Summary
ERK1/2 involvement in cell death remains unclear, although many studies have demonstrated the importance of ERK1/2 dynamics in determining cellular responses. To untangle how ERK1/2 contributes to two cell death programs, we investigated ERK1/2 signaling dynamics during hFasL-induced apoptosis and TNF-induced necroptosis in L929 cells. We observed that ERK1/2 inhibition sensitizes cells to apoptosis while delaying necroptosis. By monitoring ERK1/2 activity by live-cell imaging using an improved ERK1/2 biosensor (EKAR4.0), we reported differential ERK1/2 signaling dynamics between cell survival, apoptosis, and necroptosis. We also decrypted a temporally shifted amplitude- and frequency-modulated (AM/FM) ERK1/2 activity profile in necroptosis versus apoptosis. ERK1/2 inhibition, which disrupted ERK1/2 signaling dynamics, prevented TNF and IL-6 gene expression increase during TNF-induced necroptosis. Using an inducible cell line for activated MLKL, the final executioner of necroptosis, we showed ERK1/2 and its distinctive necroptotic ERK1/2 activity dynamics to be positioned downstream of MLKL.
Subject areas: Biological sciences, Biomolecular engineering, Cell biology
Graphical abstract
Highlights
-
•
ERK inhibition sensitizes apoptosis but delays necroptosis in L929 cells
-
•
Temporal dynamics of ERK activity distinguishes survival, apoptosis, and necroptosis
-
•
Disrupting ERK activity prevents TNF and IL6 gene expression in necroptotic L929
-
•
Expression of pro-necroptotic MLKL induces ERK activity dynamics in L929
Biological sciences; Biomolecular engineering; Cell biology
Introduction
Biological cues are processed by the cell through spatial and temporal biochemical signaling dynamics of signaling networks. The encrypted information is then decoded to elicit cellular processes. An increasing trend in cell biology reveals an additional layer of complexity in cell signaling where the cell encrypts information based on the temporal modulation of signaling activities (Levine et al., 2013; Purvis and Lahav, 2013). This signal transmission modus resides in several features such as pulse amplitude, frequency, duration, and onset. The extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) have been early examples of how signaling dynamics features drive cellular responses, such as proliferation and differentiation (Albeck et al., 2013; Aoki et al., 2013; Johnson and Toettcher, 2019; Ryu et al., 2016). While there is compelling evidence of ERK1/2 involvement in different cell death modalities, ERK1/2 controversial effects made its understanding in cell death daunting. Thus, our study seeks to clarify the complex state of the art around ERK1/2 in cell death by investigating its temporal modulation of activity in two different cell death processes. Using an improved version of the ERK1/2 Kinase Activity Reporter (EKAR4.0), we investigated at the single-cell level how ERK1/2 signaling dynamics are encoded during apoptosis and necroptosis.
Besides ERK1/2 roles in various cell survival processes (e.g., cell cycle progression, proliferation, differentiation) (Kohno and Pouyssegur, 2006; Wortzel and Seger, 2011), its involvement in different cell death modalities remains obscure. ERK1/2 activation was reported to be involved in different cell death programs, such as apoptosis, necroptosis, autophagy, and ferroptosis (Cagnol and Chambard, 2010; Yagoda et al., 2007; Zhuang and Schnellmann, 2006), either as a pro-death or a pro-survival signaling effector. In tumors, ERK1/2 was shown to promote the expression of anti-apoptotic proteins while repressing pro-apoptotic proteins, contributing to cell survival and tumorigenesis (Balmanno and Cook, 2009). In addition, Degterev et al. demonstrated that effectors of necroptosis, in this instance Receptor Interacting Protein Kinase 1 and -3 (RIPK1 and RIPK3), could directly promote inflammatory gene expression independently of lipopolysaccharides (LPS)-induced necroptosis in bone marrow-derived macrophage (BMDM) cells, and this process is mediated in part by ERK1/2 (Najjar et al., 2016). Besides, an increasing number of studies have reported pro-death functions of ERK1/2 activity in apoptosis, autophagy, and senescence depending on cell type and cell stimulation intensity [for review (Cagnol and Chambard, 2010)].
Tumor Necrosis Factor (TNF)-induced necroptosis relies on the serine-threonine kinases RIPK1 and RIPK3 (Vandenabeele et al., 2010) and the pseudo kinase Mixed-Lineage Kinase Like (MLKL) (Sun et al., 2012). Nowadays, necroptosis is defined as a RIPK1/RIPK3/MLKL-mediated cell death program (Pasparakis and Vandenabeele, 2015). Inappropriate necroptosis is linked to ischemia–reperfusion-induced injury in the brain, kidney, and heart (Oerlemans et al., 2012), inflammatory (Duprez et al., 2011; Takahashi et al., 2012, 2014), or degenerative disorders (Ofengeim et al., 2015), and tumor metastasis (Chen and Mellman, 2017). Necroptosis promotes inflammation by activating proinflammatory cytokine gene expression and releasing immunogenic molecules (Murai et al., 2018; Wallach et al., 2011; Zhu et al., 2018). Building up such a cellular stress response profoundly impacts the outcome of a pathological situation (Fulda et al., 2010). Therefore, it is crucial to identify molecular effectors that specifically modulate the signaling towards necroptosis (Vanlangenakker et al., 2010; Wallach et al., 2016).
While conflicting findings on ERK1/2 activity during necroptosis in L929 cells were reported (Cho et al., 2011; Vanlangenakker et al., 2010), we decided to mainly use the murine fibrosarcoma L929 cellular subline (L929sAhFas) since it is TNF-sensitive and stably expresses the human pro-apoptotic receptor Fas. Treatment of L929sAhFas (hereafter referred to as L929) with hFas ligand (hFasL) or TNF will specifically induce apoptosis via the caspase-8-dependent proteolytic pathway or RIPK1-dependent necroptosis, respectively (Vercammen et al., 1998). Here, we report that direct ERK1/2 inhibition while sensitizing hFasL-induced apoptosis delays TNF-induced necroptosis in L929 cells. Our results suggest that ERK1/2 is involved, in a RIPK1-kinase activity-independent manner, in necroptosis-activated cell-autonomous functions via the increase of pro-inflammatory cytokines gene expression. By fluorescent imaging using our optimized ERK1/2 Kinase Activity Reporter 4.0 (EKAR4.0), we revealed distinct amplitude- and frequency-modulated (AM/FM) ERK1/2 activity signaling dynamics in L929 depending on the triggered cellular process: survival, hFasL-induced apoptosis, or TNF-induced necroptosis. We propose a model in which the early onset of AM/FM ERK1/2 activity dynamics mediates the proinflammatory cytokine gene expression increase in TNF-induced necroptosis in L929 cells. Finally, using genetic models of RIPK1- and RIPK1/3-independent necroptosis, the results of this study propose ERK1/2 and its distinctive necroptotic ERK1/2 activity signaling dynamics to be positioned downstream of MLKL.
Results
ERK1/2 inhibition delays TNF-induced necroptosis but sensitizes hFasL-induced apoptosis
First, we investigated the impact of MAPK/ERK1/2 signaling pathway inhibition in apoptosis and necroptosis and opted for a systematic inhibition of different MAPK/ERK1/2 cascade layers using different chemical inhibitors in L929 cells (Figure S1A).
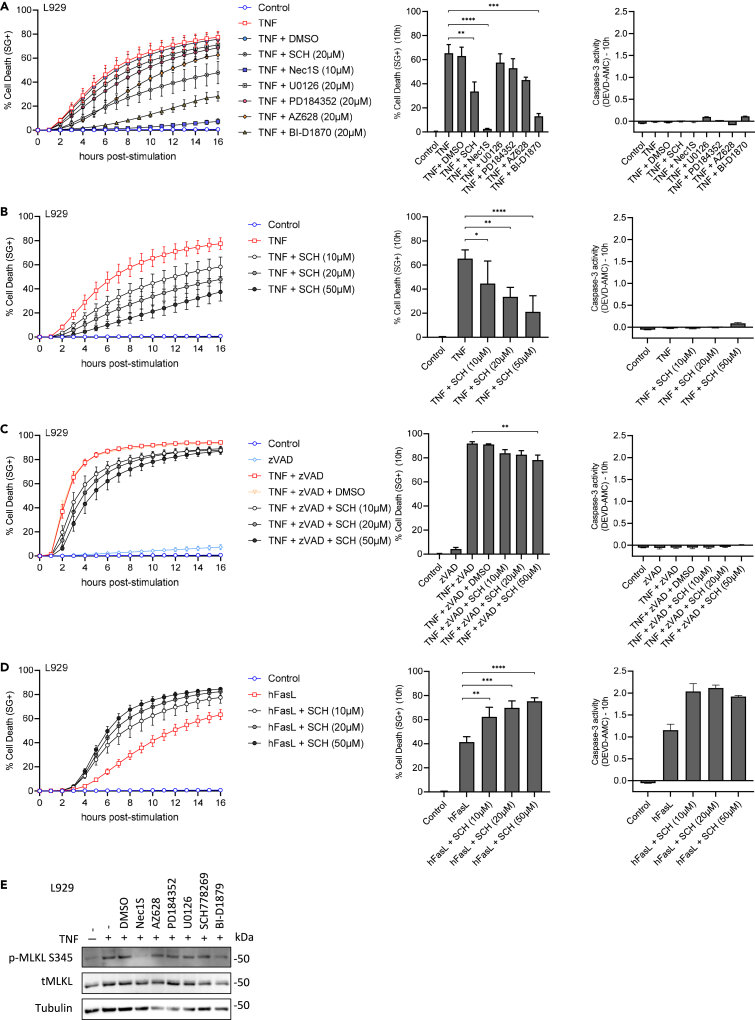
In TNF-induced necroptosis, direct inhibition of ERK1/2 (SCH772984) (Morris et al., 2013) and 90 kDa ribosomal S6 kinase (RSK) (BI-D1870) significantly delayed necroptosis (Figure 1A) in a dose-dependent manner for ERK1/2 inhibition (Figure 1B). Similar trends were observed in the presence of a pan-caspase inhibitor (TNF + zVAD) (Figure 1C) but not as pronounced when compared to TNF alone (Figure S1E vs. Figure 1A), suggesting that in necroptosis sensitizing conditions due to caspase-8 inhibition, the contribution of ERK1/2 is less outspoken. There was still a dose-dependent effect of ERK1/2 inhibition in the presence of zVAD, albeit not as big as in Figure 1B. Conversely, mitogen-activated protein kinase kinase (MEK1/2), ERK1/2, and RSK inhibition potently sensitized hFasL-induced apoptosis (Figure S1F), increasing cell death onset. In hFasL-induced apoptosis, direct ERK1/2 inhibition also sped up the process in a dose-dependent manner (Figure 1D).
Figure 1.
Effect of direct ERK1/2 inhibition in TNF-induced necroptosis and hFasL-induced apoptosis
(A) L929 wildtype cells were pretreated for 1 h with the indicated compounds, all targeting the MAPK/ERK signaling pathway (Figure S1A) or RIPK1 activity (grey curves) before TNF (red) or control (blue) treatment.
(B and C) SCH772984 dose-response in TNF-induced necroptosis, in the presence (C) or the absence (B) of the pan-caspase inhibitor zVAD-fmk (zVAD).
(D) Sensitizing effect of SCH772984 in hFas induced apoptosis. For each panel, cell death was measured as (i) function of time by SytoxGreen (SG+) positivity (A–D, left graph), and (ii) plotted at 10 h after treatment (A–D, middle graph). Caspase-3 activity was assessed by DEVD-AMC cleavage efficiency (A–D, right graph). Data are presented as mean ± SEM of at least two independent experiments. Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p < 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001.
(E) L929 cells were pre-treated or not with the indicated compounds or DMSO for 30 min before TNF treatment. After 6 h, cells were then lysed and immunoblotted as indicated on the left of each blot.
In TNF-induced necroptosis, pretreatment with a Raf inhibitor (AZ628) or two different MEK1/2 inhibitors (PD184352 and U0126) had no significant effect (Figure 1A). Of note, no cytotoxic effect was seen during ERK1/2 and RSK inhibition in the absence or presence of the pan-caspase inhibitor zVAD (Figures S1C and S1D). As a control, pretreatment of L929 with Nec1S, a specific inhibitor of RIPK1 kinase activity (Degterev et al., 2008), completely blocked RIPK1 kinase-dependent TNF-induced necroptosis (Figures 1A and 1E). Also, in all inhibitory conditions, no switch to apoptosis was observed as determined by the absence of caspase enzymatic activity (Figures 1A–1C, right panels) while the phosphorylation of MLKL was still detectable, confirming the occurrence of necroptosis (Figure 1E). Finally, we confirmed by western blot in L929 the level-by-level inhibition of the MAPK/ERK1&2 pathway using the different chemical inhibitors used in this study (Figure S1G).
While our findings concerning U0126 were consistent with an earlier study from our laboratory (Vanlangenakker et al., 2010), they were discrepant with another report showing marked protection of TNF-induced necroptosis in both L929 and Jurkat cells in the presence of 50 μM U0126 (Cho et al., 2011). To clarify this situation, we stimulated L929 cells with TNF in the presence of increasing concentrations of U0126 (10, 20, and 50 μM). Our data indicated that MEK1/2 inhibitor U0126 protects against TNF-induced necroptosis in a concentration-dependent manner and with a potent and significant inhibition at high concentration (50 μM, ∼2-fold reduction), as previously shown (Cho et al., 2011) (Figure S1B). However, such a high dose of U0126 was reported in vitro to inhibit ERK1/2 (Favata et al., 1998), which led us to use advanced and FDA-approved MEK1/2 specific inhibitors Selumetinib and Trametinib. No significant effect to either TNF-induced necroptosis or hFasL-induced apoptosis in L929 cells was observed, except for Selumetinib at 1 μM, which surprisingly lightly sensitized TNF-induced necroptosis (Figure S2A). No cytotoxic effect of Selumetinib and Trametinib was seen.
In the context of necroptosis, direct ERK1/2 inhibition in Jurkat FADD−/− cells recapitulated the same dose-dependent effect on TNF-induced necroptosis seen in L929 (Figure S2B). The same effect was observed using RSK and advanced MEK1/2 inhibitors in these experimental conditions (Figures S2B and S2C). In contrast, direct ERK1/2 inhibition (20 μM) had no significant impact on triggered necroptosis in HT29 cells (stimulated by TNF + BV6, an inhibitor of apoptosis proteins (IAP), and zVAD), while protection could be seen using Selumetinib (Figures S2D and S2E). Moreover, necroptosis in HT29 cells was still drastically reduced upon RSK inhibition (Figure S2D). Note that to induce necroptosis in HT29 we used zVAD, a known sensitizer of necroptosis, which explains why the inhibitory effect of ERK1/2 inhibition was not as marked similarly to what we showed in L929 in the presence of zVAD (Figure 1C). Again, this suggests that in necroptosis sensitizing conditions, the contribution of ERK1/2 is less outspoken. In the context of apoptosis, SCH772984 protected Jurkat FADD−/− against UV-irradiation-induced apoptosis, which engages many different apoptotic pathways besides death receptor-induced apoptosis. Finally, in TNF + BV6-induced apoptosis in HT29 while MEK1/2 inhibition had no impact, a sensitizing effect was observed in the presence of ERK1/2 inhibitor (Figure S2D) and to a lesser extent upon RSK inhibition, but overall in line with our findings in L929 cells. Altogether, our findings show a dual role for ERK1/2 in regulated cell death modalities. While ERK1/2 functions as a pro-death modulator in TNF-induced necroptosis, it mainly displays a pro-survival role in hFasL-induced apoptosis.
