Abstract
The adverse effects of stress on brain and behavior have long been known and well-studied, with abundant evidence linking stress to, among other things, mood and anxiety disorders. Likewise, many have investigated potential treatments for stress-related mood and anxiety phenotypes and demonstrated good response to standard antidepressant medications like selective serotonin reuptake inhibitors (SSRIs), as well as environmental manipulations like exercise or enrichment. However, the extent to which stress and various treatments act on overlapping pathways in the brain is less well understood. Here, we used a widely studied social defeat stress paradigm to induce a robust depression- and anxiety-like phenotype and chronic corticosterone elevation that persisted for at least 4 weeks in wild type male mice. When mice were treated with either the SSRI fluoxetine or an enriched environment, both led to similar behavioral recovery from social defeat. We then focused on the amygdala and assessed the effects of social defeat, fluoxetine, and enrichment on 168 genes broadly related to synaptic plasticity or oxidative stress. We found 24 differentially expressed genes in response to social defeat stress. Interestingly, fluoxetine led to broad normalization of the stress-induced expression pattern while enrichment led to expression changes in a separate set of genes. Together, this study provides additional insight into the chronic effects of social defeat stress on behavior and gene expression in the amygdala. The findings also suggest that, for a subset of genes assessed, fluoxetine and environmental enrichment have strikingly divergent effects on expression in the amygdala, despite leading to similar behavioral outcomes.
Keywords: Stress, Gene expression, Amygdala, Fluoxetine, Environmental enrichment, Affective disorders
Highlights
-
•
Environmental enrichment and fluoxetine result in similar recovery from stress-induced depression and anxiety-like behavior.
-
•
In the amygdala, stress-related gene expression changes are broadly normalized by fluoxetine but not environmental enrichment.
-
•
Environmental enrichment and fluoxetine lead to distinct sets of non-overlapping gene expression changes in the amygdala.
-
•
Gene expression patterns may offer insights into complex mechanisms of stress-related psychiatric disorders and treatment.
1. Introduction
The adverse effects of stress on brain and behavior are well-known and well-studied with clear evidence linking stress exposure to a number of neuropsychiatric disorders. Among the most common are depression and anxiety disorders, which, collectively, represent some of the most burdensome of all diseases (Lopez and Murray, 1998; McEwen 2007, Whiteford et al., 2013). As such, uncovering the mechanisms of stress-related disorders and the development of treatment strategies have long been urgent priorities. For many years, our understanding of affective disorders was based on two prevailing and well supported hypotheses about pathogenesis, namely, disruption of monoamine balance and neurogenesis (Hindmarch 2001, Boku et al., 2018). Understanding of the action of existing therapies and the development of new treatment approaches, therefore, has remained largely anchored to these hypotheses. For example, selective serotonin reuptake inhibitors (SSRIs) thought to target the monoamine hypothesis (Owens, 2004), and exercise/environmental enrichment thought to enhance neuroplasticity (Hotting 2013) remain mainstays of treatment for depression and anxiety. However, the over simplicity of this model was recognized early on as some noted that, for example, SSRIs have more complex effects including neurogenic and anti-inflammatory properties (Tynan et al., 2012; Gałecki et al., 2018), while exercise has been associated with modulation of oxidative stress (Leeuwenburgh et al., 2001) as well as monoamines (Lin and Kuo, 2013), and environmental enrichment has been found to have markedly different effects on different brain regions (Smail et al., 2020). Furthermore, several medications with antidepressant properties, but limited or idiosyncratic effects on monoamine systems, were identified early on, and recent development of medications like ketamine and brexanolone reinforce the likelihood that: 1) the mechanisms leading to affective disorders are likely more complicated and varied than we know, and 2) there is good reason to investigate treatment approaches that look beyond canonical pathways (Cordner et al., 2020).
In recent years, there has been increasing attention to this issue of complexity in the pathogenesis of stress-related disorders. In human studies, for example, the most recent genome-wide association analysis of depression uncovered 44 loci related to 19 different biological pathways and associated with a number of other complex traits (Wray et al., 2018). Further, recent neuroimaging studies have uncovered functional connections between brain regions that predict mood disorder diagnoses and treatment response, but the connections suggest far more complex pathophysiology than is currently understood (Osuch et al., 2018). This work has been paralleled in rodent models, which have uncovered complexity in both cellular pathways and brain circuits that appear relevant to stress-induced phenotypes. For example, a growing body of work using the chronic social defeat (SD) stress paradigm in rodents, which reliably leads to depression-like and anxiety-like behavior, has strongly implicated the dopaminergic reward circuit in stress susceptibility as well as antidepressant response (Berton et al., 2006, Cao et al., 2010; Der-Avakian et al., 2014; Mul et al., 2018). The same model has been used to elucidate transcriptional and epigenetic effects of chronic stress in the brain, which appear to impact far more molecular pathways than was previously known (Krishnan et al., 2007; Covington et al., 2009; Wilkinson et al., 2009, Wilkinson et al., 2011; Hing et al., 2018).
Despite this growing interest in the complexity of stress and related neuropsychiatric disorders, less attention has been paid to potential complexity in the effects of treatment, though there are now several studies suggesting that such investigations may prove fruitful for both understanding mechanisms of existing therapies and elucidating potentially novel approaches. For example, in one such study focused on the nucleus accumbens, the tricyclic antidepressant imipramine reversed many changes in gene expression regulation induced by chronic SD, and the effects of imipramine overlapped with patterns of regulation found in stress-resistant mice (Wilkinson et al., 2009). In another focused on the nucleus accumbens, a histone deacetylase inhibitor and fluoxetine were shown to at least partially and similarly reverse gene expression changes induced by chronic SD (Covington et al., 2009). In a zebrafish model, fluoxetine has been found to alter the expression of hundreds of genes that can lead to long-lasting suppression of cortisol synthesis (Vera-Chang et al., 2018) and can at least partially explain the drug's side effects, as well as therapeutic mechanisms (Wong et al., 2013).
Here, we assess the effects of chronic SD and treatment with either fluoxetine or environmental enrichment on behavior, plasma corticosterone levels, and the expression of 168 genes broadly related to synaptic plasticity or oxidative stress. For the gene expression analyses, we focused on the amygdala, which is highly relevant for stress-related psychiatric disorders (Campeau and Davis, 1995; Ashokan et al., 2016; Novaes et al., 2017), but less well studied than other brain regions like the nucleus accumbens and hippocampus. In particular, many have shown that chronic stress leads to persistent changes in neuronal activity, excitability, proliferation, and dendritic morphology in the amygdala (Vyas et al., 2002; McEwen 2007, Rosenkranz et al., 2010; Hing et al., 2014; Smail et al., 2020) which is in contrast with other brain regions, like the hippocampus and prefrontal cortex, which tend to undergo some degree of recovery after removal of stressors (Vyas et al., 2002). We found that SD induced a robust depression-like and anxiety-like phenotype and elevation of baseline corticosterone that persisted for at least 4 weeks after stress. Fluoxetine and enrichment resulted in similar recovery from the stress-induced phenotype, but led to markedly different patterns of gene expression in the amygdala.
2. Materials & methods
2.1. Animals
A total of 80 male C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) and 44 male CD-1 retired breeder mice (Charles River, Raleigh, NC) were used in this study. At the start of each experiment, C57BL/6J mice were 8–10 weeks old and CD-1 mice were 16–32 weeks old. Throughout the study, all mice were maintained on a 12h:12h light-dark cycle with ad libitum access to water and standard chow (Teklad Global 2018 diet, Envigo). Mice were randomly assigned to experimental groups. All protocols were approved by the Animal Care and Use Committee of the Johns Hopkins University School of Medicine and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2. Chronic social defeat (SD) stress
SD stress was conducted according to a previously published protocol (Golden et al., 2011; Hing et al., 2018). Briefly, C57BL/6J mice assigned to SD were placed in the home cage of a different CD-1 aggressor each day for 14 days. Immediately after being moved to a different aggressor's cage, each SD mouse and aggressor pair were allowed to directly interact for 10 min. Mice were then separated by a clear, ventilated barrier that allowed visual, auditory and olfactory stimulation, but no physical contact, and they remained co-housed for the next 24 h. Unstressed control (Ctrl) mice were co-housed in pairs. Each pair was separated by a clear, ventilated barrier.
2.3. Treatment
In the first cohort, mice were exposed to Ctrl conditions or SD (n = 16/group) for 14 days. Thereafter, half of the mice in each group were given access to environmental enrichment (EE) or remained in their standard home cage (n = 8/group) for 28 days prior to tissue collection. In a second cohort, mice were exposed to Ctrl conditions or SD (n = 16/group) for 14 days. Thereafter, half of the mice in each group were treated with fluoxetine (fluox) or placebo (n = 8/group) for 28 days prior to tissue collection. During the 28-day treatment phase, all mice remained individually housed.
2.3.1. Environmental enrichment (EE)
EE included a larger tub cage (104 × 56 × 48 cm) filled with extra cob bedding, nesting sheets, and polycarbonate tunnels, balls, and housing domes (Bio-Serv, Frenchtown, NJ), consistent with a previously published protocol (Cordner and Tamashiro., 2016). All other C57BL/6J mice were housed in standard cages. This resulted in four groups: Ctrl without EE (Ctrl - EE, n = 8), Ctrl with EE (Ctrl + EE, n = 8), SD without EE (SD - EE, n = 8), and SD with EE (SD + EE, n = 8).
