Summary
Transgenic reporters allow the measurement of regulatory DNA activity in vivo and consequently have long been useful tools for studying enhancers. Despite their utility, few studies have investigated the effects these reporters may have on the expression of other genes. Understanding these effects is required to accurately interpret reporter data and characterize gene regulatory mechanisms. By measuring the expression of Kruppel (Kr) enhancer reporters in live Drosophila embryos, we find reporters inhibit one another’s expression and that of a nearby endogenous gene. Using synthetic transcription factor (TF) binding site arrays, we present evidence that competition for TFs is partially responsible for the observed transcriptional inhibition. We develop a simple thermodynamic model that predicts competition of the measured magnitude specifically when TF binding is restricted to distinct nuclear subregions. Our findings underline an unexpected role of the non-homogenous nature of the nucleus in regulating gene expression.
Subject areas: Biological sciences, Molecular biology, Molecular interaction, Cell biology, Developmental biology, Mathematical biosciences
Graphical abstract

Highlights
-
•
Live tracking of transcription reveals competition between transgenic reporters
-
•
Transgenic reporters can also depress the expression of a neighboring gene
-
•
Expression inhibition is in part because of competition for transcription factors (TFs)
-
•
Competition is predicted with a model that restricts TFs to sub-nuclear “hubs”
Biological sciences; Molecular biology; Molecular interaction; Cell biology; Developmental biology; Mathematical biosciences
Introduction
An animal's ability to precisely control gene expression is dependent on the activity of enhancers. Through the binding of specific combinations of transcription factors (TFs), which can be either activating or repressive, enhancers are able to control the expression of their target genes in time and space. Enhancers control gene expression across all aspects of organismal functioning, from the immune system to the nervous system, and play a particularly important and well-studied role in the process of embryonic development (Levine, 2010; Shlyueva et al., 2014). During this period, enhancers regulate the expression of genes that determine critical cell fate decisions underlying patterning and organogenesis.
A significant amount of our understanding of enhancers and other cis-regulatory elements has come from the use of transgenic reporter lines. These transgenic animals express measurable reporters, such as fluorescent proteins or LacZ, under the control of cis-regulatory elements to enable observation of that element's activity in living organisms in different life stages, tissue types, or conditions (reviewed in Kvon, 2015; Wood, 1995). Studies of transgenic animals have enabled the discoveries of previously unknown enhancers, the modularity of enhancers, and the importance of the arrangement of TF binding sites or enhancer “grammar”, among others (Pennacchio et al., 2006; O'Kane and Gehring, 1987; Bier et al., 1989; Visel et al., 2009; Swanson et al., 2010).
Despite the remarkable utility of transgenic reporters, or perhaps in part because of it, little work has been done to look at the effect of these reporters on expression of other reporters or endogenous genes. Although reporters are exogenous regions of DNA that can originate from completely different species than the host animal, once integrated into the genome, these transgenes rely on the same pool of transcription factors, polymerases, and other molecular factors required for gene expression as endogenous genes. Given that most of these factors are present at relatively high copy numbers in the cell, for example, 250,000 Zld TF molecules per nucleus in the Drosophila embryo (Biggin, 2011; BNID 106849, Milo et al., 2009) or over 80,000 RNAP molecules per nucleus in human cells (Zhao et al., 2014; BNID 112321, Milo et al., 2009), it is commonly assumed that adding an additional enhancer would have little impact on the availability of key expression machinery. However, a couple of examples suggest that there may be competition between transgenic reporters and endogenous genes. A study by Laboulaye et al. measured the effect of three different transgenic reporters on endogenous gene expression in mice (Laboulaye et al., 2018). The authors found that the transgenic reporters all decreased the expression of the closest endogenous gene. Thompson & Gasson noted that endogenous protein levels may be slightly decreased in Saccharomyces cerevisiae and Lactococcus lactis expressing transgenic reporters, but the results were inconclusive (Thompson and Gasson, 2001). These examples suggest that transgenic reporters may decrease endogenous gene expression but leave open the questions of the mechanisms behind these decreases and whether such an effect is limited to certain organisms or reporters.
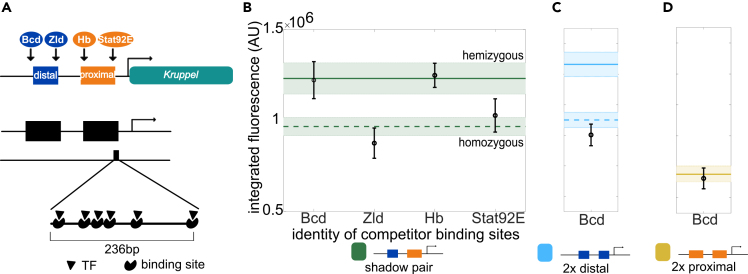
Like much of the field, we often used transgenic reporters under the assumption that they had no effect on the expression of other genes until we saw evidence to the contrary in our own data. In a study investigating gene expression noise in Drosophila embryos, we observed evidence of competition between identical copies of transcriptional reporters integrated on homologous chromosomes (Waymack et al., 2020). We were surprised to find that homozygous reporter embryos produced less mRNA per reporter allele than hemizygous embryos, with a reporter present on only one of the two homologous chromosomes (Figure 1). We suspected this could have important implications not only for the use of transgenic reporters, but also for our understanding of the balance between the supply of and demand for transcriptional machinery within the nucleus.
Figure 1.
Differences in mRNA production in homozygous and hemizygous embryos suggest competition between reporters
(A) This panel is adapted from Waymack et al. (Waymack et al., 2020). (Top) Kr expression in the early embryo is controlled by the activity of a pair of shadow enhancers, termed proximal and distal based on their location relative to the Kr promoter, that are each activated by different transcription factors (TFs). (Middle) The expression pattern driven by this pair of shadow enhancers is a stripe in the center 20% of the embryo. We use the MS2 system to image active transcription driven by enhancer reporters in living embryos. The cut out from the embryo shows a still frame of a movie where red circles are nuclei and green spots are sites of active transcription. To test whether transgenic reporters affect each other's expression, we generated embryos that are either homozygous or hemizygous for a particular reporter construct. (Bottom) Hemizygous embryos have the enhancer-MS2 reporter inserted on only one homologous chromosome and therefore display one transcriptional spot per nucleus. Homozygous embryos have the same enhancer-MS2 reporter inserted at the same location on both homologous chromosomes and therefore display two transcriptional spots per nucleus.
(B) mRNA production from homozygous reporter constructs compared to production from hemizygous constructs suggests competition between reporters. The graph shows total mRNA produced per allele in homozygous embryos as a function of total mRNA produced per allele in hemizygous embryos for the reporter construct indicated. The dashed diagonal line represents expected expression assuming independent activity of the two reporters in homozygous embryos. Points falling above this line display synergy in the activity of the two reporters in homozygotes; whereas, points falling below display competition. Error bars indicate 95% confidence intervals. Inset shows the percent higher expression in hemizygous versus homozygous embryos for each reporter construct. Error bars represent 95% confidence intervals from 1,000 rounds of bootstrapping.
(C) To rule out reporter competition being an artifact of our imaging system, we measured expression of the distal enhancer MS2 reporter in the presence of a non-transcribing distal enhancer on the homologous chromosome. The second distal enhancer is identical to the reporter, but lacks both a promoter and MS2 sequence. The graph shows expression driven by the distal enhancer reporter is significantly reduced in the presence of the non-transcribing distal enhancer (p value = 0.02, t test). The top horizontal line indicates expression driven by the distal enhancer reporter in the hemizygous configuration and bottom horizontal line indicates peak expression per allele driven in the homozygous configuration. Error bars and shading represent 95% confidence intervals. The number of embryos used for analysis of all constructs is recorded in Table S1. See also Figure S1.
Here we track the activity of multiple configurations of transgenic reporters in Drosophila embryos to assess the impact of these reporters on one another and endogenous genes. We measured live mRNA dynamics driven by the embryonic enhancers of the gap gene Kruppel (Kr) in the presence or absence of a second transcriptional reporter or a competitor TF binding array. We find that enhancer reporter expression is lower not only in the presence of a second reporter, but also in the presence of non-transcribing TF binding arrays, suggesting that there is competition for locally limited levels of certain TFs. This effect is not restricted to reporters; expression of a nearby endogenous gene is also decreased in transgenic embryos. To understand how the addition of the relatively small number of TF binding sites present in our constructs can measurably decrease reporter expression, we developed a thermodynamic model of our system. We predict reduced expression of the magnitude observed in transgenic embryos if we assume TF binding is restricted to so-called “hub” regions, but not if we assume TFs have access to the whole genome. This work reconciles the question of how tens of TF binding sites in a transgenic reporter construct can impact the available supply of tens of thousands of TF molecules. We suggest that the TF supply relevant to a particular enhancer is limited to a smaller pool of the TFs in a nucleus.
Results
Homozygous reporters display evidence of competition
To test whether transgenic reporters affect the expression of other alleles, we compared the expression output in embryos either homozygous or hemizygous for different reporter constructs. In the absence of reporter interactions, we will see the same levels of mRNA production per allele in hemizygous embryos and homozygous embryos. Conversely, if the reporters do affect one another's expression, then expression levels per allele will differ in hemizygous vs homozygous embryos, depending on the nature of this interaction. A synergistic interaction, perhaps through a mechanism such as increasing the local concentration of a key TF, would lead to higher levels of transcription in homozygous embryos than in hemizygous embryos (Figure 1B upper half). An antagonistic interaction, such as competition for a limited shared resource, would lead to lower levels of transcription in homozygous embryos than in hemizygous embryos (Figure 1B lower half).
To assess the nature of potential reporter interactions, we measured transcriptional output of different enhancers in living embryos using the MS2 reporter system. When transcribed, the MS2 sequence forms stem loops that are then bound by an MCP-GFP fusion protein expressed in the embryo, enabling us to visualize sites of nascent transcription (Figure 1A; Garcia et al., 2013). We can track these individual transcriptional spots across the time of nuclear cycle 14 (nc14), when these enhancers are most active, to measure total transcriptional output and dynamics. As a test case, we used different combinations of the two Kruppel (Kr) embryonic shadow enhancers, which we refer to as the proximal and distal enhancers. The Kr proximal and distal enhancers, individually or in combination, drive a stripe of expression in the central 20% of the embryo (Figure 1A). We generated transgenic flies with each individual enhancer, the shadow enhancer pair (which is the distal and proximal enhancer together, as in Waymack et al., 2020), or each enhancer duplicated in tandem driving an MS2 reporter (Figure 1B). Despite the similar pattern of expression driven by the two individual enhancers, the distal and proximal enhancers are each activated by different sets of TFs (Wunderlich et al., 2015). We previously showed that this separation of TF inputs plays an important role in suppressing gene expression noise (Waymack et al., 2020). Here, this separation of TF inputs allows us to investigate whether the reporter interactions we observe are influenced by specific regulatory factors or are more general consequences of having two reporters present.
