Abstract
The need for co-ordinate, high-level, and stable expression of multiple genes is essential for the engineering of biosynthetic circuits and metabolic pathways. This work outlines the functionality and design of IRES- and 2 A-peptide-based constructs by comparing different strategies for co-expression in polycistronic vectors. In particular, 2 A sequences are small peptides, mostly derived from viral polyproteins, that mediate a ribosome-skipping event such that several, different, separate proteins can be generated from a single open reading frame. When applied to metabolic engineering and synthetic gene circuits, 2 A peptides permit to achieve co-regulated and reliable expression of various genes in eukaryotic cells.
Keywords: Synthetic biology, 2A peptide, IRES, Polycistronic sequence
1. Introduction
The joint expression, from a single DNA sequence, of several genes in re-engineered eukaryotic cells has the potential to facilitate the implementation of otherwise intricated biological networks. In Synthetic Biology, genetic circuits are made of transcription units (TUs) that, in eukaryotic cells, consist of three standard biological parts [1]: promoters, coding sequences (CDSs), and terminators. Each TU produces a single protein, a small RNA molecule, or a protein domain (which, for instance, interacts with peptides from other TUs to give rise to a chimeric protein). Therefore, the expression of each CDSs demands a different promoter inside a synthetic gene circuit. This multiple-promoter architecture is commonly adopted in the design of eukaryotic genetic circuits [2]. The establishment of a polycistronic expression system i.e. a structure that allows the simultaneous transcription of multiple TUs, would bring several benefits to eukaryotic Synthetic Biology. For example, a simple biotechnological application is the construction of vectors carrying a bicistronic sequence, where one CDS encodes for a fluorescent protein and the other the protein of interest. This facilitates the screening and selection of properly genetically modified cells [3,4]. A more elaborate utilization requires longer polycistrons for the simultaneous delivery of several therapeutic agents necessary to treat human diseases [[5], [6], [7], [8]].
Different multi-gene expression systems have been proposed: (i) pre-mRNA splicing; (ii) proteolytic cleavage sites; (iii) fusion proteins; (iv) internal ribosomal entry sites (IRESs); and (v) self-cleaving 2 A peptides (2 A) [[9], [10], [11], [12]].
If, on the one hand, each of these approaches provides a way of merging a few TUs into a single one, on the other hand every strategy presents some disadvantages due to its own peculiar features. To be specific, the splicing process is the mechanism that produces two different mature mRNAs from one pre-mRNA through the cleavage at two splice sites—named splice donor and splice acceptor site—resulting in two open reading frames (ORFs) [13]. RNA splicing, however, cannot be controlled easily, and sequence rearrangements or errors in the whole procedure are relatively frequent [118,119]. A fusion protein guarantees the expression of, usually, two genes simultaneously. However, this approach may lead to losing or lowering the activity of one of the two proteins, and it may not work out well when combining proteins with different subcellular localizations [14]. The applications of proteolytic cleavage sites are limited to the expression of a limited type of proteins. For instance, furin is a ubiquitous, highly conserved eukaryotic endoprotease localized in the Golgi that can be adopted to process polyproteins containing the signal sequence Arg.X.Arg/Lys.Arg. Hence, only certain multi-proteins, e.g. those targeted by the cellular secretion pathway and, therefore, connected to the Golgi can be co-expressed and processed properly, whereas general cytoplasmic proteins cannot [15].
Among the five systems listed above, the most widely used are the IRESs and the self-cleaving 2 A peptides. For this reason, this paper is focused on the description of multicistronic constructs based on IRES and 2 A peptides, with particular attention to 2 A applications in Synthetic Biology.
2. IRESs
In 1988, an internal ribosome entry site (IRES) was discovered in the uncapped RNAs from poliovirus [16] and encephalomyocarditis virus (EMCV) [17]. IRESs are cis-acting RNA elements capable of recruiting the small ribosomal subunit and initiating translation in a 5′ cap-independent manner. In eukaryotic cells, indeed, the 7-methylguanosine cap structure at the 5’ end of the RNA is a signal for the initiation factors to recruit ribosomes and initiate the translation of the peptides encoded in the messenger RNA. IRES-mediated translation is driven by complex RNA secondary structures that interact with initiation factors, bridging RNA-binding proteins, or the small ribosomal subunit directly, leading to ribosomal positioning at, or close to, the initiation codon. They typically foster translation initiation with little or no ribosomal scanning [8,[18], [19], [20]]. After their discovery, IRES-mediated systems became well-known and were employed to build polycistronic transcription units in eukaryotic cells that expressed two or more proteins by promoting internal initiation of mRNA translation [7], [21], [22].
In particular, bicistronic constructs (Fig. 1) have been widely used for both in vitro and in vivo applications in mammalian cells, transgenic animals and plants, and yeast cells [[23], [24], [25], [26], [27], [28], [29]]. Along bicistrons, initiation of translation occurs, at the first ORF, according to the canonical 5′ cap-dependent mechanism and, at the second ORF, in an IRES-dependent manner. Importantly, one of the advantages of IRES sequences is the complete separation of the proteins coupled on the DNA, which avoids the risk of protein inactivation [7], [14]. An additional advantage is given by the fact that IRES-dependent initiation mechanism still works in situations where cap-dependent translation is shut down, such as during viral infections, growth arrest, mitosis, apoptosis, and hypoxia [27], [30], [31], [32], [33].
Fig. 1.
The translation mechanism of IRES-based bicistronic constructs.
However, several problems have been also reported in the usage of IRES elements inside multicistronic sequences. First, the considerable length of natural IRES (typically over 500 nucleotides) can reduce the plasmid packaging capacity, thus preventing the successful utilization of long and complex multicistronic vectors [5,9]. Yet, particular cellular IRESs were identified to be composed of shorter elements that displayed independent IRES activity when tested in isolation. For instance, a 9-nt-long IRES module from the 5’ UTR (untranslated region) of the homeodomain protein Gtx, which is 100% complementary to the 18S ribosomal RNA, was shown to have a considerable translation initiation activity in mouse cell line [34]. This mini-IRES efficiency can be increased up to ∼500-fold by linking 10 copies of it. They perform even better than the classical EMCV IRES. A library of short synthetic IRES elements (∼18-nt long) was also isolated in the yeast Saccharomyces cerevisiae [35]. In general, the development of a set of IRES elements spanning a broad range of translational activity would facilitate the construction of polycistronic expression cassette with a consequent substantial reduction in plasmid size and circuit design simplification.
Second, the IRES-driven translation has lower efficiency than the cap-dependent one, resulting in lower expression of genes placed downstream of an IRES [7,25,29,36]. However, in some cases, this feature can be exploited to modulate translation. For instance, most monoclonal antibodies (mAbs) consist of two identical polypeptides, the heavy (HC) and light chain (LC). An excess of LCs in the cell is known to be beneficial for the overall mAb expression [37,38]. This was achieved via the tricistronic cassette: LC-IRES–HC–IRES-selective marker [39,40]. Some improvements made in a further study [41] permitted to fix the relative expression of LC over HC, and obtain, in this way, a stable mAb expression level.
