Abstract
Using magnetic resonance diffusion tensor imaging data from 45 patients with major depressive disorder (MDD) and 41 healthy controls (HCs), network indices based on a 246‐region Brainnetcome Atlas were investigated in the two groups, and in the MDD subgroups that were subgrouped based on their duration of the disease. Correlation between the network indices and the duration of illness was also examined. Differences were observed between the MDDS subgroup (short disease duration) and the HC group, but not between the MDD and HC groups. Compared with the HCs, the clustering coefficient (CC) values of MDDS were higher in precentral gyrus, and caudal lingual gyrus; the CC of MDDL subgroup (long disease duration) was higher in postcentral gyrus and dorsal granular insula in the right hemisphere. Network resilience analyses showed that the MDDS group was higher than the HC group, representing relatively more randomized networks in the diseased brains. The correlation analyses showed that the caudal lingual gyrus in the right hemisphere and the rostral lingual gyrus in the left hemisphere were particularly correlated with disease duration. The analyses showed that duration of the illness appears to have an impact on the networking patterns. Networking abnormalities in MDD patients could be blurred or hidden by the heterogeneity of the MDD clinical subgroups. Brain plasticity may introduce a recovery effect to the abnormal network patterns seen in patients with a relative short term of the illness, as the abnormalities may disappear in MDDL.
Keywords: brain networking pattern, diffusion tensor imaging, major depressive disorder
Using magnetic resonance diffusion tensor imaging data from 45 patients with major depressive disorder (MDD) and 41 healthy controls, network indices based on a 246‐region Brainnetcome Atlas were investigated in the two groups, and in the MDD subgroups that were subgrouped based on their duration of the disease. The analyses showed that duration of the illness appears to have an impact on the networking patterns. Networking abnormalities in MDD patients could be blurred or hidden by the heterogeneity of the MDD clinical subgroups. Brain plasticity may introduce a recovery effect to the abnormal network patterns seen in patients with a relative short term of the illness, as the abnormalities may disappear in MDD with a long duration of the disease.

1. INTRODUCTION
Despite a debate over the degree of distributed processing in the human brain, there are functionally specialized regions for different cognitive processes, in addition to highly localized motor and primary sensory brain regions (Caramazza & Coltheart, 2006). Nevertheless, individual brain areas may not be working independently. Instead, multiple brain regions may coordinate in a complex network (Rubinov & Sporns, 2010; Strogatz, 2001) or a so‐called small‐world network (Bassett & Bullmore, 2006; Bassett, Edward, & Bullmore, 2017). Complex network analysis seeks to characterize brain networks using a few indices that are not only neurobiologically meaningful, but also easily computable, based on quantitative data of brain connectivity such as diffusion tensor imaging (DTI), functional magnetic resonance imaging (fMRI), electroencephalograph (EEG) or magnetoencephalograph (MEG) data. A small‐world network, one type of such complex networks, is characterized with a topology such that any pair of nodes in the network may be connected with each other via connection to just a few other nodes, even when the two nodes are not in the immediate vicinity of each other. The small‐world topology provides an infrastructure for effectively processing information, both segregated/specialized and distributed/integrated. A complex brain network typically maintains a high dynamic complexity at minimized wiring costs (Schilling, 2005), thereby supporting functions such as cognitive insight (Schilling, 2005), self‐sustained persistent activity or failure transition (Roxin, Riecke, & Solla, 2004), decision‐making (Minat, Grisoli, Seth, & Critchley, 2012), and executive functions (Satterthwaite et al., 2013) in routine life. Functional networks are further divided into sublevel networks, each of which typically consists of brain nodes with synchronizing brain activities and generally focusing on a concrete brain function. Depending on the analysis that is employed, 7–11 prevailing subnetworks are generally identified and involved (Gordon et al., 2016; Power et al., 2011; Yeo et al., 2011). Somatomotor, visual, auditory, salience, default mode networks, and fronto‐parietal control systems are some of the commonly described functional networks in brain studies (Gordon et al., 2016; Power et al., 2011; Yeo et al., 2011). These functional networks can be identified by a functional clustering characteristic, that is, functional synchronizing rhythm. Brains of individuals with various psychiatric or neurological disorders (e.g., autism, Alzheimer's disease, mild cognitive impairment, schizophrenia, pain disorder, major depression and anorexia nervosa) typically exhibit abnormalities in terms of small‐world networking properties [Nir et al., 2012; Zhou & Lui, 2013; Zhou, Yu, & Duong, 2014], functional connectivity in the networks [Sun et al., 2020; Xia et al., 2019; Zhao et al., 2017; Zhao et al., 2018], and physical rewiring in the brain [Hu et al., 2017; Li, Weissman, Posner, & Xu, 2018]).
Major depressive disorder (MDD), schizophrenia, and bipolar disorder have been conceptualized as brain network disorders (Fornito, Zalesky, & Breakspear, 2015; Gong & He, 2015). MDD has topological alterations in functional brain modules (Hu et al., 2019; Zhang et al., 2011), as do other psychiatric diseases (Liu et al., 2008; Spielberg et al., 2016; Wang et al., 2019; Zhao et al., 2018; Zhu et al., 2016). In the investigations, graph theory has been widely used assisting analyses based on anatomical, structural, and functional imaging data (Chen et al., 2017; Korgaonkar, Fornito, Williams, & Grieve, 2014; Lu et al., 2017; Sacchet, Prasad, Foland‐Ross, Thompson, & Gotlib, 2015; Tymofiyeva et al., 2017; Wang, Yuan, You, & Zhang, 2019; Zhang et al., 2011). Topological organization of whole brain networks in MDD based on resting‐state fMRI data from 30 drug‐naïve, first‐episode MDD patients and 63 healthy control (HC) subjects (Zhang et al., 2011) identified lower path length and higher global efficiency, implying a shift toward randomization in brain networks, although both groups still showed small‐world architecture in brain functional networks. Another fMRI study found disrupted functional connectivity of right posterior insula in adolescents and young adults with MDD (Hu et al., 2019). Despite reported correspondence between functional and structural measures of brain connectivity (Goni et al., 2014; Honey et al., 2009; Mišić et al., 2016), data on functional/structural correspondence in MDD are relatively scant. There had been some work on relevant topics, but either based on a specific depressive population or using methods not exactly desired. For instance, based on data from patients of remitted late‐onset depression, a DTI study using tract‐based spatial statistics and small‐world indices (path length & network efficiency) found that disrupted structural connectivity contributed to cognitive deficits in these patients (Wang, Yuan, et al., 2019). Focused on adolescents (age 16.2 ± 1.3 years) with MDD, another DTI‐based connectome analysis revealed hypoconnectivity of the right caudate using graph indices (Tymofiyeva et al., 2017). Restricted to a patient population with very short disease durations of 1–10 months, white matter structural network topology in the brain at early stage of MDD was investigated (Lu et al., 2017). The findings from these studies thus applied only to their specific patient group or age segment lacking a general consensus. A structural MRI study based on T1‐weighted data from 33 patients versus 33 matched normal controls reported alterations in morphological cortical networks in MDD when assessing whole‐brain morphological networks, a picture indicating lower global efficiency and higher modularity, suggesting impaired integration and increased segregation of brain networks in the MDD patients (Chen et al., 2017). Nevertheless, the brain networks they evaluated were constructed based on reported intra‐cortical similarities in gray matter morphology using T1‐weighted imaging data, which were actually quantified by the covariation of gray matter volume and thickness in the brain, rather than on the commonly recognized known brain functions or functional networks. In short, the physical network wiring in MDD brains using DTI has not yet been studied adequately.
Although depression is actually quite heterogeneous in its manifestation from one episode to another (Lopez‐Castroman et al., 2012; Yu et al., 2019), the biological underpinnings of this heterogeneity are largely unknown. Age of onset and disease duration is often considered in biological studies, but symptom pattern may also be relevant (Matthew et al., 2010). On the other hand, the diagnostic assessment of depression could be a potential source of heterogeneity in the patient group although all of them are diagnosed as patients with MDD. A recent study showed that different rating scales have only a moderate mean overlap of 0.41(Fried, 2016; Fried, 2020). Similarly, test–retest reliability estimates for diagnosis of DSM‐5 MDD are suboptimal (Regier et al., 2013). The heterogeneity of depression, including disease duration, thus is worthy of detailed investigation.
Using DTI, a powerful imaging tool for directly examining structural connectivity, the present work aimed to study network patterns in MDD that may correspond with the small‐world properties discussed earlier. We investigated the DTI networking patterns in MDD and HCs, and the impact of subgroup heterogeneity with respect to disease duration. We hypothesized that MDD has altered brain‐networking patterns, and explored the possibility that the clinical heterogeneity of MDD may be related to networking pattern abnormalities. We expected that this study may help to resolve some of the mixed findings due to such heterogeneity of depression.
