Summary
DNA nanotechnology offers new biosensing approaches by templating different sensor and transducer components. Here, we combine DNA origami nanoantennas with label-free antibody detection by incorporating a nanoswitch in the plasmonic hotspot of the nanoantenna. The nanoswitch contains two antigens that are displaced by antibody binding, thereby eliciting a fluorescent signal. Single-antibody detection is demonstrated with a DNA origami integrated anti-digoxigenin antibody nanoswitch. In combination with the nanoantenna, the signal generated by the antibody is additionally amplified. This allows the detection of single antibodies on a portable smartphone microscope. Overall, fluorescence-enhanced antibody detection in DNA origami nanoantennas shows that fluorescence-enhanced biosensing can be expanded beyond the scope of the nucleic acids realm.
Subject areas: Physical chemistry, Optics, Nanotechnology, Biochemistry
Graphical abstract

Highlights
-
•
Single-antibody detection with nanoswitch sensor incorporated in DNA origami structures
-
•
Fluorescence-enhanced single antibody detection in DNA origami nanoantennas
-
•
Detection of single antibodies on a portable smartphone microscope
Physical chemistry; Optics; Nanotechnology; Biochemistry
Introduction
Over the last decades, DNA nanotechnology (Seeman and Sleiman, 2017) and in particular the DNA origami technique (Rothemund, 2006; Dey et al., 2021) have emerged as an indispensable tool for designing new biosensors on the nanoscale. As introduced by Rothemund (Rothemund, 2006), DNA origami can be used to fabricate various two- or three-dimensional shapes using a long single-stranded (ss) scaffold (about 7000–8000 nucleotides (nt)) and hundreds of short staple strands (about 40 nt). Utilizing the programmable nature of DNA base pairing and functionalized staple strands, a large number of different functionalities can be introduced on the nanoscale. This unprecedented addressability of the DNA origami approach allows arranging different biosensing components, introducing new biorecognition elements and multiplexing strategies, as well as the implementation of signal transduction and amplification mechanisms. Using DNA origami, a number of biosensors have been developed capable of single-molecule detection of DNA and RNA (Ke et al., 2008; Zhang et al., 2010a; Kuzuya et al., 2011; Ochmann et al., 2017; Selnihhin et al., 2018; Funck et al., 2018; Trofymchuk et al., 2021), single nucleotide polymorphisms (Zhang et al., 2010b; Subramanian et al., 2011), specific metal ions (Ke et al., 2008; Marras et al., 2018), as well as various protein biomarkers (Rinker et al., 2008; Koirala et al., 2014; Godonoga et al., 2016; Raveendran et al., 2020) among many others (Wang et al., 2017a, 2017b, 2020; Chandrasekaran, 2017; Ke et al., 2018; Loretan et al., 2020; Dass et al., 2021).
Nevertheless, the use of these biosensors is often limited to the detection of targets that directly interact with DNA (e.g. nucleic acids or proteins with aptameric probes). The methods that are used to detect the analyte-sensor interactions also often require low-throughput, complex analytical techniques, such as atomic force microscopy, limiting the widespread application of DNA origami biosensors in clinical diagnostics. Recent advances in the development of high-throughput DNA origami-enabled optical (Domljanovic et al., 2017; Funck et al., 2018) or nanopore-based (Keyser, 2016; Raveendran et al., 2020) sensing strategies provide excellent examples of how to bridge this gap. However, general strategies on how to incorporate recognition elements for targets that go beyond nucleic acids (such as antibodies) are still highly sought after (Wang et al., 2020).
Due to its quick response, high contrast, and good sensitivity, fluorescence provides a powerful readout strategy for developing such sensing devices. Using the DNA origami approach, one can also incorporate methods to further amplify the fluorescence response (Wang et al., 2017a). This enables the detection of single target molecules on low-cost optical devices (Trofymchuk et al., 2021). A way to enhance the fluorescence intensity of a molecule is to put it in a higher electric field environment, which for example is created close to the surface of a plasmonic silver (Ag) or gold (Au) nanoparticle (NP) upon their illumination (plasmonic hotspot) (Novotny and van Hulst, 2011; Purcell, 1946). Even higher electric field enhancement can be achieved in a gap between two plasmonic NPs (Li et al., 2003). Our group has explored this signal amplification strategy to design light antennas on the nanoscale (Acuna et al., 2012; Puchkova et al., 2015; Vietz et al., 2017a) that can be used to amplify the signal of molecular assays (Ochmann et al., 2017; Trofymchuk et al., 2021). In this context, the strength of DNA nanotechnology compared to other approaches of creating plasmonic fluorescence enhancement is the possibility of targeted placement of NPs and fluorescence enhancement with respect to each other. First, we showed that the detection of DNA and RNA specific to Zika virus can be achieved with a fluorescence-quenched hairpin-based assay when positioned next to a plasmonic AgNP, allowing for around 7-fold average fluorescence enhancement (Ochmann et al., 2017). More recently, we also reported a DNA origami NanoAntenna design with a Cleared plasmonic HOtSpot (NACHOS), which allowed positioning a DNA detection assay in the hotspot of two AgNPs enabling an average fluorescence enhancement of 90-fold and the detection of single target molecules on a low-cost, portable, and battery-driven smartphone device (Trofymchuk et al., 2021).
Here, we address the question of whether DNA origami nanoantennas could be applied to larger targets beyond the realm of nucleic acids, such as antibodies. The use of specific recognition elements capable of eliciting a signal response upon binding of other non-labeled biomolecular targets, including antibodies, could provide means to expand the utility of the DNA nanoantenna, as well as other DNA-origami-based sensors for a wider range of targets and diagnostic applications. A promising strategy to combine the specific recognition of antibody targets with DNA-based biosensors that relies on a DNA nanoswitch has been developed recently (Ranallo et al., 2015, 2019). DNA containing a fluorophore-quencher pair is used as a scaffold to attach specific recognition elements and transduce target detection through fluorescence output. In absence of an antibody target, the nanoswitch adopts a stem-loop conformation that opens upon binding to the antibody target, separating the fluorophore-quencher pair and resulting in a fluorescence signal. With this class of nanoswitch, different antibody targets can be simultaneously detected (Porchetta et al., 2018). A detection limit in the nanomolar range can be achieved (Porchetta et al., 2018) and the dynamic range of the response possibly can also be tuned by varying the sequence of the stem similarly as shown for other nanoswitches (Ricci et al., 2016).
In this work, we report a single-molecule DNA origami-based sensor for antibodies by incorporating nanoswitch recognition elements into DNA origami nanostructures. First, we demonstrate the feasibility of this sensing strategy on a simple new rectangular DNA origami (NRO) structure (Rothemund, 2006; Woo and Rothemund, 2011; Li et al., 2012), showing the specific detection of anti-digoxigenin (Dig) antibodies at sub-nanomolar concentrations within few minutes. We then incorporate the nanoswitch elements in the hotspot of DNA nanoantennas showing that the signal of the nanosensor can be enhanced up to ∼60-folds. The single-molecule sensing platform reported here allows us to increase the limit of detection of the nanoswitch and combine it with signal amplification strategies. This allowed us to carry out an exemplary single antibody detection assay on a portable smartphone microscope. Additionally, the modular nature of the DNA origami approach opens exciting possibilities for even further multiplexing in rapid antibody detection.
Results
Detection of anti-dig antibodies with a nanoswitch on the NRO at the single-molecule level
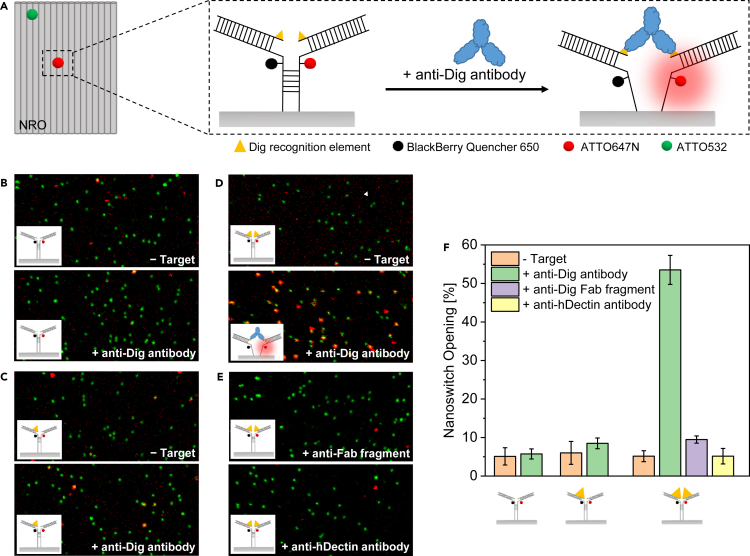
To demonstrate direct detection of antibodies on DNA origami at the single-molecule level, the NRO DNA origami was chosen as a model structure. The simple two-dimensional shape of the NRO provides an ideal platform to incorporate antibody sensing units with high accessibility (Figure 1A, Tables S1 and S2). A sensing unit, which was inspired by the nanoswitch sensor for antibodies developed by the Ricci group (Ranallo et al., 2015), was incorporated into the NRO structure during the DNA origami folding process. The nanoswitch consists of two ssDNA strands protruding from the NRO nanostructure with their 3′- and 5′-ends. Both strands contain a 7-nt long, non-complementary linker sequence followed by a 5-nt long complementary sequence which forms a stem (Figure 1A, Table S2). One of the ssDNA strands is modified with ATTO 647N and the other ssDNA strand is modified with BlackBerry Quencher 650 (BBQ-650), which forms a dye-quencher pair. The stem is followed by DNA anchors on both strands. The DNA anchors allow the hybridization of two Dig-conjugated ssDNA strands which provide binding sites for anti-Dig antibodies (McCreery, 1997). Each NRO nanostructure is equipped with six biotinylated DNA strands for surface immobilization on BSA-biotin-neutravidin coated glass coverslips. Additionally, to aid in identifying each DNA origami nanosensor position in the single-molecule fluorescence experiments, a green ATTO 532 dye is incorporated in the NRO nanostructure (Figures 1A and S1).