Lastly, homologous recombination and competition among different IRESs for translation factors are other issues [22]. IRES activity may also vary considerably with cell type. Borman and co-authors [24] have examined the translation efficiencies of six different picornaviral IRESs in a variety of human and non-human cell lines. In all tested cell lines, type II IRESs (EMCV, FMDV, and HCV) did not show any particular sensitivity to cell-type and were able to generally initiate protein synthesis effectively. Type I IRESs (PV, ECHO, and HRV) drove translation strongly in HeLa, HepG2, and FRhK4 cells, but had extremely poor performance in Neuro-2A and human neuronal cell line. Type III IRES (HAV) was relatively inefficient in any of the cell lines used in the study—including human hepatocytes. Thus, it is highly probable that IRESs have varying requirements for working in the right way in diverse cell types.
IRESs were, nevertheless, the first successful tool for the construction of eukaryotic multicistronic transcription units. Several works published during the 1990's [42], reported vectors where up to 4 genes were co-expressed via IRESs. However, the above described obstacles, such as their large size and the imbalanced gene co-expression, prompted researchers to explore other strategies adopted by viruses for multiple protein synthesis. Finally, the hurdles posed by the IRESs were partially circumvented with the discovery of the 2 A peptide sequences.
3. 2 A peptides
The first 2 A peptide sequence was identified in and characterized from the foot-and-mouth disease virus. FMDV2A (shortly, F2A), is an oligopeptide (18–22 amino acids) located between two CDSs in some members of the picornavirus family—the same where the first IRESs were discovered [[43], [44], [45], [46]]. Differentiate expression of genes separated by a 2 A peptide sequence is due to a sort of “cleavage” event that takes place at a highly conserved consensus sequence at the C terminus of the 2 A sequence. More precisely: between the glycine (Gly) and the proline (Pro) residue at the very end of the 2 A sequence that looks like: Asp-Val/Ile-Glu-X-Asn-Pro-Gly-Pro. A 2 A peptide sequence impairs the formation of a peptide bond via a “ribosomal skip” mechanism [47] (also referred to as ‘stop-go’ [48] or ‘stop carry-on’ [49]) that does not affect the translation of the downstream gene [50,51] (Fig. 2). 2 A peptide sequences have been termed CHYSELs (cis-acting hydrolase elements) too. Additional CHYSELs, different from 2 A peptides, have been found in viruses [[52], [53], [54]] but will not be discussed in this review.
Fig. 2.
The translation mechanism of 2 A-based bicistronic constructs.
Since the discovery of their ability of “self-cleavage”, a quest for more 2 A peptides has started. Several new 2 A peptide sequences have been found in different viruses such as the Porcine teschovirus-1 (P2A); the Thosea asigna virus (T2A); and the Equine rhinitis A virus (E2A) [10,45,46,55]. (Table 1) Theoretically, ‘ribosome skipping’ provides a complete mRNA cleavage. However, failures can occur in the process. Overall, there are three possible conclusions of a 2 A-mediated skipping event: (1) the skipping is successful, and two proteins are translated properly. The upstream protein is fused to the complete 2 A peptide except for the last proline residue that, in contrast, is attached to the N terminus of the protein downstream of the 2 A; (2) after a successful skipping, the ribosome drops off the mRNA such that the translation of the downstream protein is aborted (whereas the upstream protein was synthesized correctly) (3) the skipping is unsuccessful, i.e. a ribosome “readthrough” takes place, and translation results in a fusion protein [54,55]. In some cases, the presence of both cleaved and uncleaved forms can be, nevertheless, useful. For example, such a mixture was necessary in the work by Cruz-Teran et al. [56], who used F2A18 (i.e. 18 AA long) with ∼50% efficiency of ribosomal skipping, to guarantee, in S. cerevisiae, high production of a soluble protein of interest via the simultaneous cell surface display (due to ribosome readthrough) and secretion (following a successful ribosome skipping).
Table 1.
The sequences of commonly used 2 A peptide. Additional (GSG) residues can affect self-cleavage efficiency positively.
Compared to IRESs and more elaborated approaches, such as mRNA splicing, 2 A peptide sequences present some major advantages for implementing multicistronic vectors. First of all, 2 A peptides have a small size (about 60–70 nucleotides) compared to IRESs. Therefore, ORFs can be easily extended, via PCR, to contain the chosen 2 A sequence. Moreover, the size of the overall expression vector is significantly reduced, which facilitates its cloning and cell transfection/transformation [57], [58], [59]. Second, translation of multigene constructs that include 2 A peptides can be regulated to achieve a well-defined ratio of the desired proteins. This is possible just by altering gene position, and is, therefore, ideal to fine tune the co-expression of polypeptides that work cooperatively and require a precise stoichiometry [54,60,61]. Third, some of the so-far discovered 2 A peptides have shown high cleavage efficiency (defined as the percentage of correctly cleaved translation products). These three unique features together make 2 A peptides an attractive tool for the design and construction of polycistronic vectors.
To assure that protein synthesis has taken place after a successful ‘self-cleavage’, the most straightforward way is Western blotting by means of antibodies against either the target proteins or the 2 A peptides themselves [44,45,58,62,63,68]. As an alternative detection method, genes encoding for fluorescence proteins (reporters) or antibiotic resistance (in bacteria) can be linked, through a 2 A peptide sequences, downstream of the gene(s) encoding for the target protein(s) [[65], [66], [67]].
4. Designing multicistronic sequences
The cleavage efficiency of 2 A peptides depends on its amino acidic sequence. However, in this regard contrasting results are present in the literature. Szymczak et al. [10] used the T-cell receptor (TCR):CD3 complex as a test system and demonstrated that, in mammalian cells, F2A and T2A have higher activities than E2A in the multicistronic CD3δ,γ,ε,ζ (four CD3 proteins divided by 2As) and TCRα-2A-TCRβ constructs. In a later work by the same lab [53], P2A was shown to outperform F2A, T2A, and E2A. Kim and co-authors [5] used Western blotting and fluorescent protein localization analysis to show that P2A had the highest cleavage efficiency followed by T2A, E2A, and F2A (all preceded by the GSG linker) in three commonly used cell lines from human, zebra fish, and adult mice. When tested in Nannochloropsis salina (a marine microalga) [51], T2A and E2A gave better performance than P2A and F2A, with a maximum cleavage rate close to 45%. Souza-Moreira et al. [68] screened 22 2 A peptides, placed between two fluorescent proteins, in S. cerevisiae and assessed by Western blotting the cleavage levels of the above cited commonly used sequences: P2A (85%), T2A (56%), E2A (46%), F2A (43%). However, it should be noted that the best performance (almost 100% cleavage efficiency) was achieved by means of the less frequently used 2 A peptide from Equine rhinitis B virus (ERBV-1).
Additionally, Liu et al. [54] carried out a systematic analysis, in mammalian cells, of different 2 A peptides alone and combined them either in tandem or in triplets (Fig. 3) by realizing bi-, tri-, and quad-cistronic vectors. Their work showed that the P2A-T2A tandem improved the cleavage efficiency of the individual 2 A peptides since it did not generate any uncleaved products. In contrast, the P2A-T2A-E2A triplet led to a lower expression of the downstream protein. Therefore, 2As in tandem turned out to be the most performant configuration for protein co-expression. Moreover, they found that, after selecting a particular protein, its synthesis decreased as it was moved towards the end of the construct (third and fourth place). In contrast, the best results were obtained when the protein of interest occupied the second position.
Fig. 3.
Bi-cistronic constructs with different 2 A peptides alone and combined either in tandem or in triplets.
