Abstract
A duplex PCR to detect Bordetella pertussis and Bordetella parapertussis was developed with the insertion sequences IS481 (B. pertussis) and IS1001 (B. parapertussis) and evaluated with specimens from 520 consecutive patients presenting with possible pertussis. No culture-positive–PCR-negative results occurred, giving the method a sensitivity of 100%. For B. pertussis, 58 of 520 patients (11.2%) were positive by PCR compared to 17 of 520 patients positive (3.3%) by culture. For B. parapertussis, 7 of 520 patients (1.3%) were positive by PCR compared to 2 of 520 patients positive (0.4%) by culture. Two patients were positive for both B. pertussis and B. parapertussis. Patient records were reviewed to determine the validity of PCR-positive–culture-negative results. Forty-two of 49 patients who could be evaluated fulfilled the criteria for a case definition of pertussis, with 32 patients being <1 year of age and having classical pertussis symptoms. The seven patients who did not fulfil the criteria were aged 7 to 55 years and had a persistent cough for >2 weeks. The method was also used to investigate a classroom outbreak in which B. pertussis culture was positive for 5 of 28 patients. All five culture-positive specimens were confirmed by PCR, and an additional eight were positive by PCR. Of 25 patients from a suspected pertussis outbreak in a girls’ dormitory, seven of seven specimens were negative for B. pertussis, although 13 of 25 patients were positive for B. pertussis immunoglobulin M (IgM) (2 of which produced equivocal IgA results, with 23 of 25 patients being negative). Five symptomatic patients were subsequently found to be positive (by IgM and particle agglutination assays) for Mycoplasma pneumoniae, demonstrating the value of PCR in rapidly excluding B. pertussis infection in an outbreak situation. Twenty-two of 71 (30.1%) throat swabs were positive by PCR compared to 2 of 71 (2.8%) throat swabs positive by culture, indicating that a reassessment of the use of throat swabs should be considered, particularly for older patients, in contact tracing, and in situations in which specimen collection is difficult.
The striking and unique presentation of classical pertussis occurs in previously unimmunized children and does not usually present a clinical diagnostic dilemma (3). Atypical pertussis occurs in two general scenarios and offers a greater diagnostic challenge to the clinician. First, neonates and very young infants may present with apnea and seizures with no accompanying paroxysms (10). Second, mild or absent symptoms may occur in adults (6, 20) or previously vaccinated children (9, 13). It has been shown that atypical illness in adults is common, endemic, and usually unrecognized (6). The epidemiological implications of unrecognized pertussis are that exposure of unimmunized infants to individuals with pertussis places them at high risk and that pertussis remains endemic in society (16). Bordetella parapertussis also causes pertussis syndrome, usually a milder illness than that caused by Bordetella pertussis, although severe cases have been described (10). To add to the diagnostic dilemma, other agents are also thought to cause the pertussis syndrome. Adenovirus (2, 4, 11), chlamydiae, cytomegalovirus, and respiratory syncytial virus (4) have all been implicated. The role of laboratory testing is extremely important in the diagnosis of atypical disease and in the detection of other putative causative agents in classical disease.
Isolation of B. pertussis is considered to be the “gold-standard” for the diagnosis of pertussis due to its high specificity (17). The success of culture is highly dependent upon collection and laboratory techniques, the age and immune status of the patient (success of culture is high with unimmunized infants but low with older, immunized, and partially treated patients), and the stage of disease (success of culture is high at the end of incubation or start of the catarrhal phase but low after this) (17). Because of all of these factors, the sensitivity of culture is low, especially for the atypical pertussis population. Although direct fluorescent antibody testing can provide a rapid diagnosis for a patient with classical pertussis, its specificity is poor and should not replace culture or be used to detect atypical disease (14). Serological tests have been used extensively for the diagnosis of pertussis, but without a sensitive gold standard, their reliability has not yet been ascertained.