In the majority of cases, hemizygous embryos produce more mRNA per allele than homozygous embryos (Figure 1B). To calculate the mRNA produced by each reporter, we integrate the area under the fluorescence traces of activity measured during nc14 at the anterior-posterior position in the embryo of peak expression (Figure S1). The single and duplicated distal constructs produce 62% and 40%, respectively, more mRNA per allele in hemizygous embryos than in homozygous embryos. The shadow pair and proximal enhancer reporters produce 27% and 22% more mRNA per allele at their respective regions of peak expression in hemizygous embryos than in homozygous embryos. The duplicated proximal construct drives the same level of expression in hemizygous and homozygous embryos. By comparing the competition exhibited by duplicated and single enhancers, we do not find evidence that longer reporter sequences drive stronger reporter competition (Figure 1B inset). We suspect this trend may arise because duplicated enhancers with a large array of similar binding sites can recruit a larger pool of TFs (Tsai et al., 2019) or because there can be synergy between the enhancers in promoter activation (Bothma et al., 2015). In sum, when two reporters are present in the same nucleus, in most cases neither transcribes to its full potential, suggesting that there is some form of competition between the two reporters. We hypothesized that the reporters are competing for one or more molecular factors required for reporter transcription or visualization.
Reporter competition is not an artifact of imaging system
To assess whether reporter competition is the result of a biological phenomena, such as limiting levels of a TF, or an artifact of our reporter system, such as limiting levels of MCP-GFP, we measured reporter output in the presence of a second non-transcribing transgenic construct. We produced a version of our distal enhancer construct that lacks both a promoter and the MS2 cassette. This construct therefore can bind the same regulatory TFs as the original distal construct but will not drive transcription. Therefore, it should not interact with promoter-bound factors, such as RNA polymerase, or the MCP-GFP coat protein. If the enhancer-only construct has no effect on reporter expression when present on the homologous chromosome, this suggests that the observed competition is only for MCP-GFP or is dependent on transcription. Instead, if we observe a decrease in reporter expression from the presence of the enhancer-only construct, this suggests that the observed competition is at least partially for one or more regulatory factors binding the enhancer.
In the presence of the distal enhancer-only construct, the distal enhancer reporter drives 11% lower levels of expression at its region of peak expression than in the hemizygous configuration (Figure 1C). Although significant (t test p value = 0.02), this decrease is not as large as the one we see when a second transcribing distal enhancer reporter is present on the homologous chromosome. We suspect the smaller effect of the non-transcribing distal enhancer construct is because of differences in the exact composition and levels of factors that are recruited to transcriptionally active versus inactive enhancers (Savic et al., 2015; Bozek and Gompel, 2020; Li et al., 2008). The observation that a non-transcribing enhancer construct can reduce reporter activity suggests that our reporters are competing for an endogenous factor bound by the enhancer sequence itself.
Reporters are competing for transcription factors
The above results suggest that reporter competition is for an enhancer-bound factor, and the degree of competition depends on the identity of the enhancer driving reporter expression (Figure 1B). The presence of a second identical reporter has a large effect on expression driven by the shadow pair construct and an even larger effect on the duplicated distal construct, although it has no significant effect on the duplicated proximal construct (Figure 1B). Because the Kr distal and proximal enhancers are regulated by separate sets of TFs (Wunderlich et al., 2015), we hypothesized that reporters may be competing for one or more of these TFs and that this may underlie the difference in competition levels between constructs. To test this hypothesis, we measured the effect of TF binding site arrays on the activity of the reporters. As the level of competition is not significantly different between the single and duplicated enhancer constructs, we focused on the two duplicated enhancers and the shadow pair, which are of similar lengths and therefore have similar numbers of TF binding sites. We created DNA sequences consisting of six strong TF binding sites for each of the key activating TFs of the Kr enhancers and inserted them into the identical site on the homologous chromosome, opposite to one of the enhancer-MS2 reporters (Figure 2A). Critically, these TF binding site arrays lack promoter and MS2 sequences. We reasoned that these TF binding site arrays would function to sequester TF molecules without affecting factors specifically involved in transcript production (such as RNAP) or reporter visualization (i.e., MCP-GFP). Therefore, any changes in transcriptional output by the enhancer-MS2 reporter observed in the presence of a TF binding site array should stem from decreased levels of available TF, not higher demand for basal transcriptional machinery or MCP-GFP.
Figure 2.
Competitor TF binding sites on the homologous chromosome decrease reporter activity
To test whether limiting levels of one or more activating TFs contribute to the reporter competition we observe, we measured the activity of our reporters in the presence of TF binding site arrays.
(A) (Top) The Kr shadow enhancers are activated by different sets of TFs. (Bottom) A schematic of TF binding site arrays that are intended to act as sinks for TF molecules. The arrays are each 236bp long, contain six binding sites for the indicated TF, and are inserted at the same genomic site as enhancer-MS2 reporters on the homologous chromosome. The binding site arrays do not contain a promoter or MS2 sequence.
(B) The activity of the shadow pair reporter is reduced in the presence of some TF binding site arrays. Graph shows the peak expression of the shadow pair in the presence of the indicated TF binding site array on the homologous chromosome. In (B–D), the horizontal solid line indicates the peak expression level in hemizygous embryos of the indicated reporter construct and the horizontal dashed line indicates the peak expression level per allele in homozygous embryos.
(C) The activity of the duplicated distal reporter is reduced to homozygous levels when the Bcd binding array is present on the homologous chromosome.
(D) Activity of the duplicated proximal reporter, which is not activated by Bcd, is not reduced when the Bcd binding array is present on the homologous chromosome. Note that the homozygous and hemizygous peak expression levels (dashed and solid horizontal lines) overlap for the duplicated proximal reporter. Error bars and shading in B-D indicate 95% confidence intervals. See also Figure S2.
Specifically, we created four binding site arrays corresponding to the four key TF activators of the shadow pair (Bicoid (Bcd), Hunchback (Hb), Stat92E, and Zelda (Zld); Figure 2A), which each contain six binding sites for the respective TF across 236bp. As the shadow pair is the only construct known to be regulated by all four TFs, we first assessed the impact of these binding site arrays on the activity of the shadow pair reporter. We find that the binding site arrays for the Zld and Stat92E each reduce the activity of the shadow pair down to the levels seen in homozygous embryos, whereas the Bcd and Hb binding site arrays do not have a significant effect on the shadow pair's activity (Figure 2B). We suspect that the Stat92E and Zld binding arrays may have the largest effect on the shadow pair's activity because of their essential roles in early gene activation (Harrison et al., 2011; Tsurumi et al., 2011).
As we observe a stark difference in the levels of competition in the duplicated distal versus duplicated proximal constructs, we asked whether the Bcd binding site array affects the expression of either construct. Bcd is a key activator of the distal enhancer, but not the proximal enhancer (Figure 2A). In line with this, the duplicated distal reporter's activity is reduced 37% compared to hemizygous levels at their regions of peak activity in the presence of the Bcd binding site array (Figure 2C), whereas the activity of the duplicated proximal enhancer is not significantly changed (Figure 2D). The large effect that the Bcd array (from here on called Bcd_6) has on the duplicated distal reporter is striking, as the TF binding site array is less than one-fifth the size of either Kr enhancer and contains only six, albeit strong, binding sites for Bcd. The specificity of the Bcd_6 array in reducing expression only of the Bcd-activated duplicated distal reporter, but not of the duplicated proximal reporter, suggests that the effect we observe is specific to sequestering Bcd molecules, and not a general effect of inserted DNA sequences. To eliminate the possibility that reporter competition is not for Bcd but instead a cofactor bound by Bcd, we compared the extent of reporter competition in regions of the embryo with high and low Bcd levels. We found that Bcd binding site arrays have a larger effect on reporter expression levels in regions of the embryo with lower Bcd levels (Figure S2), emphasizing that the distal and shadow pair enhancers are competing for Bcd itself.
Reporters show dosage-dependent response to increasing number of Bcd competitor sites
As a whole, these experiments suggest that limiting levels of TFs play an important role in reporter competition. When comparing the effects of the Bcd_6 array across constructs, the expression of the duplicated distal reporter is dramatically reduced in the presence of the array, whereas the expression of the duplicated proximal and shadow pair constructs are unaffected. The shadow pair is activated by a wider range of TF inputs beyond Bcd than the duplicated distal reporter. Since a larger number of unique TFs can activate the shadow pair than the duplicated distal reporter, we hypothesized that the activity of the shadow pair may be less sensitive to Bcd competition. This hypothesis is corroborated by our previous work that shows the shadow pair is less sensitive to fluctuations in Bcd levels than the distal enhancer (Waymack et al., 2020). To test this hypothesis, we attempted to sequester larger amounts of Bcd and measure the effect on the shadow pair's activity. We measured the activity of the shadow pair reporter in the presence of larger binding site arrays consisting of three (Bcd_18; 3 × 6 = 18 Bcd binding sites) or six (Bcd_36; 6 × 6 = 36 Bcd binding sites) copies of the original Bcd_6 binding site array.
In line with our hypothesis, we find that the shadow pair reporter activity decreases with increasing number of Bcd binding sites in the competitor array. Peak expression is reduced 1% in the presence of the Bcd_6 array, 21% with the Bcd_18 array, and 38% with the Bcd_36 array relative to expression in hemizygotes (Figure 3A). We also measured expression of the duplicated distal enhancer with the larger Bcd binding arrays and find a non-linear effect of increasing the number of competitor Bcd binding sites (Figure S3A; Discussion). To assess whether the reduction in mRNA output in the presence of the Bcd array is specific to the Kr enhancers or a general phenomenon, we measured the expression driven by the hunchback (hb) P2 Bcd-responsive enhancer in the presence or absence of the Bcd_6 and Bcd_36 arrays. Similar to our findings with the Kr enhancers, the Bcd binding arrays decrease the expression of the hb P2 enhancer (Figure 3B). Relative to hemizygous levels, peak expression of the hb P2 enhancer is decreased 32% with the Bcd_6array and 48% with the Bcd_36 array. We suspect that the relatively modest effect of larger numbers of Bcd competitor sites on reporter activity stems from an upper limit to the amount of Bcd molecules that can be effectively sequestered away from the enhancers at our binding site arrays (Wang et al., 2005; Lee and Maheshri, 2012).
Figure 3.