2. MATERIAL AND METHODS
The data in the present study were extracted from our brain‐imaging database collected over the past 4 years from a number of studies with different scientific objectives, as described in previous publications (Chhetry et al., 2016; Rizk et al., 2017). We identified raw DTI datasets of 112 human subjects, including 63 MDD patients and 49 HC. A screening and preprocessing procedure of the DTI data was performed to identify and exclude defective datasets that showed data missing, artifacts or incompatibility and those had a disturbing medication history before acquisition of the imaging data, although patients who had a single antidepressant were originally allowed at the time of recruitment.
2.1. Participants and data acquisition
All included study protocols followed the Declaration of Helsinki and were approved by the Institutional Review Board of New York State Psychiatric Institute (NYSPI). Participants were recruited locally at Columbia University Irving Medical Center and NYSPI in New York City and from surrounding areas. Detailed procedures were as follows: All subjects provided written informed consent to participate in the research study. At study entry, the depressed adults, ages 18–65 years, met DSM‐IV criteria (A.P. Association, 1994) for a current major depressive episode in the context of MDD, without a lifetime history of psychosis, or a history of drug or alcohol abuse within the past 2 months or drug or alcohol dependence (except nicotine) within the past 6 months, based on the Structured Clinical Interview for DSM‐IV (First, Williams, Spitzer, & Gibbon, 1997), physician obtained medical history, review of systems, physical examination, and laboratory tests. Female participants also underwent pregnancy testing. Patients were not actively suicidal, had not received electroconvulsive therapy within the past 6 months, and presented with scores ≥16 on the 17‐item Hamilton Depression Rating Scale (HDRS; Hamilton, 1960; Hamilton, 1967) at study entry. MDD participants were permitted to be on a single antidepressant or were medication‐free and had no history of antipsychotic medications or mood stabilizers within 6 weeks; no special washouts were performed. A team of experienced clinical research psychologists and psychiatrists reviewed the results of SCID interviews and medical records, and achieved diagnostic consensus. HC had no history of Axis I disorders or of Axis II Cluster B disorders. Participants in both groups did not have active medical illness. All participants were also assessed for acute severity of illness within 1 week of the scan. If necessary, patients were allowed to use short‐acting benzodiazepines up to 3 days immediately prior to scans for symptomatic relief, which enhances GABA‐induced inhibition (Hollister, Müller‐oerlinghausen, Rickels, & Shader, 1993; Wise, Berger, & Stein, 1972) but was considered unlikely to alter white matter function or structure.
Both anatomical T1‐weighted (T1) and DTI MRI images were acquired on a 3.0T Signa Advantage system (GE Healthcare, Waukesha, WI), as described elsewhere (Chhetry et al., 2016). Briefly, the setting of the MRI pulse sequence for acquiring the T1 images used the following imaging parameters: echo time (TE)/repetition time (TR) = 2.8 ms/7.1 ms, field of view (FOV) = 256 mm × 256 mm, matrix size =256 × 256, slice thickness = 1 mm (voxel size = 1 mm3 isotropic), at 178 slice locations. The diffusion data were acquired using a single‐shot EPI (echo planar imaging) sequence. Its imaging parameters were: TR/TE = 14,000 ms / 82 ms, flip angle = 900, slice thickness = 3 mm, number of excitation (NEX) = 1, FOV = 240 mm × 240 mm, matrix = 256 × 256 (voxel dimensions = 0.9375 mm × 0.9375 mm × 3.0 mm anisotropic); b‐value = 1,000 s/mm2. The diffusion datasets were scanned along 25 or 30 non‐collinear directions plus five baseline images without applying diffusion gradient, depending on project‐wise designs. The scanning time was approximately 11 min per data set.
2.2. Data processing
The steps of preprocessing were as follows: (a) We checked all the available DTI datasets both visually and quantitatively, and removed those with apparent artifact, head motion (>2 mm of translation or 1° of rotation), or poor image quality. (b) Eddy current correction was performed using the FMRIB Software Library (FSL 6; http://www.fmrib.ox.ac.uk/fsl) and our own method (Liu et al., 2012), including removing head motion and registering all volumes of diffusion‐weighted imaging (DWI) data to the baseline images. (c) Nonbrain tissues were removed (skull‐stripping) using BET in the FSL software kit. (d) Tensor estimation was performed using the DSI‐Studio Toolbox (http://dsi-studio.labsolver.org/), based on the data that were already corrected. As the datasets were acquired from the same scanner along 25 or 30 noncollinear gradient directions, with a small difference in the numbers but a large redundancy far more than the required six for tolerating possible errors, no extra calibration was needed. (e) Spatial normalization of each individual brain into the standard space of the MNI152 template was accomplished using DSI‐Studio. (f) Whole‐brain deterministic tractography was performed in the template space using the same parameter settings in DSI‐Studio as reported in (Baum et al., 2017). (g) The connectivity matrix was computed based on 246 brain regions as identified in the literature (see Section 2.4. Regions of Interest, below; Fan et al., 2016). (h) For networking and modularity analysis, measuring indices, including clustering coefficient (CC), global efficiency, modular resilience, characteristic path length, and small‐worldness were calculated using the GRETNA software package (Wang et al., 2015). Figure 1 is an overview of the general procedure of our analysis.
FIGURE 1.
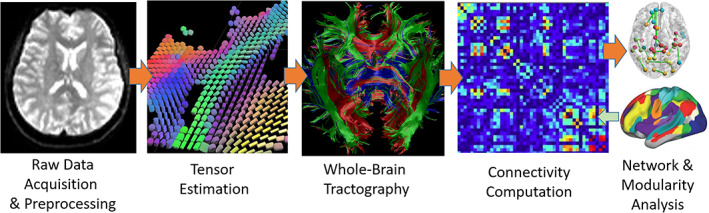
Overview of the general procedure of the analysis
2.3. Whole‐brain tractography
Using the DSI‐Studio software package, one million seeding points were cast for deterministic fiber tracking in the brain for construction of a whole‐brain fiber map. The angular threshold was 45° and the fractional anisotropy (FA) threshold was set to the default settings in DSI‐Studio. The step size of the tracking was 0.94 mm. The fiber trajectories were smoothed by averaging the propagation direction with 90% of the previous direction. Neural fiber tracts with length shorter than 10 mm or longer than 400 mm were discarded.
2.4. Regions of interest (ROI)
We adopted 246 brain regions (including 210 cortical and 36 subcortical regions) as ROIs from the human Brainnetome Atlas (http://atlas.brainnetome.org/), which is a freely available resource (Fan et al., 2016). This fine‐grained atlas has been cross validated by hundreds of independent neuroimaging studies, and it contains information on both anatomical and functional connections. It is deemed objective and stable for exploring the complex relationships between structure, connectivity, and function, and thus has been widely cited in the community.
2.5. Connectivity matrix
Based on the whole‐brain fiber tracking result, a connectivity matrix C was computed. Each element c ij (i,j = 1,…,246) in C was determined by the number k of fibers passing through both ROIs i and j divided by the total number m of fibers passing through i or j: c ij = k/m. Because c ij could differ from c ji due to the performance of the tracking algorithm on the complex neural fiber structure, the average of c ij and c ji was assigned to them so that the matrix C was forced symmetry.
2.6. Networking indices
The following indices were calculated: connectivity, CC, global efficiency, and modular resilience (Rubinov & Sporns, 2010). In addition, characteristic path length, and small‐worldness were also calculated, for comparison purposes, as these were previously examined in other studies on depressive patients (Korgaonkar et al., 2014; Lu et al., 2017; Sacchet et al., 2015; Tymofiyeva et al., 2017).
The degree of connectivity D of a node (ROI) i is denoted as D i, which is the number of nonzero elements c ij in the i‐th row in the connectivity matrix C. D i indicates how many ROIs other than itself are connected with the node i.
The CC of the entire network including all the nodes is defined as (Rubinov & Sporns, 2010; Watts & Strogatz, 1998):
| (1) |
where N is the set of all n ROIs, C i is the CC of ROI i, and t i is the number of triangles in the graph involving all the ROIs that ROI i forms with all the other nodes in N. In our case, the set N contained 246 nodes, that is, n = 246. Both CC and C i take a value range from 0 to 1. Here, C i approaching 1.0 would indicate that node i is a highly clustering hub in the network, whereas a value close to 0.0 would indicate that the node is highly segregated from other nodes. Consequently, a low value of CC means that in general the entire brain network is highly segregated.
The global efficiency E of the network is defined as
| (2) |
where E i is the efficiency of node i, and d ij is the shortest path (distance) between nodes i and j (Lo et al., 2015; Rubinov & Sporns, 2010). The global efficiency index E measures how effectively the brain regions are connected with each other in a brain network; the higher the value, the more densely connected. In depressed brains, increased efficiency may be a trace of a tendency toward a relatively more randomized network organization (Ajilore, Lamar, & Kumar, 2014).