A highly sensitive, specific, and rapid laboratory test to detect the presence of B. pertussis in clinical specimens is urgently needed for the diagnosis of acute disease, to detect atypical disease, and to assess the reliability of other testing modes such as the direct fluorescent antibody assay and serology. The use of nucleic acid amplification methods, such as PCR, are highly suited to the detection of fastidious organisms which are significant by their presence even in an asymptomatic individual. B. pertussis is such an organism, and many nucleic acid amplification-based tests have been developed over the past few years, although a suitable protocol has not yet been agreed upon. Although excellent sensitivity compared to that of culture has been achieved when B. pertussis alone has been assayed (1, 7, 8), less than optimal sensitivity compared to that of culture has been observed in large studies with methods that detect both pathogens (12, 19, 24).
In the present study, a nested duplex PCR assay was developed to detect both B. pertussis and B. parapertussis by using as a basis two previously published methods (1, 23) and international recommendations for the use of PCR in the diagnosis of pertussis (15). We chose the repetitive insertion sequences IS481 (B. pertussis) and IS1001 (B. parapertussis) as targets for our assay because of their high copy number per cell and species specificity. A sensitivity of 1 organism per reaction was obtained for both B. pertussis and B. parapertussis even in the presence of an excess amount of the other organism. In addition, the nested format reduced the chance of PCR inhibition due to the significant dilution of the primary specimen.
We applied this method to the diagnosis of pertussis in a semirural-to-rural area of Australia and compared the results to those of culture and clinical data to determine its reliability. The method was also applied to a culture-confirmed outbreak of primary B. pertussis infection and a pseudo-outbreak of pertussis later confirmed as an outbreak of Mycoplasma pneumoniae infection. We also assessed the value of using this method to diagnose infection from throat swabs, with the rationale that diagnosis of adults with atypical disease and asymptomatic carriers could be more easily achieved due to the expected greater patient compliance with this less invasive collection method.
MATERIALS AND METHODS
Bacterial strains and chromosomal DNA.
Bacterial strains used in this study are listed in Table 1. American Type Culture Collection (Remel) strains were purchased from Microdiagnostics, Brisbane, Australia. B. pertussis Tohama I and III and B. pertussis 18323 strains were kindly donated by Alison Weiss (University of Cincinnati, Ohio). Cincinnati, Patrick Blackall (Department of Primary Industries, Queensland Government, Brisbane, Australia) kindly donated DNA from Bordetella avium and Bordetella hinzii. The remainder of the strains were clinical strains from the Queensland Health Pathology Service, Toowoomba Laboratory culture collection. DNAs were extracted from cultures with an Orca IsoQuick nucleic acid extraction kit (Progen, Brisbane, Australia) as per the manufacturer’s instructions. DNA was quantitated with a GeneQuant II RNA-DNA calculator (Pharmacia Biotech).
TABLE 1.
Bacterial species (strains and DNA) used in this study
| Strain(s) or DNA | ||
|---|---|---|
| Bordetella species (no. tested) | Pseudomonas aeruginosa | |
| B. pertussis ATCC 9340, ATCC 12242 | Mycoplasma pneumoniae DNA | |
| B. pertussis Tohama I and III | Moraxella catarrhalis | |
| B. pertussis 18323 | Haemophilus influenzae | |
| B. pertussis clinical strains (16) | Neisseria lactamica | |
| B. parapertussis ATCC 15237 | Burkholderia cepacia | |
| B. parapertussis clinical strains (5) | Streptococcus anginosus | |
| B. bronchiseptica ATCC 10580 | Streptococcus intermedius | |
| B. bronchiseptica clinical strains (2) | Streptococcus constellatus | |
| B. avium DNA (1) | Streptococcus milleri | |
| B. hinzii DNA (1) | Streptococcus pneumoniae | |
| Other (1 strain tested) | Streptococcus pyogenes | |
| Alcaligenes faecalis II | Staphylococcus aureus | |
| Acinetobacter baumanii | Staphylococcus epidermidis | |
| Kingella kingae | Bacteroides fragilis | |
| Brucella suis | Fusobacterium necrophorum | |
| Shewanella putrifaciens | Chlamydia trachomatis | |
| Stenotrophomonas maltophilia | DNA | |
| Escherichia coli | Neisseria gonorrhoeae | |
Collection and processing of samples.