Competitor Bicoid binding sites decrease and shift the activity of shadow pair reporter
To assess whether limiting levels of the activating TF Bicoid (Bcd) cause the observed competition between reporters, we measured the transcriptional output of the shadow pair construct in the presence of Bcd binding site arrays of increasing length on the homologous chromosome. The Bcd_6 binding site array consists of six strong Bcd sites but lacks a promoter or MS2 cassette. The Bcd_18 and Bcd_36 binding site arrays are three and six repeats, respectively, of the Bcd_6 array and therefore contain a total of 18 and 36 strong Bcd sites, although both lack a promoter or MS2 cassette. These binding site arrays were inserted into the same location on Chromosome 2 as the enhancer reporters.
(A) Peak expression per allele driven by the shadow pair reporter decreases as the number of competitor Bcd binding sites increases. The horizontal lines mark the peak total expression per allele driven by the shadow pair reporter as hemizygotes (top solid line) or homozygotes (bottom dashed line). Shading and error bars indicate 95% confidence intervals.
(B) Competitor Bcd binding site arrays decrease the expression of an unrelated Bcd-responsive enhancer. To test if the effect of the Bcd binding arrays is specific to the Kr enhancers, we measured expression driven by the hunchback P2 (hbP2) enhancer, which is also activated by Bcd, in the presence of the Bcd_6 and Bcd_36 binding site arrays. The graph is the peak expression per allele driven by the hbP2 reporter in the presence of the indicated Bcd binding site arrays. The horizontal lines indicate the peak total expression driven by the hbP2 reporter as hemizygotes (solid line) or homozygotes (dashed line). Shading and error bars in A and B indicate 95% confidence intervals.
(C) (Left) Bcd is expressed in a gradient from the anterior of the embryo (0% egg length) to the posterior (100% egg length). The Kr expression domain is indicated by dashed vertical lines. Schematics above the embryo diagram show the Bcd_6, Bcd_18, or Bcd_36 arrays used with the enhancer reporters. (Right) Expression pattern driven by the shadow pair reporter constructs in the presence of increasing numbers of competitor Bcd binding sites. Graph shows the range of the expression pattern of each configuration to 50% of peak expression levels of the homozygous configuration, whose boundaries are indicated with dashed vertical lines. Error bars represent 95% confidence intervals found from 1,000 rounds of bootstrapping. See also Figure S3.
Because TFs can control both the level and pattern of enhancer activity, we measured how the expression boundaries of our reporters changed in response to the Bcd binding arrays. Bcd is expressed in a gradient from the anterior to the posterior of the embryo. Even though Bcd is present at high levels in the anterior of the embryo, Kr enhancers do not drive expression there because of repression by Giant (Gt), Knirps (Kni), and Hb, which can act as both an activator and a repressor (Jaeger, 2011; Vincent et al., 2018; Papatsenko and Levine, 2008). Therefore, we expected little effect on the anterior boundary by the Bcd binding site arrays. In contrast, because the posterior boundary is partially set by the decreasing levels of Bcd, if our Bcd arrays functionally reduce Bcd levels available for enhancer activation, we would expect to see a larger effect at the posterior boundary. We find that the posterior boundary of the shadow pair's expression domain moves towards the anterior in response to increasing number of competitor Bcd binding sites (Figure 3C). Relative to the homozygous configuration, the posterior border of shadow pair expression shifts anteriorly 2.5% of embryo length in the presence of the Bcd_18 array and 5% in the presence of the Bcd_36 array. Similar to peak expression levels, the Bcd_6 array does not change the expression boundaries of the shadow pair reporter. We see similar anterior shifts of the duplicated distal expression pattern with the Bcd binding arrays that qualitatively match the decrease in peak expression seen with each array (Figure S3B). We note that the anterior boundary shifts towards the posterior in the presence of the Bcd_18 and Bcd_36 arrays, which we suspect stems from the balance of activity between Bcd and the repressive TFs in this region (Kraut and Levine, 1991; Papatsenko and Levine, 2008; Small et al., 1991; Stanojevic et al., 1991).
Competition occurs at another genomic site and with an endogenous gene
Based on our findings thus far suggesting that reporter competition stems from competition for Bcd and other TFs, we reasoned that this competition should occur at other genomic insertion sites and with endogenous genes reliant on the same TFs. To first assess whether the observed reporter competition occurs at other genomic insertion sites, we measured the expression of the reporters in homozygous versus hemizygous configurations when inserted into a different chromosome (chromosome 3). Similar to our findings at the chromosome 2 insertion site (Figure 1), expression levels driven by the duplicated distal and shadow pair reporters are significantly lower in the presence of a second identical reporter on the homologous chromosome (Figures 4A and 4B). On chromosome 3, expression in homozygous embryos is 82% and 75% of expression in hemizygous embryos for the duplicated distal and shadow pair reporters, respectively. With both of these reporters, the degree of competition is consistent between the two insertion sites, indicating that the observed competition occurs independently of which pair of homologous chromosomes the reporters are inserted on (Figures 4A and B insets).
Figure 4.
Competition occurs at additional locations and genes
Based on our data suggesting that reporters are competing for limited levels of TFs, we suspected this competition would also occur at other transgenic insertion sites and with endogenous genes.
(A) Reporter competition occurs between homologous chromosomes at multiple genomic insertion sites. Graph shows the peak expression levels per allele of homozygous embryos as a function of the peak expression levels in hemizygous embryos for the shadow pair reporter inserted in either chromosome 2L or 3L. The data for chromosome 2 are the same as in Figure 1B. Diagonal line marks expected values for homozygous expression if reporters do not interact and instead display independent expression. Error bars represent 95% confidence intervals. Inset shows the peak expression levels in homozygous embryos relative to hemizygous embryos with the shadow pair reporter inserted on either chromosome 2L or chromosome 3L. Error bars in the inset represent 95% confidence intervals from 1,000 rounds of bootstrapping.
(B) The graph is as in A with the duplicated distal reporter inserted on chromosome 2 or chromosome 3.
(C) To determine the effect, if any, of transgene's use of resources on endogenous genes' expression, we compared the expression levels of three endogenous genes likely to be Bcd-regulated at increasing genetic distances from the transgenic insertion site in embryos with or without the duplicated distal transgene. Graph shows the percentage change in expression of Piezo, Mcr, and Bkt29A in embryos homozygous for the duplicated distal transgene compared to WT embryos as measured by qPCR. Error bars represent 95% confidence intervals and black circle indicates the mean. Schematic below graph shows the genetic distance of the three measured genes (indicated with a blue, red, or yellow vertical line) from the attP site (VK000002) on chromosome 2L (marked with green line and star) where all transgenic constructs, unless otherwise specified, were inserted. See also Figures S4, S5, S9, and S10.
Based on previous work in the mouse, we suspected that the reporter-induced competition would be limited to endogenous genes that are a short linear distance from the reporter insertion site (Laboulaye et al., 2018). To assess whether this is true, we measured the expression of three genes likely to be regulated by Bcd at varying linear distances from the chromosome 2 insertion site. We measured the expression of Piezo (22kb from insertion site), Mcr (58kb from insertion site), and Btk29A (160kb from insertion site) via qPCR in embryos with or without two copies of the duplicated distal transgene. All three of these genes are predicted to be regulated by Bcd, based on both their expression patterns and the observation of Bcd binding near these genes in the early embryo (Fisher et al., 2012; Hannon et al., 2017; Figure S4). In transgenic embryos, expression of the gene closest to the insertion site, Piezo, is significantly reduced to 60% of the levels seen in embryos of the same genetic background but lacking the transgene (Figure 4B). The expression levels of Mcr and Btk29A, which are further removed from the transgenic insertion site, are not significantly changed in transgenic embryos (Figure 4B). This potential distance-dependent effect of our transgene on endogenous gene expression is consistent with our finding that, in homozygous reporter embryos, there is more competition in nuclei in which the MS2 spots are physically closer together (Figure S5B).
A hub-based model of TF-enhancer interactions predicts TF competition
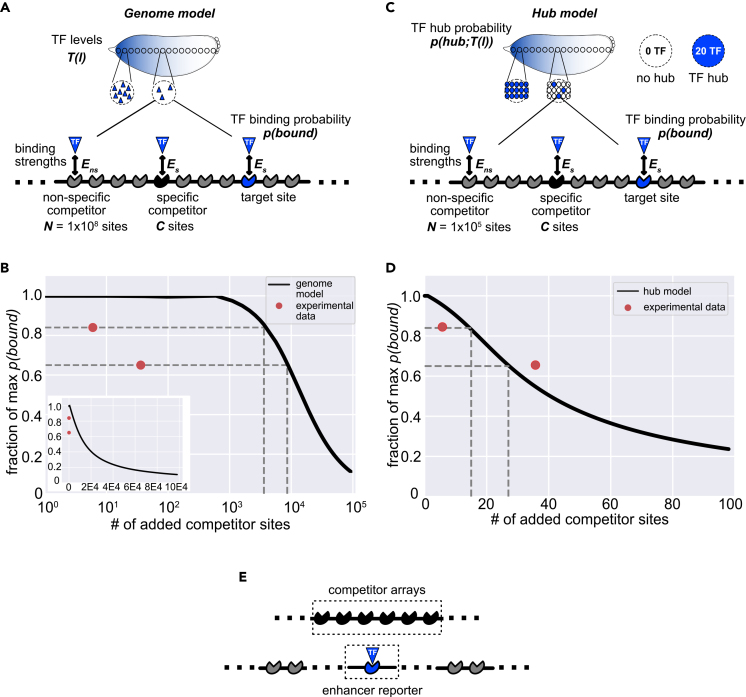
We were initially surprised to find that a reporter construct, with a length less than 0.001% of the genome, can have measurable effects on the expression of both other reporters and a nearby endogenous gene. Even more surprising is our finding that Bcd binding site arrays, which do not themselves drive any expression and have as few as six binding sites, also significantly reduce the expression of our Bcd-regulated enhancer reporters (Figures 2 and 3). This suggests that competition for Bcd can be induced by the addition of a relatively small number of binding sites, despite the fact that Bcd copy numbers vary between approximately 1,500 and 3,000 molecules per nucleus in the region of Kr expression (Biggin, 2011; Fowlkes et al., 2008). To better understand how the addition of a small sequence could induce competition for TFs, we developed a simple thermodynamic model of our system. The goal of our modeling effort is not to fit parameters such that the model precisely recapitulates our experimental data, but rather to see if our experimental observations are sensible by generating ballpark estimates of molecular competition using models that only rely on measured biophysical parameters.