Figure 1.
Detection of anti-Dig antibodies with a nanoswitch on the NRO at the single-molecule level
(A) Schematic representation of the NRO nanostructure with an incorporated nanoswitch (dimensions are not to scale). The nanoswitch consists of two DNA strands protruding from the NRO with their 3′ and 5′ ends. Both strands contain a 7-nt long, non-complementary linker sequence followed by a 5-nt long stem. The stem is modified with a dye (ATTO 647N)-quencher (BlackBerry Quencher 650) pair and followed by an ssDNA anchor on both strands. The DNA anchors allow hybridizing two Dig-modified ssDNA strands which form binding sites for Dig binding antibodies to the nanoswitch. In absence of target molecules, the stem is closed and an efficient energy transfer from the fluorophore to the quencher occurs due to their close proximity. Upon bivalent binding of an antibody to the Dig recognition elements, fluorophore and quencher are spatially separated and the fluorescence signal of ATTO 647N is increased. To localize the DNA origami structure, a green-absorbing ATTO 532 dye is incorporated. To test the specificity of the nanoswitch opening and the opening mechanism of the nanoswitch, three different nanoswitch constructs bearing one, two or no Dig recognition elements were investigated.
(B–D) Two-color fluorescence confocal images of surface immobilized NRO-nanoswitch constructs bearing no Dig recognition elements (B), one Dig recognition element (C), and two Dig recognition elements (D) before and after 20 min incubation with 100 nM anti-Dig antibodies.
(E) Two-color fluorescence images of surface immobilized NRO-nanoswitch constructs bearing two Dig recognition elements after incubation with 100 nM anti-Dig Fab fragment (upper) and anti-h-Dectin antibodies (lower), respectively. The images show a field of view of 20 μm × 10 μm. Co-localized green and red spots are attributed to functional NRO structures with an open nanoswitch.
(F) The fraction of open nanoswitches was quantified for every sample by dividing the number of green and red co-localized spots by the total number of green spots from fluorescence confocal scans. Over 300 structures from 5 different areas per sample were analyzed. Error bars represent the standard deviation of the 5 areas.
The principle of the nanoswitch is illustrated in Figure 1A. In absence of target molecules, the stem is closed and the dye-quencher pair is in close proximity so that the fluorescence of the ATTO 647N dye is quenched. Bivalent binding of anti-Dig antibodies to the Dig-recognition elements opens the stem and spatially separates fluorophore and quencher. This results in an increase of the fluorescence of ATTO 647N. The binding of the antibody can be detected in single-molecule confocal fluorescence images (Figures 1B–1E) where red-green colocalized spots are attributed to DNA origami structures with open nanoswitches, whereas only green spots indicate the presence of structures with closed nanoswitches.
Upon antibody binding, the chosen 7-nt long linker and the 5-nt long stem together with the nucleotide where the dye or the quencher are attached to, provide a 26-nt (2 x (7-nt + 5-nt + 1-nt)) long ssDNA spacer between the two Dig-moieties. This spacer length and design is comparable to the 27-nt long ssDNA spacer in the original nanoswitch design (Ranallo et al., 2015) and, based on the approximation of 0.67 nm length per ssDNA base (Chi et al., 2013), is designed to mimic the optimal ∼16 nm distance (distance with highest binding affinity) between two Dig binding moieties on DNA origami reported recently (Shaw et al., 2019).
To test the specificity of the nanoswitch opening on the NRO and to investigate the opening mechanism, we studied nanoswitch constructs bearing one, two or no Dig recognition elements. After surface immobilization, fluorescence scans of the three constructs were taken before and after 20 min incubation with 100 nM anti-Dig antibody on a confocal fluorescence microscope (Figures 1B–1D). For all three constructs, surface scans before incubation with the target antibody showed very few co-localized spots indicating only a low fraction of open nanoswitches (Figures 1B–1D). This small number of false positive signals might originate from unspecifically opened nanoswitches, mislabeled nanoswitches (structures in which the quencher was not incorporated), or nanoswitches containing a photobleached BBQ-650 quencher (Holzmeister et al., 2014; Grabenhorst et al., 2020). After incubating the different nanoswitch constructs with anti-Dig antibodies, we noted a significant increase of co-localized spots only for the nanoswitch construct bearing two Dig recognition elements (Figures 1B–1D). This confirms the specific nature of antibody binding to the Dig recognition elements and excludes an opening of the nanoswitch by monovalent binding of one anti-Dig antibody.
We quantified the efficiency of the nanoswitch opening by calculating the fraction of NRO nanostructures with an open nanoswitch (fraction of co-localized green and red spots, Figure 1F). An unpaired T test was used to compare the magnitude of changes in nanoswitch opening (see STAR Methods). For the construct bearing two Dig-recognition elements, the percentage of constructs with open nanoswitches increased from ∼5% to ∼54% upon addition of anti-Dig antibody while no significant increase was observed for both other constructs, demonstrating specific binding of the anti-Dig antibody to the Dig recognition elements.
To further investigate the opening mechanism and exclude possible cross-reactivity with other bivalent binding antibodies, we incubated the nanoswitch construct with 100 nM anti-Dig Fab fragment and 100 nM anti-hDectin-1 antibody (see Figure 1E). The anti-Dig Fab fragment is a monovalent Dig-binding protein and thus can be used to exclude an opening of the nanoswitch by monovalent binding of two antibodies. We observed only a slight increase (∼4%) in the fraction of open nanoswitches upon addition of the anti-Dig Fab fragment compared to the values obtained in presence of anti-Dig antibodies (∼49%, see Figure 1F), supporting the assumption that the nanoswitch is primarily open due to the bivalent binding of an anti-Dig antibody which is consistent with the mechanism proposed by Ranallo et al. (Ranallo et al., 2015). Anti-hDectin1 is a bivalent antibody specific for human Dectin-1. The nanoswitch opening measured in presence of anti-hDectin-1 antibodies (∼5%) (Figure 1F) was comparable to the unspecific signal gain, further demonstrating that no cross-reactivity occurs. These control experiments confirmed that the nanoswitch is specific for the target anti-Dig antibody and works according to the mechanism depicted in Figure 1A.
Enhancing the output signal of the nanoswitch in the plasmonic hotspot of nanoantennas
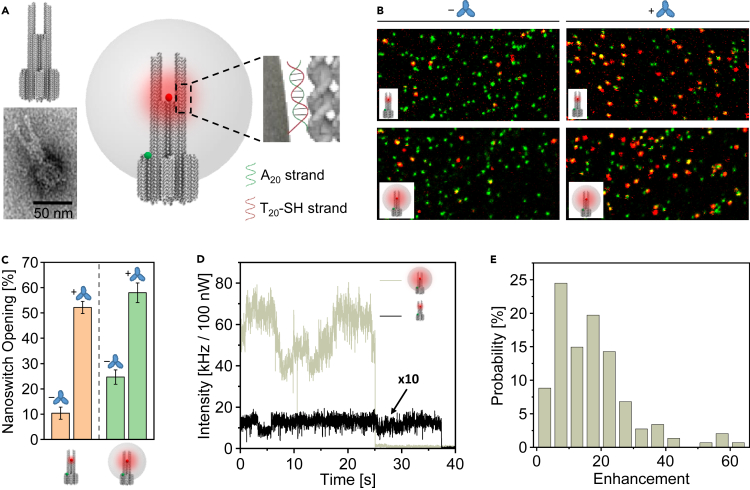
To provide physical amplification of the signal upon detection of a single antibody for possible point-of-care diagnostic applications on low-cost setups, we utilized recently developed NACHOS. NACHOS is a three-dimensional DNA origami structure consisting of two pillars each bearing six protruding staple strands (A20) which provide anchor points for 100-nm AgNPs functionalized with ssDNA strands (T20) (Figure 2A, Tables S3–S6). A plasmonic hotspot is created at the bifurcation in the gap between the two pillars and the nanoparticle (see DNA origami sketch and TEM image in Figure 2A (left) and full NACHOS structure in Figure 2A (right)). For immobilization on BSA-biotin-neutravidin coated glass coverslips, the DNA origami structure is equipped with a rigid cross-like shaped base (Figures S2 and S3) that contains six biotin-modified staple strands (Trofymchuk et al., 2021). For identifying NACHOS in single-molecule fluorescence images, a reference green dye ATTO 542 is incorporated at the base of the DNA origami structure.
