Abstract
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are among the most ancient small RNAs in all domains of life and are generated by the cleavage of tRNAs. Emerging studies have begun to reveal the versatile roles of tsRNAs in fundamental biological processes, including gene silencing, ribosome biogenesis, retrotransposition, and epigenetic inheritance, which are rooted in tsRNA sequence conservation, RNA modifications, and protein-binding abilities. We summarize the mechanisms of tsRNA biogenesis and the impact of RNA modifications, and propose how thinking of tsRNA functionality from an evolutionary perspective urges the expansion of tsRNA research into a wider spectrum, including cross-tissue/cross-species regulation and harnessing of the ‘tsRNA code’ for precision medicine.
On the ancientness of tsRNAs/tRNAs
With the wide application of high-throughput RNA sequencing (RNA-seq), tRNA-derived small RNAs (tsRNAs) (also called tDRs or tRNA-derived fragments, tRFs) are increasingly recognized as an emerging class of functional small noncoding RNAs (sncRNAs) in various fundamental biological and disease conditions [1,2]. tsRNAs have been identified in a wide range of species across all three domains of life, including Archaea, Bacteria, and some unicellular organisms (e.g., Protozoa), where other specialized small RNA pathways, such as miRNAs, siRNAs, and Piwi-interacting RNAs (piRNAs), are lacking [3–7]. These observations have positioned tsRNAs among the most ancient classes of sncRNAs for intra- and intercellular functionality and communication that may pre-date the emergence of more specialized sncRNAs.
The evolutionary conservation of tsRNAs is not entirely surprising because their tRNA precursors are among the most ancient and conserved RNA species that act as essential elements in all biological systems and were supposedly core players in the last universal common ancestor (LUCA; see Glossary) [8]. The main function of tRNAs is to decode the mRNA sequence and deliver specific amino acids from aminoacyl-tRNA synthetases (aaRSs) to the ribosome [2]. Beyond their canonical role in protein translation, tRNAs are also actively involved in other biological contexts such as being used as primers for reverse transcription (RT) of the retroviral genome [9]. Interestingly, many viral RNA genomes contain tRNA-like structures (TLSs) that can not only perform charging of amino acids but also function as telomeres for viruses, assist with viral replication, and interact with the ribosome – the translational machinery with a ribozyme at its core [10]. The intricate relationship between viruses and tRNAs (and TLSs) suggests that they may share a common evolutionary origin and have perhaps served as the drivers of early life on Earth ever since the LUCA [11] or even the ‘RNA world’ [12] (Box 1).
Box 1. The emergence, existence, and exit of the ‘RNA world’.
Ever since the discovery of ribozymes in the 1980s, the hypothesis that early life forms were dependent solely on RNA to both store genetic information and catalyze chemical reactions for self-propagation has gained momentum, making an ‘RNA world’ before DNA-protein-dominated life highly probable [118]. Recent triumphs in the RNA world hypothesis include the demonstration that the fundamental macromolecular machineries – the ribosome [119] and spliceosome [120] that are essential for protein translation and mRNA processing – both possess ribozymes at their cores. Furthermore, some ancient RNA viruses (e.g., hepatitis delta virus, HDV) and viroids are circular RNAs with ribozyme functions that replicate their genomes without a protein enzyme [121]. In vitro evolution experiments [122,123] have also generated an increasing repertoire of synthetic ribozymes with essential functions, such as a ribozyme with RNA polymerase (or replicase) activity to direct RNA-catalyzed RNA polymerization [124] and self-sustaining RNA replication through cross-catalytic reactions between different ribozymes [125]. Interestingly, tRNA-like sequences can replicate by self-assembly, and this may be a relic of self-replicating RNAs from the RNA world [126,127]. Ribozymes can also add site-specific RNA methylations [38], and the ribozyme-mediated RNA modifications may in turn modulate the functionality of ribozymes. Finally, ribozymes that can catalyze the aminoacylation of tRNA [128] may have triggered exit from the RNA world by entering into an RNA–amino acid hybrid form leading to the formation of RNPs that may outperform the function of ribozymes solely based on RNA [129], eventually moving towards protein-dominated functionality.
Pre-dating the ribozyme-dominated RNA world, earlier propagating systems could have consisted of nucleic acids other than the present-day RNA and DNA molecules [130]. Indeed, nonenzymatic template-directed RNA synthesis can be catalyzed by the coordination of Mg2+ and citrate inside fatty acid membranes – a hypothetical protocell [131] – and this provides the wider possibility that metal ions and other mineral cofactors may have assisted with the formation, polymerization, and propagation of ribose-based nucleic acid structures before the emergence of ribozymes. Notably, many active molecules in extant cellular life, including ATP, NAD+, acetyl-CoA, and vitamin B12 (Figure I), are structural derivatives of ribonucleosides. These molecules are deeply incorporated into cellular metabolism and RNA and DNA modifications [132] where they may represent chemical ‘fossils’ that were concurrent with or pre-dated the RNA world. Finally, the existence of the RNA world might also be accompanied by DNA through chemical coproduction [133], where DNA could either have a catalytic function (e.g., RNA ligation [134]) or be harnessed for genetic information storage.
Figure I.

Chemical structures of ATP, NAD+, acetyl-CoA, vitamin B12, and adenosine (A).
In the earlier mentioned evolutionary context, when tRNAs are fragmented through either nonspecific metabolic turnover or specific cleavage, they give rise to diverse forms of tsRNAs that may interfere with or enhance the role of their precursor tRNAs (or TLSs) through basic molecular interactions by competing with their target or by generating new functions via interacting with other RNA and proteins by sequence complementarity and structural effects. In other words, we argue that the fragmentation of tRNAs created a pool of tsRNA molecules for functional selection that have undergone Darwinian evolution ever since the RNA world and the LUCA, which would in theory result in heterogeneous and versatile functions of tsRNAs that penetrate fundamental biological machineries throughout evolution. Indeed, this overarching idea resonates well with the diverse tsRNA functions observed today, ranging from regulating translation, ribosome biogenesis, retrotransposition, cell–cell communication, and epigenetic inheritance, as well as with how tsRNA dysregulation is being linked to various human diseases (summarized in recent reviews [1,2]). From an evolutionary perspective, we first discuss the fundamental aspects of tsRNA biogenesis and regulation, and then propose a framework for the evolving principles of their mechanisms of action from which a wider spectrum of tsRNA functionality can be deduced. We also discuss the potential future integration and harnessing of different layers of tsRNA information (e.g., expression, cleavage patterns, and RNA modifications) for disease diagnosis and precision medicine.
tsRNA biogenesis: from degradation to regulation
From a structural perspective, the cloverleaf-shaped secondary structure of tRNA is folded into an L-shape in 3D (Figure 1A) [2]. This L-shaped structure is overall tightly condensed but has two relatively exposed sites: the anticodon at one end of the L and the tRNA elbow at the bending site of the L, where the D-loop and the T-loop meet and interact with each other. The exposed sites of the tRNA structure could be ‘points of attack’ in an ancient cellular (and perhaps early proto-cell) environment, being fragmented by either nonspecific stress signals such as radiation and reactive oxygen species (ROS), specific recognition by enzymes or ribozymes, or a combination of both.