Nasopharyngeal aspirates (NPA), throat swabs, nasal swabs, and nasopharyngeal swabs (NPS) were received by the laboratory from pediatricians, general practitioners, outlying hospitals, and other laboratories. Specimen collectors were questioned for collection protocols, and as far as can be ascertained, all specimens were collected by standard collection techniques (14). Three types of specimens were tested: (i) 532 consecutive specimens from 520 patients that were submitted to our laboratory between 1992 and 1997; (ii) 28 specimens from patients and contacts of these patients after a culture-proven outbreak, sent upon a request from another laboratory; and (iii) 7 throat swab specimens obtained from a suspected outbreak of pertussis in a girls’ dormitory in Toowoomba, Queensland, Australia. Also, blood from seven of the patients in the last group and 18 contacts of these patients was collected for serological investigation of pertussis.
All specimens were swabbed onto two prewarmed full-plate media, in the laboratory, as soon as possible after collection. The media were Regan-Lowe medium with 10% horse blood and 40 mg of cephalexin (Microdiagnostics) per liter and Moredun medium (for the selective isolation of B. parapertussis [5] Microdiagnostics). Plates were incubated at 35°C in a perspex chamber containing air with 100% humidity for up to 12 days. Throat swabs were collected into either Bordetella Transport Medium (Microdiagnostics) tubes or charcoal-based Stuart’s transport medium with a Dacron swab. Bordetella Transport Medium is identical to Regan-Lowe medium except that half-strength agar is used. Plates were examined daily under a plate microscope for suspect colonies, which were identified by phenotypic characteristics (14) and agglutination with antipertussis and antiparapertussis antibodies as per the recommendations of the manufacturer (Murex Diagnostics).
NPA were mixed well by vortexing for 30 s, and aliquots were frozen at −20°C until they were tested. Swabs were placed in 0.75 ml of sterile RNase- and DNase-free water, swirled vigorously, and wrung out, and specimens were removed before being frozen at −20°C until they were tested. All specimens were frozen before further treatment even if only overnight. On the day of testing specimens were removed from the freezer, thawed at 37°C, and then vortexed for 30 s. A 25-μl aliquot was heated to 99.9°C for 20 min in a thermal cycler, and PCR was performed directly from this specimen.
Reagents and synthetic oligonucleotides.
Oligonucleotides used in this study are listed in Table 2. They were synthesized commercially by three suppliers: Bresatech (Adelaide, South Australia), Gibco-Life Technologies (Gaithersburg, Md.), and Operon Technologies or Genemed Technologies (both in San Francisco, Calif.; oligonucleotides were purchased from Fisher-Biotech, Perth, Australia).
TABLE 2.
Oligonucleotides used in this study to detect B. pertussis and B. parapertussis
| Organism | Oligonucleotide | Sequence | Reference or Source |
|---|---|---|---|
| B. pertussis (target IS481) | Outer primers | ||
| BPIS4801 | 5′GACTTCGTCTTCGTGGCCAT3′ | ||
| BPIS4802 | 5′GTACAGCGCGCCCGATGCCT3′ | 1 | |
| Inner primers | |||
| BPNEST1 | 5′CGCGTGGCCTTCACCGACAT3′ | 1 | |
| BPNEST2 | 5′GGGCGGTAAGGTCGGGTAAA3′ | 1 | |
| B. parapertussis (target IS1001) | Outer primers | ||
| BPPA | 5′CGCCGCTTGATGACCTTGATA3′ | 23 | |
| BPPZ | 5′CACCGCCTACGAGTTGGAGAT3′ | 23 | |
| Inner primers | |||
| PARAN1 | 5′CGCTGGCTGCTGCTGCGCAA3′ | This study | |
| PARAN2 | 5′GTGGTTCCAGGCTTGTCTTG3′ | This study |
Reagents and their manufacturers and concentrations used in this study were as follows: an ultrapure deoxynucleoside triphosphate (dNTP) set (dATP, dCTP, dGTP, and dTTP, 10 mM each; Pharmacia Biotech) and Taq polymerase (5 U/μl with 10× PCR buffer and 25 mM MgCl2; Boehringer Mannheim).
