Abstract
The loop of a stem structure close to the 5′ end of the 18S rRNA is complementary to the box A region of the U3 small nucleolar RNA (snoRNA). Substitution of the 18S loop nucleotides inhibited pre-rRNA cleavage at site A1, the 5′ end of the 18S rRNA, and at site A2, located 1.9 kb away in internal transcribed spacer 1. This inhibition was largely suppressed by a compensatory mutation in U3, demonstrating functional base pairing. The U3–pre-rRNA base pairing is incompatible with the structure that forms in the mature 18S rRNA and may prevent premature folding of the pre-rRNA. In the Escherichia coli pre-rRNA the homologous region of the 16S rRNA is also sequestered, in that case by base pairing to the 5′ external transcribed spacer (5′ ETS). Cleavage at site A0 in the yeast 5′ ETS strictly requires base pairing between U3 and a sequence within the 5′ ETS. In contrast, the U3-18S interaction is not required for A0 cleavage. U3 therefore carries out at least two functionally distinct base pair interactions with the pre-rRNA. The nucleotide at the site of A1 cleavage was shown to be specified by two distinct signals; one of these is the stem-loop structure within the 18S rRNA. However, in contrast to the efficiency of cleavage, the position of A1 cleavage is not dependent on the U3-loop interaction. We conclude that the 18S stem-loop structure is recognized at least twice during pre-rRNA processing.
Eukaryotic nucleoli contain a large number of small nucleolar RNA (snoRNA) species, most of which function as guides for rRNA modifications. However, a small number of snoRNAs are required for processing of the pre-rRNA (reviewed in references 21 and 36), of which the most studied is U3. Genetic depletion of U3 in the yeast Saccharomyces cerevisiae inhibits three early pre-rRNA cleavage reactions on the pathway of 18S rRNA synthesis (Fig. 1); cleavage is inhibited at sites A0 (in the 5′ external transcribed spacer [5′ ETS]), A1 (the 5′ end of the mature 18S rRNA), and A2 (in internal transcribed spacer 1 [ITS1]) (14). In contrast, the cleavage of site A3 and sites further in the 3′ direction on the pathway of 5.8S and 25S synthesis is unaffected by depletion of U3. Depletion of the U3-associated proteins Nop1p, Sof1p, and Mpp10p leads to essentially identical phenotypes (8, 15, 37), indicating that the intact U3 small nucleolar ribonucleoprotein (snoRNP) particle is required for pre-rRNA cleavage at these sites. Depletion of U3 has also been reported to inhibit in vitro cleavage of the mouse 5′ ETS (16) and pre-rRNA processing in Xenopus oocytes (5, 30).
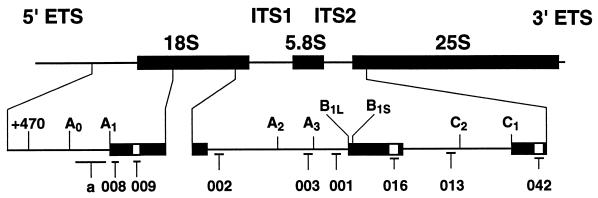
FIG. 1.
Structure of the pre-rRNA and locations of oligonucleotide hybridization probes. Thick bars represent the mature rRNAs; thin bars indicate the transcribed spacer regions, which are not drawn to scale. The 18S, 5.8S, and 25S rRNAs are flanked by the 5′ ETS and 3′ ETS and separated by ITS1 and ITS2. Probe a is a riboprobe complementary to the fragment from A0 to A1. Probes 009, 016, and 042 hybridize to the tags within the mature 18S, 5.8S, and 25S rRNAs, respectively. Probe 008 hybridizes to the mature 18S rRNA. Probes 002, 003, and 001 hybridize to ITS1 at positions 5′ to site A2, between A2 and A3, and 3′ to site A3, respectively. Probe 013 hybridizes to the 5′ region of ITS2.
In vivo psoralen cross-linking experiments identified several sites of interaction between the yeast U3 snoRNA and the pre-rRNA. One was a single-stranded region in the 5′ region of the U3 snoRNA (nucleotides [nt] 39 to 48) which exhibited a 10-nt complementarity to a region of the 5′ ETS (nt 470 to 479; approximately 140 nt 5′ to site A0 and 230 nt 5′ to site A1) (3, 4). Disruption of this base pairing blocked cleavage at sites A0, A1, and A2 and accumulation of the 18S rRNA, closely mimicking the effects of U3 depletion in trans, while compensatory mutations largely restored processing and synthesis of 18S rRNA. This indicates that U3 snoRNA interacts with the pre-rRNA at this position and is required for cleavages which lie 100, 200, and 2,000 nt distant. A second site of U3 cross-linking was with the loop of an extended stem structure that lies between sites A0 and A1 (4). The significance of this interaction remains unclear, since mutation or deletion of this loop did not detectably affect pre-rRNA processing. In Trypanosoma brucei three sites of cross-linking between U3 and the 5′ ETS have been mapped (11, 11a). As observed for yeast, these include sites required for 18S rRNA synthesis and dispensable sites. One of these sites closely resembles the yeast U3–pre-rRNA interaction site at +470. Both sites are predicted to include 10 consecutive base pairs with the hinge region of U3, are required for 18S rRNA synthesis, and are similarly located in the predicted structure of the 5′ ETS (11a). Mammalian U3 can also be cross-linked to the 5′ ETS at more than one site (20, 34, 38).