Specific spatio-temporal ERK1/2 activation profiles in TNF-induced necroptosis
Early transient and late sustained ERK1/2 phosphorylation profiles during necroptosis
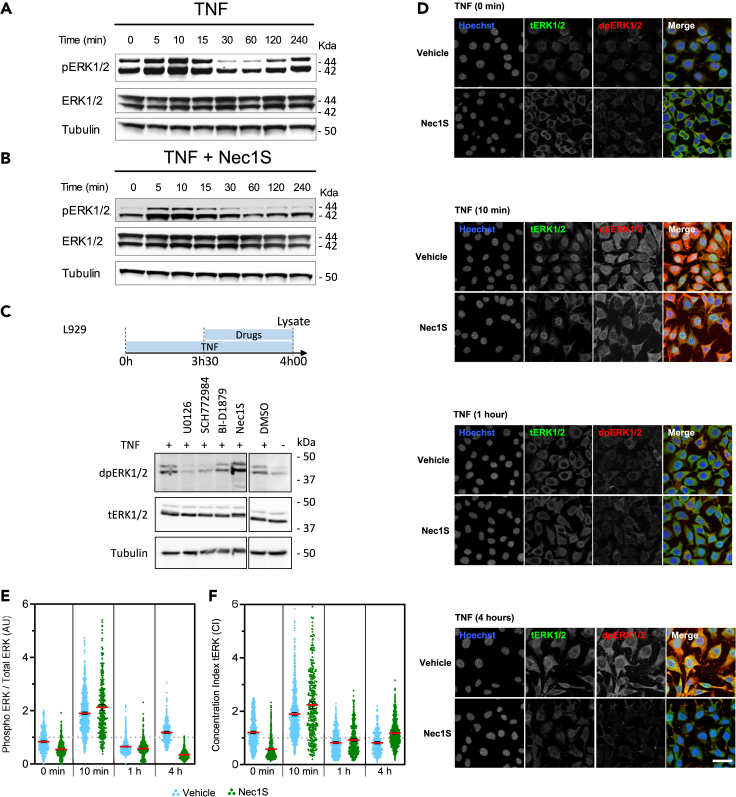
Evaluation of ERK1/2 phosphorylation patterns during TNF-induced necroptosis in L929 cells by western blot (WB) showed a biphasic phosphorylation profile of ERK1/2 (Figure 2A). First, TNF provoked a rapid and transient ERK1/2 phosphorylation between 5 and 15 min, followed by sustained ERK1/2 phosphorylation starting 2 h after TNF stimulation (Figures 2A and S3A) with no ERK1/2 isoform-specific contribution (Figure S3A). Pretreatment with Nec1S did not affect the duration of the early transient phosphorylation pattern while abolishing the late sustained ERK1/2 phosphorylation (Figure 2B). These results demonstrate that the former is RIPK1 kinase activity-independent, while the latter may directly or indirectly be associated with it. We did not observe any modification of ERK1/2 or MEK1/2 phosphorylation profiles in serum-induced ERK1/2 phosphorylation upon RIPK1 inhibition (Nec1S) (Figure S3B), suggesting that Nec1S is not interfering with the MAPK/ERK1/2 pathway. Next, to test if the sustained ERK1/2 phosphorylation depended on RIPK1 kinase activity, inhibitors were added 3h30 after TNF stimulation for a 30-min duration (Figure 2C). We observed that Nec1S did not interfere with the late sustained ERK1/2 phosphorylation while it was sensitive to both MEK1/2 and ERK1/2 inhibitions (Figure 2C), indicating that RIPK1 does not play a role directly in the late sustained ERK1/2 phosphorylation.
Figure 2.
Spatio-temporal patterns of activated ERK1/2 during TNF-induced necroptosis in L929
(A and B) L929 cells were serum-deprived (1% FCS) for 12 h and then pretreated (B) or not (A) for 30 min with Nec1S (10 μM) and subsequently stimulated with TNF for the indicated time points. Cells were then lysed and immunoblotted as indicated on the left of each blot.
(C) L929 cells were serum-deprived (1% FCS) for 12h and then treated, harvested, and lysed according to the temporal scheme (C, upper) for immunoblotting. Corresponding molecular weights are indicated on the right of each blot.
(D) L929 cells were serum-deprived (FCS 1%) for 12h and then pretreated or not with Nec1S (10 μM) for 30 min and subsequently stimulated with TNF. Cells were then fixed and processed for immunofluorescence, at the indicated time points, with an antibody against total ERK1/2 (tERK1/2, green) and di-phosphorylated activated ERK1/2 (dpERK1/2, red) and stained for DNA with Hoechst (blue). DMSO (vehicle) was added as control at a volume equivalent to 10 μM Nec1S treatment condition. Scale bar: 50 μm.
(E and F) Scatter plots showing levels of phosphorylated ERK1/2 (E) and subcellular distribution (concentration index of ERK1/2 distribution between the nucleus and the cytoplasm) of total ERK1/2 (F) at the indicated time points in cells undergoing necroptosis (DMSO, blue) or in which necroptosis was inhibited by Nec1S (green). Data are presented as mean ±95 CI of at least two independent experiments, with at least 300 single cells analyzed per condition. Statistical significance was determined using one-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; NS, not significant. Note that all differences were significant but not indicated.
Altogether, these data show that inhibition of RIPK1 activity did not affect early ERK1/2 phosphorylation but lowered the overall level of sustained ERK1/2 phosphorylation, likely by interfering with necroptosis initiation.
Biphasic and compartmentalized activated ERK1/2 profiles during TNF-induced necroptosis
To examine the subcellular localization of activated ERK1/2, immunostainings of diphosphorylated ERK1/2 (activated dpERK1/2) and total ERK1/2 (tERK1/2) in TNF-stimulated L929 cells pretreated or not with Nec1S were performed. At the basal level (TNF 0 min), ERK1/2 displayed a basal phosphorylation level evenly distributed in the cell (Figures 2D–2F). Pretreatment of non-stimulated cells with Nec1S alone produced a significant depletion of total nuclear ERK1/2 in unstimulated conditions while attenuating the ERK1/2 phosphorylation level (Figures 2D–2F), suggesting that RIPK1 kinase activity during non-stimulated conditions impacts ERK1/2 localization in the nucleus. After 10 min of TNF stimulation, phosphorylated ERK1/2 accumulated in the nucleus. One hour after TNF stimulation, ERK1/2 phosphorylation level returned to its initial level and was mainly cytoplasmic (Figure 2F). Both observations were irrespective of Nec1S pre-treatment (Figure 2F). At 4 h after TNF stimulation, ERK1/2 overall phosphorylation level increased and phosphorylated ERK1/2 remained mainly cytosolic. In contrast, Nec1S pretreatment at this time point prevented TNF-induced ERK1/2 phosphorylation level and produced an ERK1/2 localization pattern similar to that of the non-stimulated control condition (TNF 0 min). The latter observation could be expected since cells cannot undergo necroptosis in the presence of Nec1S and are essentially in a survival modus.
Together, our data show a biphasic ERK1/2 activation profile in TNF-induced necroptosis, with an early transient ERK1/2 activation wave yielding an ERK1/2 nuclear accumulation followed by a late sustained ERK1/2 activation profile mainly cytoplasmic. While both were shown to be RIPK1 independent, our results favor the late cytoplasmic signal to depend on the necroptosis process.
EKAR4.0, an improved FRET reporter for ERK1/2 activity detection
Improving KAR engineering and biosensor reversibility by MultiSite Gateway 4.0 cloning
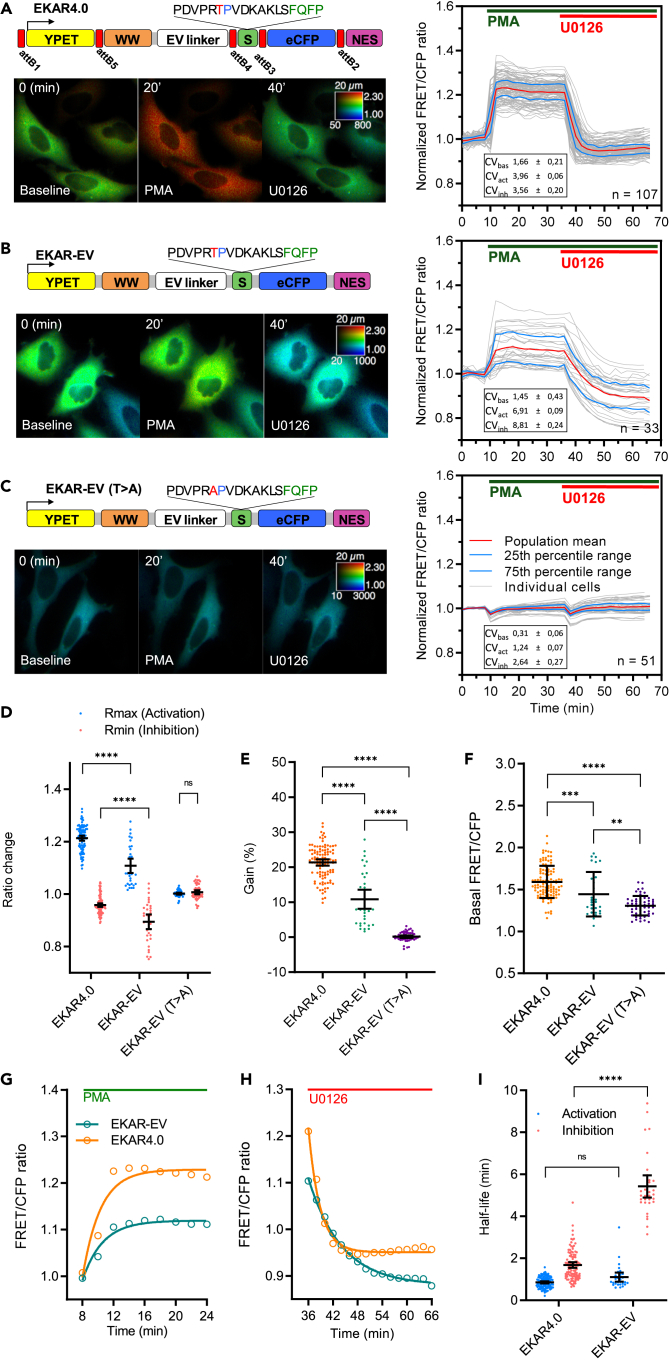
Testing two of the various versions of ERK1/2 FRET biosensors, EKAR-EV (Komatsu et al., 2011) and EKAR-EV-TVV (Vandame et al., 2014) in HeLa and L929 cells (Figures 3B and S4A), we observed that the ratio decrease reported by these biosensors upon ERK1/2 inhibition was relatively slow, and therefore poorly efficient in reporting ERK1/2 activity decrease (i.e., biosensor dephosphorylation), as already mentioned in the literature (Regot et al., 2014). We reasoned that the obligated linker composition required for KAR engineering (Table S1) resulting from the recombination cloning method (MultiSite Gateway Technology) (Jones et al., 2014) might alleviate this shortcoming by adding flexibility. We first evaluated how such modifications, using a four-molecular brick design (Figure S4B), would affect KAR performance. This approach was first tested with PKA biosensors of the AKAR family, using AKAR3-EV as a template (Komatsu et al., 2011). We constructed ‘AKAR3-EV GW4.0’ version that we named AKAR4.0 and compared them together and against the mutant version of AKAR3-EV (AKAR-EV(T > A)) (Figures S5A–S5I). We found AKAR4.0 to report almost three times faster than AKAR-EV on the decrease in PKA activity. Taken together, our data indicate that the obligated linker modifications during complete KAR engineering via the MultiSite Gateway 4.0 cloning alleviate the slowness in reporting downregulation of kinases (Regot et al., 2014).
Figure 3.
Benchmarking EKAR4.0, our improved version of EKAR_EV biosensor, in living cells
(A–C) Biosensor reference experiments: HeLa cells expressing EKAR4.0 (A), EKAR-EV (B) or its mutant version, EKAR-EV (T > A) (C) were time lapse-imaged by FRET microscopy and treated with PMA (0.5 μM) at t = 10 min and then with U0126 (20 μM) at t = 34 min. Top of each panel: Schematic representation of ERK1/2 Kinase Activity Reporter (EKAR) construct used. The small red rectangles in (A) map the positions of obligated attB linkers between each FRET biosensor domain. For the substrate peptide sequence, red letters indicate the phosphorylation site, blue letters indicate amino-acid substitutions to increase the WW domain’s affinity, and green letters indicate ERK1/2 docking site. Bottom left of each panel: Pseudo-color FRET/CFP ratio images of ERK1/2 activity at the steady-state (t = 0 min), upon PMA addition (t = 20 min) and U0126 addition (t = 40 min) are shown. Right of each panel: Graph presenting the FRET/CFP ratio values of every single cell normalized to the averaged baseline FRET/CFP ratio value throughout reference experiments. All individual cells (grey curves), as well as the mean (red curve) and the 25th and 75th percentile range (blue curves) from at least 30 cells, are plotted as a function of time. CVbas, CVact, and CVinh indicate the coefficient of variation for cells at the baseline, cells in ERK1/2 activated state, and cells in the ERK1/2 inhibited state, respectively. Imaging parameters were kept identical for the different biosensors tested. Cells were imaged for a total duration of 70 min at the rate of 1 acquisition every 2 min.
(D–I) Characterization and comparison of different EKAR FRET biosensors responses were performed based on the following parameters: ratio changes (Rmin, Rmax) (D), gain (E), basal FRET/CFP value without any stimulation (F), and fitted curves to determine the speed of activation upon ERK1/2 activation (G, I) and the reversibility of the biosensor upon MEK1/2 inhibition (H, I). Fitted curves were plotted based on Tau ½, which is the time value representing half of the measured duration of the inhibition or activation phase. Tau ½ was calculated by applying a non-linear regression curve on ration values over the complete set of individual cells during the activation or inhibition phase. The nonlinear regression was calculated between t = 8 min and t = 24 min for the activation and between t = 36 min and t = 66 min for the inhibition and plotted as fitted curves. All calculations were performed using GraphPad PRISM 8. Data are mean ± SD from at least two independent experiments. Scale bar: 20 μm. The number of cells analyzed (n) is indicated on the lower right corner of the graph on the right panel (A–C). Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; ns, not significant.
EKAR4.0, an improved FRET reporter for enhanced monitoring of ERK1/2 activity dynamics
The same KAR toolkit was used to build ‘EKAR-EV GW4.0’ aka EKAR4.0 (Figure 3A). Compared to EKAR-EV (Figures 3A–3C), we found EKAR4.0 to report three times faster on the decrease in ERK1/2 activity and exhibit a 10% increase in dynamic range (Figures 3D–3I). At the time, a comparison in HeLa cells of EKAR4.0 to others FRET biosensors for monitoring ERK1/2 activity from the same design, except EKAR3 (Sparta et al., 2015), showed EKAR4.0 to overall perform better (Figures S6A–S6G). Of note, when compared to EKAR-EV (Komatsu et al., 2011) and t-EKAR-EV-vv (Vandame et al., 2014), EKAR4.0 also performed better in L929 cells (Figure S4A).