Figure 2.
Detection of anti-Dig antibodies with a nanoswitch in the plasmonic hotspot of NACHOS nanostructures with AgNPs
(A) Sketch of the DNA origami structure used for the nanoantenna assembly. A representative class average TEM image of the DNA origami used is shown on the lower left. Schematics of nanoantennas assembly on the right: thiolated DNA-functionalized 100-nm AgNPs are attached to DNA origami nanoantennas via polyadenine (A20) binding strands in zipper-like geometry (Vietz et al., 2016).
(B) Two-color fluorescence confocal images of a reference DNA origami structure without nanoparticles (upper) and with one 100-nm AgNP (lower) before and after 20 min incubation with 100 nM anti-Dig antibodies. The scans show a field of view of 20 μm × 10 μm.
(C) Fraction of open nanoswitches quantified by dividing the number of red-green colocalized spots by the total number of all green spots. Over 400 structures from at least 4 different areas per sample were analyzed. Error bars represent the standard deviation of the 5 areas.
(D) Single-molecule fluorescence intensity transients, measured on a confocal microscope, normalized to the same excitation power of a single ATTO 647N dye incorporated in a DNA origami (black) and in a monomer NACHOS structure containing 100-nm AgNP (olive) excited at 639 nm.
(E) Fluorescence enhancement distribution of the open nanoswitch (ATTO 647N dye) measured in the NACHOS structure. The fluorescence enhancement values were calculated by comparing the intensity of the open nanoswitch (intensity of a single ATTO 647N) in the NACHOS structure to the mean intensity of the open nanoswitch in the reference structure averaged over 113 molecules. A total number of 147 NACHOS structures were analyzed.
In order to study the possible detection of antibodies in the plasmonic hotspot of NACHOS, we incorporated the nanoswitch sensing unit in NACHOS containing a 100-nm AgNP as well as in the same DNA origami nanostructure without NPs serving as a reference. The efficiency of the nanoswitch opening upon the addition of anti-Dig antibodies was then determined analogously to the measurements on the NRO nanosensors (Figure 2B and STAR Methods). In absence of anti-Dig antibodies, nanoswitch openings of ∼10% and ∼25% were recorded for the reference structure and NACHOS containing 100-nm AgNP, respectively (Figure 2C). The higher level of false-positive signals in the NACHOS structure compared with the reference structure can be related to accelerated photobleaching of BBQ-650 in the hotspot of plasmonic nanoantennas (Grabenhorst et al., 2020). After 20 min incubation with 100 nM anti-Dig antibodies, nanoswitch openings of ∼52% and ∼57% were measured for the reference structure and NACHOS, respectively. This demonstrates similar accessibility of the nanoswitch for the antibody in the NACHOS structure as in the reference structure and even is comparable to the sterically less complex two-dimensional NRO structure.
Next, we investigated the fluorescence enhancement achievable in this single-molecule antibody diagnostic assay. Single-molecule fluorescence transients of the nanoswitch (Figures 2D and S4) were recorded on a confocal microscope for the reference structure and for NACHOS containing AgNPs upon incubation with anti-Dig antibody. Fluorescence enhancement values were calculated by comparing the intensity of the open nanoswitch (intensity of a single ATTO 647N dye) in the NACHOS structure to the mean intensity of the open nanoswitch in the reference structure averaged over 113 molecules. Only transients showing single-step photobleaching (69% and 52% of all recorded transients of the open nanoswitch in the reference structures and the NACHOS structure, respectively, see STAR Methods) were included in the analysis. As shown in Figure 2E, fluorescence enhancement values up to 63-fold were achieved with the NACHOS structure (average fluorescence enhancement 17-fold).
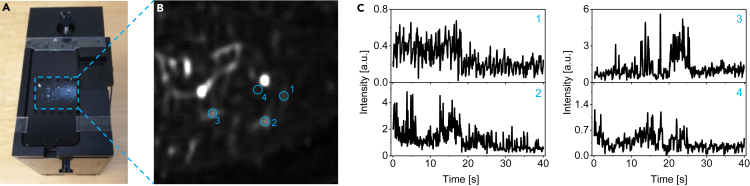
To investigate the possible detection of single antibodies on a simple setup with low-NA optics (Figure 3A) (Vietz et al., 2019; Trofymchuk et al., 2021), we recorded movies of the nanoswitch in the NACHOS nanostructure containing a 100-nm AgNP upon incubation with anti-Dig antibodies. Upon illumination, we were able to observe multiple spots on a smartphone camera (Figure 3B) which bleached upon continuous illumination with a 639-nm laser (Videos S1 and S2). Analogous to the transients recorded on a confocal microscope, most of the transients demonstrated intensity fluctuations ending with single bleaching steps (Figures 3C and S9, STAR Methods), demonstrating the detection of antibodies at the single-molecule level. To the best of our knowledge, these transients represent the first examples of the fluorescence-based detection of single antibodies on a portable smartphone microscope.
Figure 3.
Single antibody detection on a portable smartphone microscope
(A). Photograph of the portable smartphone microscope.
(B) Background corrected fluorescence image of the open nanoswitch (intensity of a single ATTO 647N dye) in NACHOS containing a 100-nm AgNP.
(C) Exemplary fluorescence transients of the nanoswitch (single ATTO 647N dye) in NACHOS containing a 100-nm AgNP measured on a portable microscope setup. Analogous to the single-molecule confocal microscopy, most of the transients demonstrated intensity fluctuations ending with a single bleaching step. The fluorescence transients shown in panel c were extracted from a single movie.
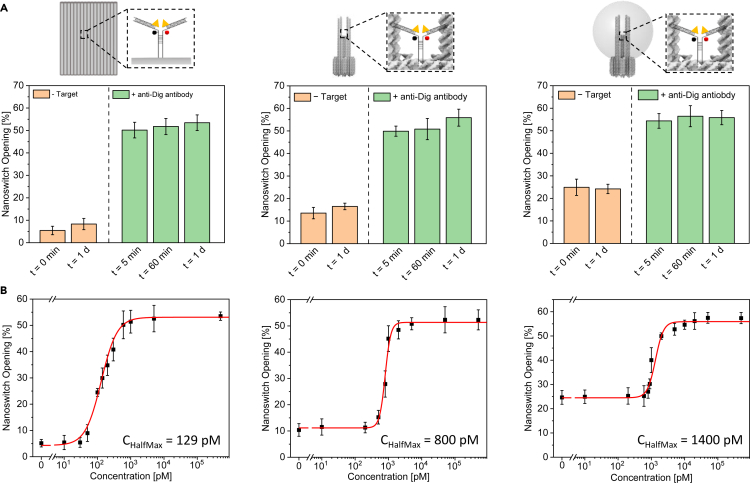
Opening kinetics and sensitive concentration range of the nanoswitch on different DNA origami nanostructures
We evaluated the time required to perform the antibody detection assay and the sensitive concentration range of the nanoswitch on the NRO, the reference DNA origami structure without NPs and NACHOS containing a 100-nm AgNP. The kinetics of the nanoswitch opening was quantified under non-diffusion limited conditions by recording confocal fluorescence scans of surface-immobilized DNA origami nanoswitches before and after different incubation times with 100 nM anti-Dig antibodies (Figure S5). To evaluate the sensitive concentration range under clinically relevant conditions, we chose an incubation time of 20 min. We measured the nanoswitch opening before and after 20 min incubation with different anti-Dig antibody concentrations (Figures S6–S8). The efficiency of nanoswitch opening was determined for each incubation time (Figure 4A) and each antibody concentration (Figure 4B), respectively. We found that under non-diffusion limited conditions the detection was equally rapid on the different DNA origami structures (for 100 nM anti-Dig antibodies), achieving the highest signal gain after just 5 min. This is in accordance with the fast nanoswitch opening kinetics in solution reported previously (Ranallo et al., 2015).
Figure 4.
Opening kinetics and sensitive concentration range of the nanoswitch on different DNA origami nanostructures
(A) Nanoswitch opening on the NRO (left), the reference DNA origami structure without NPs (middle) and NACHOS with 100-nm AgNP (right) before and after the incubation with 100 nM anti-Dig antibodies.