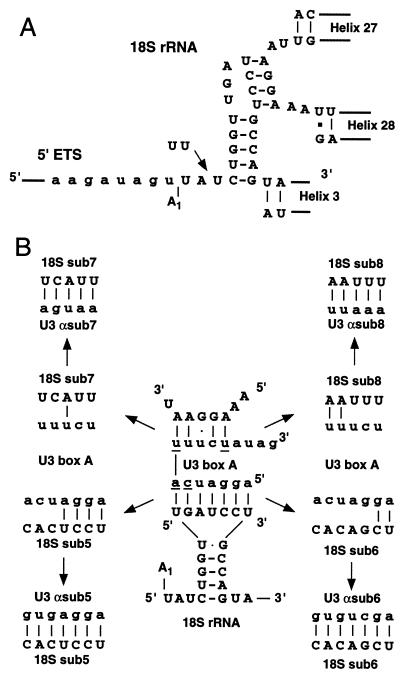
The strongest cross-linking sites in the yeast U3 molecule were in the box A region, which is conserved throughout eukaryotes (4). Box A has the potential to base pair across the central pseudoknot of the 18S rRNA (13, 22). The central pseudoknot is a universally conserved long-range interaction within the small-subunit rRNA that plays a crucial role in the overall folding of the mature rRNA (Fig. 2A). In this proposed interaction, U3 box A would form seven base pairs with the loop of the 5′ stem of the 18S rRNA and five base pairs with nt 914 to 918, which include the 3′ side of the pseudoknot in the mature rRNA (Fig. 2B). This base pairing would include the four box A nucleotides that were cross-linked to the pre-rRNA in vivo (Fig. 2B) (4). Due to the abundance of the mature rRNAs, the previous analyses would not have identified cross-linking sites in the pre-rRNA that lie within the mature 18S rRNA region.
FIG. 2.
(A) Structure of the central pseudoknot. The 5′ ETS region is shown in lowercase; the mature 18S rRNA region is shown in uppercase. Also shown is the two-U insertion present in the sub5 and sub6 pre-rRNAs. (B) Potential base pairing between the 18S rRNA region of the pre-rRNA and U3 box A in the wild-type and mutant constructs. U3 box A is shown in lowercase; the 18S rRNA sequence is shown in uppercase. Underlined nucleotides were cross-linked in vivo to the pre-rRNA.
To analyze the significance of this potential base pairing, we have expressed pre-rRNAs carrying mutations in the 18S rRNA in the presence and absence of the U3 snoRNA containing compensatory mutations in box A. Here, we demonstrate that U3 box A base pairs to the loop region of 18S rRNA and that perturbation of this base pairing inhibits processing at sites A1 and A2 without preventing cleavage at site A0.
MATERIALS AND METHODS
Strains and media.
Standard S. cerevisiae techniques were employed. The yeast strain NOY504 (MATa rpa12::LEU2 leu2-3,112 ura3-1 trp1-1 his3-11 ade2-101 CAN1-100) (26) (generously provided by M. Nomura, University of California at Irvine) was used for all the experiments. Yeast strains were grown in minimal medium containing 2% galactose and 0.67% yeast nitrogen base plus nutrients and supplemented with the required amino acids (33).
Plasmids and constructs used in this study.
A plasmid containing the entire yeast rDNA repeat fused to an inducible GAL7 promoter (pGAL::rDNA) was used as a wild-type control (12, 27, 33). Synthesis of ribosomes derived from this plasmid was monitored by hybridization to small oligonucleotide tags present within the 18S, 5.8S, and 25S rRNA sequences. A YEplac 195 plasmid (URA3 [2μ]) which does not contain a ribosomal DNA (rDNA) unit was used as a negative control (10). The pre-rRNA mutations were generated via a two-step PCR approach. For sub5 and sub6, two oligonucleotide primers, a 3′ mutagenic primer and a 5′ primer complementary to a sequence in the 5′ ETS, were used. With the tagged rDNA plasmid as template, a 200-nt fragment was amplified. This was gel purified, digested with NdeI plus HindIII, and subcloned into vector pTH66, which contains the sequences of the 5′ ETS and 18S rRNA up to the BamHI site in the tag. The 200 nt were sequenced to confirm the mutation and to eliminate any additional errors induced during amplification. Correct clones and the wild-type pGAL::rDNA were digested with BamHI, and the fragments were exchanged (39). The 18S sub7 and sub8 mutations were generated by amplifying a 200-nt fragment with an oligonucleotide including the SacI site at position 1234 within the 18S rRNA and a mutagenic primer. The resulting product was then used in a second PCR to generate a fragment extending from the SfiI site at position 646 within the 18S rRNA to the SacI site. This region was entirely sequenced, digested with SacI plus SfiI, gel purified, and exchanged with the wild-type fragment in plasmid pTH70.
