Summary
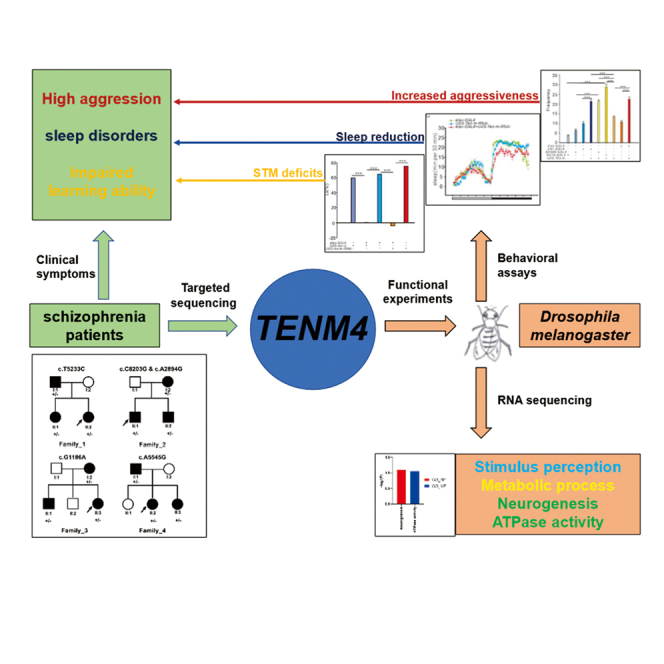
TENM4, encoding a member of the teneurin protein family, is a risk gene shared by many types of mental diseases and is implicated in neuronal plasticity and signaling. However, the role and the mechanisms of TENM4 in schizophrenia (SCZ) remain unclear. We identified possible pathogenic mutations in the TENM4 gene through target sequencing of TENM4 in 68 SCZ families. We further demonstrated that aberrant expression of Ten-m leads to lower learning ability, sleep reduction, and increased aggressiveness in animal models. RNA sequencing showed that aberrant expression of Ten-m was related to stimulus perception and metabolic process, and Gene Ontology enrichment terms were neurogenesis and ATPase activity. This study provides strong evidence that TENM4 contributes to SCZ, and its functional mutations might be responsible for the impaired neural circuits and behaviors observed in SCZ.
Subject areas: genetics, human genetics, molecular genetics, neurogenetics, psychology
Graphical abstract
Highlights
-
•
Possible pathogenic rare missense mutations in TENM4 gene contribute to SCZ
-
•
Aberrant expression of Ten-m leads to behavioral disturbances related to SCZ symptoms
-
•
Ten-m affects stimulation, metabolic process, neurogenesis, and ATPase activity
Genetics; Human genetics; Human Genetics; Molecular genetics; Neurogenetics; Psychology
Introduction
Schizophrenia (SCZ) is a severe psychiatric disorder with high heritability, genetic heterogeneity, and polygenic effect (Kavanagh et al., 2015). It has been reported that SCZ shares the same risk genes and pathways as common mental diseases, including bipolar disorder, autism, depression, Alzheimer disease, mental retardation, and epilepsy (Sullivan et al., 2012). Previous studies have confirmed that TENM4 is associated with bipolar disorder (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011) (Ikeda et al., 2018) (Ivorra et al., 2014) (Heinrich et al., 2013), autism (Iossifov et al., 2014), and essential tremor (Hor et al., 2015) (Yan et al., 2020) (Chao et al., 2016). In most genome-wide association studies (GWAS) of SCZ, the common single-nucleotide polymorphisms (SNPs) in TENM4 did not reach genome-wide significance. However, a case-control replication study showed that an SNP (rs12290811) located in TENM4 was associated with SCZ (Ivorra et al., 2014). A whole-exome sequencing study in an SCZ family also identified one missense mutation co-segregated with the SCZ in TENM4 (Xue et al., 2018). A genome-wide meta-analysis with individuals of East Asian or European ancestry identified 176 loci associated with SCZ, including TENM4 (Lam et al., 2019). These studies partly provided some genetic evidence for TENM4 as an important candidate gene for SCZ.
TENM4, also known as ODZ4, is a member of the teneurin protein family, which comprises large transmembrane proteins (Baumgartner et al., 1994). The TENM4 gene is highly conserved among species (Tucker and Chiquet-Ehrismann, 2006). The temporal and spatial expression of teneurin family proteins in Drosophila (Baumgartner et al., 1994), zebrafish (Feng et al., 2002), birds (Tucker et al., 2000), and mice (Zhou et al., 2003) showed that Tenm4 protein was highly expressed in the nervous system, particularly in the brain. In Drosophila, a study showed that Ten-m acts as a synaptic cooperative matching molecule between the axons of olfactory receptor neurons (ORNs) and the dendrites of projecting neurons (PNs) in the olfactory system, and Ten-m could also guide synapses to achieve specific matches by homophilic interactions to ensure proper matching between neurons, indicating Ten-m plays an important role in neuron orientation (Hong et al., 2012). In addition, Ten-m was required for the organization and target selection of synapses in neurons (Mosca et al., 2012). In mice, a mutation in Tenm4 suppressed oligodendrocyte differentiation and myelination of small-diameter axons (Suzuki et al., 2012). Knockdown expression of Tenm4 in Neuro-2a cell lines decreased the length of individual neurites, while overexpression of Tenm4 promoted cellular protrusion formation and neurite outgrowth (Suzuki et al., 2014). These studies indicated that aberrant expression of Ten-m might lead to mis-matched neural circuits.
Based on these previously reported findings, we hypothesized that TENM4 plays an important role in SCZ development and contributes to SCZ-related clinical symptoms, such as learning disability, sleep disorders, and violent behavior. Patients with SCZ often show violence, social withdrawal, learning and memory declines, and sleep disorders (Dowd and Barch, 2012; Cohrs, 2008; Patel et al., 2014). To verify this hypothesis, we performed targeted sequencing of TENM4 in 68 SCZ families to investigate the genetic contribution of TENM4 on SCZ. Then, to identify the role of TENM4 in SCZ, we examined the phenotypic characteristics by knockdown and overexpression of Ten-m in Drosophila and evaluated the effects of Ten-m on learning ability, sleep behavior, and aggressive behaviors. We also used mRNA sequencing to reveal the potential affected signaling pathways and molecular mechanisms of TENM4.
Results
Rare missense mutations in TENM4 are co-segregated with disease status in SCZ families
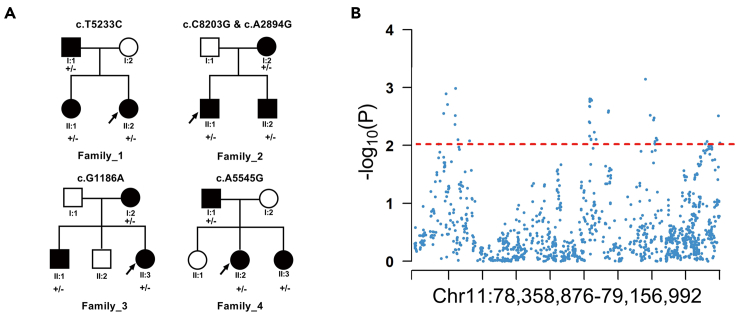
To determine whether rare mutations in TENM4 are the candidate causal variants of SCZ, we initially performed whole TENM4 exome sequencing in 351 subjects from 68 SCZ families. A total of 36 mutations in TENM4 were identified using a step-by-step depth, quality filtering method. Then, 17 noncoding and synonymous variants were filtered to remove, and 19 nonsynonymous variants were kept. The 19 variants were annotated by ANNOVAR software (2019Oct24) with dbnsfp35c, gnomad211_exome, avsnp150, and intervar_20,180,118 databases to evaluate the function and allele frequency in the population. Given the pattern of dominant inheritance in the families, only rare heterozygous variants (allele frequency<1%) shared by the patients but not found in the normal controls in the same family were considered to be candidate mutations. At last, we found 5 variants were co-segregated with SCZ in their families, which were p.S1745P in Family_1, p.Q2735E and p.N965S in Family_2, p.V396I in Family_3, and p.T1849A in Family_4 (Table 1 and Figure 1A). In addition, all the five missense mutations were predicted to be deleterious to the TENM4 protein function by no less than three software.