A structural modeling approach of EKAR4.0 favors higher flexibility of the biosensor
The successful optimization of the EKAR-EV may be attributed in part to the modification of linker composition and/or length between each of the functional elements of EKAR-EV, as reported by Griesbeck et al. (Thestrup et al., 2014). Knowing that the non-radiative energy transfer efficiency between fluorescent proteins (FPs) depends on the relative distance and orientation between the donor and acceptor FPs (Sipieter et al., 2013), we compared EKAR-EV and EKAR4.0 molecular models by using Fusion Protein MODeller (FPMOD) to predict their structure (Pham et al., 2007). In the context of EKAR4.0, the generated 3D models of this ERK1/2 biosensor suggested increased flexibility (Figure S6H) which led us to hypothesize that the increase in flexibility of the ‘FP1-PAABD’ polypeptide might explain the improvement of EKAR4.0 dynamic range. Although not modeled, linker length and composition might also affect the closed conformation of EKAR4.0, thus favoring phosphatases accessibility to the phosphorylated substrate and therefore speeding up dephosphorylation.
Early AM/FMERK1/2 activity dynamics during TNF-induced necroptosis
Distinct temporal ERK1/2 activity dynamics in hFasL- and TNF-induced cell deaths
Based on the opposite effects of direct ERK1/2 inhibition bySCH772984 observed in hFasL- and TNF-treated L929 cells (Figures 1A–1D), we hypothesized that ERK1/2 activity dynamics would be different between apoptosis and necroptosis in L929 cells. For this reason, we investigated using EKAR4.0 whether ERK1/2 would present a different temporal activity signature depending on the modality of programmed cell death. We observed isolated and stochastic bursts of ERK1/2 activity in non-stimulated cells (Figures 4A and 5B), which confirmed the previously reported behavior of ERK1/2 activity in non-stimulated cells (Albeck et al., 2013; Aoki et al., 2013). Since these variations occurred asynchronously, the mean of ERK1/2 activity at the population level displayed a steady-state level (Figure 4A), consistent with WB analysis (Figure 2A). Direct ERK1/2 inhibition by SCH772984 reduced the ERK1/2 activity means at the lower level (Figure S6A).
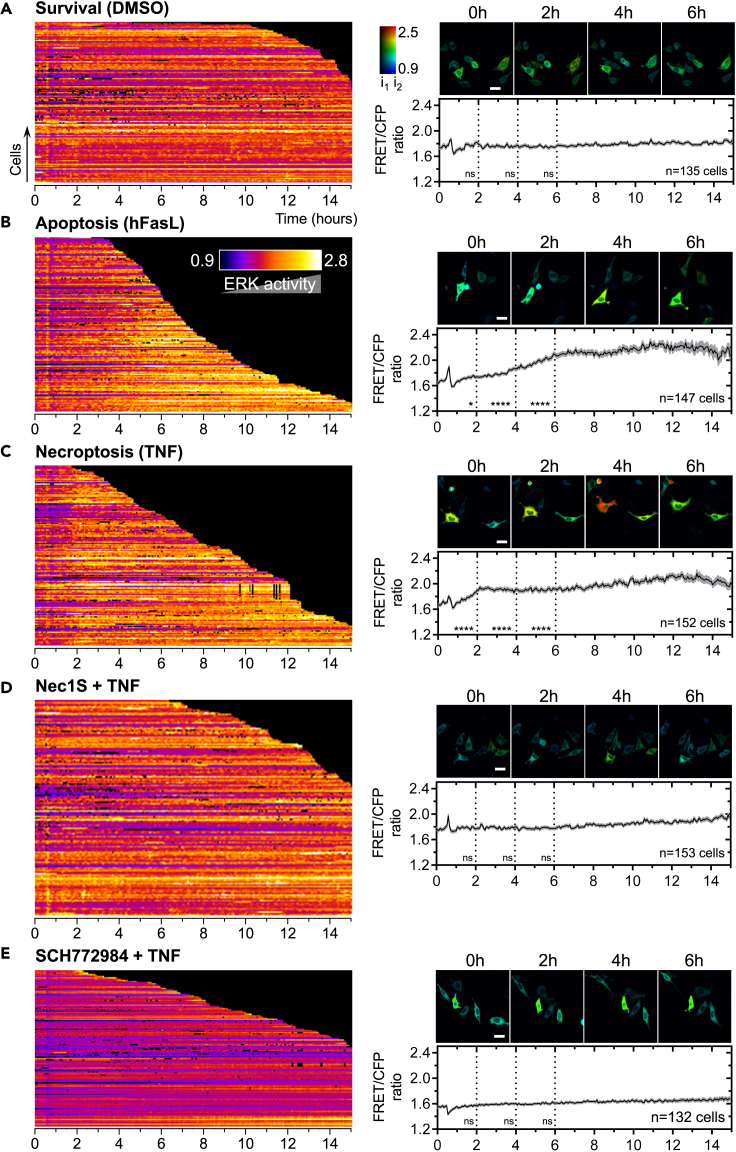
Figure 4.
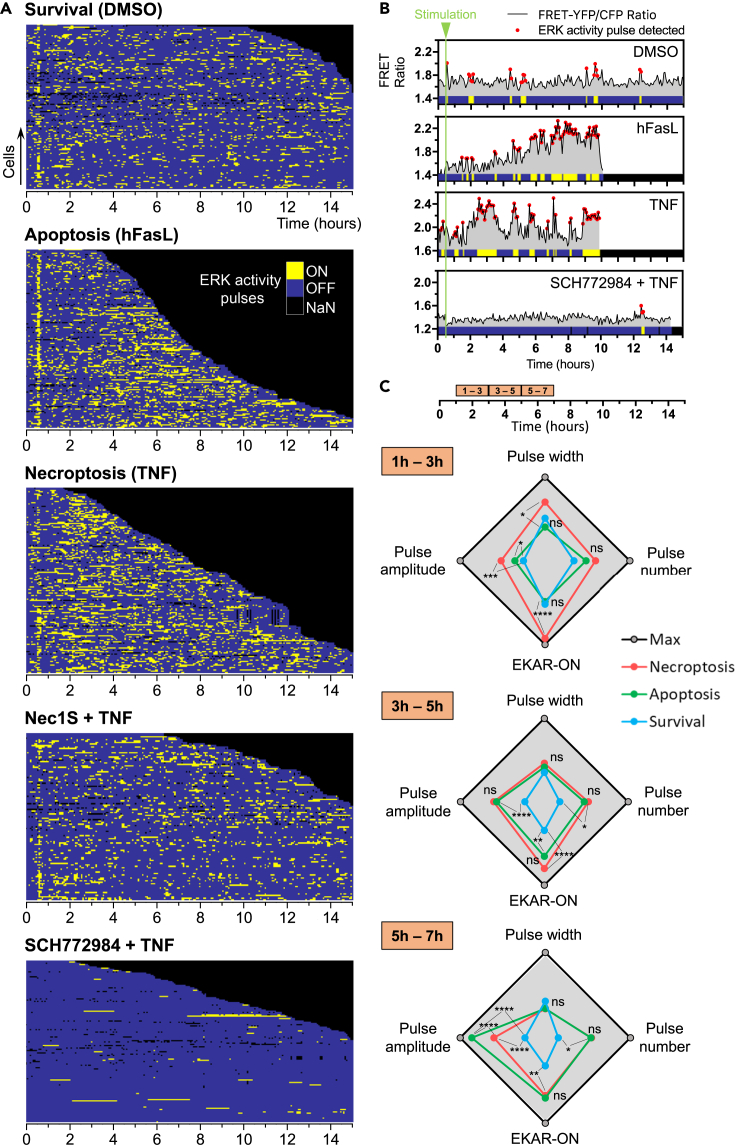
Differential temporal activation of ERK1/2 in TNF-induced necroptosis compared to hFasL-induced apoptosis
(A–E) biosensing experiments: L929 expressing EKAR4.0 were treated with DMSO as a control (A), hFasL (B), TNF (C), and Nec1S orSCH772984 (added 1h in pretreatment) + TNF (D and E, respectively). Samples were time lapse-imaged by FRET microscopy for 30 min before treatment and 14.5 h after stimulation at the rate of 1 acquisition every 4 min. The FRET/CFP ratio for at least 100 cells in each cellular context (survival, apoptosis, and necroptosis) is displayed using a kymographic representation in which each row represents one cell. Cells were ordered by the duration of cell life, from top to bottom. Data shown are from at least two independent replicates. The time scale is at the bottom of each kymograph, and the color-coded fire LUT denotes FRET ratio changes and, therefore, reflects the changes in ERK1/2 activation over time. Black color indicates dead cells or missing points during the automatic analysis. Associated time-lapse images for each condition at different time points (0, 2, 4, and 6h) are displayed. The color-coded LUT reports on both FRET ratio changes and fluorescence intensity (i1 = 50; i2 = 1300). Scale bar: 20 μm. EKAR4.0 signal in each condition was then averaged. The mean (dark curve) and the SEM (grey errors bars) are plotted as a function of time. The number of cells analyzed is reported on each graph (n).
Figure 5.
Differential patterns of EKAR4.0 pulse dynamics in TNF-induced necroptosis and hFasL-induced apoptosis compared to survival
(A) For each condition, similarly to the FRET/CFP ratio, a binary kymographic representation displays the corresponding EKAR4.0 pulse dynamics pattern.
(B) Automatic detection of ERK1/2 activity pulses was performed using a peak detection algorithm, and regions of increased ERK1/2 activity are identified by red dots on one representative FRET/CFP ratio profile for one cell and each condition, plotted against time. The green line corresponds to the time of stimulation. At the bottom of each graph, a corresponding heatmap of EKAR4.0 signal is shown with binary values (0–1 or dark blue to yellow, respectively), meaning a shift of ERK1/2 activity between the “OFF” state (EKAR4.0 FRETOFF) to the “ON” state (EKAR4.0 FRETON). Black indicates dead cells.
(C) Time-resolved quantitative analysis of EKAR4.0 pulse dynamics based on the following parameters: pulse amplitude, pulse number per time fraction, pulse duration (min), and FRETON state of ERK1/2 (calculated for each cell, normalized to the cell life duration of each cell, and expressed as a percentage of time of the cell lifespan). These parameters were calculated 1h after the start of imaging with a 2h time interval for each cell and each condition. Data are presented as radar plots in which each spot corresponds to the mean of at least two independent experiments. Dark spots and lines indicate the maximum for each parameter, and the min and max values are reported as follows: pulses width (6–10 min); pulses number (4–6); pulses amplitude (1.1–1.4); FRETON (20–35%). Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; ∗∗∗∗p< 0.0001; ns, not significant.
We then monitored ERK1/2 activity in L929 during cell death processes. At the population level, stimulation with hFasL (apoptosis condition) induced a slow and progressive significant increase in FRET/CFP ratio starting 3 h after stimulation and reaching a plateau after 6 h (Figure 4B). Looking at the single-cell profiles, almost all dying cells exhibited an increase in ERK1/2 activity to reach a plateau (Figure 4B). Of note, no ERK1/2 activity increase was observed in cells dying rapidly (Figure 4B, first cells). This observation is compatible with the observed ERK1/2 protective role noticed in hFasL-induced apoptosis in L929 cells (Figure 1D), suggesting that increased ERK1/2 activity in this condition might maintain cells alive for more extended periods following hFasL stimulation. Upon TNF stimulation, resulting in necroptosis, the ERK1/2 activity profile was temporally shifted at the population level compared to hFasL-induced apoptosis, with a significant increase in FRET/CFP ratio occurring earlier, 1h30 after TNF stimulation and ramping-up for another 1h followed by an elevated plateau (Figure 4C). Conclusively, ERK1/2 activity profiles were readily different between TNF-induced necroptosis and hFasL-induced apoptosis in L929. However, it is unclear whether these ERK activity profiles are linked to a differential ERK1/2 modulating role during necroptosis and apoptosis in L929 cells (Figures 1B and 1D).
At the population level, in cells pretreated with RIPK1 inhibitor Nec1S, stimulation with TNF did not induce sustained ERK1/2 activation beyond the initial transient burst of ERK1/2 activity (Figure 4D). These results are consistent with those of the WB analysis (Figure 2). Besides, Nec1S pretreatment did not interfere with ERK1/2 stochastic activation in cells protected from TNF-induced necroptosis (TNF + Nec1S), confirming the WB analysis (Figure S3B). Upon SCH772984 addition in TNF-treated cells, the average basal level of ERK1/2 activity was reduced (Figure 4E), and ERK1/2 activity variations were completely inhibited. This trend was comparable to control conditions in the presence of SCH772984 + DMEM (Figure S7A, bottom panel). Using the mutant version of EKAR4.0, similar results were obtained upon TNF stimulation of L929 cells (Figure S7A, top panel).
Taken together, live-cell imaging with EKAR4.0 in our cellular context confirmed that cells exhibited isolated and stochastic pulsatile ERK1/2 activation in survival conditions. Our results show that temporally distinct patterns of ERK1/2 activation are associated with the different stimulated cell death processes. Indeed, we noticed a modulation of both the temporality and the amplitude of the ERK1/2 activity signal. While an early onset of fast ramping-up of ERK1/2 activity followed by a plateau occurred in TNF-induced necroptosis, a progressive and later ramping-up of ERK1/2 activity was recorded during hFasL-induced apoptosis.
Increased amplitude- and frequency-modulated ERK1/2 activity dynamics during cell deaths
The frequency of ERK1/2 activity is a crucial determinant of cell fate decisions (Albeck et al., 2013; Aoki et al., 2013, 2017; Ryu et al., 2016). To resolve the distinct dynamical profiles for ERK1/2 activity between hFasL-induced apoptosis and TNF-induced necroptosis, we investigated the encoding of ERK1/2 activity dynamics. To determine whether the frequency and/or amplitude of ERK1/2 activity could be modulated between survival and programmed cell deaths in L929 cells, we quantified ERK1/2 activity pulses at the single-cell level with high temporal resolution from FRET oscillations data series using a robust FRET analysis workflow.
To provide analog-to-digital conversion of the biological signal which is discrete in time and amplitude, we identified ERK1/2 activity pulses (Figure 5B) using a custom-made Matlab-based semi-automated scoring routine (see Materials & Methods section). First, we determined an experimental threshold from conditions where theoretically few or no pulse of ERK1/2 activity should be detected: using the mutant version of EKAR4.0 (T > A) or EKAR4.0 in the presence of ERK1/2 inhibitor in L929 (Figure S6A). This threshold was used to generate binary kymographs of ERK1/2 activity pulses from the FRET/CFP ratio kymographs previously shown (Figures 5A and S7A).
Focusing on the pulse modulation, we observed as expected spontaneous and stochastic pulses of ERK1/2 activity in survival (Figures 5A and 5B). Analysis of the aligned profiles revealed a temporal shift in the onset of ERK1/2 pulses between apoptosis and necroptosis, which can be readily seen in the representative single-cell data (Figure 5B) and at the cell population level (Figure 5A). In contrast to survival, we found an increase of ERK1/2 activity pulses starting ∼ 1 h 30 min after TNF-induced necroptosis and ∼ 3 h after hFasL-induced apoptosis (Figures 5A and 5B). Not surprisingly, ERK1/2 pulses dynamics, reminiscent of ERK1/2 burst of stochastic activity, remained unaffected upon RIPK1 activity inhibition (Figure 5A) and comparable to survival conditions.