(B) Nanoswitch opening versus anti-Dig antibody concentration on the NRO (left), the reference DNA origami structure without NPs (middle) and NACHOS with 100-nm AgNP (right). The fraction of open nanoswitches was quantified from confocal fluorescence scans by dividing the number of red-green colocalized spots by the total number of green spots. Over 300 structures from at least 3 different areas per sample were analyzed. Error bars represent the standard deviation of the areas. For the binding curves the observed nanoswitch opening was fitted using a four-parameter logistic equation (see STAR Methods).
The target concentration at which the observed signal change is half the maximum signal change (CHalfMax) had values of 129 pM, 800 pM and 1.4 nM for the NRO, the reference DNA origami structure without NPs and the NACHOS structure with one 100-nm AgNP, respectively. We attribute the different CHalfMax values to the different accessibilities of the nanoswitch on the different DNA origami structures. One of the main challenges in designing ultrasensitive biosensors are slow binding kinetics at low target concentrations (Simon et al., 2014; Wu et al., 2019). Given that our nanoswitch opening titrations were performed starting at low pM concentrations, very long incubation times would have been required to achieve the equilibrium opening values, and we deemed it more important to report the nanosensor response (CHalfMax) under diagnostically relevant conditions (the chosen 20 min incubation time). The diffusion limited binding of the antibody at low concentrations makes the accessibility of the nanoswitch the critical criterion when studying the CHalfMax values (see STAR Methods). In the two-dimensional NRO nanostructure, the nanoswitch is expected to be easily accessible for the anti-Dig antibody binding, while the accessibility for the target antibody is expected to be lower in the three-dimensional NACHOS reference structure. Upon the attachment of 100-nm AgNP, the accessibility of NACHOS structure is even further hindered. These results emphasize that the accessibility of the nanostructure is a crucial factor to be considered when designing biosensors with low sensitive concentration ranges under diagnostically relevant incubation times.
Discussion
Rapidly increasing versatility and complexity of DNA nanostructures together with decrease in production cost of DNA that follows Moore's law (halved every 30 months) (Schmidt et al., 2015) makes them ideal platforms for the development of rapid and low-cost biosensors. While the modular and programmable DNA origami approach has been extensively used for the development of various biosensors, its scope of different targets is often limited to nucleic acids. In this work, we report a general strategy to incorporate fluorescence-based sensors for antibodies into DNA origami platforms. Building on previous work of nanoswitch sensors for antibodies in solution (Ranallo et al., 2015, 2019; Porchetta et al., 2018), we showed that these nanoswitch sensors can be successfully incorporated in different DNA origami nanostructures to provide rapid and specific detection of antibodies at the single-molecule level.
The clinically relevant concentration for detecting antibodies varies depending on the application. For the detection of antibodies used for the diagnosis of infectious diseases (Fiorentini et al., 2008; Rostenberg and Peñaloza, 1978; Barletta et al., 2004; Raulf et al., 2019), the concentration level expected to have in clinical samples (i.e. blood, serum) is – depending on the stage of infection – in the pM to low nM range. Conversely, for the detection of antibodies for other purposes (e.g. to monitor immunotherapy), the concentration level of the target antibody can be much higher (high nM low uM range) (Robert et al., 2001; Ji and Lee, 2021; Chames et al., 2009). Thus, the CHalfMax values we achieved here (Figure 4B) show sensitive concentration ranges relevant for diagnostic applications.
The biorecognition elements of the nanoswitch (antigen or small molecule) were not directly incorporated into the DNA origami design but each anchor was designed to carry a cDNA sequence to which different biorecognition elements can be bound post synthetically. This flexible and modular nature of the sensing platform allows one to easily extend it to the detection of other antibodies or biomolecular targets. This sensing approach is also not limited to the detection of bivalent antibodies as it was already demonstrated that monovalent binding of two different antibodies or proteins could also lead to the nanoswitch opening (Ranallo et al., 2015). However, as has been shown before due to the lower affinity of monovalent binding such nanoswitches would require higher antibody (substrate) concentrations to achieve similar signal responses.
On the other hand, the addressability of the DNA origami approach itself opens numbers of exciting directions as well. In this work, we demonstrated how the modular nature of DNA origami can be used to implement a strategy for signal amplification upon antibody detection, leading to an average fluorescence enhancement of 17-fold. This allows the detection on low-cost optical devices which to the best of our knowledge was so far not possible in other single-molecule approaches (Table S7) and thus is a crucial step toward the implementation of point-of-care diagnostics on the single-molecule level. An exciting future direction would be to utilize the unprecedented multiplexing capabilities of DNA origami to extend this platform or apply it for highly multiplexed detection of a large number of clinically relevant targets. It has, for example, been shown that up to 216 distinct fluorescence barcodes can be implemented on a single DNA origami nanostructure opening exciting opportunities in the development of highly multiplexed biosensing strategies (Lin et al., 2012). The nanoswitch sensor platform shown here would allow combining this multiplexing advantage with specific and sensitive (single-molecule level) detection of antibodies on one DNA origami nanostructure.
Limitations of the study
The single-molecule antibody sensing platform, however, also has some limitations, especially in combination with plasmonic signal enhancement by the nanoantenna. One obvious limitation stems from the fact that the relatively small hotspot size of the DNA nanoantennas (such as the NACHOS structure used here), required for high signal enhancement, limits the size of the antibody or the biomolecular target that can be detected. Another challenge is related to the photophysics of fluorophore-quencher pair – we have recently shown that the photobleaching of the quencher molecule (BBQ 650) is accelerated in the hotspot of dimer DNA nanoantennas, leading to an increase of acceptor (ATTO 647N) fluorescence even in absence of target molecules and giving rise to a false-positive signal (Grabenhorst et al., 2020). We also incorporated the nanoswitch in dimer nanoantennas with two NPs to achieve higher fluorescence enhancement. For NACHOS with two 100-nm AgNPs we however observed a large number of red-green colocalized spots even before the addition of target antibodies (Figure S10), which prevented us from using this structure for the detection of antibodies. The higher fraction of active red dyes in the absence the target molecules and giving the rise of false positive signals on the supposedly closed nanoswitch can have different origins. As previously shown (Grabenhorst et al., 2020), higher excitation powers in the dimer hotspot can lead to premature photobleaching of the quencher molecule (BBQ650). Another effect is that quenched dyes are enhanced more than non-quenched dyes as radiative rates are enhanced by nanoantennas so that the efficiency of BBQ650 quenching might not be strong enough (Vietz et al., 2017b). We can therefore not conclusively answer the question whether NACHOS would sterically enable antibody binding in the hotspot between two NPs. To this end, multiple and more efficient and photostable quencher molecules or replacing the molecular quencher with a small metallic NP (Swierczewska et al., 2011) might improve the assay. Nevertheless, we believe that the single-molecule DNA origami antibody sensor platform introduced here presents a useful starting point to further extending DNA origami sensors beyond the detection of nucleic acids and expanding their scope to antibodies and other sensing applications.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Dig antibodies | Thermo Fisher Scientific, USA | cat#: 700772, PRID: AB_2532342 |
| anti-Dig Fab fragment | Merck, Germany | cat#: 11214667001 |
| Chemicals, peptides, and recombinant proteins | ||
| Magnesium chloride | Sigma-Aldrich, Germany | Cat#: M8266 |
| Tris(hydroxymethyl)aminomethane (Tris) | Sigma-Aldrich, Germany | Cat#: 648314 |
| Sodium chloride | Sigma-Aldrich, Germany | Cat#: S9888 |
| Ethylendiaminetetraacetic acid (EDTA) | Sigma-Aldrich, Germany | Cat#: 03620 |
| 100-nm BioPure Silver Nanospheres | nanoComposix, USA | Cat#: HS3822 |
| Tween®20 | Thermo Fisher Scientific, USA | Cat#: 85113 |
| Monobasic potassium buffer | Merck, Germany | Cat#: 8709 |
| Dibasic potassium buffer | Merck, Germany | Cat#: 8584 |
| Phosphate buffered saline (PBS) | Sigma-Aldrich | Cat#: P4417-50TAB |
| Bovine serum albumin (BSA)-Biotin | Sigma-Aldrich, Germany | Cat#: A8549-10MG |
| Neutravidin | Thermo Fisher Scientific, USA | Cat#: 31050 |
| PEG-8000 | Sigma-Aldrich, Germany | 87006 |
| Oligonucleotides | ||
| Unmodified ssDNA strands | Eurofins Genomics GmbH, Germany | https://eurofinsgenomics.eu |
| Modified ssDNA strands (despite BBQ-lalled DNA strands and SH-labeled DNA strands) | Biomers.net GmbH, Germany | https://www.biomers.net |
| BBQ-lalled DNA strands | Eurofins Genomics GmbH, Germany | https://eurofinsgenomics.eu |
| T20-SH DNA strands | EllaBiotech, Germany | https://www.ellabiotech.com/ |
| Software and algorithms | ||
| LabView | National Instruments, USA | https://www.ni.com |
| OriginPro2020 | OriginLab, USA | https://www.originlab.com/2020 |
| ImageJ: Fiji | Schindelin et al. 2012 | https://imagej.net/software/fiji/ |
| Other | ||
| 100 kDa Amicon Ultra filters | Merck | Cat#: MPUFC510024 |
| TEM grids (Formvar/carbon, 400 mesh, Cu) | TedPella, Inc., USA | Cat#: 01702-F |
| Grace Bio-Labs hybridization chambers | Merck | Cat#: GBL623504-50EA |
| Microscope coverslips (24 mm × 60mm size and 170 μm thickness) | Carl Roth, Germany | Cat#: H878.2 |
| Transmission electron microscope | JOEL GmbH, Japan | JOEL JEM-1100 microscope |
| UV-Vis spectrometer NanoDrop 2000 | Thermo Fisher Scientific, USA | Cat#: ND-2000 |
| Inverted microscope | Olympus Corporation, Japan | IX83 inverted microscope |
| 78 MHz-pulsed white light laser | NKT Photonics A/S, Denmark | SuperK Extreme EXW-12 |
| acousto-optically tunable filter (AOTF) | NKT Photonics A/S, Denmark | Super Dual AOTF |
| Digital controller | Crystal Technology, USA | AODS 20160 8 R |
| AOTF | AA Opto-Electronic, France | AA.AOTF.ns: TN |
| Dichroic beam splitter | Chroma Technology, USA | ZT532/640rpc |
| Immersion oil objective | Olympus Corporation, Japan | UPlanSApo 100 x, NA = 1.4, WD = 0.12 mm |
| Pinhole | Linos AG, Germany | |
| Piezo-Stage | Physik Instrumente GmbH&Co. KG, Germany | P-517.3CL, E-501.00 |
| Avalanche Photodiodes | Perkin Elmer Inc., USA | SPCM, AQR 14 |
| TCSPC | PicoQuant GmbH, Germany | HydraHarp 400 |
| Spectral filter (red channel) | Semrock Inc., USA | RazorEdge 647 |
| Spectral filter (green channel) | Semrock Inc., USA | BrightLine HC 582/75 |
| Laser diode | UAB, Lithuania | 0638L-11A, Integrated Optics |
| Objective lens | UCTRONICS, USA | NA = 0.25, LS-40166, |
| Monochrome camera of the smartphone | P20 | Huawei, China |
| Filter | BrightLine HC 731/137 | Semrock Inc., USA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Philip Tinnefeld (Department of Chemistry and Center for NanoScience, Ludwig-Maximilians-Universität München, Butenandtstr. 5–13, 81,377 München, Germany. Philip.tinnefeld@cup.uni-muenchen.de).