Table 1.
Genetic analysis of TENM4 in SCZ
| FAM_ID | Position | Allele | SNP | AA_Change | AF | No. | Interpro_domain |
|---|---|---|---|---|---|---|---|
| Family_1 | 78,399,126 | A > G | Ns | p.S1745P | ns | 7 | YD repeats |
| Family_2 | 78,369,210 | G > C | rs527857070 | p.Q2735E | 1.605 × 10−5 | 6 | Tox-GHH domain |
| 78,443,605 | T > C | rs141471364 | p.N965S | 0.0053 | 3 | Carboxypeptidase-like, regulatory domain | |
| Family_3 | 78,574,076 | C > T | rs3812723 | p.V396I | 0.0003 | 3 | . |
| Family_4 | 78,383,326 | T > C | rs772977333 | p.T1849A | 0.0002 | 9 | YD repeats |
SNP, single-nucleotide polymorphism; AA, amino acid; AF, allele frequency; No. the number of software that supported the damaging effect of the missense mutation on TENM4 protein function.
Figure 1.
Genetic analysis of TENM4 in SCZ
(A) TENM4 rare mutations were identified in four SCZ families, which were p.S1745P in Family_1, p.Q2735E and p.N965S in Family_2, p.V396I in Family_3, and p.T1849A in Family_4.
(B) TENM4 showed nominally significant association with SCZ according to the previous GWAS.
Meanwhile, to confirm the relationship between TENM4 and SCZ, we re-analyzed the data from the genome-wide association analysis in a total of 50,874 SCZ cases and 83,493 controls from the Chinese and PGC2 genome-wide meta-analysis (Li et al., 2017). We analyzed 1196 SNPs in the region (chr11: 78,358,876-79,156,992) and found that 39 SNPs showed nominally significant association with SCZ (p < 0.01, Figure 1B).
Ten-m affects the learning abilities in Drosophila
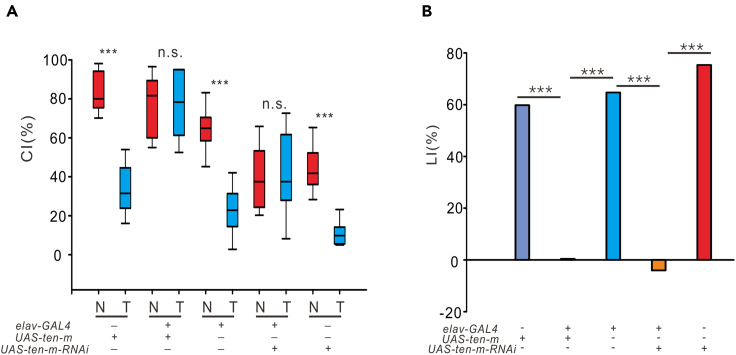
To investigate the potential contribution of TENM4 to SCZ, we examined the function of Ten-m in the Drosophila nervous system. We used the courtship conditioning assay, a behavioral assay that tests the courtship short-term memory (STM) to indicate learning ability (Siegel and Hall, 1979). After selection at the eclosion stage, male flies that had never mated were solely paired with mated females whose cuticles had already obtained cis-vaccenyl acetate (cVA), a male-specific and volatile pheromone, from males on mating (Everaerts et al., 2010). The males would be rejected by the mated females; then, the males formed STM, resulting in the suppression of courtship behavior when placed with other mated female flies over a period of time (Siwicki et al., 2005). The reduction of suppression indicates a lack of STM and the damage to learning ability. We used courtship indices (CIs), calculated as the percentage of courtship duration within 10 min, and learning indices (LIs), generated by the relative difference between the mean CIs of naive and trained male flies, to quantify the STM. Our data revealed that males of control genotypes (elav-GAL4, UAS-Ten-m, and UAS-Ten-m-RNAi) exhibited courtship suppression after training compared to naive flies. Otherwise, there was no significant difference in CIs between naive and trained experimental flies (elav > UAS-Ten-m and elav > UAS-Ten-m-RNAi) with Ten-m aberrant expression in the nervous system, indicating that pan-neuronal overexpression or downexpression of Ten-m lead to lack of the learning ability to suppress courtship behavior (Figure 2A, naive vs trained; elav-GAL4, p < 0.0001; UAS-Ten-m, p < 0.0001; UAS-Ten-m-RNAi, p < 0.0001; elav > Ten-m, p = 0.820; elav > Ten-m-RNAi, p = 1.000). In addition, compared with the control groups, flies with aberrantly expressed Ten-m pan-neuronally revealed a significant reduction in LIs, which highlighted the courtship STM deficits (Figure 2B, elav-GAL4 vs elav > Ten-m, p < 0.0001, UAS-Ten-m vs elav > Ten-m, p < 0.0001; elav-GAL4 vs elav > Ten-m-RNAi, p < 0.0001; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p < 0.0001). These results suggested that aberrant expression of Ten-m in the nervous system impairs STM.
Figure 2.
Aberrant expression of Ten-m impairs courtship-conditioning learning
(A) CIs of control males (elav-GAL4, UAS-Ten-m, UAS-Ten-m-RNAi, n = 10) and experimental males (elav > Ten-m and elav > Ten-m-RNAi, n = 10). Box-and-whisker plots of CIs show 10th, 25th, 50th, 75th, and 90th centiles and means (short dashed line). N, naive; T, trained. Experimental males showed no difference in CIs between naive and trained groups, while the CIs of control males in the trained group were significantly lower than in the naive groups.
(B) LIs of control males (elav-GAL4, UAS-Ten-m, UAS-Ten-m-RNAi, n = 10) and experimental males (elav > Ten-m and elav > Ten-m-RNAi, n = 10). LIs were compared using the permutation test with 100,000 random permutations (one-tail, H1: LI control > LI experimental males). LIs of experimental males were significantly higher than those in control males.
The Mann-Whitney U test was used for CIs. ∗∗∗p < 0.001; n.s., no significance.