Serological assays.
Sera were assayed with Panbio (Brisbane, Australia) B. pertussis immunoglobulin A (IgA) and IgM kits as per the manufacturer’s instructions. M. pneumoniae particle agglutination (Fujirebio; Australian Diagnostics) and IgM (Immunocard; Oxoid Australia) serology assays were performed per the manufacturers’ instructions.
Duplex nested PCR.
Ten microliters of treated specimen was added to 40 μl of a master mix (made immediately before use) containing 5 μl of 10× PCR buffer, 200 μM each dNTP, 10 pmol of primers IS4801 and IS4802, 12 pmol of primers BPPA and BPPZ, 2.0 U of Taq polymerase, and 1.5 mM MgCl2. Amplification was performed in a Perkin-Elmer 9600 GeneAmp thermal cycler with the following parameters: 2 min at 94°C followed by 20 cycles of 30 s at 94°C, 30 s at 57°C, and 30 s at 72°C, with the last cycle concluding with a reaction for 5 min at 72°C. Five microliters of this product was transferred to 45 μl of a new master mix containing the following: 5 μl of 10× PCR buffer; 200 μM each dNTP; 10 pmol of primers BPNEST1, BPNEST2, PARAN1, and PARAN2; 2.0 U of Taq polymerase; and 1.5 mM MgCl2. Amplification was performed in a Perkin-Elmer 9600 GeneAmp thermal cycler with the following parameters: 2 min at 94°C followed by 30 cycles of 30 s at 94°C, 30 s at 57°C, and 30 s at 72°C, with the last cycle concluding with a reaction for 5 min at 72°C. The amplified products were detected after electrophoresis through a 2% agarose gel containing 0.5 μg of ethidium bromide per ml.
RESULTS
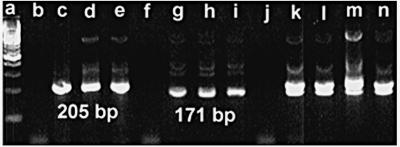
The presence of an expected 205-bp band for B. pertussis or a 171-bp band for B. parapertussis was scored as a positive result (Fig. 1). All positive results were confirmed by repeating the PCR with the primary specimen in the next run. A low-copy-number positive control (∼20 CFU per reaction mixture) for both B. pertussis and B. parapertussis was used in every run, and every third specimen was a negative control. A run was repeated with the original specimens if any negative control gave a positive result or a positive control gave a negative result.
FIG. 1.
Lanes (all values in parentheses are CFU per reaction mixture); a, 100-bp ladder; b, negative control; c to e, B. pertussis (1,000, 100, and 10, respectively); negative control; g to i, B. parapertussis (1,000, 100, and 10, respectively); j, negative control; k, B. pertussis (1,000) and B. parapertussis (1,000); l, B. pertussis (10) and B. parapertussis (1,000); m, B. pertussis (1,000) and B. parapertussis (10); n, B. pertussis (10) and B. parapertussis (10).
The specificity of the duplex nested PCR assay was 100% with 100 ng of purified DNA from each organism listed in Table 1. The clinical strains of B. pertussis tested were chosen from different geographical locations and years of isolation to minimize clonality.
Analytical sensitivity was assessed by diluting 48-h cultures in Stainer-Scholt broth (Microdiagnostics) (22) and performing standard plate counts with two fresh clinical strains, DFB102 (B. pertussis) and DFB952 (B. parapertussis), on Bordet-Gengou medium with 15% horse blood (Microdiagnostics). With log10 dilutions of these strains, treated identically to specimens, the duplex assay was able to detect 1 viable organism per reaction mixture for each organism even when an excess of the other organism was present.