Network resilience is another measure assessing the robustness of a network, by inspecting the impact of continuously losing one particular node in a network on the global efficiency of the entire network (Lo et al., 2015). In particular, all the c ij elements in the upper triangle of the connectivity matrix C will be first ranked in a descending order. At a given density level, denoted as a percentage number x%, that is, taking into consideration only the first x percentile of all the c ij's in the connectivity matrix C, resilience R is defined as the total area in the first quadrant in a two dimensional coordinate space directly under the curve of the global efficiency depending on the removal of the nodes, either in a descending order or a random order. If the global efficiency E of the network after removing one node s does not change much, node s must not be an important one, as the remaining nodes in the network may easily find alternative paths connecting with each other. In contrast, a significant reduction of E means that the removed node is a relatively important hub in the network. Removing nodes in a random order (random attack) basically regards all the nodes with equal importance and therefore would profile the baseline resilience of a brain network, while targeted attack (each time removing a node with the top rank following their descending order) would estimate the general vulnerability of the brain network. Because a targeted attack precisely aims at the most densely connected (least segregated) node in a network, the network resilience due to a targeted attack normally would be significantly lower than that of a random attack, as a network is certainly more vulnerable to a targeted attack. Moreover, in the scenario of a targeted attack, a higher resilience value of a network, which usually is closer to the baseline profile at random attack, indicates that the roles of all the nodes in the network are relatively more similar, as they are more similarly and more densely connected with each other, without anyone being particularly distinguished. In other words, higher CC, higher E, and higher R infer that the nodes in the network are generally less segregated and therefore the network is less vulnerable to each removal of the nodes. When a targeted attack occurs to a well‐segregated network (typically with a low CC value), each node removal would sharply lower the global efficiency of the remaining network and therefore its resilience is lower than that would be when the attack occurs to a less‐segregated network. In short, the resilience value of a well‐segregated network is typically low.
Characteristic path length L measures in general the average distance between different nodes in the network. L is one type of indices measuring the integration of a network, and it can be computed as follows:
| (3) |
where, L i is the average distance between node i to all other nodes in the set N of all the nodes, and d ij is the shortest path length between nodes i and j. A lower value of L indicates a relatively densely connected network (less segregated in our case), whereas a higher L value means a relatively sparsely connected network (highly segregated in our case).
Small‐worldness S of a network is defined as:
| (4) |
where, CC and L are the clustering coefficient and characteristic path length of the network, respectively; whereas and are the average values of the corresponding indices measured base on the same network's randomized version, respectively. Here, the corresponding randomized version of the same network refers to a network with the same topological configuration of nodes but the connections between the nodes are randomly assigned. The index S is usually computed based on the average CC r and L r of a few hundreds of repetitions of such a randomized network to achieve a non‐biased estimation. A network features small‐worldness typically has a value S > 1, when is much larger than 1.0 and is approximately 1.0 (Achard, Salvador, Whitcher, Suckling, & Bullmore, 2006; Bassett & Bullmore, 2006; Humphries, Gurney, & Prescott, 2006).
2.7. Group analysis
To explore the possible differences in connectivity in MDD compared with HC and the possible heterogeneity within the MDD patients, we performed two sets of analysis: (a) A general group analysis to compare all the patients with MDD versus HCs; (b) A subgroup analysis to determine whether there were heterogeneities that may have blurred the contrast between MDD patients and HCs, by comparing MDD patients with a relatively short duration of the disease (≤10 years; MDDS) versus those with a relatively long duration of the disease (>10 years; MDDL) versus HCs. The disease duration was defined as the time interval between the date of MRI data acquisition and the recorded age of the patient's first onset. Limited by the available number of patients to be subgrouped for studying possible heterogeneities with adequate statistical power in the patient cohort, the cutoff was determined based on the median years of disease duration adjusted for the counts of subjects and their sex and age in each subgroup suitable for the available MDD datasets. Two‐sample t‐tests were performed, covarying with nuisance variables (age, sex, HDRS scores). False discovery rate (FDR) correction was applied at p < .05 to control for false positives for all the tests involving all the 246 regions. In addition, we analyzed the correlation (Pearson) between the network indices and disease duration, to examine how the disease process could have affected the efficiency of brain regions and network and to identify brain areas that were the key contributors. Considering that the cutoff age of 10 years for subgrouping the patients into long and short disease durations may not exactly be an optimal choice although our samples only allowed using the 10‐year as a cut‐off age, we also performed multi‐regression analysis using disease duration as a continuous variable in the model, covariated with the nuisance variables, to double check the possible effect of disease duration on the patient cohort, thereby justifying the cutoff point of the two subgroups and the correlation analysis that was just mentioned. The model was:
| (5) |
which took the respective network indices as Y, plotting b 1, and DD was the continuous variable of disease duration.
3. RESULTS
There were no differences between the MDD and HC groups in terms of the proposed network indices. However, differences were observed between the three subgroups, that is, between the MDDS, MDDL, and HC groups.
3.1. Demographic and clinical characteristics
Eleven MDD patients and eight HCs were excluded due to imaging data deficiencies, artifacts, incompatibility of data format (pulse sequence parameters, including the schema of gradient direction, gradient strengths), or errors in spatial normalization. Another seven patients were also excluded due to their medication history, thus none of the included in this study had any antidepressant within 6 weeks prior to the MRI scanning and all were drug free for at least 3 weeks. A final sample of 45 MDD patients (22 males, 23 females) and 41 HC (15 males, 26 females) survived initial processing steps from (a) to (d) in this procedure and proceeded to the remaining steps with usable DTI data. The MDDL subgroup contained 27 participants (13 males, 14 females), and the MDDS subgroup had 18 participants (nine males, nine females). Demographic data are reported in Table 1. There were no differences across the groups or subgroups, except in the age between MDDL and HC (p = .031). Current clinical severity did not differ between the MDD subgroups (17‐item HDRS score, Beck Depression Inventory score and Beck Scale for Suicidal Ideation score).
TABLE 1.
Demographic and clinical data of the participants
| HC (N = 41) | MDD (N = 45) | MDD (N = 52) | |||
|---|---|---|---|---|---|
| MDDS (N = 18) | MDDL (N = 27) | p‐values | |||
| Age | 30.47 ± 8.08 | 33.16 ± 10.49 | 29.67 ± 8.87 | 35.48 ± 10.98 |
MDD versus HC: .183 MDDL versus HC: .031 MDDS versus HC: .729 MDDL versus MDDS: .068 |
| Sex (M:F) | 15:26 | 22:23 | 9:9 | 13:14 |
MDD versus HC: .175 MDDL versus HC: .451 MDDS versus HC: .248 MDDL versus MDDS: .572 |
| Onset age | – | 18.84 ± 9.49 | 25.90 ± 9.07 | 14.10 ± 6.35 | MDDL versus MDDS: .469 |
| HDRS‐17 | 1.87 ± 2.59 | 18.02 ± 4.79 | 17.72 ± 3.58 | 18.22 ± 5.51 | MDDL versus MDDS: .736 |
| BECK DEPN | 1.70 ± 2.81 | 23.90 ± 8.03 | 22.61 ± 6.31 | 24.81 ± 9.04 | MDDL versus MDDS: .635 |
| Suicidal ideation | – | 5.84 ± 8.34 | 4.89 ± 7.19 | 6.48 ± 9.10 | MDDL versus MDDS: .536 |
Abbreviations: BECK DEPN, Beck Depression Inventory score; F, female; HC, healthy controls; HDRS‐17:17‐item Hamilton Depression Rating Scale; M, male; MDD: patients with major depressive disorder; MDDS, MDD with a history ≤10 years; MDDL, MDD with a history >10 years.
3.2. Clustering coefficient
At all density levels, the CC of the HC group was consistently the lowest of the three subgroups numerically, and MDDS had the highest. There were no statistically significant differences between the MDD and HC groups or between the MDDL and the HC groups. However, differences were observed between the MDDS and HC groups at density levels ranging from 17 to 23%, with p‐values <.05, respectively. In particular, the difference between the MDDS and HC groups was significant at p = .0486 at density = 20% (Figure 2). Unfortunately, the significance did not survive FDR‐correction, perhaps due to limited size of the samples.
FIGURE 2.
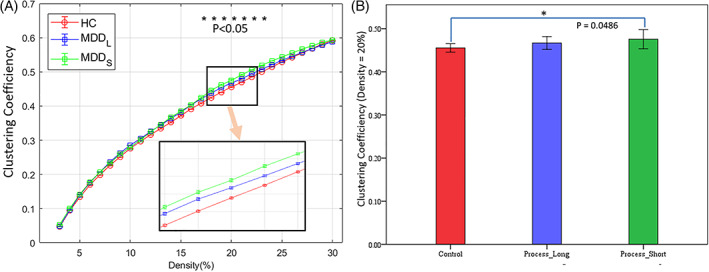
Clustering coefficients of the three groups at all density levels and their statistical difference. (a) The profile of the clustering coefficients at density levels from 2 to 30%. Among them, the differences between the MDDS and HC group showed statistical significance p < .05(*) at density levels from 17 to 23%. The light orange arrow in (a) indicates that the bottom‐right inset is a zoomed‐in view of the area outlined by the black rectangle above it. (b) The statistical significance between the MDDS and HC group was p = .0486 (uncorrected) at density = 20%
Individual brain nodes differed between groups (Table 2 and Figure 3). Comparing MDDS and HC groups (Figure 3a), MDDS group's CC values were higher (p < .01 uncorrected) in precentral gyrus (PrG) and in caudal lingual gyrus (cLinG) (p = .0011, FDR‐corrected). Compared with the HC group, MDDL had higher CC values (p < .01 uncorrected) in dorsal granular insula (dlg) and postcentral gyrus (PoG; p = .0016, FDR‐corrected) in the right hemisphere (Figure 3b). The CC values of MDDS were also higher than MDDL in cLinG (p = .0035, FDR‐corrected) in the right hemisphere and in rostral lingual gyrus (rLing; p < .01 uncorrected) in the left hemisphere (Figure 3c).