The yeast U3 genes (SNR17A and SNR17B) contain introns (25). To avoid problems with splicing efficiency, U3 mutations were constructed in an ARS-CEN plasmid carrying an ADE2 selective marker and expressing the U3 cDNA under the control of its own promoter (kindly provided by R. Fournier and C. Branlant, Nancy, France) (3). A 350-nt PCR product was amplified with a mutagenic oligonucleotide including the SalI site in the U3 5′ flanking sequence and an oligonucleotide complementary to the 3′ flanking region outside the EcoRI site, which was inserted 50 nt 3′ to the end of the mature U3 sequence (3). The PCR product was digested with SalI plus EcoRI, gel purified, and then exchanged with the wild-type fragment in pU3-wt. This region was fully sequenced to confirm the mutation and eliminate any random mutations.
Analysis of pre-rRNA processing.
The plasmids containing the mutations and the positive and negative controls were transformed into strain NOY504 by using the lithium acetate method as described previously (9). Cells were grown at 23°C to mid-log phase in minimal medium containing galactose, diluted to an optical density at 600 nm of 0.1, and shifted to 37°C for 6 h (12). Cells were harvested by centrifugation at 4°C, washed with ice-cold water, centrifuged again, and stored at −80°C. RNA was extracted from the frozen cell pellets as previously described (31). Total RNA was separated on 1.2% agarose–6% formaldehyde gels with 4 μg of total RNA per lane as previously described (35). The gel was then transferred to a Hybond N+ membrane (Amersham) with 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) as the transfer buffer. Hybridizations were performed in 6× SSPE–0.5% sodium dodecyl sulfate–5× Denhardt’s solution (29). Additionally, the filter was hybridized with a riboprobe, generated as previously described (39), to detect and distinguish the 33S from the 32S pre-rRNA precursor. This was performed in 40% formamide–5× SSPE–5× Denhardt’s solution–1% sodium dodecyl sulfate–200 μg of herring sperm DNA per ml.
Oligonucleotides used as hybridization probes had the following sequences: 001 (27SA-2), CCAGTTACGAAAATTCTTG; 002 (20S-2), GCTCTTTGCTCTTGCC; 003 (27SA-3), TGTTACCTCTGGGCCC; 008 (18S+34), CATGGCTTAATCTTTGAGAC; 009 (18S-αTAG), CGAGGATCCAGGCTTT; 013 (rna2.1), GGCCAGCAATTTCAAGTTA; 016 (5.8S-Ftag), DGDDUDCUGGCGDdGdC; 042 (25S Tag 1), ACTCGAGAGCTTCAGTACC; 062 (α18S sub6), CAGCTGTGACCAGAAATAACT; and 200 (αU3), UUAUGGGACUUGUU. Oligonucleotide 016 is largely composed of 2′-methyl RNA; D is diaminopurine. Oligonucleotide 200 (αU3) is fully composed of 2′-methyl RNA and hybridizes with yeast U3 between nt 82 and 95.
Primer extensions.
Primer extension analysis was performed as previously described (4), using 4 μg of RNA. A sequencing ladder was run in parallel by using the same oligonucleotide primer after 5′ phosphorylation with unlabeled ATP.
RESULTS
U3 base pairs to the 18S loop sequence.
To test the significance of the potential U3-stem loop base pairing, two mutations were constructed in the stem-loop structure at the 5′ end of the 18S rRNA. The sub5 mutation alters the three loop nucleotides that are not engaged in the pseudoknot interaction and is therefore predicted to be least disruptive for the overall secondary structure, while the sub6 mutation alters two additional nucleotides (Fig. 2). The positioning of site A1 is determined with respect to two signals: the sequence 5′ to the site of cleavage and the 5′ stem loop within the 18S rRNA (32, 39). The insertion of two U residues immediately 5′ to the stem allows the contribution made by each of these signals to the positioning of the cleavage site to be resolved (39). The sub5 and sub6 mutations were therefore combined with a two-U insertion, allowing the role of the putative U3-18S interaction in this positioning to be determined.
The mutant constructs were cloned into plasmids that express the entire pre-rRNA under the control of the RNA polymerase II GAL7 promoter (12). These plasmids were expressed in strain NOY504, which carries the rpa12::LEU2 mutation and is temperature sensitive (TS) for RNA polymerase I (Pol I) (27) (kindly provided by M. Nomura). When cells are shifted to 37°C for 6 h in galactose-containing medium, chromosomal rDNA synthesis is reduced to a low level, allowing the analysis of the processing of the mutant pre-rRNAs. Short oligonucleotide tags present in the mature 18S, 5.8S, and 25S rRNA sequences allowed their synthesis to be monitored (4, 12, 24). The mutant pre-rRNAs were expressed alone or together with U3 mutants U3αsub5 and U3αsub6, which contain alterations in the box A region that restore complementarity. The mutations and the predicted base pairings are shown in Fig. 2B.
