Abstract
DNA purified from clinical cerebrospinal fluid and urine specimens by a silica-guanidiniumthiocyanate procedure frequently contained an inhibitor(s) of DNA-processing enzymes which may have been introduced by the purification procedure itself. Inhibition could be relieved by the use of a novel lysis buffer containing alpha-casein. When the novel lysis buffer was used, alpha-casein was bound by the silica particles in the first step of the procedure and eluted together with DNA in the last step, after which it exerted its beneficial effects for DNA-processing enzymes. In the present study we have compared the novel lysis buffer with the previously described lysis buffer with respect to double-stranded DNA yield (which was nearly 100%) and the performance of DNA-processing enzymes.
It has been our main goal to develop reliable and sensitive PCR (19) assays for the detection of viral DNA in clinical specimens like blood, serum, cerebrospinal fluid (CSF), and urine. DNA from these specimens is routinely extracted by a silica-guanidiniumthiocyanate (GuSCN) procedure previously developed in our Clinical Virology Laboratory (5). The procedure has been acknowledged for its potency in the removal of inhibitory substances present in clinical specimens (6, 10, 11, 12, 13, 18, 20, 22, 23, 24) and is now widely used for the purification of nucleic acids from a variety of clinical specimens.
By our routine procedure for the detection of human cytomegalovirus (CMV) DNA, internal control (IC) DNA (a DNA sequence that mimics the CMV DNA sequence) is coextracted with CMV DNA from the clinical specimen. The extracted DNA is subjected to PCR, the amplimers are hybridized with probes specific for IC or CMV DNA, and the amounts of hybrids are measured by electrochemiluminescence (ECL) in the Perkin-Elmer (PE) QPCR System 5000 (5a). Due to the presence of IC DNA, the combined effects of DNA extraction efficiency and the presence of PCR inhibitors can be monitored, thus precluding false-negative reactions. Although no problems were encountered with clinical specimens like whole blood, serum, and plasma, low IC DNA signals were frequently obtained for CSF and urine. These low signals appeared to be due to the presence of inhibitors. The observation of Dreyer and Schulte-Holthausen (9) that casein enhances restriction enzyme activity prompted us to study the effects of alpha-casein.
Here we report that DNA purified from clinical specimens like CSF and urine frequently contained an inhibitor(s) of DNA-processing enzymes which was possibly introduced by the purification procedure itself. Inhibition could be relieved by the use of a novel lysis buffer containing alpha-casein. We have compared the previously described (5) lysis buffer (buffer L6) with the novel buffer containing alpha-casein (buffer L7A) with respect to double-stranded DNA yields and the performance of the DNA-processing enzymes.
MATERIALS AND METHODS
Restriction enzyme analysis and agarose gel electrophoresis.
Restriction enzyme (Boehringer Mannheim B.V., Almere, The Netherlands) digestions of 1 μg (48 kb) of phage lambda DNA (250 ng/μl; Boehringer Mannheim) were done for 1 h at 37°C in a 30-μl volume with 5 U of enzyme in the buffer systems provided by the manufacturer. DNA was electrophoresed through agarose gels (1) containing 1 μg of ethidium bromide per ml and was photographed under UV illumination. DNA was obtained from Boehringer (48-kb phage lambda DNA and HindIII-digested phage lambda DNA) and Gibco BRL, Breda, The Netherlands (100-bp DNA ladder).
Polyacrylamide gel electrophoresis (PAGE).
After reduction by 2-mercaptoethanol, proteins were electrophoresed through 15% polyacrylamide-sodium dodecyl sulfate gels as described by Laemmli (16) and were visualized by silver staining (4).
Preparation of buffer L7A.
Buffer L6 (5.25 M GuSCN, 50 mM Tris · HCl [pH 6.4], 20 mM EDTA, 1.3% [wt/vol] Triton X-100) was prepared as described previously (5). Buffer L7A was prepared from buffer L6 by the addition of alpha-casein to a final concentration of 1 mg/ml. Alpha-casein was obtained from Sigma Chemical Company (chromatographically purified to a purity of at least 85%; catalog no. C 6780; Sigma-Aldrich Chemie, Zwijndrecht, The Netherlands). Buffer L7A was stable for at least 2 months when it was stored at room temperature in the dark. Concentrated (50 times) stock solutions (50 mg of alpha-casein/ml of L6) were stored at −20°C and were stable for at least 1 year. Wash buffer L2 (5.25 M GuSCN, 50 mM Tris · HCl [pH 6.4]) was prepared as described previously (5).