Ten-m affects sleep behavior in Drosophila
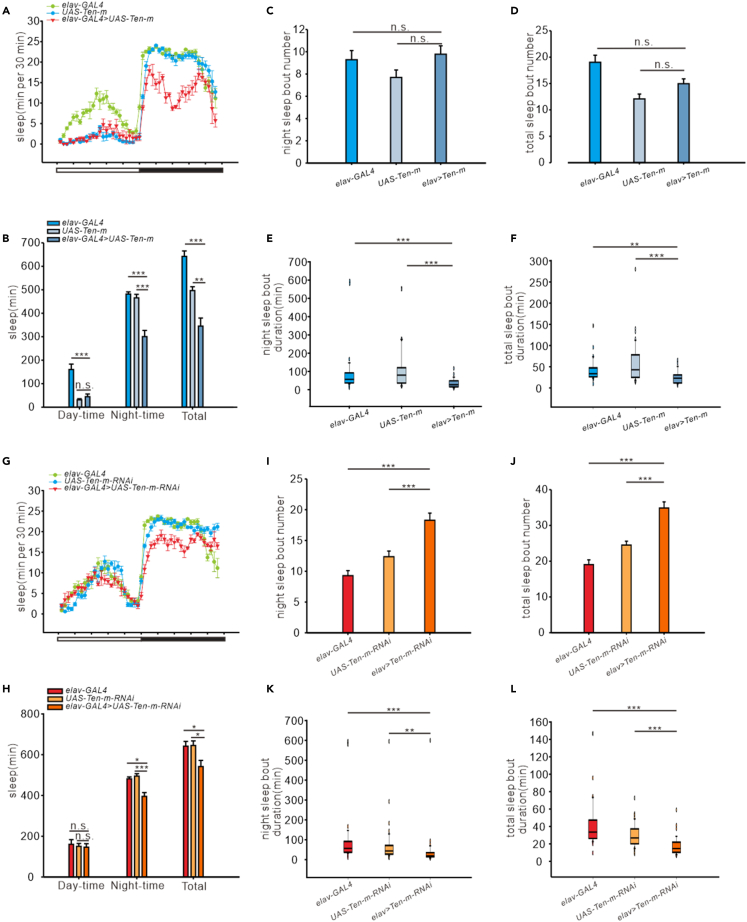
To determine whether Ten-m regulates sleep in Drosophila, we increased or decreased Ten-m expression levels using transgenes UAS-Ten-m and UAS-Ten-m-RNAi, respectively, driven by pan-neuronal elav-GAL4. Total and nighttime sleep amounts were decreased both in elav > Ten-m (Figures 3A and 3B nighttime: elav-GAL4 vs elav > Ten-m, p < 0.0001, UAS-Ten-m vs elav > Ten-m, p < 0.0001; total: elav-GAL4 vs elav > Ten-m, p < 0.0001; UAS-Ten-m vs elav > Ten-m, p = 0.002) and elav > Ten-m-RNAi files (Figures 3G and 3H, nighttime: elav-GAL4 vs elav > Ten-m-RNAi, p = 0.012; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p < 0.0001; total: elav-GAL4 vs elav > Ten-m-RNAi, p = 0.019; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p = 0.012) compared to controls. In contrast, there was no change in mutants in the daytime sleep of the mutants (Figures 3A and 3B, daytime: elav-GAL4 vs elav > Ten-m, p < 0.0001; UAS-Ten-m vs elav > Ten-m, p = 1.000; Figures 3G and 3H, elav-GAL4 vs elav > Ten-m-RNAi, p = 0.853; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p = 0.986) which might be due to a floor effect, that is, daytime sleep of the controls was minimal. Further analysis revealed that the decreased night and overall sleep in elav > Ten-m flies was due to the reduced sleep bout duration but not sleep bout number (Figure 3C, elav-GAL4 vs elav > Ten-m, p = 0.905, UAS-Ten-m vs elav > Ten-m, p = 0.154; Figure 3D, elav-GAL4 vs elav > Ten-m, p = 0.905, UAS-Ten-m vs elav > Ten-m, p = 0.121; Figure 3E, p < 0.0001; Figure 3F, elav-GAL4 vs elav > Ten-m, p = 0.005; UAS-Ten-m vs elav > Ten-m, p < 0.0001). These results indicated that over-expressing Ten-m in all neurons would impair sleep behavior in Drosophila by reducing night sleep bout duration. The night and total sleep bout numbers were significantly increased in elav > Ten-m-RNAi flies compared to the control groups (Figures 3I and 3J, p < 0.0001). Meanwhile, the night and total sleep bout durations of elav > Ten-m-RNAi groups were shorter than those of the control groups (Figure 3K, elav-GAL4 vs elav > Ten-m-RNAi, p < 0.0001; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p = 0.001; Figure 3L, p < 0.0001), suggesting the lack of Ten-m impaired sleep maintenance and caused sleep fragmentation in Drosophila. Overall, both Ten-m overexpression and downexpression impaired sleep maintenance and caused fragmented sleep.
Figure 3.
Sleep phenotypes in Ten-m overexpressed and downexpressed mutants
(A) Conventional sleep plots of controls (elav-GAL4, n = 13, UAS-Ten-m, n = 16) and experimental flies (elav > Ten-m, n = 9) in 12-hr LD conditions. White and black bars indicate 12-hr LD periods, respectively.
(B) Total sleep amount (in 24 hr) and sleep during daytime and nighttime in control and experimental flies. Data are represented as mean ± SEM.
(C and D) Histograms of the number of nighttime sleep bouts during nighttime (C) and histograms of the number of total sleep bouts (D) for controls and experimental flies. Data are represented as mean ± SEM.
(E and F) Boxplots of the nighttime sleep bout duration (E) and boxplots of the total sleep bout duration (F) for controls and experimental flies. The line inside the box indicates the median; the upper and lower box limits the 75% and 25% quantiles, respectively; vertical lines above and below the box represent the 90% and 10% quantiles, respectively; points show the 95% and 5% outliers.
(G–L) Conventional sleep plots of controls (elav-GAL4, n = 13, and UAS-Ten-m-RNAi, n = 14) and experimental flies (elav > Ten-m-RNAi, n = 13) in 12-hr LD (G). Histograms of total sleep amount and sleep during daytime and nighttime (H), nighttime sleep bout number (I), nighttime sleep bout duration (K), total sleep bout number (J), and total sleep bout duration (L) for controls and experimental flies. Data are represented as mean ± SEM.
One-way analysis of variance (ANOVA) followed by post hoc Tukey were used, except for the comparison of nonparametrically distributed data where the Kruskal-Wallis test was used. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., no significance.
Ten-m affects the aggressive behavior in Drosophila
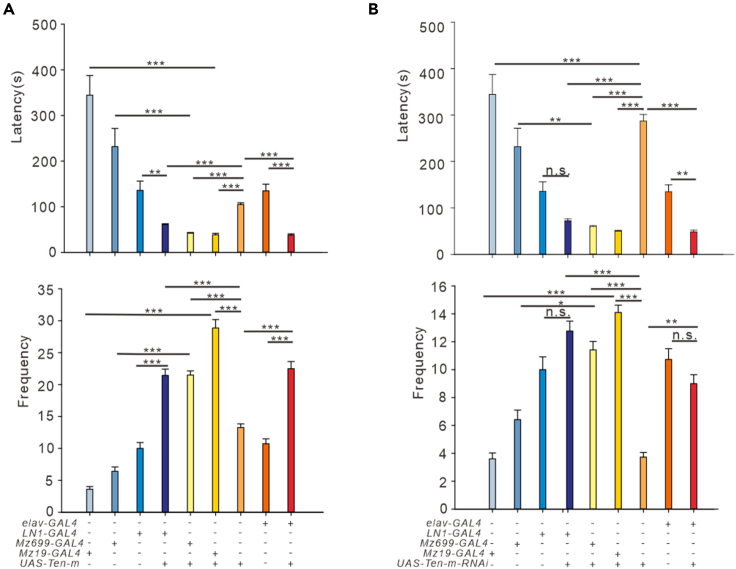
We examined the aggressive behavior of Ten-m and Ten-m-RNAi flies using different GAL4 drivers. The latency to initiate fighting was significantly reduced (Figure 4A p < 0.0001; Figure 4B, elav-GAL4 vs elav > Ten-m-RNAi, p = 0.009; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p < 0.0001), and the frequency was increased (Figure 4A, p < 0.0001; Figure 4B, elav-GAL4 vs elav > Ten-m-RNAi, p = 0.832; UAS-Ten-m-RNAi vs elav > Ten-m-RNAi, p = 0.001) in the flies with aberrantly expressed Ten-m pan-neuronally compared to the control groups. Male-male aggression is promoted by cVA in Drosophila via Or67d and Or65a receptors expressed by ORNs (Wang and Anderson, 2010). ORNs form synapses with PNs and local interneurons (LNs) in the antennal lobe (Das et al., 2008). Mz699-GAL4 is expressed in 90% of all ventral PNs, and 80% of these Mz699-PNs have been identified as GABAergic neurons, which are related to aggression (Lai et al., 2008) (Alekseyenko et al., 2019), and Mz19 PNs are inherently connected to Or67d ORNs (Hong et al., 2012). We examined the aggressive behavior of UAS-Ten-m and UAS-Ten-m-RNAi transgenic flies using Mz699-GAL4, Mz19-GAL4, and LN1-GAL4, and latency and frequency changed sharply between the experimental and the control groups (Figures 4A and 4B, p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., no significance.). These results reveal that aberrant expression of Ten-m in Drosophila would lead flies to become more aggressive.