Figure 1. The biogenesis of tRNA-derived small RNAs (tsRNAs) is rooted in tRNA structure, regulated by tRNA modifications and RNases.

(A) The 2D and 3D structures of a tRNA, showing the accessible sites at the anticodon loop (green) and the tRNA elbow [the junction of the D- (blue) and T-loops (gray)], which represent the preferred sites of tRNA fragmentation to generate diverse tsRNAs. (B) tRNA modifications and related enzymes that relate to the biogenesis of tsRNAs (TET2 [22], ALKBH1 [48], and others are reviewed in [1]). Arrows indicate enzymes that add a specific RNA modification, inhibition lines indicate the effect of demethylation. (C) Currently known RNases for tsRNA biogenesis include Dicer [17–19], RNase 1 [116], RNase P [16], and others (reviewed in [1]). Abbreviations: hm5C, 5-hydroxymethylcytosine; Me, methylation; Q, queuosine.
This simple view coincides with prevailing observations that the most abundantly detected tsRNAs are fragmented at the anticodon, and are derived from the 5′ half of the tRNA (~30 nt), whereas shorter 3′ or 5′ tsRNAs (~18–22 nt) fragmented at the T-loop or D-loop, respectively, or internal tsRNAs derived from sequences between these loops, are less abundant [13,14] (Figure 1A).
This observation may support the assumption that, in early life forms, the biogenesis of tsRNAs directly originated from tRNA degradation processes starting with these loops, perhaps including multistep degradation and the generation of different intermediates [15], accompanied by regulatory elements such as RNA modifications (Figure 1B) and specific RNases targeting these loops (Figure 1C) that emerged during evolution.
Notably, tRNA cleavage can also occur independently of the loop site, such as by targeting a specific tRNA stem position by RNase P that recognizes a specific (e.g., GC-rich) sequence [16] or, more generally, by enzymes targeting double-stranded (ds) RNA regions. For example, although it is well-known that the RNase Dicer cleaves dsRNAs to generate siRNAs and miRNAs, Dicer is also responsible for the biogenesis of some tsRNAs from tRNAs [17–19]. Since tsRNAs are present in all domains of life, whereas Dicer has not been identified in prokaryotes [20], this supports the notion that Dicer-based canonical small RNAs (i.e., siRNAs and miRNAs) emerged later than tsRNAs during evolution, and Dicer may have recognized the ds stem region in a tRNA-like structure when it first emerged. Currently known enzymes that cleave tRNA (and pre-tRNAs) to generate tsRNAs are well-summarized in a recent review [1], and are shown in Figure 1C. The list of enzymes mediating tsRNA biogenesis is expected to expand in the future [21,22].
tsRNA termini
RNA cleavage by several classes of self-cleaving ribozymes [23] produces RNA fragments with 5′-hydroxyl (5′-OH) and 2′,3′-cyclic phosphate (2′,3′-CP) termini, whereas ancient endonucleases in the protein world, including tRNA splicing endonuclease [24], RNase T2 [25], RNase L [26], and RNase A [27], also similarly generate 5′-OH and 2′,3′-CP RNAs. Interestingly, the RNases T2 [28,29] and L [26], and the vertebrate-specific angiogenin (RNase A family) [30,31], can all cleave tRNAs at the anticodon loop, resulting in their fragmentation into tsRNAs. Cleavage of tRNAs by these ribozymes and enzymes provides unique differences in tsRNAs from other sncRNAs such as those generated by Dicer, which bear a 5′-phosphate (5′-P) and a 3′-hydroxyl (3′-OH) [32], or piRNAs (and plant miRNAs) that bear 2′-O-methylation at the 3′ terminus as a result of additional enzymatic processing [33].
Notably, some tsRNA sequences can be found in the piRNA database, which could be partially due to misannotation or contamination (and not to PIWI-binding) [34]. However, some of them may represent genuine piRNAs (i.e., tRNAAspGUC and tRNAHisGUG in Bombyx) because they bind to PIWI, and have 3′ terminal 2′-O-methylation but not 2′,3′-CP; these piRNAs are further processed from tsRNAs and are termed ‘tRNA-derived piRNAs’ [35]. Although the different sncRNA termini represent the chemical properties of their processing enzymes, they may also have functional consequences. Different termini might be harnessed as intra- or intercellular sorting signals that determine protein-binding potential and subcellular compartmentalization (e.g., cytoplasmic versus nuclear), which is an exciting area that awaits further investigation.
Impact of tRNA modifications
Both tsRNAs and their precursor tRNAs are heavily modified. More than 150 types of modifications are found in tRNAs [36], and their evolutionary origin remains an interesting question. Hypothetically, in early life forms on Earth, RNA modifications might have been first added by random chemical reactions or by base-modifying ribozymes [37,38], which in turn expanded the functionality of ribozymes. In this regard, the RNA modifications on ancient RNAs, such as tRNAs, rRNAs, and spliceosomal small nuclear RNAs (snRNAs), may represent the vestiges of the RNA world at a time when RNA modifications were maximally exploited to increase the functional diversity of ribozymes, before being replaced by the emergence of functionally more versatile protein-based enzymes. The present-day tRNA modifications contribute to multiple aspects of tRNA function, including stability, amino acid charging, and translational accuracy, as well as to tsRNA biogenesis [39,40].
It has been demonstrated that DNMT2- and NSUN2-dependent addition of a 5-methylcytosine (m5C) modification to several tRNAs (e.g., tRNAAsp, tRNAVal, tRNAGly, and tRNALeu) increases tRNA stability in flies and mice, whereas deletion of Dnmt2 and/or Nsun2 abolishes m5C on these tRNAs, making them more likely to be cleaved into tsRNAs under stress conditions [41–43]. The queuosine (Q) modification by QTRT1 occurs in the wobble anticodon position of several tRNAs (tRNAHis, tRNAAsn, tRNATyr, and tRNAAsp) and protects tRNAs against cleavage into tsRNAs in human HEK293T cells [44]. Interestingly, recent reports showed that C38 Q-modified tRNA promotes DNMT2-mediated m5C on C38 of tRNAAsp [45,46]; these discoveries resonate with findings that the establishment of one RNA modification can depend on the existence of another [47]. Recent evidence also shows that deletion of Alkbh1 [48] or Alkbh3 [49] increased the levels of N1-methyladenine (m1A) in tRNAs, preventing tRNA cleavage and resulting in less tsRNA production. TRMT10A-mediated N1-methylguanine (m1G) modification also leads to increased tRNAGln stability and less production of tsRNAGln [50]. Moreover, 2′-O-methylation of C34 in human tRNAMet can prevent site-specific cleavage of tRNAMet by angiogenin and reduce tsRNA production [51].