TABLE 2.
List of brain regions (nodes) that showed statistically significant differences in clustering coefficient between the group and subgroups (p < .01 uncorrected, except #190a with p = .0011, #160 with p = .0016 and #190b with p = .0035, FDR‐corrected)
| Nodal index | MNI coordinates | Label | Left or right hemisphere |
|---|---|---|---|
| MDDS > HC | |||
| 54 | 55, −2, 33 | Precentral gyrus (PrG) | R |
| 190a | 10, −85, −9 | Caudal lingual gyrus (cLinG) | R |
| MDDL > HC | |||
| 160 | 48,‐24,48 | Postcentral gyrus (PoG) | R |
| 172 | 39, −7, 8 | Dorsal granular insula (dlg) | R |
| MDDS > MDD L | |||
| 190b | 10, −85, −9 | Caudal lingual gyrus (cLinG) | R |
| 195 | ‐17, −60, −6 | Rostral lingual gyrus (rLinG) | L |
FIGURE 3.
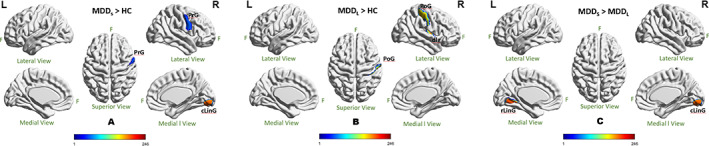
Statistical difference of clustering coefficient of individual brain nodes when comparing across groups at 20% density. (a) MDDS > HC; (b) MDDL > HC; (c) MDDS > MDDL. All the significances were thresholded at p = .01(uncorrected) except cLinG (p = .0011; FDR‐corrected) in (a), PoG (p = .0016, FDR‐corrected) in (b), and cLinG (p = .0035; FDR‐corrected) in (c). The color bar encoded the 246 individual brain regions (cLinG, caudal lingual gyrus; dlg, dorsal granular insula; F, front of the brain; L, left hemisphere; PrG, precentral gyrus; PoG, postcentral gyrus; rLing, rostral lingual gyrus; R, right hemisphere)
3.3. Global efficiency
None of the group or subgroup differences between the patients and HCs, or between the patient subgroups and the HCs survived correction. In particular, no differences were identified at the 20% density.
3.4. Network resilience
Both the MDD population and the HC had well developed, functionally segregated, brain regions and brain network, as the resilience curves of all the groups and subgroups were far below the baseline profile outlined by the random attack based on HC at all density levels, indicating targeted attack on brain hubs would easily harm the global efficiency of the brain network (Figure 4). Again, the control group was numerically consistently the lowest in resilience, with MDDL in the middle, and MDDS the highest among the three subgroups (Figure 4a). Although the MDDL group did not differ statistically significantly from the HC or MDDS group, the MDDS group showed statistical significance of either p < .05 or p < .01 at density levels from 19 to 27%, compared with the HC group (Figure 4a). In particular, the statistical significance of the difference between MDDS and the HC groups was p = .0157 (FDR‐corrected) at density = 20% (Figure 4b), which is the density level commonly used in the literature.
FIGURE 4.
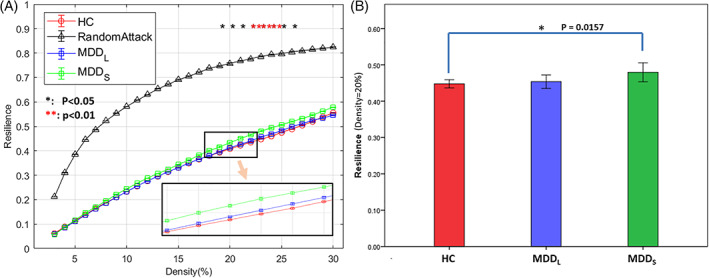
Network Resilience of the three groups at all density levels and their statistical differences. (a) The profile of the network resilience at density levels from 2 to 30%. Among them, the differences between the MDDS and HC group showed statistical significance either p < .05(*) or p < .01(**) at density levels from 19 to 27%, all FDR‐corrected. The black‐triangle profile of random attack on the top was based on the healthy controls for reference. At all density levels, the HC group was consistently the lowest while the MDDS group was the highest. The light orange arrow indicates that the bottom‐right inset is a zoomed‐in view of the area (containing the HC, MDDL, and MDDS curves) outlined by the small rectangle above it. (b) The statistical significance between the MDDS and HC group was p = .0157 (FDR‐corrected) at density = 20%
3.5. Correlations between network resilience and the duration of disease
The Pearson correlation showed that the network resilience and CCs were negatively correlated with duration of illness since onset of MDD, both generally and locally (Figure 5). In particular, the correlation coefficient between the network resilience and the duration of disease was r = −.305 at p = .041 (Figure 5a). Two brain regions showed a negative correlation with the CC, bilaterally (Figure 5b): cLinG (r = −.342, p = .021) in the right hemisphere, and rLinG (r = −.328, p = .028) in the left hemisphere.
FIGURE 5.
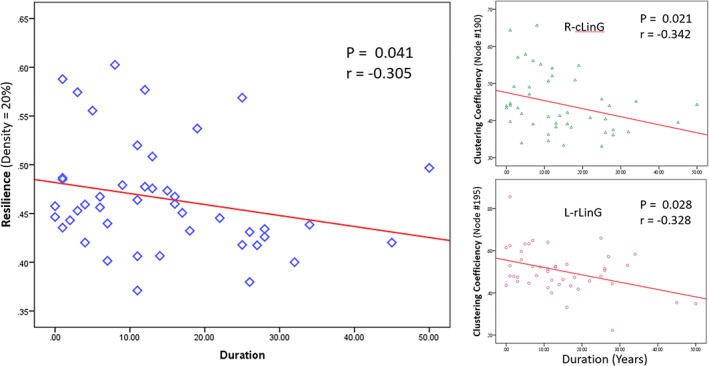
Pearson correlation analysis showed negative correlations between networking indices and the duration of MDD, represented by those at the density level of 20%. (a) The network resilience was negatively correlated with the duration of the disease: r = −.305 (p = .041). (b) Clustering coefficients of most of the 246 brain regions were not correlated with the duration of the disease except for the right side cLinG (r = −.3421, p = .021) and the left side rLinG (r = −.328, p = .028) whose clustering coefficients were negatively correlated with the disease duration (cLinG, caudal lingual gyrus in the right hemisphere; rLinG, rostral lingual gyrus in the left hemisphere)
3.6. Characteristic path length
The analyses showed that the difference was not statistically significant between the patients and HCs, or between the patient subgroups and the HCs.
3.7. Small‐worldness
All the groups demonstrated small‐worldness, and their values were between 2.0 and 3.5 (Figure 6), with HCs consistently the lowest. However, difference was significant only between MDDS and HC in the density range from 24 to 30%, not at the 20% level under observation in this study. Moreover, none of the significance survived FDR‐correction.
FIGURE 6.
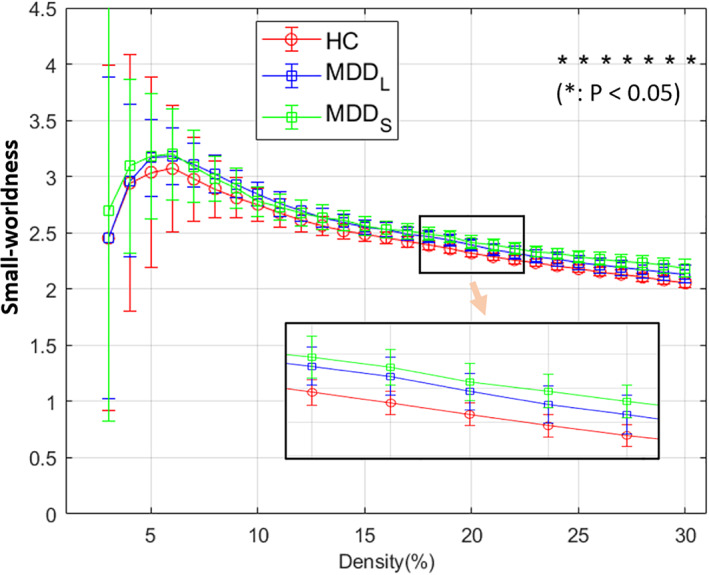
Small‐worldness of the three groups at all density levels and their statistics at density levels from 2 to 30%. Among them, MDDS and HC group showed statistical difference with p < .05(*) at density levels from 24 to 30%, however, none survived FDR‐correction. At all density levels, the HC group (red) was consistently the lowest while the MDDS group (green) was relatively the highest at all the density levels higher than 13%. The light orange arrow indicates that the bottom‐right inset is a zoomed‐in view of the area (containing the HC, MDDL, and MDDS curves) outlined by the small rectangle above it
3.8. Multiple regression using disease duration as a continuous variable
The analyses did not identify disease duration to be a factor of statistical significance for the network indices that we have examined in this work, with that for network resilience being the most prominent among them, showing a negative relationship, at p = .098 though (density level = 20%). Age and sex appeared to be insignificant factors (p = .479 and .772, respectively), either.