2.3.2. Fluoxetine (fluox)
Fluoxetine hydrochloride pellets (14 mg/pellet, 28 day sustained release) or inactive placebo pellets were implanted subcutaneously in the left lateral neck according to the manufacturer's protocol (Innovative Research of America, Sarasota, FL). This method of delivery of fluoxetine provided a dose of 17–21 mg/kg/day and an average dose of 19.8 mg/kg/day, which is consistent with other mouse studies (Covington et al., 2009; Kitahara et al., 2016). This resulted in four groups: Ctrl treated with placebo (Ctrl placebo, n = 8), Ctrl treated with fluoxetine (Ctrl fluox, n = 8), SD treated with placebo (SD placebo, n = 8), and SD treated with fluoxetine (SD fluox, n = 8).
2.4. Behavior
Behavioral testing was conducted on all mice between day 21 and 28 of treatment. No more than one behavioral assay was conducted each day. Tests were conducted in the following order: an open field test was conducted on treatment day 21, an elevated plus maze test was conducted on treatment day 22, a sucrose preference test was conducted from treatment day 23–26, and a forced swim test was conducted on treatment day 28. The open field, elevated plus maze, and forced swim tests were conducted during the middle of the light phase.
2.4.1. Open field
The open field consisted of an opaque plastic box (60 cm square chamber, 60 cm high walls) with a central zone (circle with a 35 cm diameter). Each mouse was allowed to freely explore the open field for 10 min. Horizontal activity was recorded by a computerized detection system (Omnitech Electronics, Columbus, OH).
2.4.2. Elevated plus maze
The elevated plus maze consisted of a plastic platform with four arms (10 cm × 50 cm) joined by a square intersection (10 cm × 10 cm) to form a ‘+’ shape that was elevated 50 cm above the ground. Two opposing arms had walls that were 30 cm high. The remaining two arms were open. Each mouse was placed at the center of the maze facing an open arm and allowed to freely explore for 5 min. Behavior was recorded by a digital camera and later coded by 2 blinded observers.
2.4.3. Sucrose preference test
Mice were habituated to a 1% w/v sucrose solution for 3 h on two consecutive days before the start of the test. During the preference test, mice were given ad libitum access to two bottles: one containing water, and one containing a 1% w/v sucrose solution. The test began immediately prior to onset of the dark period. Intake was measured after 12 and 24 h of access and a preference ratio was calculated.
2.4.4. Forced swim test
Each mouse was placed in a cylinder filled with water (22 cm diameter, 17 cm water height, 22 °C water temperature) for 6 min. The first 2 min were treated as a habituation period and only the last 4 min were scored. Behavior was recorded by a digital camera and later coded by 2 blinded observers.
2.5. Tissue collection
Three days after behavioral testing, mice were deeply anesthetized with isoflurane, retro-orbital blood was collected and all mice were killed by rapid decapitation. Brains were removed, immediately frozen on powdered dry ice and stored at −80°C. The amygdala was then dissected from 200 μm coronal slices. The following coordinates were used to target dissection of the basolateral complex, which has previously been implicated in affective disorders, fear learning, and the stress response (Campeau and Davis, 1995; Herringa et al., 2004; Ashokan et al., 2016; Novaes et al., 2017): Bregma −0.94 mm to −1.74 mm; dorsal/ventral 4.5–5 mm where the dorsal/ventral position of Bregma is defined as 0 mm; medial/lateral 2.5 to 3.25 in both the right and left hemisphere where the midline is defined as 0 mm (Paxinos and Franklin, 2004). The tissue was then stored at −80°C.
2.6. Plasma corticosterone
Plasma was isolated from retro-orbital blood and corticosterone (Cort) concentration was determined by radioimmunoassay according to the manufacturer's protocol (MP Biomedicals, Solon, OH).
2.7. Gene expression
RNA from the amygdala was extracted using the RNeasy Mini Kit with on-column DNase digestion (Qiagen, Valencia, CA). RNA quality and concentration were evaluated by electrophoresis (TapeStation, Agilent, CA). All samples had an RIN of 8.0 or greater. cDNA was generated using the RT2 First Strand Kit (Qiagen, Valencia, CA). Quantitative real-time PCR reactions were carried out using the RT2 Profiler PCR Array Mouse Oxidative Stress and Antioxidant Defense and the RT2 Profiler PCR Array Mouse Synaptic Plasticity with RT2 SYBR Green qPCR Mastermix, all according to the manufacturer's protocols (Qiagen, Valencia, CA). Each array assayed a set of 84 genes relevant to the respective pathway (Supplemental Table 1). Expression levels relative to the average of 5 housekeeping genes- Actb, B2m, Gapdh, Gusb, and Hsp90ab1-were determined by the -ΔΔCt method.
2.7.1. Validation of top gene expression findings
Validation was performed on the top 10 differentially expressed genes (DEGs), ranked by p-value, from the combined oxidative stress and synaptic plasticity data set. Validation was carried out in triplicate using RT2 SYBR Green qPCR Mastermix andRT2 qPCR Primer Assays against each candidate gene (Qiagen, Valencia, CA). For validation, expression levels relative to Actb were determined by the -ΔΔCt method.
2.7.2. Replication of top gene expression findings
A separate set of mice were used to replicate top gene expression findings from the initial arrays. Mice were exposed to control conditions (Ctrl, n = 4), social defeat (SD, n = 4), social defeat followed by enrichment (SD + EE, n = 4), or social defeat followed by fluoxetine (SD fluox, n = 4) as described above. Behaviors were not assessed in these mice to eliminate potential confounds introduced by behavioral testing. Instead, after 28 days of treatment, amygdala tissue was collected, RNA was isolated, and cDNA was generated as described above.
Replication was performed on the top 10 differentially expressed genes (DEGs), ranked by p-value, from the combined oxidative stress and synaptic plasticity data set. Replication was carried out in triplicate using RT2 SYBR Green qPCR Mastermix andRT2 qPCR Primer Assays against each candidate gene (Qiagen, Valencia, CA). Expression levels relative to Actb were determined by the -ΔΔCt method.
2.8. Statistical analysis
Statistical analyses were completed using Prism 8.0 (GraphPad Software, San Diego, CA) and GeneGlobe (Qiagen, Valencia, CA). For behavioral tests and Cort, differences between groups were assessed by two-way ANOVA followed by Tukey post hoc analysis to assess for ‘stress,’ ‘treatment,’ and ‘stress’ x ‘treatment’ interactions with p < 0.05 considered significant. For each gene assessed by expression array, differences between groups were assessed by one-way ANOVA; a significance threshold of p < 0.0006 was used to account for multiple comparisons. For each gene that met this threshold, Dunnett's Test was then used to compare each group to Ctrl with p < 0.05 considered significant. For validation and replication steps, differences between groups were assessed by one-way ANOVA followed by Dunnett's Test to compare each group to Ctrl, with p < 0.05 considered significant.
3. Results
3.1. Social defeat stress resulted in a phenotype that persisted for at least 28 days, but could be reversed by environmental enrichment
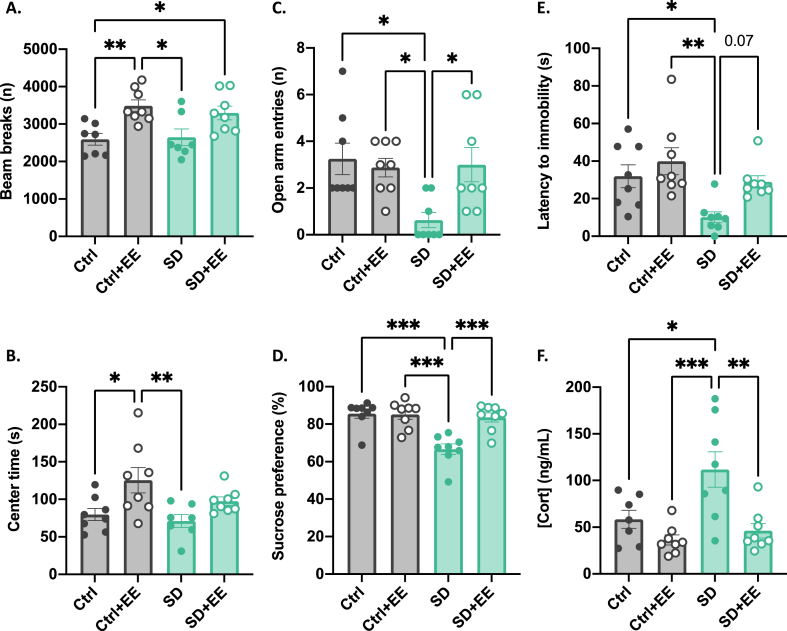
Behavioral phenotyping was conducted between recovery day 21 and 28. In the open field, we found no effect of SD. There was, however, an overall effect of EE such that mice with access to enrichment had greater total activity (EE: F(1, 27) = 18.24, p = 0.0002) (Fig. 1A) and spent more time in the center zone (EE: F(1, 27) = 10.94, p = 0.003) (Fig. 1B) suggesting that EE exposure alone may increase exploratory behavior independent of past stress exposure.
Fig. 1.
Effects of social defeat stress and environmental enrichment on behavior and plasma corticosterone.
In the open field, SD had no effect on exploratory behavior but EE increased total activity (A) and time in the center zone (B). In the elevated plus maze, SD resulted in decreased exploration of the open arms (C). SD also decreased sucrose preference (D), decreased latency to immobility in the forced swim test (E), and increased plasma Cort (F). All of the stress-induced changes were normalized by treatment with EE. Graphs indicate mean ± standard error of the mean. *p < 0.05, **<0.01, ***<0.001.