Next, we extracted quantitative parameters to decrypt the encoding of ERK1/2 activity signal. Those are automatically calculated based on the statistical analysis of ERK1/2 pulses per cell for each condition and integrated into our FRET analysis workflow (manuscript in preparation). A very similar approach was performed to characterize cell-to-cell variability in ERK1/2 pulse dynamics in cell proliferation (Albeck et al., 2013; Aoki et al., 2013). The metrics bundle includes a fraction of time spent in the “activated” ERK1/2 and “deactivated” ERK1/2 states (EKARON and EKAROFF, respectively), pulse amplitude, pulse frequency, and pulse width.
Over the entire time span of the experiment, the analysis indicated apparent differences between cell death programs and survival, while limited ones could be seen between hFasL-induced apoptosis and TNF-induced necroptosis (Figure S7B). Compared to survival, both cell death programs increased the overall EKARON fraction, peak amplitude, and peak period. Of note, only the EKARON was statistically different between survival, apoptosis, and necroptosis. Under Nec1S in TNF-induced necroptosis, all metrics reverted to values seen in survival condition. No matter the experimental condition, the ERK1/2 pulse width remained unchanged (mean pulse width: ∼10 min) except for TNF stimulation, where a slight but significant increase was observed compared with survival but not with apoptosis condition (Figure S7B).
A cellular threshold of basal ERK1/2 activity level was shown to determine cell fate decision between growth arrest and death response (Hong et al., 2018). For this reason, we asked whether the basal ERK1/2 activity level could serve a similar purpose in our context. Correlation analysis concluded on no significant relationship between basal FRET level and death time in both TNF-induced necroptosis and hFasL-induced apoptosis in L929 cells (Figures S7C and S7D). So, besides the EKARON metric, the global quantitative analysis did not show any differences in ERK1/2 pulses dynamics between apoptosis and necroptosis in L929 cells.
At the cellular level, signaling operates sequentially in a time-resolved manner with distinct phases; each phase commanding to specific but coordinated cellular output based on the integration of external cues with pulsatile dynamics at the heart of this temporal organization (Levine et al., 2013; Purvis and Lahav, 2013). Thus, we opted for time-resolved analysis of ERK1/2 activity pulses. We analyzed ERK1/2 activity pulses data in time blocks of 2 h, corresponding to the maximum value for the peak period in survival condition (Figure S7B). The analysis started 1 h after stimulation to avoid any effect due to the stimulation itself. By contrast with the complete time span analysis, the time-resolved analysis reveals differences between all the conditions (Figure 5C). At first (time frame 1 h–3 h after stimulation), we observed an early distinction of ERK1/2 pulsatile activity during necroptosis, while it remains similar between apoptosis and survival. The “active” ERK1/2 fraction was significantly higher, as were the pulse amplitude and duration (pulse width). In apoptosis, only the pulse amplitude slightly increased compared to survival. However, no significant differences were visible concerning the pulse number, indicating that pulse frequency seemed unaffected at that stage. Later in the process, between 3 h and 5 h after stimulation, ERK1/2 pulses activity profiles were similar between apoptosis and necroptosis. We measured a significantly higher EKARON fraction, pulse amplitude, and pulse number compared to survival condition. Surprisingly, it was only during the following time frame (5-7h after stimulation) that apoptosis became distinguishable from either necroptosis or survival. Indeed, while necroptosis and apoptosis shared a significant high EKARON fraction and pulse number compared to survival, pulse amplitude during cell death signaling was significantly boosted compared to survival and became significantly different between apoptosis and necroptosis.
To more carefully investigate ERK1/2 signaling dynamics features in a time window preceding the death events, dead cells identified using propidium iodide staining from experimental data presented in Figure 4 were systematically backtracked in all three cellular contexts (survival, apoptosis, and necroptosis) (Figures S8A–S8C). Of note, long-term imaging at high temporal resolution can lead to cells dying accidentally. In WB-like representation, the light red area preceding cell death (30 min) corresponds to the disintegration phase (DP) coinciding with a known burst of ROS production, as it was already observed and quantified in a previous study from our lab (Vanden Berghe et al., 2010). These data do not allow to see apparent differences between cell death programs, except a global increase in basal ERK1/2 activity in cell death programs relative to survival condition, as already shown in Figure 4. Again, as previously described (Figure S7B), no significant differences were visible between apoptosis and necroptosis for the complete time span analysis, except for pulse width (Figure S9A).
Time-resolved analysis of EKAR4.0 signal in single cells was performed in time blocks of 2 h starting 30 min before PI positivity (Figure S8D). Overall, we showed a higher pulse amplitude (Figure S8E), pulse number (Figure S8F), and EKARON fraction (Figure S8G) in apoptosis and necroptosis compared to survival condition. Interestingly, we observed a clear distinction of ERK1/2 dynamics in necroptosis compared to apoptosis in L929 cells. Comparing all temporal frames, pulse amplitude but not pulse number gradually increased in apoptosis compared to necroptosis (Figure S8E). By contrast to apoptosis, pulse number but not pulse amplitude gradually increased in necroptosis (Figure S8F). In addition, cell death observed in the control condition displayed the same ERK1/2 activity profile detected in apoptosis, reflecting the type of cell death in these cells. Altogether, ERK1/2 activity dynamics analysis centered around the time of death confirmed and refined our previous finding, emphasizing ERK1/2 activity dynamics to be more Amplitude-Modulated during hFasL-induced apoptosis and more Frequency-Modulated during TNF-induced necroptosis in L929 cells.
Frequency domain analysis confirms differences in ERK1/2 activity dynamics between necroptosis and apoptosis
While this time-resolved analysis of metrics has been applied before (Albeck et al., 2013; Aoki et al., 2013), it relies on data thresholding which could be considered biased. In addition, pulses analysis converted the signal from analog to digital binary, which narrows the resolution. Yet, another argument of a limitation of the temporal domain analysis in these circumstances is that all frequencies within the biological signal are de facto intertwined and not revealed. The higher temporal sampling resolution of our biosensing data and the nature of ERK1/2 signal allowed us to quantify the oscillatory component of ERK1/2 activity through frequency domain using Fourier analysis. Fourier analysis of such FRET biosensing data has already been used to quantify calcium oscillations (Candeo et al., 2017). To this aim, Fourier transformation was applied to our EKAR ratiometric data in order to generate different power spectral density (PSD) spectra (Figure 6A). The PSD describes the strength of the variations as a function of frequency. In other words, it shows at which frequency range (Hz or min−1) variations are strong or weak (power, dB).
Figure 6.
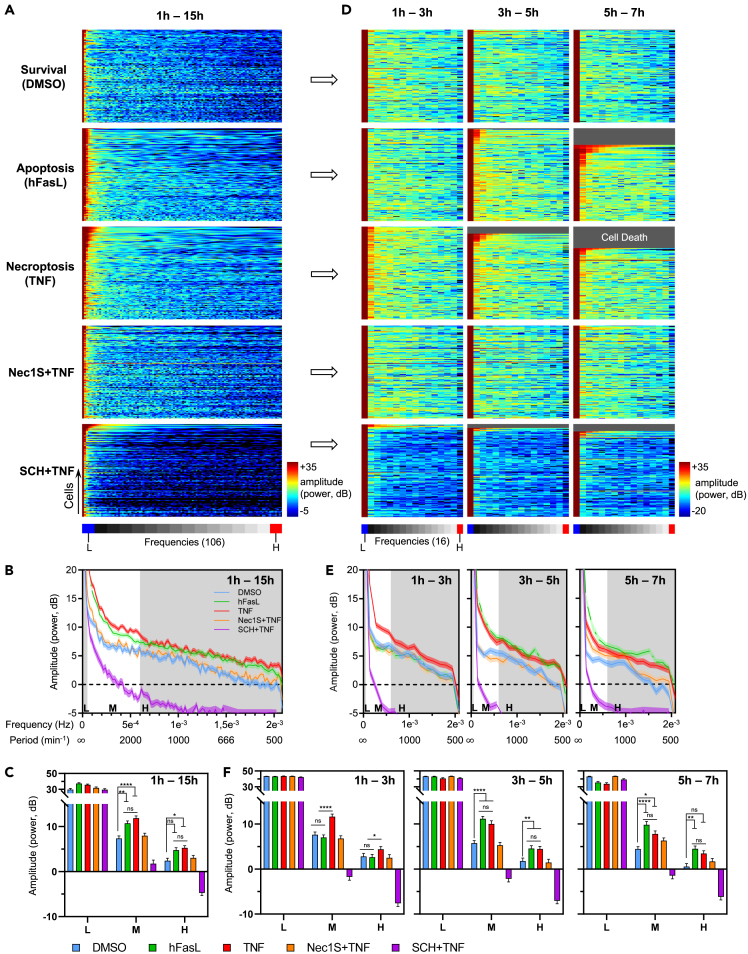
Fourier transform-based analysis helps crack a distinct ERK1/2 activity code in necroptosis in L929 cells
(A) Fourier analysis of the ERK1/2 oscillations extracted from raw FRET/CFP ratio data of Figure 4. For each condition described in Figure 4, the corresponding power spectral density (PSD) for each frequency (106) is displayed by a kymographic representation (from 1h to 15h after the beginning of imaging) in which each row represents one cell. Lower (0.000018 Hz) and higher (0.002 Hz) frequencies are indicated in blue (left) and red (right), respectively. Color-coded LUT denotes the amplitude of PSD (−5 to +35) for each frequency.
(B) For each condition, averaged data are represented where the mean PSD amplitude is plotted against frequency.
(C) For each frequency range (Low (L) f < 5 × 10−5 Hz; Medium (M); High (H) f > 6 × 10−4 Hz frequencies), the mean PSD amplitude (mean ± SEM) for each biological condition is compared.
(D) Fourier analysis of the ERK1/2 oscillations extracted from FRET/CFP ratio data was then performed on three temporal frames with a 2h time-interval for each condition: 1h–3h, 3h–5h, and 5h–7h (from left to right).
(E) For each temporal frame and for each frequency range (Low (L) f < 5 × 10−5 Hz; Medium (M); High (H) f > 6 × 10−4 Hz frequencies), and the mean PSD amplitude (mean ± SEM) for each biological condition is displayed. Grey color on the kymographs.
(F) For each frequency range (Low (L) f < 5 × 10−5 Hz; Medium (M); High (H) f > 6 × 10−4 Hz frequencies), the mean PSD amplitude (mean ± SEM) for each biological condition is compared. Lower (0.00013 Hz) and higher (0.002 Hz) frequencies are indicated in blue (left) and red (right), respectively. Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
PSD has been represented for each cell as kymographs within the entire frequency range (Figure 6A), together with the mean PSD ± standard deviation curves (Figure 6B) for each condition. At the global level, three observations can be made: (1) L929 in survival conditions (unstimulated and Nec1S + TNF) exhibited a similar PSD profile, (2) the PSD of TNF-induced necroptosis in SCH772984-pretreated L929 cells was strongly affected as expected, (3) PSD profiles of L929 cells stimulated with hFasL and TNF were comparable but higher than those in survival conditions. Note, however, that PSD in cell death programs was intertwined within the medium frequencies (Figure 6B). To appreciate more precisely these differences, we divided the entire frequency range into three main frequency bands corresponding to three different oscillatory components: low (LF, less than 0.05 mHz), which mainly represent the background noise originating from the system, medium (MF, between 0.05 and 0.6 mHz), and high (HF, more than 0.6 mHz) frequency bands (Figure 6B). Thus, the period of the MF oscillations ranged between 27 and 166 min and corresponded to the period range of ERK1/2 pulses registered for each condition as presented in Figure S7B. The mean PSD ±standard deviation histograms within each frequency band have been represented for each condition (Figure 6C). Results showed a significant difference in both the MF and HF bands in programmed cell death conditions than survival, validating the results obtained previously (Figures 5C and S7B).
Then, we performed a time-resolved analysis of PSD for each condition (Figures 6D–6F). Importantly, in the time frame 1h–3h after stimulation, PSD in both the MF and HF bands was significantly different in necroptosis from that of survival and apoptosis (Figures 6E and 6F, left panels). Therefore, in this particular time frame, we could discriminate apoptosis from necroptosis in L929 cells based on PSD analysis of frequency-modulated ERK1/2 activity. As expected, the time frame 3h–5h of TNF- and hFasL-stimulated L929 cells shared a similar PSD in both the MF and HF bands, significantly different from that of survival or TNF + Nec1S conditions. Later, in the time frame 5h–7h after stimulation, PSD results were comparable to those from the time frame 3h–5h.
In sum, spectral decomposition analysis of ERK1/2 activity pulses supported in an unbiased way our previous observations and indicated that a specific signature of ERK1/2 activity dynamics discriminates apoptosis from necroptosis in L929 cells, especially early on. TNF stimulation triggered early amplitude and frequency modulation of ERK1/2 activity mainly in two frequency bands, distinct from hFasL stimulation showing a delayed activation.
Disruption of ERK1/2 activity dynamics prevents pro-inflammatory cytokine gene expression during TNF-induced necroptosis
Several studies already proposed that temporal differences in frequency- and amplitude-modulated ERK1/2 activity could be decoded by downstream transcription factors driving, therefore, a differential gene expression pattern (Albeck et al., 2013; Muta et al., 2018; Wilson et al., 2017). Necroptosis can trigger inflammation by inducing pro-inflammatory gene expression in a cell-autonomous manner (Najjar et al., 2016; Zhu et al., 2018). Recently, Degterev et al. demonstrated in BMDM cells that this process is mediated in part by ERK1/2 (Najjar et al., 2016). Thus, we hypothesized that ERK1/2 activity could impact pro-inflammatory cytokines gene expression profiles during necroptosis in L929 cells. To this aim, we characterized the transcriptional changes in TNF-stimulated L929 cells by focusing on a small panel of genes involved in survival, inflammation, and signaling. As expected from previous studies (Martens et al., 2017), mRNA expression of nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα) and A20, known as regulators of inflammation and cell survival, were upregulated upon TNF-induced necroptosis and insensitive to ERK1/2 inhibition (Figure S10A).
All pro-inflammatory cytokines or chemokines tested – TNF, monocyte chemoattractant protein 1 (MCP1), chemokine (C-X-C motif) ligand 1 (Cxcl1), and interleukin-6 (IL-6) – showed an increase in mRNA expression upon TNF-treatment. Of those, only TNF and IL-6 were significantly sensitive to ERK1/2 inhibition 2 h after TNF stimulation. Of note, both basal (unstimulated conditions) and TNF-stimulated gene expression were impacted by ERK1/2 inhibition. Since our results showed the AM/FM ERK1/2 activity to be specifically upregulated in the time frame 1h–3h after TNF stimulation in L929 cells, we investigated the impact of ERK1/2 inhibition on TNF and IL-6 mRNA expression 4 h after TNF stimulation. Results indicated IL-6 mRNA expression to increase further, while TNF slightly decreased (Figure S10B). Next, to assess the consequence of a blockage of the upregulated AM/FM ERK1/2 activity on the expression of those particular genes, a similar experiment was performed except that ERK1/2 inhibition was performed 2 h after TNF treatment (within the time frame 1h–3h after TNF stimulation) (Figure S10C). The later TNF and IL-6 mRNA levels were again strongly reduced, similar to the ERK1/2 inhibition before TNF stimulation, indicating that the observed transient ERK1/2 activation occurring between 5 and 15 min was not implicated in TNF and IL-6 gene expression.