Previous studies [9,44,45,[69], [70], [71]] have pointed out that the substitution or mutation of the amino acids in the highly conserved “DxExNPGP” motif can affect the ribosomal skipping activity remarkably, since this sequence is regarded as the most effective one to achieve ‘stop-go’ [59]. Furthermore, the sequence of the gene immediately upstream of the 2 A may affect the interaction between the 2 A itself and the ribosome exit tunnel and, thus, its cleavage activity. A screening on the length of the 2 A peptides was performed by Minskaia et al. [72] by modifying the number of amino acids of an F2A connecting two reporter proteins: the green (GFP) and the red (mCherry) fluorescent proteins. The screening was carried out both in vitro and in vivo (HeLa cells) and indicated that F2A30 (30 AA long) was the optimal choice in terms of cleavage efficiency, which resulted completely unaffected by the sequence of the upstream gene.
In addition, other strategies have been proposed to improve the 2 A processing efficiency. One, for instance, demands the insertion of a spacer just upstream of the 2 A sequence, such as the glycine-serine linker (in the form of GSG or SGSG) [50,73], the V5 peptide tag (GKPUPNPLLGLDST) [74], or the 3xFLAG epitope tag (DYKDHDGDYKDHDIDYKDDDDK) [75]. The optimized Kozak sequence from silkworm (ACCACCATGG) [76] was placed, in contrast, after the 2 A peptide to optimize the expression of the downstream gene. On the whole, the effect on 2 A cleavage efficiency due to gene arrangement or modifications with short tags must be taken into consideration carefully for the optimized (or balanced) expression of each individual gene belonging to a multicistronic sequence [39], [44], [60], [76], [77].
Proteins expressed via a 2 A-containing multicistronic sequences can be correctly targeted to various subcellular locations via either cotranslational or posttranslational mechanisms [[83], [84], [85],87]. Trichas et al. [88] designed a bicistronic construct made of a membrane- (red) and a nuclear-localized (green) fluorescent protein separated by a 2 A sequence. Upon expression in transgenic mice, the two fluorescent proteins located in a mutually exclusive way into the two cellular compartments.
de Felipe et al. [89] showed that, in mammalian cells, when certain proteins upstream of the 2 A peptide bear an N-terminal signal sequence, the downstream protein—with or without any post-translational localization sequences—is also translocated into the endoplasmic reticulum (ER). The localization of a downstream protein into the ER is considered as a “slipstreamed” phenomenon, which means that when the upstream protein carrying a peptide signal interacts with the ribosome-translocon complex, the downstream protein also passes through the translocation pore and, then, appears in the ER lumen. The occurrence of “slipstream” translocation is due to the inhibition of the 2 A cleavage reaction—with the consequent formation of a fusion protein—by the carboxy-terminal region of the protein upstream of the 2 A when translocation into the ER occurs [73,82].
By contrast, Yan et al. [90] pointed out that the protein downstream of the 2 A requires a signal sequence for secretion or membrane-anchored expression in mammalian cells. Additionally, Roulston and co-authors [91] described a novel mechanism of dual protein localization due to a “2 A-like” N-terminal oligopeptide termed SpNLR2A (S. purpuratus NLR N-terminal 2 A-like sequences). If ‘ribosome skipping’ has not occurred, the nascent polypeptide is fused to the 2 A-like N-terminal signal sequence and targeted, as a consequence, to the exocytic pathway. In contrast, when ‘ribosome skipping’ has been successful, the downstream protein lacks the 2 A-like signal and is then localized into the cytoplasm. All the studies briefly sketched in this section provided a valuable guidance to achieve accurate protein localization when designing bi- or multi-cistronic expression vectors via 2 A peptides.
5. Applications
Multicistronic expression vectors incorporating 2 A peptide sequences have been used in a variety of applications, including gene therapy, plant biotechnology, genomic engineering, metabolic engineering, and synthetic biology.
In plant biotechnology, traditional methods for the introduction of multiple genes into cells demand sexual crossing to accumulate transgenes and successive cell transformations to integrate monocistronic expression cassettes into the genome. These techniques present several drawbacks such as time-consuming, unpredictable silencing, and unstable expression of multiple transgenes [21,29,86,92]. Thus, the “2 A polyprotein” system has become a powerful new tool for the co-expression and the controlled expression (i.e. no large variations) of several genes under a single promoter. To analyze the activity of 2 A-based biosynthetic constructs in plants, Halpin et al. [93] realized the bi-cistron CAT-F2A-GUS, where CAT stands for chloramphenicol acetyltransferase and GUT represents β-glucuronidase. This bicistronic system showed in wheatgerm lysates and transgenic tobacco plants an efficient dissociation of the two proteins that allowed a co-ordinated expression of both active CAT and GUS. This work spurred more labs to engineer plant expression vectors where multiple proteins were expressed via a polycistron, thus, co-translationally cleaved apart with high efficiency. In a similar attempt to coordinate the expression of two transgenes, Ha and co-workers [29] used both 2 A from FMDV and IRES from crucifer-infecting tobamovirus (CrTMV), separately, to link phytoene synthase (Psy)—from the herb Capsicum—and carotene desaturase (CrtI), from the bacterium Pantoea, to realize two similar bicistronic sequences (Psy-2A-CrtI and Psy-IRES-CrtI) for the synthesis of carotenoids in rice endosperm. This work not only highlighted the better performance of 2 A sequences with respect to IRESs but also paved the way for the improvement of crop breeding via a biotech approach. Hence, 2 A peptides acquired a practical significance as gene linkers for introducing novel product traits and modifying plant metabolism [81,82,94,95].
In gene therapy, monoclonal antibodies and antibody fragments (e.g. Fab fragments) are important therapeutic agents for the diagnosis and the treatment of cancer. In the early 2000s, Fang et al. [62] described a 2 A-mediated mAb delivery system where a 2 A peptide determined the equimolar expression of both heavy and light chain, thus allowing the continuous generation of full-length antibody at high levels. To further optimize 2 A-mediated mAb expression, a dual vector system (one plasmid expressing a dimer activator protein, the other the antibody) was engineered [78]. Upon induction with rapamycin, a promoter got activated and initiated the expression of the antibody heavy and light chains linked, on the DNA, by a 2 A peptide and a furin recognition site. Overall, this system guaranteed inducible, high-level expression of full-length mAbs in vivo. Chng et al. [96] compared the cleavage efficiencies of four different 2 A peptides (F2A, P2A, E2A, T2A) flanked by HC and LC. In their work, T2A preceded by a GSG linker (GT2A) exhibited the highest cleavage efficiency and mAb expression level. Beside antibody production in mammal cells [78,79,96], Mizote and colleagues [97] engineered a more complex expression cassette for the HC and LC genes (HC-GSG-furin signal-GSG-2A-LC) that exhibited high productivity of Fab fragments in Lepidoptera. Hence, this work demonstrated that bicistronic constructs using, in the appropriate order, GSG linkers, a furin cleavage site, and a 2 A peptide allow for the effective production of recombinant antibody molecules also in insect cells.
Due to its relatively small size and efficient co-expression of transgenes, self-cleaving 2 A peptites have also been successfully incorporated into many viral vectors like retroviral [10,98], lentivirus [80], and adeno-associated virus (AAV) vectors [99]. The 2 A-based multicistronic constructs have shown the ability to combine therapeutic agents and increase their effectiveness due to synergistic effects. Furthermore, the usage of multicistronic vectors in clinical trials appeared absolutely safe, which offers great opportunities for improving the treatments of human diseases [7,64].