In the elevated plus maze, there was an overall effect of SD and an interaction between SD and EE on the number of entries into the open arm such that the SD-EE group made fewer entries into the open arm, while the SD + EE group was indistinguishable from controls (SD: F(1, 28) = 4.98, p = 0.03; SD*EE: F(1, 28) = 6.03, p = 0.02) (Fig. 1C).
In the sucrose preference test, there was an overall effect of SD, and an interaction between SD and EE: the SD-EE group displayed decreased preference for a sucrose solution, while the SD + EE group was indistinguishable from controls (SD: F(1, 28) = 15.70, p = 0.0005; SD*EE: F(1, 28) = 11.26, p = 0.002) (Fig. 1D).
Likewise, in the forced swim test, we found an overall effect of SD and an interaction between SD and EE on latency to immobility (SD: F(1, 28) = 10.08, p = 0.004; SD*EE: F(1, 28) = 5.26, p = 0.01) (Fig. 1E). We observed that the SD-EE group displayed increased immobile behavior while the SD + EE group was indistinguishable from controls.
When plasma Cort was measured after 28 days of recovery, we found an overall effect of SD and an interaction between SD and EE (SD: F(1, 27) = 13.69, p = 0.001; SD*EE: F(1, 27) = 7.14, p = 0.01) (Fig. 1F), with Cort being highest in the SD-EE group, while Cort levels in the SD + EE group were indistinguishable from controls.
3.2. The social defeat-induced phenotype was also recovered by fluoxetine
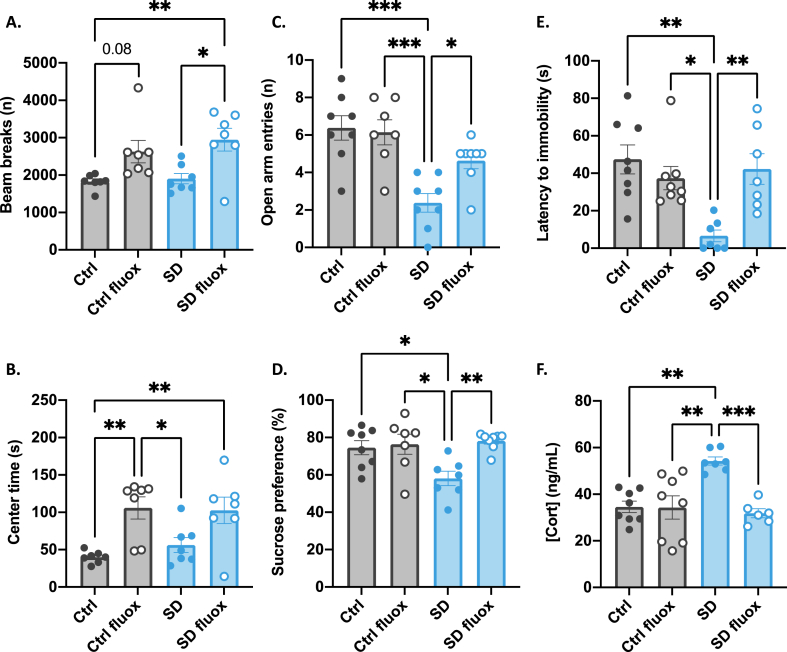
As in the first cohort, behavioral phenotyping was conducted between recovery day 21 and 28. In the open field, we again found no effect of SD. There was, however, an overall effect of fluox such that mice treated with fluox had greater total activity (fluox: F(1, 28) = 21.17, p < 0.0001) (Fig. 2A) and spent more time in the center zone (fluox: F(1, 28) = 19.73, p = 0.0002) (Fig. 2B) suggesting that, like EE, fluox exposure alone may increase exploratory behavior independent of past stress exposure.
Fig. 2.
Effects of social defeat stress and fluoxetine on behavior and plasma corticosterone.
In the open field, SD had no effect on exploratory behavior but fluox increased total activity (A) and time in the center zone (B). In the elevated plus maze, SD resulted in decreased exploration of the open arms (C). SD also decreased sucrose preference (D), decreased latency to immobility in the forced swim test (E), and increased plasma Cort (F). All of the stress-induced changes were normalized by treatment with fluox. Graphs indicate mean ± standard error of the mean. *p < 0.05, **<0.01, ***<0.001.
In the elevated plus maze, there was an overall effect of SD and an interaction between SD and fluox on the number of entries into the open arm. The SD placebo group made fewer entries into the open arm, while the SD fluox group was indistinguishable from controls (SD: F(1, 27) = 23.88, p < 0.0001; SD*fluox: F(1, 27) = 4.83, p = 0.04) (Fig. 2C).
In the sucrose preference test, there was an overall effect of SD, and an interaction between SD and fluox, with the SD placebo group displaying decreased preference for a 1% sucrose solution, while the SD fluox group was indistinguishable from controls (SD: F(1, 26) = 8.35, p = 0.008; SD*fluox: F(1, 26) = 5.81, p = 0.02) (Fig. 2D).
Likewise, in the forced swim test, we found an overall effect of SD and an interaction between SD and fluox on latency to immobility (SD: F(1, 26) = 7.22, p = 0.01; SD*fluox: F(1, 26) = 11.68, p = 0.002) (Fig. 2E). The SD placebo group displayed increased immobile behavior while the SD fluox group was indistinguishable from controls.
When plasma Cort was measured after 28 days of recovery, we found an overall effect of SD and an interaction between SD and fluox (SD: F(1, 26) = 6.74, p = 0.02; SD*fluox: F(1, 26) = 11.1, p = 0.003) (Fig. 2F) such that Cort was highest in the SD placebo group while Cort levels in the SD fluox group was indistinguishable from controls.
Taken together, these data extend the findings of other studies using similar stress models by suggesting that the depression-like and anxiety-like phenotype and HPA axis hyperactivity induced by social defeat stress persists for at least 28 days after last stress exposure. However, the phenotype is reversed by access to either EE or fluoxetine.
3.3. In the amygdala, stress-related changes in gene expression are broadly normalized by fluoxetine, but not environmental enrichment
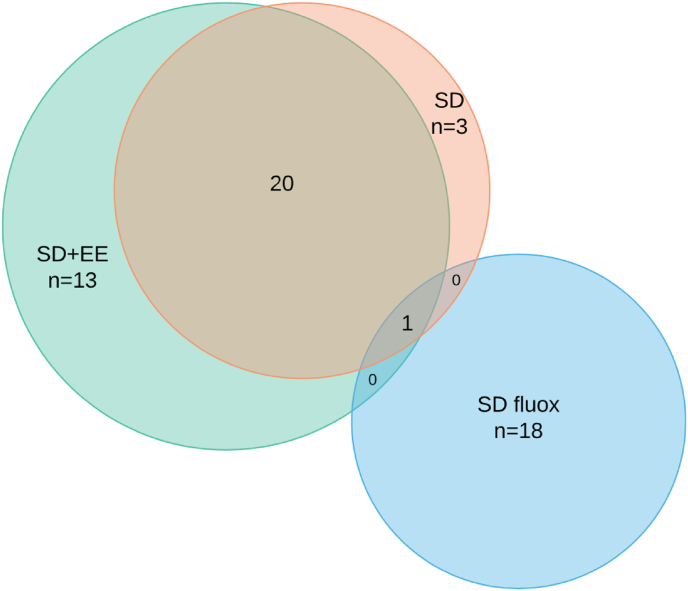
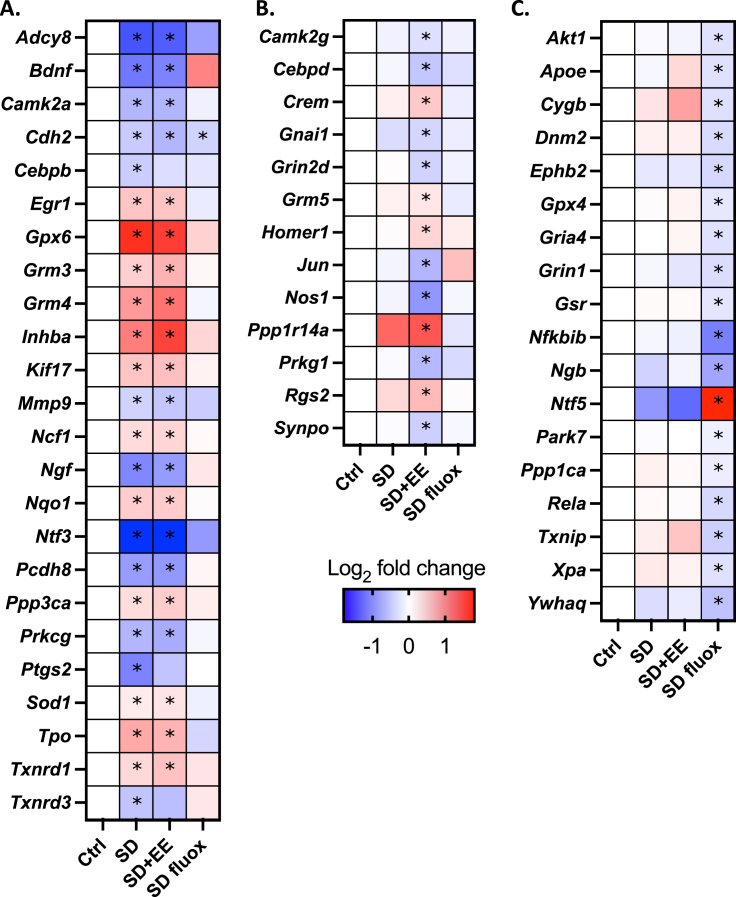
Expression of 168 genes related to synaptic plasticity or oxidative stress was then assessed in the amygdala. Among mice exposed to SD, we found a set of 24 genes that were differentially expressed compared to controls after accounting for multiple comparisons (Fig. 3) (Fig. 4A). Of those 24 genes, 23 had expression levels in the SD fluox group that were not significantly different than controls, that is, they were normalized by treatment with fluox. By contrast, only 3 of the 24 genes in the SD EE group had expression levels that were not significantly different than controls, that is., were normalized by exposure to the EE, while the remaining 21 showed the same expression change as in the SD group. A separate set of 13 genes were differentially expressed among SD + EE mice compared to controls, and a non-overlapping set of 18 genes were differentially expressed among SD fluox mice compared to controls (Fig. 3) (Fig. 4B and C).
