Abstract
Papillomaviruses, polyomaviruses and adenoviruses are collectively categorized as the small DNA tumour viruses. Notably, human adenoviruses were the first human viruses demonstrated to be able to cause cancer, albeit in non-human animal models. Despite their long history, no human adenovirus is a known causative agent of human cancers, unlike a subset of their more famous cousins, including human papillomaviruses and human Merkel cell polyomavirus. Nevertheless, seminal research using human adenoviruses has been highly informative in understanding the basics of cell cycle control, gene expression, apoptosis and cell differentiation. This review highlights the contributions of human adenovirus research in advancing our knowledge of the molecular basis of cancer.
Keywords: E1A, E1B, E4, Transformation, Hallmarks of cancer, Human adenovirus
1. Introduction
Papillomaviruses, polyomaviruses and adenoviruses are collectively categorized as the small DNA tumour viruses based on their small double-stranded DNA genome sizes and their ability to induce cancer in either experimental systems or real-world contexts. While other human tumour viruses exist, including human T-lymphotropic virus (HTLV), Epstein-Barr virus (EBV), hepatitis B and C virus (HBV and HCV), and Kaposi's sarcoma-associated herpesvirus (KSHV), these are not classified as small DNA tumour viruses. Human adenoviruses (HAdV) infect the respiratory tract, gastrointestinal tract, and the corneal epithelium. These prevalent human pathogens typically cause self-limiting infections in immunocompetent individuals; however, they have periodically been associated with outbreaks causing significant illness and mortality [1]. The first HAdV isolates were obtained in 1953 from the tonsils and adenoids of healthy individuals and in 1954 from military personnel with acute respiratory disease [2,3]. In 1962, HAdVs gained the distinction of being the first human virus demonstrated to cause cancer. These studies were originally performed in newborn hamsters, but later duplicated in other rodent models, as well as baboons [[4], [5], [6]]. Despite the notoriety of being the first known human tumour virus, no convincing relationship between HAdV infection and human cancer has been identified. Thus, unlike their more famous cousins human papillomavirus (HPV) and human polyomavirus (HPyV), HAdV is the only small DNA tumour virus without a family member directly associated with human oncogenesis [7,8]. Despite not actually causing human cancer, studies of HAdVs have provided tremendous insight into multiple cancer-relevant processes and this review will highlight some of these important advances.
1.1. HAdV-dependent oncogenesis
Early studies of HAdV-mediated tumourigenesis were based on injection of virus into newborn rodents, and these identified a range of oncogenicity between different HAdVs [9,10]. Over 80 different HAdV types infect humans and these are grouped into 7 species labeled A–G [11]. HAdVs from species A, notably HAdV-A12, -A18 and -A31, are known as the highly oncogenic types. These viruses produce tumours at high frequency within a few months at the site of injection in neonatal rodents [9,10]. Additionally, several viruses from species D, including HAdV-D9 and -D10, elicit estrogen-dependent mammary tumours in adult female rats [10]. In contrast, the vast majority of HAdVs are non-oncogenic or poorly oncogenic when directly injected into rodents [9,10]. Detailed investigation has suggested that the inability of most HAdV types to induce tumours in animal models likely reflects type-specific inabilities to evade host natural killer (NK) and T-cell based immune responses that subsequently clear virally-transformed cells [9,10]. As a consequence, when tested for oncogenic transformation activity in primary rodent cell culture-based systems that lack cell-based immunity, all tested HAdVs appear able to transform [9,10]. Additionally, rodent cells transformed in vitro by non-oncogenic HAdVs can form tumours when implanted in immunosupressed, but not syngeneic immunocompetent animals. This further supports the concept that differential sensitivity to immune surveillance is a major factor contributing to the tumourigenic potential of a given HAdV [10,12]. Regardless of their ability to induce tumours in animal models, all HAdVs are considered tumour viruses based on their transformation activities in cell-culture based systems.
Oncogenesis by HAdVs is not restricted to rodent systems. For example, HAdV-A12 was shown to induce retinal tumours in a subset of newborn baboons after intraocular injection [4]. Notably, the interior portions of the eye are considered immune privileged, and this may play a significant role in allowing HAdV-A12 oncogenesis in this system. Rare examples of transformation of primary human cells with HAdV DNA in culture exist, but an etiological role for HAdV in human oncogenesis has never been identified [13,14].
Molecular studies have shown that the major oncogenic activities within HAdV reside within the left end of the genome, encompassing early region 1 (E1), which encodes the E1A and E1B oncoproteins [15]. These sequences are consistently retained and expressed in HAdV-transformed cells and this region is both necessary and sufficient for oncogenic transformation by most HAdVs. Additional oncogenic functions are present in the right end of the HAdV genome within the E4 region, which may contribute to oncogenesis via alternative mechanisms, particularly for the induction of estrogen-dependent mammary tumours by HAdV-D9 [14,16]. For the purposes of this review, we will focus on the E1A, E1B and E4 proteins and their contributions to understanding oncogenic processes. Importantly, it is worth mentioning that much of the work involving these proteins has largely been done in the context of HAdV-C5. Therefore, much of what will be covered in the following sections will pertain to HAdV-C5, while details from other HAdV species will be mentioned where necessary. For reference, a simplified representation of the HAdV-C5 genome is depicted in Fig. 1, which is broadly representative of all HAdV species [17].
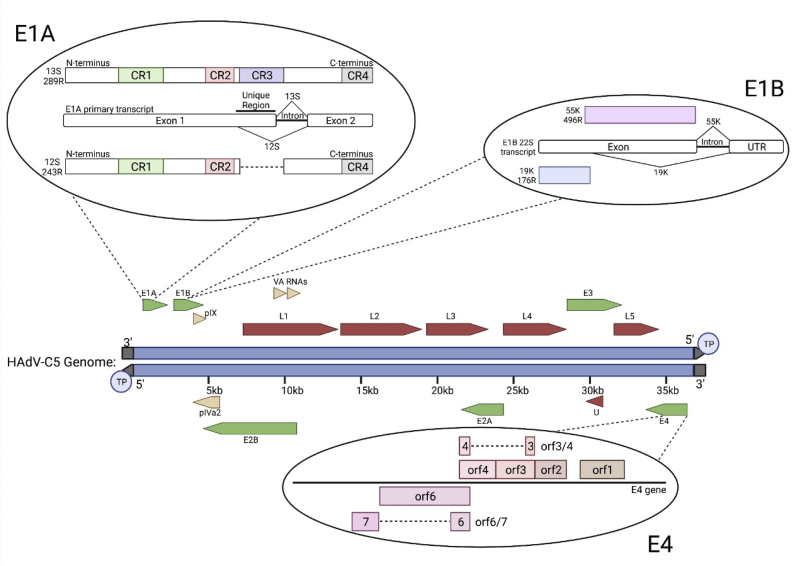
Fig. 1.
HAdV-C5 genome organization with highlighted E1A, E1B, and E4 genes. The human adenovirus (HAdV)-C5 genome, shown in blue, is approximately 36 kb of linear double-stranded DNA. Although Fig. 1 depicts the HAdV-C5 genome, all known HAdV genomes are similarly organized. However, some individual open reading frames (ORFs) may be present or absent in any given HAdV species. The viral genome is flanked by inverted terminal repeats (grey) on both strands, with the 5′ end covalently bound to the viral terminal protein (TP). Early genes E1A, E1B, E2A, E2B, E3 and E4 are shown as green arrows and are expressed prior to the onset of viral genome replication. Late genes L1-5 and μ are shown as red arrows and are expressed following the onset of viral genome replication. Intermediate early genes pIX and pVIa2, as well as the viral-associated (VA) RNAs are shown as yellow arrows. E1A, E1B and E4 genes have oncogenic properties and have been selected for more detailed description. E1A: The E1A primary transcript contains two exons separated by an intron which is spliced out for the 13S/289R E1A isoform. Exon 1 contains a unique region that is present in the 13S/289R E1A isoform, but is spliced out in the 12S/243R E1A isoform. The protein products of both major E1A isoforms contain conserved regions (CR)1, 2 and 4, while CR3 is unique to 13S/289R. Only the E1A primary transcript and two major isoforms are shown for simplicity. E1B: The E1B 22S transcript contains an exon, intron, and 3′ UTR. E1B 19K/176R is translated from the start of exon 1 and spliced in the middle of the exon to the 3′ UTR. E1B 55K/496R is translated starting from an internal translation initiation site and spliced at the intron to the 3′ UTR. Only the E1B 22S transcript and two major isoforms are shown. E4: The E4 gene and all seven major E4 protein products are shown. E4 contains multiple ORFs that encode for the following seven proteins: E4orf1, E4orf2, E4orf3, E4orf4, E4orf3/4, E4orf6 and E4orf6/7. Created with BioRender.com. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
1.2. HAdV and the hallmarks of cancer
The "Hallmarks of Cancer", as described by Weinberg and Hanahan, divides the malignant phenotype into subsets of cellular capabilities acquired during carcinogenesis [18]. The biological consequences of oncogenic alterations lead to phenotypic characteristics commonly associated with tumours, and these represent the individual ‘‘Hallmarks’’ of cancer. These hallmarks are a useful framework to organize and discern the contribution of individual HAdV genes to the oncogenic process. As previously done for the viral oncogenes of tumour viruses actually responsible for human cancers, we have framed each of the HAdV oncogenes in terms of their effects on the host cell or cellular pathways that are relevant to the hallmarks of cancer (Fig. 2) [19].
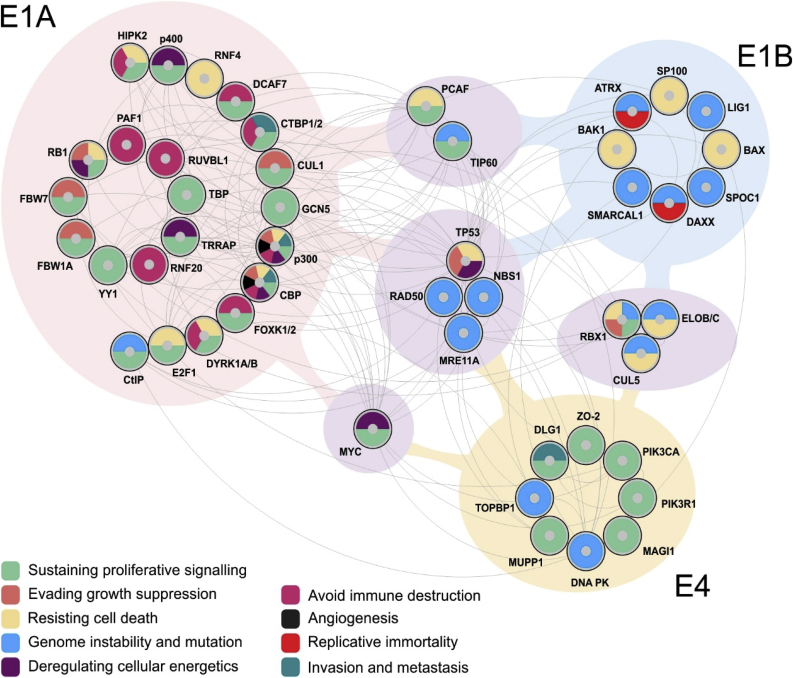
Fig. 2.
Protein interaction map of HAdV oncoproteins E1A, E1B and E4 and their connection to the hallmarks of cancer. Based on their relevance to the hallmarks of cancer, selected protein-protein interactions for E1A, E1B and E4 (including their isoforms) were visualized using Cytoscape [324]. Proteins shaded with light purple indicated a cellular target is shared by more than one of the HAdV proteins described in this review. Cancer-relevant cellular targets of each viral protein were identified from the literature and imported from the STRING database into Cytoscape. Interactions between cellular proteins were filtered to include only those with experimental evidence and high STRING confidence scores (>0.7). Targets of each viral protein have been colour coded according to their association with each respective hallmark of cancer. Importantly, each cellular target has a potential relevance to various hallmarks of cancer, even though their role in HAdV-mediated transformation may be unknown. Although an integrated analysis of E1A, E1B and E4 has yet to been performed, visualizing their interactions in this way demonstrates substantial overlap within the underlying interaction map. Notably, several cellular proteins, highlighted in purple, are targeted by multiple different viral proteins, suggesting potential synergy or antagonism on their function. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2. HAdV viral oncogenes and targets
The HAdV proteins E1A, E1B and E4 have well established roles in oncogenic processes. Interest in these regions as oncogenic entities arose from early observations demonstrating that the left end of the genome, encompassing the E1 region, was consistently retained in virus-transformed cells [20]. Alternatively, E4 has been shown to enhance transformation by conferring additional oncogenic properties, and in the case of HAdV-D9, it was shown to be the main determinant of tumourigenicity in animals [21]. For these reasons, the study of HAdV-mediated oncogenesis has been focused on the gene products of E1A, E1B and E4. A comprehensive analysis comparing each of these viral proteins between different HAdV species has not been performed; therefore, whether or not they modulate the same set of host factors has not been entirely established. However, essential interactions like that between E1A and the retinoblastoma tumour suppressor protein (pRb; see section 2.1.1 below) are assumed to be conserved based on the presence of an LxCxE binding motif within all HAdV E1A proteins [22]. Nevertheless, it is highly likely the E1A, E1B and E4 proteins from distinct HAdV species target various cellular pathways in different and nuanced ways, ultimately providing different means to a similar end with respect to regulating the viral replication cycle. Overall, these proteins can deregulate many cellular pathways, including those central to cell cycle regulation and cell death. Unsurprisingly, many of these pathways are also essential to carcinogenesis.
Despite there being no direct link between HAdV and human cancer, the repertoire of virus-host interactions these proteins facilitate is highly reminiscent to that of other well-characterized and more infamous tumour viruses. These similarities have made the study of HAdV-mediated transformation highly relevant to both viral and non-viral cancers. The roles that E1A, E1B and E4 play in transformation have mostly been studied as individual entities. However, this fails to capture the collective relevance of these viral oncoproteins to cancer as a whole. To illustrate this graphically, we constructed a protein interaction map using their well-established targets using experimentally determined, high confidence interactions. Cellular targets were colour-coded to reflect the relationship to the hallmarks of cancer. This visualization reveals the intricacy of this network and a complex cross-coupling of protein-protein interactions. Furthermore, this identifies a number of potentially shared pathways within the hallmarks of cancer framework (Fig. 2). These observations also stress the complexity and integrated nature of pathways involved in oncogenesis and may also highlight additional avenues of HAdV-mediated transformation that remain to be explored. Within this section we will discuss the contributions that E1A, E1B and E4 have made on our understanding of the molecular mechanisms underlying cancer and their connection to the hallmarks of cancer.
2.1. E1A
The E1A region of all HAdVs is the first viral gene expressed during infection and the corresponding mRNA is alternatively spliced into its two predominant isoforms that encode proteins of 289 and 243 amino acids with respect to HAdV-C5 (these proteins are also referred to as 13S and 12S respectively, which is based on the size of the fully-spliced mRNAs). The 13S-encoded isoform contains four conserved regions (CR1-4) while the shorter 12S-encoded isoform is devoid of CR3 [22] (Fig. 1). The larger E1A isoform varies in length from 249 to 289 amino acids between HAdV species, with species C encoding the largest isoforms. Despite these differences, all E1As are spliced similarly and contain all four conserved regions. Although the E1A 12S isoform has historically been used as the model oncoprotein, either 13S or 12S E1A isoforms are sufficient to immortalize primary rodent cells in culture. Furthermore, when combined with a cooperating oncogene, such as HAdV E1B or activated RAS, E1A can fully transform these cells [23]. Collectively, the multifunctional E1A proteins are largely responsible for rewiring infected host protein interaction networks into a state amenable for viral replication [24].
The E1A region is absolutely essential for viral replication. Viruses lacking this region, such as dl312, are replication deficient and can only replicate if E1A is provided in trans, such as in the case of HEK293 cells that express E1A endogenously [25]. While both the 13S-encoded and 12S-encoded E1A isoforms contribute to HAdV replication, the 13S E1A isoform appears to be responsible for modulating the expression of a wider number of HAdV-encoded viral genes [26]. HAdV that expressed only the 12S E1A isoform, but not the 13S E1A isoform, show delayed expression of a number of viral genes, lower expression of viral structural proteins, and a lower replication rate [26].
During infection, E1A functions as a viral hub protein by binding and altering the functions of several dozen cellular hubs within the human proteome [27]. Through these interactions E1A can manipulate a multitude of host processes, including transcription, epigenetic regulation, subcellular localization, metabolism, cell cycle control, and post-translational modification (PTM). In fact, many of these E1A interactions include well-established oncoproteins or tumour suppressor proteins. Given the high sequence similarity between different HAdV E1A proteins, many of these interactions are conserved across HAdV species [22,28]. However, not all E1A proteins from different HAdV species share the same repertoire of binding partners. For example, CR2 from HAdV-C5, -A12, -A18 and A31 are able to interact with the transcriptional corepressor BS69 via a PxLxP motif [29]. Intriguingly, for reasons which remain unknown, both HAdV-A12 and -A18 have evolved two BS69 binding motifs. Additionally, all but species C HAdVs possess a linker region between CR2 and CR3. Compositional differences within the linker between species exist, with the highly oncogenic species A HAdVs being particularly rich in alanine residues. These examples highlight some of the differences between E1A proteins; however, despite these differences all HAdVs tested are able to transform cells in culture with similar efficiencies [14]. Furthermore, additional evidence using replication-defective HAdV-C5 vectors lacking the E1 region could be complemented by various other HAdV species, demonstrating a high level of functional conservation within the E1 region [30,31].