Taken together, the role of ERK1/2 activation in TNF and IL-6 gene expression was confirmed since ERK1/2 inhibition strongly inhibited basal and TNF-stimulated gene expression in L929 cells. The effect of ERK1/2 inhibition in combination with TNF stimulation caused a greater decrease in TNF and IL-6 gene expression compared to ERK1/2 inhibition alone. Similar to the reported oscillations of p38 activity regulating pro-inflammatory gene expression (Tomida et al., 2015), we speculate that the AM/FM ERK1/2 activity dynamics might affect TNF and IL-6 gene expression.
Necroptotic ERK1/2 activity dynamics is recapitulated in RIPK1/3 independent necroptosis model
ERK1/2 inhibition protects against RIPK3- and MLKL-induced necroptosis
To map the position of ERK1/2 with respect to the canonical RIPK1/RIPK3/MLKL necroptotic signaling pathway, we took advantage of genetic models in which necroptosis is induced independently of any receptor activation. These models developed in our lab were derived from either L929 or the murine colon carcinoma CT26 cell line in which the expression of downstream necroptotic effectors are controlled by a doxycycline (Dox) inducible system. For RIPK1-independent necroptosis, we used BALB/c mouse-derived CT26 cells Dox-inducible RIPK3 (hereinafter referred to as CT26 mRIPK3) (Aaes et al., 2016). Concerning RIPK1/3-independent necroptosis, L929 cells were knocked out for Mlkl gene expression and reconstituted with mMLKL phospho-mimetic form (S345D), which expression can be triggered upon Dox treatment yielding the following cell line L929 Mlkl−/- Dox-inducible mMLKL(S345D)-Flag (hereinafter referred to as L929 iMLKL(S345D)).
As previously described (Figure S1A), a systematic chemical perturbation of all layers of the MAPK/ERK1/2 cascade was performed in these cell lines in the presence of Dox (Figures 7A and S11A). Importantly, direct ERK1/2 inhibition using SCH772984 showed a substantial and significant decrease in RIPK1 and RIPK1/3 independent necroptosis in CT26 mRIPK3 (Figure S11A) and L929 iMLKL(S345D) (Figure 7A), respectively. Interestingly, while similar results to that of direct ERK1/2 inhibition were observed when MEK1/2 was inhibited using PD184352, this was not the case for U0126 mediated MEK1/2 inhibition in our RIPK1-activity-independent necroptosis model. Indeed, MEK1/2 inhibition by U0126 showed no significant effect or a moderate delay of cell death, which was reminiscent of our observations in TNF-induced necroptosis in L929 (Figure 1A and (Vanlangenakker et al., 2010)). These findings advocated for ERK1/2 engagement during necroptosis signaling in CT26 and L929 and downstream of activated MLKL in the latter.
Figure 7.
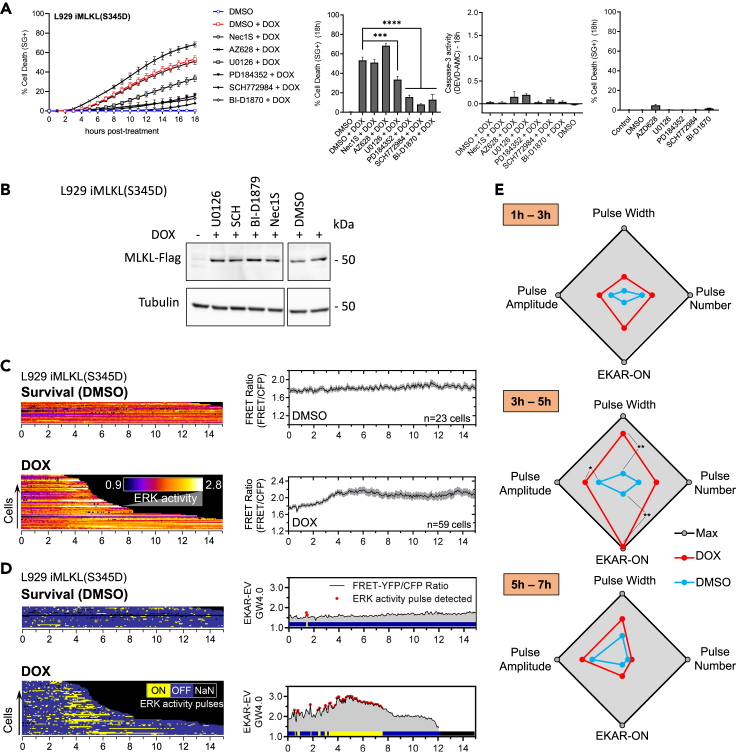
Direct ERK1/2 inhibition efficiently impairs necroptosis driven by MLKL S345D overexpression in L929 cells
(A) MLKL-deficient L929 reconstituted with Dox-inducible phosphomimetic S345D mutant MLKL expression were used. These cells were pretreated for 1 h with the indicated compounds before Dox (red) or DMEM (blue) stimulation (grey curves). Cell death was measured as a function of time by SytoxGreen (SG+) positivity (kinetic experiment, left), 18 h after Dox stimulation (A, left histogram), and caspase-3 activity was assessed by DEVD-AMC cleavage efficiency (A, middle histogram) 18 h after Dox stimulation. Drugs-induced toxicity measurements (SG+ positivity) 18 h after DMEM stimulation (A, right histogram). The cell death data are presented as mean ± SEM of at least two independent experiments. Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; ns, not significant.
(B) Cells were pretreated or not for 1 h with the indicated compounds (10 μM) and subsequently stimulated with Dox. Cells were then lysed and immunoblotted as indicated on the left of each blot. Corresponding molecular weights are reported on the right of each blot. Both left and right parts came from the same blot; a cut was performed to remove non-relevant conditions.
(C) Biosensing experiment: Cells expressing the FRET biosensor EKAR4.0 were time lapse-imaged by FRET microscopy for 30 min before treatment (control & Dox) and 14.5 h after stimulation at the rate of 1 acquisition every 4 min. From left to right, the raw FRET/CFP ratio of individual cells is displayed by a FRET/CFP ratio kymographic representation (C, left), averaged raw FRET/CFP ratio (mean ± SEM) as a function of time (C, right).
(D) Corresponding quantitative analyses of ERK1/2 pulse dynamics are displayed by a binary kymographic representation of ERKON and ERKOFF states (D, left). As described in Figure 5, ERK1/2 activity is identified by red dots on one representative FRET/CFP ratio profile for each condition, plotted against time.
(E) Time-resolved quantitative analysis of EKAR4.0 pulse dynamics based on parameters identified in Figure 5 were calculated 1h after the start of imaging with a 2-h time interval, for each cell and for each condition (blue line: survival; red: necroptosis triggered by Dox-induced S345D MLKL expression). Temporal frame data analysis is presented as radar plots similar to Figure 5C. Dark spots and lines indicate the maximum for each parameter, while the center indicated the minimum. Min and max values are reported as follows: pulses width (6–15 min); pulses number (0.5–3); pulses amplitude (1.0–1.6); FRETON (4–16%). Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; ∗∗∗∗p< 0.0001; ns, not significant. Experimental and analysis protocols, as well as graphical representations, are identical to Figures 4 and 5.
Interestingly, the RSK inhibitor BI-D1870 consistently protected against necroptosis with a 2-fold and up to a 10-fold decrease in CT26 mRIPK3 and L929 iMLKL(S345D), respectively (Figures S11A and 7A). This RSK inhibition effect was very much comparable to the one recorded previously in TNF-induced necroptosis in L929 cells (Figure 1A). Note that in all experimental conditions, neither caspase activation as expected nor chemical compounds cytotoxicity was detected (Figures 7A and S11A). Also, inhibitors used at the specified concentrations did not dampen the Dox-driven production of RIPK3 and MLKL (Figure 7B). As anticipated, RIPK1 inhibition using Nec1S did not prevent RIPK1 and RIPK1/3 independent necroptosis (Figures 7A and S11A). In sum, using our genetic models, we showed that inhibition of MEK1/2, ERK1/2, and its downstream effector RSK amounted to significant inhibition of both RIPK1- and RIPK1/3-independent necroptosis.
Necroptotic ERK1/2 activity dynamics is recapitulated downstream of MLKL
Based on our previous results using genetic models of necroptosis, we hypothesized that TNF-stimulated AM/FM ERK1/2 activity dynamics could also be observed in cells undergoing necroptosis independent of RIPK1 and RIPK3 activities. We first imaged L929 iMLKL(S345D) expressing EKAR4.0 to report on ERK1/2 activity without stimulation. ERK1/2 activity was maintained at basal level with the detection of isolated and stochastic bursts of ERK1/2 activity (Figure 7C), comparable to the unperturbed condition in L929 cells (Figure 4A), confirming thereby that knocking out MLKL did not impact basal ERK1/2 activity (Yoon et al., 2017). Interestingly, in comparison to Figure 4C, ERK1/2 activity profile in necroptosis was temporally changed with an increase in FRET/CFP ratio occurring about 2 h after Dox stimulation and mainly followed by a sustained ERK1/2 activity level for the remainder of the experiment as shown by ERK1/2 activity mean average population representation (Figure 7C). Of note, RIPK1 inhibition did not affect ERK1/2 activity dynamics in these experimental conditions (data not shown), reinforcing the notion that the AM/FM ERK1/2 activity in L929 cells is independent of RIPK1 activity.
Applying the metrics bundle analysis strategy over the entire time span of this experiment again indicated apparent differences between MLKL(S345D)-driven necroptosis and survival. Importantly, we found that the AM/FM ERK1/2 activity signature observed in TNF-induced necroptosis in L929 cells (Figure 5) was recapitulated in Dox-stimulated L929 iMLKL(S345D) (Figures 7D, S7B and S11B). Indeed, the EKARON fraction and the frequency of ERK1/2 activity pulses almost doubled, and pulse amplitude was also raised (Figure S11B). Next, time-resolved analysis of ERK1/2 activity pulses in time blocks of 2 h revealed a distinct ERK1/2 activity signature in Dox-stimulated cells compared to survival only in the time frame 3h–5h after stimulation (Figure 7E). Metrics of ERK1/2 activity are similar to those observed in TNF-induced necroptosis in L929 cells in the time frame 1h–3h, confirming the previously observed temporal shift of the AM/FM ERK1/2 activity upon Dox-induced MLKL(S345D) production. Taking together, these results in RIPK1/3 independent necroptosis in L929 iMLKL(S345D) are reminiscent of our findings in TNF-induced necroptosis in L929 cells (Figure 5D). Therefore, these observations suggest that necroptosis is associated with a specific temporally regulated AM/FM ERK1/2 activity signature in L929 cells, which appears to be part of the execution process, and more precisely, downstream of MLKL.
Discussion
This study has identified and exemplified ERK1/2 modulator function in programmed cell death. Our findings indicate that direct ERK1/2 inhibition delayed TNF-induced necroptosis while it sensitized hFasL-induced apoptosis in L929 cells. Biochemical data revealed a RIPK1-activity independent biphasic trend of ERK1/2 activity upon TNF stimulation at the population level, with an early nuclear transient and a late cytoplasmic sustained ERK1/2 activity profile.
Using live-cell imaging with a much-improved FRET biosensor to monitor ERK1/2 activity, we have reported that in TNF-induced necroptosis and hFasL-induced apoptosis, ERK1/2 activity dynamics display amplitude and frequency-modulation contrasting with the ERK1/2 activity dynamics recorded in survival. Using a robust FRET analysis workflow, our measurements shed light on ERK1/2 activity implication to discriminate among necroptosis and apoptosis in L929 cells both at early and late stages of cell death. By comparison with hFasL, TNF-stimulated cells underwent a rapid change in ERK1/2 pulsatile activity characterized by modulation of pulses amplitude and duration. At later time points, ERK1/2 pulse modulation between necroptosis and apoptosis remained comparable and could only be discriminated by a higher amplitude modulation during apoptosis, clearly indicating an increase in signal strength. Also, time-resolved analysis together with a spectral decomposition of ERK1/2 pulsatile activity demonstrated that an early onset of AM- and FM- ERK1/2 activity pulses are characteristic of TNF-induced necroptosis by comparison to hFasL-induced apoptosis in L929 cells.
Beyond the encoding phase that links cell death stimulus to a specific ERK1/2 activity profile, we also investigated the decoding phase that ties the ERK1/2 signal to a particular cellular outcome. ERK1/2 activity recorded in TNF-induced necroptosis in L929 cells was found to be associated with an increase of pro-inflammatory cytokines gene expression. However, only TNF and IL-6 were significantly sensitive to ERK1/2 inhibition 2 h after TNF stimulation. Besides, using genetic models of RIPK1- and RIPK1/3-independent necroptosis, our results advocated for an MEK/ERK/RSK axis engagement, actually downstream of activated MLKL. Moreover, we found that the uncovered AM/FM ERK1/2 activity signature was recapitulated in a genetic model of RIPK1/3-independent necroptosis. Based on our results, we could hypothesize that a distinct AM/FM ERK1/2 activity signature would regulate pro-inflammatory cytokines gene expression downstream of activated MLKL during necroptosis in L929 cells.
Uncovering pulsatile dynamics of signaling molecules has provided unprecedented insights into the biochemical signal encoding towards cell fate. In survival conditions, ERK1/2 activity pulses occur spontaneously and are mainly due to autocrine epidermal growth factor (EGF) signaling (Aoki et al., 2013, 2017; Hiratsuka et al., 2015). In apoptotic and necroptotic L929 cells, these pulses increased in duration and decreased spacing because individual bursts in ERK1/2 activity were no longer distinguishable. While pulse amplitude was reported in EGF-stimulated conditions (Albeck et al., 2013; Aoki et al., 2013), ERK1/2 pulse amplitude varied over time in hFasL- and TNF-stimulated L929 cells. As far as we know, quantitative measurements of amplitude-modulated ERK1/2 activity dynamics have solely been shown experimentally in the context of MEK1/2 inhibition in a dose-dependent manner (Albeck et al., 2013).
Fourier analysis of ERK1/2 oscillations within three main frequency bands (named LF, MF, and HF) offered the possibility to perform unbiased statistical analysis of ERK1/2 pulsatile activity. Fourier based frequency decomposition can be cast as a relevant approach in our field to decrypt the context-specific ERK1/2 signaling code and can be applied to other kinases. Spectral decomposition analysis of ERK1/2 kinase activity dynamics might reveal an additional layer of biochemical signal encoding, which could help rational pharmacological targeting of diseases associated with MAPK/ERK1/2- and other kinases aberrant signaling.
Specificity in signal transduction is determined by the cell’s computational capacity to encode and decode a signal and is likely applicable to most signaling pathways (Madsen and Vanhaesebroeck, 2020). Next, we explored the functional relevance of this AM/FM ERK1/2 activity in TNF-induced necroptosis in L929 cells. Necroptosis promotes proinflammatory cytokine gene expression in a cell-autonomous manner (Najjar et al., 2016; Zhu et al., 2018) and the release of immunogenic molecules (Murai et al., 2018). Pursuing this paradigm, we found ERK1/2 to be a transcriptional activator of pro-inflammatory cytokine gene expressions such as TNF and IL-6. This proposed function of ERK1/2 in the context of TNF-induced necroptosis is associated with specific ERK1/2 activity dynamics. Our results align with that of Najjar et al. exposing ERK1/2 as a critical regulator promoting inflammatory gene expression in LPS-induced necroptosis in BMDM cells (Najjar et al., 2016).