In genome editing (GE) technology, three major custom-designed nuclease tool kits have been developed. They are based on zinc finger proteins (ZFPs), transcription activator-like effectors (TALEs), and CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated proteins) systems. They can all produce DNA double-strand breaks (DSBs) at specific sites with varying degrees of specificity and efficiency, thus enabling a broad range of genetic modifications in various organisms [[100], [101], [102]]. For instance, cells characterized by 2 A peptide-coupled co-expression of a fluorescent protein and a nuclease were analysed via fluorescence-activated cell sorting (FACS), to screen for cell populations with high nuclease expression levels [3,55,103]. Duda et al. [104] used this approach to isolate nuclease-positive cells, which led to increased genome editing rates in the presence of both ZFNs and the CRISPR-Cas9 system, with low off-target mutagenesis. Ding et al. [105] demonstrated that 2 A-coupled co-expression of a nuclease and a fluorescent protein worked well also for TALENs. Since they contain FokI nuclease, TALENs need to dimerize in order to carry out DNA cleavage [106]. As monomers, TALENS are usually expressed from different ORFs. Mariano et al. [107] developed a scaffold where two TALENs could be co-expressed from a single transcript at similar concentrations by using a 2 A peptide, which permitted, also, to achieve higher genome editing efficiencies. CRISPR-Cas technology and 2 A peptides were employed together to realize multicistronic constructs as components of complex, regulatory synthetic gene networks like genetic switches [108], logic gates [109], and dynamic pulse response circuit [110].
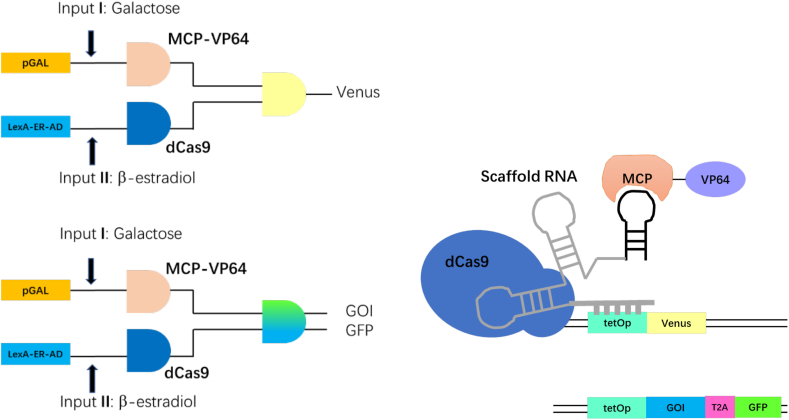
In metabolic engineering and synthetic biology, viral 2 A sequences in combination with other tools—such that in vivo assembly to generate large constructs—permitted the production of complex molecules, in heterologous eukaryotic chassis or the implementation of novel transcriptional regulation systems [[120], [121], [122], [123]] Enzymatic systems requiring several cofactors have been repurposed, by means of 2 A peptides, both in Schizosaccharomyces pombe and S. cerevisie. [[113], [114], [115]], The baker's yeast, moreover, is commonly treated as a cell factory to host heterologous pathways (such as that for β-carotene biosynthesis [60,112]) and as a chassis for synthetic networks that make use of the interactions between transcription factors derived from nuclease-deficient Cas protein and their antagonist anti-CRISPR macromolecules (see Fig. 4) [109,111]. Beekwilder and coworkers [112] introduced in S. cerevisiae a polycistronic vector that carried three β-carotene biosynthesis genes: crtI, crtE, and crtYB, to engineer a carotenoid pathway. The three genes were separated by two T2As, which displayed a well detectable protein release and hardly any wrong fusion proteins. Interestingly, they modified the two T2A peptide sequences and made them differ for 18 out of the 60 encoding nucleotides to avoid loss of productivity due to homologous recombination of T2A themselves. To explore carotenoid metabolic pathway further, Jiao and colleagues [60] considered the effects of the gene order in a polycistronic 2 A construct. Hence, they reconstituted the carotenoid production system by using three codon-optimized carotenogenic genes (GGPPS, CARB, and CARRP) linked by P2A sequences. The highest β-carotene yield was achieved with CARB in the first position within the polycistronic construct regardless of the location of GGPPS, CARRP downstream. Hence, gene order alteration in a polycistronic construct may contribute to optimize heterologous expression pathways, thus increase the yield of nature products.
Fig. 4.
The AND gate based on dCas9-mediated transcription activation. There are two different reporter systems: single reporter system using a yellow fluorophore (Venus) as reporter and double reporter system containing a combination of GOI (gene of interest) and GFP via T2A peptide. The pGAL and LexA-ER (estrogen receptor)-AD (activation domain) were activated by inducers such as galactose and β-estradiol, respectively, and led to the expression of MCP-VP64 and dCas9. Only in the presence of both input molecules reporter (and GOI) gene expression takes place [109].
To sum up, the potential of 2 A-mediated co-expression systems is clearly illustrated by all the applications mentioned above. To the best of our knowledge, the work by Geier et al. [116], where nine genes were successfully expressed from a single polycistronic transcript via T2A peptides in the yeast Pichia pastoris, represents, at the moment, the construct with the highest number of genes expressed in a coordinated fashion. As we mentioned in the Introduction, the usage of 2 A sequences could reduce, drastically, the complexity of circuit schemes with respect to the multiple-promoter architecture. Nevertheless, as we have described in this work, gene expression via 2 A sequences (or similar tools such as IRESs) is difficult to control. In contrast, many synthetic promoters of different, well-characterized strength, were developed over the last four decades, at least. They permit to tune, rather precisely, the amounts of proteins in a circuit through various expedients such as: assembling multiple UASs (upstream activating sequences) with different core sequences [124]; nucleosome removal [125]; insertion of intron sequences along the 5′UTR [126]; mutations in the TATA box sequence; and varying the distance between the same TATA box and either the TSS (transcription start site) or the UASs [127]. Moreover, longer polycistronic sequences might be obtained in the future by combining 2 A peptides with bidirectional synthetic promoters [116,117].
6. Conclusions
Metabolic engineering often demands the introduction of multiple genes into a heterologous host. Differently from bacteria, eukaryotes are not equipped with polycistrons. Therefore, the reconstruction of a metabolic pathway in eukaryotic cells requires the assembly and insertion of multiple transcription units, each of them delimited by a promoter and a terminator. Self-cleaving 2 A peptides appear to be the best genetic tool to reduce the number of transcription units in pathways construction since they allow the building of polycistronic sequences for gene co-expression in eukaryotes. In this way, potentially, an entire pathway or a complete synthetic gene circuit could be placed into a single transcription unit, with a significant reduction in the number of promoters and terminators involved in the synthetic network.
Although, so far, the use of 2 A-based multicistronic vectors was limited since a full understanding and characterization of the working of 2 A peptides in different organisms is missing, we have seen, in recent years, a renewed interest in this particular DNA sequences. We believe that future works will be mainly focused on the development of new approaches to regulate the cleavage efficiency in 2 A-based constructs and expand their usage in Synthetic Biology using different kinds of eukaryotic cells as chassis.
Author contribution
XW Writing original draft MAM Supervision; Writing review and editing.