4. DISCUSSION
Our examination of network clustering efficiency, global efficiency, network resilience, characteristic path length, and small‐worldness based on 45 MDD patients and 41 demographically matched HC did not identify differences between the two general groups. However, taking into account the duration of MDD illness, by subgrouping MDD into those who had this disorder for more than or less than 10 years, we found heterogeneity in terms of networking patterns characterized by the indices of a small‐world network.
In the clustering efficiency analysis of the 246 brain regions, we observed that the HC group exhibited a higher level of baseline functional segregation when compared with the MDD group, based on a lower numerical CC of the HC group, indicating that the brain regions had relatively fewer inter‐regional connections. However, only the difference between MDDS and HC reached statistical significance, as the subgroup heterogeneity could have blurred the difference between the general MDD group and the HC. Brains undergoing normal development are progressively more functionally and structurally segregated (Baum et al., 2017). Resting fMRI data of patients with MDD have found greater clustering when compared with normal controls (Zhang et al., 2011). Our observation that the CC values of MDDS were higher than the HC group suggests that these network patterns may represent a structural change occurring around illness onset, or early in the course of illness (Figure 2). In contrast, CC values of the MDDL were not different from the HC group, suggesting that this anatomic pattern does not persist with more chronic illness states, possibly due to homeostatic responses, functional and structural neuroplasticity (Liu et al., 2017) or treatment effects (Abdallah et al., 2017). MDDS and MDDL groups did not differ in terms of current depression severity, ruling that out as an explanation of the group differences.
A similar pattern of subgroup heterogeneity related to illness duration was observed in relation to the network resilience analysis, where values in MDDS were higher compared with the HC group (Figure 4). This is consistent with fMRI findings in MDD showing higher global efficiency in the literature, representing relatively more randomized brain networks in MDD (Zhang et al., 2011). Using network resilience, an index derived from network connectivity and global efficiency, but profiling the efficiency globally in a dynamic fashion, we were able to measure the vulnerability of the network‐clustering pattern. We have found that the difference in MDD from HCs was confined to MDDS. This observation suggests that MDD earlier in its course (≤10 years) involves a more severe structural abnormality in the brain than later in the course of the disorder. This surprising finding requires replication in an independent data set.
While all the groups and subgroups showed small‐worldness, our analyses again found the HC group was the lowest among the groups. Although the differences were not significant, the relativity of the groups was in line with the findings in the literature, which saw a similar situation (HC smaller than MDD, yet difference not significant; Sacchet et al., 2015). Moreover, MDDS again was the highest and MDDL in the middle, indicating a short term tendency in the MDDS developed toward randomization and in a long run MDDL's returning to HC. The small‐worldness measure was thus also in line with our findings in the analyses of CC and network resilience. In the literature, investigators reported a potential scarring effect suggesting that a depressive episode may leave lasting changes in personality and self‐concept (Christensen & Kessing, 2006; Kaltenboeck & Harmer, 2018; Rohde, Lewinsohn, & Seeley, 1990; Zautra et al., 2007), and a 911 study also showed multimodal neuroimaging evidences of stress‐related changes in healthy adult brains (Ganzel, Kim, Glover, & Temple, 2008). However, such arguments remain largely inconclusive, psychologically, biologically and cognitively (Wichers, Geschwind, van Os, & Peeters, 2010). For example, a study based on a large sample of 350 participants using a daily diary design found that the scar hypothesis was not clearly supported (O'Grady, Tennen, & Armeli, 2010). Moreover, these studies mostly focused on personality traits. Yet, how the psychological effect may reflect in brain imaging, is still mysterious. Our findings based on the current patient cohort demonstrated a curing tendency in neuroimaging in a long run, which in fact does not conflict with the possible scarring effect—just like a cut on the skin will cure anyway whether or not a scar would present and last later. Nevertheless, the curing effect found in our study could have resulted from the heterogeneity that may exist in the depressive patients, because, as mentioned in the text, depression is actually quite heterogeneous in its manifestation from one episode to another (Lopez‐Castroman et al., 2012; Yu et al., 2019). Future studies should conduct a more precise and detailed work to clarify all the variables that may have contributed to the heterogeneity.
Our data identified no group or subgroup difference concerning the characteristic path length between MDD and HC, or MDDS/MDDL and HC. Studies in the literature actually reported conflicted results. For example, one DTI study on a remitted geriatric depression population (35 geriatric depression patients vs. 30 HC) found increased path length in the patient group compared with the HCs at p < .05, uncorrected (Bai et al., 2012). A second DTI Study on MDD (95 MDD vs. 102 HC) found that the patient group was slightly higher than the controls (p‐value = .048, uncorrected; Korgaonkar et al., 2014), and another structural MRI study (93 MDD vs. 151 HC) also had a similar result (p < .05; Singh et al., 2013). The statistical significances in these studies were generally weak and did not survive correction, although the latter two studies involved relatively large sizes of samples. Moreover, the second DTI study focused mostly on a part of the brain, that is, the nodes of two networks (the default mode network and frontal‐thalamo‐caudate regions), which may not reflect the whole‐brain status like what we have done in the current study. The third study again constructed the network based on volumetric correspondence in the cortices, which was very different from the ways of constructing either the conventional fMRI network or the DTI network. In contrast, studies on depression using graph theory based on functional MRI data (resting state function MRI data) reported abnormally low values of the characteristic path length and consequently higher global efficiency in the patient group, which contradicted with the findings from the aforementioned imaging studies either using DTI or structural imaging data (Zhang et al., 2011). Although people have tried to explain the inconsistency, lack of consensus is factual. We attribute the disagreement to the heterogeneity in the employed imaging modalities and the analysis methods, and to the possible heterogeneity in the sample of MDD patients as well. Our study resulted in no difference in the measures of characteristic path length and global efficiency was therefore reasonable. The unsettled argument shall be further studied with better design to comprehend,
In general, many MDD studies comparing MDD and matched HC employed graph theory, and many of them identified no or only weak global network abnormalities in the brains with MDD. This perhaps is partly due to their strategy that used all the MDD participant as one group, without taking into consideration of the possible heterogeneity that may exist (Ajilore et al., 2014; Korgaonkar et al., 2014; Sacchet et al., 2015). The DTI study using a support vector machine (SVM) on MDD (Sacchet et al., 2015) investigated the respective relationship between nine graph indices (including CC, global efficiency, characteristic path length, and small‐worldness) in MDD against normal controls and identified uncorrected weak significance from only one index (global flow coefficient), but none survived FDR‐correction. However, its analysis on the SVM employed in this work showed that small‐worldness and global efficiency were the two leading variables that contributed to the accuracy of classifying MDD out from normal healthy people. Yet, directly analyzing the correlation showed that none of the clinical variables under investigation (disease duration, age of onset, Beck Depression Inventor‐II, Global Assessment of Function) was correlated to small‐worldness with statistical significance. Our work in the current study that subgrouped the MDD patients thus may be an encouraging effort, although our analyses based on the small sized samples revealed group difference only in a part of the nine indices.
The insula plays a key role in emotional‐cognitive integration (Augustine, 1996). The brain connectivity pattern in the right insula region is abnormal in people at high familial risk for depression (Li et al., 2018). In the current study, we found an abnormal connectivity pattern in the right insula in MDD compared with the HC group, however only in the MDDL subgroup and not in the MDDS subgroup; perhaps, because the deficits of emotional‐cognitive functions integrated with this particular brain region in MDD need an extensively long time to develop.
Numerous imaging studies on MDD versus HC have found abnormalities in the PoG area, including those studying functional connectivity, regional homogeneity, local brain volume, cortical thickness, cortical area, or serotonin 1A receptor binding, using MRI or PET (Drevets et al., 1999; Jaworska et al., 2014; Li et al., 2010; Mao et al., 2020; Shen et al., 2015; The‐ENIGMA‐MDD‐Working‐Group, 2017; Vasic, Walter, Höse, & Wolf, 2008; Wehry et al., 2015). The PoG is the primary sensorimotor area and receives the majority of the somatic sensory relay information from the thalamus. Functional imaging studies have previously found that PoG is associated with higher ReHo in MDD patients with somatic symptoms including pain (Liang et al., 2020). We also identified an abnormality in this region in MDD. Interestingly, the significant difference was only detected in MDDL. We do not know whether this difference means the distinctive connectivity pattern in PoG explains difference in somatic symptoms in MDD or a difference in brain between MDDL and MDDS groups. Future studies must address this question.