Figure 4.
Ten-m modulates aggression
(A) The latency and frequency of male flies overexpressing Ten-m in all neurons (elav > Ten-m, n = 14) or in specific neurons such as Mz699, Mz19, and LN1 (Mz699>Ten-m, n = 14, Mz19>Ten-m, n = 15, LN1>Ten-m, n = 9) compared to the controls (elav-GAL4, n = 15, UAS-Ten-m, n = 11, Mz699-GAL4, n = 12, Mz19-GAL4, n = 10, LN1-GAL4, n = 10). Fighting latencies of experimental flies were significantly longer than those in controls, and fighting frequencies of experimental flies were significantly higher than those in controls. Data are represented as mean ± SEM.
(B) The latency and frequency of male flies downexpressing Ten-m in all neurons (elav > Ten-m-RNAi, n = 12) or in specific neurons such as Mz699, Mz19, and LN1 (Mz699>Ten-m-RNAi, n = 12, Mz19>Ten-m-RNAi, n = 10, LN1>Ten-m-RNAi, n = 13) compared to the controls (elav-GAL4, n = 15, UAS-Ten-m-RNAi, n = 11, Mz699-GAL4, n = 12, Mz19-GAL4, n = 10, LN1-GAL4, n = 10). Fighting latencies of experimental flies were significantly longer than those of controls, and fighting frequencies of experimental flies were significantly higher than those of controls. Data are represented as mean ± SEM.
One-way ANOVA followed by post hoc Tukey were used, except for the comparison of nonparametrically distributed data where the Kruskal-Wallis test was used. ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., no significance.
RNA sequencing reveals the potential molecular mechanism
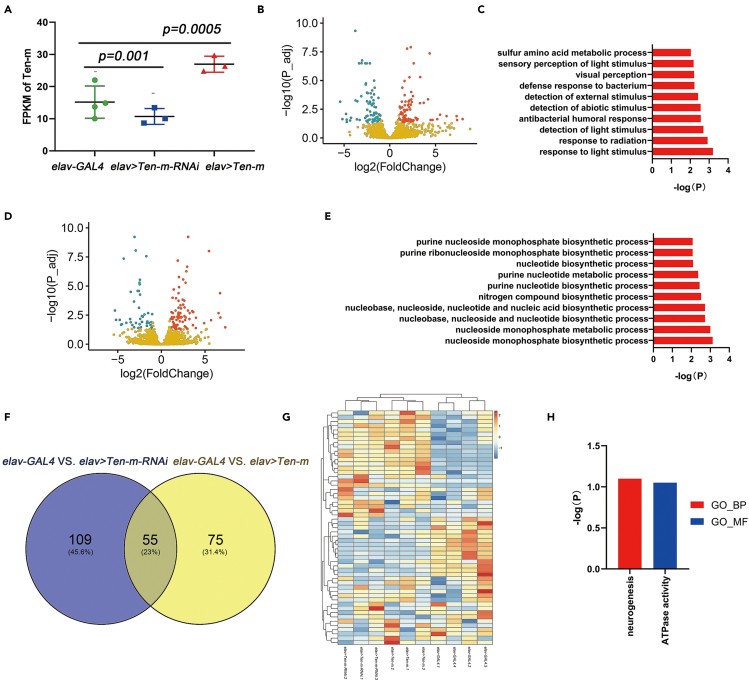
Through RNA sequencing of brain tissues in Drosophila, differentially expressed genes (DEGs) under the conditions of Ten-m overexpression or downexpression were compared (Figure 5A), and the functional classification of these DEGs was carried out. A total of 164 significant DEGs were identified between elav > Ten-m-RNAi and elav-GAL4 groups, including 83 upregulated genes and 81 downregulated genes (Figure 5B, Table S3). Among DEGs in the elav > Ten-m-RNAi vs elav-GAL4, Gene Ontology (GO) enrichment analysis was related to stimulation, such as biological process (BP) terms linked to response to light stimulus and detection of external stimulus (Figure 5C, Table S5). We also revealed that 130 genes were differentially expressed in elav > Ten-m vs elav-GAL4 groups, and 85 genes were upregulated, and 45 genes were downregulated (Figure 5D, Table S2). In the elav > Ten-m vs elav-GAL4, the significantly enriched GO terms were those involved in metabolic processes such as the nucleoside monophosphate metabolic process (Figure 5E, Table S4). As shown in the Venn diagram (Figure 5F), we found there were 55 overlapped DEGs with both overexpressed and decreased expression of Ten-m. The clustering heatmap analysis of 55 overlapped significant DEGs indicated a contrasting gene expression profile between experimental groups. elav > Ten-m-RNAi and elav > Ten-m had a very similar profile of gene expression, revealing the same high and low expression trends while showing a different profile against elav-GAL4 (Figure 5G). GO enrichment analysis of overlapped significant DEGs showed that the only BP term was neurogenesis, and the only molecular function (MF) term was ATPase activity (Figure 5H, Table S6).
Figure 5.
RNA sequencing of brain tissues with aberrant expression of Ten-m in Drosophila
(A) FPKM of controls (elav-GAL4) and experimental flies (elav > Ten-m and elav > Ten-m-RNAi). Data are represented as mean ± SD. The Kruskal-Wallis test was used for comparison. Compared to the control groups, the experimental groups had increased or decreased the expression of Ten-m.
(B) Volcano plots of DEGs from elav > Ten-m-RNAi vs elav-GAL4 groups. Red denotes high expression while blue denotes low expression. In total, 164 significant DEGs were identified, including 83 upregulated genes and 81 downregulated genes.
(C) GO enrichment analysis of DEGs in the elav > Ten-m-RNAi group. The DEGs were enriched in stimulation-associated pathways, including light stimulus and detection of external stimulus.
(D) Volcano plots of DEGs from elav > Ten-m vs elav-GAL4 groups. Red denotes high expression while blue denotes low expression. In total, 130 significant DEGs were identified, including 85 upregulated genes and 45 downregulated genes.
(E) GO enrichment analysis of DEGs in the elav > Ten-m group. The DEGs were enriched in metabolism-associated pathways, such as the nucleoside monophosphate metabolic process.
(F) Venn diagram showing the intersection of significant DEGs observed in elav > Ten-m and elav > Ten-m-RNAi groups. There were 55 overlapped DEGs for both decreased and overexpressed Ten-m.
(G) Heatmap of overlapped significant DEGs. Hierarchical clustering was performed to highlight the DEGs, indicating a contrasting gene expression profile between experimental groups. Red denotes high expression while blue denotes low expression.
(H) GO enrichment analysis of overlapped significant DEGs in elav > Ten-m and elav > Ten-m-RNAi groups. The only BP term was neurogenesis, and the only MF term was ATPase activity.