Interestingly, ERK1/2 functioned in LPS-induced necroptosis in BMDM cells as an uncoupling modulator that directly mediated RIPK1/3 dependent inflammation, independently of MLKL. Could there be a correlation between specific ERK1/2 pulsatile activity dynamics and pro-inflammatory cytokine gene expression patterns? This has been demonstrated before by Saito et al. in 2014 reporting on the role of interleukin-1 beta (IL-1β)-stimulated p38 pulsatile activity in controlling pro-inflammatory cytokines gene expression profile (Tomida et al., 2015). They found the p38 pulsatile activity to control efficient expression of genes such as IL-6, interleukin 8 (IL-8), and cyclooxygenase-2 (COX-2).
Considering that AM/FM ERK1/2 activity dynamics would mediate the increase in pro-inflammatory cytokines gene expression profile in programmed cell death, what could be the pulse generator? In 2013, Aoki et al. showed Raf activity as a generator of ERK1/2 activity pulses in NRK-52E cells by impacting pulses width. Here, the pulse width was significantly impacted in both TNF- and MLKL-induced necroptosis in L929, and at the same time, inhibition of Raf did not significantly modulate neither necroptosis nor hFasL-induced apoptosis in L929 cells. Despite the RIPK1 kinase activity-independent nature of AM/FM ERK1/2 activity in TNF-induced necroptosis, but based on the fact that Najjar et al. mapped ERK1/2 downstream of RIPK1/RIPK3 platform upon LPS-induced necroptosis in BMDM cells (Najjar et al., 2016), we tested RIPK1 as a potential candidate for the generation of ERK1/2 activity pulses. Nec1S alone or Nec1S together with TNF did not affect ERK1/2 activity pulses width, excluding RIPK1 as a pulse generator. In line with this, we found the impact of ERK1/2 inhibition recapitulated in RIPK1/3 independent necroptosis in L929 cells. Since the specific AM/FM ERK1/2 activity signature could be monitored in a model of MLKL-induced necroptosis, we came to consider other potential scenarios downstream of MLKL that could explain our findings.
MLKL depletion was shown to affect endosomal transport independently of RIPK3 (Yoon et al., 2017) and to provoke a slowdown of TNF and TNF receptor (TNFR) degradation, and this was also true for EGF and EGF receptor (EGFR). In both cases, this boosts either TNF or EGF signaling activities, including enhanced gene induction. During necroptosis, phosphorylated MLKL significantly affects plasma membrane integrity, and this process is now known to increase the load on the endosomal trafficking (Vandenabeele et al., 2017). The endosomal sorting complexes required for transport (ESCRT)-III machinery, which acts downstream of MLKL, manages necroptotic cell death to allow for pro-inflammatory cytokine production and extracellular-vesicle (EV) production and release towards emergency signaling purposes before cell demise (Gong et al., 2017). Like the impact of MLKL depletion, overloading of the endosomal machinery during necroptosis could amount to the slowdown of EGFR recycling with an enhancement of EGF signaling. On the opposite, sustained ERK1/2 signaling could equally be caused by enhanced recycling of endocytosed EGFR [for review (Mattissek and Teis, 2014)]. The low expression level of the ESCRT-I protein Tsg101 was reported to cause rapid recycling of EGFR to the cell surface and a subsequent increase in EGFR signaling associated with a sustained ERK1/2 phosphorylation (Babst et al., 2000). A later report indicated that only knock-down of ESCRT subunits −0 and −I proteins would contribute to this effect (Raiborg et al., 2008). Finally, depending on the stressor, EGFR receptor recycling has been shown to potentiate cell death or promote cell survival (Tomas et al., 2014).
Previous observations reported that targeted inhibition of EGFR (using Gefitinib) reduces ERK1/2 activity pulses frequency while inhibition of MEK1/2 reduces their amplitude (Albeck et al., 2013). Including results from this study, further investigation would be required to determine whether the interference with endosomal trafficking downstream of MLKL during necroptosis leads to aberrant recycling of EGFR, which acts as a pulse generator, boosting AM/FM ERK1/2 activity dynamics towards increasing pro-inflammatory cytokine gene expression.
In a follow-up study, it might be possible to monitor necroptosis at the single-cell level, thanks to Nakano et al. who recently filled a challenging gap in functional live-cell monitoring of necroptosis (Murai et al., 2018). They produced a sensor for MLKL activation by RIPK3 based on FRET (SMART) biosensor (Murai et al., 2018; Nakano et al., 2019). Using this SMART biosensor in future experiments would help correlate the engagement of MLKL with AM/FM ERK1/2 activity profile during either TNF- or MLKL-induced necroptosis in L929 cells. Simultaneous recording of dual kinase activities in the same cell could be performed using our recently published method (Demeautis et al., 2017). While this would enlighten us about the coincidence of MLKL activation and ERK1/2 pulsatile activity, it would not provide a strong causality link between necroptosis initiation and AM/FM ERK1/2 activity-mediated pro-inflammatory cytokine gene expression increase in TNF-stimulated L929 cells. Instead, this will be addressed by generating controlled activation of ERK1/2 by repetitive pulsatile TNF stimulations using a microfluidic system (Ryu et al., 2015; Tomida et al., 2015) and/or by applying synthetic ERK1/2 activity pulses using optogenetic (Aoki et al., 2013; Zhou et al., 2017).
Limitations of the study
This report provided an accurate correlation between specific ERK1/2 signaling dynamics features during TNF-induced necroptosis and hFasL-induced apoptosis in L929. However, this study does not provide a causality link: (1) between ERK signaling dynamics features and programmed cell death; (2) between necroptosis initiation and AM/FM ERK1/2 activity-mediated pro-inflammatory cytokine gene expression increase in TNF-stimulated L929 cells. In addition, while we observed a modulation of MAPK/ERK pathways in several cell lines using chemical inhibitors, ERK activity signatures were only recorded in the L929 cellular model.
This study does not link molecular signaling events. Cell death is asynchronous, and the time to death varies across the cell population. The lack of fluorogenic tools to monitor in real-time necroptosis hallmarks such as caspase FRET biosensors in apoptosis hampered the detection of necroptotic specific signaling events at the single living cells level. Therefore, beyond the known morphological features of necroptosis, only the time to death using PI has been used as default confirmation of necroptosis in this report.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ERK1 + ERK2 antibody | Abcam | Cat# ab17942 |
| anti-β-actin | Santa Cruz Biotechnology | Cat# I-19 |
| anti-RIPK1 | BD-Biosciences | Cat# 610459 |
| anti-β-tubulin | Abcam | Cat# ab6046 |
| anti-β-tubulin-HRP | Abcam | Cat# ab21058 |
| Anti-MAP Kinase, Activated (Diphosphorylated ERK-1&2) antibody | Sigma Aldrich | Clone MAPK-YT, M9692 |
| anti-MLKL | Merck Millipore | Cat# MABC604, Clone 3H1 |
| Anti-phospho-MLKL (Ser345) | Merck Millipore | Cat# MABC1158, clone 7C6.1 |
| Phospho-MEK1/2 (Ser217/221) | Cell Signalling | Cat# 9121 |
| Anti-MEK1/2 | Cell Signalling | Cat# 9122 |
| anti-RIPK3 | Santa Cruz | Cat# sc-374639 |
| anti-RSK1/2/3 | Cell Signalling | Cat# 9355, clone 32D7 |
| anti-phspho-p90-RSK (Ser380) | Cell Signalling | Cat# 9341 |
| anti-GSK3β | Cell Signalling | Cat# 12456, clone D5C5Z-XP |
| anti-phospho-GSK-3α/β | Cell Signalling | Cat# 9331 |
| anti-mouse Alexa Fluor 488 | Thermo Fischer | Cat# A-10667 |
| HRP-conjugated anti-rabbit IgG | Santa Cruz Biotechnology | Cat# sc-2004 |
| HRP-conjugated anti-mouse IgG | Santa Cruz Biotechnology | Cat# sc-2005 |
| HRP-conjugated anti-goat IgG | Santa Cruz Biotechnology | Cat# sc-2020 |
| HRP-conjugated anti-rat- IgG | GE Healthcare | Cat# NA935 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant mouse TNFα | In house production | N/A |
| Anti-hFas ligand | Upstate | Cat# 05-201Clone CH11 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma Aldrich | Cat# P8139 |
| U0126 | Sigma Aldrich | Cat# U120 |
| PD184352 | Sigma Aldrich | Cat# PZ0181 |
| AZ628 | Sigma Aldrich | Cat# SML0785 |
| BI-D1878 | Sigma Aldrich | Cat# 559286 |
| Forskoline | Tocris | Cat# 1099/10 |
| SCH772984 | Selleckchem | Cat# S7101 |
| Selumetinib (AZD6244) | Selleckchem | Cat# S1008 |
| Trametinib (GSK1120212) | Selleckchem | Cat# S2673 |
| Necrostatin-1S (Nec1S) | Laboratory of Medicinal Chemistry; University of Antwerp | Cat# UAMC-02197 |
| Propidium Iodide | Sigma Aldrich | Cat# P-4170 |
| Sytox Green | Thermo Fischer Scientific | Cat# S7020 |
| Doxycycline Hyclate (Dox) | Sigma Aldrich | Cat# D9891 |
| Rneasy Mini Kit | Qiagen | Cat# 74104 |
| Qiashredder | Qiagen | Cat# 79654 |
| DNA-free™ DNA Removal Kit | Invitrogen | Cat# AM1906 |
| SuperScript® III First-Strand Synthesis System for RT-PCR | Invitrogen | Cat# 18080-51 |
| qPCR MasterMix Plus for SYBR® Green I No ROX | Eurogentec | Cat# RT-SN2X-03+NR |
| Ac-DEVD-AMC | Pepta Nova | Cat# 3171-V |
| Bio-Gel P-6 Micro Bio-Spin chromatography columns | Biorad | Cat# 732-6222 |
| plasmid purification columns | Qiagen | Cat# 28106, 28706, and 27106 |
| PCR colony screening kit | 5Prime | Cat# MasterTaq Kit |
| Q5 DNA polymerase | NEB | Cat# M0491 |
| Taq DNA polymerase | NEB | Cat# M0273 |
| Klenow fragment | NEB | Cat# M0212 |
| T4 Polynucleotide Kinase (PNK) | NEB | Cat# M0201 |
| T4 DNA ligase | NEB | Cat# M0202 |
| dNTPs | NEB | Cat# N0447 |
| ATP | NEB | Cat# P0756 |
| Restriction Enzymes, endonucleases | NEB | N/A |
| four-fragment Multisite Gateway Pro Plus system | Life Technologies | Cat# 12537-100 |
| EDTA-free Complete protease inhibitor cocktail tablets | Roche Diagnostics Belgium | Cat# 11873580001 |
| phosphatase inhibitor cocktail tablets | Roche Diagnostics Belgium | Cat# 04906837001, PhosSTOP |
| Luminata Classico Western HRP Chemiluminescence Detection Reagents | Millipore | Cat# WBLUC0500 |
| Experimental models: Cell lines | ||
| HeLa | ECACC | Cat# 93021013 |
| HEK 293T | ATCC | Cat# CRL-11268 |
| L929sAhFas | Generated in our unit | N/A |
| HT-29 | ATCC | Cat# ATCC-HTB-38 |
| inducible mRIPK3-CT26 | Generated in our unit | Aaes et al. (2016) |
| Fadd-/-Jurkat | Generated in our unit | N/A |
| Mlkl-/- L929sAhFas | Generated in our unit | N/A |
| inducible mMLKL(S345D)-L929sAhFas | Generated in our unit | N/A |
| Recombinant DNA | ||
| AKAR4.0 | This study | N/A |
| EKAR4.0 | This study | N/A |
| AKAR3-EV | Kindly provided by the Michiyuki Matsuda laboratory (Laboratory of Bioimaging and Cell Signaling, Graduate School of Biostudies, Kyoto University, Kyoto, Japan) | Komatsu, N., Aoki, K., Yamada, M., Yukinaga, H., Fujita, Y., Kamioka, Y., and Matsuda, M. (2011). Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656. |
| AKAR-EV (T>A) | This study | N/A |
| EKAR-EV | Kindly provided by the Michiyuki Matsuda laboratory (Laboratory of Bioimaging and Cell Signaling, Graduate School of Biostudies, Kyoto University, Kyoto, Japan) | Komatsu, N., Aoki, K., Yamada, M., Yukinaga, H., Fujita, Y., Kamioka, Y., and Matsuda, M. (2011). Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656. |
| EKAR-EV (T>A) | This study | N/A |
| EKAR2G-WT | A gift from Olivier Pertz | Addgene plasmid # 39813 ; http://n2t.net/addgene:39813 ; RRID:Addgene_39813 |
| EKAR2G-105/WT | A gift from Olivier Pertz | Addgene plasmid # 39818 ; http://n2t.net/addgene:39818 ; RRID:Addgene_39818 |
| EKAR-EV-VV | Previously developed | Vandame, P., Spriet, C., Riquet, F., Trinel, D., Cailliau-Maggio, K., and Bodart, J.-F. (2014). Optimization of ERK activity biosensors for both ratiometric and lifetime FRET measurements. Sensors (Basel). 14, 1140–1154. |
| pDG2-flag-rtTA3-puro-mRIPK3 | Previously generated in our unit | Aaes et al. (2016) |
| pDG2-flag-rtTA3-puro-mMLKL(S345D) | Generated in our unit | N/A |
| Software and algorithms | ||
| Statistical analyses | GraphPad Prism | PRISM 8 software, RRID: SCR_002798 |
| qPCR analysis | Biogazelle | qBasePLUS, RRID: SCR_003370 |
| FRET analysis (short experiment) | IGOR Pro - Wavemetrics | Ratioscope_8.31, DOI:10.25493/5G5V-HBC |
| FRET analysis (long experiment) | Matlab, MathWorks | CellTraFIC code, RRID: SCR_001622 |
| CellTraFIC (Cellular Tracking in FRET Image aCquisition) | APP Paris | IDDN.FR.001.230009.000.S.P. 2017.000.21000 |
| CellTraFIC (Cellular Tracking in FRET Image aCquisition) | GitHub | https://github.com/ayleray/CellTraFIC |
| Cell Death Analysis | Fiji | RRID: SCR_002285 |
| Immunofluorescence quantification | Cell Profiler | RRID: SCR_007358 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Franck. B. Riquet (franck.riquet@univ-lille.fr).
Materials availability
Plasmids generated in this study are available upon request to the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell lines
HeLa cells (ECACC 93021013), HEK 293T (ATCC CRL-11268), L929sAhFas, HT-29, and CT26 cells were cultured at 37°C under 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS, qualified, heat-inactivated, EU-approved, South America Origin), Na-Pyruvate (400 mM), 2 mM L-Glutamine and 100 U/mL Penicillin/Streptomycin (Gibco, Thermo Fisher Scientific). Jurkat FADD−/− were cultured similarly but in Roswell Park Memorial Institute (RPMI) medium. All cell lines were regularly tested against mycoplasma contamination. L929sAhFas (referred to as L929) were previously described (Vercammen et al., 1998). Mlkl−/- L929sAhFas cells were generated by CRISPR/Cas9. These deficient cells were transduced with pDG2-flag-rtTA3-puro-mMLKL(S345D), and clones were obtained by limited dilution. The inducible RIPK3 murine colon carcinoma CT26 was produced as described previously (Aaes et al., 2016). Briefly, CT26 cells were transduced with pDG2-flag-rtTA3-puro-mRIPK3, and clones were obtained by limited dilution. Both constructions were cloned into a Flag-containing, doxycycline-inducible pLenti6 derived vector, already described elsewhere (De Groote et al., 2016). Both inducible cell lines were stimulated with doxycycline (1 μg/mL, Dox) to induce necroptosis.