Declaration of competing interest
The authors Xuekun Wang and Mario Andrea Marchisio declare no conflict of interest.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xuekun Wang, Email: wxk_11@tju.edu.cn.
Mario Andrea Marchisio, Email: mario@tju.edu.cn, mamarchisio@yahoo.com.
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438(7067):449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Marchisio M.A., Colaiacovo M., Whitehead E., Stelling J. Modular, rule-based modeling for the design of eukaryotic synthetic gene circuits. BMC Syst Biol. 2013;7(1):42. doi: 10.1186/1752-0509-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luke G.A., Ryan M.D. Using the 2A protein coexpression system: multicistronic 2A vectors expressing gene(s) of interest and reporter proteins. Methods Mol Biol. 2018;1755:31–48. doi: 10.1007/978-1-4939-7724-6_3. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Q., Zhang X., Yang H., Xie J., Xie Y., Chen J., Yu C., Zhong C. Internal ribosome entry site dramatically reduces transgene expression in hematopoietic cells in a position-dependent manner. Viruses. 2019;11(10):920. doi: 10.3390/v11100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J.H., Lee S.-R., Li L.-H., Park H.-J., Park J.-H. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One. 2011;6(4) doi: 10.1371/journal.pone.0018556. https://doi:10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels R.W., Rossano A.J., Macleod G.T., Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PloS One. 2014;9(6) doi: 10.1371/journal.pone.0100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaimardanova Alisa A., Chulpanova Daria S., Kitaeva Kristina V. Production and application of multicistronic constructs for various human disease therapies. Pharmaceutics 2019. 2019;11(11):580. doi: 10.3390/pharmaceutics11110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Curr Opin Biotechnol. 1999;10(5):458–464. doi: 10.1016/s0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 9.Minskaia E., Ryan M.D. Protein coexpression using FMDV 2A: effect of "linker" residues. BioMed Res Int. 2013 doi: 10.1155/2013/291730. 2013, 291730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., Vignali D.A. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 11.Szymczak A.L., Vignali D.A. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expet Opin Biol Ther. 2005;5(5):627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y., Feuer G., Day S.L., Wrzesinski S., Planelles V. Multigene lentiviral vectors based on differential splicing and translational control. Mol Ther: the journal of the American Society of Gene Therapy. 2001;4(4):375–382. doi: 10.1006/mthe.2001.0469. [DOI] [PubMed] [Google Scholar]
- 13.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 14.de Felipe P. Polycistronic viral vectors. Curr Gene Ther. 2002;2(3):355–378. doi: 10.2174/1566523023347742. [DOI] [PubMed] [Google Scholar]
- 15.Gäken J., Jiang J., Daniel K., van Berkel E., Hughes C., Kuiper M., Darling D., Tavassoli M., Galea-Lauri J., Ford K., Kemeny M., Russell S., Farzaneh F. Fusagene vectors: a novel strategy for the expression of multiple genes from a single cistron. Gene Ther. 2000;7(23):1979–1985. doi: 10.1038/sj.gt.3301341. [DOI] [PubMed] [Google Scholar]
- 16.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 17.Jang S.K., Kräusslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62(8):2636–2643. doi: 10.1128/JVI.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts L., Wieden H.J. Viruses, IRESs, and a universal translation initiation mechanism. Biotechnol Genet Eng Rev. 2018;34(1):60–75. doi: 10.1080/02648725.2018.1471567. [DOI] [PubMed] [Google Scholar]
- 19.Baird S.D., Turcotte M., Korneluk R.G., Holcik M. Searching for IRES. RNA. 2006;12(10):1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebauer F., Hentze M.W. Genetics. IRES unplugged. Science. 2016;351(6270) doi: 10.1126/science.aad8540. 228–228. [DOI] [PubMed] [Google Scholar]
- 21.Francois I.E.J.A., Broekaert W.F., Cammue B.P.A. Different approaches for multi-transgene-stacking in plants. Plant Sci. 2002;163:281–295. doi: 10.1016/S0168-9452(02)00130-9. [DOI] [Google Scholar]
- 22.Douin V., Bornes S., Creancier L., Rochaix P., Favre G., Prats A.C., Couderc B. Use and comparison of different internal ribosomal entry sites (IRES) in tricistronic retroviral vectors. BMC Biotechnol. 2004;4:16. doi: 10.1186/1472-6750-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mountford P.S., Smith A.G. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet: TIG (Trends Genet) 1995;11(5):179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borman A.M., Le Mercier P., Girard M., Kean K.M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25(5):925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennecke M., Kwissa M., Metzger K., Oumard A., Kröger A., Schirmbeck R., Reimann J., Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29(16):3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouabe H., Fässler R., Heesemann J. Improvement of reporter activity by IRES-mediated polycistronic reporter system. Nucleic Acids Res. 2008;36(5):e28. doi: 10.1093/nar/gkm1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W., Edelman G.M., Mauro V.P. Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98(4):1531–1536. doi: 10.1073/pnas.98.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenfeld A.B., Racaniello V.R. Hepatitis C virus internal ribosome entry site-dependent translation in Saccharomyces cerevisiae is independent of polypyrimidine tract-binding protein, poly(rC)-binding protein 2, and La protein. J Virol. 2005;79(16):10126–10137. doi: 10.1128/JVI.79.16.10126-10137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha Sun-Hwa, Liang Ying Shi, Jung Harin, etal Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant biological Journal. 2010;8(8):928–938. doi: 10.1111/j.1467-7652.2010.00543.x. October 2010. [DOI] [PubMed] [Google Scholar]
- 30.Johannes G., Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4(12):1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein I., Itin A., Einat P., Skaliter R., Grossman Z., Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18(6):3112–3119. doi: 10.1128/MCB.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs A.B. Cell cycle-dependent translation initiation: IRES elements prevail. Cell. 2000;101(3):243–245. doi: 10.1016/s0092-8674(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 33.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Gene Dev. 2001;15(13):1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 34.Chappell S.A., Edelman G.M., Mauro V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. U.S.A. 2000;97(4):1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W., Edelman G.M., Mauro V.P. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae. Proc Natl Acad Sci Unit States Am. 2003;100(8):4457–4462. doi: 10.1073/pnas.0437993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan H.Y., Sivakamasundari V., Xing X., Kraus P., Yap S.P., Ng P. Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PloS One. 2011;6(12) doi: 10.1371/journal.pone.0028885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlatter S., Stansfield S.H., Dinnis D.M., Racher A.J., Birch J.R., James D.C. On the optimal ratio of heavy to light chain genes for efficient recombinant antibody production by CHO cells. Biotechnol Prog. 2005;21(1):122–133. doi: 10.1021/bp049780w. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Zhang C., Jostock T., Dübel S. Analysis of IgG heavy chain to light chain ratio with mutant Encephalomyocarditis virus internal ribosome entry site. Protein Eng Des Sel: PEDS. 2007;20(10):491–496. doi: 10.1093/protein/gzm038. [DOI] [PubMed] [Google Scholar]