DNA purification.
DNA was purified from 200 μl of CSF, serum, plasma, urine, and water as described previously (5) with 20 μl of size-fractionated silica particles by using either buffer L6 or buffer L7A. Elution was in 100 μl of TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]).
PCR.
Primers (purified by high-pressure liquid chromatography) were from PE (Nieuwerkerk a/d IJssel, The Netherlands). The primer pair used for amplification consisted of CMV-531 (5′-ACA AGG TGC TCA CGC ACA TTG ATC-3′; nucleotide [nt] positions 2034 to 2057) and Bio-CMV-1107 (5′-CAC TGG CTC AGA CTT GAC AGA CAC-3′; 5′ biotinylated; nt positions 2588 to 2611); the nt numbering was that of Akrigg et al. (2). The primer pair amplifies a 578-bp fragment from exon 4 of the major immediate-early gene of human CMV or a fragment of identical size and with the same GC content from internal control DNA (see below). The final reaction mixture (50 μl) contained 20 μl of eluate, 28 pmol of each primer, 2.5 U of Amplitaq DNA polymerase (PE), 0.5 U of Amperase (PE), 5 μg of bovine serum albumin (Boehringer), 10 mM Tris · HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, dATP, dGTP, and dCTP each at a concentration of 200 μM, and 400 μM dUTP (PE). PCRs were done in a PE 9600 thermocycler, as follows: 2 min at 50°C and 5 min at 95°C, followed by 35 cycles each consisting of 20 s at 95°C, 20 s at 63°C, and 1 min at 72°C; this was followed by 5 min at 72°C.
IC DNA.
A 597-bp amplimer was generated by PCR from CMV AD169 DNA (Advanced Biotechnologies Inc, Columbia, Md.) with primers CMV-517 (5′-GAT GAG GAG AGA GAC AAG GTG C-3′; nt positions 2021 to 2042) and CMV-1113 (5′-CTC AGA CAC TGG CTC AGA CTT G 3′; nt positions 2596 to 2617). The amplimer was digested with AocI, and a 227-bp fragment (nt positions 2021 to 2248) was purified from the gel. The amplimer was also digested with AlwI, and a 296-bp fragment (nt positions 2321 to 2617) was purified from the gel. A double-stranded DNA sequence was obtained from in vitro-synthesized single-stranded DNAs (5′ phosphorylated; PE) by hybridization of the complementary sequences 5′-TGA CCT TAT CAG TGT AAT GAA CCG CCG CAT TGA GGA GAT CTG CAC CCT TTA CAT CTT TCT GAA GTA GGG G-3′ and 5′-CCC CCT ACT TCA GAA AGA TGT AAA GGG TGC AGA TCT CCT CAA TGC GGC GGT TCA TTA CAC TGA TAA GG-3′. This double-stranded DNA sequence contained DNA overhangs and was ligated to the AocI site of the 227-bp fragment and to the AlwI site of the 296-bp fragment, generating a 597-bp fragment. This fragment was purified from the gel and amplified by PCR with primers CMV-517 and CMV-1113, and the amplimer was cloned into a plasmid vector (PCRII; 3,932 bp; Promega, Leiden, The Netherlands), resulting in a plasmid pCMV marker which served as the IC DNA. Relative to the CMV sequence (2), the AocI site (5′-CCTGAGG-3′; nt positions 2246 to 2252) has been replaced by the sequence 5′-CCTGACC-3′, the HhaI site (5′-GCGC-3′; nt positions 2269 to 2272) has been replaced by the sequence 5′-CCGC-3′, and the CMV sequence at nt positions 2292 to 2316 has been replaced by the sequence 5′-CCC TTT ACA TCT TTC TGA AGT AGG G-3′ to serve as a probe area. These modifications allowed discrimination between the CMV and IC amplimers resulting from the PCR described above by the restriction enzymes HhaI and AocI and by probes specific for either CMV DNA- or IC DNA-specific areas (see below). The pCMV marker was purified from bacterial cultures as described previously (14); the plasmid was linearized with HindIII and was used as input for PCR.
Hybridization and measurement by ECL.