The lingual gyrus is relevant for anxiety‐depression severity in young adults (Couvy‐Duchesne et al., 2018), but its volume may impact depression/anxiety severity and antidepressant response (Jung et al., 2014). We found that part of the right lingual gyrus (cLinG) of the MDDS brains was more clustered compared with MDDL. The longer the MDD course, the closer the clustering pattern resembled HC (Figures 2 and 5). This effect was most prominent in the lingual gyri in both hemispheres, which suggests that these brain regions may be more plastic (Papmeyer et al., 2015; Peng et al., 2015). Cognitive therapy, pharmacotherapy (SSRI, ketamine, etc.) and even religious beliefs could influence cognitive and structural renormalization of the brain (Abdallah et al., 2017; Godlewska, Norbury, Selvaraj, Cowen, & Harmer, 2012; Hasler & Northoff, 2011; Joffe, Levitt, Sokolov, & Young, 1996; Li et al., 2018; Simons, Garfield, & Murphy, 1984), although not specifically the lingual gyri. For example, abnormal brain activity patterns reversed after MDD recovery (Phillips, Drevets, Rauch, & Lane, 2003). While our patients were drug‐free at least 3 weeks and antidepressant‐free at least 6 weeks at the time of the MRI scans in this study, many had received antidepressant treatments previously. However, due to the variation in types of medication, duration of usage and time since the last dose, we were unable to statistically account for the potential effect of such past lifetime medication or treatment in our analyses. We speculate that such prior treatment could have contributed to time‐dependent recovery in terms of brain connectivity in MDD.
To address possible concerns on the 10‐year cutoff to subgroup the patients, we also performed additional regression analyses using disease duration as a continuous variable to check the effect of this variable, although we have explained that using this particular cutoff was an optimal choice in the current study. Unfortunately, no significance was detected for most of the network indices we examined in this study (Section 3.8), perhaps due to the modest size of our subgroups. Nevertheless, the negative correlationship between disease duration and the network resilience was the most prominent among all the network indices, although its p‐value did not reach a conventionally significant level, at only p = .098. This observation demonstrated somewhat agreement with our correlation analysis between the index of network resilience and disease duration that showed a negative correlation with statistical significance (Section 3.5 & Figure 5), inferring that disease duration is an important factor in studying MDD. While the latter analysis was basically a discrete version of using the variable of disease duration, the slight discrepancy between the results of the two strategies encouraged using larger sample sizes in the fitting process for a better estimation of the relationship.
The current work has limitations. First, 246 brain regions employed in the current study were mostly DTI‐based, with the nodes centered at the voxel with a local peak FA value. The regions thus did not have an exact correspondence with those in the functional neuroimaging studies, where brain regions were identified based on functional activity. Therefore, structural findings based on DTI could differ from those in functional neuroimaging studies. Similar structural functional network analyses should be employed in future studies combining fMRI and DTI. Second, we studied all the 246 regions within one small‐world network, while functional neuroimaging studies typically study the brain based on a number of functional neural networks identified based on their synchronized rhythms of the blood‐oxygen‐level‐dependency signals. The network indices in this current study thus were all used as global measures to provide an overview of the networking status based on DTI data, but did not follow exactly the conventional approach of using them at separated nodal or global levels. The interpretations thus were made with caution from a slightly different angle of view. Third, the sample sizes were modest. While this study involved nearly 100 participants, we divided the MDD cohort into two subgroups. Future studies demand larger sample sizes to permit more detailed clinical subgrouping approaches to investigate the possible heterogeneities of networking patterns in MDD.
In summary, our study employed DTI to investigate the networking patterns in MDD and possible heterogeneity in this patient population. Our findings showed that duration of the illness has an impact on the networking patterns. MDD patients have networking abnormalities compared with HCs; however, such abnormalities could be blurred or hidden by the heterogeneity of the MDD clinical subgroups, such as duration of illness. Brain plasticity may introduce a recovery effect to the abnormal network patterns seen in patients with a relative short term of the illness, as these abnormalities are not observable in those whose onset of MDD occurred longer than 10 years prior to scanning. Future studies should recruit larger sample sizes to examine longitudinal clinical effects in more detail, including the effects of treatment on network patterns.
CONFLICT OF INTERESTS
The authors declare no conflict of interest pertaining to the content of this study. Also the authors would like to thank Dr. Jun Liu for helpful discussions on the statistical analyses.
ACKNOWLEDGMENT
This project was supported in part by NIMH grants 5R01MH108032‐02 (J. Mann; PI) and R01 MH 5P50MH090964(J. Mann; PI). G. Xu and Z. Zhao were supported by a scholarship sponsored by Chinese Scholarship Council, and in part by an award by China National Key R&D Program (No. 2017YFC1308500).
Xu, D., Xu, G., Zhao, Z., Sublette, M. E., Miller, J. M., & Mann, J. J. (2021). Diffusion tensor imaging brain structural clustering patterns in major depressive disorder. Human Brain Mapping, 42(15), 5023–5036. 10.1002/hbm.25597
Funding information NIMH, Grant/Award Numbers: 5R01MH108032‐02, R01 MH 5P50MH090964; Chinese Scholarship Council, Grant/Award Number: 2017YFC1308500
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available upon request. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- American Psychiatric Association . (1994). Diagnostic and statistical manual of mental disorders (4th ed.). Washington: American Psychiatric Association. [Google Scholar]
- Abdallah, C. G., Jackowski, A., Salas, R., Gupta, S., Sato, J. R., Mao, X., … Mathew, S. J. M. (2017). The nucleus accumbens and ketamine treatment in major depressive disorder. Neuropsychopharmacology, 42, 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, S., Salvador, R., Whitcher, B., Suckling, J., & Bullmore, E. (2006). A resilient, low‐frequency, small‐world human brain functional network with highly connected association cortical hubs. Journal of Neuroscience, 26(1), 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajilore, O., Lamar, M., & Kumar, A. (2014). Association of brain network efficiency with aging, depression, and cognition. The American Journal of Geriatric Psychiatry, 22(2), 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine, J. R. (1996). Circuitry and functional aspects of the insular lobe in primates including humans. Brain Research Reviews, 22, 229–244. [DOI] [PubMed] [Google Scholar]
- Bai, F., Shu, N., Yuan, Y., Shi, Y., Yu, H., Wu, D., … Zhang, Z. (2012). Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. The Journal of Neuroscience, 32(12), 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett, D. S., & Bullmore, E. (2006). Small‐world brain networks. The Neuroscientist, 12(6), 512–523. [DOI] [PubMed] [Google Scholar]
- Bassett, D. S., Edward, E. T., & Bullmore, T. (2017). Small‐world brain networks revisited. Neuroscientist, 23(5), 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, G. L., Ciric, R., Roalf, D. R., Betzel, R. F., Moore, T. M., Shinohara, R. T., … Satterthwaite, T. D. (2017). Modular segregation of structural brain networks supports the development of executive function in youth. Current Biology, 27, 1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramazza, A., & Coltheart, M. (2006). Cognitive neuropsychology twenty years on. Cognitive Neuropsychology, 23(1), 3–12. [DOI] [PubMed] [Google Scholar]
- Chen, T., Kendrick, K. M., Wang, J., Wu, M., Li, K., Huang, X., … Gong, Q. (2017). Anomalous single‐subject based morphological cortical networks in drug‐naive, first‐episode major depressive disorder. Human Brain Mapping, 38(5), 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetry, B. T., Hezghia, A. M., Jeffrey, M., Lee, S., Rubin‐Falcone, H., Cooper, T. B., … Sublette, M. E. (2016). Omega‐3 polyunsaturated fatty acid supplementation and white matter changes in major depression. Journal of Psychiatric Research, 75, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, M. V., & Kessing, L. V. (2006). Do personality traits predict first onset in depressive and bipolar disorder? Nordic Journal of Psychiatry, 60, 79–88. [DOI] [PubMed] [Google Scholar]
- Couvy‐Duchesne, B., Strike, L. T., de Zubicaray, G. I., McMahon, K. L., Thompson, P. M., Hickie, I. B., … Wright, M. J. (2018). Lingual gyrus surface area is associated with anxiety‐depression severity in young adults: A genetic clustering approach. eNeuro, 5(1), 1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets, W. C., Frank, E., Price, J. C., Kupfer, D. J., Holt, D., Greer, P. J., … Mathis, C. (1999). PET imaging of serotonin 1A receptor binding in depression. Biological Psychiatry, 46(10), 1375–1387. [DOI] [PubMed] [Google Scholar]