Fig. 3.
Differential gene expression in the amygdala.
Of 168 genes assessed in the amygdala, SD resulted in differential expression of 24. Of those, 23 were normalized by treatment with fluox and 3 were normalized by treatment with EE. EE and fluox also led to separate, non-overlapping sets of 13 and 18 DEGs, respectively.
Fig. 4.
Differentially expressed genes in response to SD and treatment with EE or fluox.
24 genes were differentially expressed in response to SD, of which 23 were normalized by treatment with fluox and 3 were normalized by treatment with EE (A). 13 genes were differentially expressed only in response to treatment with EE (B), and a non-overlapping set of 18 genes were differentially expressed only in response to treatment with fluox (C). Within each set, genes are listed alphabetically. Data expressed as a heatmap indicating log2 of the fold change relative to Ctrl. *p < 0.05 relative to Ctrl.
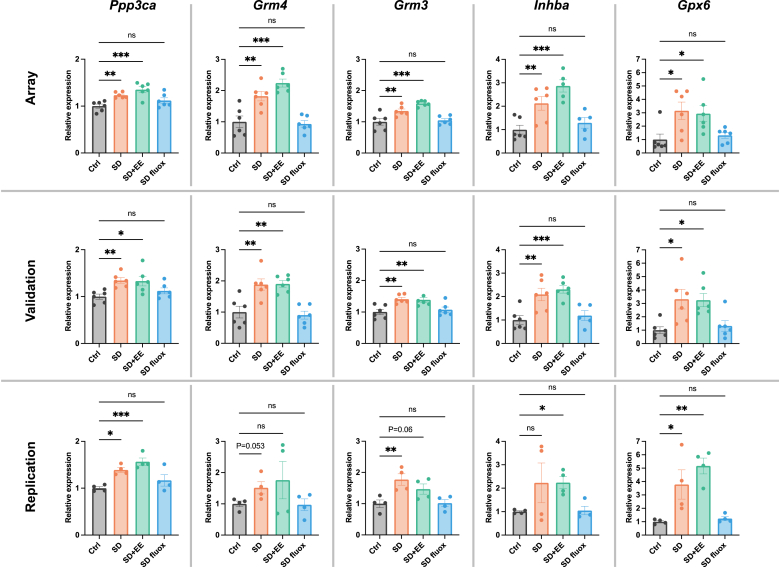
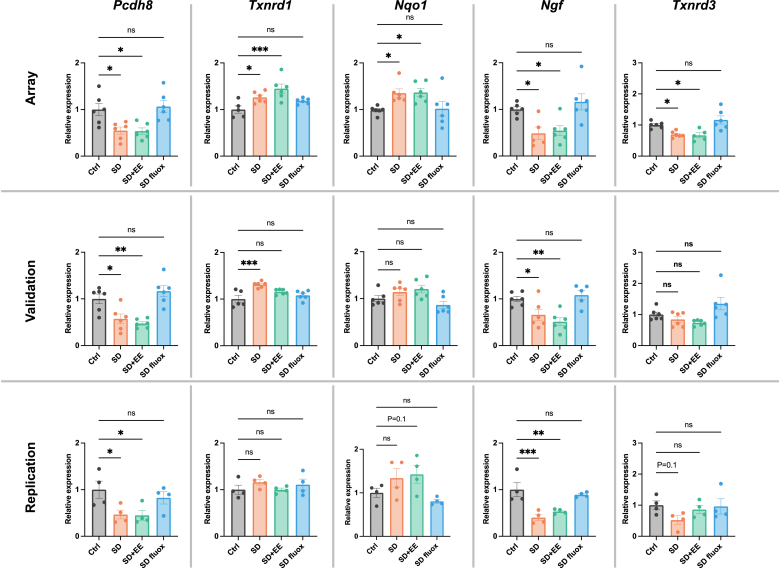
Validation was then performed on the top 10 differentially expressed genes, ranked by p-value. Expression levels of Ppp3ca, Grm4, Grm3, Inhba, Gpx6, Pcdh8, Txnrd1, Nqo1, Ngf, and Txnrd3 were again assessed in the amygdala. The expression differences were validated in 8 of 10 genes. Only Nqo1, and Txnrd3 failed to validate (Fig. 5). Validation generally confirmed the pattern of fluoxetine, but not EE, having the effect of normalizing stress-related gene expression changes.
Fig. 5.
Validation and replication of differential gene expression.
Validation was performed on the top 10 DEGs, ranked by p-value. The expression patterns of Ppp3ca, Grm4, Grm3, Inhba, Gpx6, Pcdh8, Txnrd1, and Ngf (the 1st through 7th and 9th ranked DEGs) were consistent with array data. The expression patterns of Nqo1 and Txnrd3 (the 8th and 10th ranked DEGs) failed to validate findings from the array, though the trends were similar. Replication attempted using amygdala tissue from a separate set of mice revealed similar patterns of gene expression except for Txnrd1, where no significant differences or trends were found. Graphs indicate mean ± standard error of the mean. *p < 0.05, **<0.01, ***<0.001.
Replication of the top 10 differentially expressed genes ranked by p-value was also attempted using a separate set of mice that were not exposed to behavioral testing. Expression levels of Ppp3ca, Grm4, Grm3, Inhba, Gpx6, Pcdh8, Txnrd1, Nqo1, Ngf, and Txnrd3 were assessed in the amygdala. Statistically significant expression differences were replicated for Ppp3ca, Grm3, Inhba, Gpx6, Pcdh8, and Ngf. Non-significant trends resembling the expression patterns from the initial array were found for Grm4, Nqo1, and Txnrd3 (Fig. 5). Broadly, the pattern of fluoxetine, but not EE, having the effect of normalizing stress-related gene expression changes was replicated.
4. Discussion
In this study, we first assessed effects of two distinctly different treatment approaches – one behaviorally-based (environmental enrichment) and the second a pharmacological treatment (the SSRI fluoxetine) – on a chronic social stress-related phenotype. We found that 2 weeks of social stress resulted in depression-like and anxiety-like behaviors and was associated with elevated basal plasma Cort that persisted for at least 4 weeks after the withdrawal of stressors. This is consistent with other studies using a similar chronic social stress model (Martinez et al., 1998; Krishnan et al., 2007), though the long-term persistence of the phenotype has not been as well-characterized. When mice were treated with either environmental enrichment or fluoxetine for 4 weeks after stress, both treatments resulted in similar normalization of behavior and plasma Cort levels.
Regarding the effects of enrichment, another study found that socially defeated mice housed in an enriched environment display less depression- and anxiety-like behaviors than mice housed in an impoverished environment, though unstressed controls were not included for comparison (Schloesser et al., 2010). The same group also found that exposure to enrichment prior to SD confers resilience to the effects of stress (Lehmann and Herkenham, 2011). Enrichment has also been previously shown to decrease baseline stress hormone levels among unstressed rats (Belz et al., 2003) and, consistent with our findings, reverse HPA-axis hyperactivity induced by early life stress (Francis et al., 2002; Morley‐Fletcher et al., 2003). Like enrichment, fluoxetine has been shown to reverse the behavioral and endocrine effects of social stress in mice (Beitia 2005, Razzoli et al., 2011), though at least one study has shown no effect of treatment with fluoxetine alone (Ma et al., 2016).
We next assessed the effects of stress and treatment on the expression of candidate genes related to synaptic plasticity and oxidative stress in the amygdala, an area with high relevance for stress-related psychiatric disorders, though one that has been less well studied than others like the hippocampus, prefrontal cortex, and reward circuits. We focused on the amygdala based on well-supported observations that chronic stress results in persistent changes in dendritic morphology accompanied by increased neuronal activity, excitability, and proliferation in this brain region (Vyas et al., 2002; McEwen 2007, Rosenkranz et al., 2010; Hing et al., 2014; Smail et al., 2020). These changes, and the amygdala's resistance to recovery from stress, offer interesting contrasts to other brain regions, like the hippocampus and prefrontal cortex, that demonstrate some degree of recovery after removal of stressors (Vyas et al., 2002). Thus, examination of transcriptional changes in the amygdala could provide novel insights into how chronic stress has persistent negative effects on affective behavior and help to elucidate mechanisms through which behavioral or pharmacological treatment exerts therapeutic effects.