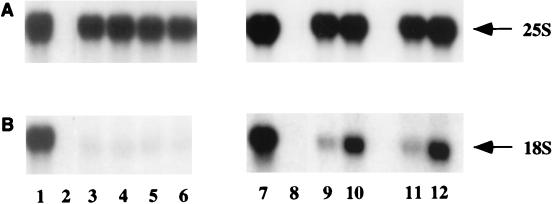
Compared to the tagged but otherwise wild-type pre-rRNA (Fig. 3, lane 1), the sub5 mutation substantially reduced 18S rRNA accumulation (Fig. 3, lanes 3 and 5), while 25S rRNA accumulation was unaffected. Coexpression of U3αsub5 did not restore 18S rRNA accumulation (Fig. 3, lanes 4 and 6). In contrast, the larger sub6 mutation also strongly inhibited synthesis of the 18S rRNA (Fig. 3, lanes 9 and 11), which was partially restored by coexpression of the compensatory U3αsub6 (Fig. 3, lanes 10 and 12). Synthesis of the 25S rRNA (Fig. 3) was unaffected by the mutation in the 18S rRNA or the compensatory U3 mutation. No signal was observed in a strain lacking the tagged rDNA plasmid (Fig. 3, lane 2).
FIG. 3.
Effects of compensatory mutations between U3 and the 18S rRNA 5′ stem-loop on rRNA synthesis. RNA was extracted from strains carrying plasmids expressing the mutant and wild-type pre-rRNAs and analyzed by Northern hybridization. Lanes: 1 and 7, wild-type rDNA; 2 and 8, plasmid lacking the rDNA sequence; 3 and 5, sub5 rDNA; 4 and 6, sub5 rDNA with coexpression of U3αsub5; 9 and 11, sub6 rDNA; 10 and 12, sub6 rDNA with coexpression of U3αsub6. For the mutant pre-rRNAs, the analysis of two independent transformants is presented. (A) Probe 042, complementary to the 25S rRNA tag; (B) probe 009, complementary to the 18S rRNA tag.
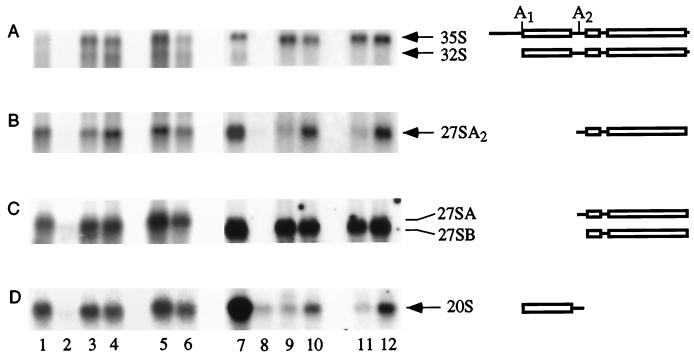
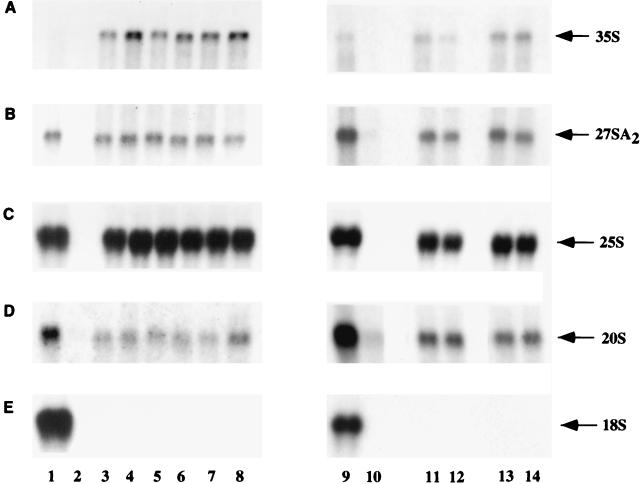
The effects of the mutations on pre-rRNA processing were assessed by Northern hybridization (Fig. 4). The sub5 mutation did not strongly inhibit pre-rRNA processing, and no clear alterations in the levels of the 20S or 27SA2 pre-rRNAs were observed. Pre-rRNA processing at sites A1 and A2 was strongly inhibited by the sub6 mutation. The products of cleavage at these sites, the 27SA2 and 20S pre-rRNAs, were reduced to levels close to those of the −rDNA negative control (Fig. 4B and D; compare lanes 9 and 11 with lane 8). The levels of both the 27SA2 and 20S pre-rRNAs were largely restored by coexpression of U3αsub6 (Fig. 4, lanes 10 and 12). No aberrant processing intermediates were detectably synthesized from the sub6 pre-rRNA. The levels of the 27SA3 and 27SB pre-rRNAs were unaffected by the sub6 mutation or the expression of U3αsub6 (data not shown), indicating that processing at later steps on the pathway of 25S and 5.8S rRNA synthesis was unaffected.