Methods details
Antibodies, cytokines, and reagents
Recombinant mouse TNFα (TNF) produced in our laboratory has a specific biological activity of 6,12x109 IU/mL and was used at 1000 IU/mL to stimulate L929 cells. Anti-Fas (hFasL, human, activating, clone CH11, #05-201) was purchased from Upstate and used at a final concentration of 500 ng/mL. Phorbol myristate acetate (PMA), MEK1/2 inhibitors U0126 and PD184352, Raf inhibitor, and RSK inhibitor (BI-D1870) were purchased from Sigma Aldrich, Forskolin (Fsk), MEK1/2 (Trametinib and Selumetinib), and ERK1/2 inhibitor (SCH772984) from Selleckchem. All MEK1/2 and ERK1/2 inhibitors were used at 20 μM unless indicated otherwise. RIPK1 inhibitor, necrostatin-1S (Nec1S) (UAMC-02197, synthesised by the Laboratory of Medicinal Chemistry; University of Antwerp) was used at 10 μM. Propidium iodide (PI) (Sigma Aldrich) and SytoxGreen (Thermo Fisher Scientific) were used at 3 μM and 5 μM, respectively. Doxycycline Hyclate (Dox, Sigma Aldrich) was used at 1 μg/mL. The following antibodies were used for western blot: anti-ERK1/2 (Abcam, ab17942, 1:1000), anti-β-actin (Santa Cruz Biotechnology, I-19, 1:1000), anti-RIPK1 (BD Biosciences, 610459, 1:1000), anti-β tubulin (Abcam, ab6046, 1:1000), anti- β-tubulin-HRP (Abcam, ab21058), anti-activated MAPK/ERK1&2 (diphosphorylated ERK1/2) antibody (clone MAPK-YT, Sigma Aldrich, M9692, 1:2000), anti-phosphoMEK1/2 (Cell Signalling, 9121), anti-MEK1/2 (Cell Signalling, 9122), anti-RIPK3 (Santa-Cruz, sc-374639), anti-MLKL (Merck Millipore, MABC604, Clone 3H1), anti-phospho-S345-MLKL (Merck Millipore, MABC1158, clone 7C6.1), anti-RSK1/2/3 (Cell Signalling, 32D7, 9355), anti-phspho-p90-RSK (Ser380) (Cell Signalling, 9341), anti-GSK3β (Cell Signalling, D5C5Z-XP, 12456) and anti-phospho-GSK-3α/β (Ser21/9) (Cell Signalling, 9331). HRP-conjugated secondary antibodies were anti-rabbit IgG, anti-mouse IgG, or anti-goat IgG (Santa Cruz Biotechnology, sc-2004, sc-2005, sc-2020 respectively, 1:10000).
DNA constructs
FRET biosensor plasmids AKAR3-EV and EKAR-EV were kindly provided by the Michiyuki Matsuda laboratory (Laboratory of Bioimaging and Cell Signaling, Graduate School of Biostudies, Kyoto University, Kyoto, Japan). EKAR2G-WT (mTFP wild type and Venus wild type) and EKAR2G-105/WT (mTFP 105 and Venus wild type) were a gift from Olivier Pertz (Addgene plasmid # 39813 ; http://n2t.net/addgene:39813 ; RRID:Addgene_39813 and Addgene plasmid # 39818 ; http://n2t.net/addgene:39818 ; RRID:Addgene_39818 respectively) (Fritz et al., 2013). t-EKAR-EV-VV was previously described (Vandame et al., 2014).
EKAR3-EV GW4.0 and AKAR3-EV GW4.0 were developed starting from the ERKAR3-EV and AKAR3-EV, respectively, using the four-fragment Multisite Gateway Pro Plus system (Life Technologies). Briefly, this system allows four separate FRET biosensor bricks to be simultaneously transferred to a Destination vector using Gateway BP and LR recombination reactions. The extended method is described in the Supplementary material. A schematic overview of the cloning strategy is presented in Figure S3B. Briefly, the destination vector was generated starting from the original biosensors. The sensing domain was excised and replaced by a reading frame (Rf) Gateway cassette. The pEntr vectors containing the fluorescent proteins and the PAABD + linker were generated by PCR amplification of the elements from the original vectors, followed by a BP recombination with the appropriate pDonr vectors. Specific sequences named attB were included in the design of the PCR primers. Concerning the substrate, an oligonucleotide cassette was inserted in the dedicated pEntr vector. Finally, to generate the pEx vector coding for the FRET biosensor, we performed a quadruple LR reaction containing the different pEntr vectors containing the donor and acceptor fluorescent proteins, the PAABD with its linker, and the substrate, according to the manufacturer’s recommendations.
The identity of all recombined vectors was verified by restriction digestion profiling or by PCR colony screening (MasterTaq Kit, 5Prime), and then validated by sequencing (Genoscreen, France). Restriction endonucleases, Q5 and Taq DNA polymerase, Klenow fragment, T4 Polynucleotide Kinase (PNK), T4 DNA ligase, dNTPs, ATP, and specific buffers were all purchased from New England Biolabs. All oligonucleotides were synthesized by Eurogentec (Belgium). Each complementary oligonucleotide designed to create double-stranded cassettes was 5’-phosphorylated by T4 PNK and then purified using Bio-Gel P-6 Micro Bio-Spin chromatography columns (#732-6222, Biorad). DNA fragments were purified on Qiagen plasmid purification columns (#28106, #28706, and # 27106, Qiagen).
Western blotting
After TNF stimulation, at specified time intervals, cells were washed twice in ice-cold PBS and scraped using ice-cold RIPA lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; 0.5% sodium deoxycholate; 1% Triton X-100 and 0.1% SDS) freshly supplemented with EDTA-free Complete protease inhibitor cocktail tablets (no. 11873580001) and phosphatase inhibitor cocktail tablets (PhosSTOP, no. 04906837001, Roche Diagnostics Belgium N.V.). Extracted proteins were separated on 12% SDS polyacrylamide gels or 4-12% Bis-Tris Plus Gels (ThermoFisher) and then transferred onto nitrocellulose membranes (Amersham Bioscience). Membranes were blocked using TBS with 0.05% Tween20 (TBS-T) containing 5% non-fat dry milk (Biorad) or in 2% BSA for phospho-antibodies. Luminata Classico Western HRP Chemiluminescence Detection Reagents (Millipore) were used for antibody detection.
Immunostaining
At specified time intervals upon TNF stimulation, L929 cells were fixed with 4% paraformaldehyde in PBS for 10 min. Afterward, cells were rinsed three times with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 min. Cells were incubated with a blocking solution composed of 2% FBS, 5% normal goat serum, and 2% BSA in PBS for 1h at room temperature and then further incubated with primary antibodies in blocking solution at 4°C overnight. The antibodies used were anti-MAPK activated (Sigma Aldrich, 1:500). After washing steps, cells were incubated in a blocking solution containing anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific, A-10667, 1:500) for 1h at room temperature in the dark. Finally, cells were incubated for 10 min in Hoechst 33258 (10 μg/mL), and then slides were mounted in ProLong Gold anti-fading reagent (Thermo Fisher Scientific, P36930). Immunofluorescence imaging was performed at room temperature with an inverted confocal Leica TCS SP5 X microscope (DMI6000, Leica Microsystems). A 40X/1.25NA oil immersion objective was used. Image size was 1024 x 1024 pixels, and the zoom factor was 1, for a pixel size of 0.3788 μm. Lasers source was a white light laser (Koheras). Image quantification was performed using CellProfiler.
Cell death analysis
Cell death was determined by measuring Sytox Green (5 μM, Life Technologies) and Ac-DEVD-AMC (20 μM, Pepta Nova) intensity using a FLUOstar Omega fluorescence plate reader (BMG Labtech) with excitation/emission filters of 485/520 nm and 360/460 nm, respectively. Triton X-100 at a final concentration of 0.1% was added at the end of time-lapse experiments for 1 h to obtain the maximal fluorescence intensity, corresponding to 100%. Cell death was also detected by monitoring Hoechst and PI fluorescence intensity using video-microscopy with excitation filters 395/25 and 575/25 and emission filters 440/40 and 632/60, respectively. Quantification was performed using Image J.
Live cell imaging
Cells were seeded on 8-well dishes ibiTreat (Ibidi) to reach 60% confluence at the time of transfection. For live-cell imaging, cells were cultured in Fluorobrite DMEM (Gibco, Thermo Fisher Scientific). Transfections were performed using JetPrime reagent (Polyplus) according to the manufacturer’s instructions. The medium was refreshed 4 h after transfection to reduce cytotoxicity. Cells were serum-deprived with a medium containing 1% FBS for 12 h before experiments began. For long-term experiments, Hoechst 33342 (1 μM) was added in order to track the cells. On the day of microscopy experiments, cells were placed directly inside the microscope stage chamber under controlled temperature, CO2, and humidity (top stage incubator, Okolab) 1 h before starting cellular imaging to equilibrate. Wide-field images were captured with a Nikon TiE inverted microscope with a 20X/0.5NA objective and a DS-Qi2 CMOS camera (Nikon). Images were acquired at intervals of 2 min (biosensor reference experiment) or 4 min (biosensor imaging) with the Nikon NIS-Elements acquisition software using JOBS module (Nikon). A Lumencor Spectra X LED Light Engine (Lumencor) provided the excitation light source. Ratio imaging used a 440/30 excitation filter, a t440/510/575rpc multi-band dichroic mirror, and two emission filters (ET480/40M (CFP) and AT545/30M (FRET)). Lumencor provided excitation filters, and all dichroic mirrors and emission filters were obtained from Chroma Technology. An automated emission filter wheel Lambda 10-B Smart Shutter (Sutter Instrument) was used.
Image data analysis and data representation
Analysis of the ERK1/2 and PKA biosensors comparison was performed with custom routines written in the IGOR Pro environment (Wavemetrics, available at https://doi.org/10.25493/5G5V-HBC). Long time-lapse FRET biosensing measurements in the context of cell death study were analyzed using custom Matlab (MathWorks, available at github.com/ayleray/CellTraFIC) routines for segmentation (with a standard connected components algorithm), tracking (by comparing as a function of time the Euclidean distance of the coordinates of the centroids of segmented cells) and quantification of ratiometric FRET data. Based on Hoechst staining for nuclear segmentation and cell tracking, we monitored ERK1/2 activity over time. FRET intensity was calculated as a ratio of the CFP/YFP signal for each pixel on background-subtracted images. Pseudocolor images represent both the ratio and the fluorescence intensity values, where warmer colors denote high FRET and cooler colors low FRET efficiency. The calibration bar shows the fluorescence intensity horizontally and the ratio values vertically. The micron-scale of the images corresponds to the size of the calibration bar.
We borrowed the semi-automated image analysis protocol previously described for enhanced FRET variations representation purpose and optimal reporting on both single-cell and population-level responses (Sparta et al., 2015). We displayed a kymographic representation of ERK1/2 activity shown as a color range, representative FRET pictures, and population-level trends. Kymographs represent a dynamic process on a single panel and correspond to a FRET ratio-time scan, where the FRET ratio values are plotted along the timeline for all images of the time-lapse stack. One line corresponds to one cell, and one column corresponds to one time point. These lines are then stacked along the x-axis (time) for all the cells (y-axis). Dead cells were detected by incubating cells using propidium iodide. When red fluorescence was detected in the red channel, image analysis was stopped. The cells were ordered from top to bottom based on their living time span.
The variations of ERK1/2 activity (pulses) were detected using a Matlab-based, semi-automated scoring routine. We first determined an experimental threshold (T=0,15) corresponding to the condition where little or no pulse of ERK1/2 activity should be detected, theoretically. Alternatively, results obtained with the mutated version of ERK1/2 biosensor (T>A) could also be used and processed for pulse analysis to define the threshold value. By systematically applying the threshold obtained following the ERK1/2 pulses analysis from the SCH772984 stimulated cell condition, we generated binary kymographs of ERK1/2 activity pulses over time for all remaining experimental conditions.
3D modeling & prediction of FRET efficiency changes
The different protein domains of the biosensor were modeled separately. A BLAST search against all Protein Data Bank (PDB) structures was performed for YPET, WW domain, ERK1/2 substrate, and eCFP. Multiple templates were selected for homology modeling with Modeller, and 10 models were generated for each protein domain. The best model was selected by comparing scores (molpdf, DOPE score, GA341 score) and Ramachandran plot assessment. Fusion Protein MODeller (FPMOD) was used to predict FRET efficiency changes by sampling conformational space through rigid body rotation using the pdb files of the modeled proteins as input and defining flexible linkers. Multiple conformations were calculated for the N-terminal part (YPET+WW domain) and the C-terminal part (ERK+eCFP) using different linkers.
Power spectral density (PSD) analysis
ERK1/2 peaks were detected with a standard algorithm for finding local maxima based on the derivative of the signal; the corresponding amplitude and time of each maximum were recorded. In order to facilitate the visualization, we represent only the mean in Figure 6. At low frequency (f<0.1mHz), the PSD curves decrease proportionally to 1/f2, corresponding to a Brownian noise (decrease of 20dB per decade) which is produced by Brownian motion (or random walk process). For high frequency, we observe that PSD curves decrease with a slope of 10 dB per decade, corresponding to the signature of pink noise (also called 1/f noise). This kind of noise occurs in numerous biological systems (e.g., brain or heart) because it originates from the ion channel’s opening-closing process.
Gene expression analysis
According to the manufacturer’s protocol, RNA was extracted from L929 cells using the RNeasy Mini Kit (Qiagen, 74104) and Qiashredder (Qiagen, 79654). The DNA-free™ DNA Removal Kit (Invitrogen, AM1906) was used to remove contaminating DNA and added DNase from the RNA samples. The first-strand cDNA of L929 cells was synthesized from total RNA using SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen, 18080-51). Plate-based real-time PCR amplification and detection of the synthesized cDNA was done on the Lightcycler 480 (Roche) with qPCR MasterMix Plus for SYBR® Green I No ROX (Eurogentec, RT-SN2X-03+NR) according to the manufacturer’s guidelines. mRNA levels of murine A20, IκB-α, Tnf, Mcp1, Il-6, Cxcl-1 were normalized to multiple reference genes (Actb, Cdc40, Htatsf) using qbase+ software and primers listed in Table S2.
Quantification and statistical analysis
Results are presented as means ± SEM unless indicated otherwise in figure captions. Statistical analyses were performed using PRISM 8 software (GraphPad). One-way and two-way ANOVA tests were performed, followed by Tukey’s post hoc test. Alpha was set at 0,05. For qPCR, statistical significance was determined using an unpaired t-test. Significance between samples is indicated as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant.