We focused on a set of 168 candidate genes that are broadly involved in synaptic function or oxidative stress and found that chronic social stress led to differential expression of 24 genes. Closer inspection shows that these genes are involved in diverse functions and signaling pathways, and may provide some insight into the complexity of the cellular response to chronic stress. For example, stress exposure was associated with down regulation of the neurotrophins Bdnf and Ngf, which have previously been implicated in stress-related affective disorders and the mechanisms of antidepressants (Mondal and Fatima, 2019). Though far better studied in the cortex and hippocampus, a few prior studies have also found decreased NGF (von Richthofen et al., 2003) and BDNF (Reus 2013) in the amygdala in response to stress. Additionally, we found upregulation of genes coding for the group II/III metabotropic glutamate receptors Grm3 and Grm4, which are thought to have primarily inhibitory roles (Krystal et al., 2010). Several lines of evidence potentially link these changes to depression- and anxiety-like phenotypes. Specifically, group II metabotropic glutamate receptor antagonists have been shown to exert anxiolytic effects in an outbred mouse model (Shimazaki et al., 2004) as well as anxiolytic and antidepressant effects in a rat model of learned helplessness (Yoshimizu et al., 2006). In a human study, GRM4 was upregulated among depressed subjects, and this effect was moderated by antidepressant treatment (Lopez et al., 2014). Another postmortem study also found increased expression of GRM4, as well as several other glutamate receptors, in the cortex of depressed female subjects (Gray et al., 2015). That said, at least one study in mice exposed to chronic restraint stress found decreased expression of Grm3 and Grm4 in the hippocampus (Sathyanesan et al., 2017), suggesting potential brain region-specific or stress-paradigm-specific effects.
In our chronic stress model, we also found increased expression of the immediate early gene Egr1, which codes for the early growth response protein 1, also commonly referred to as nerve growth factor-induced protein A (NGF-IA) or zing finger protein 268 (ZNF268). EGR1 is thought to be broadly critical for post-natal epigenetic regulation of gene transcription by direct recruitment of the demethylase, TET1 (Sun et al., 2019). Stimulated by a range of factors including neuronal activity and stress, Egr1 expression is usually increased transiently and is thought to ultimately play critical roles in consolidation of fear-related memories (Revest et al., 2010) as well as maintenance of long-term potentiation (Jones et al., 2001). The mechanisms through which Egr1 acts in response to stress are complex, but include epigenetic remodeling, direct binding of regulatory elements, and interactions with other transcription factors (Dulcot et al., 2017). Interestingly, EGR1 appears to directly regulate transcription of Nr3c1, the gene coding for the glucocorticoid receptor (Moser et al., 2007; Vandevyver et al., 2014; Weaver et al., 2014), while activation of the glucocorticoid receptor by stress hormone binding leads to upregulation of Egr1 (Revest et al., 2005, Revest et al., 2010). While acute stress exposure robustly increases Egr1 throughout the brain, it is interesting to note that chronic stress and depression are associated with decreased Egr1 in the cortex as well as the hippocampus and these changes have been subsequently associated with impaired learning and synaptic plasticity (Duclot et al., 2017). In the amygdala, one study found enhanced long-term fear memory as well as persistently increased expression of Egr1 and another immediate early gene, Arc, among rats exposed to chronic corticosterone (Monsey et al., 2014). Generally, little work has been done to investigate the long-term trajectory of Egr1 expression in the amygdala and its potential role in perpetuating stress-induced affective phenotypes. However, complementary data from Egr1 knock out mice, which have impaired long term memory (Jones et al., 2001) as well as decreased anxiety-like behaviors (Ko 2005), and mice overexpressing Egr1, which have enhanced spatial learning (Penke et al., 2014) and reduced extinction of aversive learning (Baumgartel et al., 2008), suggest that such studies may prove useful. Ultimately, though much work remains to be done to validate and expand upon the changes we observed, the current findings may provide new insights into the complex transcriptional response to chronic stress and how different therapeutic approaches can have varied influence on gene expression, but lead to the same behavioral consequence.
When we then assessed the effects of two different treatment approaches, fluoxetine and environmental enrichment, on stress-induced gene expression changes in the amygdala, strikingly different transcriptional patterns emerged despite similar effects on behavior. Fluoxetine normalized 23 out of 24 genes that were differentially expressed in response to stress, whereas enrichment normalized only 3 of 24 in the amygdala. Regarding the broadly normalizing effects of fluoxetine treatment following chronic defeat stress, others have reported a similar pattern in the nucleus accumbens (Covington et al., 2009). In the hippocampus, the SSRI paroxetine normalized the behavior and broadly altered gene expression in an inbred mouse model with high anxiety-like behaviors (Sillaber et al., 2008). Though limited work has been done in the amygdala, one study focused on immediate early gene expression found that fluoxetine normalizes changes induced by chronic corticosterone exposure, including normalization of Egr1 (Monsey et al., 2014), which is consistent with our findings. Less work has been done to assess effects of environmental enrichment on gene expression in the context of stress, or in translational models for affective disorders. Two studies focused on individual gene products in the rat amygdala have found that enrichment can normalize chronic restraint stress-induced reduction in Bdnf (Ashokan et al., 2016), and can prevent acute restraint stress-induced increase in Egr1 (Novaes et al., 2017). In our study in mice, which used different stress and enrichment paradigms, we found stress-related changes in Bdnf and Egr1, but neither were normalized by enrichment.
Apart from the normalizing effects of treatment, we separately found that 18 genes were differentially expressed in response to fluoxetine, and 13 genes were differentially expressed in response to environmental enrichment, but were not affected by stress. Surprisingly, these sets of 18 and 13 genes were non-overlapping. Thus, these gene expression changes may reflect unique actions of fluoxetine and environmental enrichment treatment that are separate from any stress-normalizing effects. However, it is not currently known whether any of these changes cause alterations in behavior or represent unrelated, off-target effects of treatment. Regardless, these changes also suggest a substantial degree of complexity and diversity in the transcriptional response to fluoxetine and environmental enrichment, despite similar behavioral outcomes. In particular, we found that fluoxetine decreased expression of several genes whose products are closely interconnected to the AKT/protein kinase B pathway including Akt1 itself as well as two elements of the NFKB complex, Rela, and Nfkbib. We also found decreased expression of an NMDA receptor subunit Grin1 and an AMPA receptor subunit Gria4 that are both activated by the AKT pathway (Leonard and Hell, 1997; Nuriya et al., 2005) and involved in synaptic plasticity and memory through their function at excitatory glutamatergic synapses (Chen et al., 2017; Martin et al., 2017). Though only speculative, one potential outcome of these changes could be modulation of neuronal hyperactivity and proliferation in the amygdala following chronic stress. Intriguingly, while this may seem counter to their often-cited neurogenic effects in the hippocampus (Malberg, 2000), the possibility that SSRIs may have varying effects on the AKT pathway is supported by studies suggesting that AKT signaling is activated by fluoxetine in a neuron-like pheochromacytoma cell line (Zeng et al., 2016), but suppressed by fluoxetine in liver (Yang et al., 2020). In astrocytes cultured from mouse cortex, fluoxetine increased AKT activity at low dose but inhibited activity at high dose (Bai et al., 2017), further supporting the possibility that SSRI effects on the AKT pathway are robust and context dependent. Environmental enrichment, on the other hand, altered several other distinct pathways. For example, treatment with environmental enrichment increased expression of the excitatory glutamate receptor Grm5 and the gene coding for its scaffolding protein, Homer. Additionally, environmental enrichment decreased neuronal nitric oxide synthase Nos1 and the transcription factor Jun, which is upregulated by Nos1 (Srivastava et al., 2017) and plays critical roles in cell cycle progression, inhibition of apoptosis, and regulation of inflammatory states (Wisdom et al., 1999). Perhaps related, environmental enrichment also led to decreased expression of the transcription factor Cebpd, which is itself upregulated by Jun (Liu et al., 2007) and glucocorticoids, and plays important roles in cellular differentiation and pro-inflammatory responses (Ko et al., 2015). A relatively small number of studies have assessed the transcriptional effects of environmental enrichment on more than single genes of interest, and fewer still have focused on models of stress or affective disorders. In one, treatment of Bdnf-deficient mice with environmental enrichment led to numerous changes in the expression of neurotransmitter receptors in the cortex and hippocampus, including several glutamate receptors, though Grm5 was not specifically changed (Dong et al., 2020). Two other studies have assessed broad transcriptional effects of environmental enrichment in naïve, wild type rodents. In the first, which used cortical tissue from mice, environmental enrichment lasting between 3 h and 14 days altered the expression of approximately 100 transcripts involved in diverse processes including RNA synthesis, protein processing, synaptic signaling, and neuronal structure. More specifically, 14 days of environmental enrichment was found to decrease expression of the cell cycle regulator C-fos, while also altering several genes involved in glutamatergic signaling and nitric oxide synthesis (Rampon et al., 2000), which offer some correlations with our findings. Another study focused on rat hippocampus and sensorimotor cortex found that 2 weeks of environmental enrichment altered similar, and similarly diverse pathways (Keyvani 2004). Finally, with regard to the lack of overlap between genes differentially expressed by fluoxetine and environmental enrichment, our findings appear novel as direct comparisons of these two treatments have not been done. However, one study did find that treating unstressed rats with fluoxetine, the tricyclic antidepressant amitriptyline, or the neurotrophin NGF led to largely non-overlapping expression in amygdala and hippocampal tissue (McGeary et al., 2011). Another found that treating socially defeated mice with fluoxetine or the histone deacetylase inhibitor MS-275 led to similar behavioral response, and similar normalization of stress-induced gene expression changes, but also a large number of non-overlapping gene expression changes in the nucleus accumbens (Covington et al., 2009). These studies, along with data presented here, clearly suggest that directly comparing the actions of different treatments may provide unique insights into the mechanisms of existing therapies and help identify potentially novel targets.