FIG. 4.
Effects of compensatory mutations between U3 and the 18S rRNA 5′ stem-loop on pre-rRNA processing. RNA was extracted from strains carrying plasmids expressing the mutant and wild-type pre-rRNAs and analyzed by Northern hybridization. Lanes: 1 and 7, wild-type rDNA; 2 and 8, plasmid lacking the rDNA sequence; 3 and 5, sub5 rDNA; 4 and 6, sub5 rDNA with coexpression of U3αsub5; 9 and 11, sub6 rDNA; 10 and 12, sub6 rDNA with coexpression of U3αsub6. For the mutant pre-rRNAs, the analysis of two independent transformants is presented. Panels A through D show different hybridizations of the same gels. (A and B) Probe 003, hybridizing in ITS1 between A2 and A3; (C) probe 013, hybridizing to ITS2; (D) probe 002, hybridizing in ITS1 5′ to A2. Lanes 7 through 12 show different hybridizations of the same Northern blot.
The coexpression of the sub6 pre-rRNA and U3αsub6 did not support growth of the Pol I TS strain at 37°C, indicating that the mutant ribosomes are not functional. Several other mutations tested in the 18S stem-loop structure all prevented the synthesis of functional ribosomes (32). This region of the ribosome is highly conserved in evolution (2), and it appears that changes are poorly tolerated.
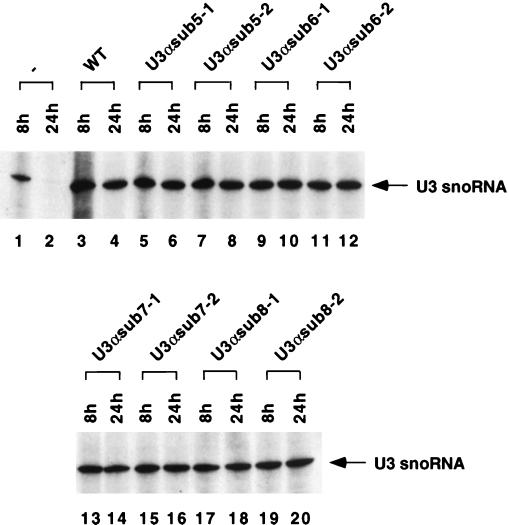
To test the accumulation of the mutant U3 snoRNAs, U3αsub5 and U3αsub6 were expressed in a GAL::U3 strain, JH84 (12a, 14). Both were expressed at wild-type levels (Fig. 5), consistent with the report that the 5′ region of U3 is not required for its accumulation (28). Surprisingly, both mutant U3 constructs were able to complement the GAL::U3 mutation for growth on glucose medium at 18, 30, and 37°C, although the U3αsub6 strain was mildly cold sensitive. A detailed analysis of the effects of these and other mutations in the 5′ region of U3 on pre-rRNA processing will be published elsewhere.
FIG. 5.
Expression of the mutant forms of U3. Plasmids expressing the wild-type and mutant U3 cDNAs were transformed into a strain in which the chromosomal U3 expression is under GAL regulation. Lanes: 1 and 2, nontransformed strain; 3 and 4, strain transformed with the wild-type U3 cDNA; 5 through 20, strains transformed with the U3 constructs indicated. RNA was extracted 8 and 24 h after transfer of the strains to glucose medium and analyzed by Northern analysis with probe 200, which hybridizes 3′ to the mutated regions.
We conclude that base pairing between U3 and the 5′ loop within the 18S rRNA is required for pre-rRNA processing at sites A1 and A2. However, even when the complementary U3 is present, not all loop sequences are tolerated for 18S rRNA accumulation.
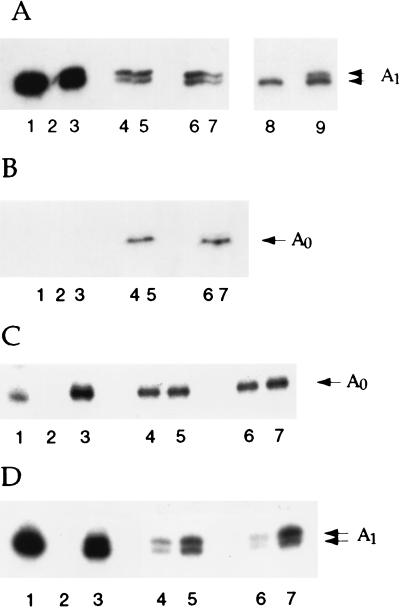
The site of A1 cleavage is positioned with respect to both the 5′ flanking sequence and the stem loop within the 18S rRNA (32, 39). The contributions made by these two signals can be resolved by the insertion 5′ to the stem of two uracil residues (32, 39), which are present in the sub5 and sub6 pre-rRNAs (Fig. 2A). Both the sub5 and sub6 mutations reduced the use of the 3′ A1 site relative to that of the 5′ A1 site (compare Fig. 6A and D, lanes 4 and 6, with Fig. 6A, lane 9), showing that they partially inhibited the positioning of the cleavage site with respect to the stem-loop structure. However, coexpression of U3αsub5 or U3αsub6 did not affect the relative utilization of the two sites (Figs. 6A and D, lanes 5 and 7). We conclude that recognition of the loop region of the stem-loop structure to position the site of A1 cleavage is independent of U3 base pairing.