Our model predicts the probability of a TF being bound to a target site, such as one of the binding sites that exist in an enhancer (Figure 5). For simplicity, we assume that TF binding at the target site is proportional to enhancer activity (Bintu et al., 2005; Phillips et al., 2019). In reality, enhancer activity depends on the combined occupancy of many TF binding sites (Levine, 2010; Kazemian et al., 2013). The simplifying assumption that enhancer activity is proportional to binding site occupancy allows us to avoid the need to test multiple models with different components, such as cooperative TF binding or activation behavior. In addition to the target site, TF molecules can bind to specific or non-specific competitor sites. Because most TFs have sequence-independent affinity for DNA (Slattery et al., 2014), the number of non-specific binding sites, N, is set to 1 × 108, roughly the size of the Drosophila melanogaster genome. The number of specific competitor sites, C, is varied. To maintain the simplicity of the model, the binding energy of specific competitor sites, Es, is equal to the binding energy of the target site, whereas the binding energy of all non-specific sites is represented as Ens. Because the specific binding energy is representing that of multiple binding sites, which may differ in their affinities, we vary the difference between specific and non-specific binding energies. Lastly, to allow for comparisons with our experimental data and to measure the effect of TF levels on binding, we vary the levels of our input TF T as a function of embryo length, l, in accordance with the measured Bcd gradient (Fowlkes et al., 2008). In this way, we can look at how the probability of TF binding to a single target site, p(bound; T(l)), changes as a function of number of specific competitor sites, binding strength relative to non-specific binding, and TF abundance.
| (Equation 1) |
Figure 5.
Modeling the impact of competitor binding sites on TF-enhancer binding
To understand how small transgenic sequences could induce the competition for TFs we observe, we created a thermodynamic model of TF binding at a single site as a function of TF levels, competitor sites, and binding strengths.
(A) Schematic of the parameters of the genome model where the whole genome is considered for TF binding. The probability of a TF molecule being bound at the target site, p(bound), is determined by the parameters shown. The number of available TFs, T, varies as a function of embryo position l to match the measured Bcd gradient. We assume that all TF molecules are bound and can be bound to either the target site or competitor sites, which are divided into specific and non-specific sites. The number of non-specific competitor sites, N, is held constant at 1 × 108 while the number of specific competitor sites, C, is varied. TF molecules bind the target site and specific competitor sites with binding energy Es, and bind non-specific competitor sites with binding energy Ens. Ens is held constant at zero and Es is varied. With each set of parameters, p(bound) is calculated using Equation 1 from the text.
(B) The fraction of maximum p(bound) as a function of the number of added specific competitor sites using the genome model. Es is held constant at 10 and l is held constant at 27% embryo length. Model predictions are in black. Experimental data of the fraction of maximum hemizygous hbP2 reporter expression as a function of the number of Bcd binding sites in the transgene on the homologous chromosome is shown in red. Data points indicate the fraction of maximum hbP2 expression with the Bcd_6 or Bcd_36 arrays measured at 27% egg length. Dashed lines indicate the number of additional competitor sites predicted by the genome model to be required to produce the experimentally observed decrease in expression. The inset shows the same data on a linear x axis.
(C) Schematic of the parameters of the hub model where TF binding is assumed to only occur within nuclear subregions. Each nucleus is divided into 1,000 equally-sized regions, one of which contains the target site. As in the genome model, the output of the model is the probability of a TF molecule being bound at the target site, p(bound). Based on previous measurements, the number of available TFs, T, is held constant at 20 for hub regions and 0 for non-hub regions (Mir et al., 2018). Instead, the probability that a region in a nucleus is a hub is a function of embryo position l to match the Bcd gradient and we call this probability p(hub; T(l)) (Equation 2 in the text). As in the genome model, we assume all TFs are bound at the target site, competitor sites, or non-specific sites. In each region, the number of non-specific competitor sites, N, is 100,000 while the number of specific competitor sites, C, varies. The binding strength parameters Es and Ens are the same as those used in the genome model. p(bound) is calculated as in the genome model using Equation 1 from the text and multiplying the resulting value by p(hub;T(l)). This product is the final p(bound) value.
(D) Results of the hub model. The graph is as in (B), with the fraction of maximum p(bound) as a function of the number of added competitor sites where the black line is the prediction of the hub model. As in B, Es is held at 10 and l is held constant at 27% egg length. Red points are the same experimental data as in (B) Dashed lines indicate the number of additional competitor sites predicted by the hub model to be needed to produce the experimentally observed decrease in expression.
(E) Model results can be compared to experimentally measured decreases in reporter expression. The schematic shows how our models relate to our experimental system. The target site in the models is analogous to the enhancer-MS2 reporters in our experimental system. The added specific competitor sites of the models represent the TF binding site arrays or second reporter introduced on the homologous chromosome opposite the enhancer reporter. Although the exact relationship is not known, TF binding at enhancers is related to enhancer activity so the p(bound) output of our models is related to our measured enhancer reporter activity. See also Figure S6.
As we vary the parameters, we find that p(bound; T(l)) changes in a qualitatively intuitive way. The probability of the target site being bound, p(bound; T(l)) decreases as a function of increasing competitor sites, decreasing difference in specific and non-specific binding strength, and decreasing TF levels (Figure S6). To test the accuracy of our model, we compared our experimental measurements of expression changes as a function of additional Bcd binding sites to predicted changes in p(bound; T(l)) as a function of added competitor sites. In our model, we assume there are 2000 specific competitor sites, based on experimental measurements of genome-wide Bcd binding in nc14 embryos (Hannon et al., 2017), and then add additional competitor sites to mimic the addition of a reporter of Bcd binding site array. Our findings are similar even if we do not assume these “background” sites exist (Figure S6). We recognize that the relationship between TF binding at an enhancer and gene transcription is complex (Grossman et al., 2017; Chen et al., 2020; Liu and Tjian, 2018) and do not expect our predicted p(bound; T(l)) values to exactly predict gene expression levels. Still, gene expression is dependent on TF binding (Mir et al., 2018; Shariati et al., 2019) and so our p(bound; T(l)) values provide useful ballpark estimates of how gene expression is expected to change as new competitor sites are introduced.
We compared our model predictions to the experimentally measured changes in activity of the hbP2 reporter. The hbP2 enhancer is a well-studied, Bcd-responsive enhancer and therefore makes a useful point of comparison for our model of Bcd binding (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989; Chen et al., 2012). We compared our experimental data and model predictions at one position in the embryo, 27% egg length, where the hbP2 enhancer drives peak levels of expression in homozygous embryos. This means we hold l, and consequently T, constant and therefore refer to our model output as p(bound) from here on. To observe the effect specifically of introducing new specific competitor binding sites, we also used experimental measurements to estimate the difference between Es and Ens and held this constant (see STAR Methods). In our experimental data, we see a 16% reduction in activity driven by the hbP2 reporter when the Bcd_6 array is present on the homologous chromosome. In contrast, our model predicts a 0.0003% decrease in p(bound) from the addition of 6 specific competitor sites. With this model, 3,600 competitor sites are needed to get a 16% reduction in p(bound) (Figure 5C). Similarly, although we observe a 35% decrease in hbP2 expression in the presence of the Bcd_36 array, which contains 36 Bcd binding sites, our model predicts only a 0.002% decrease in p(bound) with this number of added competitor sites. Based on model predictions, 8,600 specific competitor sites need to be added to achieve a 35% reduction in p(bound). Thus, a simple thermodynamic model of molecular competition produces estimates a couple of orders of magnitude different from experimental measurements.
We suspected that the large discrepancy between our measured decreases in reporter activity and our model's predictions of decrease in p(bound) are partially because of the model's assumption that any Bcd molecule in the nucleus can bind the target site. Growing evidence indicates that TFs and other pieces of the transcriptional machinery are not distributed evenly throughout the nucleus, but instead tend to cluster in regions of high density, called hubs, separated by low density regions (Tsai et al., 2019; Mir et al., 2018; Cho et al., 2018; Boehning et al., 2018; Tsai et al., 2017). This non-homogenous distribution seems functional, as transcription itself is also associated with these hubs (Tsai et al., 2019; Chong et al., 2018; Cho et al., 2018). Compared to the whole nucleus, hubs have a higher concentration of TFs and a lower number of specific and non-specific binding sites. We predicted that the addition of a small number of binding sites, similar to the numbers found in our reporter constructs, may have a sizable impact on p(bound) in the context of individual TF hubs.
To test this, we modified our previous model (genome model) to look at the probability of TF binding at the same target site, assuming all TF binding happens within hubs (hub model). In our hub model, we divide the nucleus into 1,000 hub-sized regions, based on the size of Drosophila embryonic nuclei and previous estimates of the distance between enhancers associated with the same TF hub (Tsai et al., 2019; see STAR Methods). Based on the measured distribution of distances between transcriptional spots in homozygous embryos, it is likely that reporters and TF binding site arrays transiently co-localize to the same hub-sized region (Figure S7). Binding times of TFs to specific DNA sites are short compared to the time scale of our imaging (<10s compared to 30s for each time point of our data; Swift and Coruzzi, 2017) so these transient co-localizations may be enough to induce competition between reporters. Within each region, we assume there are 100,000 non-specific binding sites, which is the number of non-specific sites in the genome model (1 × 108) divided by 1,000. The number of specific competitor sites is varied from 0 to 100. Based on previous measurements,the number of Bcd TFs present in a hub, Thub, is held constant at 20 molecules per hub, but the number of total Bcd molecules per nucleus, T(l), follows the Bcd gradient along the embryo (Mir et al., 2018). Regions that are not a hub are assumed to have 0 Bcd molecules. Consequently, the p(bound) value in our hub model is found by multiplying the p(bound) value calculated using the same formula as the genome model (Equation 1) by the probability that a given region is a TF hub (p(hub; T(l)); Equation 2). As in our genome model, we varied the difference between specific and non-specific binding energies.
| (Equation 2) |
In comparison to the genome model, the hub model shows far better agreement with our experimental data. As with the genome model, we assume that some specific competitor sites already exist and ask how p(bound) changes as additional specific competitor sites are added. In the hub model, we assume the 2000 specific competitor sites of the genome model are evenly distributed throughout the genome and consequently two specific competitor sites are present in each sub-region of the nucleus. We again focus on one position in the embryo, 27% egg length, and therefore hold T(l) constant. Experimentally, we see a 16% decrease in the activity of the hbP2 reporter with the addition of the Bcd_6 array. The hub model predicts a 5% decrease in p(bound) from the addition of the 6 Bcd binding sites in the Bcd_6 array (Figure 5D). Unlike the genome model, which requires 3,600 competitor sites for a 16% reduction in p(bound), this magnitude of reduction is achieved by 15 competitor sites in the hub model. With 36 competitor sites, the number of Bcd binding sites in the Bcd_36 array, the hub model predicts a 46% decrease in p(bound) compared to the 35% decrease in hbP2 reporter expression we measure in the presence of the Bcd_36 array.