- Fan, L., Li, H., Zhuo, J., Zhang, Y., Wang, J., Chen, L., … Jiang, T. (2016). The human brainnetome atlas: A new brain atlas based on connectional architecture. Cerebral Cortex, 26, 3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First, M., Williams, J., Spitzer, R., & Gibbon, M. (1997). Structured clinical interview for DSM‐IV Axis I disorders. Washington D.C: American Psychiatric Publishing, Inc. [Google Scholar]
- Fornito, A., Zalesky, A., & Breakspear, M. (2015). The connectomics of brain disorders. Nature Reviews. Neuroscience, 16, 159–172. [DOI] [PubMed] [Google Scholar]
- Fried, E. I. (2016). The 52 symptoms of major depression: Lack of content overlap among seven common depression scales. Journal of Affective Disorders, 208, 191–197. [DOI] [PubMed] [Google Scholar]
- Fried, E. I. (2020). Corrigendum to “the 52 symptoms of major depression: Lack of content overlap among seven common depression scales”, [journal of affective disorders, 208, 191–197]. Journal of Affective Disorders, 260, 744. [DOI] [PubMed] [Google Scholar]
- Ganzel, B. L., Kim, P., Glover, G. H., & Temple, E. (2008). Resilience after 9/11: Multimodal neuroimaging evidence for stress related change in the healthy adult brain. NeuroImage, 40, 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewska, B. R., Norbury, R., Selvaraj, S., Cowen, P. J., & Harmer, C. J. (2012). Short‐term SSRI treatment normalises amygdala hyperactivity in depressed patients. Psychological Medicine, 42(12), 2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Q., & He, Y. (2015). Depression, neuroimaging and connectomics: A selective overview. Biological Psychiatry, 77, 223–235. [DOI] [PubMed] [Google Scholar]
- Goni, J., van den Heuvel, M. P., Avena‐Koenigsberger, A., de Mendizabal, N. V., Betzel, R. F., Griffa, A., … Sporns, O. (2014). Resting‐brain functional connectivity predicted by analytic measures of network communication. Proceedings of the National Academy of Sciences, 111, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, E. M., Laumann, T. O., Adeyemo, B., Huckins, J. F., Kelley, W. M., & Petersen, S. E. (2016). Generation and evaluation of a cortical area parcellation from resting‐state correlations. Cerebral Cortex, 26, 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M. (1960). A rating scale for depression. Journal of Neurology, Neurosurgery, and Psychiatry, 23, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M. (1967). Development of a rating scale for primary depressive illness. The British Journal of Social and Clinical Psychology, 6, 278–296. [DOI] [PubMed] [Google Scholar]
- Hasler, G., & Northoff, G. (2011). Discovering imaging endophenotypes for major depression. Molecular Psychiatry, 16, 604–619. [DOI] [PubMed] [Google Scholar]
- Hollister, L., Müller‐oerlinghausen, B., Rickels, K., & Shader, R. (1993). Clinical uses of benzodiazepines. Journal of Clinical Psychopharmacology, 13(6), 25. [PubMed] [Google Scholar]
- Honey, C. J., Sporns, O., Cammoun, L., Gigandet, X., Thiran, J. P., Meuli, R., & Hagmann, P. (2009). Predicting human resting‐state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences, 106, 2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L., Xiao, M., Ai, M., Wang, W., Chen, J., Tan, Z., … Kuang, L. (2019). Disruption of resting‐state functional connectivity of right posterior insula in adolescents and young adults with major depressive disorder. Journal of Affective Disorders, 257, 23–30. [DOI] [PubMed] [Google Scholar]
- Hu, S. F., Feng, H., Xu, T., Zhang, H., Zhao, Z., Lai, J., … Xu, Y. (2017). Altered microstructure of brain white matter in females with anorexia nervosa: A diffusion tensor imaging study. Scientific Reports, 13, 2829–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, M. D., Gurney, K., & Prescott, T. J. (2006). The brainstem reticular formation is a small‐world, not scale‐free, network. Proceedings of the Biological Sciences, 273(1585), 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska, N., MacMaster, F. P., Gaxiola, I., Cortese, F., Goodyear, B., & Ramasubbu, R. (2014). A preliminary study of the influence of age of onset and childhood trauma on cortical thickness in major depressive disorder. BioMed Research International, 2014 (Translational Neuroimaging of the Mood and Anxiety Disorders), 410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe, R. T., Levitt, A. J., Sokolov, S. T., & Young, L. T. (1996). Response to an open trial of a second SSRI in major depression. The Journal of Clinical Psychiatry, 57(3), 114–115. [PubMed] [Google Scholar]
- Jung, J., Kang, J., Won, E., Nam, K., Lee, M.‐S., Tae, W. S., & Ham, B.‐J. (2014). Impact of lingual gyrus volume on antidepressant response and neurocognitive functions in major depressive disorder: A voxel‐based morphometry study. Journal of Affective Disorders, 169, 179–197. [DOI] [PubMed] [Google Scholar]
- Kaltenboeck, A., & Harmer, C. (2018). The neuroscience of depressive disorders: A brief review of the past and some considerations about the future. Brain and Neuroscience Advances, 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar, M. S., Fornito, A., Williams, L. M., & Grieve, S. M. (2014). Abnormal structural networks characterize major depressive disorder: A connectome analysis. Biological Psychiatry, 76(7), 567–574. [DOI] [PubMed] [Google Scholar]
- Li, C.‐T., Lin, C.‐P., Chou, K.‐H., Chen, I.‐Y., Hsieh, J.‐C., Wu, C.‐L., … Su, T.‐P. (2010). Structural and cognitive deficits in remitting and non‐remitting recurrent depression: A voxel‐based morphometric study. NeuroImage, 50(1), 347–356. [DOI] [PubMed] [Google Scholar]
- Li, X., Weissman, M. M., Posner, J., & Xu, D. (2018). A diffusion tensor imaging study of altered brain microstructure by religiosity and spirituality underlying familial risk for depression. Brain and Behavior, 9(2), e01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J., Liao, H., Li, X., Xu, C., Xu, Z., Yu, Y., … Xie, G. (2020). Functional abnormalities in first‐episode major depressive disorder with somatic pain. Journal of Affective Disorders, 2, 100029. [Google Scholar]
- Liu, W., Ge, T., Leng, Y., Pan, Z., Fan, J., Yang, W., & Cui, R. (2017). The role of neural plasticity in depression: From hippocampus to prefrontal cortex. Neural Plasticity, 2017, 6871089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Liu, X., Yang, G., Zhou, Z., Zhou, Y., Li, G., … Xu, D. (2012). Improving the correction of Eddy current induced distortion in diffusion‐weighted imaging data by recognizing and excluding ventricles. Computerized Medical Imaging and Graphics, 36, 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Liang, M., Zhou, Y., He, Y., Hao, Y., Song, M., … Jiang, T. (2008). Disrupted small‐world networks in schizophrenia. Brain, 131, 945–961. [DOI] [PubMed] [Google Scholar]
- Lo, C.‐Y. Z., Su, T.‐W., Hung, C.‐C. X., Hung, C.‐C., Chen, W.‐L., Lan, T.‐H., … Bullmore, E. T. (2015). Randomization and resilience of brain functional networks as systems‐level endophenotypes of schizophrenia. PNAS, 112(29), 9123–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Castroman, J., Galfalvy, H., Currier, D., Stanley, B., Blasco‐Fontecilla, H., Baca‐Garcia, E., … Oquendo, M. A. (2012). Personality disorder assessments in acute depressive episodes: Stability at follow‐up. The Journal of Nervous and Mental Disease, 200(6), 526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Shen, Z., Cheng, Y., Yang, H., He, B., Xie, Y., … Han, H. (2017). Alternations of white matter structural networks in First episode untreated major depressive disorder with short duration. Frontiers in Psychology, 8, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, N., Che, K., Chu, T., Li, Y., Wang, Q., Liu, M., … Ji, H. (2020). Aberrant resting‐state brain function in adolescent depression. Frontiers in Psychology, 11, 1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew, R. V. P., Milak, S., Oquendo, M. A., Kevin, M., Malone, J., & Mann, J. (2010). Regional brain metabolic correlates of self‐reported depression severity contrasted with clinician ratings. Journal of Affective Disorders, 126(1–2), 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minat, L., Grisoli, M., Seth, A. K., & Critchley, H. D. (2012). Decision‐making under risk: A graph‐based network analysis using functional MRI. NeuroImage, 60(4), 2191–2205. [DOI] [PubMed] [Google Scholar]
- Mišić, B., Betzel, R. F., de Reus, M. A., van den Heuvel, M. P., Berman, M. G., McIntosh, A. R., & Sporns, O. (2016). Network‐level structurefunction relationships in human neocortex. Cerebral Cortex, 26, 3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir, T., Jahanshad, N., Jack, C. R., Weiner, M. W., Toga, A. W., & Thompson, P. M. (2012). Small world network measures predict white matter degeneration in patients with early‐stage mild cognitive impairment, in 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). Barcelona. 1405–1408. [DOI] [PMC free article] [PubMed]
- O'Grady, M. A., Tennen, H., & Armeli, S. (2010). Depression history, depression vulnerability and the experience of everyday negative events. Social and Clinical Psychology, 29(9), 949–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papmeyer, M., Giles, S., Sussmann, J. E., Kielty, S., Stewart, T., Lawrie, S. M., … McIntosh, A. M. (2015). Cortical thickness in individuals at high familial risk of mood disorders as they develop major depressive disorder. Biological Psychiatry, 78(1), 58–66. [DOI] [PubMed] [Google Scholar]