FIG. 6.
Effects of compensatory mutations between U3 and the 18S rRNA 5′ stem on processing at sites A1 and A0. RNA was extracted from strains carrying plasmids expressing the mutant and wild-type pre-rRNAs and analyzed by primer extension. For the mutant pre-rRNAs, the analysis of two independent transformants is presented. (A) Primer extension through site A1 from oligonucleotide 009. Lanes: 1 and 8, wild-type rDNA; 2, plasmid lacking the rDNA sequence; 3 and 9, A1+2U rDNA; 4 and 6, sub5 rDNA; 5 and 7, sub5 rDNA with coexpression of U3αsub5. (B) Primer extension through site A0 from oligonucleotide 062 (α18S sub6), specific for the mutant pre-rRNA. Lanes: 1, wild-type rDNA; 2, plasmid lacking the rDNA sequence; 3, A1+2U rDNA; 4 and 6, sub6 rDNA; lanes 5 and 7, sub6 rDNA with coexpression of U3αsub6. (C) Primer extension through site A0 from oligonucleotide 008. Lane order is as described for panel B. (D) Primer extension through site A1 from oligonucleotide 009. Lane order is as described for panel B.
With the Pol I TS system, pre-rRNA processing cannot be analyzed at other temperatures. However, the sub6 mutation also inhibited 18S rRNA synthesis at 25°C, and suppression by coexpression of U3αsub6 was similar to that observed at 37°C (data not shown).
The potential interaction between U3 snoRNA and the sequence on the 3′ side of the central pseudoknot was also tested. Mutation sub7 substituted all five nucleotides involved in the proposed interaction, while sub8 altered only three of these nucleotides (Fig. 2B). The presence of either the sub7 or sub8 mutation in the pre-rRNA greatly reduced accumulation of the mature 18S rRNA (Fig. 7E), and 20S pre-rRNA accumulation was also reduced, without effect on 25S rRNA synthesis (Fig. 7C). No suppression was observed upon coexpression of U3 carrying the compensatory mutations U3αsub7 and U3αsub8. Both U3 mutants were shown to be expressed at wild-type levels in a GAL::U3 strain (Fig. 5). These data are consistent with strong effects of the sub7 and sub8 mutations on 18S rRNA stability and probably also on 20S pre-rRNA stability but provide no support for the base pairing of U3 across the central pseudoknot.
FIG. 7.
Compensatory mutations between the U3 snoRNA and the sequence on the 3′ side of the central pseudoknot. RNA was extracted from strains carrying plasmids expressing the mutant and wild-type pre-rRNAs and analyzed by Northern hybridization. Lanes: 1 and 9, wild-type rDNA; 2 and 10, plasmid lacking the rDNA sequence; 3, 5, and 7, sub7 rDNA; 4, 6, and 8, sub7 rDNA with coexpression of U3αsub7; 11 and 13, sub8 rDNA; 12 and 14, sub8 rDNA with coexpression of U3αsub8. For the mutant pre-rRNAs, the analysis of three (sub7) or two (sub8) independent transformants is presented. (A and B) Probe 003, hybridizing in ITS1 between A2 and A3; (C) probe 042, complementary to the 25S rRNA tag; (D) probe 002, hybridizing in ITS1 5′ to A2; (E) probe 009, complementary to the 18S rRNA tag.
Cleavage at site A0 is unaffected by the mutations in the 5′ loop of 18S rRNA.
Previous analyses have shown that the U3 snoRNA base pairs with the 5′ ETS region of the 35S pre-rRNA (4). Compensatory mutations in this region established that this base pairing is strictly required for the pre-rRNA cleavages at sites A0, A1, and A2 (3). However, the lack of clear accumulation of the 35S pre-rRNA or 23S RNA in the sub6 mutant suggested that the U3-18S interaction might not be required for cleavage at site A0. The 33S pre-rRNA, which is the normal product of cleavage at site A0, cannot readily be detected by Northern hybridization, and its abundance was therefore assessed by primer extension. Primer extension with oligonucleotide 008, complementary to the 5′ region of the 18S rRNA, revealed that the level of the 33S pre-rRNA, shown by the stop at A0, was unaffected by the sub6 mutation with or without coexpression of U3αsub6 (Fig. 6C), in contrast to the stop at A1 (Fig. 6D). To confirm this result, an oligonucleotide that hybridizes specifically to the sub6 mutant pre-rRNA was used. This oligonucleotide (α18S sub6) did not give a signal on the wild-type pre-rRNA (Fig. 6B, lane 1) and confirmed that processing of the sub6 pre-rRNA at site A0 was unaffected by coexpression of U3αsub6. Both primer extensions showed that A0 cleavage occurs at the correct nucleotide.