Materials availability
This study did not generate new unique reagents.
Method details
Synthesis of DNA origami
DNA origami structures were designed using the open-source software caDNAno2 (Douglas et al., 2009) and assembled and purified using published protocols (Wagenbauer et al., 2017). For the exact sequences of all unmodified and modified DNA staple strands used to fold the DNA origami structures see Tables S1 and S2 (NRO) and Tables S3 and S4 (DNA origami used to build NACHOS). The BBQ650-labeled staple strands were purchased from Biomers.net GmbH (Germany). All other staples were purchased from Eurofins Genomics GmbH (Germany).
For the DNA origami structure used to build NACHOS, 25 μL of p8064 scaffold (produced in-house) at 100 nM were mixed with 18 μL of unmodified staples pooled from 100 μM original concentration and 2 μL of modified staples pooled from 100 μM original concentration. For DNA origami folding, 5 μL of 10x FoB20 folding buffer (200 mM MgCl2, 50 mM Tris, 50 mM NaCl, 10 mM EDTA) were added and the mixture was subjected to a thermal annealing ramp (Table S5). Folded DNA origamis were purified from excessive staple strands using 100 kDa Amicon Ultra filters (Merk, Germany) with 6 washing steps with a lower ionic strength buffer (5 mM MgCl2, 5 mM Tris, 5 mM NaCl, 1 mM EDTA) at 10 krcf for 5 min and 20°C. To extract the purified DNA origamis, the filter was inverted in a new Eppendorf tube and the sample was recovered by spinning for 2 min at 1 krcf and 20°C.
For the NRO, 10 μL of p7249 scaffold (produced in-house) at 100 nM were mixed with 18 μL of unmodified staples pooled from 100 μM original concentration and 4 μL of modified staples pooled from 100 μM original concentration. Briefly, 10 μL of 10x folding buffer (125 mM MgCl2, 400 mM Tris, 200 mM acetic acid, 10 mM EDTA) were added and the mixture was heated to 65°C in a thermocycler. The solution was kept at this temperature for 15 min before being cooled down to 25°C with a temperature gradient of – 1°C min−1. Samples were purified from excess staple strands by PEG-precipitation. The reaction mixture was mixed 1:1 (v:v) with precipitation buffer (15% PEG-8000, 10 mM MgCl2, 250 mM NaCl) and spinned at 16,000 g for 45 min at 4°C. The supernatant was discarded and the pellet was re-dissolved in 100 μL storage buffer (12.5 mM MgCl2, 40 mM Tris, 20 mM acetic acid, 10 mM EDTA). The precipitation procedure was carried out 3 times. Finally, the pellet was dissolved in 20 μL storage buffer.
Functionalization of AgNPs
100 nm AgNP (100-nm BioPure Silver Nanospheres (Citrate), nanoComposix, USA) were functionalized with polythymidine (T20) ssDNA strands with a thiol modification at the 3ʼ-end (Ella Biotech GmbH, Germany) based on previously described procedures (Trofymchuk et al., 2021). For the fabrication of T20-functionalized AgNPs, 2 mL of 0.025 mg/mL nanoparticle solution in ultra-pure water (Sigma Aldrich, Germany) was heated to 40°C under permanent stirring at 550 rpm. Briefly, 20 μL of 10% Tween20 (Thermo Fisher Scientific, USA), 20 μL of a 4:5 (v:v) mixture of 1 M monobasic and dibasic potassium phosphate buffers (P8709 and P8584 Sigma Aldrich, Germany) and 20 μL of a 2 nM polythymidine ssDNA strands (T20-SH-3ʼ) were added to the nanoparticle solution and stirred at 40°C for 1 h. Then, different amounts of 1x PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) containing 3.3 M NaCl were added stepwise every three minutes to the mixture, until a final concentration of 750 mM NaCl was reached – for the exact salting procedure see Table S6. Afterward, the solution was centrifuged for 12 min at 2800 g and 20°C. The supernatant was discarded and the pellet, in which the particles were concentrated, was re-suspended in PBS10 buffer (147 mM NaCl, 2.7 mM KCl, 10mM Na2HPO4, 3.9 mM KH2PO4, 2.9 mM K2HPO4, 2.5 mM EDTA, 0.01% Tween20). The washing step was carried out six times. Finally, the NPs were diluted in 1x TE buffer (10 mM Tris, 1 mM EDTA) containing 750 mM NaCl to an absorption of 0.1 (0.1 mm path length) at the excitation maxima on a UV-Vis spectrometer (NanoDrop, 2000; Thermo Fisher Scientific, USA).
Transmission electron microscopy (TEM) measurements
TEM grids (Formvar/carbon, 400 mesh, Cu, TedPella, Inc., USA) were Ar-plasma cleaned and incubated with 5 μL of ∼2–10 nM DNA origami sample for 60 s. Grids were washed with 5 μL 2% uranyl formate solution and incubated afterward again with 5 μL 2% uranyl formate solution for staining. TEM imaging was performed on a JOEL JEM-1100 microscope (JEOL GmbH, Japan) with an acceleration voltage of 80 kV.
Sample preparation on the coverslip for single-molecule confocal measurements
Adhesive SecureSeal™ Hybridization Chambers (2.6 mm depth, Grace Bio-Labs, USA) were glued on microscope coverslips of 24 mm × 60 mm size and 170 μm thickness (Carl Roth GmbH, Germany). The created wells were incubated with 1 M KOH for 1 h and washed three times with 1×PBS buffer. After surface passivation by incubation with BSA-Biotin (0.5 mg/mL, Sigma Aldrich, USA) for 15 min, the surface was again washed three times with 1× PBS buffer. 100 μL neutravidin (0.25 mg/mL, Thermo Fisher, USA) was incubated for 10 min and then washed three times with 1× PBS buffer. The DNA origami solution was diluted with 1× TE buffer containing 750 mM NaCl to a concentration of ∼10–100 pM and then immobilized on the biotin-neutravidin surface via biotin-neutravidin interactions. For this, 100 μL of the DNA origami sample solution was added and incubated for 3 min. Residual unbound DNA origami was removed by washing the wells three times with 1x TE buffer containing 750 mM NaCl. The density of DNA origami on the surface suitable for single-molecule measurements was checked on a confocal microscope. Nanoantenna samples were then incubated with 150 μL of the T20-functionalized AgNPs in 1x TE buffer containing 750 mM NaCl overnight at room temperature. Unbound NPs were removed by washing the samples three times with 1x TE buffer containing 750 mM NaCl. To prevent the evaporation of the samples, wells were glued with tapes. The samples were then imaged either directly or after performing a sensing procedure in antibody binding buffer (150 mM NaCl, 50 mM Na2HPO4, pH 7).