Five hundred twenty patients (532 specimens) were assayed for the presence of B. pertussis and B. parapertussis by the nested duplex PCR and culture. The results of this study are given in Table 3. No culture-positive–PCR-negative results were obtained, giving the PCR assay 100% sensitivity; however, a large number of PCR-positive–culture-negative specimens were obtained. To determine the validity of these results, patient records were reviewed to determine if clinical data supported the positive PCR result. A PCR-positive–culture-negative result was considered a true positive if a National Health and Medical Research Council (NHMRC) case definition was fulfilled (18), that is, if (i) B. pertussis was isolated from a clinical specimen; (ii) specific IgA was found in the serum of a patient with a history of a clinically compatible illness; or (iii) a patient having an illness lasting 2 weeks or more had one of the following: paroxysms of coughing, inspiratory whoop without other causes, or posttussive vomiting. An illness characterized by a cough lasting at least 2 weeks in a patient who is epidemiologically related to a patient with a laboratory-confirmed case of pertussis was also considered a true-positive case by NHMRC guidelines. Sufficient clinical data were available to assess 49 of 51 positive patients, with 42 of 49 fulfilling the NHMRC criteria for confirmed pertussis. From this group of 42 patients, 32 were infants of <1 year of age with classical pertussis symptoms. The seven patients which did not fulfill NHMRC criteria were aged 7, 7, 9, 11, 27, 34, and 55 years, and all had had a persistent cough for >2 weeks. Table 4 shows the distribution of results by specimen type.
TABLE 3.
Results of pertussis and parapertussis PCR versus results of culturea
| Culture result | No. of patients (n = 520)
|
|||
|---|---|---|---|---|
| Pertussis PCR
|
Parapertussis PCR
|
|||
| Positive | Negative | Positive | Negative | |
| Positive | 17 | 0 | 2 | 0 |
| Negative | 41b | 462 | 5b | 513 |
The percentage of patients who were PCR positive for pertussis was 11.2%; that of patients who were culture positive for pertussis was 3.3%. The percentage of patients who were PCR positive for parapertussis was 1.3%; that of patients who were culture positive for parapertussis was 0.4%.
Includes two patients whose specimens were both positive by PCR and negative by culture for both B. pertussis and B. parapertussis.
TABLE 4.
Distribution of results by specimen type
| Specimen type | Total no. of specimens (n = 532)a | No. of specimens (% positive)
|
|
|---|---|---|---|
| PCR positive | Culture positive | ||
| NPA | 445 | 41 (9.2) | 14 (3.1) |
| Throat swab | 71 | 22 (30.1) | 2 (2.8) |
| Nasal swab | 5 | 2 | 0 |
| Sputum | 9 | 1 | 1 |
| Lung tissue | 2 | 1 | 0 |
Number of patients, 520.
Twenty-eight nasopharyngeal specimens from 28 patients from the same school class were sent to us, from another laboratory at our request, upon our learning that five of these patients were culture positive. We repeated the culture and obtained the same five positive results. All five culture-positive specimens were positive by PCR. Eight of the 23 culture-negative specimens were also positive by PCR.
Two of the 25 patients from the suspected pertussis outbreak in the girls’ dormitory gave an equivocal result for pertussis IgA. The remaining 23 patients were negative for pertussis IgA. Thirteen of the 25 patients were positive for pertussis IgM. However, the throat swabs collected from all seven symptomatic patients tested negative by PCR. The main presenting symptoms were described by the clinician, as a dry persistent cough and a sore throat. We performed mycoplasma serology on these specimens, and five of the symptomatic patients tested positive by both particle agglutination and IgM serology.
DISCUSSION
The duplex-nested-PCR assay described in this paper was shown to be both highly sensitive and specific when it was tested with B. pertussis, B. parapertussis, commonly encountered respiratory organisms, other bordetellae, and genetically similar organisms. A sensitivity of 100% was achieved for both organisms when the method was compared to culture, representing a significant advantage over previously published methods. When applied clinically, and assessed against culture and clinical data, the method proved to be more sensitive than culture.