2.1.1. E1A induces cell cycle through modulation of the retinoblastoma protein pathway
The role E1A plays in transformation has been extensively studied and reviewed in the past [23,[32], [33], [34], [35], [36]]. Since E1A has no direct DNA-binding or enzymatic ability, its participation in oncogenic transformation is achieved through direct protein-protein interactions as well as secondary interactions within the network of its primary targets [24]. The region of E1A encoded by exon 1 contains both CR1 and CR2 and is primarily responsible for transformation (Fig. 1) [33]. Interestingly, either the extreme N-terminal region up to the end of CR1 (N-terminus/CR1) or CR2 alone is sufficient to induce S phase in primary rodent cells; however, both regions are required to carry cells into mitosis [37]. Historically, the first cellular protein identified to bind E1A directly was pRb [38]. This interaction has historical significance; it was recognized as the first example of a viral oncogene interacting with a cellular tumour suppressor. While the interaction between the simian polyomavirus 40 (SV40) large T antigen and the p53 tumour suppressor protein predates this observation, at the time p53 was mistakenly characterized as an oncogene and it was not until after the E1A-pRb interaction was identified that p53 was correctly classified as a tumour suppressor [[39], [40], [41], [42]].
Using a conserved LxCxE amino acid motif located in CR2, E1A from all HAdVs so far tested can directly bind pRb and its family members p107 and p130 with high affinity [43]. Once bound through CR2, a lower affinity binding region within CR1 can displace E2F transcription factors from pRb [44,45]. Note that the use of two separate motifs within E1A to bind a given target is actually quite common, as other cellular factors including p300, TBP and PCAF are known to bind both CR1 and CR3 [[46], [47], [48]]. The interaction with pRb functionally inactivates it, freeing E2Fs of pRb and its associated repressive chromatin modifying enzymes, resulting in derepression of cell cycle genes [49,50]. Indeed, functional inactivation of pRb is frequently observed in cancers and is also a target of several infamous viral oncogenes, such as HPV E7 and HPyV large T antigen [43,51,52]. In the context of non-virally induced cancers, the inactivation of pRb contributes to sustained proliferative signalling as well as evasion of growth suppression [53]. The ability of E1A and other viral oncogenes to inactivate pRb contributes to both of these hallmarks.
2.1.2. E1A promotes transformation and cell cycle entry through binding chromatin modification complexes
The mechanism by which E1A induces S phase through CR2 and pRb is relatively straight forward compared to the N-terminus/CR1 region. This region of E1A is densely packed with protein interaction motifs, which creates difficulty in assigning causality of E1A's function to a specific E1A-interacting partner [24,48]. Despite the difficulty in unraveling specific interactions and their functions within this region, it has been established that E1A's N-terminus/CR1 interacts with a plethora of chromatin modifiers, many of which have recognized roles in oncogenesis (Fig. 2) [35].
Involvement of E1A's N-terminus/CR1 in transformation has largely been attributed to the interaction with p300 and its homologue CREB-binding protein (CBP) [34]. The discovery of p300, originally named E1A-associated 300-kDa protein, was identified through study of E1A's growth controlling functions [[54], [55], [56]]. It was later discovered that E1A also targeted CBP, and that interactions with p300/CBP were necessary for transformation [34,35,48,57].
Both p300 and CBP are lysine acetyltransferases that function as transcriptional cofactors involved in chromatin modification and play important roles in transformation [58,59]. Precisely how p300 contributes to E1A-mediated transformation is not fully understood; however, several consequences of this interaction are apparent. First, early studies of E1A and p300 demonstrated that they form a multimeric complex with pRb, suggesting a proximal association of the three proteins could be important [60]. This led to the concomitant discovery of p300-mediated acetylation of pRb at K873/K874 during S phase and that E1A enhances pRb acetylation through this multimeric complex [61]. Acetylation within this region of pRb was also shown to impede pRb phosphorylation as well as promote dissociation of E2F1 from pRb during the DNA damage response (DDR) [61,62]. These observations are interesting, but somewhat paradoxical for several reasons. Unlike other E2Fs, E2F1 has a unique role in apoptosis and DNA repair, as well as an additional pRb binding region within its C-terminal domain [[63], [64], [65]]. Notably, E1A-mediated transformation involves disruption of pRb-E2F complexes; however, the pRb-E2F1 complex is resistant to E1A-mediated disruption during HAdV infection, which was suggested to promote cell cycle progression while avoiding apoptosis [66]. Based on these findings, E1A appears to enhance p300-mediated pRb acetylation without causing E2F1 dissociation. Furthermore, E2F1 is also known to be acetylated by p300/CBP and its associated factor PCAF [67]. In vitro acetylation of E2F1, in the presence of pRb and p300, was decreased in response to increasing E1A protein levels [61]. E2F1 is acetylated in response to DNA damage; however, how E1A simultaneously enhances pRb and reduces E2F1 acetylation in an in vitro system is unclear. Regardless, these observations demonstrate a complex interplay between various cellular factors and highlight the utility of using E1A as a molecular tool.
Analysis of host gene expression during infection demonstrated the E1A-p300 interaction is responsible for the majority of E1A-mediated repression of host gene expression [68]. Indeed, a number of genes involved in chromatin condensation are regulated by the multimeric E1A-pRb-p300 complex. Mechanistically, it appears that E1A relocalizes pRb to chromatin-bound p300. Here, E1A-mediated enhancement of pRb acetylation via p300 impedes pRb phosphorylation, presumably locking pRb in a hypophosphorylated repressive state. By hijacking p300 and pRb along with its associated repressive chromatin modifiers, E1A can inhibit expression of genes which would otherwise inhibit cell cycle or promote apoptosis [50,68].
E1A has also been reported to alter global H3K18 acetylation through its interaction with p300/CBP. These epigenetic changes affect the promoters for thousands of genes, including those involved in the antiviral response and cell cycle control. Importantly, point mutants of p300/CBP that fail to induce H3K18 hypoacetylation and subsequent antiviral gene repression, severely impact E1A's transforming abilities, suggesting a link between H3K18 hypoacetylation and transformation [69]. Similarly, a global reduction in H3K18 acetylation was also observed in cells expressing SV40 large T antigen, potentially raising the question if this is a common strategy among DNA tumour viruses [69]. Furthermore, E1A has been shown to induce gene expression of c-Myc through derepression of a p300-YY1-HDAC3 complex [70]. This complex is involved in repressing c-Myc transcription and thereby preventing aberrant S phase entry [71]. By disrupting the p300 interaction with YY1, E1A alleviates c-Myc repression to induce S phase [72]. Notably, the induction of c-Myc expression by a viral oncoprotein involving p300 was first shown with E1A and subsequently demonstrated with the SV40 large T antigen soon after [73,74]. The similar function of these two viral oncoproteins highlights the benefits of using viruses as tools to study the molecular mechanisms behind cancer. Taken together, these findings demonstrate that E1A is able to modulate p300/CBP-mediated acetylation of both histone and non-histone proteins, resulting in dramatic changes in cell host gene expression.
In addition to p300/CBP, the E1A N-terminus, including CR1, interacts with a variety of other cellular proteins involved in chromatin modification and transcriptional regulation that are important for transformation [48]. Although p300/CBP appears to be the major determinant for transformation via the N-terminus, several other cellular factors that bind within this region are also important, such as PCAF, p400, TBP, GCN5 and TRRAP (Fig. 2) [48]. Moreover, these proteins function together in larger multimeric complexes which E1A can also modulate. For example, p300/CBP associates with the histone acetyltransferase PCAF and acts as a cofactor for several transcription factors, including p53 [75,76]. Fascinatingly, E1A has been shown to both disrupt binding of PCAF to p300/CBP as well as bind PCAF directly to sequester its activity [77].
The complexes E1A forms with multimeric histone acetyltransferases that involve the TRRAP adaptor protein are particularly important in E1A-mediated transformation [78]. One such TRRAP complex involves c-Myc, which when depleted reduces the transforming abilities of E1A [79]. E1A also interacts with the NuA4/TIP60 complex (containing TRRAP, p400, DMAP1, TIP60, RUBL2 and RUVBL1) and enhances its association with c-Myc [80]. While earlier studies demonstrated p400 was essential for E1A-mediated transformation, these studies used deletions of E1A (Δ26–35) that also prevented TRRAP binding, making it difficult to determine which protein E1A is binding directly [80,81]. E1A also targets the TRRAP complex involving GCN5 which is associated with c-Myc directed transcriptional activation [82]. Proteomic analysis of E1A's N-terminus suggested a preference for binding TRRAP-NuA4/TIP60 complexes, possibly suggesting that E1A can target two different TRRAP complexes [80]. It remains to be determined if E1A differentially regulates c-Myc mediated transcription depending on which TRRAP complex it binds, and whether this is context dependent or a result of different E1A proteoforms. Nevertheless, E1A may be able to modulate c-Myc-driven transcription of cellular genes essential for transformation through these interactions [83].
Overall, it is important to consider the complexity of interactions taking place within the N-terminus. For example, even though p300 has intrinsic histone acetylase activity, it also behaves as a hub protein with an interaction network spanning hundreds of cellular proteins [84]. Therefore, through binding p300, E1A has access to an astonishing number of p300-associated proteins. Even within the interaction map (Fig. 2), p300 and CBP interact with nearly half of all other proteins depicted. It is possible that on a genome-wide scale, promoter-bound p300 is found within a variety of different complexes. Some of these complexes are bound by E1A, which may act to either disrupt, stabilize, or reorganized their composition. Further adding to this complexity is that p300 and CBP are often treated interchangeably in the context of viral and cellular protein interactions; however, this is not necessarily true. Divergent functions between the two proteins exist, given that neither can compensate entirely for the other [85]. Whether or not E1A uses the overlapping functions of p300 and CBP to achieve a common goal, or if their unique functions are differentially exploited by E1A remains to be determined.
The role of p300/CBP as a tumour-suppressor has been previously reviewed and is widely accepted [59,86]. Other viral oncoproteins including SV40 large T antigen and HPV E6 and E7 are also able to bind and modulate the activity of p300/CBP in a variety of ways [19,[87], [88], [89]]. The interaction between E1A and p300/CBP clearly points towards cell cycle progression and sustained proliferation; however, when considering the diverse cellular roles p300/CBP partakes in, the E1A-p300/CBP interaction can be categorized into several hallmarks of cancer (some of which are discussed in the following sections).
2.1.3. E1A promotes transformation through modulating the ubiquitin-proteosome system
E1A's multi-faceted approach to targeting both tumour suppressors and oncogenes has also been observed through fine tuning PTMs resulting in ubiquitin-mediated proteasomal degradation. One protein complex implicated in this process is the Skp-Cullin-F-box (SCF) complex. Within the SCF complex, Skp1 acts as an adapter that binds a variety of F-box proteins — which provide substrate specificity — and Cul1 that functions as a scaffold by binding both Rbx1 and Skp1 [90]. Fascinatingly, E1A has been shown to target SCF complexes containing two different F-box proteins: Fbw7 (SCFFbw7) and Fbw1/β-TrCP (SCFβ-TrCP) [91,92]. In targeting SCFFbw7, E1A was shown to directly bind Roc1/Rbx1 and Cul1, resulting in an attenuated turnover of SCFFbw7 targets including the proto-oncogenes c-Myc, c-Myb, c-Jun and Cyclin E [91,93]. This pathway may be a common target among tumour viruses, as SV40 large T antigen has also been observed to interfere with Fbw7 [94]. Given the role of Fbw7 in regulating diverse cellular processes, which includes regulating the activities of several proto-oncogenes, it's role in E1A-mediated oncogenesis is most likely an important factor.
Molecular details regarding the ability of E1A to target SCFβ-TrCP are less characterized. Observations of HAdV-A12 transformed cells revealed that the cellular REST protein was functionally defective and depleted within the nucleus [95]. These observations were later extended mechanistically through experiments showing that E1A induces expression of SCFβ-TrCP which leads to ubiquitination and degradation of nuclear REST [92]. REST is involved in the regulation of a wide array of genes and has both oncogenic and tumour suppressive properties [96]. Interestingly, depending on the cell cycle phase, SCFβ-TrCP and SCFFbw7 can exert different effects of c-Myc stability [97]. β-TrCP-mediated ubiquitination was shown to antagonize Fbw7-mediated turnover of c-Myc resulting in accelerated cell cycle progression. Intriguingly, the relationship between E1A and Fbw7/β-TrCP has not been explored in this context. Concomitant E1A-mediated stabilization of c-Myc through SCFFbw7 and increased SCFβ-TrCP expression may be another important, and yet uncharacterized, factor in E1A-mediated transformation.
Furthermore, SCFβ-TrCP targets additional E1A interactors, such as the DYRK1A/B kinases, through the adapter protein DCAF7 [98,99]. Intriguingly, SCFβ-TrCP-mediated degradation of DYRK1A has been shown to be essential for cell cycle progression in HEK293 cells, which endogenously express E1A and require it for growth [100,101]. Given that cells expressing E1A mutants that are deficient in binding DYRK1A/B show increased proliferation and transformation, a link between E1A and DYRK1A degradation by SCFβ-TrCP in the nucleus might exist [102]. Furthermore, the degron motif identified within the N-terminus of DYRK1A does not appear to exist in DYRK1B, raising additional questions as to how E1A could regulate these proteins functions in the context of proliferation and or transformation.
Under normal cellular conditions, ubiquitin-mediated degradation of proteins provides powerful negative-feedback mechanisms that attenuate proliferative signalling; however, in the context of cancer, disruption of this system is often a contributing factor to sustained proliferation [18,103,104]. Cellular protein complexes like SCFFbw7 and SCFβ-TrCP regulate a multitude of oncoproteins and tumour suppressors, making them important factors involved in non-virally induced transformation [93,96]. E1A-mediated disruption of these pathways likely contributes to transformation in an analogous fashion.
2.1.4. Transformation by E1A's C-terminus is context dependent
The C-terminal region of E1A encompassing exon 2, including CR4, also consists of multiple overlapping protein interaction motifs like the N-terminus/CR1. This is exemplified by binding motifs for DCAF7 and CtBP1/2 which overlap with the bipartite nuclear localization sequence [98,105]. Additionally, a region preceding CR4 is responsible for binding FOXK1/2 in a phosphorylation-dependent manner. Paradoxically, the C-terminus of E1A is required for transformation in cells co-transformed with E1B; however, in the context of activated RAS it functions to suppress transformation [106,107]. The molecular mechanism by which E1A exploits these cellular proteins to promote transformation remains unclear.
CtBP1/2 was originally identified via its role as a negative regulator of E1A-mediated transformation [108]. CtBP1/2 functions as a transcriptional corepressor by interacting with chromatin modifying complexes and has well recognized roles in cancer [109,110]. Both CtBP1 and CtBP2 isoforms bind E1A through a highly conserved PxDLS amino acid motif initially identified within E1A, and this is necessary for transformation in cooperation with E1B [98,111]. However, whether CtBP1/2 is necessary for transformation with activated RAS remains obscure, as different studies have shown conflicting outcomes [98,112].
Importantly, a number of cellular proteins contain CtBP1/2 binding motifs that may compete with E1A for binding. Specifically, E1A has been shown to compete with the tumour suppressor CtBP interacting protein (CtIP) for binding to CtBP1/2 [111]. Characterization of CtIP has revealed a primary role in DNA repair and genome stability through interacting with Nbs1 within the Mre11-Rad50-Nbs1 (MRN) DNA damage sensor protein complex [113,114]. Additionally, CtIP has also been shown to regulate cell cycle progression by binding pRb and regulating E2F target genes [115]. CtIP binds pRb and CtBP through an LxCxE and PxDLS motif, respectively, suggesting E1A can compete with CtIP for binding to both targets using it's own highly similar motifs [116]. Intriguingly, even though E1A can outcompete CtIP binding to CtBP, E1A has also been shown to bind CtIP directly via the N-terminus [117]. The functional consequence of this interaction with E1A is unknown. Additionally, an interesting observation may be drawn between E1A and the MRN complex: CtIP and TRRAP both bind the E1A N-terminus and are associated with the MRN complex [114,118]. Furthermore, both HAdV E4 and E1B proteins have been shown to interact with the MRN complex (Fig. 2) resulting in its degradation (discussed in sections 2.2.2, 2.3.5); however, a direct link with E1A has not been established [119].
Preceding E1A's CtBP1/2 binding motif is a region responsible for binding DCAF7, which acts as an adapter for binding DYRK1A/B and HIPK2, as well as the FOXK1/2 binding region [98,99]. Point mutants within E1A that disrupt the interaction with DYRK1A/B and HIPK2, likely through inhibiting the association with DCAF7, demonstrated that both of these interactions are important for transformation with E1B. However, in the context of activated RAS, only certain DYRK1A/B and HIPK2 binding mutants exhibit suppressed transformation, suggesting the involvement of additional factors or that some of these interactions are qualitatively different than others [98,112,120].
Transformation with E1B using E1A mutants unable to bind FOXK1/2 has not been tested; however, when tested with activated RAS it appears this interaction is responsible for repressing transformation [102]. FOXK1/2 was also shown to bind the cutaneous low-risk HPV14 and HPV21 E6 proteins and this was similarly associated with suppressed transformation [102]. Collectively, these data show that the C-terminus of E1A influences transformation in a context-dependent fashion. In cooperation with E1B, the C-terminus complements the N-terminal oncogenic functions; however, with RAS, the C-terminus elicits tumour suppressive properties. Nevertheless, it seems that the C-terminus has clear functions involved in suppressing proliferation in the context of activated RAS [102]. In this context, these observations make the C-terminus an “anti” hallmark of cancer with respect to sustained proliferation.
Overall, the implications for each of these interactions, alone or in combination with one another, with respect to transformation require further investigation and may provide additional insight into the mechanisms of their anti-proliferative functions. Several consequences of the CtBP1/2 interaction have been established and will be addressed in the following section. Although speculative, potential consequences of binding DYRK1A/B, FOXK1/2 and HIPK2 will also be addressed in additional sections. Nevertheless, the consequences of E1A interacting with each of these targets likely results in either re-tasking their functions or in sequestering them to prevent their activity.