In addition to preventing tRNA cleavage, some RNA modifications can also promote tsRNA biogenesis. For example, PUS7-mediated pseudouridine (Ψ) at the U8 position has been shown to affect tsRNA biogenesis in stem cells, where deletion of Pus7 leads to a decreased levels of several types of 5′-tsRNAs with terminal oligo(G), suggesting that (Ψ) at the U8 position increases the cleavage of these tRNAs to generate tsRNAs [52]. In another example in yeast, 5-methoxycarbonylmethyl-2-thiouridine (mcm5S2) at the anticodon wobble position can promote the cleavage of tRNA into tsRNAs [53].
In an intriguing scenario, when bacterial tRNAs (tRNAAsp and tRNAArg) are cleaved in half by ribotoxins that recognize the Q-containing anticodon of tRNA [54], the newly generated two tsRNA halves can be reunited/repaired into a full tRNA by a Pnkp/Hen1 heterotetramer while adding a 2′-O-methylation to the previous RNA cleavage site [55]. The added 2′-O-methylation can protect the repaired tRNA from being recut by ribotoxins. This example shows how dynamic RNA modifications are harnessed to balance tRNA stability and tsRNA biogenesis, and also raises the possibility that tsRNAs can gain additional modifications after being cleaved from precursor tRNAs.
Importantly, it should be noted that the changes in tRNA modifications not only affect tsRNA biogenesis but may also affect the function of the resulting tsRNAs owing to altered modification status [56]. In fact, the RNA modifications of tsRNAs have posed challenges for RNA-seq library preparation and for functional studies of tsRNAs (Box 2), and this will need to be resolved to permit new waves of tsRNA research.
Box 2. Technical challenges centered on tsRNA modifications.
Fully understanding the impact of RNA modifications on the function of each tsRNA remains experimentally challenging, as synthetic tsRNAs may not fully mimic the modified tsRNAs that exist in tissues/cells. Two recent reports have provided valuable protocols to isolate individual tsRNAs in vivo [135,136], enabling the future possibility of comparing the functions of modified tsRNAs versus synthetic unmodified tsRNAs – this might permit the engineering of individual tsRNAs by adding site-specific modifications to mimic or enhance their functions. In addition, when performing functional studies (such as cellular transfection) of either synthetic tsRNAs or isolated tsRNAs in vivo, quantitative and stoichiometric factors should aim to reflect the intracellular situation. Blockade of tsRNAs using complementary sequences such as antisense oligonucleotides (ASOs) or locked nucleic acids (LNAs) should be carefully validated so as to not affect the levels of precursor tRNAs [72].
When profiling tsRNAs using RNA-seq, the specific tsRNA termini (e.g., 3′-phosphate and 2′,3′-cyclic phosphate) and RNA modifications (e.g., m1A, m3C, m1G, and N2-dimethylguanosine, m22G) have created challenges to prevent efficient and complete conversion of tsRNAs into cDNA libraries during standard RNA-seq protocols, resulting in biased detection and quantitation during deep sequencing [14]. Recently improved RNA-seq methods such as panoramic RNA display overcoming RNA modification aborted sequencing (PANDORA)-seq [14] and Cap-Clip acid pyrophosphatase, PNK, and AlkB (CPA)-seq [137] aim to overcome these problems by removing key RNA modifications that block adapter ligation and RT by using consecutive enzymatic treatment; these techniques await further optimization such as combination with other strategies focusing on the 5′-termini [138].
Finally, some RNA modifications of tsRNAs/tRNAs may affect probe binding efficiency during northern blot (NB) examination, which may lead to incorrect interpretations when an altered NB signal is associated with altered levels of tRNA/tsRNA modifications. For example, in a cell/tissue with altered RNA modifications caused by mutation or specific environmental stimuli, changes in the observed NB signal may reflect altered tsRNA levels, or altered probe hybridization efficiency owing to RNA modification changes, or both. In addition, in studies involving the knockdown or knockout of tRNA modification enzymes, the resulting phenotypes will represent a mixed effect of altered levels of tRNAs, tsRNAs, and potentially other molecules. These challenging situations in tsRNA research await clarification and resolution.
Evolving tsRNA functionalities
Research on the functions of tsRNAs continues to expand, and roles have been reported in regulating stem cell maintenance [57,58], cancer [59–61], viral infection [9,62], neurological diseases [63,64], epigenetic inheritance [42,65–68], and symbiosis [69]. Unlike other canonical sncRNAs such as miRNAs, siRNAs, and piRNAs, which usually function through strict molecular mechanisms by binding to Argonaute family proteins, the mechanisms of action of tsRNAs appear to be more diverse. Instead of listing known tsRNA functions and mechanisms that have been well summarized elsewhere [1,2,70], we propose to streamline these tsRNA functions and mechanisms in a new narrative to dissect/propose three possible major steps regarding how the function of tsRNAs has emerged and evolved: (i) tsRNAs function via mimicry/replacement of tRNAs with sequence/structure effects (Figure 2A), (ii) tsRNAs function via association with ribonucleoproteins (RNPs) (Figure 2B), and (iii) tsRNAs function via Argonaute proteins (i.e., AGO, PIWI) (Figure 2C). In addition, tsRNAs (and other mobile RNAs) can travel between cells and organisms, enabling long-distance regulation inside and between organisms, and even mediate cross-kingdom regulation between prokaryotes and eukaryotes (Figure 3).
Figure 2. The evolving principles of tRNA-derived small RNA (tsRNA) functionality from an evolutionary perspective.

We propose three possible major steps regarding how tsRNA function has emerged and evolved; the arrow at the bottom shows evolution of tsRNA functionality towards more specialized roles. (A) tsRNA function by tRNA mimicry/displacement and structural effects. (B) tsRNA function via cytosolic and nuclear ribonucleoproteins (RNPs). (C) tsRNA function through binding to prokaryotic or eukaryotic Argonaute proteins. Abbreviations: ERV, endogenous retrovirus; RT, reverse transcription.
Figure 3. Mechanisms and routes of tRNA-derived small RNA (tsRNA) movement between cells, individual organisms, and species.

(A) Extracellular tsRNAs can be encapsulated in extracellular vesicles (EVs), bind to ribonucleoproteins (RNPs), or form specific secondary/3D structures with the aid of RNA modifications. (B) Known and predicted (?) movement of tsRNA between somatic cells or between somatic and germ cells in a single individual, or between organisms/species (e.g., between microbiomes, grafted plants), where tsRNAs may mediate cross-kingdom regulation such as between bacteria and plants/animals.