Antibody detection assay
For the detection of anti-Dig antibodies, DNA origami bearing a nanoswitch were immobilized on a surface via biotin-neutravidin interactions and NPs were attached to DNA origami samples in analogous way to the previous section. anti-Dig antibodies (Rb Monoclonal, Thermo Fisher Scientific, USA, cat#: 700,772, PRID: AB_2532342) were diluted to 0.01–500 nM in antibody binding buffer. DNA origami samples were incubated 20 min (unless stated otherwise in the text) with 150 μL of the anti-Dig antibody solution at room temperature before imaging.
Confocal measurements and data analysis
For detection of single-molecule fluorescence, a home-build confocal microscope was used. The setup was based on an inverted microscope (IX-83, Olympus Corportation, Japan) and a 78 MHz-pulsed white light laser (SuperK Extreme EXW-12, NKT Photonics A/S, Denmark) with selected wavelengths of 532 nm and 639 nm. The wavelengths were selected via an acousto-optically tunable filter (AOTF, SuperK Dual AOTF, NKT Photonics A/S, Denmark). This was controlled by a digital controller (AODS, 20160 8 R, Crystal Technology, USA) via a computer software (AODS, 20160 Control Panel, Crystal Technology, Inc., USA). A second AOTF (AA.AOTF.ns: TN, AA Opto-Electronic, France) was used to alternate 532 nm and 639 nm wavelengths if required, as well as to further spectrally clean the laser beam. It was controlled via home-made LabVIEW software (National Instruments, USA). A neutral density filter was used to regulate the laser intensity, followed by a linear polarizer and a λ/4 plate to obtain circularly polarized excitation. A dichroic beam splitter (ZT532/640rpc, Chroma Technology, USA) and an immersion oil objective (UPlanSApo 100×, NA = 1.4, WD = 0.12 mm, Olympus Corporation, Japan) were used to focus the excitation laser onto the sample. Micropositioning was performed using a Piezo-Stage (P-517.3CL, E−501.00, Physik Instrumente GmbH&Co. KG, Germany). The excitation powers at 639 nm were set to 500 nW for the reference samples and to 100 nW for the nanoantennas for the recording of fluorescence transients. For the confocal scans, 1 μW at 532 nm and 1 μW at 639 nm were used for all samples. Emitted light was collected by the same objective and filtered from the excitation light by a dichroic beam splitter. The light was later focused on a 50 μm pinhole (Linos AG, Germany) and detected using avalanche photodiodes (SPCM, AQR 14, PerkinElmer, Inc., USA) registered by an TCSPC system (HydraHarp 400, PicoQuant GmbH, Germany) after additional spectral filtering (RazorEdge 647, Semrock Inc., USA for the red channel and BrightLine HC 582/75, Semrock Inc., USA for the green channel). Custom-made LabVIEW software (National Instruments, USA) was used to process the acquired raw data. Background correction was carried out individually for each transient. The extracted data was analyzed in OriginPro2020.
Sample preparation for single-molecule measurements on the smartphone microscope
Microscope cover slides of 22 mm × 22 mm size and 170 μm thickness (Carl Roth GmbH, Germany) were cleaned with UV-Ozone cleaner (PSD-UV4, Novascan Technologies, USA) for 30 min at 100°C. After this a silicon mask was glued on a coverslip to create a chamber for surface functionalization, DNA origami immobilization (20 pM) and NACHOS assembly. Then the antibody detection assay was performed analogously as described above. The silicon mask was removed, and a double-sided tape was glued on both sides of the cover slide. The cover slides were covered with 76 mm × 26 mm microscope slides (1 mm thickness, Carl Roth GmbH, Germany) which were priory cleaned with UV-Ozone cleaner for 30 min at 100°C. Chambers were sealed before imaging.
Single-molecule measurements and analysis on the smartphone
Single antibody detection measurements on the smartphone were performed using a home-built portable setup (Trofymchuk et al., 2021). The 638 nm laser diode (0638L-11A, Integrated Optics, UAB, Lithuania) with an output power 180 mW that can be driven by various (portable) voltage sources was focused (f = 50 mm) in 45° angle onto the sample. The fluorescence of the molecules was collected using an objective lens (NA = 0.25, LS-40166, UCTRONICS, USA) and detected using a monochrome camera of the smartphone (P20, Huawei, China) after filtering out the excitation light (BrightLine HC 731/137, Semrock Inc., USA). Movies were recorded via FreeDCam application and analyzed with ImageJ (FIJI) equipped with FFMPEG plugin using a home written macro to convert MP4 format of the acquired movies to a TIFF format and find the single-molecule signals and extract the fluorescence intensity as a function of illumination time. The extracted data were analyzed in OriginPro2020.
Quantification and statistical analysis
Statistical analysis on nanoswitch opening
An unpaired t test was used to compare the magnitude of changes in nanoswitch opening on the NRO before and after the addition of 100 nM antibodies. The changes in nanoswitch opening before and after incubation with 100 nM anti-Dig antibodies were significant only for the nanoswitch construct bearing two Dig recognition elements (p < 0.05). While the changes in nanoswitch opening before and after incubation with anti-hDectin-1 antibody is not significant (p > 0.05), we observed a significant increase in nanoswitch opening before and after incubation with anti-Dig Fab fragment (p < 0.05). However, the difference before and after incubation with anti-Dig Fab fragment was very small (∼4%) even at 100 nM anti-Dig Fab fragment concentration. Given that the sensitive concentration range of the nanoswitch is way below this (Figure 3B), monovalent binding of two antibodies should not contribute much to the nanoswitch opening in the performed experiments.
Additionally, an unpaired t test was used to compare the changes in nanoswitch opening in NACHOS containing a 100-nm AgNP as well as in the same DNA origami nanostructure without NPs. The changes in nanoswitch opening before and after incubation with 100 nM anti-Dig antibodies was significant for the NACHOS containing a 100-nm AgNP and the DNA origami nanostructure without NPs (p < 0.05).
Impact of photophysics on the antibody assay
The single-molecule fluorescence transients of the nanoswitch in both the reference structure and the NACHOS structure upon incubation with anti-Dig antibody show short time blinking and also fluctuations between multiple intensity states. The intensity fluctuations could report on unbinding/rebinding of one of the two binding sites, distance fluctuations between dye and quencher due to the flexibility of ssDNA strands or could also be caused by photophysical processes unrelated to both (e.g. dim and dark states of the ATTO 647N dye, quencher blinking, fluctuations in the interaction between the dye and the Ag nanoparticle (e.g. orientation change of the transition dipole moment).
The photophysics of the ATTO 647N dye also were used to confirm the presence of single molecules in the assay. Only transients showing single-step photobleaching – transients with short time blinking and reversible intensity fluctuations between a bright and a dark or dim state – were included in the fluorescence enhancement analysis (69% and 52% of all recorded transients of the open nanoswitch in the reference structures and the NACHOS structure, respectively, Figure 2E). Multi-step photobleaching as observed for some nanostructures can be caused by a spectral shift of the ATTO 647N dye(Vogelsang et al., 2008; Glembockyte et al., 2016) – visible in the transients as an irreversible intensity fluctuation between a bright state and a dim state before photobleaching – or aggregated DNA origami structures. Furthermore, NACHOS transients without any photobleaching step can be related to nanoparticle aggregates on the surface.
Sensitive concentration ranges
For the binding curves in Figure 3B the observed nanoswitch opening (NO) was fitted using the following four parameter logistic equation: NO = NOmin + (NOmax – NOmin) [[Ab]nH/([Ab]nH + CHalfMaxnH)]. Here, NOmin and NOmax are the minimum and maximum nanoswitch opening values, CHalfMax is the antibody concentration at half-maximum signal after 20 min incubation, nH is the apparent Hill coefficient, and [Ab] is the concentration of the anti-Dig antibody added. This function usually is used for fitting dose-response functions and allows to study cooperativity if all data is collected in equilibrium limit. As we aimed to validate the assay under conditions used for diagnostic applications, we did not perform it in the equilibrium limit but with an incubation time of only 20 min for each target concentration. For this incubation time, binding of the antibody is most probably diffusion limited at low concentrations (Simon et al., 2014; Wu et al., 2019). This becomes apparent when fitting the binding curves as apparent Hill coefficients larger than one are found for all three DNA origami nanostructures. Thus, the performed measurements do not allow determining the KD value (antibody concentration at half maximum signal gain und equilibrium conditions) but show the sensitive concentration range achievable under clinically relevant conditions, e.g. short incubation times. The diffusion limitation shifts the CHalfMax to higher values than the KD value. They thus represent the upper KD limit.