2.1.5. E1A influences tumour cell invasion and metastasis (anoikis/epithelialization)
The process of invasion and metastasis refers to tumour cell detachment, migration and secondary colonization [121]. From a molecular point of view, this process must be preceded by cellular changes that permit cancer cell detachment and survival. Since carcinomas are epithelial in nature, these cells have a physical barrier to surpass in order to migrate to distal sites: epithelial cells are anchored in place by cell-cell adhesion and contact with the extracellular matrix (ECM) [122]. To surpass this barrier, carcinoma cells must undergo an epithelial-mesenchymal transition (EMT) to become migratory, avoid death, and invade distal tissues. Through the study of HAdV-C5 E1A, we have learned many lessons on the EMT process. While E1A was originally described as an oncogene for its ability to induce cell cycle progression and cell growth, it has also been defined as a tumour suppressor gene based on its ability to trigger mesenchymal-epithelial transition (MET), the reverse of EMT. The earliest studies on this topic showed that E1A expression in cancer cells from different origins would cause their conversion to an epithelial phenotype. Specifically, E1A induces expression of cell-cell adhesion and junction molecules, cell polarization, repression of other cell type-specific programing, anchorage dependency and anoikis sensitivity [123]. Anoikis is a type of apoptosis that occurs when epithelial cells lose contact with neighbouring cells and the ECM, which serves to prevent aberrant epithelial cell detachment and dissemination [124]. Anoikis-resistance is an important characteristic of carcinomas that allows them to survive detachment from the ECM [18]. Indeed, seminal studies of E1A done by Frisch and Francis in 1994 led to the identification and naming of this specialized death program [124].
The epithelial cell program can be considered to be the default cell type because the transcription factors responsible for epithelial programming are ubiquitous and must be repressed by other cell type-specific factors to induce differentiation [125]. In order to convert cells to the epithelial default programming, E1A uses two strategies: relieve the repression of epithelial factors and remove the activation of other cell type-specific factors. As mentioned previously (section 2.1.4), E1A interacts with CtBP1/2, which is a key antagonist of the epithelial phenotype [126]. CtBP interacts with other repressive factors like δEF1/ZEB1 to repress E-cadherin expression, which is considered a primary suppressor of motility and is responsible for the majority of epithelial cell connections [122,124,127]. E1A sequesters CtBP1/2, preventing its antagonism of E-cadherin and the epithelial program. This interaction with CtBP1/2 is also linked to E1A's ability to sensitize cells to anoikis [23]. E1A binds to a set of nuclear acetylases, namely p300, CBP, and PCAF, to prevent these proteins from activating cell type-specific programs [126]. E1A has taught us much about MET, which could be taken as an anti-hallmark role, but the true biological picture is more complex. Both EMT and MET are important components in the invasion and metastasis hallmark. While tumour cells must initially undergo EMT to disseminate from their primary tumour location, they must subsequently use MET to seed and colonize distal sites [18]. The lessons E1A has taught us about engaging cells to undergo MET or epithelialization illustrate its usefulness in studying pathways related to the hallmarks and anti-hallmarks of cancer.
2.1.6. E1A affects angiogenesis
Tumour cells frequently exist in a state of hypoxia. Hypoxic conditions lead to stabilization of the transcription factor HIF1α, which can then activate transcription of genes involved in the hypoxia stress response. Hypoxia induces genes that promote the development and branching of new vasculature in the tumour microenvironment, a process termed angiogenesis, which is another hallmark of cancer. The primary mediator of angiogenesis is vascular endothelial growth factor (VEGF), which acts on vascular epithelial cells by binding VEGF receptors on their cell surface [128]. While the benefits of disrupting angiogenic signalling for HAdV replication remain unclear, E1A's reach does extend to this process. Some evidence suggests that E1A may be able to indirectly provide anti-angiogenic signals to neighbouring cells through a bystander effect [129]. Aside from this interesting possibility, E1A specifically disrupts expression of VEGF and factor VIII, an important angiogenic marker [130,131]. Through its interaction with p300/CBP, E1A mediates the repression of VEGF expression [132]. These experiments with E1A and p300/CBP were among the earliest to show these coactivator proteins interact with HIF1α to promote angiogenesis, contributing to our knowledge of the induction of angiogenesis as a hallmark of cancer [133]. Indeed, the HIF1α-p300/CBP interaction is now considered an important aspect of angiogenic signalling during hypoxia-related stress [134]. These findings implicate p300/CBP in yet another hallmark of cancer, further highlighting E1A as a powerful tool for studying cellular pathways, even in those seemingly unrelated to the HAdV replicative cycle.
2.1.7. E1A influences p53 signalling
Many of the functions carried out by E1A, such as aberrantly forcing cells to enter the cell cycle, can be viewed as an apoptosis-inducing stress. As a result of deregulating cellular proteins like pRb and p300, apoptosis is an impending certainty for the cell [35]. Indeed, E1A expression has been associated with the induction of p53-dependent apoptosis, but this is largely mitigated by the HAdV E1B proteins during infection [35]. Originally, p53-dependent apoptosis was thought to primarily occur via the DDR pathway [135]. HAdVs, along with many other viruses, provoke the DDR pathway in a variety of ways that predominately lead to apoptotic signalling through p53 [136]. Alternatively, p53-dependent apoptosis can be induced through oncogenic signalling by a separate pathway involving mouse tumour suppressor p19ARF (or p14ARF in humans). In fact, some of the earlier work on p19ARF-induced stability of p53 was facilitated through studying E1A [135]. Specifically, E1A-mediated inactivation of pRb and release of E2Fs induces gene expression of p19ARF which prevents MDM2 from targeting p53 for ubiquitin-mediated destruction [35,135]. Interestingly, p19ARF can disrupt the YY1-MDM2 complex that is important for ubiquitinating p53 for proteasomal degradation [137]. As mentioned previously, E1A disrupts the interaction between p300 and YY1 (section 2.1.2); however, it is unknown if this has any effect on p53-mediated apoptosis. Additionally, E1A may also promote apoptosis through the p19ARF-p53 pathway by binding pRb and a p400-containing TRRAP complex [138]. While the molecular mechanisms behind E1A's interaction with p53 have not been fully elucidated, it is clear E1A is heavily involved in the anti-hallmark process of promoting cell death.
Additionally, the interaction between E1A and p300/CBP may also connect to p53 signalling. E1A binding to p300/CBP induces acetylation of pRb, which promotes its interaction with MDM2 [61,139]. While the direct consequences of this interaction remain unclear, it is possible that E1A is modifying p53-dependent transactivation, while leaving apoptotic signalling unabated [35]. This possibility raises the question of whether the functions of p53 can be attenuated during oncogenesis without full loss of function. While cases have been reported of p53 attenuation through disruption of MDM2, MDM4, and p19ARF, this is an area for future research that could benefit from HAdV E1A as a molecular tool [140].
In addition, both DYRK1A and HIPK2, kinases targeted by E1A's C-terminus, have established roles in regulating p53 function through PTMs [141]. Depending on the severity of DNA damage, p53 can direct responses towards cell cycle arrest or apoptosis, which is generally reflected in the phosphorylation status of p53 [141]. Specifically, phosphorylation of p53 at S15 and S20 are observed under mild genotoxic stress and favour cell cycle arrest. Conversely, under severe genotoxic stress, phosphorylation of S46 can be observed and is an important marker for apoptosis. Intriguingly, S15 and S46 are targets of DYRK1A and HIPK2, respectively [142,143]. These observations may suggest that E1A, which binds these targets indirectly via DCAF7, is also modulating p53 activity and its associated hallmarks of cancer with regards to cell cycle arrest and apoptosis [99].
2.1.8. E1A influences autophagy
E1A has evidently contributed to our understanding of the underlying processes of apoptotic cell death (section 2.1.7). Beyond apoptosis, other forms of cell death and decomposition are becoming increasingly relevant in the field of cancer research [18]. Over the past decade there has been a growing body of evidence that implicates autophagy in the development of cancer [144,145]. Interestingly, this complex process involves both tumour suppressive and tumour promoting aspects. During infection, some viruses make use of autophagy to fuel their own replicative cycles through virally-induced autophagy. Since HAdVs infect lung epithelial cells, which are under constant hyperoxia that induces adaptive autophagy, it is unsurprising that HAdVs have evolved to make use of this scenario [146,147]. Adaptive autophagy leads to increased E1A expression and viral replication of HAdV-C2 [147]. While the mechanistic details surrounding the involvement of autophagy in the HAdV replicative cycle are not fully understood, it is hypothesized to be beneficial for escape of viral particles from early endosomes. Autophagosomal fusion with early endosomes creates amphisomes that appear to be beneficial to viral particle release [147]. One possible connection could be the interaction between E1A and the FOXK1/2 proteins, which have been implicated as regulators of autophagy [148]. Conversely, autophagy might be more harmful than helpful for HAdV replication, as it could be used by the host as a form of nutritional immunity [149]. Regardless of whether the virus benefits from these pathways or not, there is an apparent relationship that could be exploited to further study autophagy and how it relates to cancer.
2.1.9. E1A helps avoid immune detection and destruction
Like all viral infections, HAdVs are subject to detection by cellular immune surveillance mechanisms. Thus, HAdV must be equipped with tools to either avoid immune detection or to disrupt immune system-mediated destruction. Likewise, the immune system imposes comparable constraints on cancer cells, making the process of avoiding immune-mediated destruction an important hallmark of cancer [18]. Unsurprisingly, viral and cancer immune evasion strategies have much in common [150].
Specifically, HAdV infections trigger innate immune responses like interferon (IFN) and other pro-inflammatory cytokines [146]. Toll-like receptors (TLR) represent the conventional immune sensory molecule for detecting HAdV infections, with TLR4 and TLR9 being of particular importance. TLR4 binds to complement-bound HAdV particles and initiates an IL1β response. Intracellularly, TLR9 senses the HAdV genome and activates factors like MyD88, NFκB, MAPKs, and IRFs to mediate the anti-HAdV response. HAdV induces the cGAS/STING pathway during infection, which activates TBK1 and IRF3 to ultimately mount an IFNβ response [151]. Since the HAdV genome is an open-ended, linear, double-stranded DNA molecule, DDR sensors are activated during infection. These sensors identify the genome as a double-stranded DNA break and aim to repair it through non-homologous end joining (NHEJ). Similarly, with the TP protein primer bound to the 5’ ends of the HAdV genome, the cell is triggered to repair an inappropriate DNA-protein hybrid [151]. Fascinatingly, an additional mechanism used to suppress the host inflammatory response during viral infection involves the HAdV protein VII, which associates with cellular chromatin by acting as a viral histone. This results in the retention of HMGB1 on cellular chromatin, thereby preventing proinflammatory HMGB1-related danger signals [152,153].
Many HAdV proteins have some form of antiviral function to aid in avoiding immune recognition and killing, but E1A specifically works through transcriptional regulation of immune surveillance molecules and IFN effectors. E1A's approach to blocking the type I IFN response is multi-faceted. First, by interacting with and sequestering a number of histone acetyltransferases which are capable of modifying the cellular chromatin program, E1A can efficiently suppress the expression of IFN-stimulated genes (ISGs) [151]. As mentioned previously (section 2.1.2), through p300/CBP, E1A reduces H3K18 acetylation, which subsequently impairs ISG expression [69]. E1A also interacts with hBre1/RNF20 to block mono-ubiquitination of H2BK120, again causing a reduction in ISG production [154]. E1A makes use of this interaction to promote viral gene expression by repurposing hBre1 to act as a scaffold for hPaf1 recruitment [155]. It is interesting that HAdV E1A interacts with hBre1 to mediate suppression of ISG expression, since hBre1 has been reported to have tumour suppressive functions, such as upregulating p53 and downregulating oncogenes such as c-Myc and c-Fos [156].
While the N-terminus of E1A interacts with a number of histone acetyltransferases to mediate immune suppression, the C-terminal CR4 region has a different role in preventing immune-mediated destruction. Here, E1A interacts with RuvBL1 to prevent it from activating ISG promoters [157]. E1A's C-terminus also creates an important complex between FOXK1/2, DCAF7 and CtBP1/2. This complex allows E1A to downregulate a unique set of ISGs that are induced later during infection than a typical TLR-mediated IFN response [158]. Interestingly, it appears that the portion of E1A's C-terminus which engages in this complex is also involved in suppressing the Ras signalling pathway [158]. Overall, E1A seems to be suppressing the IFN response at the transcriptional level to weaken pathogen detection and antiviral response. IFN suppression is also an essential aspect of oncogenesis, as IFN pathways are capable of suppressing tumour growth and development [159]. Furthermore, many of the viruses that are truly oncogenic in humans rely on strong immune evasion strategies to avoid detection and destruction, ultimately contributing to their oncogenic potential [19].
Important lessons on how HAdV avoids immune detection can be taken from the highly oncogenic HAdV-A12. While the majority of HAdV species are inefficient at oncogenic transformation and incapable of producing tumours in immunocompetent rodents, HAdV-A12 is the exception. Using a chimeric E1A from HAdV-C5 and -A12, the alanine rich linker region located between CR2 and CR3 was suggested to be one of the determinants for HAdV-A12 oncogenicity. The underlying reason for this association is not fully understood; however, given this region's low sequence complexity and the failure to identify any protein targets of this region, it may simply function as a spacer between CR2 and CR3. This could subsequently influence steric effects involving interactions within these two adjacent regions, ultimately enhancing their unique functions [160].
Essential to its high oncogenic potential, HAdV-A12 E1A is able to repress the expression of genes involved in MHC class I antigen processing and presentation, allowing the virus to evade T-cell and NK-mediated killing. Investigating the differences between the highly oncogenic HAdV-A12 and its non-oncogenic counterparts has taught us about molecular mechanisms contributing to tumour evasion of the immune system. Similarly, HPVs can be classified as oncogenic or non-oncogenic, and their effects on the immune system may contribute to differences in their oncogenic potential [161].
HAdV-A12 infected cells have been shown to express lower levels of cell surface MHC-I. Specifically, HAdV-A12 disrupts MHC-I presentation by repressing expression of MHC as well as antigen processing genes. This can be attributed to E1A disrupting MHC transcriptional regulation via multiple distinct regulatory sites. Importantly, these sites belong to class I regulatory elements (CRE), which is a key point of control for MHC-I expression that also has implications in cancer [162]. HAdV-A12 E1A disrupts NFκB binding to CRE by disrupting the NFκB cytoplasmic/nuclear equilibrium such that NFκB is relocalized to the cytoplasm. HAdV-A12 E1A also promotes binding of nuclear factors like COUP-TF and N-CoR to the second region of the CRE, which is associated with repression of MHC transcription. Besides MHC itself, HAdV-A12 also represses expression of genes involved in processing and presenting antigens like TAP1, TAP2, LMP2, and LMP7. Notably, TAP1 and LMP2 share a bidirectional promoter that is normally regulated by NFκB. Thus, the impact of E1A on subcellular localization of NFκB, a transcription factor that promotes expression of proinflammatory pathways, has multiple transcriptional effects that promote immune evasion. Finally, foreign antigenic peptides present in the E1A C-terminus are much less abundant on MHC-I molecules in HAdV-A12 infected cells, possibly due to its inefficient processing. This could partially explain why this species is more likely to be oncogenic, as it has a reduced capacity to be detected by immune surveillance.
2.1.10. E1A impacts cell metabolism
Deregulating cellular energetics is another hallmark of cancer that can be induced by HAdV [18]. In particular, HAdV induces a metabolic phenotype reminiscent of the Warburg effect, otherwise known as aerobic glycolysis [163,164]. The Warburg effect involves an upregulation of glycolysis with a concurrent downregulation of cellular respiration despite ample oxygen that would otherwise favour cellular respiration [165]. In addition, it is becoming increasingly appreciated that cancers still perform a degree of metabolic activity related to cellular respiration, including the tricarboxylic acid cycle and oxidative phosphorylation [166]. HAdV-infected cells have a similar phenotype in this respect, which further emphasizes the utility of studying HAdV-mediated transformation [167].
E1A is a strong candidate for mediating metabolic reprogramming during HAdV infection as it is capable of interacting with a wide number of host-cell regulatory proteins, many of which can influence metabolism [24]. Some of the host-cell regulatory proteins that can interact with both E1A and regulate metabolic gene expression include c-Myc, p53 and pRb/E2F [168]. It is possible that interactions between E1A and c-Myc, mediated through E1A-TRRAP-c-Myc, E1A-p300-TRRAP-c-Myc, or E1A-p400-c-Myc complexes, are responsible for some of the metabolic changes observed during HAdV infection, although a direct interaction between E1A and c-Myc is also possible [[169], [170], [171]]. Likewise, a variety of interactions between E1A and pRb could also modulate metabolism [24,68,164,172,173]. The ability of E1A to activate p53 activity and the implications this may have on metabolic regulation, such as a theoretical increase in cellular respiration and decrease in glycolysis due to p53 induction of SCO2 and TIGAR, which serves to promote cellular respiration and inhibit glycolysis, respectively, remains to be fully understood [[174], [175], [176]]. However, little research has been done exploring how the interactions between these proteins and E1A can directly influence metabolism.
Recently, the E1A 13S-encoded isoform of HAdV-C5 was shown to be capable of regulating the expression of transcripts involved in a variety of metabolic pathways to a greater extent than the 12S isoform of E1A during infection [177]. These include an upregulation of genes involved in glycolysis and the tricarboxylic acid cycle along with a downregulation of genes involved in cellular respiration. Additionally, A549 cells that constitutively express 13S E1A have a drastically increased functional rate of glycolysis, which is not mirrored in A549 cells with constitutive expression of 12S E1A. It is possible that upregulation of SLC2A3, which encodes the GLUT3 transporter, is responsible for the increased utilization of glucose that occurs in 13S E1A expressing A549 cells. In addition, cells expressing the 13S E1A isoform, but not the 12S E1A isoform also have decreased functional rates of cellular respiration. No other study has yet shown a similar effect of E1A from other HAdV species on metabolism, but this does not rule out the possibility that other HAdVs may also regulate metabolism through E1A.