Mimicry/displacement of tRNAs
Since the 1960s it has been reported that a tRNA fragment from tRNAfMet can interact with ribosomal subunits in a fashion similar to that of its precursor mature tRNAfMet [71]; this suggests that tsRNAs may naturally compete with the function of tRNAs under some circumstances. The tRNA: tsRNA ratio may reach an equilibrium under normal cellular conditions, whereas the waxing and waning of this ratio under changing environments (e.g., stress conditions that cleave more tRNAs) enables regulatory effects. Supporting this idea, recent evidence has shown that, in Archaea (e.g., Haloferax volcanii), a stress-induced 5′tsRNAVal can interact with the rRNA of the ribosomal subunit, and interferes with the loading of mRNAs into ribosomes, thus inhibiting global protein synthesis [4,5] (Figure 2A, top). By contrast, in another example, in Trypanosoma brucei, a unicellular protozoan, a stress-induced 3′tsRNAThr can be incorporated into ribosomes to enhance mRNA loading during stress recovery, and thus increases global translation [6]. These opposing examples suggest that tsRNAs can function flexibly in a context-dependent manner, possibly depending on where they interact with the ribosome and how they might alter the local structure of the rRNAs. In another example, 3′tsRNALeu can bind to specific ribosomal protein mRNAs (e.g., RPS28 and RPS15) to enhance their translation by changing their mRNA structure [72,73]; depletion of 3′tsRNALeu causes decreased translation of RPS28, which impairs pre-18S ribosomal RNA processing and ribosome biogenesis [72].
In addition to tsRNA–ribosome interactions, many RNA viruses and retrotransposons (e.g., endogenous retroviruses, ERVs) can duplicate themselves by using the 3′ terminus of mature tRNA as primers for their RT [9] (Figure 2A, top). In a recent study, transfection of exogenous 18 nt 3′tsRNAs led to competition with the mature tRNAs for the primer binding sequence (PBS) of the ERVs, leading to RT blockade and thus impeding retroviral cDNA synthesis [74]. A similar tsRNA blockade effect at the PBS site may also take place in HIV-infected T cells [75]. By contrast, in other scenarios, tsRNAs can be directly used as primers for RT in human T cell leukemia virus type 1 (HTLV-1) [76] and copia retrovirus-like particles [16]. These opposite roles of tsRNAs in retroviral RT processes suggest adaptation of the viral world to specific tsRNAs that either block or mimic the function of tRNAs.
In another interesting case, ATE1-mediated post-translational arginylation of proteins directly competes with protein translation involving the use of arginine-charged tRNAArg [77]. In this context, it was found that when arginine-charged tRNAArg (Arg-tRNAArg) is fragmented into tsRNA, Arg remains bound to the 3′tsRNAArg (Arg-3′tsRNAArg) and the translation-incompetent Arg-3′tsRNAArg can still be efficiently used as a donor for arginylation by the ATE1 enzyme [77] (Figure 2A, bottom). Although this is only one piece of evidence, these data suggest a scenario in which the balance between translation and arginylation might be regulated by the tsRNAArg:tRNAArg ratio.
We anticipate the discovery of more tsRNA functions under the principle of mimicking/displacing precursor tRNAs because we believe this scenario may represent the most ancient regulatory mode of tsRNAs – simply based on sequence/structural competition.
Forming RNPs
Another well-studied mode of tsRNA functionality is through the formation of tsRNA-RNP complexes. For example, under stress conditions, angiogenin-induced tsRNAs can induce stress granule (SG) formation in a portion (~4%) of cells and the tsRNAs could be incorporated into SGs [78,79] (Figure 2B, top). The ability of tsRNAs to induce SG formation requires the action of YB1 [79], and the role of tsRNAs in suppressing translation is dependent on their binding with eukaryotic initiation factors (eIFs) to sequester them from the translational machinery [79,80]. The formation of this tsRNA-RNP complex involves tsRNA secondary structures such as intermolecular RNA G-quadruplexes (RNA-G4) [81], and RNA modifications such as PUS7-mediated Ψ modifications at U8 [52]. Angiogenin-induced tsRNA has also been shown to form an RNP complex with cytochrome c that is released from the mitochondria, where the tsRNA-mediated sequestration of cytochrome c protects the cells from apoptosome formation and apoptosis under cell stress [82]. Interestingly, the binding of tsRNAs to cytochrome c to inhibit apoptosis resembles the function of some tRNAs [83].
In addition to functioning with cytosolic RNPs, tsRNA can also interact with spliceosomal proteins (e.g., hnRNPF/H) in the nuclei, where hnRNPF/H contributes to the formation of the Cajal body [84] (Figure 2B, bottom). Cajal bodies are known to regulate the production of nuclear small RNAs, including small nucleolar RNAs (snoRNAs), small Cajal body-specific RNAs (scaRNAs), and snRNAs. These nuclear small RNAs, along with tsRNAs, may form other nuclear RNP complexes that contribute to the processing [85] and modification [86] of rRNAs, resulting in ribosome heterogeneity that leads to translational specificity for a selected pool of mRNAs to direct the cell to a specific functional state [87]. To date, the possible nuclear function of tsRNAs remains mysterious but intriguing, especially in early embryo development. Recent evidence has shown that tsRNAs in mouse sperm are responsive to environmental stimuli [65,66], and tsRNAs are enriched at a higher level in the sperm head where the nucleus is condensed [14]. Importantly, pronuclear zygotic injection of tsRNA-containing sperm RNA fractions from high-fat diet-exposed males can induce metabolic phenotypes in the resulting offspring, suggesting that tsRNAs might function through nuclear events that drive butterfly effects throughout embryo development since the zygote stage, possibly by regulating the translational specificity of the early embryo [88].
Binding to Argonaute
tsRNA binding to Argonaute family proteins has been reported to mediate post-transcriptional silencing (either by RNA degradation or mRNA translational inhibition) in multiple cases [18,69,89–91], mostly in an RNAi-like fashion using complementary sequences binding to the targeted mRNAs (Figure 2C). However, the Argonaute-dependent RNAi systems only prevail in eukaryotes and are believed to have emerged primarily to defend against viruses, represented by either the AGO-centered antiviral machinery in the cytoplasm or the PIWI-centered function in the nuclei to suppress endogenous viruses (e.g., transposons) [20].
Unlike in eukaryotes, prokaryotic Argonaute proteins primarily use small DNA as a guide to cleave DNA, a process known as DNA interference [92], to inhibit the propagation of foreign plasmids and infection by phages [93] (functionally similar to the CRISPR/Cas system but mechanistically analogous to RNAi). We believe that this evidence from prokaryotic systems suggests an ancient origin of Argonaute proteins that pre-dates the emergence of RNAi. Since tsRNAs are widely present in prokaryotes [4,5,7], it is an intriguing hypothesis that tsRNAs might be used by prokaryotic Argonaute as an alternative substrate. Indeed, in a seminal study exploring the binding of bacterial Argonaute to RNAs, it was found that the 3′ halves of two types of tsRNAMet are abundantly found as the majority of 45 nt reads [94], although their biological functions remain unresolved. It is our hypothesis that other tsRNAs might also bind to the prokaryotic Argonaute, which awaits to be confirmed by improved library construction protocols that consider the specific tsRNA termini and modifications [14].