Comparison with other single-antibody assays
The nanoswitch-based single-antibody detection assay was compared with three previously reported single-molecule antibody detection assays (Table S7). Criteria with possible relevance for point-of-care clinical applications were selected.
Acknowledgments
The authors thank Prof. Tim Liedl/Prof. Joachim Rädler (Ludwig-Maximilians-Universität, Department für Physik, Munich, Germany) for providing access to their facilities especially to the transmission electron microscope. P.T. acknowledges the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy — EXC 2089/1–390776260 for funding. P.T. gratefully acknowledges financial support from the DFG (Grant Nos. INST 86/1904-1 FUGG and TI 329/9-2) and BMBF (Grants POCEMON, 13N14336, and SIBOF, 03VP03891). V.G. acknowledges the support by Humboldt Research Fellowships from the Alexander von Humboldt Foundation and European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie. K.T. acknowledges the support by Humboldt Research Fellowships from the Alexander von Humboldt Foundation. S.R. is supported by European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement n. 843179 (“DNA-NANO-AB”). The work was also supported by Associazione Italiana per la Ricerca sul Cancro, AIRC (project n. 21965, F.R.) and by the European Research Council, ERC (Consolidator Grant project n. 819160, F.R.). We thank Luna for her support with the smartphone measurements.
Author contributions
M.P., K.T., V.G., and S.R. developed the concept of combining the nanoswitch and DNA origami platforms. M.P., K.T., and V.G. prepared samples, performed, and analyzed the measurements. M.P., K.T., V.G., F.S., and F.C. performed and analyzed the measurements on the smartphone microscope. P.T. and F.R. supervised the project. All authors have written, read, and approved the final manuscript.
Declaration of interests
P.T. is an inventor on an awarded patent of the described bottom-up method for fluorescence enhancement in molecular assays, EP1260316.1, 2012. The remaining authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103072.
Contributor Information
Viktorija Glembockyte, Email: viktorija.glembockyte@cup.lmu.de.
Philip Tinnefeld, Email: philip.tinnefeld@cup.uni-muenchen.de.
Supporting Citations
The following references appear in the Supplemental information: Fahie et al., 2015; Li et al., 2002; Rissin et al., 2010.
Supplemental information
Data and code availability
-
•
All raw data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Acuna G.P., Moller F.M., Holzmeister P., Beater S., Lalkens B., Tinnefeld P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science. 2012;338:506–510. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- Barletta J.M., Edelman D.C., Constantine N.T. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am. J. Clin. Pathol. 2004;122:20–27. doi: 10.1309/529T2WDNEB6X8VUN. [DOI] [PubMed] [Google Scholar]
- Chames P., Van Regenmortel M., Weiss E., Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br. J. Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A.R. DNA nanobiosensors: an outlook on signal readout strategies. J. Nanomater. 2017;2017:2820619. doi: 10.1155/2017/2820619. [DOI] [Google Scholar]
- Chi Q., Wang G., Jiang J. The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Phys. A Stat. Mech. Appl. 2013;392:1072–1079. doi: 10.1016/j.physa.2012.09.022. [DOI] [Google Scholar]
- Dass M., Gür F.N., Kołątaj K., Urban M.J., Liedl T. DNA origami-enabled plasmonic sensing. J. Phys. Chem. C. 2021;125:5969–5981. doi: 10.1021/acs.jpcc.0c11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S., Fan C., Gothelf K.V., Li J., Lin C., Liu L., Liu N., Nijenhuis M.A.D., Saccà B., Simmel F.C. DNA origami. Nat. Rev. Methods Primers. 2021;1:13. doi: 10.1038/s43586-020-00009-8. [DOI] [Google Scholar]
- Domljanovic I., Carstens A., Okholm A., Kjems J., Nielsen C.T., Heegaard N.H.H., Astakhova K. Complexes of DNA with fluorescent dyes are effective reagents for detection of autoimmune antibodies. Sci.Rep. 2017;7:1925. doi: 10.1038/s41598-017-02214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahie M., Chisholm C., Chen M. Resolved single-molecule detection of individual species within a mixture of anti-biotin antibodies using an engineered monomeric nanopore. ACS Nano. 2015;9:1089–1098. doi: 10.1021/nn506606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini S., Marsico S., Becker P.D., Iaria M.L., Bruno R., Guzmán C.A., Caruso A. Synthetic peptide AT20 coupled to KLH elicits antibodies against a conserved conformational epitope from a major functional area of the HIV-1 matrix protein p17. Vaccine. 2008;26:4758–4765. doi: 10.1016/j.vaccine.2008.06.082. [DOI] [PubMed] [Google Scholar]
- Funck T., Nicoli F., Kuzyk A., Liedl T. Sensing picomolar concentrations of RNA using switchable plasmonic chirality. Angew. Chem. Int. Ed. Engl. 2018;57:13495–13498. doi: 10.1002/anie.201807029. [DOI] [PubMed] [Google Scholar]
- Glembockyte V., Lin J., Cosa G. Improving the photostability of red- and green-emissive single-molecule fluorophores via Ni2+ mediated excited triplet-state quenching. J. Phys. Chem. B. 2016;120:11923–11929. doi: 10.1021/acs.jpcb.6b10725. [DOI] [PubMed] [Google Scholar]
- Godonoga M., Lin T.-Y., Oshima A., Sumitomo K., Tang M.S.L., Cheung Y.-W., Kinghorn A.B., Dirkzwager R.M., Zhou C., Kuzuya A. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci. Rep. 2016;6:21266. doi: 10.1038/srep21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenhorst L., Trofymchuk K., Steiner F., Glembockyte V., Tinnefeld P. Fluorophore photostability and saturation in the hotspot of DNA origami nanoantennas. Methods Appl. Fluoresc. 2020;8:024003. doi: 10.1088/2050-6120/ab6ac8. [DOI] [PubMed] [Google Scholar]
- Holzmeister P., Wünsch B., Gietl A., Tinnefeld P. Single-molecule photophysics of dark quenchers as non-fluorescent FRET acceptors. Photochem. Photobiol. Sci. 2014;13:853–858. doi: 10.1039/C3PP50274K. [DOI] [PubMed] [Google Scholar]
- Ji E., Lee S. Antibody-based therapeutics for atherosclerosis and cardiovascular diseases. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22115770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Castro C., Choi J.H. Structural DNA nanotechnology: artificial nanostructures for biomedical research. Annu. Rev. Biomed. Eng. 2018;20:375–401. doi: 10.1146/annurev-bioeng-062117-120904. [DOI] [PubMed] [Google Scholar]
- Ke Y., Lindsay S., Chang Y., Liu Y., Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- Keyser U.F. Enhancing nanopore sensing with DNA nanotechnology. Nat. Nanotechnol. 2016;11:106–108. doi: 10.1038/nnano.2016.2. [DOI] [PubMed] [Google Scholar]
- Koirala D., Shrestha P., Emura T., Hidaka K., Mandal S., Endo M., Sugiyama H., Mao H. Single-molecule mechanochemical sensing using DNA origami nanostructures. Angew. Chem. Int. Ed. 2014;53:8137–8141. doi: 10.1002/anie.201404043. [DOI] [PubMed] [Google Scholar]
- Kuzuya A., Sakai Y., Yamazaki T., Xu Y., Komiyama M. Nanomechanical DNA origami 'single-molecule beacons' directly imaged by atomic force microscopy. Nat. Commun. 2011;2:449. doi: 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Stockman M.I., Bergman D.J. Self-similar chain of metal Nanospheres as an efficient nanolens. Phys. Rev. Lett. 2003;91:227402. doi: 10.1103/PhysRevLett.91.227402. [DOI] [PubMed] [Google Scholar]
- Li L., Chen S., Oh S., Jiang S. In situ single-molecule detection of Antibody−Antigen binding by tapping-mode atomic force microscopy. Anal. Chem. 2002;74:6017–6022. doi: 10.1021/ac0258148. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang L., Yan H., Liu Y. Effect of DNA hairpin loops on the twist of planar DNA origami tiles. Langmuir. 2012;28:1959–1965. doi: 10.1021/la2037873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Jungmann R., Leifer A.M., Li C., Levner D., Church G.M., Shih W.M., Yin P. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loretan M., Domljanovic I., Lakatos M., Rüegg C., Acuna G.P. DNA origami as emerging Technology for the engineering of fluorescent and plasmonic-based biosensors. Materials. 2020;13:2185. doi: 10.3390/ma13092185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras A.E., Shi Z., Lindell M.G., Patton R.A., Huang C.-M., Zhou L., Su H.-J., Arya G., Castro C.E. Cation-activated avidity for rapid reconfiguration of DNA nanodevices. ACS Nano. 2018;12:9484–9494. doi: 10.1021/acsnano.8b04817. [DOI] [PubMed] [Google Scholar]