Furthermore, E1A plays a major role in regulating the expression of other HAdV proteins, such as E4orf1 (discussed in section 2.3.2), which has been implicated in HAdV metabolic regulation [24,178]. This pathway may be another way in which E1A can modulate host-cell metabolism during infection. The ability of E4orf1 to modulate metabolism during HAdV infection will be discussed below.
2.2. E1B
Adjacent to E1A in early region 1 is E1B, which encodes two major protein products, E1B–19K and E1B–55K (Fig. 1). Like E1A, much of our knowledge regarding E1B has been derived from studies using HAdV-C5 and -C2. However, as mentioned previously, viruses lacking the E1 region can be complemented by other HAdVs, indicating a level of functional conservation across different HAdV species [31]. During E1A-mediated transformation, cells will eventually enter a pro-apoptotic state that requires suppression of apoptosis and evasion of growth suppressors for efficient transformation [33,179]. This is perhaps best exemplified by the observation that E1A can fully transform primary fibroblasts from p53-deficient mice [180]. Both E1B–55K and E1B–19K demonstrate the ability to antagonize growth arrest and apoptosis, albeit with different molecular mechanisms and through different targets [181]. Interestingly, both E1B–55K and −19K can independently induce transformation with E1A [181].
Similar to viruses lacking E1A, E1B-deficient viruses also display severely crippled growth [25]. Infection of HeLa or KB cells with loss-of-function E1B–19K mutant viruses produces extremely large plaques, a result of increased apoptosis/enhanced cytopathic effects and degradation of both host cell and viral DNA, known as the cyt and deg phenotypes, respectively [182,183]. Much of what we know regarding E1B–19K has come from studies involving species C HAdVs; however, based on experiments using HAdV-A12, E1B–19K mutants similarly display large plaques as well as the cyt and deg phenotype [184]. Furthermore, HAdV-C2 E1B–19K is able to complement the cyt and deg phenotype of HAdV-A12, further suggesting functional similarity between E1B–19K from different HAdV species tested [185]. Infecting HeLa or KB cells with an E1B–55K mutant virus displays a prominent phenotype of late structural gene expression inhibition [186]. Additionally, E1B–55K mutants that could not bind p53 were unable to transform baby rat kidney (BRK) cells in conjunction with E1A [187]. E1B–55K mutant viruses have been investigated as anti-tumour agents based off of the relationship between E1B–55K and p53, though these studies were heavily critiqued and had varying degrees of success [188,189].
These seminal observations helped to lay the framework for what would be a detailed molecular understanding of E1B and its functions over the past decades. Regardless, investigation of the effect of E1B on apoptosis, cell cycle regulation, degradation of host proteins, and repression of gene transcription in the context of HAdV infection has generated important insights into how these processes impact transformation and tumourigenesis. Similar to E1A, the oncogenic activities of E1B contribute to a number of the hallmarks of cancer (Fig. 2).
2.2.1. E1B–55K targets p53 to inhibit apoptosis and cell cycle arrest
In order to complete transformation in concert with E1A, E1B must counteract E1A-induced growth arrest and pro-apoptotic signalling [[190], [191], [192]]. This is mainly achieved though counteracting the activities of p53 [191]. In particular, the role of E1B–55K-mediated inhibition of p53 has been discussed in other excellent reviews that supplement this one [14,193].
The p53 protein was first discovered through its association with E1B–55K and SV40 T antigen [39,194]. Under normal conditions p53 regulates various cellular activities, including cell cycle check points in response to DNA damage; thus, p53 inhibition by E1B is important for transformation and oncogenesis [179,195]. Mechanistically, E1B–55K has evolved a variety of strategies to inhibit or repress p53 function. These include localizing to p53-responsive promoters to make direct interactions with the N-terminus of p53, thereby repressing its transactivation; relocalization of p53 to perinuclear bodies to inhibit its transcriptional activity; forming an E3 ubiquitin-ligase complex with E4orf6 to polyubiquitinate p53, leading to subsequent proteasomal degradation; and lastly, regulation of p53 PTMs to modulate p53 activity [[196], [197], [198], [199], [200], [201]].
E1B–55K from various HAdV species, with the exception of HAdV-E4, are able to repress the transactivation activity of p53 [187,202]. HAdV E1B–55K proteins also differed in their ability to relocalize p53, with HAdV-C5 and HAdV-B16 E1B–55K relocalizing p53 to juxtanuclear aggresomes and HAdV-B34 and HAdV-F40 relocalizing p53 to distinct nuclear dots [202]. HAdV-A12 E1B–55K displayed some to no ability to relocalize p53 to aggresome-like structures; however, p53 relocalization to the cytoplasm has been observed [202]. Further investigation into HAdV-E4 E1B–55K also demonstrated its inability to transform BRK cells in conjunction with E1A [202]. Given that HAdV-E4 E1B–55K cannot repress p53 transactivation these findings highlight the importance of this interaction in the transformation process.
With respect to PTMs, E1B–55K influences both the acetylation and SUMOylation of p53. Acetylation of p53 by cellular factors, including PCAF and p300/CBP, is known to enhance p53 sequence-specific DNA binding [203]. Through simultaneously binding PCAF and p53, E1B–55K inhibits p53 acetylation by PCAF resulting in reduced DNA binding [203]. E1B–55K can also act as an E3 SUMO1-p53 ligase to SUMOylate p53 resulting in its relocalization to promyelocytic leukemia (PML) nuclear bodies (PML-NB) and subsequent nuclear export [201]. Intriguingly, HIPK2 (discussed in 2.1.7 as a target of E1A) is also known to localize to PML-NBs and form a complex with p53 that is important for HIPK2-mediate phosphorylation of S46 [204]. However, whether E1A and E1B are playing a concerted role in regulating p53 activity via HIPK2/p53 near or within PML-NBs is unknown.
Targeting p53 is an ideal strategy for E1B and this is made evident by the involvement of p53 in multiple hallmarks of cancer, including evading growth suppressors, resisting cell death, and genome instability and mutation [18]. By subverting p53, the primary functions of E1B result in evasion of growth suppression and prevention of cell death [18,179,195]. E1B's strategy to inhibit the tumour suppressive functions of p53 mirrors what is observed in cancer cells to evade or limit apoptosis [18]. Furthermore, even though a direct link between E1B and genome instability has not been observed, viruses frequently co-opt the DDR pathway [136]. Given these similarities, it's likely E1B utilizes p53s role in the DDR pathway in a parallel manner to what is observed in cancer. Importantly, the p53 pathway is targeted by a wide variety of viral oncoproteins, including HPV E6, HBV Hbx, HTLV Tax, KSHV LANA and SV40 large T antigen [19].
2.2.2. E1B 55K and the E1B-E4orf6 E3 ubiquitin-ligase complex target a variety of cellular proteins to promote transformation
Many of E1B–55K and E1B-E4orf6's shared targets, some of which are also shared with E1A (Fig. 2), that are important for viral replication can be found at PML-NBs. These factors include concentrated amounts of the tumour suppressor protein PML, as well as other regulatory proteins such as Sp100A, CBP, pRb, RAD51, Mre11, Daxx and p53 [205,206]. Several targets of this complex also include components of DDR pathways that are commonly activated upon viral infection [207].
The E1B-E4orf6 E3 ubiquitin-ligase complex, which includes Cul5, Elongins B/C and Rbx1, is able to target a set of cellular proteins, including p53, for polyubiquitination and subsequent proteasomal degradation [198,208]. In addition to p53, this complex targets the DNA damage sensor MRN complex (Mre11, Rad50, Nbs1), DNA damage effectors DNA ligase I, SPOC1, and Tip60, as well as DNA damage repair protein SMARCAL1 [[209], [210], [211], [212], [213]]. Depending on the HAdV species, the E1B-E4orf6 E3 ubiquitin-ligase complex may incorporate either Cul2 or Cul5, or both in the case of HAdV-A12, allowing them to differentially target cellular proteins for degradation [214,215]. For example, all HAdV species appear to degrade DNA ligase IV, whereas others preferentially degrade p53 and/or Mre11, among others [215]. Furthermore, there are examples of substrates that efficiently bind to E1B–55K and are not degraded, suggesting additional factors, like substrate orientation within the complex, may be critical [202]. Regardless of these differences, E1B–55K was shown to be the major substrate recognition component of the E4orf6/E1B55K E3 ubiquitin ligase across the different HAdV species [202].
Defects within these pathways have been noted to contribute to the hallmark of genome instability. Notably, Tip60 has also been shown to be targeted for degradation by low- and high-risk HPV E6 proteins [216]. Mre11 has been shown to suppress E1A/E1B-mediated transformation in BRK cells, demonstrating it is an important target that inhibits cell transformation [217]. Additionally, PML itself has also been shown to suppress transformation of BRK cells [218]. E1B–55K can interact with two PML isoforms, PML-IV and PML-V, the latter of which is able to recruit and modulate p53 and possibly contribute to transformation [219]. Furthermore, this strategy may be common among tumour viruses considering that PML is also targeted for degradation by HPV18 E6 and virtually every other DNA virus perturbs PML bodies in some way or another [220,221].
E1B–55K also targets the tumour suppressor Sp100A, which can activate p53-depedent transcription and reduce E1A/E1B–55K-mediated transformation [222]. E1B–55K recruits and binds both Sp100A and p53, thereby preventing Sp100A's activation of p53 [222]. Additionally, E1B–55K can induce the SUMOylation of Sp100A, relocalizing it to the nuclear matrix and cytoplasmic inclusion bodies where it cannot carry out its suppressive effects [222].
Death-associated protein Daxx plays a role in transcriptional regulation, intrinsic antiviral response, chromatin remolding, and apoptosis [223]. E1B in conjunction with cellular SUMO-targeted ubiquitin ligase RNF4 polyubiquinates Daxx, targeting it for proteasomal degradation [223]. Degradation of Daxx is required for efficient transformation of primary rodent cells via an unknown mechanism that appears to be p53-independent [206]. More recently, HPV16 E6 was shown to target Daxx to deregulate its activity in order to inhibit apoptosis [224]. Daxx is also known to act as a histone H3.3 chaperone when complexed with ATRX, which is also targeted by E1B-E4orf6 for degradation [225,226]. Cellular Daxx/ATRX has been suggested to be an important factor for HAdV gene expression by counteracting chromatin remodeling complexes [226]. Intriguingly, inactivating mutations in ATRX or Daxx are observed in a variety of cancers which exhibit an increased DNA damage response and other hallmarks of cancer including alternative lengthening of telomeres and genomic instability [227,228]. However, whether or not these changes are observed as a result of Daxx/ATRX degradation via by E1B-E4orf6 is unknown.
2.2.3. E1B–19K suppression of apoptosis
E1B–19K can suppress E1A-induced apoptosis by p53-dependent and -independent pathways [229]. In response to apoptotic stress during HAdV infection, small amounts of p53 are targeted to the mitochondria to promote apoptosis by causing mitochondrial dysfunction [230]. E1B–19K is also targeted to the mitochondria, where it enacts most of its anti-apoptotic function [230]. E1B–19K was shown to interact with p53 to rescue BRK cells from p53-induced apoptosis [230]. E1B–19K regulates p53's function by alleviating p53-mediated transcriptional repression, rather than blocking p53-mediated transactivation [231].
Investigation into E1B–19K's ability to block E1A-induced apoptosis led to its identification as a Bcl-2 homologue [179]. This discovery was significant as it represented one of the first identified viral Bcl-2 homologues [[232], [233], [234]]. Bcl-2 is an anti-apoptotic oncoprotein that was identified in human B-cell follicular lymphoma [235]. Despite a low level of sequence homology between Bcl-2 and E1B–19K, mutational analyses revealed conserved residues essential for E1B–19K function [229]. Bcl-2 can functionally substitute E1B–19K, as Bcl-2 comparably rescued infected cells from E1A-induced apoptosis and cooperated with E1A to induce transformation [232,236,237]. These findings were significant, as they cemented the role of both cellular and viral Bcl-2 family members in apoptosis and epithelial transformation [238]. Based on these properties of E1B–19K, a novel protein, Bcl-2/Adenovirus E1B 19 KDa Protein-Interacting Protein 2 (BNIP-2), was discovered through a yeast-two hybrid screen with E1B–19K and is currently of interest for its role in breast cancer evasion and metastasis [236,239,240].
E1B–19K likely inhibits apoptosis during transformation through the TNFα and Fas ligand cell death pathways via its interaction with pro-apoptotic proteins BAX and BAK [229,[241], [242], [243]]. Other viral oncogenes, such as HPV E6 have also been shown to target BAX, leading to its degradation thereby preventing its pro-apoptotic effects [244]. Like Bcl-2, E1B–19K can also act as a BH3 receptor [242]. BAX and BAK are activated by caspase-8-cleaved BID, triggering a conformational change leading to exposure of their BH3 domain [245,246]. E1B–19K can then bind to the BH3 domain, blocking BAK and BAX oligomerization, subsequent caspase-9 activation, the release of pro-apoptotic mitochondrial proteins, and ultimately, apoptosis [179,246,247]. However, there are conflicting results regarding the necessity of E1B–19K for efficient transformation, as HAdV-A12 and HAdV-C5 E1B–19K was not required for efficient oncogenic transformation in both primary BRK and baby mouse kidney cells [184,248,249].
Overall, E1B–19K can inhibit apoptosis by p53-dependent and -independent pathways and promote the growth of epithelial carcinomas. Studies of viral oncogenes such as E1B–19K have implicated BAX/BAK, as well as p53, as critical targets involved in apoptosis and epithelial tumourigenesis, thus contributing to our knowledge of cellular strategies involved in the resisting cell death hallmark of cancer [179].
2.3. E4
The HAdV E4 gene, located at the right end of the genome, is involved in lytic infection and oncogenesis [21]. The E4 region encodes for one precursor RNA that is alternatively spliced to generate multiple polypeptides with a variety of functions that aid in viral replication [250]. With the exception of species F HAdVs, the E4 region encodes for up to 7 polypeptides which are named E4orf1 through E4orf6/7 (Fig. 1). Similar to E1A and E1B, our understanding of the E4 region has predominately been from studies involving HAdV-C5 and -C2. Furthermore, this region is homologous among different HAdV species and shows a similar sequence organization [250]. Studies, predominately using HAdV-C5, have demonstrated the entire E4 region is indispensable for viral replication. However, the expression of only one or a few of the E4 proteins can compensate for the absence of the others; for example, E4orf6 was capable of compensating for the absence of all other E4 proteins to promote successful viral replication [251]. Four gene products of the E4 region, namely E4orf1, E4orf3, E4orf4 and E4orf6, have been implicated in oncogenic pathways that are relevant to several hallmarks of cancer (collectively summarized as E4 in Fig. 2) [14,208]. These E4 proteins operate by interacting with key viral and cellular regulatory components involved in cell cycle control, transcription, apoptosis and DNA repair.
2.3.1. E4orf1 sustains proliferative signalling
HAdV-D9 E4orf1 can promote proliferative signalling and survival by selectively activating phosphatidylinositol 3-kinase (PI3K) [252]. The C-terminus of E4orf1 contains a functional PDZ domain-binding motif that can interact with several PDZ domain-containing proteins involved in signal transduction [252]. These proteins include the multi-PDZ protein (MUPP1) and three membrane-associated guanylate kinase family proteins: DLG1, MAGI-1 and ZO-2 [[253], [254], [255], [256]]. HAdV-D9 E4orf1 was among the first viral oncoproteins identified to have a PDZ domain-binding motif, alongside other viral oncoproteins including HTLV-1 Tax and high-risk HPV E6 [254,257,258]. The interaction of E4orf1 with these PDZ proteins causes the localization of signalling complexes, including assembled receptors and cytosolic factors, to the plasma membrane for the selective activation of PI3K [252]. The PI3K family of enzymes are involved in cellular functions such as cell growth, proliferation, differentiation and survival; therefore, the activation of PI3K is crucial for the oncogenic potential of HAdV-D9 E4orf1 to promote proliferative signalling and survival [259].
Although E4orf1 proteins from HAdV species A, B and C stimulate PI3K, they have limited transforming potential compared to species D [259,260]. The unique oncogenic activity of HAdV-D9 E4orf1 is in its ability to selectively interact with the tumour suppressor protein ZO-2, causing its sequestration within the cytoplasm [256]. Furthermore, HAdV-D9 E4orf1 has been shown to directly bind the p85 regulatory and p110 catalytic PI3K subunits to form a cytoplasmic heterocomplex. This complex is recruited to the plasma membrane in a DLG1-dependent fashion to stimulate PI3K catalytic activity and promote oncogenic cellular transformation [261]. Thus, the serotype-specific oncogenic activity of E4orf1 may be a result of the DLG1-dependent stimulation of PI3K catalytic activity that leads to sustained proliferative signalling.
Interestingly, the observation that E4orf1 activates PI3K in a DLG1-dependent manner has provided valuable insight into viral carcinogenesis. A study conducted on the activation of the PI3K pathway by E4orf1 identified that the increase in PI3K activity was not a result of DLG1 inactivation, as previously thought, but that E4orf1 promotes DLG1 activity to direct PI3K activation [262]. This observation not only elucidated a mechanism to promote proliferative signalling by E4orf1 but revealed a previously unrecognized DLG1 oncogenic function. This oncogenic function of DLG1 may be important given that high-risk HPV E6 proteins require DLG1 to promote invasive properties in cervical carcinoma cells [263,264].