The IC-specific probe (TBR-CMV-2; 5′-CCC TTT ACA TCT TTC TGA AGT AGG G-3′) contained a single TBR [Tris (2,2′-bipyridine) ruthenium (II) chelate] label at the 5′ end and was obtained from PE. Probes were diluted in 1× PCR II buffer to 1 ng/μl. To remove excess primers, 40 μl of PCR product was added to a mixture of 1 ml of buffer L6 and 20 μl of silica particle suspension (5); after 10 min the tube was centrifuged (at 12,000 × g for 1 min) and the supernatant was removed. The quasiempty tube was centrifuged again, the supernatant was removed, and the pellet was washed with 1 ml of acetone. After centrifugation the supernatant was removed and the pellets were dried (10 min at 56°C). DNA was eluted in 100 μl of 1× PCR II buffer (10 min at 56°C), followed by centrifugation. To 30 μl of the purified PCR product, 20 μl of probe was added and hybridization was done (2 min at 95°C, followed by 5 min at 56°C). Ten microliters of streptavidin-coated magnetic beads (PE) was added, and the mixture was incubated for 15 min at 56°C. Forty microliters of the bead-hybrid suspension was added to 400 μl of QPCR assay buffer (PE), and the ECL signal, expressed in luminosity units, was measured in the QPCR System 5000 (PE).
RESULTS
Inhibition of restriction enzyme activity by L6 eluate and relief of inhibition by L7A eluate.
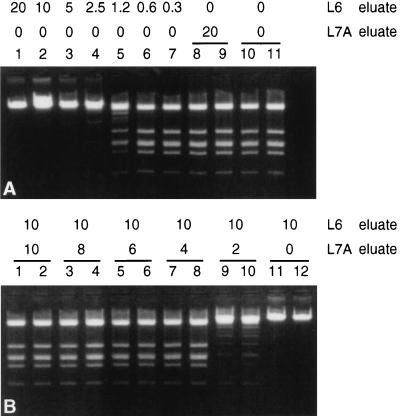
To monitor for inhibitors of restriction enzymes present in eluates, we performed extractions with different input materials. Subsequently, phage lambda DNA was added to the eluates and the activities of the restriction enzymes were determined. These experiments permitted us to make conclusions about the presence of inhibitors of restriction enzymes. Figure 1A (lanes 1 to 7) shows that an inhibitor of EcoRI was present in the eluate resulting from an extraction of pure water with L6 buffer (L6 eluate) since dilution of the eluate restored EcoRI activity. This experiment suggested that inhibitors were introduced by the DNA extraction procedure itself. Figure 1A (lanes 8 and 9) shows that inhibition of EcoRI was not observed when extractions were done with buffer L7A, which contains alpha-casein. Figure 1B (lanes 1 to 10) shows that the addition of the L7A eluate to the L6 eluate relieved the inhibition of EcoRI, suggesting that the inhibitor in the L6 eluate was neutralized by a factor present in the L7A eluate.
FIG. 1.
Phage lambda DNA was added to eluates resulting from water extractions, and DNA was digested with EcoRI in a constant 30-μl reaction volume (after adjustment with TE buffer). (A) Decreasing amounts of L6 eluate (20, 10, 5, 2.5, 1.25, 0.6, and 0.3 μl; lanes 1 to 7, respectively). Control digestions contained 20 μl of L7A eluate (lanes 8 and 9) and TE buffer only (lanes 10 and 11). (B) To a constant amount of L6 eluate (10 μl), decreasing amounts of L7A eluate were added. Lanes 1 and 2, 10 μl of L7A eluate; lanes 3 and 4, 8 μl of L7A eluate; lanes 5 and 6, 6 μl of L7A eluate; lanes 7 and 8, 4 μl of L7A eluate; lanes 9 and 10, 2 μl of L7A eluate, lanes 11 and 12, no L7A eluate.
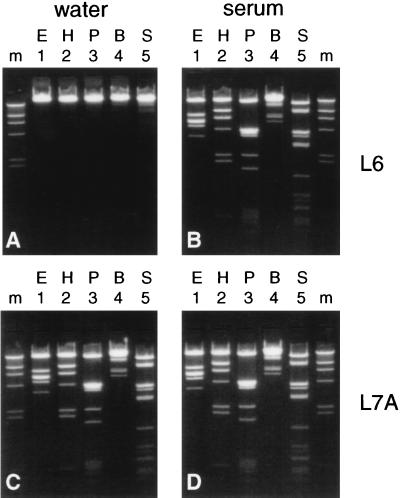
Inhibition of restriction enzymes is not observed after serum extraction, and the inhibition observed after water extraction is relieved by buffer L7A.