In a large vaccine efficacy trial in Germany (555 patients), the sensitivity of PCR was shown to be fourfold greater than that of culture (21). In that study 26 of 28 culture-positive specimens were positive by PCR and an additional 82 specimens were positive by PCR and negative by culture. The PCR-positive–culture-negative patients had good clinical correlation with pertussis. Importantly, an excellent culture protocol was used for this study, including the use of transport media in outlying areas, but the culture positivity rate was only 25% of the PCR positivity rate. This trend has been observed in all other published studies, including the present study, particularly in the schoolroom epidemic we described above, from which eight extra positive results were obtained over the number of positive results obtained by culture. Clearly, PCR provides a useful adjunct to conventional testing methodologies and should be used wherever possible.
Despite impressive results from all published studies, agreement on a standard methodology has not been forthcoming. No method which fulfils all of the proposed recommendations has been offered (15). Specimen preparation is critical to the success of any PCR method. Elaborate methods to extract DNA can result in loss of DNA. Conversely, the chance of PCR inhibition increases with a decrease in the purity of the specimen. Simple specimen preparation is preferred for pertussis (15); however, many NPA are mucoid and are therefore difficult to process. In contrast to results of other studies, a sensitivity of 100% compared to that of culture was observed in our study. This result shows that the efficacy of simple sample processing can be as high or higher than more elaborate methods. We did not use an inhibition detection method in our study, however. We felt that this was probably not necessary in a nested method due to the great dilution of the sample and the requirement of the first round to provide only a small number of targets; i.e., unless complete inhibition has occurred, the reaction should be successful.
Investigations into the reliability of serological testing for the diagnosis of pertussis have been hampered due to the lack of a reliable diagnostic test for comparison. It seems from our pseudo-outbreak that the role of IgM testing in pertussis diagnosis needs urgent attention. Although a negative IgA test result is of little use with infants, due to delayed antibody production, it has proven useful in the diagnosis of atypical pertussis in older children and adults. Certainly in our pseudo-outbreak the IgA proved to be more reliable. Interestingly, three adult patients in our study complained of a chronic cough for greater than 3 months. All three patients were repeatedly equivocal for pertussis IgA during this period and were PCR positive for pertussis by throat swab (one culture-positive specimen). The method described here provides an opportunity to investigate the strengths and weaknesses of serological testing and the complex epidemiology of pertussis, areas needing intensive research efforts.
In our patients, only 1.3% of PCR-diagnosed pertussis was caused by B. parapertussis. This low rate of infection is similar to those of two previous studies in which rates were reported to be 4% (24) and 1.6% (21). The value of detecting B. parapertussis when it causes infection at such low rates can be questioned on the basis of economics. It may be, however, that the incidence of B. parapertussis is underestimated because it usually results in a less severe disease and specimen collection may not be deemed necessary. Clearly, further research is needed in the epidemiology of infections due to B. parapertussis, and the method described here has sufficient sensitivity and specificity for this purpose.
Throat swabs are not recommended for culture of B. pertussis and B. parapertussis, because although these organisms colonize the entire upper respiratory tract, culture of specimens from this site has low sensitivity (14). In Table 4, our data also show the low yield of positive specimens observed with throat swab cultures, with only 2 of the 22 PCR-positive specimens being positive by culture. Indeed, the high percentage (30.1%, n = 71) of PCR-positive specimens from throat swabs is surprising, as the value is higher than that for NPA (9.2%, n = 445). Although this difference is significant, we do not feel it justifies the replacement of NPA for throat swabs, as several other factors may cause a bias. For instance, many of the NPA were collected during the winter months from infants with respiratory syncytial virus (RSV) infection and the number of pertussis PCR-positive specimens in this group was low in comparison to the total number (data not shown). Conversely, outside of the “RSV season” the number of pertussis PCR-positive results was high in comparison to the total number collected (data not shown). Throat swabs were generally collected only when pertussis (and not RSV) was suspected, often from general practitioners or pediatricians who were reasonably confident of a clinical diagnosis of pertussis and could not collect an NPA or did not want to subject the patient to the more invasive NPS. Indeed, most older patients agreed to a throat swab but not to an NPS.