Discussion
To date, hundreds of genes related to SCZ genes have been identified from GWAS, exome sequencing studies, transcriptomic studies, and epigenetic studies according to the SCZ database (SZDB, http://www.szdb.org/SZDB/index.html) (Sanders and Gill, 2007; Khavari and Cairns, 2020; Xu et al., 2011; Chen et al., 2015; Farrell et al., 2015). Previous studies have also provided amount of evidence for the polygenic architecture of SCZ (Smeland et al., 2020). Many common genetic variants and rare functional mutations make up the polygenic architecture of SCZ (Henriksen et al., 2017). In SCZ, common and rare genetic variants are associated with disorders of biological processes related to neurodevelopment, neuroexcitability, synaptic function, and the immune system (Ukkola-Vuoti et al., 2019; Boyajyan et al., 2015; Jiang et al., 2020). In this study, we focused on rare functional mutations with minor allele frequency (MAF) < 0.01 in the candidate gene, TENM4. We identified five rare functional mutations of TENM4 gene in SCZ families, suggesting TENM4 contributes to SCZ. Interestingly, common SNPs in TENM4 only presented a nominally significant association with SCZ and did not reach the level of genome-wide significance, which might be because most mutations identified in TENM4 were missense mutations. Unlike the loss of function mutations, most missense mutations have a reduced effect on gene function, and some mutations might even not affect gene function. It is not easily to distinguish these mutations from the true functional missense mutations, but the GWAS results revealed that there might still be a genetic influence of TENM4 on SCZ. We have not performed the sequencing in patients with sporadic SCZ because the limited sample size would affect the results a lot, and the very big sample size may find the association between rare functional mutations.
TENM4 is known to be involved in mental illness and cognition, but there has been no behavioral analysis of TENM4 in animal models. Thus, we used the Drosophila model to demonstrate the potential mechanism of SCZ caused by TENM4 mutations. We observed reduced STM with abnormal neuronal expression of Ten-m. Intellectual disability and learning disabilities are prevalent in patients with SCZ. SCZ is also associated with a substantial cognitive impairment that affects working memory, processing speed, attention, and verbal memory (Dickinson et al., 2007). It is also difficult for patients with SCZ to learn from positive feedback (Dowd and Barch, 2012). Patients with SCZ often reveal social withdrawal, and the possibility of violence is increased (Lamberti, 2007). Sleep and circadian rhythm disorders occur in many psychiatric patients, including SCZ (Wulff et al., 2010). Furthermore, sleep disturbances occur frequently in patients with SCZ (Cohrs, 2008), including shorter duration, sleep fragmentation, insomnia, or other sleeping disorders (Monti and Monti, 2004). We found that the aberrant expression of Ten-m could cause sleep reduction and increased aggressiveness, which exhibit an SCZ-like phenotype. Thus, we inferred that TENM4 is a promising candidate gene, which might be related to some SCZ symptoms in SCZ, such as high aggression levels, sleep disorders, and learning ability related to cognitive deficits.
Overexpression and downexpression of Ten-m both caused night-sleep reduction in Drosophila, but the effect was not the same. Overexpression of Ten-m caused reduced sleep duration, and both overexpression and downexpression of Ten-m caused sleep fragmentation. We speculated that these effects were caused by different mechanisms, which need to be studied. The GWAS of actigraphic sleep phenotypes indicated that wake after sleep onset is associated with TENM4 (Spada et al., 2016).
SCZ is related to severe abnormal mutations in genes essential for synaptic plasticity (Gulsuner et al., 2020). Neural circuits of the human brain are composed of approximately 100 million neurons intercommunicated by synapses which are eventually responsible for behavior and brain function (Li et al., 2016). Ten-m affects the synapses morphological structure in Drosophila, suggesting that the aberrant expression of TENM4 may cause changes in synapse morphological structure which could lead to SCZ.
RNA sequencing showed that the effect of Ten-m on stimulation and the metabolic process might cause behavioral abnormalities. The sensitivity to cVA in males plays an important role in learning, memory, and aggressive behavior, while the sensory perception of light stimulation affects sleep in Drosophila. In addition, the imbalance of the nucleoside pathway and metabolism significantly affects the neuromodulatory function significantly (Ipata et al., 2011). The GO enrichment analysis of overlapped significant DEGs revealed neurogenesis and ATPase activity. Moreover, hippocampal neurogenesis and synaptic functions are crucial for learning and memory (Nabavi et al., 2014). Aberrant subcortical neurogenesis often occurs in SCZ (Hoffmann et al., 2019). The abnormal regulation of energy metabolism leads to changes in the synthesis and release of ATP. Studies have shown that ATP is the main stimulant of microglial and can be used as a signal to mediate the growth of microglial and the communication of nerve cells, which would affect the function of neurons and synapses (Davalos et al., 2005). Thus, the molecular mechanism of TENM4 on SCZ might be mainly through affecting neurogenesis and regulating ATPase activity.
Limitations of the study
There are still some limitations in our study. For example, we need a big sample size of sporadic SCZ patients to perform the target sequencing to find the association between rare functional mutations. More functional experiments should be set to find more direct pathological evidence in SCZ. The molecular mechanism of TENM4 on SCZ remains unclear.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human blood | SJTU BIO-X psychiatric sample bank | http://www.bio-x.cn/ |
| Deposited data | ||
| Raw data of Target sequencing | This paper | Mendeley Data, V2, https://doi.org/10.17632/6w5zmn6j8h.2. |
| mRNA sequencing analyzed data | This paper | Mendeley Data, V1, https://doi.org/10.17632/hy3bh2z8x8.1 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: elav-GAL4 | Bloomington Drosophila Stock Center | flybase_FBst0000458 |
| D. melanogaster: Mz19-GAL4 | Bloomington Drosophila Stock Center | flybase_FBal0155865 |
| D. melanogaster: Mz699-GAL4 | Bloomington Drosophila Stock Center | flybase_FBal0066073 |
| D. melanogaster: UAS-Ten-m | Bloomington Drosophila Stock Center | flybase_FBst0041569 |
| D. melanogaster: UAS-Ten-m-RNAi | Bloomington Drosophila Stock Center | flybase_FBtp0052660 |
| D. melanogaster: w;Cyo/sco;MKRS/Tb | Kyoto Stock Center | flybase_FBst0305835 |
| D. melanogaster: LN1-GAL4 | Kyoto Stock Center | flybase_FBst0302813 |
| Oligonucleotides | ||
| Primers for TENM4, see supplemental information Such as: GACCCTGTTTGT GGATGTGGA |
This paper | N/A |
| Software and algorithms | ||
| Burrows-Wheeler Aligner (BWA) | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| Samtools | Li et al. (2009) | http://samtools.sourceforge.net/ |
| Genome Analysis Toolkit (GATK) | McKenna et al. (2010) | http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit |
| ANNOVAR | Wang et al. (2010) | https://annovar.openbioinformatics.org/en/latest/ |
| IGV | Robinson et al. (2011) | http://www.igv.org/ |
| Trimmomatic | Bolger et al. (2014) | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| TopHat2 | Kim et al. (2013) | http://ccb.jhu.edu/software/tophat/manual.shtml |
| Cufflinks | Trapnell et al. (2012) | http://cole-trapnell-lab.github.io/cufflinks/ |
| Bowtie2 | Langmead and Salzberg (2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| HTSeq | Anders et al. (2015) | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al. (2014) | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daizhan Zhou (zhoudaizhan@sjtu.edu.cn).
Materials availability
Materials used or generated in this study will be available upon reasonable request, and a material transfer agreement may be required.
Experimental model and subject details
Patients and samples
In this study, all individuals with SCZ were interviewed by at least two independent psychiatrists. Diagnoses were made strictly according to the Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV criteria, and patients with other neurological, mental, or psychiatric disorders, or with history of drug use were excluded. All participants provided written informed consent. Approval for our study was received from the Ethics Committee of Human Genetic Resources of Shanghai Jiao Tong University and local hospitals.
For the family-based sequencing study, 68 SCZ families with no less than 3 patients each were selected, and 351 samples were collected for targeted sequencing. There are 183 males and 168 females, and the average age was 39. All blood samples were collected during 1998-2005 in Shanghai, Shandong, Hebei, Anhui, Gansu, Heilongjiang and Guangxi provinces in China.
Fly lines
The fly lines used can be found in Table S1. Other genotypes were obtained by fly crossing. All procedures were approved by the Ethics Committee of Animal Research of Shanghai Jiao Tong University.