Discussion
Since the discovery of enhancers 40 years ago (Banerji et al., 1981; Moreau et al., 1981), transgenic reporters have been invaluable tools to study the principles governing cis-regulatory regions. With a few exceptions, it has largely been assumed that transgenic reporters do not meaningfully affect the expression of other genes. Here we challenge this assumption and investigate the observed competition between transgenic transcriptional reporters in developing Drosophila embryos. Using reporters controlled by different configurations of the Kruppel shadow enhancers, we show that expression of a single reporter is decreased in the presence of a second identical reporter. We further show that this effect is not limited to transgenic reporters, but that the expression of a nearby endogenous gene is also decreased in transgenic embryos. Using non-transcribing arrays of TF binding sites, we find evidence that decreased reporter expression is due in part to decreased availability of key activating TFs of the Kr enhancers. Focusing on enhancer competition for the TF Bcd, we show that competitor Bcd binding arrays specifically affect the expression of Bcd-regulated enhancers, have a dosage-dependent effect on these enhancers, and shrink the width of the expression pattern of the enhancers. By developing a simple thermodynamic model, we predicted that the introduction of tens of additional Bcd binding sites can appreciably decrease gene expression, but only when TF binding is assumed to be limited to nuclear subregions.
Transgenic reporters can affect the expression of other reporters and an endogenous gene
Because of the widespread use of transgenic reporters, we were surprised to find that our small reporters reduce the expression of not only other reporters, but a nearby endogenous gene. A deeper search of literature revealed that Laboulaye et al., also found a distance-dependent decrease in endogenous gene expression in mice that is similar to our own results (Laboulaye et al., 2018). Although further systematic investigation is needed, these similar results in these distantly related organisms suggest that decreased endogenous gene expression may be a common consequence of transgenic reporters.
At first glance, our findings are also reminiscent of the transgene silencing previously reported in Drosophila (Kassis et al., 1991; Kassis, 1994; Pal-Bhadra et al., 2002). Pioneering studies found that flies containing multiple copies of a transgene showed reduced expression of the transgene as well as the corresponding endogenous gene (Pal-Bhadra et al., 1997; Pal-Bhadra et al., 1999). This silencing was shown to depend on Polycomb-mediated repression in the case of transgenes, and post-transcriptional RNAi mechanisms in the case of the endogenous gene (Pal-Bhadra et al., 2002). Although our findings share some similarities with transgene silencing, and may well rely on related mechanisms, numerous differences lead us to believe we are observing a distinct phenomenon. Unlike these previous studies, our transgenic flies, which contain a mini-white marker, do not show lighter eye color in homozygotes compared to hemizygotes (Figure S8). This suggests that overall expression from our transgenic insertion sites is not being ubiquitously repressed. If our observations were only the consequence of silencing mechanisms, triggered by increased amounts of transgenic DNA, we would expect to see larger reporter competition effects with our duplicated enhancer constructs compared to the single enhancer versions. Instead, for both the distal and proximal enhancers, we see a trend of larger decreases in homozygous expression levels compared to hemizygous expression levels with the single enhancer constructs (Figure 1B). Further, unlike the findings of Pal-Bhadra, our transgenic reporter decreases expression of an endogenous gene with which it does not share sequence homology (Pal-Bhadra et al., 2002).
Reporters and non-transcribed DNA sequences can induce competition for TFs
In addition to the studies described above, which describe how a transgene can alter the expression of genes, there are several studies that describe how the presence of non-transcribing pieces of DNA can alter expression. Work in flies and mouse cells showed that highly repetitive genomic sequences can alter gene expression, likely by binding and sequestering TF molecules away from their target genes (Liu et al., 2007; Janssen et al., 2000). In yeast, repetitive sequences of “decoy” tetO TF binding sites can change the relationship between tetO levels and the expression of a gene regulated by tetO (Lee and Maheshri, 2012), and individual competitor binding sites in bacteria also have a similar effect (Brewster et al., 2014). These studies underscore the regulatory importance of repetitive non-coding DNA sequences, which make up the majority of many genomes, by titrating available TF levels.
The repetitive sequences investigated in these studies are much longer (6Mb of major satellite DNA in mice, 7Mb of satellite V DNA in flies) than our transgenic constructs, which all contain less than 5kb of regulatory sequence and 10s of TF binding sites (Guenatri et al., 2004; Janssen et al., 2000). The large effects our transgenic constructs have on gene expression levels are therefore initially surprising. It is easier to imagine how very large, repetitive DNA sequences could sequester meaningful amounts of TFs than small sequences containing as few as six TF binding sites. In particular, this competition is surprising because we observe it even in the peak regions of the reporter expression patterns, where we expect activating TF levels to be high. For example, there are 250,000 molecules of the TF Zld per nucleus in the embryo (Biggin, 2011; BNID 106849, Milo et al., 2009), yet we see evidence of competition by introducing only six new strong Zld binding sites.
We suspect that the effect of our reporters and binding site arrays on expression levels partially stems from the non-uniform distribution of TFs in the nucleus. Although heterogeneity in the nucleus has been long observed with DNA (Nagele et al., 1999; Manuelidis, 1984), recent studies revealed that TFs and other pieces of the transcriptional machinery are also distributed unevenly throughout the nucleus (Tsai et al., 2019; Mir et al., 2018; Cho et al., 2018; Boehning et al., 2018; Tsai et al., 2017). There are several potential consequences of the organization of TFs into hubs. First, if our competitive binding arrays end up outside of a so-called TF “hub” with the enhancer reporter (or nearby endogenous genes), then the TF levels functionally available to the enhancer may be low enough to disrupt reporter activity. Second, if our binding arrays and reporters are found in the same hub, they may be competing for a fairly small pool of TFs. Previous measurements suggest there are roughly 20 Bcd molecules per hub (Mir et al., 2018). Lastly, the presence of a binding array may affect the properties of the hubs themselves. Another study showed that the deletion of TF-recruiting enhancers can decrease TF hub size and therefore lower gene expression (Tsai et al., 2019). In addition, Zld plays a key role in the formation of Bcd hubs (Mir et al., 2018), suggesting that our Zld binding site arrays may sequester both Zld and Bcd molecules.
Two aspects of our data support the hypothesis of local competition. First, reporters that spend more time in close physical proximity in the nucleus compete more than reporters that are further apart (Figure S5). Similarly, the endogenous gene Piezo, whose expression is decreased in the presence of the duplicated distal reporter, is within the same topologically associating domain (TAD) as the insertion site of the transgene during nc14 (Hug et al., 2017; Figure S9). This suggests that Piezo and the reporter likely inhabit the same nuclear subregion and have access to the same local pool of TFs.
Thermodynamic model of TF binding implicates TF hubs in competition
In line with our experimental data, our modeling results suggest that local competition for TFs is consistent with the observed decrease in expression levels. To rationalize how the addition of a small number of competitor TF binding sites could meaningfully decrease expression levels, we developed two simple thermodynamic models of TF binding. Our hub model, which assumes all TF binding is restricted to nuclear subregions, matches our experimental data more closely than the genome model, which assumes that all TF molecules have access to the whole genome. Our findings suggest an unexplored consequence of TF hubs. Previous studies have shown that TF hubs help to increase local concentration of TFs to increase gene expression (Tsai et al., 2019; Chong et al., 2018). Here, we show the flip side of this coin -- the non-uniform distribution of TFs can also induce competition among binding sites. We note that we have used only strong binding sites in our competitor arrays and plan to test the effect of binding arrays consisting of non-optimal TF binding sites, as enhancers containing sub-optimal binding sites have been shown to be important for establishing TF hubs (Tsai et al., 2019). There may be a balance between sequestering TFs, as we see here, and recruiting TFs to a local region that could depend on the affinity of the binding sites present.
Our goal in developing a simple model of our system was to generate ballpark predictions about the behavior of the system, using experimentally measured parameters and minimal assumptions. Although our hub model better matches our experimental findings than the genome model, we recognize that it is a simplification of reality and as such cannot fully describe our system. For example, we assume that existing specific competitor sites are evenly distributed throughout the genome, but in reality, chromatin accessibility and the clustering of TF binding sites in cis-regulatory regions alter the distribution of available binding sites (Berman et al., 2002; Li et al., 2011). The “true” number of specific competitor sites in any given sub-nuclear region will vary and consequently p(bound) of a given target site will depend on the surrounding sequences in the same region (Figure S10). With these simplifying assumptions, comes an incomplete ability to explain some experimental data. We find that expression levels driven by the duplicated distal reporter significantly decrease in the presence of the smallest and largest of our Bcd binding site arrays, but, unexpectedly, are not affected by the presence of the intermediate sized array (Figure S3). We do not fully understand this observation, but suspect that it has to do with the exact recruitment of TFs and other molecular factors mediated by this combination of DNA sequences.
Implications for transgenic reporters and underlying biology
Our work adds to the evidence that transgenic reporters can have measurable effects on endogenous gene expression (Laboulaye et al., 2018; Pal-Bhadra et al., 1997) and also builds on our understanding of the mechanisms behind this phenomenon. We note that our transgenic fly lines develop without any gross phenotypic defects in ideal laboratory conditions, making it tempting to assume that any effects of transgenic reporters are negligible. Although much about the mechanisms and effects of reporters on endogenous gene expression remains to be discovered, our findings provide some practical lessons for using transgenic reporters. First, investigators should use caution in interpreting changes in expression levels or patterns when comparing assays using one reporter to those using multiple reporters simultaneously. Controls must be used to determine whether changes in expression are because of additional reporters or the variable of interest. We find clear evidence that our reporters compete with one another when present in the same nucleus and as this seems to be mediated by competition for TFs, we suspect this finding is true beyond our specific reporters and system. In addition, potential effects of reporters on nearby endogenous gene expression should be considered in study design and data interpretation. In these cases, experiments that compare the effect of multiple transgenes with similar expression outputs may distinguish between the general impacts of inserting trangenes from the specific effect of a given transgene of interest. Our study again emphasizes the importance of taking into account the full genetic background of an organism to enable useful comparisons between studies (Chandler et al., 2014; Montagutelli, 2000).
Beyond the implications for the use of transgenic reporters, our findings suggest that the distribution of TF binding sites, both in the genome and in 3D space, is a potential tuning mechanism for dose-response relationships between TF levels and target genes. Previous studies in bacteria and yeast have shown that competitor TF binding sites can modulate the dose-response relationship of TF levels and gene expression, and that this modulation depends on the relative affinity of competitor versus gene-regulating TF binding sites (Brewster et al., 2014; Lee and Maheshri, 2012). This effect may generate an unappreciated selection pressure to either retain or eliminate TF binding sites that are not directly regulating a specific target gene. The observations of TF sequestration across a wide range of organisms suggest that this phenomenon is conserved and likely plays a functional role in regulating gene expression.