In fact, because tRNA/tsRNA sequences are conserved across species during evolution, tsRNAs might be harnessed under some circumstances to defend against foreign DNA/RNA invasions as a first-frontier responding molecules in a fashion similar to the innate immune system, given their rapid elevation upon active infection [95] and that tsRNAs activate Toll-like receptors (TLRs) [96]. This is in contrast to the siRNA/piRNA systems that more resemble the adaptive immune system in that they involve de novo generation of perfectly matched sncRNAs customized to the invading DNA/RNA [12].
Importantly, although the earlier mentioned tsRNA mechanisms of action have been discussed separately and from an evolutionary perspective, different mechanisms can coexist or function synergistically to achieve common outcomes. For example, the suppression of retrotransposons could be achieved by both tsRNA displacement of tRNA-mediated RT [74] and RNAi-like post-transcriptional suppression [74,97]. tsRNA-mediated translational regulation can be achieved by tsRNA–ribosome interaction, tsRNA-RNP-based eIF sequestration, and Argonaute-dependent RNAi effects [98]. In addition, it has been shown in Tetrahymena that tsRNA can bind to an Argonaute family protein to form nuclear RNPs, which may facilitate nuclear rRNA processing and cell growth [85,99].
tsRNAs on the move
In addition to functioning in the cytoplasm and nuclei, tsRNAs are also abundant in body fluids [100] and can be delivered between somatic cells or between somatic and germ cells [66], and even across species in which bacterial tsRNAs are essential for prokaryote–eukaryote communication to ensure microbiome–plant symbiosis for soybean growth [69] (Figure 3). Intercellular communication by tsRNAs could be either dependent or independent of extracellular vesicles (EVs) [101] (Figure 3A), which resonates with the wisdom of Charles Darwin – who proposed in 1868 that each cell emits small particles or molecules, called ‘gemmules’, that diffuse and move between both somatic cells and germ cells [102], a concept that is now gaining new momentum [103].
In fact, the phenomenon of membrane vesicle release in Bacteria, Archaea, and Eukaryotes is an evolutionarily conserved mechanism [104] that might enable robust horizontal information exchange and facilitate the selection and propagation of traits under pressure. Whether and how tsRNAs are selectively or randomly encapsulated into EVs as cargo and specifically delivered to another cell or organism to exert their function remains largely unknown and certainly deserves intense investigation.
Importantly, tsRNAs have been observed outside the cell without being encapsulated into EVs [95,105,106]. In these cases, tsRNA could travel with extracellular RNPs or depend on specific RNA structures (e.g., dimers, RNA G4 tetraplexes [107,108]) or modifications, which increase their stability. Interestingly, engineered mRNAs harboring TLSs with predicted stem–bulge–stem–loop structures are sufficient to mediate systemic mRNA transport in plants and move through graft junctions [109]. Thus, specific tRNA-related and tsRNA-mediated structures may represent RNA mobility motifs that facilitate EV-independent RNA transfer, and their entry into a target cell may or may not rely on specific cell-membrane receptors. This type of extracellular tsRNA transfer could have significant implications because it might be involved in crosstalk between food-derived RNAs and the gut microbiome, or enable the direct transport of food-derived small RNAs into animal cells [110] (Figure 3B).
Concluding remarks
As summarized here, the studies on the biogenesis and functions of tsRNAs have advanced greatly and now constitute an emerging field in RNA biology; as a result, the association and causative roles of tsRNAs in various disease conditions are also being revealed. In particular, tsRNAs are found in all types of body fluids [100] and are dynamically regulated under pathophysiological conditions [95,106], which we argue potentially makes tsRNAs excellent candidates as liquid biopsy biomarkers for clinical translational applications, especially when augmented with the power of machine learning to reveal the multifaceted nature of the ‘tsRNA code’ (Box 3).
Box 3. Harnessing the ‘tsRNA code’ for precision medicine.
tsRNAs are relatively stable in body fluids, and this may be due in part to the fact that they are heavily modified. RNA modifications also provide additional layers of information compared with other RNA-based biomarkers (such as mRNAs and miRNAs). More specifically, a complete tsRNA profile should include not only the expression level of each tsRNA sequence but also the cleavage site/pattern from which they are derived, the overall spectrum of RNA modifications, and the site-specific RNA modifications on each tsRNA. This complex nature of tsRNA profiles, in theory, would generate astronomical permutations when considering each layer of information [2], making the following data analyses high-dimensional, sparse, and possibly nonlinear and thus difficult to precisely describe using traditional statistical approaches. Furthermore, we believe that, given the emerging application of artificial intelligence in biomedical research, the complex layers of information embedded in tsRNA profiles provides an excellent research/clinical opportunity to analyze tsRNA signatures with deep learning algorithms such as artificial neural networks (Figure I) [139]. In our opinion, the level of complexity buried in the tsRNA signature, once fully deciphered with deep learning programs, holds great promise to provide superior biomarkers with better resolution for the diagnosis and prognosis of complex diseases that were previously difficult to distinguish at the molecular level, facilitating the future development of precision medicine [140].
Figure I. Schematics of the use of artificial neural networks to analyze multiple layers of tRNA-derived small RNA (tsRNA) profiles.