2.3.2. Metabolic changes associated with E4orf1
The first HAdV protein identified to directly influence host-cell metabolism and induce a cancer-like metabolic phenotype was E4orf1 [265,266]. In two separate studies, HAdV-C5 E4orf1 was responsible for mediating increases in both glycolysis and glutaminolysis [265,266]. This upregulation occurs through an interaction between E4orf1 and c-Myc, which leads to the transcription of genes related to both metabolic pathways. E4orf1 was found to be sufficient on its own to induce these metabolic changes without the presence of any other HAdV proteins. In addition to glycolysis, genes involved in nucleotide metabolism, especially the pentose phosphate pathway are also upregulated due to E4orf1 [266]. Interestingly, this metabolic phenotype bears a striking resemblance to the metabolic reprogramming that occurs in cancer, namely an upregulation of glycolysis and glutaminolysis with a concurrent decrease in cellular respiration [163]. E4orf1 is not solely responsible for the full extent of metabolic changes that occur during HAdV infection, since it does not appear to cause decreased cellular respiration [266]. However, other HAdV proteins, such as E1A, are possible candidates that may contribute to the full extent of metabolic reprogramming during HAdV infection in conjunction with E4orf1 (discussed in 2.1.10) [177]. As for other HAdV species, HAdV-D36 E4orf1 has been implicated in modulating a wide array of metabolic pathways in infected animal models, some of which are linked to obesity [267]. Considering the link between cancer and obesity, it is interesting to consider parallels between all three disease states [268]. The role of E4orf1 in metabolic regulation of other HAdV species besides C and D have not yet been characterized. However, HAdV-F types do not contain E4orf1, which could point to a larger role for E1A or another HAdV-F viral protein in regulating metabolism during infection [269]. Like E1A, E4orf1 could contribute to the cancer-like metabolic reprogramming of infected host-cell metabolism which phenotypically resembles the metabolic changes that lead to aerobic glycolysis, which is also considered to be a hallmark of cancer [18,163].
2.3.3. E4orf3 and E4orf6 redundantly target p53 tumour suppressor activity
Upon activation, p53 functions as a transcription factor, regulating the expression of numerous genes to mediate its anticancer and antiviral activities [270,271]. As p53 activation plays an important role in genome stability and regulating cell death, inhibition of p53 activity can promote oncogenesis.
E4orf3 and E4orf6 proteins redundantly target p53 through different mechanisms to prevent pro-apoptotic pathways. E4orf6 works alongside E1B–55K, forming an E3 ubiquitin ligase complex to degrade p53 and inhibit its transcriptional activity [198,209,272,273]. In this E3 ligase complex, E1B–55K provides the substrate recognition function, while E4orf6 assembles the specific cellular components needed for proteasomal degradation of p53 [198]. In comparison, E4orf3 inactivates p53 independently of E1B–55K by forming a scaffold within the nucleus that directs H3K9 methylation at p53 target promoters. This H3K9 methylation causes heterochromatin assembly to prevent p53 binding and activation [274,275]. As p53 is a critical tumour suppressor, inhibition of its transcriptional activity by either E4orf6/E1B–55K or E4orf3 may inhibit pro-apoptotic pathways and contribute to the resistance of the cell death hallmark of cancer. This activity would support the transforming functions of these viral proteins, as observed with HAdV-C5 E4orf6/E1B–55K [276].
2.3.4. E4orf3 targets PML-NBs
The E4orf3 protein may promote proliferative signalling by targeting PML-NBs. PML-NBs are nuclear structures within the host cell that are targeted by DNA viruses in the early stages of an infection due to their role in viral detection and subsequent cellular response [277,278]. E4orf3 of HAdV-C5 and -C2 can disrupt the cellular regulatory functions of PML-NBs by specifically targeting the PML-II isoform to form elongated track-like structures distributed throughout the nucleus [279,280]. This ability of E4orf3 to inhibit PML-NBs through the formation of unusual track-like nuclear structures is conserved among a variety of HAdVs [281]. As PML-NBs have several cellular regulatory functions, including induction of apoptosis, maintenance of genome stability, senescence and the interferon response, abrogation of PML activities have been implicated in a variety of cancers to sustain proliferative signalling [277]. As a result, inhibition of PML-NBs by the E4orf3 HAdV protein is likely to promote proliferative signalling, contributing to its transforming properties [218]. The diverse functions of PML-NBs are likely a consequence of a variety of protein populations known to associate with PML-NBs, including the MRN DNA repair complex, which are also inhibited by the activity of E4 gene products [119].
2.3.5. E4orf3, E4orf4 and E4orf6 redundantly target DNA damage repair pathways
The E4orf3, E4orf4 and E4orf6 proteins redundantly target cellular pathways involved in the DDR pathway, including the MRN complex, which is also targeted by E1B and potentially E1A (Fig. 2) [119]. The MRN complex can detect double-stranded breaks (DSB) and signal through ATM kinase to phosphorylate several downstream proteins involved in cell cycle control and DDR [[282], [283], [284]]. Compared to wild-type, mutant HAdV infections containing deletions in their E4 region demonstrated intense activation of cellular ATM/ATR-mediated DDR pathways [119,285]. Additionally, deletions of the E4 region result in viral genomes forming concatemers due to the inability to inactivate the MRN complex involved in the DDR pathway [119,286]. Consequently, viral genomes are too large to be packaged and this results in abortive infection. As the MRN complex is involved in activation of ATM signalling, these data indicate that E4 early proteins are crucial for suppression of host DDR through inhibition of the MRN complex which also prevents concatemerized viral genomes. Interestingly, signalling through ATM in response to genotoxic stress ultimately leads to p53-mediated apoptosis via HIPK2 phosphorylation of S46 [141]. How E1A, E1B and E4 may converge on p53 signalling (Fig. 2) and if this involves HIPK2 is not fully understood and warrants further investigation.
Although E4orf6 and E4orf3 both target the MRN complex in addition to E1B, the mechanism by which they inhibit this DDR pathway differs. E4orf3 inhibits the activation of the MRN complex by relocalizing it to PML tracks and excluding it from sites of viral replication [119]. By sequestering the MRN complex away from viral genomes, this prevents activation of the DDR pathway [119]. Alternatively, degradation of the MRN complex by E4orf6 is dependent on its interaction with E1B–55K and formation of the E3 ubiquitin ligase complex to target these proteins for proteolytic degradation [287]. Interestingly, the ability of E4orf3 to relocalize the MRN complex is specifically a feature of HAdV-C5, meaning DSB repair (DSBR) is redundantly inhibited by E4orf3 and E4orf6 in a species-specific manner [288,289]. Relocalization or degradation of the MRN complex to promote efficient viral replication, inhibits its DSBR activity and as a result, may contribute to genomic instability and mutation that could contribute to the so called ‘hit-and-run’ transformation [119].
E4orf3 and E4orf6 also inhibit the cellular DDR pathway through DNA-dependent protein kinase (DNA PK) [290]. DNA PK is a member of the PI3K-related kinase family and is best known for its involvement in DSBR through NHEJ [291]. DNA PK is also involved in the immune system V(D)J recombination, as well as cell cycle progression and telomere maintenance [[292], [293], [294]]. E4orf3 and E4orf6 have been shown to bind DNA PK, preventing DNA PK-dependent DSBR [295]. The redundant inhibition of DNA PK by E4orf3 and E4orf6 may also contribute to genome instability and mutation through prevention of DSBR.
E4orf4, together with protein phosphatase 2A (PP2A), has recently been shown to reduce phosphorylation of ATM and ATR substrates, inhibiting the DDR pathway [296]. Reduction of ATM and ATR signalling by E4orf4 is also dependent on its biphasic functional interaction with DNA PK [297]. The interaction between E4orf4 and DNA PK is required for reduction of ATM and ATR signalling during early HAdV infection; however, during the later stages of infection DNA PK activity is inhibited by E4orf4 to promote viral replication [297]. Although deficiencies in DDR pathways have been shown to contribute to tumour development [298,299], manipulation of the remaining DDR pathways by E4orf4 in malignant cells with existing mutations in DDR genes may promote cell death [297].
Another aspect of HAdV-A12's unique oncogenicity (discussed in 2.1.9) is the ability of E4orf6 to inhibit topoisomerase IIβ-binding protein 1 (TopBP1) to further suppress the DDR pathway, independent of E1B–55K. TopBP1 is involved in numerous cell processes, including DDR and cell cycle checkpoint control [[300], [301], [302], [303], [304]]. This involvement is through activation of the ATR/Chk1 signalling pathway [305]. HAdV-A12 E4orf6 degrades TopBP1, thereby challenging ATR signalling and inhibiting DDR [306]. Interestingly, HAdV-C5 has demonstrated no measurable effect on TopBP1, further indicating that different HAdVs may target the DDR in host cells through different mechanisms [306]. As TopBP1 is involved in both DDR and cell cycle checkpoint control, its inhibition by HAdV-A12 E4orf6 likely promotes genome instability and mutation, and possibly proliferative signalling.
The E4 region and its potentially oncogenic gene products E4orf1, E4orf3 and E4orf6, target specific cellular proteins and pathways to promote HAdV replication that may also contribute to the hallmarks of cancer. Studying the mechanisms by which E4 regulates cell cycle control, DDR, and apoptosis has revealed novel oncogenic functions of cellular proteins as well as mechanisms by which oncogenic viruses may induce transformation. Therefore, the study of the E4 has provided valuable insight in understanding the molecular basis of cancer.
3. Why doesn't HAdV cause human cancer?
Given the large body of evidence discussed above, which clearly links many different HAdV functions to cancer-relevant pathways, why doesn't HAdV cause actual human malignancies? Unlike a subset of HPVs and HPyVs, which do cause human cancers, HAdVs only cause tumours in heterologous animal models. The mechanisms behind the inability of HAdVs to cause human cancer is not entirely understood. Simplistically, tumourigenesis post-infection with HAdV is only observed in animal hosts that the virus is either unable to replicate in, or replicates poorly in [307]. The inability to complete a lytic infection ending in the death of the host cell may provide the time and pressure necessary to allow viral genome integration and acquire the additional mutations to develop a tumour. Although likely a contributing factor, HAdV infections in humans can be persistent, suggesting that the time interval between infection and cell death is not solely responsible for the lack of HAdV-mediated tumourigenesis in humans [308]. Furthermore, studies of HAdV transformation using viral DNA or vectors expressing HAdV oncogenes generally suggest that oncogenic transformation of primary human kidney cells is fundamentally less efficient than parallel experiments using primary rodent kidney cells [309,310]. These results suggest that there may be inherent molecular mechanisms in human cells that limit the ability of HAdV to mediate full-blown oncogenic transformation.
In addition, HAdV-induced tumourigenesis is generally restricted to neonatal animals with underdeveloped immune systems or at immune privileged sites [4,12,20]. This suggests that HAdV-transformed cells are quite susceptible to immune clearance. Given that animals with short gestation periods (i.e. mice, rats and hamsters) have relatively immature immune systems at birth compared to humans, it could be argued that humans are simply never in an immunological state susceptible to HAdV-mediated transformation [311]. Again, although likely a contributing factor to the inability of HAdV to induce human tumours, no evidence has been discovered to support a role for HAdV in tumourigenesis in immunocompromised humans.
Finally, it is important to understand that no virus, at least to our knowledge, has evolved to specifically cause cancer. With that in mind, it stands to reason that there are far more similarities between HAdV and other more famous tumour viruses, many of which we have outlined throughout this review. These differences in oncogenicity suggest that even though HAdVs possess the repertoire of tools to promote carcinogenesis (Fig. 2), it's true oncogenic potential is likely overshadowed by additional factors within the replicative cycle. Given the molecular similarities between HAdV and other tumour viruses, studying HAdV-mediated transformation has significantly contributed to our knowledge of carcinogenesis.
3.1. What about zoonotic adenovirus infections and human cancer?
In his 2008 Nobel lecture, Professor Harold zur Hausen pointed out the possibility that animal polyomaviruses and papillomaviruses are sufficiently thermostable such that they can survive heating in meat prepared medium or rare without significant loss of infectivity [312]. Like HAdV, these viruses are typically non-oncogenic in their natural animal hosts, but can exhibit carcinogenicity in heterologous species, raising the possibility that their ingestion could play an as yet undescribed role in the increased human colorectal, lung and breast carcinogenesis associated with red meat consumption [312]. Like the more famous members of the small DNA tumour viruses, adenoviruses are similarly resistant to inactivation by heating and multiple members have been identified in cattle and other domestic or wild meat animals, at least some of which are oncogenic in newborn hamsters [[313], [314], [315]]. Thus, any of the small DNA tumour viruses could represent viral agents contributing to human cancers associated with red meat consumption. Despite this possibility, comprehensive investigations of transcriptomic and genomic datasets from thousands of human cancers from dozens of cancer types did not identify sequences from adenoviruses from any species [316,317].
4. Conclusions
At this point, it seems unlikely that HAdV has a causal role in any appreciable subset of human cancers. However, studies of HAdV have a long history of teaching us about cellular biology and cancer. As examples, splicing was originally described by Nobel Prize winner Dr. Phillip Sharp based on his work with HAdV, and HAdV was the first human virus shown to be oncogenic [318,319]. As illustrated by specific examples in this review, studies of HAdV have provided a wealth of knowledge relevant to understanding oncogenesis, including the regulation of cell cycle control, DNA replication, DNA damage response, transcription, mRNA processing, metabolism, immunological responses and apoptosis, many of which are central to the hallmarks of cancer. Studies of HAdV oncoproteins like E1A, E1B and E4 have identified interactions with cellular regulators and identified protein interaction motifs that could have useful applications in protein engineering, protein network manipulation, metabolic reprogramming and synthetic biology [24,27,320,321]. In addition, the well-studied and readily manipulated HAdV genome is the foundation for vectors for gene therapy, oncolytic viruses and current vaccines [322,323].
CRediT authorship contribution statement
T.M.T: Conceptualization, Writing – Original Draft, Writing – Review and Editing. M.J.D: Conceptualization, Writing – Original Draft, Writing – Review and Editing. K.M.M: Writing – Review and Editing. A.M.E: Writing – Review and Editing. M.A.P: Writing – Review and Editing. J.S.M: Conceptualization, Writing – Review and Editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a grant from the Canadian Institute of Health Research (MOP-148689) to J.S.M. M.J.D. and K.M.M. were supported RGE Murray Scholarships and M.J.D currently holds and Ontario Graduate Scholarship. We apologize to the many colleagues whose excellent research was not cited directly in this review. In many cases, it was necessary to cite comprehensive reviews as the volume of relevant research was simply too extensive to cite directly.