- 39.Ho S.C., Bardor M., Li B., Lee J.J., Song Z., Tong Y.W., Goh L.T., Yang Y. Comparison of internal ribosome entry site (IRES) and Furin-2A (F2A) for monoclonal antibody expression level and quality in CHO cells. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0063247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho S.C., Bardor M., Feng H., Mariati Tong Y.W., Song Z., Yap M.G., Yang Y. IRES-mediated Tricistronic vectors for enhancing generation of high monoclonal antibody expressing CHO cell lines. J Biotechnol. 2012;157(1):130–139. doi: 10.1016/j.jbiotec.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Yeo J., Mariati, Yang Y. An IRES-mediated tricistronic vector for efficient generation of stable, high-level monoclonal antibody producing CHO DG44 cell lines. Methods Mol Biol. 2018:335–349. doi: 10.1007/978-1-4939-8648-4_17. 1827. [DOI] [PubMed] [Google Scholar]
- 42.Fussenegger M. The impact of mammalian gene regulation concepts on functional genomic research, metabolic engineering, and advanced gene therapies. Biotechnol Prog. 2001;17(1):1–51. doi: 10.1021/bp000129c. [DOI] [PubMed] [Google Scholar]
- 43.Ryan M.D., King A.M., Thomas G.P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol. 1991;72(Pt 11):2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 44.Donnelly M.L. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 45.Donnelly M., Hughes L.E., Luke G., Mendoza H., Ten Dam E., Gani D., Ryan M.D. The 'cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring '2A-like' sequences. J Gen Virol. 2001;82(Pt 5):1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 46.Luke G.A., de Felipe P., Lukashev A., Kallioinen S.E., Bruno E.A., Ryan M.D. Occurrence, function and evolutionary origins of '2A-like' sequences in virus genomes. J Gen Virol. 2008;89(Pt 4):1036–1042. doi: 10.1099/vir.0.83428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan M.D., Donnelly M.L., Lewis A. A model for nonstoichiometric, Co-translational protein scission in eukaryotic ribosomes. Bioorg Chem. February 1999;27:pp55–79. ISSN:0045-2068. [Google Scholar]
- 48.Atkins J.F. Rna13, 803–810. 2007. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go)http://www.rnajournal.org/cgi/doi/10.1261/rna.487907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doronina V.A., de Felipe P., Wu C., Sharma P., Sachs M.S., Ryan M.D., Brown J.D. Dissection of a co-translational nascent chain separation event. Biochem Soc Trans. 2008;36(Pt 4):712–716. doi: 10.1042/BST0360712. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y., Wang F., Wang R., Zhao P., Xia Q. 2A self-cleaving peptide-based multi-gene expression system in the silkworm Bombyx mori. Sci Rep. 2015;5:16273. doi: 10.1038/srep16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koh H.G., Kang N.K., Kim E.K., Jeon S., Shin S.E., Lee B., Chang Y.K. Advanced multigene expression system for Nannochloropsis salina using 2A self-cleaving peptides. J Biotechnol. 2018;278:39–47. doi: 10.1016/j.jbiotec.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 52.De Felipe P. Skipping the co-expression problem: the new 2A "CHYSEL" technology. Genet Vaccine Ther. 2004;2:13. doi: 10.1186/1479-0556-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szymczak-Workman A.L., Vignali K.M., Vignali D.A. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb Protoc. 2012:199–204. doi: 10.1101/pdb.ip067876. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z., Chen O., Wall J., Zheng M., Zhou Y., Wang L., Vaseghi H.R., Qian L., Liu J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep. 2017;7(1):2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luke G.A., Ryan M.D. Therapeutic applications of the 'NPGP' family of viral 2As. Rev Med Virol. 2018;28(6) doi: 10.1002/rmv.2001. [DOI] [PubMed] [Google Scholar]
- 56.Cruz-Teran C.A., Tiruthani K., Mischler A., Rao B.M. Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae. ACS Synth Biol. 2017;6(11):2096–2107. doi: 10.1021/acssynbio.7b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szymczak-Workman A.L., Vignali K.M., Vignali D.A. Generation of 2A-linked multicistronic cassettes by recombinant PCR. Cold Spring Harb Protoc. 2012;(2):251–254. doi: 10.1101/pdb.prot067884. 2012. [DOI] [PubMed] [Google Scholar]
- 58.Szymczak-Workman A.L., Vignali K.M., Vignali D.A. Verification of 2A peptide cleavage. Cold Spring Harb Protoc. 2012;(2):255–257. doi: 10.1101/pdb.prot067892. 2012. [DOI] [PubMed] [Google Scholar]
- 59.Yang X., Zeng Q., Wang M., Cheng A., Pan K., Zhu D., Liu M., Jia R., Yang Q., Wu Y., Chen S., Zhao X., Zhang S., Liu Y., Yu Y., Zhang L. DHAV-1 2A1 peptide - a newly discovered Co-expression tool that mediates the ribosomal "skipping" function. Front Microbiol. 2018;9:2727. doi: 10.3389/fmicb.2018.02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang Jiao, Sun Wenyi, Zhang Yue. Exchanging the order of carotenogenic genes linked by porcine teschovirus-1 2A peptide enable to optimize carotenoid metabolic pathway in Saccharomyces cerevisiae†. RSC Adv. 2018;8:34967–34972. doi: 10.1039/C8RA06510A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwirz J., Yan Y., Franta Z., Schetelig M.F. Bicistronic expression and differential localization of proteins in insect cells and Drosophila suzukii using picornaviral 2A peptides. Insect Biochem Mol Biol. 2020;119:103324. doi: 10.1016/j.ibmb.2020.103324. [DOI] [PubMed] [Google Scholar]
- 62.Fang J., Qian J., Yi S. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- 63.Arnaud Ahier, Jarriault Sophie. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. GENETICS March. 2014;196 doi: 10.1534/genetics.113.160846. 3 605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luke G., Escuin H., De Felipe P., Ryan M. 2A to the fore - research, technology and applications. Biotechnol Genet Eng Rev. 2010;26:223–260. doi: 10.5661/bger-26-223. [DOI] [PubMed] [Google Scholar]
- 65.Li Y.M., Wang M., Wang T.Y., Wei Y.G., Guo X., Mi C.L., Zhao C.P., Cao X.X., Dou Y.Y. Effects of different 2A peptides on transgene expression mediated by tricistronic vectors in transfected CHO cells. Mol Biol Rep. 2020;47(1):469–475. doi: 10.1007/s11033-019-05153-3. [DOI] [PubMed] [Google Scholar]
- 66.Goh J., Ianiri G., Heitman J., Dawson T.L., Jr. Expression of a malassezia codon optimized mCherry fluorescent protein in a bicistronic vector. Frontiers in cellular and infection microbiology. 2020;10:367. doi: 10.3389/fcimb.2020.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S., AlAbed D., Whitteck J.T., Chen W., Bennett S., Asberry A., Wang X., DeSloover D., Rangasamy M., Wright T.R., Gupta M. A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn. Plant Mol Biol. 2015;87(4–5):341–353. doi: 10.1007/s11103-015-0281-6. [DOI] [PubMed] [Google Scholar]
- 68.Souza-Moreira T.M., Navarrete C., Chen X., Zanelli C.F., Valentini S.R., Furlan M., Nielsen J., Krivoruchko A. Screening of 2A peptides for polycistronic gene expression in yeast. FEMS Yeast Res. 2018;18(5) doi: 10.1093/femsyr/foy036. 10.1093/femsyr/foy036. [DOI] [PubMed] [Google Scholar]