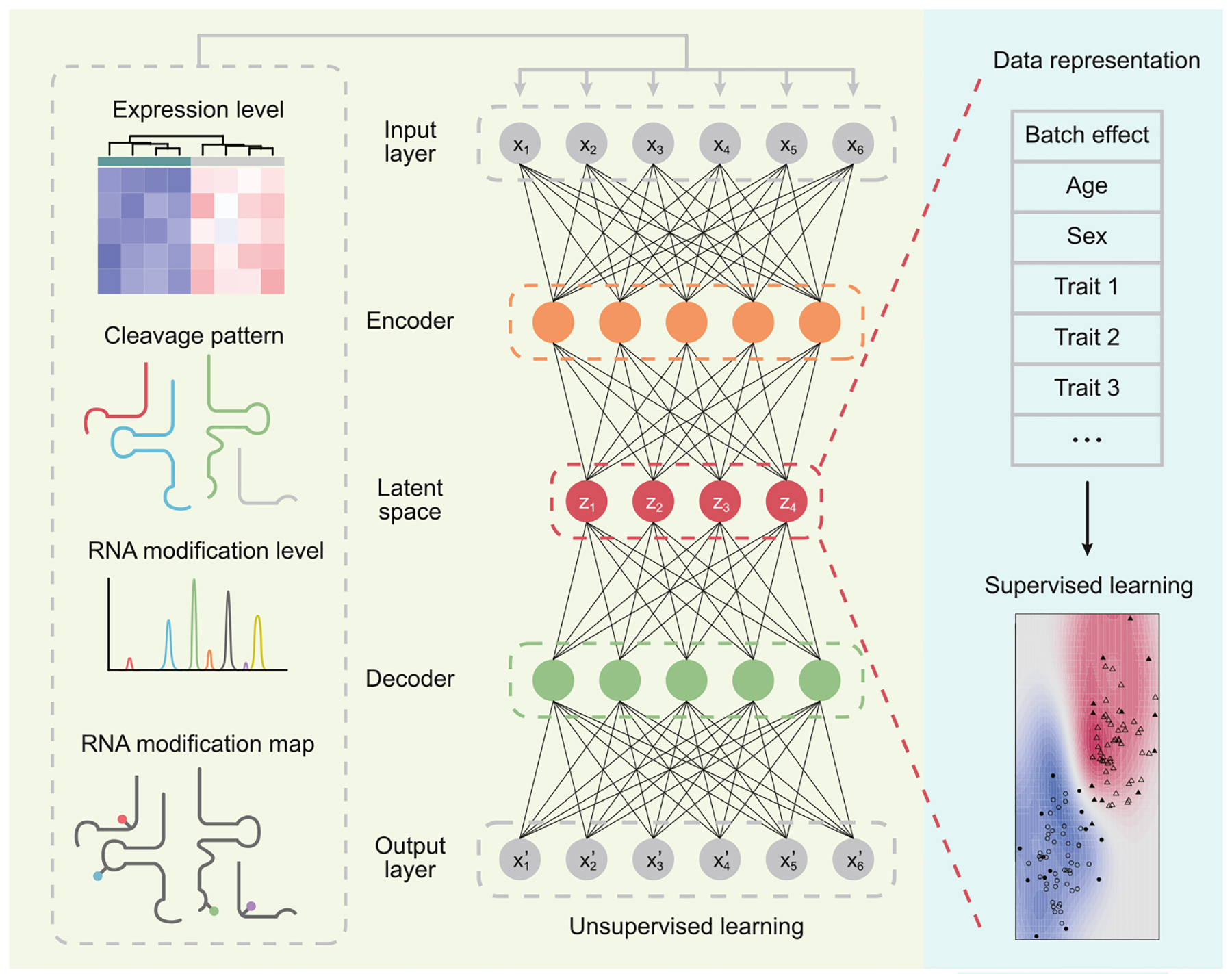
The input tsRNA data are processed by an unsupervised deep learning algorithm, Autoencoder. With multiple hidden layers, the encoder maps the input tsRNA data (X) into a lower-dimensional latent space (Z) with fewer nodes, which captures a compressed knowledge representation (supposedly containing biological/clinical features such as age, sex, disease status, batch effect, etc.) from the original input and forms the basis for reconstructing the input in the output layer (X’) via the multiple hidden layers in the decoder. In turn, the latent representation can be fed into a supervised machine learning algorithm that will aid diagnosis, prognosis, and preventive and therapeutic interventions in precision medicine.
Looking forward, reports of other noncanonical small RNAs, such as rRNA-derived small RNAs (rsRNAs) and YRNA-derived small RNAs (ysRNAs), are also emerging as new analytical tools become available [14,111]. The biogenesis and function of rsRNAs and ysRNAs are less understood but may follow the same general principles as tsRNAs because they are similarly fragmented and processed from ancient RNA species (i.e., rRNA, YRNA) that exist in all domains of life. For example, RNase T2 can cleave both rRNA and tRNA to generate rsRNAs/tsRNAs [28,29], and both tsRNA and rsRNAs are sensitive to environmental exposures such as oxidative stresses, diet, and inflammation [42,112–115]. YRNAs/tRNAs can also be cleaved inside EVs by RNases (e.g., RNase L, RNase 1) to generate ysRNAs/tsRNAs, and may contribute to circulating tsRNAs/ysRNAs in the serum and other biofluids [26,116]. Interestingly, recent data have shown that tsRNAs/rsRNAs/ysRNAs can together form a ‘disease RNA code’ that can distinguish human diseases such as lung cancer and tuberculosis, and this can outperform miRNA-based signatures [117], suggesting that their complex forms in nature (e.g., versatile cleavage patterns) enabled more permutations and superior information capacity and specificity that can be harnessed to distinguish between and subcategorize complex diseases for translational medicine.
Outstanding questions.
How many more (modified) tsRNAs can be discovered with improved RNA sequencing methods?
Do RNA modifications regulate tsRNA intracellular sorting, compartmentalization, and functionality in conjunction with associated RNA-binding proteins?
Is the principle of the functionality of each tsRNA universal or context-dependent?
Could tsRNAs mediate cross-species regulation between food and the microbiome, or between the microbiome and the host?
To what extent do RNA modifications and 3D RNA structures affect the functions of tsRNAs, in addition to their sequences?
How can tsRNA levels be experimentally manipulated without affecting their tRNA precursors?
Can multiple layers of information be extracted and integrated from tsRNAs (e.g., expression level, cleavage patterns, RNA modifications) for deep learning and clinical translational applications?
Highlights.
tRNA-derived small RNAs (tsRNAs) are present across all three domains of life: Archaea, Bacteria, and Eukarya.
The biogenesis of tsRNAs involves specific cleavage of mature tRNAs or tRNA precursors at preferred loci followed by further processing.
RNA modifications are essential for both tsRNA biogenesis and functionality.
tsRNA functionality is intricately linked to basic life elements including the ribosomal machinery and viral replication, in addition to binding to ribonucleoprotein and Argonaute proteins (i.e., AGO and PIWI).
tsRNAs can move across tissues/cells and may mediate intergenerational phenotype transmission and pathogen–host interplay and symbiosis.
tsRNAs and their RNA modifications can serve as disease biomarkers.
Acknowledgments
M.H. is in part supported by MOST (2018YFC1004500). Research in the laboratory of Q.C. is in part supported by the National Institutes of Health (NIH; R01HD092431 and R01ES032024). The laboratory of T.Z. is in part supported by the NIH (R01ES032024).
Glossary
- Aminoacyl-tRNA synthetases (aaRSs)
also termed tRNA ligases, aaRSs are enzymes that catalyze the acylation of a specific amino acid or its precursor to one of its cognate tRNAs to form an aminoacyl-tRNA.
- Apoptosome
a large quaternary protein structure formed in the process of apoptosis, the formation of which is triggered by the release of cytochrome c from the mitochondria.
- Arginylation
a post-translational modification mediated by arginyl-tRNA–protein transferase (ATE1) that transfers arginine from tRNA onto proteins.