Figures 2A and C show that the beneficial effect of buffer L7A was not restricted to the activity of EcoRI but was also observed for the activities of the other restriction enzymes tested when the L6 eluates resulting from a water extraction were tested. Inhibition by the L6 eluates was not restricted to phage lambda DNA but was also observed for other DNA sources like plasmids purified by a standard (14) procedure (data not shown).
FIG. 2.
Extractions were done with buffer L6 (A and B) and buffer L7A (C and D) with either water (A and C) or serum (B and D) as the input for extraction. Phage lambda DNA was added to 20 μl of the eluates, and DNA was digested with restriction enzymes. Lanes 1, EcoRI (E); lanes 2, HindIII (H); lanes 3, PvuII (P); lanes 4, BamHI (B); lanes 5, SspI (S); lanes m, length marker (HindIII digest of phage lambda DNA [500 ng]).
A completely different situation was encountered when serum rather than water was used as the input for extraction since no inhibition of restriction enzymes was observed with either buffer (Fig. 2B and D). This phenomenon was not restricted to a particular serum specimen since 10 other serum specimens gave similar results (data not shown).
Inhibition of PCR is not observed after serum extraction, and the inhibition of PCR observed after water extraction is relieved by buffer L7A.
To monitor for inhibitors of Taq DNA polymerase (8), 50 molecules of IC DNA were added to the L6 and L7A eluates resulting from the water and serum extractions for the procedures whose results are presented in Fig. 2. PCRs were done, and the amount of PCR product was determined by hybridization followed by ECL. The PCR product amounts were inhibited to a level of 24% compared with the amounts from the uninhibited controls (TE buffer) for the L6 eluate resulting from the water extraction, whereas no inhibition was observed for the L7A eluate. On the other hand, when extraction was done with serum as input for extraction, no inhibition was found, regardless of the buffer used.
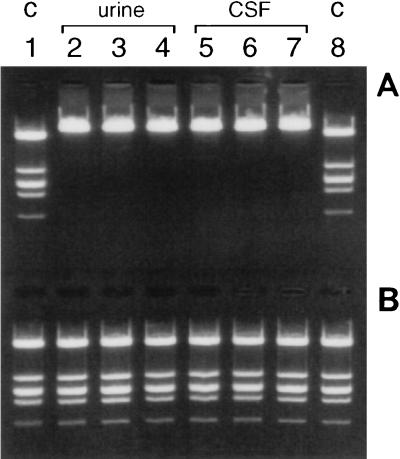
Inhibition of Taq DNA polymerase and restriction enzymes after extraction from urine and CSF is not observed with buffer L7A.
Although no significant inhibition of restriction enzymes and Taq DNA polymerase was found when the extraction was performed with serum or plasma with L6 as the buffer, other clinical materials like urine or CSF behaved like water; that is, L6 eluates inhibited restriction enzymes, whereas no inhibition was observed with L7A eluates. These results are shown for three CSF specimens and three urine specimens in Fig. 3. When the same eluates were tested for the presence of PCR inhibitors, a moderate increase (two- to threefold) in the amount of PCR product was observed when buffer L7A rather than buffer L6 was used. The amounts of PCR products obtained with the L7A eluates reached a mean level of 93% of the amounts reached in the uninhibited control reactions, suggesting that no significant inhibition of the PCR had occurred in L7A eluates derived from CSF and urine specimens. Additional experiments have shown that up to 15-fold increases in PCR products could be obtained for some CSF specimens when buffer L7A rather than buffer L6 is used.
FIG. 3.
Three different urine specimens and three different CSF specimens were used as input for extraction with buffer L6 or buffer L7A. Phage lambda DNA was added to 20 μl of the eluates, and DNA was digested with EcoRI. (A) Buffer L6; (B) buffer L7A. Lanes 1 and 8, control (C) digests in TE buffer; lanes 2 to 4, urine specimens; lanes 5 to 7, CSF specimens.