- 69.Doronina V.A., Wu C., de Felipe P., Sachs M.S., Ryan M.D., Brown J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol. 2008;28(13):4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kjær J., Belsham G.J. Modifications to the foot-and-mouth disease virus 2A peptide: influence on polyprotein processing and virus replication. J Virol. 2018;92(8) doi: 10.1128/JVI.02218-17. e02218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kjær J., Belsham G.J. Selection of functional 2A sequences within foot-and-mouth disease virus; requirements for the NPGP motif with a distinct codon bias. RNA. 2018;24(1):12–17. doi: 10.1261/rna.063339.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minskaia E., Nicholson J., Ryan M.D. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 2013;13:67. doi: 10.1186/1472-6750-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Felipe P., Luke G.A., Brown J.D., Ryan M.D. Inhibition of 2A-mediated 'cleavage' of certain artificial polyproteins bearing N-terminal signal sequences. Biotechnol J. 2010;5(2):213–223. doi: 10.1002/biot.200900134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang S., Cohen C.J., Peng P.D., Zhao Y., Cassard L., Yu Z., Zheng Z., Jones S., Restifo N.P., Rosenberg S.A., Morgan R.A. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 2008;15(21):1411–1423. doi: 10.1038/gt.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan Y., Liang H., Chen A., Guo X. Coexpression of double or triple copies of the rabies virus glycoprotein gene using a 'self-cleaving' 2A peptide-based replication-defective human adenovirus serotype 5 vector. Biologicals : J Int Assoc Buddhist Stud. 2010;38(5):586–593. doi: 10.1016/j.biologicals.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y., Wang F., Xu S., Wang R., Chen W., Hou K., Tian C., Wang F., Zhao P., Xia Q. Optimization of a 2A self-cleaving peptide-based multigene expression system for efficient expression of upstream and downstream genes in silkworm. Mol Genet Genom: Mol Gen Genet. 2019;294(4):849–859. doi: 10.1007/s00438-019-01534-2. [DOI] [PubMed] [Google Scholar]
- 77.Lengler J., Holzmüller H., Salmons B., Günzburg W.H., Renner M. FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1-reporter fusion protein. Anal Biochem. 2005;343(1):116–124. doi: 10.1016/j.ab.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Fang J., Yi S., Simmons A. An antibody delivery system for regulated expression of therapeutic levels of monoclonal antibodies in vivo. Mol Ther. June 2007;15(Issue 6):1153–1159. doi: 10.1038/sj.mt.6300142. [DOI] [PubMed] [Google Scholar]
- 79.Ebadat S., Ahmadi S., Ahmadi M., Nematpour F., Barkhordari F., Mahdian R., Davami F., Mahboudi F. Evaluating the efficiency of CHEF and CMV promoter with IRES and Furin/2A linker sequences for monoclonal antibody expression in CHO cells. PloS One. 2017;12(10) doi: 10.1371/journal.pone.0185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chinnasamy D., Milsom M.D., Shaffer J., Neuenfeldt J., Shaaban A.F., Margison G.P., Fairbairn L.J., Chinnasamy N. Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol J. 2006;3:14. doi: 10.1186/1743-422X-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun H., Lang Z., Zhu L., Huang D. Acquiring transgenic tobacco plants with insect resistance and glyphosate tolerance by fusion gene transformation. Plant Cell Rep. 2012;31(10):1877–1887. doi: 10.1007/s00299-012-1301-5. [DOI] [PubMed] [Google Scholar]
- 82.Sun H., Zhou N., Wang H., Huang D., Lang Z. Processing and targeting of proteins derived from polyprotein with 2A and LP4/2A as peptide linkers in a maize expression system. PloS One. 2017;12(3) doi: 10.1371/journal.pone.0174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B., Rapolu M., Kumar S., Gupta M., Liang Z., Han Z., Williams P., Su W.W. Coordinated protein co-expression in plants by harnessing the synergy between an intein and a viral 2A peptide. Plant biotechnology journal. 2017;15(6):718–728. doi: 10.1111/pbi.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Felipe P., Hughes L.E., Ryan M.D., Brown J.D. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J Biol Chem. 2003;278(13):11441–11448. doi: 10.1074/jbc.M211644200. [DOI] [PubMed] [Google Scholar]
- 85.El Amrani A., Barakate A., Askari B.M., Li X., Roberts A.G., Ryan M.D., Halpin C. Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol. 2004;135(1):16–24. doi: 10.1104/pp.103.032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luke G. In: In innovations in biotechnology. Agbo E., editor. InTech; Rijeka, Croatia: 2012. Translating 2A research into practice. (161–186). [Google Scholar]
- 87.De Felipe P., Luke G.A., Hughes L.E., Gani D., Halpin C., Ryan M.D. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 88.Trichas G., Begbie J., Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Felipe P., Ryan M.D. Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic. 2004;5(8):616–626. doi: 10.1111/j.1398-9219.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 90.Yan J., Wang H., Xu Q., Jain N., Toxavidis V., Tigges J., Yang H., Yue G., Gao W. Signal sequence is still required in genes downstream of "autocleaving" 2A peptide for secretary or membrane-anchored expression. Anal Biochem. 2010;399(1):144–146. doi: 10.1016/j.ab.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 91.Roulston C., Luke G.A., de Felipe P., Ruan L., Cope J., Nicholson J., Sukhodub A., Tilsner J., Ryan M.D. '2A-Like' signal sequences mediating translational recoding: a novel form of dual protein targeting. Traffic. 2016;17(8):923–939. doi: 10.1111/tra.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Halpin C., Barakate A., Askari B.M., Abbott J.C., Ryan M.D. Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol. 2001;47(1–2):295–310. [PubMed] [Google Scholar]
- 93.Halpin C., Cooke S.E., Barakate A., El Amrani A., Ryan M.D. Self-processing 2A-polyproteins--a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J: for cell and molecular biology. 1999;17(4):453–459. doi: 10.1046/j.1365-313x.1999.00394.x. [DOI] [PubMed] [Google Scholar]
- 94.Park S., Kang K., Kim Y.S. Endosperm-specific expression of tyramine N-hydroxycinnamoyltransferase and tyrosine decarboxylase from a single self-processing polypeptide produces high levels of tyramine derivatives in rice seeds. Biotechnol Lett. 2009;31:911–915. doi: 10.1007/s10529-009-9951-2. [DOI] [PubMed] [Google Scholar]
- 95.Quilis J., López-García B., Meynard D., Guiderdoni E., San Segundo B. Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant biotechnology journal. 2014;12(3):367–377. doi: 10.1111/pbi.12143. [DOI] [PubMed] [Google Scholar]
