3K/AKT pathway by upregulating DAB2
The overexpression of the DAB2 interactive protein inhibits the activation of the PI3K/AKT pathway (23). Targeting the PI3K/AKT/mTOR pathway with small molecule inhibitors may lead to decreased toxicity and therapeutic effects in AML patients (24). Thus, it was hypothesized that miR-93/DAB2 regulates the PI3K/AKT pathway to manipulate AML cell growth. Western blot analysis was performed to determine protein levels and phosphorylation of AKT and PI3K in each group. The levels of p-AKT and p-PI3K were elevated in the THP-1 cells overexpressing miR-93, but were downregulated following DAB2 overexpression. There were no notable differences in the protein levels of AKT and PI3K in the HL60 cells transfected with anti-miR-93, but the levels of p-AKT and p-PI3K were notably decreased (all P<0.05; Fig. 7A).
Figure 7.
miR-93 promotes the activation of PI3K/AKT pathway via downregulating DAB2. (A) Protein bands and relative protein levels of AKT, p-AKT, PI3K and p-PI3K were measured by western blot analysis. (B) Representative images of fluorescence expression of p-AKT by immunofluorescence assay. FITC-labeled goat anti-rabbit IgG (H + L) exhibited a green emission light (i.e., expression and localization of p-Akt protein immunofluorescence), and DAPI-stained cells exhibited a blue emission light (i.e., nuclear staining). Each experiment was performed 3 times, independently. The data are presented as the means ± standard deviation. Data in (A) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC or anti-NC group; &P<0.05, compared with the oe-miR-93 group. miR-93, microRNA-93; NC, negative control; DAB2, disabled 2; AKT, protein kinase B; PI3K, phosphatidylinositol-3 kinase.
The fluorescence expression of p-AKT was visualized using an immunofluorescence assay. In contrast to the control and NC groups, the fluorescence expression of p-AKT in the THP-1 cells was notably increased in the oe-miR-93 group (P<0.05). Compared with the oe-miR-93 group, the fluorescence expression of p-AKT in the oe-miR-93 + DAB2 group decreased, while the expression of p-AKT in HL60 cells transfected with anti-miR-93 was greatly decreased (all P<0.05; Fig. 7B). The results revealed that miR-93 promoted the activation of the PI3K/AKT pathway by targeting DAB2 to participate in the growth of AML cells.
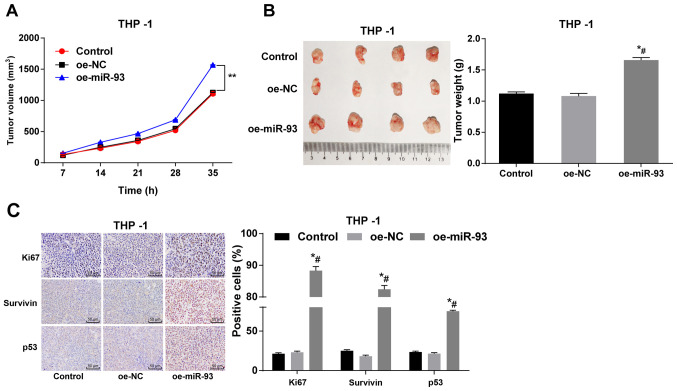
Anti-miR-93 inhibits xenograft tumors in vivo
THP-1 cells overexpressing miR-93 were subcutaneously injected into nude mice, and the tumor volume and weight were measured. No significant differences were observed in tumor volume and weight between the control and NC groups (P>0.05). Compared with the control and NC groups, the injection of THP-1 cells overexpressing miR-93 significantly increased tumor volume and weight (P<0.05; Fig. 8A and B). Immunohistochemistry was also employed to measure the expression of Ki67, survivin and p53. Compared with the control and NC groups, the numbers of Ki67-, surviving- and p53-positive THP-1 cells transfected with oe-miR-93 were notably increased (all P<0.05; Fig. 8C). Taken together, these findings demonstrate that miR-93 overexpression promotes tumor growth in nude mice.