Taken together, the data presented here suggest that chronic social stress induces long-lasting depressive and anxiety-like behaviors that are reversed by treatment with either fluoxetine or environmental enrichment. In the amygdala, which is less well studied in models for affective disorders than several other brain regions, chronic stress leads to complex changes in the expression of several genes broadly related to synaptic function or oxidative stress, and some of these changes implicate pathways that are understudied in response to stress. When treated after stress exposure, fluoxetine, but not environmental enrichment broadly normalizes these gene expression changes. Furthermore, both fluoxetine and enrichment lead to separate sets of non-overlapping expression changes that point to several pathways whose involvement in antidepressant treatment responses are not well understood.
There are several limitations that should be acknowledged. First, we chose the social defeat stress model given its wide use and robust, reproducible phenotype. However, the standard protocol can only reliably be applied to males, which presents a substantial limitation. Future studies should utilize alternative stress paradigms to assess for sex differences. Regarding gene expression, we chose to focus on a set of genes that, while reflecting a broad range of synaptic and antioxidant functions, is still narrowly targeted, thus largely limiting our analysis and potential findings to pathways whose roles in brain function are mostly well described. Given the unexpected differential effects of stress and treatment reported here, future work utilizing unbiased approaches is warranted. Further, it is important to acknowledge that the expression results presented here represent just a starting point in that they identify specific genes and implicate several pathways whose roles in stress neurobiology, affective phenotypes, and antidepressant response are currently poorly understood. However, additional studies involving targeted manipulation of these genes and pathways are required to determine the extent to which they are causally involved in the behavioral effects of stress and treatment reported here. Regarding treatments, we chose fluoxetine and environmental enrichment given that they represent distinctly different approaches, but result in similar behavioral effects. Considering that we found nearly non-overlapping changes in the amygdala in response to fluoxetine and enrichment, future work may seek to identify specific cellular populations responsible for these differences, as the current study was limited to analysis of tissue with heterogenous cell composition dissected from the basolateral complex. Moreover, evaluating the effects of fluoxetine and enrichment on unstressed brains represents an important future direction as the current study was not designed to assess stress-by-treatment interactions on gene expression, which is a limitation. Additionally, comparing these findings to other brain regions and other treatment approaches - including novel antidepressant strategies like ketamine or psilocybin - would be of great interest as such work may provide novel insights into the complex mechanisms of stress-related psychiatric disorders as well as the complex mechanisms of treatment. Finally, it is important to note that while affective disorders like major depression are uniquely human diseases, translational models may be used to advance our understanding of the impact of stress on neurobiology and behavior as well as for screening of potential novel treatment approaches, as others have recently discussed (Zhang et al., 2020; Bale et al., 2019; Gordon 2019).
Funding sources
This work was supported by National Institutes of Health (NIH) T32 MH15330, the Greif Family Scholar Fund, and the EOS Foundation Trust Fund (ZAC), NIH R01 MH090595 (JBP), NIH R21 MH108944, and Dalio Philanthropies (KLKT).
These funding sources had no role in study design, collection, analysis, or interpretation of data, preparation of the report, or decision to submit the article for publication.
CRediT authorship contribution statement
Zachary A. Cordner: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Isaiah Marshall-Thomas: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Gretha J. Boersma: Investigation, Methodology, Writing – review & editing. Richard S. Lee: Investigation, Methodology, Resources, Supervision, Writing – review & editing. James B. Potash: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. Kellie L.K. Tamashiro: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
ZAC, IMT, GJB, RSL, JBP, and KLKT have no relevant conflicts of interest to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100392.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ashokan A., Hegde A., Mitra R. Short-term environmental enrichment is sufficient to counter stress-induced anxiety and associated structural and molecular plasticity in basolateral amygdala. Psychoneuroendocrinology. 2016 Jul 1;69:189–196. doi: 10.1016/j.psyneuen.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Bai Q., Song D., Gu L., Verkhratsky A., Peng L. Bi-phasic regulation of glycogen content in astrocytes via Cav-1/PTEN/PI3K/AKT/GSK-3β pathway by fluoxetine. Psychopharmacology. 2017 Apr 1;234(7):1069–1077. doi: 10.1007/s00213-017-4547-3. [DOI] [PubMed] [Google Scholar]
- Bale T.L., Abel T., Akil H., Carlezon W.A., Jr., Moghaddam B. The critical importance of basic animal research for neuropsychiatric disorders. Neuropsychopharmacology. 2019 Jul;44(8):1349–1353. doi: 10.1038/s41386-019-0405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtel K., Genoux D., Welzl H., Tweedie-Cullen R.Y., Koshibu K., Livingstone-Zatchej M., Mamie C., Mansuy I.M. Control of the establishment of aversive memory by calcineurin and Zif268. Nat. Neurosci. 2008 May;11(5):572–578. doi: 10.1038/nn.2113. [DOI] [PubMed] [Google Scholar]
- Beitia G., Garmendia L., Azpiroz A., Vegas O., Brain P.F., Arregi A. Time-dependent behavioral, neurochemical, and immune consequences of repeated experiences of social defeat stress in male mice and the ameliorative effects of fluoxetine. Brain Behav. Immun. 2005 Nov 1;19(6):530–539. doi: 10.1016/j.bbi.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Belz E.E., Kennell J.S., Czambel R.K., Rubin R.T., Rhodes M.E. Environmental enrichment lowers stress-responsive hormones in singly housed male and female rats. Pharmacol. Biochem. Behav. 2003 Dec 1;76(3–4):481–486. doi: 10.1016/j.pbb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Berton O., McClung C.A., DiLeone R.J., Krishnan V., Renthal W., Russo S.J., Graham D., Tsankova N.M., Bolanos C.A., Rios M., Monteggia L.M. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006 Feb 10;311(5762):864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Boku S., Nakagawa S., Toda H., Hishimoto A. Neural basis of major depressive disorder: beyond monoamine hypothesis. Psychiatr. Clin. Neurosci. 2018 Jan;72(1):3–12. doi: 10.1111/pcn.12604. [DOI] [PubMed] [Google Scholar]
- Campeau S., Davis M. Involvement of the central nucleus and basolateral complex of the amygdala in fear conditioning measured with fear-potentiated startle in rats trained concurrently with auditory and visual conditioned stimuli. J. Neurosci. 1995 Mar 1;15(3):2301–2311. doi: 10.1523/JNEUROSCI.15-03-02301.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J.L., Covington H.E., Friedman A.K., Wilkinson M.B., Walsh J.J., Cooper D.C., Nestler E.J., Han M.H. Mesolimbic dopamine neurons in the brain reward circuit mediate susceptibility to social defeat and antidepressant action. J. Neurosci. 2010 Dec 8;30(49):16453–16458. doi: 10.1523/JNEUROSCI.3177-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Shieh C., Swanger S.A., Tankovic A., Au M., McGuire M., Tagliati M., Graham J.M., Madan-Khetarpal S., Traynelis S.F., Yuan H. GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J. Hum. Genet. 2017 Jun;62(6):589–597. doi: 10.1038/jhg.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordner Z.A., Tamashiro K.L. Effects of chronic variable stress on cognition and Bace1 expression among wild-type mice. Transl. Psychiatry. 2016 Jul;6(7):e854. doi: 10.1038/tp.2016.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordner Z.A., MacKinnon D.F., DePaulo J.R., Jr. The care of patients with complex mood disorders. Focus. 2020 Apr;18(2):129–138. doi: 10.1176/appi.focus.20200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington H.E., Maze I., LaPlant Q.C., Vialou V.F., Ohnishi Y.N., Berton O., Fass D.M., Renthal W., Rush A.J., Wu E.Y., Ghose S. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 2009 Sep 16;29(37):11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der-Avakian A., Mazei-Robison M.S., Kesby J.P., Nestler E.J., Markou A. Enduring deficits in brain reward function after chronic social defeat in rats: susceptibility, resilience, and antidepressant response. Biol. Psychiatr. 2014 Oct 1;76(7):542–549. doi: 10.1016/j.biopsych.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.E., Chen H., Sakata K. BDNF deficiency and enriched environment treatment affect neurotransmitter gene expression differently across ages. J. Neurochem. 2020 Jul;154(1):41–55. doi: 10.1111/jnc.15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclot F., Kabbaj M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 2017 Mar 6;11:35. doi: 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D.D., Diorio J., Plotsky P.M., Meaney M.J. Environmental enrichment reverses the effects of maternal separation on stress reactivity. J. Neurosci. 2002 Sep 15;22(18):7840–7843. doi: 10.1523/JNEUROSCI.22-18-07840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gałecki P., Mossakowska-Wójcik J., Talarowska M. The anti-inflammatory mechanism of antidepressants–SSRIs. SNRIs. Progr. Neuro-Psychopharmacol. Biol. Psychiatr. 2018 Jan 3;80:291–294. doi: 10.1016/j.pnpbp.2017.03.016. [DOI] [PubMed] [Google Scholar]