Limitations of the study
In this study, we showed that a second reporter as well as competitive binding arrays for key activating TFs decreases the activity of transcriptional reporters. Our findings strongly suggest decreased reporter activity is because of competition for these key TFs, but we did not simultaneously track subnuclear TF levels and reporter activity. Such experiments would allow us to observe whether competition is more prominent when genes are co-localized to the same TF hub and how the size or dynamics of TF hubs may be affected by the associated DNA. We also demonstrated that the expression levels of Bcd-regulated genes genetically close to our transgenic reporter insertion site are reduced in the presence of the duplicated distal reporter. This suggests that these endogenous genes can compete with the reporter for Bcd activation. This finding would be complemented by measuring the effect of the transgene on the expression levels of genes that are not regulated by Bcd, but the experiment was not possible at this genomic location because such genes were not located sufficiently close to the transgene insertion site. Lastly, our simplified thermodynamic models provide ballpark estimates of the degree of TF competition, and these models indicate our results are consistent with the organization of TF into hubs. However, we cannot rule out other unknown factors or mechanisms that may also explain our results.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: organisms/strains | ||
| D. melanogaster: y[1] w[1118]; PBac{y[+]-attP-3B}VK00002; + (Used for chromosome II insertions) | Bloomington Drosophila Stock Center | BDSC:9723 FLYB: FBti0076425 |
| D. melanogaster: yw; Kr distal - MS2; + (distal enhancer) | Waymack et al., 2020 | |
| D. melanogaster: yw; Kr proximal - MS2; + (proximal enhancer) | Waymack et al., 2020 | |
| D. melanogaster: yw; Kr duplicated distal - MS2; + (duplicated distal enhancer) | Waymack et al., 2020 | |
| D. melanogaster: yw; Kr duplicated proximal - MS2; + (duplicated proximal enhancer) | Waymack et al., 2020 | |
| D. melanogaster: yw; Kr shadow enhancer pair - MS2; + (shadow pair enhancer) | Waymack et al., 2020 | |
| D. melanogaster: yw; Kr distal; + (distal enhancer without promoter/MS2) | This study | |
| D. melanogaster: yw; Hb array; + (Hb binding site array) | This study | |
| D. melanogaster: yw; Zld array; + (Zld binding site array) | This study | |
| D. melanogaster: yw; Stat92E array; + (Stat92E binding site array) | This study | |
| D. melanogaster: yw; Bcd_6 array; + (Bcd_6 binding site array) | This study | |
| D. melanogaster: yw; Bcd_18 array; + (Bcd_18 binding site array) | This study | |
| D. melanogaster: yw; Bcd_36 array; + (Bcd_36 binding site array) | This study | |
| D. melanogaster: yw; Hb P2-MS2; + (Hb P2 enhancer) | Garcia et al., 2013 | |
| D. melanogaster: yw; +; Kr duplicated distal-MS2 (Duplicated distal enhancer on chromosome III) | This study | |
| D. melanogaster: yw; +; Kr shadow pair-MS2 (Shadow pair enhancer on chromosome III) | This study | |
| D. melanogaster: y[1] w[1118]; +; PBac{y[+]-attP-3B}VK00033 (Used for Chromosome III insertions) | Bloomington Drosophila Stock Center | BDSC:9750 FLYB: FBst0009750 |
| D. melanogaster: yw; His-RFP; MCP-GFP | Garcia et al. (2013) Curr Bio | |
| Oligonucleotides | ||
| Taqman Fam probe targeting RpII140 | Thermo Fisher | DM02134593_g1 |
| Taqman Fam probe targeting Piezo | Thermo Fisher | DM01803576_g1 |
| Taqman Fam probe targeting Mcr | Thermo Fisher | Dm01825813_g1 |
| Taqman Fam probe targeting Btk29A | Thermo Fisher | Dm01803642_g1 |
| Recombinant DNA | ||
| Plasmids used for generating transgenic fly lines | This study - available at https://benchling.com/zebabw/f/lib_co7oEHfC-tf-competition_public/ | |
| Software and algorithms | ||
| Matlab code for tracking MS2 transcriptional spots | Garcia et al. (2013) Curr Biohttps://github.com/GarciaLab/mRNADynamics | |
| Matlab code for analysis in Figures 1, 2, 3, and 4 | https://github.com/WunderlichLab/TFCompetitionCode | DOI: 10.5281/zenodo.5138138 |
| Python code for model in Figure 5 | https://github.com/WunderlichLab/TFCompetitionCode | DOI: 10.5281/zenodo.5138138 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zeba Wunderlich (zeba@bu.edu).
Materials availability
The plasmids and fly lines generated in this study are available upon request.
Experimental model and subject details
The experimental model used in this study is Drosophila melanogaster. All individuals used in this study were embryos imaged as described below during the first 3 hours of development. Adult flies were housed in cages at 25C and collected embryos were allowed to develop at room temperature. Embryo sex is not reported as it is not believed to affect the data reported here.
Generation of transgenic fly lines
Transgenic fly lines containing an enhancer-MS2 reporter were generated by phiC31-mediated insertion into the second or third chromosome, as described in Waymack et al., (2020). Unless otherwise indicated, all reporter constructs and TF binding site arrays were integrated into the same site on the second chromosome via phiC31-mediated integration. These constructs were injected into y[1] w[1118]; PBac{y[+]-attP-3B}VK00002 (BDRC stock #9723) embryos by BestGene Inc (Chino Hills, CA). For the reporter constructs inserted into chromosome 3, plasmids were injected into y[1] w[1118]; PBac{y[+]-attP-3B}VK00033 (BDRC stock #9750) embryos by BestGene Inc (Chino Hills, CA). The Kruppel enhancer reporters contained a single, duplicated, or shadow enhancer pair and the Kruppel promoter upstream of 24 MS2 repeats and a yellow reporter gene cloned into the pBphi vector (Garcia et al., 2013). These are the same enhancer-MS2 reporters as used and described in Waymack et al., (2020). The hunchback P2 enhancer reporter is that used in Garcia et al., (2013) and consists of the hunchback P2 enhancer and P2 promoter upstream of 24 MS2 repeats and a lacZ reporter (Garcia et al., 2013). TF binding site arrays were inserted into the same integration site on the second chromosome as described above. Exact genomic sequences used in each reporter construct are given in Data S1.
Hemizygous embryos were generated by crossing male flies homozygous for an enhancer-MS2 reporter to females expressing RFP-tagged histones and MCP-GFP (Garcia et al., 2013). Homozygous embryos were generated by crossing virgin females of the F1 hemizygous offspring just described with males homozygous for the same enhancer-MS2 reporter. Embryos with one copy of an enhancer reporter and one copy of a TF binding site array were generated by crossing the virgin female hemizygous offspring (i.e. containing one enhancer-MS2 reporter allele) with males homozygous for the corresponding TF binding site array.
Method details
Generation of TF binding site arrays
To generate our competitor binding site arrays for the four different TFs investigated, we started with the sequence of the hb P2 enhancer, which is well known to be Bcd responsive and contains six Bcd binding sites (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989; Chen et al., 2012). This 236bp sequence was our Bcd_6 array and the starting point for our other binding site arrays. To generate the Hb, Zld, and Stat92E arrays used we modified the six Bcd binding sites of the hb P2 enhancer to be the consensus motif for the corresponding TF (while retaining the same 236bp total length of the array) and had these sequences synthesized by Integrated DNA Technologies Inc (San Diego, CA). Previously defined consensus motifs were used for Zld (Xu et al., 2014), Hb (Stanojevic et al., 1991), and Stat92E (Yan et al., 1996). To generate the Bcd_18 and Bcd_36 arrays we performed Golden Gate assembly to ligate three or six copies of the Bcd_6 array together with 10bp random sequences between each repeat, to avoid potential repeat removal during transformations. All of the described TF binding site arrays were inserted into the same plasmid backbone, which was a modified version of the pBphi vector used for our enhancer-MS2 reporters, which lacks any enhancers, promoters, or MS2 sequence. We generated this vector by removing the Kr distal enhancer, Kr promoter, MS2 cassette, yellow sequence, and termination sequence from our distal MS2 reporter through digestion with NotI and XbaI. We then used Gibson assembly to ligate in the appropriate TF binding site array to this backbone. Sequences for the TF binding site arrays are provided in Data S1.
Embryo preparation and image acquisition
Living embryos were collected and dechorinated before being mounted onto a permeable membrane in halocarbon 27 oil and placed under a glass coverslip as in Garcia et al., (2013). Individual embryos were then imaged as described in Waymack et al., on a Nikon A1R point scanning confocal microscope using a 60X/1.4 N.A. oil immersion objective and laser settings of 40uW for 488nm and 35uW for 561nm (Waymack et al., 2020). To track transcription, 21 slice Z-stacks, at 0.5um steps, were taken throughout the length of nc14 at roughly 30s intervals. To identify the imaged position in the embryo, the whole embryo was imaged after nc14 prior to gastrulation at 20X using the same laser power settings. This whole embryo image was used to assign each transcription spot into one of 42 bins across the anterior-posterior (AP) axis of the embryo. The first bin corresponds to the anterior end of the embryo.
Measurement of transcriptional reporter activity
Tracking of nuclei and MCP-GFP bound MS2 transcriptional spots was done using the image analysis Matlab pipeline described in Garcia et al. (2013), which can be accessed at the Garcia lab Github (https://github.com/GarciaLab/mRNADynamics). Calling of transcriptional bursts to use for analysis was done as in Waymack et al. (2020). In short, transcriptional traces captured during nc14 consisting of at least three points were used for analysis. To measure total mRNA produced by all of our reporter configurations, we integrated the area under the curve of the transcriptional spot's fluorescence across the time of nc14 (Figure S1).