The larvae and adults were kept on a standard sugar-yeast-cornmeal medium under a 12-hour light/dark (LD) cycle at 25°C with 60% humidity. A list of fly lines used can be found in Table S1. Two genotypes of fly stocks were obtained by crossing other genotypes, elav>Ten-m and elav>Ten-m-RNAi. We used balanced lethal genotype flies: w;Cyo/sco;MKRS/Tb. The genotype includes four types of lethal alleles, which cause the death of the organism that carries them. To obtain elav>Ten-m flies, firstly, UAS-Ten-m flies were crossed with w;Cyo/sco;MKRS/Tb flies. We selected presenting Cyo and MKRS phenotypes of male flies from the offspring to cross with female elav;+;+ flies, then selected elav;Cyo/+;UAS-Ten-m/+ males. Simultaneously, female elav;+;+ flies were crossed with w;Cyo/sco;MKRS/Tb flies, and we selected presenting Cyo and MKRS phenotypes of male flies from the offspring to cross with female elav;+;+ flies, then selected elav;Cyo/+;MKRS/+ females to cross with the elav;Cyo/+;UAS-Ten-m/+ males. Finally, we obtained elav>Ten-m flies. The same methods were used to obtain elav>Ten-m-RNAi flies. Other fly genotypes were obtained by crossing parental flies. More than five generations with the w1118 strain outcrossing were performed to standardize the background.
In crossing experiment, we used 20 mature females and 10 mature males. In courtship conditioning assay, we used 10 males and 10 females which were selected at the eclosion stage. In aggression assay, we used only males which were kept separately for 7 days after eclosion stage. In sleep and activity assay, we used only virgin females which were collected immediately after eclosion stage.
Method details
Targeted sequencing of TENM4
Primer3 was used to design 70 target-specific primer pairs for all exons and exon-intron boundaries of TENM4 (Untergasser et al., 2012). The primers were synthesized with common adapter sequences at their 5’ ends as previously described (Forshew et al., 2012). All 70 primer pairs were divided into 6 multiplex primer pools. The library construction was performed essentially as previously described, including pre-amplification of the tagged gene amplicons, generation of a barcoded DNA library, quantification and clean-up (Forshew et al., 2012). The final DNA libraries were determined by Qubit 2100 (Thermo Scientific, USA) and Bioanalyzer (Illumina, USA) and then sequenced on the Illumina HiSeq X-Ten System (Illumina, USA) (Wu et al., 2017).
The targeted next-generation sequencing data were filtered by quality analysis and PCR primers and splices, and the paired 150 bp-long reads were mapped to hg19 human reference genome using the Burrows-Wheeler Aligner (BWA) software (Li and Durbin, 2009). Samtools was used for sequence sorting, gene location labeling, and format transformation (Li et al., 2009). Genome Analysis Toolkit (GATK) was used to identify mutation, filter and select comment to generate a VCF file (McKenna et al., 2010) . All variants were annotated by ANNOVAR software (2019Oct24) (Wang et al., 2010), and the potential effects on the protein function were predicted using 9 software (SIFT, PolyPhen2, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, MetaSVM, and MetaLR). All functional mutations were visually confirmed one-by-one using IGV software (v2.8.6) (Robinson et al., 2011).
Courtship conditioning assay
A courtship conditioning assay was used to test courtship behavior and learning memory in Drosophila. Males and females were selected at the eclosion stage and raised in Drosophila standard culture medium. Male flies were kept separately and females were kept in groups of 10-15 flies. Before the experiments, selected females were pre-mated by housing with 15-20 w1118 males per group for 18 hours. Males were divided into naive group and trained group. Male flies in trained groups aged up to 4-7 days after eclosion were solely paired with two mated females 1h for training, then the mated females were removed for half an hour before the test. Individual males from the control or trained groups were placed into the test chamber, 15 mm in diameter by 7 mm deep, and paired with two mated females. The period of experiments lasted for 10 minutes and were recorded by a video camera. The courtship index (CI) was calculated as the percentage of male courtship duration within 10 minutes. The learning index (LI) was calculated using the mean CIs, LI = [CI naive – CI trained]/CI naive (Keleman et al., 2012). As the distributions of LIs are different between groups, LIs were used for the permutation test with 100,000 random permutations to compare between groups, and the data were processed with R software. One-tail testing against the null hypothesis was used in the comparison. The Mann–Whitney U test was used for CIs.
Aggression assay
Pairs of male flies were introduced into a fighting chamber, which was 13 mm in diameter by 7 mm in height. We used 20 mm × 20 mm glass slides to cover the chamber. Fly behaviors were observed and recorded for 15 minutes (Zhou et al., 2008).The aggressive behaviors of male flies were evaluated in two aspects: latency and frequency. Latency is the duration between putting experimental flies into the chambers and the start of the first agonistic encounter while frequency is the number of aggressive behaviors of male flies, such as lunges, within 15 minutes. (Chen et al., 2002).
Sleep and activity assay
All experimental flies were virgin females collected immediately after eclosion stage; 15 flies of each genotype were required for parallel experiments. The sleep and activity of the experimental flies were monitored by the Drosophila Activity Monitor (DAM) system (TriKinetics, USA). The selected flies were raised in standard food vials until they were monitored, then they were moved into 65 mm × 5 mm glass tubes (Wuhan Yihong, China) containing solely 5% sucrose (Sangon Biotech, China) and 2% agar (Sangon Biotech, China). Subsequently, glass tubes with experimental flies were placed in the DAM system which was set in 12-hour LD conditions for the test. LD conditions were light-on at 9 am and light-off at 9 pm. Data collection started after the flies were acclimated to the environment for 3 to 5 days(Song et al., 2017). Data records of experimental activities were binned in one-minute periods in LD for three days. Flies were regarded to be sleeping when they were inactive for at least 5 minutes. Flies were implied to have died when their whole period of activity was less than 10 minutes, and the data were excluded. Flies’ sleep duration and parameters related to sleep behavior were analyzed by R software. Conventional sleep plots were portrayed by calculating the average amount of sleep duration within 30 minutes of experimental flies.
RNA extraction and sequencing
The flies were placed in 1 mL TRIzol (Invitrogen, USA), and we used forceps to rapidly break down the brains of the adult flies rapidly. Almost 20 heads of each genotype were saved in a 1.5 mL microcentrifuge tube with 200 μL of TRIzol. There were 3-4 replicates for each genotype. RNA extraction and purification were performed using TRIzol. The RNA was eluted with nuclease-free water, then a NanoDrop 2000 (Thermo Scientific, USA) was used to evaluate of RNA purity and concentration.
RNA-sequencing was performed on 3-4 replicates of RNA samples from each of elav-GAL4, elav>Ten-m-RNAi, and elav>Ten-m groups. The sample libraries were prepared according to the standard process of the TruSeq RNA Sample Prep Kit v2 (Illumina, USA). The final libraries were determined by Qubit 2100 (Thermo Scientific, USA) and Bioanalyzer (Illumina, USA) and paired-end sequencing was performed on an Illumina HiSeq X-Ten System (Illumina, USA), with a read length of 2 × 150 bp.
Adapters and low-quality reads were removed from the raw data using Trimmomatic (v0.36) (Bolger et al., 2014). TopHat (v2.1.1) was used to align the RNA-seq reads to a reference genome (BDGP5.25) with the short-read aligner Bowtie2 (v2.3.4) (Kim et al., 2013) (Langmead and Salzberg, 2012). Cufflinks (v2.2.1) was used to analyze the BAM files, including assembling the transcripts from the aligned reads and estimating their abundances using a Fragments Per Kilobase of transcript per Million mapped reads (FPKM)-based algorithm (Trapnell et al., 2012). HTSeq (v0.10) was used to generate the read count files for each gene (Anders et al., 2015). Then, DESeq2 (v1.36.0) was used to identify DEGs, which were selected with log2 (fold change) >1 or log2 (fold change) <-1 and with statistical significance (adjusted p-value <0.05) (Love et al., 2014). GO and KEGG pathway enrichment analysis of DEGs was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation tool (v6.8).