Figure 8.
Anti-miR-93 inhibits xenograft tumors in vivo. THP-1 cells overexpressing miR-93 were injected subcutaneously into nude mice. (A) Relative tumor volume. (B) Relative tumor weight. (C) Positive expression of Ki67, survivin and p53 in tumors measured by immunohistochemistry (n=6). Data are presented as the means ± standard deviation. Data in (A-C) were analyzed by one-way ANOVA, followed by Tukey's multiple comparisons test. *P<0.05, compared with the control group; #P<0.05, compared with the oe-NC group. miR-93, microRNA-93; NC, negative control; AML, acute myeloid leukemia.
Discussion
Currently, one feasible treatment option exists for patients with recurrent or refractory leukemia, namely allogeneic hematopoietic stem cell transplant, with cure rates of only ~10% using standard chemotherapy (34). With a deeper understanding of the pathogenesis of AML, new drugs for various operational targets can be developed (35). miRs are involved in normal hematopoiesis, suggesting that the dysregulation of miRs can contribute to leukemogenesis (36). The present study assessed the mechanisms of miR-93 and DAB2 in the biological processes of AML with the involvement of the PI3K/AKT pathway. Consequently, it was found that the silencing miR-93 suppressed AML cell growth by negatively regulating DAB2 and inactivating the PI3K/AKT pathway.
The first major result was that miR-93 expression was upregulated in patients with AML and in AML cells. The unfavorable role of miR-93 has been identified in other diseases. For instance, miR-93 has been shown to be markedly upregulated in nasopharyngeal carcinoma and breast cancer (37,38). A previous study demonstrated that miR-93 upregulation was positively associated with clinicopathologic grades, but negatively with overall survival time in patients with gliomas (25). Meenhuis et al confirmed that miR-93 was abundantly expressed in human and mouse hematopoietic progenitor cells, and that its expression significantly decreased during bone marrow differentiation (39). Convincingly, Verboon et al stated that the miR-106b-25 cluster, to which miR-93 belongs, was upregulated in relapsed mixed lineage leukemia-rearranged pediatric AML (40). In the present study, the area under the curve was 0.8143 (sensitivity, 0.6786; specificity, 0.8333), indicating that miR-93 expression had certain clinical significance for the diagnosis of AML. Thus, the present study investigated miR-93 expression in the patients with AML and the authors aim to perform further studies in the future on this matter as well.
Transfection with anti-miR-93 vectors inhibited AML cell proliferation, resistance to apoptosis and tumor growth. In line with the results of the present study, the downregulation of miR-93 has been shown to effectively attenuate the proliferation, migration and metastasis of A549, H1975 and H1299 cells (41). Another study revealed that the inhibition of miR-93 suppressed hepatocellular carcinoma cell proliferation and migratory ability (42). In addition, the knockdown of miR-93 has been shown to suppress gastric tumor growth in vivo (43). Cancer-derived miR-93 knockout inhibits leukemia development by reducing the number of stem cells, inhibiting cell proliferation, and promoting cell differentiation and apoptosis (14), which is consistent with the present findings. Moreover, a previous study demonstrated that AML-derived mesenchymal stem cells exhibited a low miR-93 expression, which could promote the production of antioxidant metabolites to protect cells from oxidative stress injury in the leukemia microenvironment (44).