- Argonaute family proteins
evolutionarily conserved proteins in both prokaryotes and eukaryotes that play an essential role in gene silencing. These can be subdivided into the AGO subfamily and the PIWI subfamily according to their phylogenetic classification.
- Artificial neural network
algorithms that aim to recognize underlying relationships in a dataset through a process that mimics the way the human brain operates.
- Cytochrome c
a small hemoprotein that is loosely associated with the inner membrane of the mitochondrion, and which triggers cell apoptosis on being released into the cytosol in response to apoptotic stimuli.
- Deep learning
a part of the broader family of machine learning methods.
- Hepatitis delta virus (HDV)
the smallest virus that is known to infect animals; HDV has a single-stranded circular RNA genome.
- Last universal common ancestor (LUCA)
the most recent common ancestor (most likely a single-celled organism) of all current life on Earth.
- Protocell
a self-organized, endogenously ordered, spherical collection of lipids that has been proposed as a stepping-stone toward the origin of life.
- Ribosome heterogeneity
ribosomes of distinct compositions within a cell or across tissues that generate biased translational specificity for different mRNA subpopulations.
- Ribotoxins
extracellular RNases that can enter host cells and cleave essential RNAs such as rRNAs and tRNAs.
- Ribozyme
RNA molecules that catalyze specific biochemical reactions similarly to the action of protein enzymes.
- Stress granules (SGs)
ribonucleoprotein (RNP) granules in the cytoplasm which aggregate stalled translational preinitiation complexes and nontranslating mRNAs that accumulate during stress.
- tRNA elbow
a conserved structure arising from the interaction between the D- and T-loops of a tRNA.
- Viroids
the smallest known infectious pathogens which are composed solely of a circular, single-stranded RNA that has no protein coating.
Footnotes
Declaration of interests
The authors declare no conflicts of interest.
References
- 1.Su Z et al. (2020) Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu. Rev. Genet 54, 47–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimmel P (2018) The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol 19, 45–58 [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Silva MR et al. (2014) Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res 113, 285–304 [DOI] [PubMed] [Google Scholar]
- 4.Gebetsberger J et al. (2012) tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebetsberger J et al. (2017) A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 14, 1364–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fricker R et al. (2019) A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun 10, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar P et al. (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss MC et al. (2018) The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 14, e1007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunes A et al. (2020) Emerging roles of tRNAs in RNA virus infections. Trends Biochem. Sci 45, 794–805 [DOI] [PubMed] [Google Scholar]
- 10.Dreher TW (2010) Viral tRNAs and tRNA-like structures. Wiley Interdiscip. Rev. RNA 1, 402–414 [DOI] [PubMed] [Google Scholar]
- 11.Krupovic M et al. (2020) The LUCA and its complex virome. Nat. Rev. Microbiol 18, 661–670 [DOI] [PubMed] [Google Scholar]
- 12.Broecker F and Moelling K (2019) What viruses tell us about evolution and immunity: beyond Darwin? Ann. N. Y. Acad. Sci 1447, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P et al. (2015) tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J et al. (2021) PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol 23, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng H et al. (2012) A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 22, 1609–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi Y et al. (1990) Cleavage of tRNA within the mature tRNA sequence by the catalytic RNA of RNase P: implication for the formation of the primer tRNA fragment for reverse transcription in copia retrovirus-like particles. Proc. Natl. Acad. Sci. U. S. A 87, 8105–8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole C et al. (2009) Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haussecker D et al. (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinsborough CW et al. (2019) BCDIN3D regulates tRNAHis 3′ fragment processing. PLoS Genet. 15, e1008273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shabalina SA and Koonin EV (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol 23, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama Y et al. (2019) Multiple ribonuclease A family members cleave transfer RNAs in response to stress. BioRxiv Published online October 21, 2019. 10.1101/811174 [DOI] [Google Scholar]
- 22.He C et al. (2020) TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol 28, 62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diener TO (1989) Circular RNAs: relics of precellular evolution? Proc. Natl. Acad. Sci. U. S. A 86, 9370–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trotta CR et al. (1997) The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858 [DOI] [PubMed] [Google Scholar]
- 25.Luhtala N and Parker R (2010) T2 family ribonucleases: ancient enzymes with diverse roles. Trends Biochem. Sci 35, 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donovan J et al. (2017) Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons SM et al. (2017) RNA biology of angiogenin: current state and perspectives. RNA Biol. 14, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen KL and Collins K (2012) Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol. Biol. Cell 23, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson DM and Parker R (2009) The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol 185, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamasaki S et al. (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol 185, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu H et al. (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP (2018) Metazoan microRNAs. Cell 173, 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji L and Chen X (2012) Regulation of small RNA stability: methylation and beyond. Cell Res. 22, 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tosar JP et al. (2018) Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda S et al. (2017) The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 45, 9108–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boccaletto P et al. (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poudyal RR et al. (2017) Nucleobase modification by an RNA enzyme. Nucleic Acids Res. 45, 1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheitl CPM et al. (2020) Site-specific RNA methylation by a methyltransferase ribozyme. Nature 587, 663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan T (2018) Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki T (2021) The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol Published online March 3, 2021. 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- 41.Tuorto F et al. (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol 19, 900–905 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y et al. (2018) Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol 20, 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer M et al. (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X et al. (2018) Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuorto F et al. (2018) Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller M et al. (2015) Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 43, 10952–10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barraud P et al. (2019) Time-resolved NMR monitoring of tRNA maturation. Nat. Commun 10, 3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rashad S et al. (2020) The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 17, 1092–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z et al. (2019) Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47, 2533–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosentino C et al. (2018) Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 46, 10302–10318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitali P and Kiss T (2019) Cooperative 2′-O-methylation of the wobble cytidine of human elongator tRNA(Met)(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev. 33, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guzzi N et al. (2018) Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216 [DOI] [PubMed] [Google Scholar]
- 53.Lu J et al. (2008) Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res. 36, 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogawa T et al. (2006) Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res. 34, 6065–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan CM et al. (2009) Reconstituting bacterial RNA repair and modification in vitro. Science 326, 247. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X et al. (2016) Small RNA modifications: integral to function and disease. Trends Mol. Med 22, 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanco S et al. (2016) Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krishna S et al. (2019) Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 20, e47789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honda S et al. (2015) Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U. S. A 112, E3816–E3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodarzi H et al. (2015) Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balatti V et al. (2017) tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U. S. A 114, 8071–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q et al. (2013) Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther 21, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X et al. (2020) Small RNA modifications in Alzheimer’s disease. Neurobiol. Dis 145, 105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hogg MC et al. (2019) Elevation in plasma tRNA fragments precede seizures in human epilepsy. J. Clin. Invest 129, 2946–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Q et al. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 [DOI] [PubMed] [Google Scholar]