For every tracked spot of transcription, background fluorescence at each time point is estimated as the offset of fitting the 2D maximum projection of the Z-stack image centered around the transcriptional spot to a gaussian curve, using Matlab lsqnonlin. This background estimate is subtracted from the raw spot fluorescence intensity. The resulting fluorescence traces across the time of nc14 are then subject to smoothing by the LOWESS method with a span of 10%. The smoothed traces were used to measure transcriptional parameters and noise. Traces consisting of fewer than three time frames were removed from calculations. The area under each smoothed transcriptional trace is integrated using the Matlab trapz function, which gives the total integrated fluorescence value for that transcriptional spot. This integrated fluorescence is proportional to the number of transcripts produced by an enhancer reporter (Garcia et al., 2013; (Lammers et al., 2020). We group all transcriptional spots of a given reporter configuration by AP bin (position in the embryo) and calculate the average total integrated fluorescence value in each AP bin. For each reporter configuration we identify the AP bin with the highest average integrated fluorescence value as the region of peak expression. In the text, unless otherwise indicated, the integrated fluorescence or peak expression values correspond to the average integrated fluorescence value at this AP bin (Figure S1).
qRT-PCR to measure expression of endogenous genes in varying genetic backgrounds
Flies were allowed to lay eggs on molasses plates for 2.5 hours, so that most embryos collected were in nc14. Flies were either homozygous for the duplicated distal reporter on chromosome 2 or of the same genetic background (BDSC #9723) but did not have the transgene. The embryos collected from each plate were pooled and total RNA was extracted and purified using TRIzol (Thermo Fisher Scientific) and the Direct-zol RNA Miniprep kit (Zymo Research). cDNA was generated using SuperScript III RT Kit (Thermo Fisher Scientific). qPCR amplification was then done using the TaqMan Gene Expression Master Mix (Thermo Fisher Scientific). The data for each group, transgenic or non-transgenic, is from three separate biological replicates (i.e. the colored circles in Figure 4C are biological replicates), each done in technical triplicates. Relative RNA levels of each measured gene was calculated using the 2-ddC(t) method, using RpII140 as the reference gene. The TaqMan FAM probes used for each gene were DM01803576_g1 for Piezo, Dm01825813_g1 for Mcr, Dm01803642_g1 for Btk29A, and DM02134593_g1 for RpII140.
Description of the genome model of TF binding
We developed a simple thermodynamic model that looks at the probability of a TF molecule being bound at a single target site. For simplicity, we assume all TF molecules are bound either specifically or non-specifically. The probability of TF being bound at the target site, p(bound), is then determined by the number of TF molecules, T(l), the number of non-specific competitor binding sites, N, the number of specific competitor sites, C, and the difference in specific versus non-specific binding energies, Es and Ens respectively. The number of TF molecules, T(l), follows the Bcd gradient (Fowlkes et al., 2008) and is determined by position in the embryo, l, with a maximum value of 20,000 at the anterior tip of the modeled embryo (Biggin, 2011). For ease of comparison with our experimental data, we only consider binding probability at one position (l = 27% egg length) and thereby hold T constant, unless otherwise indicated. We hold the number of non-specific binding sites, N, constant at 1 × 108. Using statistical mechanics, we first enumerated the possible states of our system and their associated Boltzman weights (Bintu et al., 2005). In these states, x indicates the number of TFs that are bound at specific competitor sites.
| TF binding configuration | State | Statistical weight |
|---|---|---|
| Non-specific binding only | ||
| Competitor sites and non-specific binding | ||
| Target site, competitor sites and non-specific binding |
With all of the possible states of the system and the associated statistical weights, we can calculate p(bound) by dividing the statistical weight of the state with TF bound at the target site by the combined statistical weights of all possible states:
| (Equation 1) |
With this equation we can calculate the probability of a TF molecule being bound at the target site for a given number of TF molecules, specific competitor sites, and difference in binding affinity at specific vs non-specific sites. In the main text (Figure 5), we assume 2,000 background competitor sites already exist and span a range of 0–100,000 added specific competitor sites. To facilitate comparison with our experimental data, we looked at binding probability at one position in the embryo by holding l and consequently T constant. We focus on the effect of adding specific competitor binding sites and as such hold the difference between Ens and Es, and T(l) constant. Ens is held at 0kBT and Es is held at −10kBT, based on previously measured differences in specific vs non-specific DNA binding (Jung et al., 2018; Bintu et al., 2005). Specifically, the formula was applied to the measured binding affinities, from Jung et al., of TFs to their consensus sequence and highly mutated consensus sequences (Jung et al., 2018). T(l) is held at 5468, corresponding to 27% egg length in the modeled embryo. In Figure S6, we explore how p(bound) changes as a function of our other parameters (i.e. T(l), the difference between Ens and Es, or the number of background specific competitor sites).
Description of the hub model of TF binding
Calculation of p(bound) in the hub model is similar to the genome model but assumes all TF molecules are restricted to “hub” regions (sub-regions of the nucleus containing a high concentration of TFs) and therefore takes into account the probability that the nuclear sub-region containing the target site is a TF hub. In the hub model, each nucleus is divided into 1,000 equal-sized regions with a radius of 256nm. This estimate for the size of nuclear regions was based on the average distance between interacting loci found by Tsai et al. (Tsai et al., 2019), of approximately 360nm, the approximate volume of Drosophila embryonic nuclei of 70um3, and an estimation of the amount of DNA contained within a TF hub of the size seen in Tsai et al. The nuclear volume of 70um3 was reached by estimating the nucleus to be a sphere and using the formula V = 4/3πr3 with r = 2.5um (estimated from imaging data).
As the nucleus is divided into 1,000 hub regions, we set the number of non-specific binding sites, N, to 100,000, which is 1/1000th of the value in the genome model (108). For simplicity, we assume that DNA is distributed uniformly in the nucleus and as such the amount of DNA in each region is the same, which also allows us to have the same number of total non-specific binding sites per nucleus as in the genome model (105 x 1,000 hubs = 108). To maintain the same number of total specific competitor sites as the genome model, we assume 2 background specific competitor sites per region and vary the number of added specific competitor sites per region from 0 to 100. These values were reached by dividing the number of background or added specific competitor sites from the genome model by 1,000 (2,000/1,000 = 2 and 100,000/1,000 = 100). Based on the observation of Mir et al., (Mir et al., 2018), that the number of Bcd molecules per hub did not change along the Bcd gradient, we hold T constant at 20 if a region is a TF hub or 0 if it is not a TF hub. To account for this additional condition of whether the target site is within a TF hub or not, we calculate the probability that the region containing the target site is a TF hub:
| (Equation 2) |
where Thub is 20 TF molecules found in a hub and T(l) is the total number of TFs in the nucleus, as determined by the Bcd gradient (Fowlkes et al., 2008). To obtain the final p(bound) value from the hub model the p(hub) value of Equation 4 is multiplied by the value obtained using the above parameters in Equation 3.
Plotting p(bound) as a function of added specific competitor sites
To best simulate our experimental system, where we add transgenic constructs containing Bcd binding sites into a genome that already contains Bcd binding sites, we focused on how p(bound) changes as a function of added specific competitor sites. We therefore hold l, and consequently nuclear TF levels T, constant. Similarly, we hold constant the difference in binding energy between specific and non-specific sites by setting Es to −10 and Ens to 0. To account for specific Bcd binding sites that already exist in the Drosophila genome, we set the p(bound) value when a set number of “background” competitor sites exist as our reference maximum p(bound) value. We estimated the total number of true Bcd binding sites in nc14 embryos to be 2,000 based on the number of genome-wide Bcd ChIP-seq peaks reported by Hannon et al. (Hannon et al., 2017). Therefore for the genome model, the graph shown in Figure 5B depicts how p(bound) changes as a function of additional specific competitor sites beyond 2,000. For the hub model, we assume that these 2,000 Bcd binding sites are equally distributed throughout the genome and consequently there are two specific Bcd binding sites per nuclear region (2,000/1,000 = 2). The graph in Figure 5D shows how p(bound) changes as a function of additional specific competitor sites per nuclear region beyond the baseline two.
Quantification and statistical analysis
To determine statistical differences in levels of competition (Figures 1 and 4) and expression boundaries (Figures 3 and S3) between reporters we performed bootstrapping to estimate 95% confidence intervals. To do so, we randomly sampled with replacement the integrated fluorescence values of all of the transcriptional spots tracked in the AP bin of peak expression for both the hemizygous and homozygous configurations of the respective enhancer reporters. We averaged this value for the hemizygous configuration and for the homozygous configuration and then divided this average homozygous expression by the hemizygous expression to get our competition value (i.e. % hemizygous expression). This was done 1,000 times and each time the difference between the competition value found using the original real data set and that found using the randomly resampled data was calculated. We then took the top and bottom 2.5 percentiles of these differences as our upper and lower error bounds, respectively.
We estimated the error in expression boundaries in a similar fashion. We again perform 1,000 rounds of bootstrapping by randomly sampling, with replacement, the integrated fluorescence values from rows of transcriptional spots along the AP embryo axis for a given enhancer construct. Each column corresponds to a single AP bin in the embryo. We randomly sampled rows equal to the total number of rows in the original data set and using these found the anterior-most and posterior-most AP bins that produce greater than or equal to 50% of the maximum expression measured in the hemizygous configuration of that reporter. Empirical 95% confidence intervals were calculated as above by finding the 2.5th and 97.5th percentiles of the distribution of differences between the expression boundaries found using the original data and those found using each iteration of resampled data.
To determine whether the distal enhancer reporter produces significantly lower expression levels in the presence of a non-transcribing distal enhancer (Figure 1C) we performed a t-test comparing the integrated fluorescence values recorded in the region of peak expression of the two configurations.
The number of embryos and transcriptional spots per reporter construct used for all analyses are given in Table S1.
Acknowledgments
We thank Hernan Garcia for flies containing MCP-GFP and His-RFP transgenes, as well as for useful discussion of our observations. We thank Lily Li for helpful suggestions on the model and all members of the Wunderlich lab for feedback on the project. We also thank German Enciso, Anthony Long, Thomas Schilling, and Rahul Warrior for helpful feedback and suggestions on the project. This study was funded by an NIH grant (R01HD095246) to ZW.
Author contributions
RW: conceptualization, software, formal analysis, investigation, writing - original draft, writing - review & editing, visualization. MG: Investigation, resources, writing - review & editing. ZW: conceptualization, resources, writing - review & editing, supervision, funding acquisition.
Declaration of interests
The authors declare no competing interests.
Published: ▪▪ ▪▪, ▪▪
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103034.