Furthermore, the data of the present study support the hypothesis that miR-93 targets DAB2 and that the overexpression of DAB2 inhibits AML cell proliferation and facilitates apoptosis. miR-93 has previously been reported as an effective inhibitor of DAB2 expression, and the overexpression of miR-93 is associated with the lower expression of DAB2 in lung cancer (20). The tumor suppressor gene, DAB2, exerts potent inhibitory functions in the growth of solid tumor cells and regulates the AKT pathway (45). Dynamic DAB2 expression in vascular and microglial progenitor cells strongly indicates that DAB2 is implicated in the development of bone marrow cells (46). The decreased expression of DAB2 in patients with leukemia is related to the aggravation of the disease (33). As previously described, the restoration of DAB2 interactive proteins prevents the proliferation, colony-forming ability, migration, subcutaneous tumor growth and pulmonary metastasis of nasopharyngeal carcinoma cells by inactivating the PI3K/AKT pathway (23). Additionally, the present study verified that anti-miR-93 inactivated the PI3K/AKT pathway by targeting DAB2. A previous study revealed that PI3K activity regulates the proliferation and apoptosis of erythroid progenitor cells, and that the PI3K/AKT pathway is activated in 50% of AML cases (47). The downregulation of miR-93 reduces the levels of p-PI3K and p-AKT, thus inactivating the PI3K/AKT pathway in nasopharyngeal carcinoma (38). Similarly, miR-22 overexpression promotes chronic lymphocytic leukemia progression by activating the PI3K/AKT pathway (48).
In conclusion, the present study demonstrates that the inhibition of miR-93 suppresses AML cell growth by negatively regulating DAB2 and inactivating the PI3K/AKT pathway. These results are promising in the prospect of promoting future individualized treatments for patients with AML. Although the current findings provide therapeutic implications for the treatment of AML, the experimental results and their translation into the effective application in clinical practice warrant further in-depth investigations. The authors aim to focus on the effects and mechanism of AML cell differentiation in future research and to improve the experimental design to explore the mechanisms of oe-miR-93 and anti-miR-93 in experimental cell lines. The authors also aim to further investigate the effects of miR-93 expression on the prognosis of patients with AML in future experiments, as well as to use core blood cells for experiments, analyze mature myeloid markers, and conduct animal experiments in an intravenous manner. However, it is hoped that the findings of the present study may aid future research on AML.
Acknowledgements
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Authors' contributions
All authors are the guarantors of the integrity of the entire study. JH, RX, XW and ZF contributed to the study concept, study design and definition of intellectual content. XW, ZF, JH, RX, ZF and BK contributed to the manuscript preparation, editing and reviewing. JL and BK contributed to the clinical experiments. MY and LZ contributed to the experiments and data acquisition. JW contributed to data analysis and statistical analysis. JH and RX confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved and supervised by the Ethics Committee of the Third Affiliated Hospital of Sun Yat Sen University. All subjects signed an informed consent form. All animal experiments involving nude mice were approved by the Laboratory Animal Ethics Committee of the Third Affiliated Hospital of Sun Yat Sen University. Significant efforts were made to minimize both the number of animals and their suffering.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Juliusson G, Hough R. Leukemia. Prog Tumor Res. 2016;43:87–100. doi: 10.1159/000447076. [DOI] [PubMed] [Google Scholar]