- Peng, D., Shi, F., Li, G., Fralick, D., Shen, T., Qiu, M., … Fang, Y. (2015). Surface vulnerability of cerebral cortex to major depressive disorder. PLoS One, 10(3), e0120704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, M. L., Drevets, W. C., Rauch, S. L., & Lane, R. (2003). Neurobiology of emotion perception II: Implications for major psychiatric disorders. Biological Psychiatry, 54, 515–528. [DOI] [PubMed] [Google Scholar]
- Power, J. D., Cohen, A. L., Nelson, S. M., Wig, G. S., Barnes, K. A., Church, J. A., … Petersen, S. E. (2011). Functional network organization of the human brain. Neuron, 72, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier, D. A., Narrow, W. E., Clarke, D. E., Kraemer, H. C., Kuramoto, S. J., Kuhl, E. A., & Kupfer, D. J. (2013). DSM‐5 field trials in the United States and Canada, part II: Test‐retest reliability of selected categorical diagnoses. The American Journal of Psychiatry, 170(1), 59–70. [DOI] [PubMed] [Google Scholar]
- Rizk, M. M., Rubin‐Falcone, H., Keilp, J., Miller, J. M., Sublette, M. E., Burke, A., … Mann, J. J. (2017). White matter correlates of impaired attention control in major depressive disorder and healthy volunteers. Journal of Affective Disorders, 222(2017), 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, P., Lewinsohn, P. M., & Seeley, J. R. (1990). Are people changed by the experience of having an episode of depression? A further test of the scar hypothesis. Journal of Abnormal Psychology, 99(3), 264–271. [DOI] [PubMed] [Google Scholar]
- Roxin, A., Riecke, H., & Solla, S. A. (2004). Self‐sustained activity in a small‐world network of excitable neurons. Physical Review Letters, 92(19), 198101. [DOI] [PubMed] [Google Scholar]
- Rubinov, M., & Sporns, O. (2010). Complex network measures of brain connectivity: Uses and interpretations. NeuroImage, 52(2010), 1059–1069. [DOI] [PubMed] [Google Scholar]
- Sacchet, M. D., Prasad, G., Foland‐Ross, L. C., Thompson, P. M., & Gotlib, I. H. (2015). Support vector machine classification of major depressive disorder using diffusion‐weighted neuroimaging and graph theory. Frontiers in Neuroscience, 6 (Article 21), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite, T. D., Wolf, D. H., Erus, G., Ruparel, K., Elliott, M. A., Gennatas, E. D., … Gur, R. E. (2013). Functional maturation of the executive system during adolescence. The Journal of Neuroscience, 33, 16249–16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling, M. A. (2005). A "small‐world" network model of cognitive insight. Creativity Research Journal, 17(2–3), 131–154. [Google Scholar]
- Shen, T., Li, C., Wang, B., Yang, W.‐m., Zhang, C., Wu, Z., … Peng, D.‐h. (2015). Increased cognition connectivity network in major depression disorder: A fMRI study. Psychiatry Investigation, 12(2), 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, A. D., Garfield, S. L., & Murphy, G. E. (1984). The process of change in cognitive therapy and pharmacotherapy for depression. Changes in mood and cognition. Archieves of General Psychiatry, 41, 45–51. [DOI] [PubMed] [Google Scholar]
- Singh, M. K., Kesler, S. R., Hosseini, S. M. H., Kelley, R. G., Amatya, D., Hamilton, J. P., … Gotlibb, I. H. (2013). Anomalous gray matter structural networks in major depressive disorder. Biological Psychiatry, 74(10), 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberg, J. M., Beall, E. B., Hulvershorn, L. A., Altinay, M., Karne, H., & Anand, A. (2016). Resting state brain network disturbances related to hypomania and depression in medication‐free bipolar disorder. Neuropsychopharmacology, 13(30), 3016–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz, S. H. (2001). Exploring complex networks. Nature, 410, 268–276. [DOI] [PubMed] [Google Scholar]
- Sun, F., Zhao, Z., Lan, M., Xu, Y., Huang, M., & Xu, D. (2020). Abnormal dynamic functional network connectivity of the mirror neuron system network and the mentalizing network in patients with adolescent onset, first‐episode, drug‐naïve schizophrenia. Neuroscience Research, 19, 30439–30440. [DOI] [PubMed] [Google Scholar]
- The‐ENIGMA‐MDD‐Working‐Group . (2017). Structural T1‐weighted brain magnetic resonance imaging (MRI) scans from 2148 MDD patients and 7957 healthy controls were analysed with harmonized protocols at 20 sites around the world. Molecular Psychiatry, 22, 900–909 Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva, O., Connolly, C. G., Ho, T. C., Sacchet, M. D., Blom, E. H., LeWinn, K. Z., … Yang, T. T. (2017). DTI‐based connectome analysis of adolescents with major depressive disorder reveals hypoconnectivity of the right caudate. Journal of Affective Disorders, 207, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasic, N., Walter, H., Höse, A., & Wolf, R. C. (2008). Gray matter reduction associated with psychopathology and cognitive dysfunction in unipolar depression: A voxel‐based morphometry study. Journal of Affective Disorders, 109(1–2), 107–116. [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, X., Xia, M., Liao, X., Evans, A., & He, Y. (2015). GRETNA: A graph theoretical network analysis toolbox for imaging connectomics. Frontiers in Human Neuroscience, 9, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Tao, F., Zuo, C., Kanji, M., Hu, M., & Wang, D. (2019). Disrupted resting frontal‐parietal attention network topology is associated with a clinical measure in children with attention‐deficit/hyperactivity disorder. Frontiers in Psychiatry, 10, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Yuan, Y., You, J., & Zhang, Z. (2019). Disrupted structural brain connectome underlying the cognitive deficits in remitted late‐onset depression. Brain Imaging and Behavior, 14(5), 1600–1611. [DOI] [PubMed] [Google Scholar]
- Watts, D. J., & Strogatz, S. H. (1998). Collective dynamics of ‘small‐world’ networks. Nature, 393(1998), 440–442. [DOI] [PubMed] [Google Scholar]
- Wehry, A. M., McNamara, R. K., Adler, C. M., Eliassen, J. C., Croarkin, P., Cerullo, M. A., … Strawn, J. R. (2015). Neurostructural impact of co‐occurring anxiety in pediatric patients with major depressive disorder: A voxel‐based morphometry study. Journal of Affective Disorders, 171, 54–59. [DOI] [PubMed] [Google Scholar]
- Wichers, M., Geschwind, N., van Os, J., & Peeters, F. (2010). Scars in depression: Is a conceptual shift necessary to solve the puzzle? Psychological Medicine, 40, 359–365. [DOI] [PubMed] [Google Scholar]
- Wise, C. D., Berger, B. D., & Stein, L. (1972). Benzodiazepines: Anxiety‐reducing activity by reduction of serotonin turnover in the brain. Science, 177(4044), 180–183. [DOI] [PubMed] [Google Scholar]
- Xia, M., Womer, F. Y., Chang, M., Zhu, Y., Zhou, Q., Edmiston, E. K., … Fei, W. (2019). Shared and distinct functional architectures of brain networks across psychiatric disorders. Schizophrenia Bulletin, 45(2), 450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, B. T. T., Krienen, F. M., Sepulcre, J., Sabuncu, M. R., Lashkari, D., Hollinshead, M., … Buckner, R. L. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106, 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., Linn, K. A., Shinohara, R. T., Oathes, D. J., Cook, P. A., Duprat, R., … Sheline, Y. I. (2019). Childhood trauma history is linked to abnormal brain connectivity in major depression. PNAS, 116(17), 8582–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zautra, A. J., Parrish, B. P., Van Puymbroeck, C. M., Tennen, H., Davis, M. C., Reich, J. W., & Irwin, M. (2007). Depression history, stress, and pain in rheumatoid arthritis patients. Journal of Behavioral Medicine, 30(3), 187–197. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Wang, J., Wu, Q., Kuang, W., Huang, X., He, Y., & Gong, Q. (2011). Disrupted brain connectivity networks in drug‐naive, first‐episode major depressive disorder. Biological Psychiatry, 70(4), 334–342. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., Li, X., Feng, G., Shen, Z., Li, S., Xu, Y., … Xu, D. (2018). Altered effective connectivity in the default network of the brains of first‐episode, drug‐naïve schizophrenia patients with auditory verbal hallucinations. Frontiers in Human Neuroscience, 12, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Huang, T., Tang, C., Ni, K., Pan, X., Yan, C., … Luo, Y. (2017). Altered resting‐state intra‐ and inter‐network functional connectivity in patients with persistent somatoform pain disorder. PLos ONE, 12(3), e0176494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., & Lui, Y. W. (2013). Small‐world properties in mild cognitive impairment and early Alzheimer's disease: A cortical thickness MRI study. ISRN Geriatrics, 2013, 542080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Yu, F., & Duong, T. (2014). Multiparametric MRI characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS One, 9(6), e90405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Zhuo, C., Liu, F., Qin, W., Xu, L., & Yu, C. (2016). Distinct disruptions of resting‐state functional brain networks in familial and sporadic schizophrenia. Scientific Reports, 6, 23577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available upon request. The data are not publicly available due to privacy or ethical restrictions.