References
- 1.Dodge M.J., MacNeil K.M., Tessier T.M., Weinberg J.B., Mymryk J.S. Emerging antiviral therapeutics for human adenovirus infection: recent developments and novel strategies. Antivir. Res. 2021;188:105034. doi: 10.1016/j.antiviral.2021.105034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilleman M.R., Werner J.H. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 1954;85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 3.Rowe W.P., Huebner R.J., Gilmore L.K., Parrott R.H., Ward T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 4.Mukai N., Kalter S.S., Cummins L.B., Matthews V.A., Nishida T., Nakajima T. Retinal tumor induced in the baboon by human adenovirus 12. Science. 1980;210:1023–1025. doi: 10.1126/science.7434012. [DOI] [PubMed] [Google Scholar]
- 5.Yabe Y., Samper L., Bryan E., Taylor G., Trentin J.J. Oncogenic effect of human adenovirus type 12, in mice. Science. 1964;143:46–47. doi: 10.1126/science.143.3601.46. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi S., Mukai N. Retinoblastoma-like tumors induced by human adenovirus type 12 in rats. Canc. Res. 1974;34:1646–1651. [PubMed] [Google Scholar]
- 7.Schwarz, E.; Freese, U.K.; Gissmann, L.; Mayer, W.; Roggenbuck, B.; Stremlau, A.; zur Hausen, H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 1985 314, 111–114, doi:10.1038/314111a0. [DOI] [PubMed]
- 8.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams J.F., Zhang Y., Williams M.A., Hou S., Kushner D., Ricciardi R.P. E1A-based determinants of oncogenicity in human adenovirus groups A and C. Curr. Top. Microbiol. Immunol. 2004;273:245–288. doi: 10.1007/978-3-662-05599-1_8. [DOI] [PubMed] [Google Scholar]
- 10.Endter C., Dobner T. Cell transformation by human adenoviruses. Curr. Top. Microbiol. Immunol. 2004;273:163–214. doi: 10.1007/978-3-662-05599-1_6. [DOI] [PubMed] [Google Scholar]
- 11.Greber U.F., Flatt J.W. Adenovirus entry: from infection to immunity. Annu. Rev. Virol. 2019;6:177–197. doi: 10.1146/annurev-virology-092818-015550. [DOI] [PubMed] [Google Scholar]
- 12.Bernards R., Van der Eb A.J. Adenovirus: transformation and oncogenicity. Biochim. Biophys. Acta. 1984;783:187–204. doi: 10.1016/0167-4781(84)90029-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhou R., Caspi R.R. Ocular immune privilege. F1000 Biol. Rep. 2010;2 doi: 10.3410/B2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ip W.H., Dobner T. Cell transformation by the adenovirus oncogenes E1 and E4. FEBS Lett. 2020;594:1848–1860. doi: 10.1002/1873-3468.13717. [DOI] [PubMed] [Google Scholar]
- 15.Graham F.L., Rowe D.T., McKinnon R., Bacchetti S., Ruben M., Branton P.E. Transformation by human adenoviruses. J. Cell. Physiol. Suppl. 1984;3:151–163. doi: 10.1002/jcp.1041210418. [DOI] [PubMed] [Google Scholar]
- 16.Thomas D.L., Shin S., Jiang B.H., Vogel H., Ross M.A., Kaplitt M., Shenk T.E., Javier R.T. Early region 1 transforming functions are dispensable for mammary tumorigenesis by human adenovirus type 9. J. Virol. 1999;73:3071–3079. doi: 10.1128/JVI.73.4.3071-3079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison A.J., Benkő M., Harrach B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Mesri E.A., Feitelson M.A., Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham F.L. The Adenoviruses. 1984. Transformation by and oncogenicity of human adenoviruses; pp. 339–398. [Google Scholar]
- 21.Täuber B., Dobner T. Adenovirus early E4 genes in viral oncogenesis. Oncogene. 2001;20:7847–7854. doi: 10.1038/sj.onc.1204914. [DOI] [PubMed] [Google Scholar]
- 22.Avvakumov N., Kajon A.E., Hoeben R.C., Mymryk J.S. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004;329:477–492. doi: 10.1016/j.virol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Frisch S.M., Mymryk J.S. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 24.King C.R., Zhang A., Tessier T.M., Gameiro S.F., Mymryk J.S. Hacking the cell: network intrusion and exploitation by adenovirus E1A. MBio. 2018;9 doi: 10.1128/mBio.00390-18. e00390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones N., Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 26.Radko S., Jung R., Olanubi O., Pelka P. Effects of adenovirus type 5 E1A isoforms on viral replication in arrested human cells. PloS One. 2015;10 doi: 10.1371/journal.pone.0140124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelka P., Ablack J.N.G., Fonseca G.J., Yousef A.F., Mymryk J.S. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avvakumov N., Wheeler R., D'Halluin J.C., Mymryk J.S. Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J. Virol. 2002;76:7968–7975. doi: 10.1128/JVI.76.16.7968-7975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang A., Tessier T.M., Galpin K.J.C., King C.R., Gameiro S.F., Anderson W.W., Yousef A.F., Qin W.T., Li S.S.C., Mymryk J.S. The transcriptional repressor BS69 is a conserved target of the E1A proteins from several human adenovirus species. Viruses. 2018;10 doi: 10.3390/v10120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rademaker H.J., El Hassan M.A.A., Versteeg G.A., Rabelink M.J.W.E., Hoeben R.C. Efficient mobilization of E1-deleted adenovirus type 5 vectors by wild-type adenoviruses of other serotypes. J. Gen. Virol. 2002;83:1311–1314. doi: 10.1099/0022-1317-83-6-1311. [DOI] [PubMed] [Google Scholar]
- 31.Brusca J.S., Chinnadurai G. Transforming genes among three different oncogenic subgroups of human adenoviruses have similar replicative functions. J. Virol. 1981;39:300–305. doi: 10.1128/JVI.39.1.300-305.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayley S.T., Mymryk J.S. Adenovirus e1a proteins and transformation (review) Int. J. Oncol. 1994;5:425–444. doi: 10.3892/ijo.5.3.425. [DOI] [PubMed] [Google Scholar]
- 33.Berk A.J. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 34.Turnell A.S., Mymryk J.S. Roles for the coactivators CBP and p300 and the APC/C E3 ubiquitin ligase in E1A-dependent cell transformation. Br. J. Canc. 2006;95:555–560. doi: 10.1038/sj.bjc.6603304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallimore P.H., Turnell A.S. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene. 2001;20:7824–7835. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- 36.Chinnadurai G. Opposing oncogenic activities of small DNA tumor virus transforming proteins. Trends Microbiol. 2011;19:174–183. doi: 10.1016/j.tim.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe J.A., Bayley S.T. Effects of Ad5 E1A mutant viruses on the cell cycle in relation to the binding of cellular proteins including the retinoblastoma protein and cyclin A. Virology. 1992;186:15–24. doi: 10.1016/0042-6822(92)90057-v. [DOI] [PubMed] [Google Scholar]
- 38.Whyte P., Buchkovich K.J., Horowitz J.M., Friend S.H., Raybuck M., Weinberg R.A., Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 39.Lane D.P., Crawford L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 40.Kress M., May E., Cassingena R., May P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 1979;31:472–483. doi: 10.1128/JVI.31.2.472-483.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linzer D.I., Levine A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 42.Baker S.J., Fearon E.R., Nigro J.M., Hamilton S.R., Preisinger A.C., Jessup J.M., VanTuinen P., Ledbetter D.H., Barker D.F., Nakamura Y. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 43.Helt A.-M., Galloway D.A. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- 44.Fattaey A.R., Harlow E., Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol. Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267-7277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21:2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelka P., Ablack J.N.G., Shuen M., Yousef A.F., Rasti M., Grand R.J., Turnell A.S., Mymryk J.S. Identification of a second independent binding site for the pCAF acetyltransferase in adenovirus E1A. Virology. 2009;391:90–98. doi: 10.1016/j.virol.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Ablack J.N.G., Pelka P., Yousef A.F., Turnell A.S., Grand R.J.A., Mymryk J.S. Comparison of E1A CR3-dependent transcriptional activation across six different human adenovirus subgroups. J. Virol. 2010;84:12771–12781. doi: 10.1128/JVI.01243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasti M., Grand R.J.A., Mymryk J.S., Gallimore P.H., Turnell A.S. Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J. Virol. 2005;79:5594–5605. doi: 10.1128/JVI.79.9.5594-5605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 50.Longworth M.S., Dyson N.J. pRb, a local chromatin organizer with global possibilities. Chromosoma. 2010;119:1–11. doi: 10.1007/s00412-009-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burkhart D.L., Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Canc. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeCaprio J.A. How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology. 2009;384:274–284. doi: 10.1016/j.virol.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Knudsen E.S., Nambiar R., Rosario S.R., Smiraglia D.J., Goodrich D.W., Witkiewicz A.K. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun. Biol. 2020;3:158. doi: 10.1038/s42003-020-0873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckner R., Ewen M.E., Newsome D., Gerdes M., DeCaprio J.A., Lawrence J.B., Livingston D.M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 55.Harlow E., Whyte P., Franza B.R., Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol. Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579-1589.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howe J.A., Mymryk J.S., Egan C., Branton P.E., Bayley S.T. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arany Z., Newsome D., Oldread E., Livingston D.M., Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 58.Goodman R.H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 59.Iyer N.G., Ozdag H., Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 60.Wang H.G., Moran E., Yaciuk P. E1A promotes association between p300 and pRB in multimeric complexes required for normal biological activity. J. Virol. 1995;69:7917–7924. doi: 10.1128/JVI.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan H.M., Krstic-Demonacos M., Smith L., Demonacos C., La Thangue N.B. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 62.Markham D., Munro S., Soloway J., O'Connor D.P., La Thangue N.B. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Julian L.M., Palander O., Seifried L.A., Foster J.E.G., Dick F.A. Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene. 2008;27:1572–1579. doi: 10.1038/sj.onc.1210803. [DOI] [PubMed] [Google Scholar]
- 64.Munro S., Carr S.M., La Thangue N.B. Diversity within the pRb pathway: is there a code of conduct? Oncogene. 2012;31:4343–4352. doi: 10.1038/onc.2011.603. [DOI] [PubMed] [Google Scholar]
- 65.Dick F.A., Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell. 2003;12:639–649. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 66.Seifried L.A., Talluri S., Cecchini M., Julian L.M., Mymryk J.S., Dick F.A. pRB-E2F1 complexes are resistant to adenovirus E1A-mediated disruption. J. Virol. 2008;82:4511–4520. doi: 10.1128/JVI.02713-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martínez-Balbás M.A., Bauer U.M., Nielsen S.J., Brehm A., Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrari R., Gou D., Jawdekar G., Johnson S.A., Nava M., Su T., Yousef A.F., Zemke N.R., Pellegrini M., Kurdistani S.K. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe. 2014;16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horwitz G.A., Zhang K., McBrian M.A., Grunstein M., Kurdistani S.K., Berk A.J. Adenovirus Small e1a Alters Global Patterns of Histone Modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. (80-.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J.S., Galvin K.M., See R.H., Eckner R., Livingston D., Moran E., Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 71.Sankar N., Baluchamy S., Kadeppagari R.-K., Singhal G., Weitzman S., Thimmapaya B. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene. 2008;27:5717–5728. doi: 10.1038/onc.2008.181. [DOI] [PubMed] [Google Scholar]
- 72.Kadeppagari R.-K., Sankar N., Thimmapaya B. Adenovirus transforming protein E1A induces c-Myc in quiescent cells by a novel mechanism. J. Virol. 2009;83:4810–4822. doi: 10.1128/JVI.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baluchamy S., Sankar N., Navaraj A., Moran E., Thimmapaya B. Relationship between E1A binding to cellular proteins, c-myc activation and S-phase induction. Oncogene. 2007;26:781–787. doi: 10.1038/sj.onc.1209825. [DOI] [PubMed] [Google Scholar]
- 74.Singhal G., Kadeppagari R.K., Sankar N., Thimmapaya B. Simian virus 40 large T overcomes p300 repression of c-Myc. Virology. 2008;377:227–232. doi: 10.1016/j.virol.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lill N.L., Grossman S.R., Ginsberg D., DeCaprio J., Livingston D.M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 76.Yang X.J., Ogryzko V.V., Nishikawa J., Howard B.H., Nakatani Y.A. p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 77.Reid J.L., Bannister A.J., Zegerman P., Martínez-Balbás M.A., Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deleu L., Shellard S., Alevizopoulos K., Amati B., Land H. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- 79.Vijayalingam S., Subramanian T., Zhao L.-J., Chinnadurai G. The cellular protein complex associated with a transforming region of E1A contains c-MYC. J. Virol. 2016;90:1070–1079. doi: 10.1128/JVI.02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao L.-J., Loewenstein P.M., Green M. Ad E1A 243R oncoprotein promotes association of proto-oncogene product MYC with the NuA4/Tip60 complex via the E1A N-terminal repression domain. Virology. 2016;499:178–184. doi: 10.1016/j.virol.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 82.Lang S.E., Hearing P. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene. 2003;22:2836–2841. doi: 10.1038/sj.onc.1206376. [DOI] [PubMed] [Google Scholar]
- 83.Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedford D.C., Kasper L.H., Fukuyama T., Brindle P.K. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 86.Attar N., Kurdistani S.K. Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eckner R., Ludlow J.W., Lill N.L., Oldread E., Arany Z., Modjtahedi N., DeCaprio J.A., Livingston D.M., Morgan J.A. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell Biol. 1996;16:3454–3464. doi: 10.1128/MCB.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patel D., Huang S.M., Baglia L.A., McCance D.J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bernat A., Avvakumov N., Mymryk J.S., Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. doi: 10.1038/sj.onc.1206896. [DOI] [PubMed] [Google Scholar]
- 90.Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 91.Isobe T., Hattori T., Kitagawa K., Uchida C., Kotake Y., Kosugi I., Oda T., Kitagawa M. Adenovirus E1A inhibits SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 2009;284:27766–27779. doi: 10.1074/jbc.M109.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guan H., Ricciardi R.P. Transformation by E1A oncoprotein involves ubiquitin-mediated proteolysis of the neuronal and tumor repressor REST in the nucleus. J. Virol. 2012;86:5594–5602. doi: 10.1128/JVI.06811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng Y., Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Canc. Metastasis Rev. 2012;31:75–87. doi: 10.1007/s10555-011-9330-z. [DOI] [PubMed] [Google Scholar]
- 94.Welcker M., Clurman B.E. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 2005;280:7654–7658. doi: 10.1074/jbc.M413377200. [DOI] [PubMed] [Google Scholar]
- 95.Guan H., Williams J.F., Ricciardi R.P. Induction of neuronal and tumor-related genes by adenovirus type 12 E1A. J. Virol. 2009;83:651–661. doi: 10.1128/JVI.01538-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5 doi: 10.4161/cc.5.17.2982. 1929–35. [DOI] [PubMed] [Google Scholar]
- 97.Popov N., Schülein C., Jaenicke L.A., Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat. Cell Biol. 2010;12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 98.Cohen M.J., Yousef A.F., Massimi P., Fonseca G.J., Todorovic B., Pelka P., Turnell A.S., Banks L., Mymryk J.S. Dissection of the C-terminal region of E1A redefines the roles of CtBP and other cellular targets in oncogenic transformation. J. Virol. 2013;87:10348–10355. doi: 10.1128/JVI.00786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glenewinkel F., Cohen M.J., King C.R., Kaspar S., Bamberg-Lemper S., Mymryk J.S., Becker W. The adaptor protein DCAF7 mediates the interaction of the adenovirus E1A oncoprotein with the protein kinases DYRK1A and HIPK2. Sci. Rep. 2016;6:28241. doi: 10.1038/srep28241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Q., Tang Y., Chen L., Liu N., Lang F., Liu H., Wang P., Sun X. E3 ligase SCFβTrCP-induced DYRK1A protein degradation is essential for cell cycle progression in HEK293 cells. J. Biol. Chem. 2016;291:26399–26409. doi: 10.1074/jbc.M116.717553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zemke N.R., Gou D., Berk A.J. Dedifferentiation by adenovirus E1A due to inactivation of Hippo pathway effectors YAP and TAZ. Genes Dev. 2019;33:828–843. doi: 10.1101/gad.324814.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Komorek J., Kuppuswamy M., Subramanian T., Vijayalingam S., Lomonosova E., Zhao L.-J., Mymryk J.S., Schmitt K., Chinnadurai G. Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous beta-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors. J. Virol. 2010;84:2719–2731. doi: 10.1128/JVI.02119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mészáros B., Kumar M., Gibson T.J., Uyar B., Dosztányi Z. Degrons in cancer. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aak9982. [DOI] [PubMed] [Google Scholar]
- 104.Duan S., Pagano M. Ubiquitin ligases in cancer: functions and clinical potentials. Cell Chem. Biol. 2021 doi: 10.1016/j.chembiol.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cohen M.J., King C.R., Dikeakos J.D., Mymryk J.S. Functional analysis of the C-terminal region of human adenovirus E1A reveals a misidentified nuclear localization signal. Virology. 2014;468–470:238–243. doi: 10.1016/j.virol.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 106.Subramanian T., La Regina M., Chinnadurai G. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene. 1989;4:415–420. [PubMed] [Google Scholar]
- 107.Douglas J.L., Quinlan M.P. Efficient nuclear localization and immortalizing ability, two functions dependent on the adenovirus type 5 (Ad5) E1A second exon, are necessary for cotransformation with Ad5 E1B but not with T24ras. J. Virol. 1995;69:8061–8065. doi: 10.1128/JVI.69.12.8061-8065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schaeper U., Boyd J.M., Verma S., Uhlmann E., Subramanian T., Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stankiewicz T.R., Gray J.J., Winter A.N., Linseman D.A. C-terminal binding proteins: central players in development and disease. Biomol. Concepts. 2014;5:489–511. doi: 10.1515/bmc-2014-0027. [DOI] [PubMed] [Google Scholar]