Previous recommendations (15) on specimen collection may need to be reviewed in light of the data presented in this paper. Our data support the use of throat swabs if a suitably sensitive PCR is available under the following circumstances: (i) an NPA is unable to be collected (often the case outside of a hospital setting), (ii) an NPS proves difficult to obtain either due to lack of compliance (an important consideration with older children and adults) or an unwillingness on the part of the physician to collect the specimen (an important consideration with infants in situations where the clinical diagnosis is high), and (iii) epidemiological investigations such as contact tracing of asymptomatic individuals are carried out.
Disadvantages of the method we have described arise from the inherent problems of nested-PCR assays, in particular the increased risk of carryover of amplified DNA. In addition, the carryover cannot be minimized by the use of the uracil-N-glycosalase carryover prevention system. Because of these problems, this method should be performed only in laboratories with staff experienced in nested-PCR techniques and is not suitable for routine laboratory use. We are currently adapting the method to a format more suitable for the routine laboratory and are attempting to fulfil the remaining recommended criteria (15).
In summary, we have described a sensitive and specific PCR method for the diagnosis of pertussis. Although further modifications are needed to allow routine use, this method further demonstrates the value of this technology for this disease. The method described is being used as a reference method to evaluate simpler methods, to investigate the reliability of serological testing, and to study the epidemiology of pertussis. Throat swabs were shown to be a useful specimen when they are used in conjunction with PCR and should be considered when specimen collection is problematic and in contact tracing.
ACKNOWLEDGMENTS
This study was funded in part by the Private Practice Trust Fund, Toowoomba Health Region.
The support of the following is gratefully appreciated: pediatricians and general practitioners from the Darling Downs region; specimens from Drs. Sullivan and Nicolaides & Partners Laboratories, Brisbane and Lismore (Lynne Wright), Australia; and cultures from J. Faoagali, M. Nolan, J. Bates, R. Reed, R. McDougall, C. Coulter, Pat Blackall, and Alison Weiss.
REFERENCES
- 1.Bäckman A, Johansson B, Olcén P. Nested PCR optimized for detection of Bordetella pertussis in clinical nasopharyngeal samples. J Clin Microbiol. 1994;32:2544–2548. doi: 10.1128/jcm.32.10.2544-2548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barraff L J, Wilkins J, Wehrle P F. The role of antibiotics, immunisations, and adenoviruses in pertussis. Pediatrics. 1978;61:224–230. [PubMed] [Google Scholar]
- 3.Cherry J D. Historical review of pertussis and the classical vaccine. J Infect Dis. 1996;174(Suppl. 3):S259–S263. doi: 10.1093/infdis/174.supplement_3.s259. [DOI] [PubMed] [Google Scholar]
- 4.Connor J D. Evidence for an etiological role of adenoviral infection in pertussis syndrome. N Engl J Med. 1970;283:390–394. doi: 10.1056/NEJM197008202830802. [DOI] [PubMed] [Google Scholar]
- 5.Connor K M, Porter J F, Quirie M M, Donachie W. Moredun Bordetella Medium, an improved selective medium for isolation of Bordetella parapertussis. J Clin Microbiol. 1996;34:638–640. doi: 10.1128/jcm.34.3.638-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deville J G, Cherry J D, Christenson P D, Pineda E, Leach C T, Kuhls T L, Viker S. Frequency of unrecognized Bordetella pertussis infections in adults. Clin Infect Dis. 1995;21:639–642. doi: 10.1093/clinids/21.3.639. [DOI] [PubMed] [Google Scholar]