- McCreery T. Digoxigenin labeling. Mol. Biotechnol. 1997;7:121–124. doi: 10.1007/bf02761747. [DOI] [PubMed] [Google Scholar]
- Novotny L., Van Hulst N. Antennas for light. Nat. Photon. 2011;5:83–90. doi: 10.1038/nphoton.2010.237. [DOI] [Google Scholar]
- Ochmann S.E., Vietz C., Trofymchuk K., Acuna G.P., Lalkens B., Tinnefeld P. Optical nanoantenna for single molecule-based detection of Zika virus nucleic acids without molecular multiplication. Anal. Chem. 2017;89:13000–13007. doi: 10.1021/acs.analchem.7b04082. [DOI] [PubMed] [Google Scholar]
- Porchetta A., Ippodrino R., Marini B., Caruso A., Caccuri F., Ricci F. Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids. J. Am. Chem. Soc. 2018;140:947–953. doi: 10.1021/jacs.7b09347. [DOI] [PubMed] [Google Scholar]
- Puchkova A., Vietz C., Pibiri E., Wunsch B., Sanz Paz M., Acuna G.P., Tinnefeld P. DNA origami nanoantennas with over 5000-fold fluorescence enhancement and single-molecule detection at 25 muM. Nano Lett. 2015;15:8354–8359. doi: 10.1021/acs.nanolett.5b04045. [DOI] [PubMed] [Google Scholar]
- Purcell E.M. Proceedings of the American physical society. Phys. Rev. 1946;69:674. doi: 10.1103/PhysRev.69.674.2. [DOI] [Google Scholar]
- Ranallo S., Porchetta A., Ricci F. DNA-based scaffolds for sensing applications. Anal. Chem. 2019;91:44–59. doi: 10.1021/acs.analchem.8b05009. [DOI] [PubMed] [Google Scholar]
- Ranallo S., Rossetti M., Plaxco K.W., Vallée-Bélisle A., Ricci F. A modular, DNA-based beacon for single-step fluorescence detection of antibodies and other proteins. Angew. Chem. Int. Ed. Engl. 2015;54:13214–13218. doi: 10.1002/anie.201505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulf M., Joest M., Sander I., Hoffmeyer F., Nowak D., Ochmann U., Preisser A., Schreiber J., Sennekamp J., Koschel D. Update of reference values for IgG antibodies against typical antigens of hypersensitivity pneumonitis. Allergo J. Int. 2019;28:192–203. doi: 10.1007/s40629-019-0099-x. [DOI] [Google Scholar]
- Raveendran M., Lee A.J., Sharma R., Wälti C., Actis P. Rational design of DNA nanostructures for single molecule biosensing. Nat. Commun. 2020;11:4384. doi: 10.1038/s41467-020-18132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci F., Vallée-Bélisle A., Simon A.J., Porchetta A., Plaxco K.W. Using nature’s “tricks” to rationally tune the binding properties of biomolecular receptors. Acc. Chem. Res. 2016;49:1884–1892. doi: 10.1021/acs.accounts.6b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker S., Ke Y., Liu Y., Chhabra R., Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand–protein binding. Nat. Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Ezekiel M.P., Spencer S.A., Meredith R.F., Bonner J.A., Khazaeli M.B., Saleh M.N., Carey D., Lobuglio A.F., Wheeler R.H. Phase I study of anti--epidermal growth factor receptor antibody cetuximab in combination with radiation therapy in patients with advanced head and neck cancer. J. Clin. Oncol. 2001;19:3234–3243. doi: 10.1200/jco.2001.19.13.3234. [DOI] [PubMed] [Google Scholar]
- Rostenberg I., Peñaloza R. Serum IgG and IgD and levels in some infectious and noninfectious diseases. Clin. Chim. Acta. 1978;85:319–321. doi: 10.1016/0009-8981(78)90310-8. [DOI] [PubMed] [Google Scholar]
- Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.L., Beliveau B.J., Uca Y.O., Theilmann M., Da Cruz F., Wu C.-T., Shih W.M. Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat. Commun. 2015;6:8634. doi: 10.1038/ncomms9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N.C., Sleiman H.F. DNA nanotechnology. Nat. Rev. Mater. 2017;3:17068. doi: 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Selnihhin D., Sparvath S.M., Preus S., Birkedal V., Andersen E.S. Multifluorophore DNA origami beacon as a biosensing platform. ACS Nano. 2018;12:5699–5708. doi: 10.1021/acsnano.8b01510. [DOI] [PubMed] [Google Scholar]
- Shaw A., Hoffecker I.T., Smyrlaki I., Rosa J., Grevys A., Bratlie D., Sandlie I., Michaelsen T.E., Andersen J.T., Högberg B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 2019;14:184–190. doi: 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.J., Vallée-Bélisle A., Ricci F., Plaxco K.W. Intrinsic disorder as a generalizable strategy for the rational design of highly responsive, allosterically cooperative receptors. Proc. Natl. Acad. Sci. U S A. 2014;111:15048. doi: 10.1073/pnas.1410796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian H.K.K., Chakraborty B., Sha R., Seeman N.C. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 2011;11:910–913. doi: 10.1021/nl104555t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczewska M., Lee S., Chen X. The design and application of fluorophore-gold nanoparticle activatable probes. Phys. Chem. Chem. Phys. : PCCP. 2011;13:9929–9941. doi: 10.1039/c0cp02967j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofymchuk K., Glembockyte V., Grabenhorst L., Steiner F., Vietz C., Close C., Pfeiffer M., Richter L., Schütte M.L., Selbach F. Addressable nanoantennas with cleared hotspots for single-molecule detection on a portable smartphone microscope. Nat. Commun. 2021;12:950. doi: 10.1038/s41467-021-21238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietz C., Kaminska I., Sanz Paz M., Tinnefeld P., Acuna G.P. Broadband fluorescence enhancement with self-assembled silver nanoparticle optical antennas. ACS Nano. 2017;11:4969–4975. doi: 10.1021/acsnano.7b01621. [DOI] [PubMed] [Google Scholar]
- Vietz C., Lalkens B., Acuna G.P., Tinnefeld P. Functionalizing large nanoparticles for small gaps in dimer nanoantennas. New J. Phys. 2016;18:045012. [Google Scholar]
- Vietz C., Lalkens B., Acuna G.P., Tinnefeld P. Synergistic combination of unquenching and plasmonic fluorescence enhancement in fluorogenic nucleic acid hybridization probes. Nano Lett. 2017;17:6496–6500. doi: 10.1021/acs.nanolett.7b03844. [DOI] [PubMed] [Google Scholar]
- Vietz C., Schütte M.L., Wei Q., Richter L., Lalkens B., Ozcan A., Tinnefeld P., Acuna G.P. Benchmarking smartphone fluorescence-based microscopy with DNA origami nanobeads: reducing the gap toward single-molecule sensitivity. ACS Omega. 2019;4:637–642. doi: 10.1021/acsomega.8b03136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelsang J., Kasper R., Steinhauer C., Person B., Heilemann M., Sauer M., Tinnefeld P. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. Int. Ed. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- Wang D., Vietz C., Schröder T., Acuna G., Lalkens B., Tinnefeld P. A DNA walker as a fluorescence signal amplifier. Nano Lett. 2017;17:5368–5374. doi: 10.1021/acs.nanolett.7b01829. [DOI] [PubMed] [Google Scholar]
- Wagenbauer K.F., Engelhardt F.A.S., Stahl E., Hechtl V.K., Stömmer P., Seebacher F., Meregalli L., Ketterer P., Gerling T., Dietz H. How we make DNA origami. ChemBioChem. 2017 doi: 10.1002/cbic.201700377. [DOI] [PubMed] [Google Scholar]
- Wang P., Meyer T.A., Pan V., Dutta P.K., Ke Y. The beauty and utility of DNA origami. Chem. 2017;2:359–382. doi: 10.1016/j.chempr.2017.02.009. [DOI] [Google Scholar]
- Wang S., Zhou Z., Ma N., Yang S., Li K., Teng C., Ke Y., Tian Y. DNA origami-enabled biosensors. Sensors. 2020;20:6899. doi: 10.3390/s20236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S., Rothemund P.W. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 2011;3:620–627. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- Wu Y., Tilley R.D., Gooding J.J. Challenges and solutions in developing ultrasensitive biosensors. J. Am. Chem. Soc. 2019;141:1162–1170. doi: 10.1021/jacs.8b09397. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang Y., Fan C., Li C., Li Y., Qian L., Fu Y., Shi Y., Hu J., He L. Asymmetric DNA origami for spatially addressable and index-free solution-phase DNA chips. Adv. Mater. 2010;22:2672–2675. doi: 10.1002/adma.201000151. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zeng D., Ma H., Feng G., Hu J., He L., Li C., Fan C. A DNA-Origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement. Small. 2010;6:1854–1858. doi: 10.1002/smll.201000908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.