Quantification and statistical analysis
In courtship conditioning assay, the courtship index (CI) was calculated as the percentage of male courtship duration within 10 minutes. The learning index (LI) was calculated using the mean CIs, LI = [CI naive – CI trained]/CI naive (Keleman et al., 2012). As the distributions of LIs are different between groups, LIs were used for the permutation test with 100,000 random permutations to compare between groups, and the data were processed with R software. One-tail testing against the null hypothesis was used in the comparison. The Mann–Whitney U test was used for CIs.
In aggression assay and sleep assay, all behavior data analyses were performed using Sigma Plot (v14.0) and SPSS (v22.0). For multiple comparisons, one-way ANOVA followed by post hoc Tukey were used, except for the comparison of nonparametrically distributed data where the Kruskal–Wallis test was used.
Acknowledgments
We acknowledge the altruism of the participants and their families and support staff at each of the participating sites for their contributions to this study. This work was supported by grants from the National Key Research and Development Program (2016YFC0906400 and 2016YFC1201700), the National Natural Science Foundation of China (grant 8142106, 81970999, 82071262 and 81872297), Innovation Funding in Shanghai (20JC1418600), the Shanghai Leading Academic Discipline Project (B205), and Shanghai Rising-Star Program (19QA1404900).
Author contributions
D.Z., Y.P., G.H., and L.H. designed the study and interpreted the results. X.L., D.C., L.W., X.W., F.Y., and X.C. collected the samples and performed the primary experiments. X.Y. and H.D. conducted targeted sequencing, and D.Z. performed statistical analysis. X.Y. and M.L. performed the animal experiments. X.Y. and D.Z. performed the RNA-seq and GO analysis. D.Z., Y.P., X.Y., and M.L. verified the underlying data. X.Y. and M.L. drafted the manuscript, and all authors contributed to the final version of the paper.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103063.
Contributor Information
Lin He, Email: helin@bio-x.cn.
Yong Ping, Email: yoping@sjtu.edu.cn.
Daizhan Zhou, Email: zhoudaizhan@sjtu.edu.cn.
Supplemental information
Data and code availability
The target sequencing data of TENM4 and the RNA sequencing data have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alekseyenko O.V., Chan Y.B., Okaty B.W., Chang Y., Dymecki S.M., Kravitz E.A. Serotonergic modulation of aggression in Drosophila involves GABAergic and cholinergic opposing pathways. Curr. Biol. 2019;29:2145–2156 e5. doi: 10.1016/j.cub.2019.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S., Martin D., Hagios C., Chiquet-Ehrismann R. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 1994;13:3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyajyan A., Zakharyan R., Atshemyan S., Chavushyan A., Mkrtchyan G. Schizophrenia-associated risk and protective variants of c-Fos encoding gene. Recent Adv. DNA Gene Seq. 2015;9:51–57. doi: 10.2174/2352092209666150223113334. [DOI] [PubMed] [Google Scholar]
- Chao Y.X., Lin Ng E.Y., Tio M., Kumar P., Tan L., Au W.L., Yih Y., Tan E.K. Essential tremor linked TENM4 mutation found in healthy Chinese individuals. Parkinsonism Relat. Disord. 2016;31:139–140. doi: 10.1016/j.parkreldis.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Chen J., Cao F., Liu L., Wang L., Chen X. Genetic studies of schizophrenia: an update. Neurosci. Bull. 2015;31:87–98. doi: 10.1007/s12264-014-1494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Lee A.Y., Bowens N.M., Huber R., Kravitz E.A. Fighting fruit flies: a model system for the study of aggression. Proc. Natl. Acad. Sci. U S A. 2002;99:5664–5668. doi: 10.1073/pnas.082102599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohrs S. Sleep disturbances in patients with schizophrenia : impact and effect of antipsychotics. CNS Drugs. 2008;22:939–962. doi: 10.2165/00023210-200822110-00004. [DOI] [PubMed] [Google Scholar]
- Das A., Sen S., Lichtneckert R., Okada R., Ito K., Rodrigues V., Reichert H. Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene. Neural Dev. 2008;3:33. doi: 10.1186/1749-8104-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., Gan W.B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Dickinson D., Ramsey M.E., Gold J.M. Overlooking the obvious: a meta-analytic comparison of digit symbol coding tasks and other cognitive measures in schizophrenia. Arch. Gen. Psychiatry. 2007;64:532–542. doi: 10.1001/archpsyc.64.5.532. [DOI] [PubMed] [Google Scholar]
- Dowd E.C., Barch D.M. Pavlovian reward prediction and receipt in schizophrenia: relationship to anhedonia. PLoS One. 2012;7:e35622. doi: 10.1371/journal.pone.0035622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaerts C., Farine J.P., Cobb M., Ferveur J.F. Drosophila cuticular hydrocarbons revisited: mating status alters cuticular profiles. PLoS One. 2010;5:e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M.S., Werge T., Sklar P., Owen M.J., Ophoff R.A., O'donovan M.C., Corvin A., Cichon S., Sullivan P.F. Evaluating historical candidate genes for schizophrenia. Mol. Psychiatry. 2015;20:555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K., Zhou X.H., Oohashi T., Morgelin M., Lustig A., Hirakawa S., Ninomiya Y., Engel J., Rauch U., Fassler R. All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J. Biol. Chem. 2002;277:26128–26135. doi: 10.1074/jbc.M203722200. [DOI] [PubMed] [Google Scholar]
- Forshew T., Murtaza M., Parkinson C., Gale D., Tsui D.W., Kaper F., Dawson S.J., Piskorz A.M., Jimenez-Linan M., Bentley D. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- Gulsuner S., Stein D.J., Susser E.S., Sibeko G., Pretorius A., Walsh T., Majara L., Mndini M.M., Mqulwana S.G., Ntola O.A. Genetics of schizophrenia in the South African Xhosa. Science. 2020;367:569–573. doi: 10.1126/science.aay8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich A., Lourdusamy A., Tzschoppe J., Vollstadt-Klein S., Buhler M., Steiner S., Bach C., Poustka L., Banaschewski T., Barker G. The risk variant in ODZ4 for bipolar disorder impacts on amygdala activation during reward processing. Bipolar Disord. 2013;15:440–445. doi: 10.1111/bdi.12068. [DOI] [PubMed] [Google Scholar]
- Henriksen M.G., Nordgaard J., Jansson L.B. Genetics of schizophrenia: overview of methods, findings and limitations. Front. Hum. Neurosci. 2017;11:322. doi: 10.3389/fnhum.2017.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Ziller M., Spengler D. Progress in iPSC-based modeling of psychiatric disorders. Int. J. Mol. Sci. 2019;20:4896. doi: 10.3390/ijms20194896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., Mosca T.J., Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor H., Francescatto L., Bartesaghi L., Ortega-Cubero S., Kousi M., Lorenzo-Betancor O., Jimenez-Jimenez F.J., Gironell A., Clarimon J., Drechsel O. Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum. Mol. Genet. 2015;24:5677–5686. doi: 10.1093/hmg/ddv281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Takahashi A., Kamatani Y., Okahisa Y., Kunugi H., Mori N., Sasaki T., Ohmori T., Okamoto Y., Kawasaki H. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry. 2018;23:639–647. doi: 10.1038/mp.2016.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I., O'roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipata P.L., Camici M., Micheli V., Tozz M.G. Metabolic network of nucleosides in the brain. Curr. Top Med. Chem. 2011;11:909–922. doi: 10.2174/156802611795347555. [DOI] [PubMed] [Google Scholar]