- 66.Sharma U et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarker G et al. (2019) Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc. Natl. Acad. Sci. U. S. A 116, 10547–10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida K et al. (2020) ATF7-dependent epigenetic changes are required for the intergenerational effect of a paternal low-protein diet. Mol. Cell 78, 445–458 [DOI] [PubMed] [Google Scholar]
- 69.Ren B et al. (2019) Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922 [DOI] [PubMed] [Google Scholar]
- 70.Magee R and Rigoutsos I (2020) On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 48, 9433–9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rudland PS and Dube SK (1969) Specific interaction of an initiator tRNA fragment with 30 s ribosomal subunits. J. Mol. Biol 43, 273–280 [DOI] [PubMed] [Google Scholar]
- 72.Kim HK et al. (2017) A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim HK et al. (2019) A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell Rep. 29, 3816–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schorn AJ et al. (2017) LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeung ML et al. (2009) Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37, 6575–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruggero K et al. (2014) Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J. Virol 88, 3612–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Avcilar-Kucukgoze I et al. (2020) tRNA(Arg)-derived fragments can serve as arginine donors for protein arginylation. Cell Chem. Biol 27, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emara MM et al. (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem 285, 10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lyons SM et al. (2016) YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 44, 6949–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ivanov P et al. (2011) Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lyons SM et al. (2017) Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun 8, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saikia M et al. (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol 34, 2450–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mei Y et al. (2010) tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 37, 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boskovic A et al. (2020) Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev. 34, 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Couvillion MT et al. (2012) A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell 48, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poole AR et al. (2017) Regulatory RNPs: a novel class of ribonucleoproteins that potentially contribute to ribosome heterogeneity. Biol. Open 6, 1342–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Genuth NR and Barna M (2018) The discovery of ribosome heterogeneity and Its implications for gene regulation and organismal life. Mol. Cell 71, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y et al. (2019) Sperm RNA code programmes the metabolic health of offspring. Nat. Rev. Endocrinol 15, 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luo S et al. (2018) Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 46, 5250–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maute RL et al. (2013) tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U. S. A 110, 1404–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuscu C et al. (2018) tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swarts DC et al. (2014) DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuzmenko A et al. (2020) DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637 [DOI] [PubMed] [Google Scholar]
- 94.Olovnikov I et al. (2013) Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell 51, 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y et al. (2014) Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol 6, 172–174 [DOI] [PubMed] [Google Scholar]
- 96.Pawar K et al. (2020) Infection-induced 5′-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 18, e3000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez G et al. (2017) tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 45, 5142–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi J et al. (2019) tsRNAs: the Swiss Army Knife for translational regulation. Trends Biochem. Sci 44, 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Couvillion MT et al. (2010) A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 24, 2742–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Godoy PM et al. (2018) Large differences in small RNA composition between human biofluids. Cell Rep. 25, 1346–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tosar JP and Cayota A (2020) Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 17, 1149–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Darwin C (1868) The Variation of Animals and Plants under Domestication, John Murray; [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y and Chen Q (2018) 150 years of Darwin’s theory of intercellular flow of hereditary information. Nat. Rev. Mol. Cell Biol 19, 749–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deatherage BL and Cookson BT (2012) Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun 80, 1948–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tosar JP et al. (2020) Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res. 48, 12874–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dhahbi JM et al. (2013) 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 14, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tosar JP et al. (2018) Dimerization confers increased stability to nucleases in 5′ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 46, 9081–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ivanov P et al. (2014) G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U. S. A 111, 18201–18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang W et al. (2016) tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28, 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou Z et al. (2015) Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 25, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi J et al. (2018) SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genomics Proteomics Bioinformatics 16, 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thompson DM et al. (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Natt D et al. (2019) Human sperm displays rapid responses to diet. PLoS Biol. 17, e3000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chu C et al. (2017) A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation. J. Mol. Cell Biol 9, 256–259 [DOI] [PubMed] [Google Scholar]
- 115.Lee SR and Collins K (2005) Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem 280, 42744–427749 [DOI] [PubMed] [Google Scholar]
- 116.Nechooshtan G et al. (2020) Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 48, 8035–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gu W et al. (2020) Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer 19, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Higgs PG and Lehman N (2015) The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet 16, 7–17 [DOI] [PubMed] [Google Scholar]
- 119.Cech TR (2000) The ribosome is a ribozyme. Science 289, 878–879 [DOI] [PubMed] [Google Scholar]
- 120.Shi Y (2017) Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol 18, 655–670 [DOI] [PubMed] [Google Scholar]
- 121.Lai MM (2005) RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol 79, 7951–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Beaudry AA and Joyce GF (1992) Directed evolution of an RNA enzyme. Science 257, 635–641 [DOI] [PubMed] [Google Scholar]
- 123.Green R and Szostak JW (1992) Selection of a ribozyme that functions as a superior template in a self-copying reaction. Science 258, 1910–1915 [DOI] [PubMed] [Google Scholar]
- 124.Johnston WK et al. (2001) RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 [DOI] [PubMed] [Google Scholar]
- 125.Lincoln TA and Joyce GF (2009) Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maizels N and Weiner AM (1994) Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. U. S. A 91, 6729–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuhnlein A et al. (2021) tRNA sequences can assemble into a replicator. Elife 10, e63431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee N et al. (2000) Ribozyme-catalyzed tRNA aminoacylation. Nat. Struct. Biol 7, 28–33 [DOI] [PubMed] [Google Scholar]
- 129.Schimmel P and Kelley SO (2000) Exiting an RNA world. Nat. Struct. Biol 7, 5–7 [DOI] [PubMed] [Google Scholar]
- 130.Joyce GF and Szostak JW (2018) Protocells and RNA self-replication. Cold Spring Harb. Perspect. Biol 10, a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Adamala K and Szostak JW (2013) Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342, 1098–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yarus M (2011) Getting past the RNA world: the initial Darwinian ancestor. Cold Spring Harb. Perspect. Biol 3, a003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu J et al. (2020) Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature 582, 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scheitl CPM et al. (2020) New deoxyribozymes for the native ligation of RNA. Molecules 25, 3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Akiyama Y et al. (2020) Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol. 17, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Drino A et al. (2020) Production and purification of endogenously modified tRNA-derived small RNAs. RNA Biol. 17, 1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang H et al. (2021) CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov. 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kugelberg U et al. (2020) 5′XP sRNA-seq: efficient identification of transcripts with and without 5 phosphorylation reveals evolutionary conserved small RNA. RNA Biol. Published online December 31, 2020. 10.1080/15476286.2020.1861770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang J et al. (2020) Denoising Autoencoder, a deep learning algorithm, aids the identification of a novel molecular signature of lung adenocarcinoma. Genomics Proteomics Bioinformatics Published online December 18, 2020. http://dx.do.org/10.1016/j.gpb.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Topol EJ (2019) High-performance medicine: the convergence of human and artificial intelligence. Nat. Med 25, 44–56 [DOI] [PubMed] [Google Scholar]