- Gray A.L., Hyde T.M., Deep-Soboslay A., Kleinman J.E., Sodhi M.S. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol. Psychiatr. 2015 Sep;20(9):1057–1068. doi: 10.1038/mp.2015.91. [DOI] [PubMed] [Google Scholar]
- Golden S.A., Covington H.E., Berton O., Russo S.J. A standardized protocol for repeated social defeat stress in mice. Nat. Protoc. 2011 Aug;6(8):1183–1191. doi: 10.1038/nprot.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J.A. From neurobiology to novel medications: a principled approach to translation. Am. J. Psychiatr. 2019 Jun 1;176(6):425–427. doi: 10.1176/appi.ajp.2019.19040386. [DOI] [PubMed] [Google Scholar]
- Herringa R.J., Nanda S.A., Hsu D.T., Roseboom P.H., Kalin N.H. The effects of acute stress on the regulation of central and basolateral amygdala CRF-binding protein gene expression. Mol. Brain Res. 2004 Nov 24;131(1–2):17–25. doi: 10.1016/j.molbrainres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Hindmarch I. Expanding the horizons of depression: beyond the monoamine hypothesis. Hum. Psychopharmacol. Clin. Exp. 2001 Apr;16(3):203–218. doi: 10.1002/hup.288. [DOI] [PubMed] [Google Scholar]
- Hing B., Gardner C., Potash J.B. Effects of negative stressors on DNA methylation in the brain: implications for mood and anxiety disorders. Am. J. Med. Genet. Part B: Neuropsychiatric Genetics. 2014 Oct;165(7):541–554. doi: 10.1002/ajmg.b.32265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hing B., Braun P., Cordner Z.A., Ewald E.R., Moody L., McKane M., Willour V.L., Tamashiro K.L., Potash J.B. Chronic social stress induces DNA methylation changes at an evolutionary conserved intergenic region in chromosome X. Epigenetics. 2018 Jun 3;13(6):627–641. doi: 10.1080/15592294.2018.1486654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hötting K., Röder B. Beneficial effects of physical exercise on neuroplasticity and cognition. Neurosci. Biobehav. Rev. 2013 Nov 1;37(9):2243–2257. doi: 10.1016/j.neubiorev.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Jones M.W., Errington M.L., French P.J., Fine A., Bliss T.V., Garel S., Charnay P., Bozon B., Laroche S., Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 2001 Mar;4(3):289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Keyvani K., Sachser N., Witte O.W., Paulus W. Gene expression profiling in the intact and injured brain following environmental enrichment. J. Neuropathol. Exp. Neurol. 2004 Jun 1;63(6):598–609. doi: 10.1093/jnen/63.6.598. [DOI] [PubMed] [Google Scholar]
- Kitahara Y., Ohta K., Hasuo H., Shuto T., Kuroiwa M., Sotogaku N., Togo A., Nakamura K.I., Nishi A. Chronic fluoxetine induces the enlargement of perforant path-granule cell synapses in the mouse dentate gyrus. PloS One. 2016 Jan 20;11(1) doi: 10.1371/journal.pone.0147307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C.Y., Chang W.C., Wang J.M. Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J. Biomed. Sci. 2015 Dec;22(1):1–8. doi: 10.1186/s12929-014-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S.W., Hu-Shan A.O., Gallitano-Mendel A., Chang-Shen Q.I., Feng W.E., Milbrandt J., Min Z.H. Transcription factor Egr-1is required for long-term fear memory and anxiety. Acta Physiol. Sin. 2005 Aug 25;57(4):421–432. [PubMed] [Google Scholar]
- Krishnan V., Han M.H., Graham D.L., Berton O., Renthal W., Russo S.J., LaPlant Q., Graham A., Lutter M., Lagace D.C., Ghose S. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007 Oct 19;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Krystal J.H., Mathew S.J., D'Souza D.C., Garakani A., Gunduz-Bruce H., Charney D.S. Potential psychiatric applications of metabotropic glutamate receptor agonists and antagonists. CNS Drugs. 2010 Aug;24(8):669–693. doi: 10.2165/11533230-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Leeuwenburgh C., Heinecke J.W. Oxidative stress and antioxidants in exercise. Curr. Med. Chem. 2001 Jun 1;8(7):829–838. doi: 10.2174/0929867013372896. [DOI] [PubMed] [Google Scholar]
- Lehmann M.L., Herkenham M. Environmental enrichment confers stress resiliency to social defeat through an infralimbic cortex-dependent neuroanatomical pathway. J. Neurosci. 2011 Apr 20;31(16):6159–6173. doi: 10.1523/JNEUROSCI.0577-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A.S., Hell J.W. Cyclic AMP-dependent protein kinase and protein kinase C phosphorylate N-methyl-D-aspartate receptors at different sites. J. Biol. Chem. 1997 May 2;272(18):12107–12115. doi: 10.1074/jbc.272.18.12107. [DOI] [PubMed] [Google Scholar]
- Lin T.W., Kuo Y.M. Exercise benefits brain function: the monoamine connection. Brain Sci. 2013 Mar;3(1):39–53. doi: 10.3390/brainsci3010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.W., Chen C.C., Wang J.M., Chang W.C., Huang Y.C., Chung S.Y., Chen B.K., Hung J.J. Role of transcriptional factors Sp1, c-Rel, and c-Jun in LPS-induced C/EBPδ gene expression of mouse macrophages. Cell. Mol. Life Sci. 2007 Dec;64(24):3282–3294. doi: 10.1007/s00018-007-7375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.D., Murray C.C. The global burden of disease, 1990–2020. Nat. Med. 1998;4:1241–1243. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- Lopez J.P., Lim R., Cruceanu C., Crapper L., Fasano C., Labonte B., Maussion G., Yang J.P., Yerko V., Vigneault E., El Mestikawy S. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 2014 Jul;20(7):764–768. doi: 10.1038/nm.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Ren Q., Yang C., Zhang J.C., Yao W., Dong C., Ohgi Y., Futamura T., Hashimoto K. Adjunctive treatment of brexpiprazole with fluoxetine shows a rapid antidepressant effect in social defeat stress model: role of BDNF-TrkB signaling. Sci. Rep. 2016 Dec 19;6(1):1–2. doi: 10.1038/srep39209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malberg J.E., Eisch A.J., Nestler E.J., Duman R.S. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J. Neurosci. 2000 Dec 15;20(24):9104–9110. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Chamberlin A., Shinde D.N., Hempel M., Strom T.M., Schreiber A., Johannsen J., Ousager L.B., Larsen M.J., Hansen L.K., Fatemi A. De novo variants in GRIA4 lead to intellectual disability with or without seizures and gait abnormalities. Am. J. Hum. Genet. 2017 Dec 7;101(6):1013–1020. doi: 10.1016/j.ajhg.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Calvo‐Torrent A., Pico‐Alfonso M.A. Social defeat and subordination as models of social stress in laboratory rodents: a review. Aggress. Behav.: Off. J. Int. Soc. Resear. Aggression. 1998;24(4):241–256. [Google Scholar]
- McEwen B.S. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol. Rev. 2007 Jul;87(3):873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- McGeary J.E., Gurel V., Knopik V.S., Spaulding J., McMichael J. Effects of nerve growth factor (NGF), fluoxetine, and amitriptyline on gene expression profiles in rat brain. Neuropeptides. 2011 Oct 1;45(5):317–322. doi: 10.1016/j.npep.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Mondal A.C., Fatima M. Direct and indirect evidences of BDNF and NGF as key modulators in depression: role of antidepressants treatment. Int. J. Neurosci. 2019 Mar 4;129(3):283–296. doi: 10.1080/00207454.2018.1527328. [DOI] [PubMed] [Google Scholar]
- Monsey M.S., Boyle L.M., Zhang M.L., Nguyen C.P., Kronman H.G., Ota K.T., Duman R.S., Taylor J.R., Schafe G.E. Chronic corticosterone exposure persistently elevates the expression of memory-related genes in the lateral amygdala and enhances the consolidation of a Pavlovian fear memory. PloS One. 2014 Mar 11;9(3) doi: 10.1371/journal.pone.0091530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley‐Fletcher S., Rea M., Maccari S., Laviola G. Environmental enrichment during adolescence reverses the effects of prenatal stress on play behaviour and HPA axis reactivity in rats. Eur. J. Neurosci. 2003 Dec;18(12):3367–3374. doi: 10.1111/j.1460-9568.2003.03070.x. [DOI] [PubMed] [Google Scholar]
- Moser D., Molitor A., Kumsta R., Tatschner T., Riederer P., Meyer J. The glucocorticoid receptor gene exon 1-F promoter is not methylated at the NGFI-A binding site in human hippocampus. World J. Biol. Psychiatr. 2007 Jan 1;8(4):262–268. doi: 10.1080/15622970701429862. [DOI] [PubMed] [Google Scholar]
- Mul J.D., Soto M., Cahill M.E., Ryan R.E., Takahashi H., So K., Zheng J., Croote D.E., Hirshman M.F., la Fleur S.E., Nestler E.J. Voluntary wheel running promotes resilience to chronic social defeat stress in mice: a role for nucleus accumbens ΔFosB. Neuropsychopharmacology. 2018 Aug;43(9):1934–1942. doi: 10.1038/s41386-018-0103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaes L.S., Dos Santos N.B., Batalhote R.F., Malta M.B., Camarini R., Scavone C., Munhoz C.D. Environmental enrichment protects against stress-induced anxiety: role of glucocorticoid receptor, ERK, and CREB signaling in the basolateral amygdala. Neuropharmacology. 2017 Feb 1;113:457–466. doi: 10.1016/j.neuropharm.2016.10.026. [DOI] [PubMed] [Google Scholar]