- 7.Glare E M, Paton J C, Premier R R, Lawrence A J, Nisbet I T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990;28:1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Q, Mertsola J, Soini H, Viljanen M K. Sensitive and specific polymerase chain reaction assays for detection of Bordetella pertussis in nasopharyngeal specimens. J Pediatr. 1994;124:421–426. doi: 10.1016/s0022-3476(94)70365-5. [DOI] [PubMed] [Google Scholar]
- 9.He Q, Viljanen M K, Nikkari S, Lyytikäinen R, Mertsola J. Outcomes of Bordetella pertussis infection in different age groups of an immunised population. J Infect Dis. 1994;170:873–877. doi: 10.1093/infdis/170.4.873. [DOI] [PubMed] [Google Scholar]
- 10.Heininger U, Stehr K, Cherry J D. Serious pertussis overlooked in infants. Eur J Pediatr. 1992;151:342–343. doi: 10.1007/BF02113254. [DOI] [PubMed] [Google Scholar]
- 11.Keller M A, Aftandelians R, Connor J D. Etiology of pertussis syndrome. Pediatrics. 1980;66:50–55. [PubMed] [Google Scholar]
- 12.Li Z, Jansen D L, Finn T M, Halperin S A, Kasina A, O’Connor S P, Aoyama T, Manclark C R, Brennan M J. Identification of Bordetella pertussis infection by shared-primer PCR. J Clin Microbiol. 1994;32:783–789. doi: 10.1128/jcm.32.3.783-789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long S S, Lischner H W, Deforest A, Clark J L. Serologic evidence of subclinical pertussis in immunised children. Pediatr Infect Dis J. 1990;9:700–705. doi: 10.1097/00006454-199010000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Marcon M J. Bordetella, P. 566–573. In: Murray P J, Barron E J, Pfaller M J, Tenover E C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 15.Meade B D, Bollen A. Recommendations for the use of the polymerase chain reaction in the diagnosis of Bordetella pertussis infections. J Med Microbiol. 1994;41:51–55. doi: 10.1099/00222615-41-1-51. [DOI] [PubMed] [Google Scholar]
- 16.Mortimer E A., Jr Pertussis and its prevention: a family affair. J Infect Dis. 1990;161:473–479. doi: 10.1093/infdis/161.3.473. [DOI] [PubMed] [Google Scholar]
- 17.Müller F C, Hoppe J E, Wirsing von König C H. Laboratory diagnosis of pertussis: state of the art in 1997. J Clin Microbiol. 1997;35:2435–2443. doi: 10.1128/jcm.35.10.2435-2443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Health and Medical Research Council. Surveillance case definitions. Canberra, Australia: National Health and Medical Research Council; 1994. [Google Scholar]
- 19.Reizenstein E, Lindberg L, Möllby R, Hallander H O. Validation of nested Bordetella PCR in pertussis vaccine trial. J Clin Microbiol. 1996;34:810–815. doi: 10.1128/jcm.34.4.810-815.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal S, Strebel P, Cassiday P, Sanden G, Brusuelas K, Wharton M. Pertussis infection among adults during the 1993 outbreak in Chicago. J Infect Dis. 1995;171:1650–1652. doi: 10.1093/infdis/171.6.1650. [DOI] [PubMed] [Google Scholar]
- 21.Schläpfer G, Cherry J D, Heininger U, Überall M, Schmitt-Grohé S, Laussucq S, Just M, Stehr K. Polymerase chain reaction identification of Bordetella pertussis infections in vaccinees and family members in a pertussis vaccine efficacy trial in Germany. Pediatr Infect Dis J. 1995;14:209–214. doi: 10.1097/00006454-199503000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Stainer D W, Scholte M J. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1971;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 23.van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi F R. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993;31:2134–2140. doi: 10.1128/jcm.31.8.2134-2140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Zee A, Agterberg C, Peeters M, Mooi F, Schellekens J. A clinical validation of Bordetella pertussis and Bordetella parapertussis polymerase chain reaction: comparison with culture and serology using samples from patients with suspected whooping cough from a highly immunised population. J Infect Dis. 1996;174:89–96. doi: 10.1093/infdis/174.1.89. [DOI] [PubMed] [Google Scholar]