- Ivorra J.L., Rivero O., Costas J., Iniesta R., Arrojo M., Ramos-Rios R., Carracedo A., Palomo T., Rodriguez-Jimenez R., Cervilla J. Replication of previous genome-wide association studies of psychiatric diseases in a large schizophrenia case-control sample from Spain. Schizophr. Res. 2014;159:107–113. doi: 10.1016/j.schres.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Jiang S., Zhou D., Wang Y.Y., Jia P., Wan C., Li X., He G., Cao D., Jiang X., Kendler K.S. Identification of de novo mutations in prenatal neurodevelopment-associated genes in schizophrenia in two Han Chinese patient-sibling family-based cohorts. Transl. Psychiatry. 2020;10:307. doi: 10.1038/s41398-020-00987-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kavanagh D.H., Tansey K.E., O'donovan M.C., Owen M.J. Schizophrenia genetics: emerging themes for a complex disorder. Mol. Psychiatry. 2015;20:72–76. doi: 10.1038/mp.2014.148. [DOI] [PubMed] [Google Scholar]
- Keleman K., Vrontou E., Kruttner S., Yu J.Y., Kurtovic-Kozaric A., Dickson B.J. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489:145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- Khavari B., Cairns M.J. Epigenomic dysregulation in schizophrenia: in search of disease etiology and biomarkers. Cells. 2020;9:1837. doi: 10.3390/cells9081837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.L., Awasaki T., Ito K., Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Lam M., Chen C.Y., Li Z., Martin A.R., Bryois J., Ma X., Gaspar H., Ikeda M., Benyamin B., Brown B.C. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti J.S. Understanding and preventing criminal recidivism among adults with psychotic disorders. Psychiatr. Serv. 2007;58:773–781. doi: 10.1176/ps.2007.58.6.773. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wilkinson B., Clementel V.A., Hou J., O'dell T.J., Coba M.P. Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Sci. Signal. 2016;9:rs8. doi: 10.1126/scisignal.aaf6716. [DOI] [PubMed] [Google Scholar]
- Li Z., Chen J., Yu H., He L., Xu Y., Zhang D., Yi Q., Li C., Li X., Shen J. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 2017;49:1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., Depristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti J.M., Monti D. Sleep in schizophrenia patients and the effects of antipsychotic drugs. Sleep Med. Rev. 2004;8:133–148. doi: 10.1016/S1087-0792(02)00158-2. [DOI] [PubMed] [Google Scholar]
- Mosca T.J., Hong W., Dani V.S., Favaloro V., Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S., Fox R., Proulx C.D., Lin J.Y., Tsien R.Y., Malinow R. Engineering a memory with LTD and LTP. Nature. 2014;511:348–352. doi: 10.1038/nature13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.R., Cherian J., Gohil K., Atkinson D. Schizophrenia: overview and treatment options. P T. 2014;39:638–645. [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J., Gill M. Unravelling the genome: a review of molecular genetic research in schizophrenia. Ir. J. Med. Sci. 2007;176:5–9. doi: 10.1007/s11845-007-0004-3. [DOI] [PubMed] [Google Scholar]
- Siegel R.W., Hall J.C. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. U S A. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki K.K., Riccio P., Ladewski L., Marcillac F., Dartevelle L., Cross S.A., Ferveur J.F. The role of cuticular pheromones in courtship conditioning of Drosophila males. Learn Mem. 2005;12:636–645. doi: 10.1101/lm.85605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeland O.B., Frei O., Dale A.M., Andreassen O.A. The polygenic architecture of schizophrenia - rethinking pathogenesis and nosology. Nat. Rev. Neurol. 2020;16:366–379. doi: 10.1038/s41582-020-0364-0. [DOI] [PubMed] [Google Scholar]
- Song Q., Feng G., Huang Z., Chen X., Chen Z., Ping Y. Aberrant axonal arborization of PDF neurons induced by abeta42-mediated JNK activation underlies sleep disturbance in an Alzheimer's model. Mol. Neurobiol. 2017;54:6317–6328. doi: 10.1007/s12035-016-0165-z. [DOI] [PubMed] [Google Scholar]
- Spada J., Scholz M., Kirsten H., Hensch T., Horn K., Jawinski P., Ulke C., Burkhardt R., Wirkner K., Loeffler M. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J. Sleep Res. 2016;25:690–701. doi: 10.1111/jsr.12421. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Daly M.J., O'donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Fukushi M., Kosaki K., Doyle A.D., De Vega S., Yoshizaki K., Akazawa C., Arikawa-Hirasawa E., Yamada Y. Teneurin-4 is a novel regulator of oligodendrocyte differentiation and myelination of small-diameter axons in the CNS. J. Neurosci. 2012;32:11586–11599. doi: 10.1523/JNEUROSCI.2045-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Numakawa T., Chou J., De Vega S., Mizuniwa C., Sekimoto K., Adachi N., Kunugi H., Arikawa-Hirasawa E., Yamada Y., Akazawa C. Teneurin-4 promotes cellular protrusion formation and neurite outgrowth through focal adhesion kinase signaling. FASEB J. 2014;28:1386–1397. doi: 10.1096/fj.13-241034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R.P., Chiquet-Ehrismann R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev. Biol. 2006;290:237–245. doi: 10.1016/j.ydbio.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Tucker R.P., Martin D., Kos R., Chiquet-Ehrismann R. The expression of teneurin-4 in the avian embryo. Mech. Dev. 2000;98:187–191. doi: 10.1016/s0925-4773(00)00444-5. [DOI] [PubMed] [Google Scholar]
- Ukkola-Vuoti L., Torniainen-Holm M., Ortega-Alonso A., Sinha V., Tuulio-Henriksson A., Paunio T., Lonnqvist J., Suvisaari J., Hennah W. Gene expression changes related to immune processes associate with cognitive endophenotypes of schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;88:159–167. doi: 10.1016/j.pnpbp.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Anderson D.J. Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature. 2010;463:227–231. doi: 10.1038/nature08678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhou D.Z., Berki D., Geisz A., Zou W.B., Sun X.T., Hu L.H., Zhao Z.H., Zhao A.J., He L. No significant enrichment of rare functionally defective CPA1 variants in a large Chinese idiopathic chronic pancreatitis cohort. Hum. Mutat. 2017;38:959–963. doi: 10.1002/humu.23254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K., Gatti S., Wettstein J.G., Foster R.G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010;11:589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- Xu B., Roos J.L., Dexheimer P., Boone B., Plummer B., Levy S., Gogos J.A., Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C.B., Xu Z.H., Zhu J., Wu Y., Zhuang X.H., Chen Q.L., Wu C.R., Hu J.T., Zhou H.S., Xie W.H. Exome sequencing identifies TENM4 as a novel candidate gene for schizophrenia in the SCZD2 locus at 11q14-21. Front. Genet. 2018;9:725. doi: 10.3389/fgene.2018.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.P., Xu C.Y., Gu L.Y., Zhang B., Shen T., Gao T., Tian J., Pu J.L., Yin X.Z., Zhang B.R., Zhao G.H. Genetic testing of FUS, HTRA2, and TENM4 genes in Chinese patients with essential tremor. CNS Neurosci. Ther. 2020;26:837–841. doi: 10.1111/cns.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Rao Y., Rao Y. A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 2008;11:1059–1067. doi: 10.1038/nn.2164. [DOI] [PubMed] [Google Scholar]
- Zhou X.H., Brandau O., Feng K., Oohashi T., Ninomiya Y., Rauch U., Fassler R. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr. Patterns. 2003;3:397–405. doi: 10.1016/s1567-133x(03)00087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The target sequencing data of TENM4 and the RNA sequencing data have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.