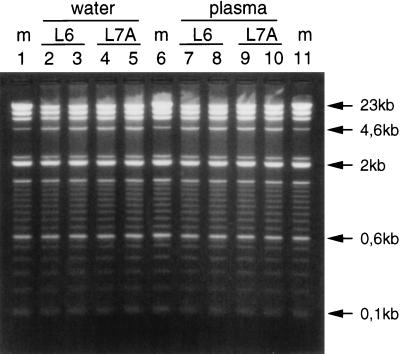
DNA extraction efficiency.
In the experiments described above, inhibition of DNA-processing enzymes was studied by adding pure DNA to the eluates resulting from extractions with buffer L6 or buffer L7A. Therefore, these experiments did not give information on the DNA yields obtained with buffer L7A. To establish the performance of buffer L7A with respect to the DNA yields with relatively high DNA inputs, plasma or water was mixed with double-stranded DNA, and DNA was extracted from these mixtures. Figure 4 shows that the yields were the same with either buffer in the range of 23 kb to 100 bp; visual comparison with the 100% recovery marker suggested that double-stranded DNA yields were nearly 100%.
FIG. 4.
HindIII-digested phage lambda DNA (4 μg) and a 100-bp DNA ladder (2 μg) were added to water or plasma, DNA was extracted from these mixtures with either buffer L6 or buffer L7A, and 25 μl of the eluates was electrophoresed through a 1.5% agarose gel. Lanes 1, 6, and 11, 100% recovery markers (m); lanes 2 to 5, extraction from water; lanes 2 and 3, extraction with buffer L6; lanes 4 and 5, extraction with buffer L7A; lanes 7 to 10, extraction from plasma; lanes 7 and 8, extraction with buffer L6; lanes 9 and 10, extraction with buffer L7A.
Alpha-casein is bound by silica particles and elutes in the last step of the procedure.
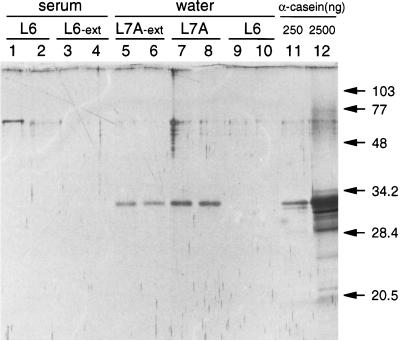
The data presented in Fig. 1 suggested that when buffer L7A was used, the eluate contained a factor which could relieve inhibition of the restriction enzymes. Since this factor most likely was alpha-casein (9), we did extractions with buffer L6 or L7A, with either water or serum as input, and studied the eluates for the presence of proteins by PAGE and silver staining. Figure 5 shows that a 32-kDa protein was isolated when buffer L7A was used for extraction (Fig. 5, lanes 7 and 8). The 32-kDa protein comigrated with the major band in the alpha-casein preparation (Fig. 5, lanes 11 and 12). The apparent molecular mass of the protein is in accordance with the published molecular mass (32 kDa) of alpha-casein (17). Since this result might also have been obtained when alpha-casein was bound to the walls of the reaction tubes, we performed additional extractions in which the silica particles were extensively washed (five washes with buffer L2) and were transferred to a new reaction tube after each of the wash steps. The data show that alpha-casein was still recovered (Fig. 5, lanes 5 and 6). In contrast, no proteins were observed in the L6 eluate when serum, which is very rich in proteins, was used as the input for extraction (Fig. 5, lanes 3 and 4), suggesting that protein binding is not a general property of the silica particles.
FIG. 5.
Extractions were done (in duplicate) with serum (lanes 1 to 4) or water (lanes 5 to 10) as input for extraction with buffer L6 (lanes 1 to 4, 9, and 10) or buffer L7A (lanes 5 to 8). In addition, in some extractions the silica particles were extensively (Ext) washed (lanes 3 to 6). Elution was with 60 μl of TE buffer; 10 μl was electrophoresed through a 15% polyacrylamide gel followed by silver staining. Marker lanes contained 250 ng (lane 11) and 2,500 ng (lane 12) of alpha-casein. Molecular masses (in kilodaltons) are indicated.
DISCUSSION
We have shown that when pure water was used as input material in the silica-GuSCN extraction procedure (5) with the original buffer, buffer L6, an inhibitor(s) of DNA-processing enzymes was present in the eluate. This inhibitor(s) was apparently introduced by the purification procedure itself. A similar inhibitory effect was observed when CSF and urine were used as input materials for extraction. Whereas inhibition was moderate for Taq DNA polymerase, strong inhibition was observed for many restriction enzymes. The nature of the inhibitor(s) is not known, but additional experiments have suggested that the inhibitor originated from the size-fractionated silica particle suspension and appeared to be tightly bound to the silica particles (4a). The inhibitor(s) remained bound throughout the purification procedure and was eluted from the silica particles in the last step of the procedure.