We conclude that base pairing between the 5′ loop of the 18S rRNA and box A in the U3 snoRNA is required for cleavage at sites A1 and A2 but not for cleavage at A0, in marked contrast to the U3-5′ ETS interaction.
DISCUSSION
U3 box A base pairs with the 5′ end of the 18S rRNA.
Cleavage of the pre-rRNA at sites A0, A1, and A2 requires the U3 snoRNP; cleavage is blocked by depletion of the U3 snoRNA or the U3-associated proteins Nop1p, Nop58p, Sof1p, and Mpp10p (8, 15, 18, 37, 40). U3 base pairs to the 5′ ETS region of the pre-rRNA, but this interaction is insufficient to account for the in vivo cross-links detected between the box A region of U3 and the pre-rRNA (4). U3 box A can also be drawn base paired to the 5′ loop region in the 18S rRNA, and this interaction would involve two nucleotides in U3 known to interact with the 35S pre-rRNA (Fig. 2B). To investigate this potential base pairing, two compensatory mutations were tested. sub5 was a substitution of the three nucleotides that are not engaged in the pseudoknot interaction, while sub6 altered two additional nucleotides (Fig. 2B). The sub5 mutation had little effect on pre-rRNA processing but strongly reduced 18S rRNA accumulation, presumably due to destabilization of the mature rRNA; this phenotype was not suppressed upon coexpression of U3αsub5. The sub6 mutation strongly inhibited pre-rRNA processing at sites A1 and A2, and this phenotype was largely suppressed by coexpression of U3αsub6. Accumulation of 18S rRNA from the sub6 pre-rRNA was also partially restored by U3αsub6. The difference between the effects of the suppressed sub5 and sub6 mutations on 18S rRNA accumulation most likely reflects the tolerance of the mature rRNA structure for the different sequences.
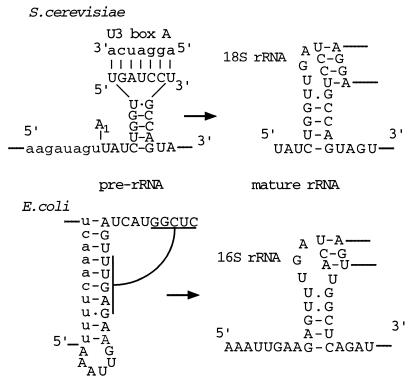
In the E. coli pre-rRNA, the 5′ end of the 16S rRNA is engaged in a base pair interaction with the 3′ region of the 5′ ETS (6). This interaction must be broken in order for the 5′ end of 16S rRNA to assume its mature conformation. As in yeast, the loop of the 5′ stem-loop structure in E. coli is involved in a long-range interaction with nucleotides around position 917, termed the central pseudoknot (Fig. 8). In yeast, base pairing of the U3 snoRNA to the 18S rRNA is also mutually exclusive with formation of the central pseudoknot. This structure is likely to play a crucial role in the overall folding of the rRNA, and in both cases the alternative structure may prevent the formation of this interaction until the correct stage in the assembly process. It is notable that this structural isomerization involves interactions in cis in E. coli and in trans in yeast. It was previously postulated that the snoRNA–pre-rRNA interactions in trans are functionally related to interactions in cis within the bacterial pre-rRNA (7, 23), and the present data are consistent with this proposal.
FIG. 8.
Comparison of the predicted structures of pre-rRNAs and mature rRNAs in E. coli and S. cerevisiae. The 5′ ETS regions and the U3 box A region are shown in lowercase; the mature rRNAs are shown in uppercase.
The box A region of U3 can also be drawn base paired to the sequence that forms the 3′ side of the central pseudoknot, in an interaction that would involve the other two box A nucleotides that were cross-linked to the pre-rRNA in vivo (4). In contrast to the strong support for the interaction of U3 box A with the loop sequence that includes the 5′ side of the central pseudoknot, the analysis of compensatory mutations provided no support for the potential interaction with the 3′ sequence. The sub7 and sub8 mutations reduced the accumulation of the 20S pre-rRNA and greatly reduced the level of mature 18S rRNA. However, no suppression was observed upon coexpression of U3αsub7 or U3αsub8. A number of more subtle mutations in the 3′ side of the pseudoknot structure also reduced 18S rRNA accumulation (data not shown). Again, coexpression of the compensatory U3 mutants had no obvious effect. While these negative data do not constitute proof that U3 does not base pair with the 3′ side of the central pseudoknot, they certainly give no support for the model. If box A does not base pair with this sequence it must interact with some other as-yet-unidentified region(s) of the rRNA, as shown by the U3-35S pre-rRNA cross-linking data.