- Nuriya M., Oh S., Huganir R.L. Phosphorylation‐dependent interactions of α‐Actinin‐1/IQGAP1 with the AMPA receptor subunit GluR4. J. Neurochem. 2005 Oct;95(2):544–552. doi: 10.1111/j.1471-4159.2005.03410.x. [DOI] [PubMed] [Google Scholar]
- Osuch E., Gao S., Wammes M., Théberge J., Williamson P., Neufeld R.J., Du Y., Sui J., Calhoun V. Complexity in mood disorder diagnosis: fMRI connectivity networks predicted medication‐class of response in complex patients. Acta Psychiatr. Scand. 2018 Nov;138(5):472–482. doi: 10.1111/acps.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens M.J. Selectivity of antidepressants: from the monoamine hypothesis of depression to the SSRI revolution and beyond. J. Clin. Psychiatr. 2004;65(4):5–10. [PubMed] [Google Scholar]
- Paxinos G., Franklin K.B. second ed. Academic Press; 2004. The Mouse Brain in Stereotaxic Coordinates, Compact. [Google Scholar]
- Penke Z., Morice E., Veyrac A., Gros A., Chagneau C., LeBlanc P., Samson N., Baumgärtel K., Mansuy I.M., Davis S., Laroche S. Zif268/Egr1 gain of function facilitates hippocampal synaptic plasticity and long-term spatial recognition memory. Phil. Trans. Biol. Sci. 2014 Jan 5;369(1633):20130159. doi: 10.1098/rstb.2013.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C., Jiang C.H., Dong H., Tang Y.P., Lockhart D.J., Schultz P.G., Tsien J.Z., Hu Y. Effects of environmental enrichment on gene expression in the brain. Proc. Natl. Acad. Sci. Unit. States Am. 2000 Nov 7;97(23):12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzoli M., Carboni L., Andreoli M., Michielin F., Ballottari A., Arban R. Strain-specific outcomes of repeated social defeat and chronic fluoxetine treatment in the mouse. Pharmacol. Biochem. Behav. 2011 Jan 1;97(3):566–576. doi: 10.1016/j.pbb.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Réus G.Z., Dos Santos M.A., Abelaira H.M., Ribeiro K.F., Petronilho F., Vuolo F., Colpo G.D., Pfaffenseller B., Kapczinski F., Dal-Pizzol F., Quevedo J. Imipramine reverses alterations in cytokines and BDNF levels induced by maternal deprivation in adult rats. Behav. Brain Res. 2013 Apr 1;242:40–46. doi: 10.1016/j.bbr.2012.11.044. [DOI] [PubMed] [Google Scholar]
- Revest J.M., Di Blasi F., Kitchener P., Rougé-Pont F., Desmedt A., Turiault M., Tronche F., Piazza P.V. The MAPK pathway and Egr-1 mediate stress-related behavioral effects of glucocorticoids. Nat. Neurosci. 2005 May;8(5):664–672. doi: 10.1038/nn1441. [DOI] [PubMed] [Google Scholar]
- Revest J.M., Kaouane N., Mondin M., Le Roux A., Rougé-Pont F., Vallée M., Barik J., Tronche F., Desmedt A., Piazza P.V. The enhancement of stress-related memory by glucocorticoids depends on synapsin-Ia/Ib. Mol. Psychiatr. 2010 Dec;15(12):1140–1151. doi: 10.1038/mp.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz J.A., Venheim E.R., Padival M. Chronic stress causes amygdala hyperexcitability in rodents. Biol. Psychiatr. 2010 Jun 15;67(12):1128–1136. doi: 10.1016/j.biopsych.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanesan M., Haiar J.M., Watt M.J., Newton S.S. Restraint stress differentially regulates inflammation and glutamate receptor gene expression in the hippocampus of C57BL/6 and BALB/c mice. Stress. 2017 Mar 4;20(2):197–204. doi: 10.1080/10253890.2017.1298587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloesser R.J., Lehmann M., Martinowich K., Manji H.K., Herkenham M. Environmental enrichment requires adult neurogenesis to facilitate the recovery from psychosocial stress. Mol. Psychiatr. 2010 Dec;15(12):1152–1163. doi: 10.1038/mp.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki T., Iijima M., Chaki S. Anxiolytic-like activity of MGS0039, a potent group II metabotropic glutamate receptor antagonist, in a marble-burying behavior test. Eur. J. Pharmacol. 2004 Oct 6;501(1–3):121–125. doi: 10.1016/j.ejphar.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sillaber I., Panhuysen M., Henniger M.S., Ohl F., Kühne C., Pütz B., Pohl T., Deussing J.M., Paez-Pereda M., Holsboer F. Profiling of behavioral changes and hippocampal gene expression in mice chronically treated with the SSRI paroxetine. Psychopharmacology. 2008 Nov;200(4):557–572. doi: 10.1007/s00213-008-1232-6. [DOI] [PubMed] [Google Scholar]
- Smail M.A., Smith B.L., Nawreen N., Herman J.P. Differential impact of stress and environmental enrichment on corticolimbic circuits. Pharmacol. Biochem. Behav. 2020 Jul 11:172993. doi: 10.1016/j.pbb.2020.172993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Saqib U., Naim A., Roy A., Liu D., Bhatnagar D., Ravinder R., Baig M.S. The TLR4–NOS1–AP1 signaling axis regulates macrophage polarization. Inflamm. Res. 2017 Apr 1;66(4):323–334. doi: 10.1007/s00011-016-1017-z. [DOI] [PubMed] [Google Scholar]
- Sun Z., Xu X., He J., Murray A., Sun M.A., Wei X., Wang X., McCoig E., Xie E., Jiang X., Li L. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 2019 Aug 29;10(1):1–2. doi: 10.1038/s41467-019-11905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan R.J., Weidenhofer J., Hinwood M., Cairns M.J., Day T.A., Walker F.R. A comparative examination of the anti-inflammatory effects of SSRI and SNRI antidepressants on LPS stimulated microglia. Brain Behav. Immun. 2012 Mar 1;26(3):469–479. doi: 10.1016/j.bbi.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Vandevyver S., Dejager L., Libert C. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr. Rev. 2014 Aug 1;35(4):671–693. doi: 10.1210/er.2014-1010. [DOI] [PubMed] [Google Scholar]
- Vera-Chang M.N., St-Jacques A.D., Gagné R., Martyniuk C.J., Yauk C.L., Moon T.W., Trudeau V.L. Transgenerational hypocortisolism and behavioral disruption are induced by the antidepressant fluoxetine in male zebrafish Danio rerio. Proc. Natl. Acad. Sci. Unit. States Am. 2018 Dec 26;115(52):E12435–E12442. doi: 10.1073/pnas.1811695115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Richthofen S., Lang U.E., Hellweg R. Effects of different kinds of acute stress on nerve growth factor content in rat brain. Brain Res. 2003 Oct 17;987(2):207–213. doi: 10.1016/s0006-8993(03)03338-9. [DOI] [PubMed] [Google Scholar]
- Vyas A., Mitra R., Rao B.S., Chattarji S. Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. J. Neurosci. 2002 Aug 1;22(15):6810–6818. doi: 10.1523/JNEUROSCI.22-15-06810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver I.C., Hellstrom I.C., Brown S.E., Andrews S.D., Dymov S., Diorio J., Zhang T.Y., Szyf M., Meaney M.J. The methylated-DNA binding protein MBD2 enhances NGFI-A (egr-1)-mediated transcriptional activation of the glucocorticoid receptor. Phil. Trans. Biol. Sci. 2014 Sep 26;369(1652):20130513. doi: 10.1098/rstb.2013.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford H.A., Degenhardt L., Rehm J. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382:1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.B., Xiao G., Kumar A., LaPlant Q., Renthal W., Sikder D., Kodadek T.J., Nestler E.J. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J. Neurosci. 2009 Jun 17;29(24):7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.B., Dias C., Magida J., Mazei-Robison M., Lobo M., Kennedy P., Dietz D., Covington H., Russo S., Neve R., Ghose S. A novel role of the WNT-dishevelled-GSK3β signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J. Neurosci. 2011 Jun 22;31(25):9084–9092. doi: 10.1523/JNEUROSCI.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Johnson R.S., Moore C. c‐Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999 Jan 4;18(1):188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.Y., Oxendine S.E., Godwin J. Behavioral and neurogenomic transcriptome changes in wild-derived zebrafish with fluoxetine treatment. BMC Genom. 2013 Dec;14(1):1–3. doi: 10.1186/1471-2164-14-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M., Bacanu S.A. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018 May;50(5):668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Cao Q., Xiong X., Zhao P., Shen D., Zhang Y., Zhang N. Fluoxetine regulates glucose and lipid metabolism via the PI3K-AKT signaling pathway in diabetic rats. Mol. Med. Rep. 2020 Oct 1;22(4):3073–3080. doi: 10.3892/mmr.2020.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T., Shimazaki T., Ito A., Chaki S. An mGluR2/3 antagonist, MGS0039, exerts antidepressant and anxiolytic effects in behavioral models in rats. Psychopharmacology. 2006 Jul;186(4):587–593. doi: 10.1007/s00213-006-0390-7. [DOI] [PubMed] [Google Scholar]
- Zeng B., Li Y., Niu B., Wang X., Cheng Y., Zhou Z., You T., Liu Y., Wang H., Xu J. Involvement of PI3K/Akt/FoxO3a and PKA/CREB signaling pathways in the protective effect of fluoxetine against corticosterone-induced cytotoxicity in PC12 cells. J. Mol. Neurosci. 2016 Aug;59(4):567–578. doi: 10.1007/s12031-016-0779-7. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xiao X., Li T., Li M. Translational genomics and beyond in bipolar disorder. Mol. Psychiatr. 2020 May 18:1–7. doi: 10.1038/s41380-020-0782-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.