- 110.Dcona M.M., Morris B.L., Ellis K.C., Grossman S.R. CtBP- an emerging oncogene and novel small molecule drug target: advances in the understanding of its oncogenic action and identification of therapeutic inhibitors. Canc. Biol. Ther. 2017;18:379–391. doi: 10.1080/15384047.2017.1323586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schaeper U., Subramanian T., Lim L., Boyd J.M., Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 112.Subramanian T., Zhao L.-J., Chinnadurai G. Interaction of CtBP with adenovirus E1A suppresses immortalization of primary epithelial cells and enhances virus replication during productive infection. Virology. 2013;443:313–320. doi: 10.1016/j.virol.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Makharashvili N., Paull T.T. CtIP: a DNA damage response protein at the intersection of DNA metabolism. DNA Repair. 2015;32:75–81. doi: 10.1016/j.dnarep.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 114.Williams R.S., Dodson G.E., Limbo O., Yamada Y., Williams J.S., Guenther G., Classen S., Glover J.N.M., Iwasaki H., Russell P. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu F., Lee W.-H. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell Biol. 2006;26:3124–3134. doi: 10.1128/MCB.26.8.3124-3134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.You Z., Bailis J.M. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bruton R.K., Rasti M., Mapp K.L., Young N., Carter R.Z., Abramowicz I.A., Sedgwick G.G., Onion D.F., Shuen M., Mymryk J.S. C-terminal-binding protein interacting protein binds directly to adenovirus early region 1A through its N-terminal region and conserved region 3. Oncogene. 2007;26:7467–7479. doi: 10.1038/sj.onc.1210551. [DOI] [PubMed] [Google Scholar]
- 118.Robert F., Hardy S., Nagy Z., Baldeyron C., Murr R., Déry U., Masson J.-Y., Papadopoulo D., Herceg Z., Tora L. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell Biol. 2006;26:402–412. doi: 10.1128/MCB.26.2.402-412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stracker T.H., Carson C.T., Weitzman M.D. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 120.Boyd J.M., Subramanian T., Schaeper U., La Regina M., Bayley S., Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fidler I.J. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat. Rev. Canc. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 122.Cavallaro U., Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Canc. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 123.Frisch S.M. E1a induces the expression of epithelial characteristics. J. Cell Biol. 1994;127:1085–1096. doi: 10.1083/jcb.127.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Frisch S.M., Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Frisch S.M. The epithelial cell default-phenotype hypothesis and its implications for cancer. Bioessays. 1997;19:705–709. doi: 10.1002/bies.950190811. [DOI] [PubMed] [Google Scholar]
- 126.Grooteclaes M.L., Frisch S.M. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 127.Berx G., van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb. Perspect. Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 129.Shao R., Xia W., Hung M.C. Inhibition of angiogenesis and induction of apoptosis are involved in E1A-mediated bystander effect and tumor suppression. Canc. Res. 2000;60:3123–3126. [PubMed] [Google Scholar]
- 130.Deng J., Kloosterbooer F., Xia W., Hung M.-C. The NH(2)-terminal and conserved region 2 domains of adenovirus E1A mediate two distinct mechanisms of tumor suppression. Canc. Res. 2002;62:346–350. [PubMed] [Google Scholar]
- 131.Zhou Z., Zhou R.-R., Guan H., Bucana C.D., Kleinerman E.S. E1A gene therapy inhibits angiogenesis in a Ewing's sarcoma animal model. Mol. Canc. Therapeut. 2003;2:1313–1319. [PubMed] [Google Scholar]
- 132.Saito Y., Sunamura M., Motoi F., Abe H., Egawa S., Duda D.G., Hoshida T., Fukuyama S., Hamada H., Matsuno S. Oncolytic replication-competent adenovirus suppresses tumor angiogenesis through preserved E1A region. Canc. Gene Ther. 2006;13:242–252. doi: 10.1038/sj.cgt.7700902. [DOI] [PubMed] [Google Scholar]
- 133.Arany Z., Huang L.E., Eckner R., Bhattacharya S., Jiang C., Goldberg M.A., Bunn H.F., Livingston D.M. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kung A.L., Wang S., Klco J.M., Kaelin W.G., Livingston D.M. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 135.de Stanchina E., McCurrach M.E., Zindy F., Shieh S.Y., Ferbeyre G., Samuelson A.V., Prives C., Roussel M.F., Sherr C.J., Lowe S.W. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luftig M.A. Viruses and the DNA damage response: activation and antagonism. Annu. Rev. Virol. 2014;1:605–625. doi: 10.1146/annurev-virology-031413-085548. [DOI] [PubMed] [Google Scholar]
- 137.Sui G., Affar E.B., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 138.Samuelson A.V., Narita M., Chan H.-M., Jin J., de Stanchina E., McCurrach M.E., Narita M., Fuchs M., Livingston D.M., Lowe S.W. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 2005;280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]
- 139.Hsieh J.K., Chan F.S., O'Connor D.J., Mittnacht S., Zhong S., Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 140.Wasylishen A.R., Lozano G. Attenuating the p53 pathway in human cancers: many means to the same end. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liebl M.C., Hofmann T.G. Cell fate regulation upon DNA damage: p53 serine 46 kinases pave the cell death road. Bioessays. 2019;41 doi: 10.1002/bies.201900127. [DOI] [PubMed] [Google Scholar]
- 142.Park J., Oh Y., Yoo L., Jung M.-S., Song W.-J., Lee S.-H., Seo H., Chung K.C. Dyrk1A phosphorylates p53 and inhibits proliferation of embryonic neuronal cells. J. Biol. Chem. 2010;285:31895–31906. doi: 10.1074/jbc.M110.147520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.D'Orazi G., Cecchinelli B., Bruno T., Manni I., Higashimoto Y., Saito S., Gostissa M., Coen S., Marchetti A., Del Sal G. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 144.Yun C.W., Lee S.H. The roles of autophagy in cancer. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chavez-Dominguez R., Perez-Medina M., Lopez-Gonzalez J.S., Galicia-Velasco M., Aguilar-Cazares D. The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity. Front. Oncol. 2020;10:578418. doi: 10.3389/fonc.2020.578418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hendrickx R., Stichling N., Koelen J., Kuryk L., Lipiec A., Greber U.F. Innate immunity to adenovirus. Hum. Gene Ther. 2014;25:265–284. doi: 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeng X., Carlin C.R. Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells. J. Virol. 2013;87:2307–2319. doi: 10.1128/JVI.02014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bowman C.J., Ayer D.E., Dynlacht B.D. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat. Cell Biol. 2014;16:1202–1214. doi: 10.1038/ncb3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rodriguez-Rocha H., Gomez-Gutierrez J.G., Garcia-Garcia A., Rao X.-M., Chen L., McMasters K.M., Zhou H.S. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology. 2011;416:9–15. doi: 10.1016/j.virol.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wong J., Choi S.Y.C., Liu R., Xu E., Killam J., Gout P.W., Wang Y. Potential therapies for infectious diseases based on targeting immune evasion mechanisms that pathogens have in common with cancer cells. Front. Cell. Infect. Microbiol. 2019;9:25. doi: 10.3389/fcimb.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sohn S.-Y., Hearing P. Adenoviral strategies to overcome innate cellular responses to infection. FEBS Lett. 2019;593:3484–3495. doi: 10.1002/1873-3468.13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.King C.R., Tessier T.M., Mymryk J.S. Color me infected: painting cellular chromatin with a viral histone Mimic. Trends Microbiol. 2016;24:774–776. doi: 10.1016/j.tim.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 153.Avgousti D.C., Herrmann C., Kulej K., Pancholi N.J., Sekulic N., Petrescu J., Molden R.C., Blumenthal D., Paris A.J., Reyes E.D. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature. 2016;535:173–177. doi: 10.1038/nature18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fonseca G.J., Thillainadesan G., Yousef A.F., Ablack J.N., Mossman K.L., Torchia J., Mymryk J.S. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 155.Fonseca G.J., Cohen M.J., Nichols A.C., Barrett J.W., Mymryk J.S. Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., Tarcic G. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Olanubi O., Frost J.R., Radko S., Pelka P. Suppression of type I interferon signaling by E1A via RuvBL1/Pontin. J. Virol. 2017;91 doi: 10.1128/JVI.02484-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zemke N.R., Berk A.J. The adenovirus E1A C terminus suppresses a delayed antiviral response and modulates RAS signaling. Cell Host Microbe. 2017;22:789–800. doi: 10.1016/j.chom.2017.11.008. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 160.Blair G.E., Blair-Zajdel M.E. Evasion of the immune system by adenoviruses. Curr. Top. Microbiol. Immunol. 2004;273:3–28. doi: 10.1007/978-3-662-05599-1_1. [DOI] [PubMed] [Google Scholar]
- 161.Egawa N., Doorbar J. The low-risk papillomaviruses. Virus Res. 2017;231:119–127. doi: 10.1016/j.virusres.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 162.Jongsma M.L.M., Guarda G., Spaapen R.M. The regulatory network behind MHC class I expression. Mol. Immunol. 2019;113:16–21. doi: 10.1016/j.molimm.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 163.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Prusinkiewicz M.A., Mymryk J.S. Metabolic reprogramming of the host cell by human adenovirus infection. Viruses. 2019;11 doi: 10.3390/v11020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 166.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dyer A., Schoeps B., Frost S., Jakeman P., Scott E.M., Freedman J., Jacobus E.J., Seymour L.W. Antagonism of glycolysis and reductive carboxylation of glutamine potentiates activity of oncolytic adenoviruses in cancer cells. Canc. Res. 2019;79:331–345. doi: 10.1158/0008-5472.CAN-18-1326. [DOI] [PubMed] [Google Scholar]
- 168.Berk A.J. Functions of adenovirus E1A. Canc. Surv. 1986;5:367–387. [PubMed] [Google Scholar]
- 169.Löhr K., Hartmann O., Schäfer H., Dobbelstein M. Mutual interference of adenovirus infection and myc expression. J. Virol. 2003;77:7936–7944. doi: 10.1128/jvi.77.14.7936-7944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao L.-J., Loewenstein P.M., Green M. Enhanced MYC association with the NuA4 histone acetyltransferase complex mediated by the adenovirus E1A N-terminal domain activates a subset of MYC target genes highly expressed in cancer cells. Genes Canc. 2017;8:752–761. doi: 10.18632/genesandcancer.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tworkowski K.A., Chakraborty A.A., Samuelson A.V., Seger Y.R., Narita M., Hannon G.J., Lowe S.W., Tansey W.P. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6103–6108. doi: 10.1073/pnas.0802095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pelka P., Miller M.S., Cecchini M., Yousef A.F., Bowdish D.M., Dick F., Whyte P., Mymryk J.S. Adenovirus E1A directly targets the E2F/DP-1 complex. J. Virol. 2011;85:8841–8851. doi: 10.1128/JVI.00539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Moran E. Interaction of adenoviral proteins with pRB and p53. FASEB J. 1993;7:880–885. doi: 10.1096/fasebj.7.10.8344487. Off. Publ. Fed. Am. Soc. Exp. Biol. [DOI] [PubMed] [Google Scholar]
- 174.Matoba S., Kang J.-G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 175.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N.C., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 176.Teodoro J.G., Shore G.C., Branton P.E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 177.Prusinkiewicz M.A., Tu J., Dodge M.J., MacNeil K.M., Radko-Juettner S., Fonseca G.J., Pelka P., Mymryk J.S. Differential effects of human adenovirus E1A protein isoforms on aerobic glycolysis in A549 human lung epithelial cells. Viruses. 2020;12 doi: 10.3390/v12060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nevins J.R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981;26:213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- 179.White E. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 2006;13:1371–1377. doi: 10.1038/sj.cdd.4401941. [DOI] [PubMed] [Google Scholar]
- 180.Lowe S.W., Jacks T., Housman D.E., Ruley H.E. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McLorie W., McGlade C.J., Takayesu D., Branton P.E. Individual adenovirus E1B proteins induce transformation independently but by additive pathways. J. Gen. Virol. 1991;72(Pt 6):1467–1471. doi: 10.1099/0022-1317-72-6-1467. [DOI] [PubMed] [Google Scholar]
- 182.White E., Stillman B. Expression of adenovirus E1B mutant phenotypes is dependent on the host cell and on synthesis of E1A proteins. J. Virol. 1987;61:426–435. doi: 10.1128/JVI.61.2.426-435.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Subramanian T., Vijayalingam S., Chinnadurai G. Genetic identification of adenovirus type 5 genes that influence viral spread. J. Virol. 2006;80:2000–2012. doi: 10.1128/JVI.80.4.2000-2012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Edbauer C., Lamberti C., Tong J., Williams J. Adenovirus type 12 E1B 19-kilodalton protein is not required for oncogenic transformation in rats. J. Virol. 1988;62:3265–3273. doi: 10.1128/JVI.62.9.3265-3273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumai H., Takemori N., Hashimoto S. Role of adenovirus type 2 early region 1B 19K protein stability in expression of the cyt and deg phenotypes. J. Gen. Virol. 1989;70(Pt 8):1975–1986. doi: 10.1099/0022-1317-70-8-1975. [DOI] [PubMed] [Google Scholar]
- 186.Flint S.J., Gonzalez R.A. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 2003;272:287–330. doi: 10.1007/978-3-662-05597-7_10. [DOI] [PubMed] [Google Scholar]
- 187.Yew P.R., Berk A.J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 188.Bischoff J.R., Kirn D.H., Williams A., Heise C., Horn S., Muna M., Ng L., Nye J.A., Sampson-Johannes A., Fattaey A. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 189.Cheng P.-H., Wechman S.L., McMasters K.M., Zhou H.S. Oncolytic replication of E1b-deleted adenoviruses. Viruses. 2015;7:5767–5779. doi: 10.3390/v7112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.White E. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene. 2001;20:7836–7846. doi: 10.1038/sj.onc.1204861. [DOI] [PubMed] [Google Scholar]
- 191.Debbas M., White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 192.Lowe S.W., Ruley H.E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 193.Hidalgo P., Ip W.H., Dobner T., Gonzalez R.A. The biology of the adenovirus E1B 55K protein. FEBS Lett. 2019;593:3504–3517. doi: 10.1002/1873-3468.13694. [DOI] [PubMed] [Google Scholar]
- 194.Sarnow P., Ho Y.S., Williams J., Levine A.J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 195.Hutton F.G., Turnell A.S., Gallimore P.H., Grand R.J. Consequences of disruption of the interaction between p53 and the larger adenovirus early region 1B protein in adenovirus E1 transformed human cells. Oncogene. 2000;19:452–462. doi: 10.1038/sj.onc.1203316. [DOI] [PubMed] [Google Scholar]
- 196.Kao C.C., Yew P.R., Berk A.J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 197.Martin M.E., Berk A.J. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 1998;72:3146–3154. doi: 10.1128/JVI.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Querido E., Blanchette P., Yan Q., Kamura T., Morrison M., Boivin D., Kaelin W.G., Conaway R.C., Conaway J.W., Branton P.E. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nevels M., Rubenwolf S., Spruss T., Wolf H., Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zantema A., Schrier P.I., Davis-Olivier A., van Laar T., Vaessen R.T., van der Eb A.J. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol. Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084-3091.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pennella M.A., Liu Y., Woo J.L., Kim C.A., Berk A.J. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 2010;84:12210–12225. doi: 10.1128/JVI.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cheng C.Y., Gilson T., Wimmer P., Schreiner S., Ketner G., Dobner T., Branton P.E., Blanchette P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J. Virol. 2013;87:6232–6245. doi: 10.1128/JVI.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu Y., Colosimo A.L., Yang X.J., Liao D. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell Biol. 2000;20:5540–5553. doi: 10.1128/MCB.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Möller A., Sirma H., Hofmann T.G., Rueffer S., Klimczak E., Dröge W., Will H., Schmitz M.L. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Canc. Res. 2003;63:4310–4314. [PubMed] [Google Scholar]
- 205.Bernardi R., Pandolfi P.P. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene. 2003;22:9048–9057. doi: 10.1038/sj.onc.1207106. [DOI] [PubMed] [Google Scholar]
- 206.Schreiner S., Wimmer P., Groitl P., Chen S.-Y., Blanchette P., Branton P.E., Dobner T. Adenovirus type 5 early region 1B 55K oncoprotein-dependent degradation of cellular factor Daxx is required for efficient transformation of primary rodent cells. J. Virol. 2011;85:8752–8765. doi: 10.1128/JVI.00440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Weitzman M.D., Fradet-Turcotte A. Virus DNA replication and the host DNA damage response. Annu. Rev. Virol. 2018;5:141–164. doi: 10.1146/annurev-virology-092917-043534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schreiner S., Wimmer P., Dobner T. Adenovirus degradation of cellular proteins. Future Microbiol. 2012;7:211–225. doi: 10.2217/fmb.11.153. [DOI] [PubMed] [Google Scholar]
- 209.Harada J.N., Shevchenko A., Shevchenko A., Pallas D.C., Berk A.J. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 2002;76:9194–9206. doi: 10.1128/jvi.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Baker A., Rohleder K.J., Hanakahi L.A., Ketner G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 2007;81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mund A., Schubert T., Staege H., Kinkley S., Reumann K., Kriegs M., Fritsch L., Battisti V., Ait-Si-Ali S., Hoffbeck A.-S. SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response. Nucleic Acids Res. 2012;40:11363–11379. doi: 10.1093/nar/gks868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gupta A., Jha S., Engel D.A., Ornelles D.A., Dutta A. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene. 2013;32:5017–5025. doi: 10.1038/onc.2012.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nazeer R., Qashqari F.S.I., Albalawi A.S., Piberger A.L., Tilotta M.T., Read M.L., Hu S., Davis S., McCabe C.J., Petermann E. Adenovirus E1B 55-kilodalton protein targets SMARCAL1 for degradation during infection and modulates cellular DNA replication. J. Virol. 2019;93 doi: 10.1128/JVI.00402-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Forrester N.A., Sedgwick G.G., Thomas A., Blackford A.N., Speiseder T., Dobner T., Byrd P.J., Stewart G.S., Turnell A.S., Grand R.J.A. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. J. Virol. 2011;85:2201–2211. doi: 10.1128/JVI.01748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cheng C.Y., Gilson T., Dallaire F., Ketner G., Branton P.E., Blanchette P. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J. Virol. 2011;85:765–775. doi: 10.1128/JVI.01890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jha S., Vande Pol S., Banerjee N.S., Dutta A.B., Chow L.T., Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol. Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Härtl B., Zeller T., Blanchette P., Kremmer E., Dobner T. Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription. Oncogene. 2008;27:3673–3684. doi: 10.1038/sj.onc.1211039. [DOI] [PubMed] [Google Scholar]
- 218.Nevels M., Täuber B., Kremmer E., Spruss T., Wolf H., Dobner T. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 1999;73:1591–1600. doi: 10.1128/JVI.73.2.1591-1600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wimmer P., Berscheminski J., Blanchette P., Groitl P., Branton P.E., Hay R.T., Dobner T., Schreiner S. PML isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functionally inhibiting the tumor-suppressor p53. Oncogene. 2016;35:69–82. doi: 10.1038/onc.2015.63. [DOI] [PubMed] [Google Scholar]