The U3 snoRNA carries out functionally distinct interactions with the pre-rRNA.
Comparison of the effects of mutations in the U3 binding site within the 18S rRNA with mutations in the U3 binding site at +470 in the 5′ ETS reveals a striking difference. The U3-ETS interaction is strictly required for pre-rRNA cleavage at sites A0, A1, and A2. In contrast, the U3-18S interaction is required for cleavage at sites A1 and A2 but not for cleavage at A0. We therefore conclude that the interactions of U3 with the 5′ ETS and the 18S rRNA are functionally distinct. It may be that the U3-5′ ETS interaction is specifically required to target the endonuclease responsible for A0 cleavage, while the U3-18S interaction functions independently as a chaperone in the folding of the 5′ region of the 18S rRNA. Cleavage at A0 was initially thought to be performed by Rnt1p (1), but more recent studies have revealed that cleavage continues in the complete absence of Rnt1p (17), demonstrating that this enzyme is not required for cleavage. Another site of U3-5′ ETS interaction was detected around position 645 (4) in the loop of an extended stem that lies between sites A0 and A1 (site A0 is at 610 and site A1 is at 699). Mutation of this region had no clear effect on pre-rRNA processing (39), indicating that U3 forms at least three functionally distinct interactions with the pre-rRNA.
Mutations in the U3-associated protein Mpp10p (8) can also uncouple the requirement for the U3 snoRNP in cleavage at site A0 and at sites A1 and A2. C-terminal-truncation mutations lead to the appearance of the 22S RNA that extends from site A0 to A3, clearly showing that processing at A0 is less inhibited than processing at A1 and A2 (19). This suggests that, like the U3 snoRNA itself, Mpp10p carries out different interactions during processing at A0 and at A1 and A2.
Processing of the pre-RNA involves multiple, complex interactions with the U3 snoRNA. It remains to be determined whether these occur simultaneously or in succession and whether one or more U3 molecules are involved.
ACKNOWLEDGMENTS
We thank M. Nomura for strain NOY504, Phil Mitchell and Christine Allmang for critical reading of the manuscript, and Elisabeth Petfalski for expert technical assistance.
K.S. was the recipient of a fellowship from the Boehringer-Ingelheim Foundation for Biomedical Research, and D.T. was partially funded by the Wellcome Trust.
REFERENCES
- 1.Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 2.Alksne L E, Anthony R A, Liebman S W, Warner J R. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrame M, Tollervey D. Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borovjagin A V, Gerbi S A. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol. 1999;286:1347–1363. doi: 10.1006/jmbi.1999.2527. [DOI] [PubMed] [Google Scholar]
- 6.Dammel C S, Noller H F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 7.Dennis P P, Russell A G, Moniz De Sa M. Formation of the 5′ end pseudoknot in small subunit ribosomal RNA: involvement of U3-like sequences. RNA. 1997;3:337–343. [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2554. doi: 10.1093/nar/26.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Hartshorne, T. 1999. Personal communication.
- 12.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Hughes, J. 1997. Personal communication.
- 13.Hughes J M X. Functional base-pairing interactions between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 14.Hughes J M X, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nuclear ribonucleoprotein functions at the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 17.Kufel, J., B. Dichtl, and D. Tollervey. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA, in press. [DOI] [PMC free article] [PubMed]
- 18.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for stability of the snoRNAs. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S J, Baserga S J. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci USA. 1997;94:13536–13541. doi: 10.1073/pnas.94.25.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maser R L, Calvet J P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5′ external transcribed spacer of pre-ribosomal RNA. Proc Natl Acad Sci USA. 1989;86:6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 22.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 23.Morrissey J P, Tollervey D. Birth of the snoRNPs—the evolution of RNase MRP and the eukaryotic pre-rRNA processing system. Trends Biochem Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 24.Musters W, Venema J, van der Linden G, van Heerikhuizen H, Klootwijk J, Planta R J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989;9:551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myslinski E, Ségault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 26.Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants defective in RNA polymerase I. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samarsky D A, Fournier M J. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma K, Fabre E, Tekotte H, Hurt E C, Tollervey D. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol. 1996;16:294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma K, Venema J, Tollervey D. The 5′ end of the 18S rRNA can be positioned from within the mature rRNA. RNA. 1999;5:678–686. doi: 10.1017/s1355838299990052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F, Fink G R, Hicks J B N. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 34.Stroke I L, Weiner A M. The 5′ end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989;210:497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- 35.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 37.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyc K, Steitz J A. A new interaction between the mouse 5′ external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res. 1992;20:5375–5382. doi: 10.1093/nar/20.20.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venema J, Henry Y, Tollervey D. Two distinct recognition signals define the site of endonucleolytic cleavage at the 5′ end of yeast 18S rRNA. EMBO J. 1995;14:4883–4892. doi: 10.1002/j.1460-2075.1995.tb00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]