- 2.Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 3.Cammarata G, Augugliaro L, Salemi D, Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A, Bica MG, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 4.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76. doi: 10.1016/j.blre.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Crump C, Sundquist J, Sieh W, Winkleby MA, Sundquist K. Perinatal risk factors for acute myeloid leukemia. Eur J Epidemiol. 2015;30:1277–1285. doi: 10.1007/s10654-015-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fircanis S, Merriam P, Khan N, Castillo JJ. The relation between cigarette smoking and risk of acute myeloid leukemia: An updated meta-analysis of epidemiological studies. Am J Hematol. 2014;89:E125–E132. doi: 10.1002/ajh.23744. [DOI] [PubMed] [Google Scholar]
- 8.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 9.Kadia TM, Ravandi F, Cortes J, Kantarjian H. New drugs in acute myeloid leukemia. Ann Oncol. 2016;27:770–778. doi: 10.1093/annonc/mdw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes BC, Rueff J, Rodrigues AS. MicroRNAs and cancer drug resistance. Methods Mol Biol. 2016;1395:137–162. doi: 10.1007/978-1-4939-3347-1_9. [DOI] [PubMed] [Google Scholar]
- 11.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al. miR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352–4365. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, Matsuoka M, Jeang KT. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y, Meng M, Zhang G, Han H, Zhou Q. Oncogenic microRNAs in the genesis of leukemia and lymphoma. Curr Pharm Des. 2014;20:5260–5267. doi: 10.2174/1381612820666140128211724. [DOI] [PubMed] [Google Scholar]
- 15.Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W, Ling Y, Wang R, Yang Y, Liu Y. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8:e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Zhu L, Wang Y, Zhou D, Zhu J, Xie W, Ye X. Profiling of microRNAs in AML cells following overexpression or silencing of the VEGF gene. Oncol Lett. 2017;13:105–110. doi: 10.3892/ol.2016.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Jiang M, Yuan W, Tang H. Prognostic value of miR-93 overexpression in resectable gastric adenocarcinomas. Acta Gastroenterol Belg. 2012;75:22–27. [PubMed] [Google Scholar]
- 18.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, Xu J, Wu Y, Chen Q, Zheng W, Lu X, Zhou C, Jiao D. Identification of microRNA-93 as a functional dysregulated miRNA in triple-negative breast cancer. Tumour Biol. 2015;36:251–258. doi: 10.1007/s13277-014-2611-8. [DOI] [PubMed] [Google Scholar]
- 20.Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E, Suraokar M, Wistuba I, Minna JD, Pertsemlidis A. miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;33:4307–4315. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkielstein CV, Capelluto DG. Disabled-2: A modular scaffold protein with multifaceted functions in signaling. Bioessays. 2016;38(Suppl 1):S45–S55. doi: 10.1002/bies.201670907. [DOI] [PubMed] [Google Scholar]
- 22.Tseng CP, Huang CL, Cheng JC. Serine 24 phosphorylation of Disabled-2 negatively regulates K562 cell adhesion to fibrinogen. Proceedings of the The 95th Annual Meeting of American Association for Cancer Research (AACR); AACR, Orlando, FL. 2004. [Google Scholar]
- 23.Wang B, Gu Q, Li J. DOC-2/DAB2 interactive protein regulates proliferation and mobility of nasopharyngeal carcinoma cells by targeting PI3K/Akt pathway. Oncol Rep. 2017;38:317–324. doi: 10.3892/or.2017.5704. [DOI] [PubMed] [Google Scholar]
- 24.Martelli AM, Evangelisti C, Chiarini F, Grimaldi C, Manzoli L, McCubrey JA. Targeting the PI3K/AKT/mTOR signaling network in acute myelogenous leukemia. Expert Opin Investig Drugs. 2009;18:1333–1349. doi: 10.1517/14728220903136775. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, Wang C, Lei F, Zhang L, Zhang X, Liu A, Wu G, Zhu J, Song L. miR-93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget. 2015;6:8286–8299. doi: 10.18632/oncotarget.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Shen T. Blood disease diagnosis and efficacy standards. 3rd edition. Science Press; Beijing: 2007. [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36(Database Issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Lu Q, Zhu J, Fu J, Chen YX. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn Pathol. 2014;9:76. doi: 10.1186/1746-1596-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardoni R, Giordani G, Signorino E, Monticelli S, Messa F, Pradotto M, Rosso V, Bracco E, Giangrande A, Perini G, et al. A new BCR-ABL1 Drosophila model as a powerful tool to elucidate the pathogenesis and progression of chronic myeloid leukemia. Haematologica. 2019;104:717–728. doi: 10.3324/haematol.2018.198267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose P, Vachhani P, Cortes JE. Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol. 2017;18:17. doi: 10.1007/s11864-017-0456-2. [DOI] [PubMed] [Google Scholar]
- 35.Saygin C, Carraway HE. Emerging therapies for acute myeloid leukemia. J Hematol Oncol. 2017;10:93. doi: 10.1186/s13045-017-0463-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang H, Hong M, Jiang T, Jiang Q, Lu J, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia. 2012;26:769–777. doi: 10.1038/leu.2011.273. [DOI] [PubMed] [Google Scholar]
- 37.Deng ZQ, Qian J, Liu FQ, Lin J, Shao R, Yin JY, Tang Q, Zhang M, He L. Expression level of miR-93 in formalin-fixed paraffin-embedded tissues of breast cancer patients. Genet Test Mol Biomarkers. 2014;18:366–370. doi: 10.1089/gtmb.2013.0440. [DOI] [PubMed] [Google Scholar]
- 38.Lyu X, Fang W, Cai L, Zheng H, Ye Y, Zhang L, Li J, Peng H, Cho WC, Wang E, et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13:51. doi: 10.1186/1476-4598-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, et al. miR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verboon LJ, Obulkasim A, de Rooij JD, Katsman-Kuipers JE, Sonneveld E, Baruchel A, Trka J, Reinhardt D, Pieters R, Cloos J, et al. MicroRNA-106b~25 cluster is upregulated in relapsed MLL-rearranged pediatric acute myeloid leukemia. Oncotarget. 2016;7:48412–48422. doi: 10.18632/oncotarget.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pignataro D, Francia S, Zanetta F, Brenna G, Brandini S, Olivieri A, Torroni A, Biamonti G, Montecucco A. A missense MT-ND5 mutation in differentiated Parkinson Disease cytoplasmic hybrid induces ROS-dependent DNA damage response amplified by DROSHA. Sci Rep. 2017;7:9528. doi: 10.1038/s41598-017-09910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu D, He XX, Chang Y, Sun SZ, Xu CR, Lin JS. Downregulation of miR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology. 2012;59:2367–2373. doi: 10.5754/hge12458. [DOI] [PubMed] [Google Scholar]
- 43.Liang H, Wang F, Chu D, Zhang W, Liao Z, Fu Z, Yan X, Zhu H, Guo W, Zhang Y, et al. miR-93 functions as an oncomiR for the downregulation of PDCD4 in gastric carcinoma. Sci Rep. 2016;6:23772. doi: 10.1038/srep23772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruvolo PP, Jacamo R, McQueen T, Pan R, Samudio I, Konopleva M, Kornblau SM, Garzon R, Croce CM, Andreeff M. Suppression of Mir-93 may regulate anti-oxidant metabolism in mesenchymal stromal cells derived from acute myeloid leukemia patients. Blood. 2012;120:2354. doi: 10.1182/blood.V120.21.2354.2354. [DOI] [Google Scholar]
- 45.Yan ZX, Wu LL, Xue K, Zhang QL, Guo Y, Romero M, Leboeuf C, Janin A, Chen SJ, Wang L, Zhao WL. MicroRNA187 overexpression is related to tumor progression and determines sensitivity to bortezomib in peripheral T-cell lymphoma. Leukemia. 2014;28:880–887. doi: 10.1038/leu.2013.291. [DOI] [PubMed] [Google Scholar]
- 46.Cheung KK, Mok SC, Rezaie P, Chan WY. Dynamic expression of Dab2 in the mouse embryonic central nervous system. BMC Dev Biol. 2008;8:76. doi: 10.1186/1471-213X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, Green A, Mayeux P, Lacombe C, Bouscary D. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica. 2010;95:819–828. doi: 10.3324/haematol.2009.013797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palacios F, Abreu C, Prieto D, Morande P, Ruiz S, Fernandez-Calero T, Naya H, Libisch G, Robello C, Landoni AI, et al. Activation of the PI3K/AKT pathway by microRNA-22 results in CLL B-cell proliferation. Leukemia. 2015;29:115–125. doi: 10.1038/leu.2014.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.