Acknowledgments
We thank the VIB BioImaging Core for training, support and access to the instrument park. This collaborative work was encouraged by the CNRS Groupement de recherche (GDR) “Imabio” scientific community via the biosensor workgroup initiative. FR acknowledges Nikon BELUX partnership. The authors would like to thank Anne Schepers for technical assistance with gene expression study, Davy Gaillot, and John Albeck for helpful discussions and for critical reading of the manuscript.
At the time of the project, FS was a postdoctoral fellow paid by Methusalem grant and FWO projects (BOF16/MET_V/007, FWO G.0607.13N, FWO G.0C37.14N) and BC held a Ph.D. fellowship from the FWO and was paid by FWO projects (FWO G.0C76.18N), both are a member of the Death Dynamics Team (DDT) in the Vandenabeele unit. Research in the Vandenabeele group is supported by EOS MODEL-IDI (FWO Grant 30826052), FWO research grants (G.0E04.16N, G.0C76.18N, G.0B71.18N, G.0B96.20N), Methusalem (BOF16/MET_V/007), iBOF20/IBF/039 ATLANTIS, Foundation against Cancer (F/2016/865, F/2020/1505), CRIG and GIGG consortia, and VIB. FR is a full associate professor at Lille University, a visiting research professor at Ghent University, and DDT leader. FR acknowledges funding from Oseo - Ministère de l'enseignement supérieur et de la recherche via the national contest 2013 d'aide à la création d'entreprise de technologies innovantes catégorie émergence in the context of the KiBioS spin-off project. This research was supported by the Agence Nationale pour la Recherche (ANR) via the G2Progress program (ANR-13- BSV2-0016-02) to FR.
Author contributions
FS, BC, EDS performed experiments, and JB made the L929 iMLKL(S345D) cell line. FS, BC, AL, EDS, GVC, PH, JB, WD, LH, PVi, PVa, and FR contributed reagents. FS, BC, AL, EDS, PH, PVi, and FR analyzed the data. AL and PVi developed FRET analysis software. FS, BC, and FR wrote the manuscript. PVa and PVi gave extensive feedback on the advanced version of the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103074.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
Custom computer code used to analyze FRET data set from long-term live-cell imaging experiments has been deposited at the APP (Agence pour la Protection des Programmes) under the following terms: CellTraFIC (Cellular Tracking in FRET Image aCquisition), IDDN.FR.001.230009.000.S.P.2017.000.21000 and on GitHub (https://github.com/ayleray/CellTraFIC). In addition, FRET data set from short-term live-cell imaging experiments has been analyzed using Ratioscope_8.31, running on IGOR Pro environment (DOI: 10.25493/5G5V-HBC.)
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Aaes T.L.L., Kaczmarek A., Delvaeye T., De Craene B., De Koker S., Heyndrickx L., Delrue I., Taminau J., Wiernicki B., De Groote P. Vaccination with necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep. 2016;15:274–287. doi: 10.1016/j.celrep.2016.03.037. [DOI] [PubMed] [Google Scholar]
- Albeck J.G., Mills G.B., Brugge J.S. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Kumagai Y., Sakurai A., Komatsu N., Fujita Y., Shionyu C., Matsuda M. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Aoki K., Kondo Y., Naoki H., Hiratsuka T., Itoh R.E., Matsuda M. Propagating wave of ERK activation orients collective cell migration. Dev. Cell. 2017;43:305–317.e5. doi: 10.1016/j.devcel.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Babst M., Odorizzi G., Estepa E.J., Emr S.D. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Balmanno K., Cook S.J. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16:368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- Cagnol S., Chambard J.-C.C. ERK and cell death: mechanisms of ERK-induced cell death - apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- Candeo A., Doccula F.G., Valentini G., Bassi A., Costa A. Light sheet fluorescence microscopy quantifies calcium oscillations in root hairs of Arabidopsis thaliana. Plant Cell Physiol. 2017;58:1161–1172. doi: 10.1093/pcp/pcx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Cho Y., McQuade T., Zhang H., Zhang J., Chan F.K.M. RIP1-dependent and independent effects of necrostatin-1 in necrosis and T cell activation. PLoS ONE. 2011;6:e23209. doi: 10.1371/journal.pone.0023209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A., Hitomi J., Germscheid M., Ch’en I.L., Korkina O., Teng X., Abbott D., Cuny G.D., Yuan C., Wagner G. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeautis C., Sipieter F., Roul J., Chapuis C., Padilla-Parra S., Riquet F.B., Tramier M. Multiplexing PKA and ERK1&2 kinases FRET biosensors in living cells using single excitation wavelength dual colour FLIM. Sci. Rep. 2017;7:41026. doi: 10.1038/srep41026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L., Takahashi N., Van Hauwermeiren F., Vandendriessche B., Goossens V., Vanden Berghe T., Declercq W., Libert C., Cauwels A., Vandenabeele P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Favata M.F., Horiuchi K.Y., Manos E.J., Daulerio A.J., Stradley D.A., Feeser W.S., Van Dyk D.E., Pitts W.J., Earl R.A., Hobbs F. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Fritz R.D., Letzelter M., Reimann A., Martin K., Fusco L., Ritsma L., Ponsioen B., Fluri E., Schulte-Merker S., van Rheenen J. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci. Signal. 2013;6:rs12. doi: 10.1126/scisignal.2004135. [DOI] [PubMed] [Google Scholar]
- Fulda S., Gorman A.M., Hori O., Samali A. Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010;2010:1–23. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Guy C., Olauson H., Becker J.U., Yang M., Fitzgerald P., Linkermann A., Green D.R. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell. 2017;169:286–300.e16. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote P., Grootjans S., Lippens S., Eichperger C., Leurs K., Kahr I., Tanghe G., Bruggeman I., De Schamphelaire W., Urwyler C. Generation of a new gateway-compatible inducible lentiviral vector platform allowing easy derivation of co-transduced cells. Biotechniques. 2016;60:252–259. doi: 10.2144/000114417. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T., Fujita Y., Naoki H., Aoki K., Kamioka Y., Matsuda M. Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. eLife. 2015;4:e05178. doi: 10.7554/eLife.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.K., Wu P.K., Park J.I. A cellular threshold for active ERK1/2 levels determines Raf/MEK/ERK-mediated growth arrest versus death responses. Cell. Signal. 2018;42:11–20. doi: 10.1016/j.cellsig.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.E., Toettcher J.E. Signaling dynamics control cell fate in the early drosophila embryo. Dev. Cell. 2019;48:361–370.e3. doi: 10.1016/j.devcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Danielson J.A.h, Manojkumar S.N., Lanquar V., Grossmann G., Frommer W.B. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife. 2014;3:e01741. doi: 10.7554/eLife.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno M., Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann. Med. 2006;38:200–211. doi: 10.1080/07853890600551037. [DOI] [PubMed] [Google Scholar]
- Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y., Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.H., Lin Y., Elowitz M.B. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen R.R., Vanhaesebroeck B. Cracking the context-specific PI3K signaling code. Sci. Signal. 2020;13:1–14. doi: 10.1126/scisignal.aay2940. [DOI] [PubMed] [Google Scholar]
- Martens S., Jeong M., Tonnus W., Feldmann F., Hofmans S., Goossens V., Takahashi N., Bräsen J.H., Lee E.W., Van der Veken P. Sorafenib tosylate inhibits directly necrosome complex formation and protects in mouse models of inflammation and tissue injury. Cell Death Dis. 2017;8:e2904. doi: 10.1038/cddis.2017.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattissek C., Teis D. The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis. Mol. Membr. Biol. 2014;31:111–119. doi: 10.3109/09687688.2014.894210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A., Carr D., Deng Y., Jin W., Black S. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- Murai S., Yamaguchi Y., Shirasaki Y., Yamagishi M., Shindo R., Hildebrand J.M., Miura R., Nakabayashi O., Totsuka M., Tomida T. A FRET biosensor for necroptosis uncovers two different modes of the release of DAMPs. Nat. Commun. 2018;9:1–17. doi: 10.1038/s41467-018-06985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muta Y., Fujita Y., Sumiyama K., Sakurai A., Taketo M.M., Chiba T., Seno H., Aoki K., Matsuda M., Imajo M. Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat. Commun. 2018;9:1–16. doi: 10.1038/s41467-018-04527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar M., Saleh D., Zelic M., Nogusa S., Shah S., Tai A., Finger J.N., Polykratis A., Gough P.J., Bertin J. RIPK1 and RIPK3 kinases promote cell-death-independent inflammation by toll-like receptor 4. Immunity. 2016;45:46–59. doi: 10.1016/j.immuni.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Murai S., Yamaguchi Y., Shirasaki Y., Nakabayashi O., Yamazaki S. Development of novel methods that monitor necroptosis and the release of DAMPs at the single cell resolution. Cell Stress. 2019;3:66–69. doi: 10.15698/cst2019.02.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerlemans M.I.F.J., Liu J., Arslan F., Den Ouden K., Van Middelaar B.J., Doevendans P.A., Sluijter J.P.G. Inhibition of RIP1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Res. Cardiol. 2012;107:270. doi: 10.1007/s00395-012-0270-8. [DOI] [PubMed] [Google Scholar]
- Ofengeim D., Ito Y., Najafov A., Zhang Y., Shan B., DeWitt J., Ye J., Zhang X., Chang A., Vakifahmetoglu-Norberg H. Activation of necroptosis in multiple sclerosis. Cell Rep. 2015;10:1836–1849. doi: 10.1016/j.celrep.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- Pham E., Chiang J., Li I., Shum W., Truong K. A computational tool for designing fret protein biosensors by rigid-body sampling of their conformational space. Structure. 2007;15:515–523. doi: 10.1016/j.str.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Purvis J.E., Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Malerød L., Pedersen N.M., Stenmark H. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 2008;314:801–813. doi: 10.1016/j.yexcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Regot S., Hughey J.J., Bajar B.T., Carrasco S., Covert M.W. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Chung M., Dobrzyński M., Fey D., Blum Y., Lee S.S., Peter M., Kholodenko B.N., Jeon N.L., Pertz O. Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 2015;11:838. doi: 10.15252/msb.20156458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Chung M., Dobrzyński M., Fey D., Blum Y., Lee S.S., Peter M., Kholodenko B.N., Jeon N.L., Pertz O. Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 2016;12:866. doi: 10.15252/msb.20166982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipieter F., Vandame P., Spriet C., Leray A., Vincent P., Trinel D., Bodart J.-F., Riquet F.B., Héliot L. From FRET imaging to practical methodology for kinase activity sensing in living cells. Prog. Mol. Biol. Transl. Sci. 2013;113:145–216. doi: 10.1016/B978-0-12-386932-6.00005-3. [DOI] [PubMed] [Google Scholar]
- Sparta B., Pargett M., Minguet M., Distor K., Bell G., Albeck J.G. Receptor level mechanisms are required for epidermal growth factor (EGF)-stimulated extracellular signal-regulated kinase (ERK) activity pulses. J. Biol. Chem. 2015;290:24784–24792. doi: 10.1074/jbc.M115.662247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wang H., Wang Z., He S., Chen S., Liao D., Wang L., Yan J., Liu W., Lei X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Duprez L., Grootjans S., Cauwels A., Nerinckx W., DuHadaway J.B., Goossens V., Roelandt R., Van Hauwermeiren F., Libert C. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 2012;3:e437. doi: 10.1038/cddis.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Vereecke L., Bertrand M.J.M., Duprez L., Berger S.B., Divert T., Gonçalves A., Sze M., Gilbert B., Kourula S. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature. 2014;513:95–99. doi: 10.1038/nature13706. [DOI] [PubMed] [Google Scholar]
- Thestrup T., Litzlbauer J., Bartholomäus I., Mues M., Russo L., Dana H., Kovalchuk Y., Liang Y., Kalamakis G., Laukat Y. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods. 2014;11:175–182. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- Tomas A., Futter C.E., Eden E.R. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida T., Takekawa M., Saito H. Oscillation of p38 activity controls efficient pro-inflammatory gene expression. Nat. Commun. 2015;6:8350. doi: 10.1038/ncomms9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandame P., Spriet C., Riquet F., Trinel D., Cailliau-Maggio K., Bodart J.-F. Optimization of ERK activity biosensors for both ratiometric and lifetime FRET measurements. Sensors (Basel) 2014;14:1140–1154. doi: 10.3390/s140101140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T., Vanlangenakker N., Parthoens E., Deckers W., Devos M., Festjens N., Guerin C.J., Brunk U.T., Declercq W., Vandenabeele P. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ. 2010;17:922–930. doi: 10.1038/cdd.2009.184. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P., Declercq W., Van Herreweghe F., Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal. 2010;3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P., Riquet F., Cappe B. Necroptosis: (last) message in a bubble. Immunity. 2017;47:1–3. doi: 10.1016/j.immuni.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Vanlangenakker N., Vanden Berghe T., Bogaert P., Laukens B., Zobel K., Deshayes K., Vucic D., Fulda S., Vandenabeele P., Bertrand M.J.M.M. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 2010;18:656–665. doi: 10.1038/cdd.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D., Beyaert R., Denecker G., Goossens V., Van Loo G., Declercq W., Grooten J., Fiers W., Vandenabeele P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach D., Kovalenko A., Kang T.B. Necrosome’-induced inflammation: must cells die for it? Trends Immunol. 2011;32:505–509. doi: 10.1016/j.it.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Wallach D., Kang T.-B., Dillon C.P., Green D.R. Programmed necrosis in inflammation: toward identification of the effector molecules. Science. 2016;352:aaf2154. doi: 10.1126/science.aaf2154. [DOI] [PubMed] [Google Scholar]
- Wilson M.Z., Ravindran P.T., Lim W.A., Toettcher J.E. Tracing information flow from erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell. 2017;67:757–769.e5. doi: 10.1016/j.molcel.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I., Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N., von Rechenberg M., Zaganjor E., Bauer A.J., Yang W.S., Fridman D.J., Wolpaw A.J., Smukste I., Peltier J.M., Boniface J.J. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Kovalenko A., Bogdanov K., Wallach D. MLKL, the protein that mediates necroptosis, also regulates endosomal trafficking and extracellular vesicle generation. Immunity. 2017;47:51–65.e7. doi: 10.1016/j.immuni.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Zhou X.X., Fan L.Z., Li P., Shen K., Lin M.Z. Optical control of cell signaling by single-chain photoswitchable kinases. Science. 2017;355:836–842. doi: 10.1126/science.aah3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Liang W., Ma Z., Xu D., Cao S., Lu X., Liu N., Shan B., Qian L., Yuan J. Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression article. Cell Death Dis. 2018;9:500. doi: 10.1038/s41419-018-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang S., Schnellmann R.G. A death-promoting role for extracellular signal-regulated kinase. J. Pharmacol. Exp. Ther. 2006;319:991–997. doi: 10.1124/jpet.106.107367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
Custom computer code used to analyze FRET data set from long-term live-cell imaging experiments has been deposited at the APP (Agence pour la Protection des Programmes) under the following terms: CellTraFIC (Cellular Tracking in FRET Image aCquisition), IDDN.FR.001.230009.000.S.P.2017.000.21000 and on GitHub (https://github.com/ayleray/CellTraFIC). In addition, FRET data set from short-term live-cell imaging experiments has been analyzed using Ratioscope_8.31, running on IGOR Pro environment (DOI: 10.25493/5G5V-HBC.)
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.