Inhibition could be relieved by using a novel lysis buffer, buffer L7A, which was prepared from original lysis buffer L6 by the addition of alpha-casein. Alpha-casein appeared to be bound to the silica particles, remained bound throughout the washing steps, and coeluted with DNA, exerting its beneficial effects in subsequent enzymatic reactions.
In contrast to CSF, urine, and water, no inhibition was found with the original buffer L6 when serum was used as input material for DNA extraction. One way to explain this difference might be that components of serum like albumin, which is a known scavenger of PCR inhibitors (15), prevented copurification of the inhibitor. Alternatively, alpha-casein-like proteins might be copurified from serum and relieve the inhibition.
Buffer L7A was also examined with respect to the DNA yields relative to the DNA yields achieved with the original buffer L6. When tested with inputs in the microgram range, double-stranded DNA was isolated at the same yields (nearly 100%) for either buffer. High yields were also observed with extremely low DNA inputs; thus, the absolute yields of IC DNA purified from plasma with buffer L7A were about 100% with an input of only 4 molecules per 200 μl of plasma (5a).
A modified lysis buffer may not only affect the DNA yield but may also affect lysis of the target organism in the first step of the procedure. At present we have no indication that there are gross differences in the lysing properties of buffers L6 and L7A for the viruses studied thus far by quantitative methods (human CMV, adenovirus, and human rotavirus).
In addition to DNA, alpha-casein was bound by silica particles in the presence of the chaotropic agent GuSCN. Protein binding was apparently not a general property of silica particles since no serum proteins were detected, and in addition, alpha-casein was selectively isolated from the mixture of proteins comprising the alpha-casein preparation. It may be speculated that the binding of alpha-casein to silica particles is mediated by phosphorylated serine residues, which are abundant in alpha-casein. Especially, stretches of phosphorylated serine residues (in the motif SerP-SerP-SerP-Glu-Glu) present in bovine alpha-casein (21) might mimic DNA and mediate binding by phosphate groups to silica particles. These phosphorylated regions give alpha-casein a high chelating power toward positively charged calcium, magnesium, iron, and copper ions (3, 7, 21) and might well be involved in the enhancing properties of alpha-casein described here by capturing metal ions which otherwise might act as inhibitors of DNA-processing enzymes. Alternatively, alpha-casein might induce conformational changes in the DNA or the DNA-processing enzyme, as speculated previously (9).
In summary, we have compared the performance of a novel lysis buffer (buffer L7A) containing alpha-casein with the performance of the lysis buffer (buffer L6) originally described in the silica-GuSCN DNA purification procedure (5). Alpha-casein was copurified with DNA by binding to silica particles and relieved the inhibition of the DNA-processing enzymes which was observed when DNA was extracted from urine, CSF, and water when the original buffer was used. No inhibition was observed for either buffer after purification of DNA from serum. Double-stranded DNA was isolated at a very high efficiency (about 100%). Together, the data suggest that for clinical specimens like CSF and urine, the casein-containing buffer will perform better than the original buffer.
ACKNOWLEDGMENTS
We thank F. P. de Vries for expert assistance with PAGE and silver staining; Ans van Strien, Fokla Zorgdrager, and John Dekker for providing PCR-grade materials; Monique de Boer, Yvette Gerrits, and Alex van Breda for technical assistance; the members of the Section of Clinical Virology for providing the clinical specimens; and Wim van Est for fine artwork.
REFERENCES
- 1.Aaij C, Borst P. The gel electrophoresis of DNA. Biochim Biophys Acta. 1972;269:192–200. doi: 10.1016/0005-2787(72)90426-1. [DOI] [PubMed] [Google Scholar]
- 2.Akrigg A, Wilkinson G W G, Oram J D. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985;2:107–121. doi: 10.1016/0168-1702(85)90242-4. [DOI] [PubMed] [Google Scholar]
- 3.Bernos E, Girardet J M, Humbert G, Linden G. Role of the O-phosphoserine clusters in the interaction of the bovine milk alpha s1-, beta-, kappa-caseins and the PP3 component with immobilized iron (III) ions. Biochim Biophys Acta. 1997;1337:149–159. doi: 10.1016/s0167-4838(96)00159-8. [DOI] [PubMed] [Google Scholar]
- 4.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–98. [Google Scholar]
- 4a.Boom, R. Unpublished data.