- 96.Chng J., Wang T., Nian R., Lau A., Hoi K.M., Ho S.C., Gagnon P., Bi X., Yang Y. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. mAbs. 2015;7(2):403–412. doi: 10.1080/19420862.2015.1008351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mizote Y., Masumi-Koizumi K., Katsuda T., Yamaji H. Production of an antibody Fab fragment using 2A peptide in insect cells. J Biosci Bioeng. 2020;130(2):205–211. doi: 10.1016/j.jbiosc.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 98.Hofacre A., Yagiz K., Mendoza D., Lopez Espinoza F., Munday A.W., Burrascano C., Singer O., Gruber H.E., Jolly D.J., Lin A.H. Efficient therapeutic protein expression using retroviral replicating vector with 2A peptide in cancer models. Hum Gene Ther. 2018;29(4):437–451. doi: 10.1089/hum.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woodley E., Osmon K., Thompson P., Richmond C., Chen Z., Gray S.J., Walia J.S. Efficacy of a bicistronic vector for correction of sandhoff disease in a mouse model. Molecular therapy. Methods & clinical development. 2018;12:47–57. doi: 10.1016/j.omtm.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gupta R.M., Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124(10):4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ul Ain Q., Chung J.Y., Kim Y.H. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J Contr Release: official journal of the Controlled Release Society. 2015;205:120–127. doi: 10.1016/j.jconrel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 103.Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S., Wu W.Y., Scott D.A., Severinov K., van der Oost J., Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duda K., Lonowski L.A., Kofoed-Nielsen M., Ibarra A., Delay C.M., Kang Q., Yang Z., Pruett-Miller S.M., Bennett E.P., Wandall H.H., Davis G.D., Hansen S.H., Frödin M. High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 2014;42(10):e84. doi: 10.1093/nar/gku251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ding Q., Lee Y.K., Schaefer E.A., Peters D.T., Veres A., Kim K., Kuperwasser N., Motola D.L., Meissner T.B., Hendriks W.T., Trevisan M., Gupta R.M., Moisan A., Banks E., Friesen M., Schinzel R.T., Xia F., Tang A., Xia Y., Figueroa E., Cowan C.A. A TALEN genome-editing system for generating human stem cell-based disease models. Cell stem cell. 2013;12(2):238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mariano A., Xu L., Han R. Highly efficient genome editing via 2A-coupled co-expression of two TALEN monomers. BMC Res Notes. 2014;7:628. doi: 10.1186/1756-0500-7-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moore R., Spinhirne A., Lai M.J., Preisser S., Li Y., Kang T., Bleris L. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 2015;43(2):1297–1303. doi: 10.1093/nar/gku1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hofmann A., Falk J., Prangemeier T., Happel D., Köber A., Christmann A., Koeppl H., Kolmar H. A tightly regulated and adjustable CRISPR-dCas9 based AND gate in yeast. Nucleic Acids Res. 2019;47(1):509–520. doi: 10.1093/nar/gky1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakamura M., Srinivasan P., Chavez M., Carter M.A., Dominguez A.A., La Russa M., Lau M.B., Abbott T.R., Xu X., Zhao D., Gao Y., Kipniss N.H., Smolke C.D., Bondy-Denomy J., Qi L.S. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat Commun. 2019;10(1):194. doi: 10.1038/s41467-018-08158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu L., Marchisio M.A. Saccharomyces cerevisiae synthetic transcriptional networks harnessing dCas12a and type V-A anti-CRISPR proteins. ACS Synth Biol. 2021;10(4):870–883. doi: 10.1021/acssynbio.1c00006. [DOI] [PubMed] [Google Scholar]
- 112.Beekwilder J., van Rossum H.M., Koopman F., Sonntag F., Buchhaupt M., Schrader J., Hall R.D., Bosch D., Pronk J.T., van Maris A.J., Daran J.M. Polycistronic expression of a β-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to β-ionone production. J Biotechnol. 2014;192:383–392. doi: 10.1016/j.jbiotec.2013.12.016. Pt B. [DOI] [PubMed] [Google Scholar]
- 113.Efimova V.S., Isaeva L.V., Makeeva D.S., Rubtsov M.A., Novikova L.A. Expression of cholesterol hydroxylase/lyase system proteins in yeast S. cerevisiae cells as a self-processing polyprotein. Mol Biotechnol. 2017;59(9–10):394–406. doi: 10.1007/s12033-017-0028-5. [DOI] [PubMed] [Google Scholar]
- 114.Durairaj P., Fan L., Du W., Ahmad S., Mebrahtu D., Sharma S., Ashraf R.A., Liu J., Liu Q., Bureik M. Functional expression and activity screening of all human cytochrome P450 enzymes in fission yeast. FEBS Lett. 2019;593(12):1372–1380. doi: 10.1002/1873-3468.13441. [DOI] [PubMed] [Google Scholar]
- 115.Efimova V.S., Isaeva L.V., Orekhov P.S., Bozdaganyan M.E., Rubtsov M.A., Novikova L.A. Using a viral 2A peptide-based strategy to reconstruct the bovine P450scc steroidogenic system in S. cerevisiae: bovine P450scc system expression using 2A peptides. J Biotechnol. 2021;325:186–195. doi: 10.1016/j.jbiotec.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 116.Geier M., Fauland P., Vogl T., Glieder A. Compact multi-enzyme pathways in P. pastoris. Chem Commun. 2015;51(9):1643–1646. doi: 10.1039/c4cc08502g. [DOI] [PubMed] [Google Scholar]
- 117.Vogl T., Kickenweiz T., Pitzer J., Sturmberger L., Weninger A., Biggs B.W., Köhler E.M., Baumschlager A., Fischer J.E., Hyden P., Wagner M., Baumann M., Borth N., Geier M., Ajikumar P.K., Glieder A. Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat Commun. 2018;9(1):3589. doi: 10.1038/s41467-018-05915-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reiser J., Lai Z., Zhang X.Y., Brady R.O. Development of multigene and regulated lentivirus vectors. J Virol. 2000;74(22):10589–10599. doi: 10.1128/jvi.74.22.10589-10599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garg K., Green P. Differing patterns of selection in alternative and constitutive splice sites. Genome Res. 2007;17(7):1015–1022. doi: 10.1101/gr.6347907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Unkles S.E., Valiante V., Mattern D.J., Brakhage A.A. Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem Biol. 2014;21(4):502–508. doi: 10.1016/j.chembiol.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 121.Mol A.A., Groher F., Schreiber B., Rühmkorff C., Suess B. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module. Nucleic Acids Res. 2019;47(20):e132. doi: 10.1093/nar/gkz753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tian Y., Li Y., Zhao F., Meng C. Engineered Pichia pastoris production of fusaruside, a selective immunomodulator. BMC Biotechnol. 2019;19(1):37. doi: 10.1186/s12896-019-0532-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaczmarek M.B., Struszczyk-Swita K., Xiao M., Szczęsna-Antczak M., Antczak T., Gierszewska M., Steinbüchel A., Daroch M. Polycistronic expression system for Pichia pastoris composed of chitino- and chitosanolytic enzymes. Front. Bioeng. Biotechnol. 2021;9:710922. doi: 10.3389/fbioe.2021.710922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blazeck J., Liu L., Redden H., Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77(22):7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Curran K.A., Crook N.C., Karim A.S., Gupta A., Wagman A.M., Alper H.S. Design of synthetic yeast promoters via tuning of nucleosome architecture. Nat Commun. 2014;5:4002. doi: 10.1038/ncomms5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Myburgh M.W., Rose S.H., Viljoen-Bloom M. Evaluating and engineering Saccharomyces cerevisiae promoters for increased amylase expression and bioethanol production from raw starch. FEMS Yeast Res. 2020;20(6):foaa047. doi: 10.1093/femsyr/foaa047. [DOI] [PubMed] [Google Scholar]
- 127.Feng X., Marchisio M.A. Novel S. cerevisiae hybrid synthetic promoters based on foreign core promoter sequences. Int J Mol Sci. 2021;22(11):5704. doi: 10.3390/ijms22115704. [DOI] [PMC free article] [PubMed] [Google Scholar]