Supplemental information
Table where each row corresponds to a transgenic reporter (defined in column 42) and columns 1–41 indicate AP bins across the embryo, reporting the number of transcriptional spots analyzed in each bin for a given construct. Column 43 records the number of embryos that were analyzed for each construct
Fasta file containing the sequences of all enhancer sequences used in reporters as well as the sequences of the TF binding site arrays.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at https://github.com/WunderlichLab/TFCompetitionCode and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Banerji J., Rusconi S., Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-X. [DOI] [PubMed] [Google Scholar]
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. U S A. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., Barbel S., Ackerman L., Carretto R., Uemura T., Grell E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/J.DEVCEL.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Bintu L., Buchler N.E., Garcia H.G., Gerland U., Hwa T., Kondev J., Phillips R. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 2005 doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P., Darzacq X. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018;25:833–840. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- Bothma J.P., Garcia H.G., Ng S., Perry M.W., Gregor T., Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife. 2015;4 doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek M., Gompel N. Developmental transcriptional enhancers: a subtle interplay between accessibility and activity. BioEssays. 2020;42:1900188. doi: 10.1002/bies.201900188. [DOI] [PubMed] [Google Scholar]
- Brewster R.C., Weinert F.M., Garcia H.G., Song D., Rydenfelt M., Phillips R. The transcription factor titration effect dictates level of gene expression. Cell. 2014;156:1312–1323. doi: 10.1016/j.cell.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler C., Chari S., Tack D., Dworkin I. Causes and consequences of genetic background effects illuminated by integrative genomic analysis. Genetics. 2014;196:1321–1336. doi: 10.1534/GENETICS.113.159426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Zheng R., Tokheim C., Dong X., Fan J., Wan C., Tang Q., Brown M., Liu J.S., Meyer C.A., Liu X.S. Determinants of transcription factor regulatory range. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-16106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xu Z., Mei C., Yu D., Small S. A system of repressor gradients spatially organizes the boundaries of bicoid-dependent target genes. Cell. 2012;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cisse I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science (New York, N.Y.) 2018;361:412. doi: 10.1126/SCIENCE.AAR4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Fisher, B., Weiszmann, R., Frise, E., Hammonds, A., Tomancak, P., Beaton, A., Berman, B., Quan, E., Shu, S., Lewis, S., et al (2012). BDGP Insitu Homepage
- Fowlkes C.C., Hendriks C.L.L., Keränen S.V., Weber G.H., Rübel O., Huang M.Y., Chatoor S., DePace A.H., Simirenko L., Henriquez C., Beaton A. A quantitative spatiotemporal atlas of gene expression in the drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Garcia H.G., Tikhonov M., Lin A., Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol. 2013;23:2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S.R., Zhang X., Wang L., Engreitz J., Melnikov A., Rogov P., Tewhey R., Isakova A., Deplancke B., Bernstein B.E., Mikkelsen T.S. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc. Natl. Acad. Sci. U S A. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon C.E., Blythe S.A., Wieschaus E.F. Concentration dependent chromatin states induced by the bicoid morphogen gradient. eLife. 2017;6 doi: 10.7554/eLife.28275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.M., Li X.-Y., Kaplan T., Botchan M.R., Eisen M.B. Zelda binding in the early drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C.B., Grimaldi A.G., Kruse K., Vaquerizas J.M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169:216–228.e19. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- Jaeger J. The gap gene network. Cell Mol. Life Sci. 2011;68:243–274. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen S., Durussel T., Laemmli U.K. Chromatin opening of DNA satellites by targeted sequence-specific drugs. Mol. Cell. 2000;6:999–1011. doi: 10.1016/S1097-2765(00)00099-X. [DOI] [PubMed] [Google Scholar]
- Jung C., Bandilla P., Von Reutern M., Schnepf M., Rieder S., Unnerstall U., Gaul U. True equilibrium measurement of transcription factor-DNA binding affinities using automated polarization microscopy. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-03977-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J.A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J.A., VanSickle E.P., Sensabaugh S.M. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemian M., Pham H., Wolfe S.A., Brodsky M.H., Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucl. Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R., Levine M. Spatial regulation of the gap gene giant during drosophila development. Development. 1991;111 doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- Kvon E.Z. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015 doi: 10.1016/j.ygeno.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Laboulaye M.A., Duan X., Qiao M., Whitney I.E., Sanes J.R. Mapping transgene insertion sites reveals complex interactions between mouse transgenes and neighboring endogenous genes. Front. Mol. Neurosci. 2018;11 doi: 10.3389/fnmol.2018.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers N.C., Galstyan V., Reimer A., Medin S.A., Wiggins C.H., Garcia H.G. Multimodal transcriptional control of pattern formation in embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:836–847. doi: 10.1073/pnas.1912500117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.H., Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol. Syst. Biol. 2012;8:576. doi: 10.1038/msb.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010 doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., MacArthur S., Bourgon R., Nix D., Pollard D.A., Iyer V.N., Hechmer A., Simirenko L., Stapleton M., Hendriks C.L.L., Chu H.C. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., Thomas S., Sabo P.J., Eisen M.B., Stamatoyannopoulos J.A., Biggin M.D. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu B., Szary J., Kofoed E.M., Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J. Biol. Chem. 2007;282:20868–20876. doi: 10.1074/jbc.M702547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Tjian R. Visualizing transcription factor dynamics in living cells. J. Cell Biol. 2018 doi: 10.1083/jcb.201710038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Different central nervous system cell types display distinct and nonrandom arrangements of satellite DNA sequences. Proc. Natl. Acad. Sci. U S A. 1984;81:3123–3127. doi: 10.1073/pnas.81.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., Jorgensen P., Moran U., Weber G., Springer M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2009;38:D750. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M., Stadler M.R., Ortiz S.A., Hannon C.E., Harrison M.M., Darzacq X., Eisen M.B. Dynamic multifactor hubs interact transiently with sites of active transcription in drosophila embryos. eLife. 2018;7 doi: 10.7554/eLife.40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli X. Effect of the genetic background on the phenotype of mouse mutations. J. Am. Soc. Nephrol. 2000;11:S101–S105. doi: 10.1681/ASN.V11SUPPL_2S101. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M.P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucl. Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele R.G., Freeman T., McMorrow L., Thomson Z., Kitson-Wind K., Lee H.y. Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J. Cell Sci. 1999;112:525–535. doi: 10.1242/jcs.112.4.525. [DOI] [PubMed] [Google Scholar]
- O’Kane C.J., Gehring W.J. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra U., Birchler J.A. Cosuppression in Drosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/S0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra U., Birchler J.A. Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell. 1999;99:35–46. doi: 10.1016/S0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra U., Birchler J.A. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 2002;9:315–327. doi: 10.1016/S1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Papatsenko D., Levine M.S. Dual regulation by the Hunchback gradient in the Drosophila embryo. Proc. Natl. Acad. Sci. U S A. 2008;105:2901–2906. doi: 10.1073/pnas.0711941105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Plajzer-Frick I. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Phillips R., Belliveau N.M., Chure G., Garcia H.G., Razo-Mejia M., Scholes C. Figure 1 theory meets figure 2 experiments in the study of gene expression. Annu. Rev. Biophys. 2019;48:121–163. doi: 10.1146/annurev-biophys-052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D., Roberts B.S., Carleton J.B., Partridge E.C., White M.A., Cohen B.A., Cooper G.M., Gertz J., Myers R.M. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/enhancer-binding protein beta binding sites. Genome Res. 2015;25:1791–1800. doi: 10.1101/gr.191593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati S.A., Dominguez A., Xie S., Wernig M., Qi L.S., Skotheim J.M. Reversible disruption of specific transcription factor-DNA interactions using CRISPR/Cas9. Mol. Cell. 2019;74:622–633.e4. doi: 10.1016/j.molcel.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordân R., Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014 doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Kraut R., Hoey T., Warrior R., Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Stanojevic D., Small S., Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Struhl G., Struhl K., Macdonald P.M. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Swanson C.I., Evans N.C., Barolo S. Structural rules and complex regulatory circuitry constrain expression of a notch- and EGFR-regulated eye enhancer. Dev. Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Coruzzi G. A matter of time - how transient transcription factor interactions create dynamic gene regulatory networks. Biochim. Biophys. Acta. 2017;1860:75. doi: 10.1016/J.BBAGRM.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A., Gasson M.J. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001;67:3434–3439. doi: 10.1128/AEM.67.8.3434-3439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A., Alves M.R.P., Crocker J. Multi-enhancer transcriptional hubs confer phenotypic robustness. eLife. 2019;8 doi: 10.7554/eLife.45325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A., Muthusamy A.K., Alves M.R.P., Lavis L.D., Singer R.H., Stern D.L., Crocker J. Nuclear microenvironments modulate transcription from low-affinity enhancers. eLife. 2017;6 doi: 10.7554/eLife.28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi A., Xia F., Li J., Larson K., LaFrance R., Li W.X. STAT is an essential activator of the zygotic genome in the early drosophila embryo. PLoS Genet. 2011;7:e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B.J., Staller M.V., Lopez-Rivera F., Bragdon M.D., Pym E.C., Biette K.M., Wunderlich Z., Harden T.T., Estrada J., DePace A.H. Hunchback is counter-repressed to regulate even-skipped stripe 2 expression in Drosophila embryos. PLoS Genet. 2018;14:e1007644. doi: 10.1371/journal.pgen.1007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Akiyama J.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A. Functional autonomy of distant-acting human enhancers. Genomics. 2009;93:509–513. doi: 10.1016/j.ygeno.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tegenfeldt J.O., Reisner W., Riehn R., Guan X.J., Guo L., Golding I., Cox E.C., Sturm J., Austin R.H. Single-molecule studies of repressor–DNA interactions show long-range interactions. Proc. Natl. Acad. Sci. U S A. 2005;102:9796–9801. doi: 10.1073/PNAS.0502917102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waymack R., Fletcher A., Enciso G., Wunderlich Z. Shadow enhancers can suppress input transcription factor noise through distinct regulatory logic. eLife. 2020;9:1–57. doi: 10.7554/ELIFE.59351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.V. Marker proteins for gene expression. Curr. Opin. Biotechnol. 1995;6:50–58. doi: 10.1016/0958-1669(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z., Bragdon M.D.J., Vincent B.J., White J.A., Estrada J., DePace A.H. Krüppel expression levels are maintained through compensatory evolution of shadow enhancers. Cell Rep. 2015;12:1740–1747. doi: 10.1016/j.celrep.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Chen H., Ling J., Yu D., Struffi P., Small S. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes and Development. 2014;28:608–621. doi: 10.1101/gad.234534.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Small S., Desplan C., Dearolf C.R., Darnell J.E. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/S0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Zhao Z.W., Roy R., Gebhardt J.C.M., Suter D.M., Chapman A.R., Xie X.S. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflectedlight-sheet superresolution microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:681–686. doi: 10.1073/pnas.1318496111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table where each row corresponds to a transgenic reporter (defined in column 42) and columns 1–41 indicate AP bins across the embryo, reporting the number of transcriptional spots analyzed in each bin for a given construct. Column 43 records the number of embryos that were analyzed for each construct
Fasta file containing the sequences of all enhancer sequences used in reporters as well as the sequences of the TF binding site arrays.
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at https://github.com/WunderlichLab/TFCompetitionCode and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.