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Boom, R., et al. Submitted for publication.
- 6.Brisson-Noël A, Aznar C, Chureau C, Nguyen S, Pierre C, Bartoli M, Bonete R, Pialoux G, Gicquel B, Garrigue G. Diagnosis of tuberculosis by DNA amplification in clinical practice evaluation. Lancet. 1991;338:364–366. doi: 10.1016/0140-6736(91)90492-8. [DOI] [PubMed] [Google Scholar]
- 7.Chruscinska E, Dyba M, Micera G, Ambroziak W, Olczak J, Zabrocki J, Kozlowski H. Binding ability of Cu2+ ions by opiate-like fragments of bovine casein. J Inorg Biochem. 1997;66:19–22. doi: 10.1016/s0162-0134(96)00147-x. [DOI] [PubMed] [Google Scholar]
- 8.Cone R W, Hobson A C, Huang M W. Coamplified positive control detects inhibition of polymerase chain reactions. J Clin Microbiol. 1992;30:3185–3189. doi: 10.1128/jcm.30.12.3185-3189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyer K, Schulte-Holthausen H. Casein is a potent enhancer of restriction enzyme activity. Nucleic Acids Res. 1991;19:4295. doi: 10.1093/nar/19.15.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyer J R, Gilliam B L, Eron J J, Jr, Grosso L, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA™ with Amplicor™ reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 11.Fidler H M, Rook G A, Johnson N M, McFadden J. Mycobacterium tuberculosis DNA in tissue affected by sarcoidosis. Br Med J. 1993;306:546–549. doi: 10.1136/bmj.306.6877.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale A D, Green J, Brown D W G. Comparison of four RNA extraction methods for the detection of small round viruses in faecal specimens. J Virol Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- 13.Handt O, Höss M, Krings M, Pääbo S. Ancient DNA: methodological challenges. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 14.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreader C A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen L K, Hojrup P, Petersen T E. Disulphide arrangement in bovine caseins: localization of intrachain disulphide bridges in monomers of kappa- and alpha s2-casein from bovine milk. J Dairy Res. 1994;61:485–493. doi: 10.1017/s0022029900028417. [DOI] [PubMed] [Google Scholar]
- 18.Revello M G, Baldanti F, Sarasini A, Zavattoni M, Torsellini M, Gerna G. Prenatal diagnosis of rubella virus infection by direct detection and semiquantitation of viral RNA in clinical samples by reverse transcriptase-PCR. J Clin Microbiol. 1997;35:708–713. doi: 10.1128/jcm.35.3.708-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 20.Secchiero P, Carrigan D R, Asano Y, Benedetti L, Crowley R W, Komaroff A L, Gallo R C, Lusso P. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J Infect Dis. 1995;171:273–280. doi: 10.1093/infdis/171.2.273. [DOI] [PubMed] [Google Scholar]
- 21.Swaisgood H E. Chemistry of the caseins. In: Fox P F, editor. Advanced dairy chemistry. Vol. 1. London, United Kingdom: Elsevier Applied Science; 1992. pp. 63–110. [Google Scholar]
- 22.Vernazza P L, Dyer J R, Fiscus S A, Eron J J, Cohen M S. HIV-1 viral load in blood, semen and saliva. AIDS. 1997;11:1058–1059. [PubMed] [Google Scholar]
- 23.Wall S, Kunze Z M, Saboor S, Soufleri J, Seechurn P, Chiodini R, McFadden J J. Identification of spheroplast-like agents isolated from tissues of patients with Crohn’s disease and control tissues by polymerase chain reaction. J Clin Microbiol. 1993;31:1241–1245. doi: 10.1128/jcm.31.5.1241-1245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zipeto D, Baldanti F, Zella D, Furione M, Cavicchini A, Milanesi G, Gerna G. Quantification of human cytomegalovirus DNA in peripheral blood polymorphonuclear leukocytes of immunocompromised patients by the polymerase chain reaction. J Virol Methods. 1993;44:45–56. doi: 10.1016/0166-0934(93)90006-d. [DOI] [PubMed] [Google Scholar]