- 220.Guccione E., Lethbridge K.J., Killick N., Leppard K.N., Banks L. HPV E6 proteins interact with specific PML isoforms and allow distinctions to be made between different POD structures. Oncogene. 2004;23:4662–4672. doi: 10.1038/sj.onc.1207631. [DOI] [PubMed] [Google Scholar]
- 221.Everett R.D. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 222.Berscheminski J., Brun J., Speiseder T., Wimmer P., Ip W.H., Terzic M., Dobner T., Schreiner S. Sp100A is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation. Oncogene. 2016;35:3178–3189. doi: 10.1038/onc.2015.378. [DOI] [PubMed] [Google Scholar]
- 223.Müncheberg S., Hay R.T., Ip W.H., Meyer T., Weiß C., Brenke J., Masser S., Hadian K., Dobner T., Schreiner S. E1B-55K-Mediated regulation of RNF4 SUMO-targeted ubiquitin ligase promotes human adenovirus gene expression. J. Virol. 2018;92 doi: 10.1128/JVI.00164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tang S., Ding S., Yu L., Shen H., Wan Y., Wu Y. Effects of HPV16 E6 protein on Daxx-induced apoptosis in C33A cells. Cell. Mol. Biol. Lett. 2020;25:38. doi: 10.1186/s11658-020-00230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lewis P.W., Elsaesser S.J., Noh K.-M., Stadler S.C., Allis C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schreiner S., Bürck C., Glass M., Groitl P., Wimmer P., Kinkley S., Mund A., Everett R.D., Dobner T. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013;41:3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Watson L.A., Goldberg H., Bérubé N.G. Emerging roles of ATRX in cancer. Epigenomics. 2015;7:1365–1378. doi: 10.2217/epi.15.82. [DOI] [PubMed] [Google Scholar]
- 228.Wang X., Zhao Y., Zhang J., Chen Y. Structural basis for DAXX interaction with ATRX. Protein Cell. 2017;8:767–771. doi: 10.1007/s13238-017-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.White E., Sabbatini P., Debbas M., Wold W.S., Kusher D.I., Gooding L.R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol. Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570-2580.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lomonosova E., Subramanian T., Chinnadurai G. Mitochondrial localization of p53 during adenovirus infection and regulation of its activity by E1B-19K. Oncogene. 2005;24:6796–6808. doi: 10.1038/sj.onc.1208836. [DOI] [PubMed] [Google Scholar]
- 231.Sabbatini P., Chiou S.K., Rao L., White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol. Cell Biol. 1995;15:1060–1070. doi: 10.1128/MCB.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chiou S.K., Tseng C.C., Rao L., White E. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J. Virol. 1994;68:6553–6566. doi: 10.1128/JVI.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Henderson S., Huen D., Rowe M., Dawson C., Johnson G., Rickinson A. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Neilan J.G., Lu Z., Afonso C.L., Kutish G.F., Sussman M.D., Rock D.L. An African swine fever virus gene with similarity to the proto-oncogene bcl-2 and the Epstein-Barr virus gene BHRF1. J. Virol. 1993;67:4391–4394. doi: 10.1128/JVI.67.7.4391-4394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Vaux D.L., Cory S., Adams J.M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 236.Boyd J.M., Malstrom S., Subramanian T., Venkatesh L.K., Schaeper U., Elangovan B., D’Sa-Eipper C., Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 237.Rao L., Debbas M., Sabbatini P., Hockenbery D., Korsmeyer S., White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cuconati A., White E. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 2002;16:2465–2478. doi: 10.1101/gad.1012702. [DOI] [PubMed] [Google Scholar]
- 239.Pan M., Chew T.W., Wong D.C.P., Xiao J., Ong H.T., Chin J.F.L., Low B.C. BNIP-2 retards breast cancer cell migration by coupling microtubule-mediated GEF-H1 and RhoA activation. Sci. Adv. 2020:6. doi: 10.1126/sciadv.aaz1534. eaaz1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Low B.C., Lim Y.P., Lim J., Wong E.S., Guy G.R. Tyrosine phosphorylation of the Bcl-2-associated protein BNIP-2 by fibroblast growth factor receptor-1 prevents its binding to Cdc42GAP and Cdc42. J. Biol. Chem. 1999;274:33123–33130. doi: 10.1074/jbc.274.46.33123. [DOI] [PubMed] [Google Scholar]
- 241.White E., Cipriani R. Role of adenovirus E1B proteins in transformation: altered organization of intermediate filaments in transformed cells that express the 19-kilodalton protein. Mol. Cell Biol. 1990;10:120–130. doi: 10.1128/mcb.10.1.120-130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Han J., Sabbatini P., Perez D., Rao L., Modha D., White E. The E1B 19K protein blocks apoptosis by interacting with and inhibiting the p53-inducible and death-promoting Bax protein. Genes Dev. 1996;10:461–477. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- 243.Farrow S.N., White J.H., Martinou I., Raven T., Pun K.T., Grinham C.J., Martinou J.C., Brown R. Cloning of a bcl-2 homologue by interaction with adenovirus E1B 19K. Nature. 1995;374:731–733. doi: 10.1038/374731a0. [DOI] [PubMed] [Google Scholar]
- 244.Thomas M., Banks L. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 1999;80(Pt 6):1513–1517. doi: 10.1099/0022-1317-80-6-1513. [DOI] [PubMed] [Google Scholar]
- 245.Perez D., White E. TNF-alpha signals apoptosis through a bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol. Cell. 2000;6:53–63. [PubMed] [Google Scholar]
- 246.Sundararajan R., Cuconati A., Nelson D., White E. Tumor necrosis factor-alpha induces Bax-Bak interaction and apoptosis, which is inhibited by adenovirus E1B 19K. J. Biol. Chem. 2001;276:45120–45127. doi: 10.1074/jbc.M106386200. [DOI] [PubMed] [Google Scholar]
- 247.Sundararajan R., White E. E1B 19K blocks Bax oligomerization and tumor necrosis factor alpha-mediated apoptosis. J. Virol. 2001;75:7506–7516. doi: 10.1128/JVI.75.16.7506-7516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Williams J., Williams M., Liu C., Telling G. Assessing the role of E1A in the differential oncogenicity of group A and group C human adenoviruses. Curr. Top. Microbiol. Immunol. 1995;199(Pt 3):149–175. doi: 10.1007/978-3-642-79586-2_8. [DOI] [PubMed] [Google Scholar]
- 249.Zhang S., Mak S., Branton P.E. Overexpression of the E1B 55-kilodalton (482R) protein of human adenovirus type 12 appears to permit efficient transformation of primary baby rat kidney cells in the absence of the E1B 19-kilodalton protein. J. Virol. 1992;66:2302–2309. doi: 10.1128/JVI.66.4.2302-2309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Täuber B., Dobner T. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene. 2001;278:1–23. doi: 10.1016/s0378-1119(01)00722-3. [DOI] [PubMed] [Google Scholar]
- 251.Weitzman M.D. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front. Biosci. 2005;10:1106–1117. doi: 10.2741/1604. [DOI] [PubMed] [Google Scholar]
- 252.Sheng M., Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 253.Lee S.S., Glaunsinger B., Mantovani F., Banks L., Javier R.T. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 2000;74:9680–9693. doi: 10.1128/jvi.74.20.9680-9693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lee S.S., Weiss R.S., Javier R.T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Glaunsinger B.A., Lee S.S., Thomas M., Banks L., Javier R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene. 2000;19:5270–5280. doi: 10.1038/sj.onc.1203906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Glaunsinger B.A., Weiss R.S., Lee S.S., Javier R. Link of the unique oncogenic properties of adenovirus type 9 E4-ORF1 to a select interaction with the candidate tumor suppressor protein ZO-2. EMBO J. 2001;20:5578–5586. doi: 10.1093/emboj/20.20.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rousset R., Fabre S., Desbois C., Bantignies F., Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 258.Kiyono T., Hiraiwa A., Fujita M., Hayashi Y., Akiyama T., Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Frese K.K., Lee S.S., Thomas D.L., Latorre I.J., Weiss R.S., Glaunsinger B.A., Javier R.T. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene. 2003;22:710–721. doi: 10.1038/sj.onc.1206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Weiss R.S., Lee S.S., Prasad B.V., Javier R.T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J. Virol. 1997;71:1857–1870. doi: 10.1128/JVI.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kong K., Kumar M., Taruishi M., Javier R.T. The human adenovirus E4-ORF1 protein subverts discs large 1 to mediate membrane recruitment and dysregulation of phosphatidylinositol 3-kinase. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Frese K.K., Latorre I.J., Chung S.-H., Caruana G., Bernstein A., Jones S.N., Donehower L.A., Justice M.J., Garner C.C., Javier R.T. Oncogenic function for the Dlg1 mammalian homolog of the Drosophila discs-large tumor suppressor. EMBO J. 2006;25:1406–1417. doi: 10.1038/sj.emboj.7601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Roberts S., Delury C., Marsh E. The PDZ protein discs-large (DLG): the “Jekyll and Hyde” of the epithelial polarity proteins. FEBS J. 2012;279:3549–3558. doi: 10.1111/j.1742-4658.2012.08729.x. [DOI] [PubMed] [Google Scholar]
- 264.Krishna Subbaiah V., Massimi P., Boon S.S., Myers M.P., Sharek L., Garcia-Mata R., Banks L. The invasive capacity of HPV transformed cells requires the hDlg-dependent enhancement of SGEF/RhoG activity. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Thai M., Thaker S.K., Feng J., Du Y., Hu H., Ting Wu T., Graeber T.G., Braas D., Christofk H.R. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat. Commun. 2015;6:8873. doi: 10.1038/ncomms9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Thai M., Graham N.A., Braas D., Nehil M., Komisopoulou E., Kurdistani S.K., McCormick F., Graeber T.G., Christofk H.R. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metabol. 2014;19:694–701. doi: 10.1016/j.cmet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Na H.-N., Dubuisson O., Hegde V., Nam J.-H., Dhurandhar N.V. Human adenovirus Ad36 and its E4orf1 gene enhance cellular glucose uptake even in the presence of inflammatory cytokines. Biochimie. 2016;124:3–10. doi: 10.1016/j.biochi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 268.Calle E.E., Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Canc. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 269.Guissoni A.C.P., Soares C.M.A., Badr K.R., Ficcadori F.S., Parente A.F.A., Parente J.A., Baeza L.C., Souza M., Cardoso D., das D.P. Proteomic analysis of A-549 cells infected with human adenovirus 40 by LC-MS. Virus Gene. 2018;54:351–360. doi: 10.1007/s11262-018-1554-3. [DOI] [PubMed] [Google Scholar]
- 270.Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018;25:133–143. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Laptenko O., Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 272.Steegenga W.T., Riteco N., Jochemsen A.G., Fallaux F.J., Bos J.L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 273.Blanchette P., Cheng C.Y., Yan Q., Ketner G., Ornelles D.A., Dobner T., Conaway R.C., Conaway J.W., Branton P.E. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell Biol. 2004;24:9619–9629. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Soria C., Estermann F.E., Espantman K.C., O'Shea C.C. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ou H.D., Kwiatkowski W., Deerinck T.J., Noske A., Blain K.Y., Land H.S., Soria C., Powers C.J., May A.P., Shu X. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Nevels M., Spruss T., Wolf H., Dobner T. The adenovirus E4orf6 protein contributes to malignant transformation by antagonizing E1A-induced accumulation of the tumor suppressor protein p53. Oncogene. 1999;18:9–17. doi: 10.1038/sj.onc.1202284. [DOI] [PubMed] [Google Scholar]
- 277.Bernardi R., Pandolfi P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 278.Ishov A.M., Stenberg R.M., Maul G.G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Carvalho T., Seeler J.S., Ohman K., Jordan P., Pettersson U., Akusjärvi G., Carmo-Fonseca M., Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Doucas V., Ishov A.M., Romo A., Juguilon H., Weitzman M.D., Evans R.M., Maul G.G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 281.Evans J.D., Hearing P. Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 2003;77:5295–5304. doi: 10.1128/jvi.77.9.5295-5304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lavin M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 283.Williams R.S., Williams J.S., Tainer J.A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell. Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 284.Tisi R., Vertemara J., Zampella G., Longhese M.P. Functional and structural insights into the MRX/MRN complex, a key player in recognition and repair of DNA double-strand breaks. Comput. Struct. Biotechnol. J. 2020;18:1137–1152. doi: 10.1016/j.csbj.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Carson C.T., Schwartz R.A., Stracker T.H., Lilley C.E., Lee D.V., Weitzman M.D. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Weiden M.D., Ginsberg H.S. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U. S. A. 1994;91:153–157. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Schwartz R.A., Lakdawala S.S., Eshleman H.D., Russell M.R., Carson C.T., Weitzman M.D. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 2008;82:9043–9055. doi: 10.1128/JVI.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Stracker T.H., Lee D.V., Carson C.T., Araujo F.D., Ornelles D.A., Weitzman M.D. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 2005;79:6664–6673. doi: 10.1128/JVI.79.11.6664-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pancholi N.J., Weitzman M.D. Serotype-specific restriction of wild-type adenoviruses by the cellular Mre11-Rad50-Nbs1 complex. Virology. 2018;518:221–231. doi: 10.1016/j.virol.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Smith G.C., Jackson S.P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 291.Shrivastav M., De Haro L.P., Nickoloff J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 292.Björkman A., Du L., Felgentreff K., Rosner C., Pankaj Kamdar R., Kokaraki G., Matsumoto Y., Davies E.G., van der Burg M., Notarangelo L.D. DNA-PKcs is involved in Ig class switch recombination in human B cells. J. Immunol. 2015;195:5608–5615. doi: 10.4049/jimmunol.1501633. [DOI] [PubMed] [Google Scholar]
- 293.Ruis B.L., Fattah K.R., Hendrickson E.A. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Jette N., Lees-Miller S.P. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Boyer J., Rohleder K., Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 296.Brestovitsky A., Nebenzahl-Sharon K., Kechker P., Sharf R., Kleinberger T. The adenovirus E4orf4 protein provides a novel mechanism for inhibition of the DNA damage response. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Nebenzahl-Sharon K., Shalata H., Sharf R., Amer J., Khoury-Haddad H., Sohn S.-Y., Ayoub N., Hearing P., Kleinberger T. Biphasic functional interaction between the adenovirus E4orf4 protein and DNA-PK. J. Virol. 2019;93 doi: 10.1128/JVI.01365-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Goode E.L., Ulrich C.M., Potter J.D. Polymorphisms in DNA repair genes and associations with cancer risk. Canc. Epidemiol. Biomark. Prev. 2002;11:1513–1530. a Publ. Am. Assoc. Cancer Res. cosponsored by Am. Soc. Prev. Oncol. [PubMed] [Google Scholar]
- 299.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability--an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 300.Mordes D.A., Glick G.G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 302.Leimbacher P.-A., Jones S.E., Shorrocks A.-M.K., de Marco Zompit M., Day M., Blaauwendraad J., Bundschuh D., Bonham S., Fischer R., Fink D. MDC1 interacts with TOPBP1 to maintain chromosomal stability during mitosis. Mol. Cell. 2019;74:571–583. doi: 10.1016/j.molcel.2019.02.014. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Moudry P., Watanabe K., Wolanin K.M., Bartkova J., Wassing I.E., Watanabe S., Strauss R., Troelsgaard Pedersen R., Oestergaard V.H., Lisby M. TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity. J. Cell Biol. 2016;212:281–288. doi: 10.1083/jcb.201507042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Liu Y., Cussiol J.R., Dibitetto D., Sims J.R., Twayana S., Weiss R.S., Freire R., Marini F., Pellicioli A., Smolka M.B. TOPBP1(Dpb11) plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1(Rad9) J. Cell Biol. 2017;216:623–639. doi: 10.1083/jcb.201607031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Saldivar J.C., Cortez D., Cimprich K.A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Blackford A.N., Patel R.N., Forrester N.A., Theil K., Groitl P., Stewart G.S., Taylor A.M.R., Morgan I.M., Dobner T., Grand R.J.A. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12251–12256. doi: 10.1073/pnas.0914605107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Schlesinger R.W. Adenoviruses. The nature of the virion and of controlling factors in productive or abortive infection and tumorigenesis. Adv. Virus Res. 1969;14:1–61. doi: 10.1016/s0065-3527(08)60556-4. [DOI] [PubMed] [Google Scholar]
- 308.King C.R., Zhang A., Mymryk J.S. The persistent mystery of adenovirus persistence. Trends Microbiol. 2016;24:323–324. doi: 10.1016/j.tim.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 309.Graham F.L., Smiley J., Russell W.C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 310.Johnson J.T. Pseudohyperparathyroidism associated with metastatic adenocarcinoma of undetermined origin in the dog. J. Am. Vet. Med. Assoc. 1978;173:82–83. [PubMed] [Google Scholar]
- 311.Kimura M., Shirakura K., Hasegawa A., Kobayashi Y., Udagawa E. Anatomy and pathophysiology of the popliteal tendon area in the lateral meniscus: 2. Clinical investigation. Arthrosc. J. Arthrosc. Relat. Surg. 1992;8:424–427. doi: 10.1016/0749-8063(92)90002-s. Off. Publ. Arthrosc. Assoc. North Am. Int. Arthrosc. Assoc. [DOI] [PubMed] [Google Scholar]
- 312.zur Hausen H. The search for infectious causes of human cancers: where and why (Nobel lecture) Angew. Chem., Int. Ed. Engl. 2009;48:5798–5808. doi: 10.1002/anie.200901917. [DOI] [PubMed] [Google Scholar]
- 313.Ishibashi M., Yasue H. The Adenoviruses. 1984. Adenoviruses of animals; pp. 497–562. [Google Scholar]
- 314.Lelie P.N., Reesink H.W., Lucas C.J. Inactivation of 12 viruses by heating steps applied during manufacture of a hepatitis B vaccine. J. Med. Virol. 1987;23:297–301. doi: 10.1002/jmv.1890230313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Harrach B., Tarján Z.L., Benkő M. Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett. 2019;593:3660–3673. doi: 10.1002/1873-3468.13687. [DOI] [PubMed] [Google Scholar]
- 316.Cantalupo P.G., Katz J.P., Pipas J.M. Viral sequences in human cancer. Virology. 2018;513:208–216. doi: 10.1016/j.virol.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zapatka M., Borozan I., Brewer D.S., Iskar M., Grundhoff A., Alawi M., Desai N., Sültmann H., Moch H., Cooper C.S. The landscape of viral associations in human cancers. Nat. Genet. 2020;52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Trentin J.J., Yabe Y., Taylor G. The quest for human cancer viruses. Science. 1962;137:835–841. doi: 10.1126/science.137.3533.835. [DOI] [PubMed] [Google Scholar]
- 319.Berget S.M., Moore C., Sharp P.A. Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Yousef A.F., Fonseca G.J., Cohen M.J., Mymryk J.S. The C-terminal region of E1A: a molecular tool for cellular cartography. Biochem. Cell. Biol. 2012;90:153–163. doi: 10.1139/o11-080. [DOI] [PubMed] [Google Scholar]
- 321.Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wold W.S.M., Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013;13:421–433. doi: 10.2174/1566523213666131125095046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Sun M. Acid aerosols called health hazard. Science. 1988;240:1727. [PubMed] [Google Scholar]
- 324.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]