Abstract
Background
One-third of patients with hormone receptor (HR)-positive breast cancers fail to respond to hormone therapy, and some patients even progress within two years of adjuvant endocrine therapy (ET) toward primary endocrine resistance. However, there is no effective way to predict endocrine resistance.
Objective
To build a model that incorporates the radiomic signature of pretreatment magnetic resonance imaging (MRI) with clinical information to predict endocrine resistance.
Methods
Clinical data of non-metastatic breast cancer patients diagnosed between May 1, 2015 and December 31, 2018 and preoperative dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) were retrospectively collected from three hospitals in China. The significant clinicopathological characteristics and radiomic signatures were included in multivariable logistic regression to establish a combined model to predict endocrine resistance in the training set, and validate the internal and external validation set.
Results
A total of 744 female non-metastatic breast cancer patients from three hospitals in China were included. In the training cohort, the AUC of the Radiomic-Clinical combined model to predict endocrine resistance was 0.975, which was higher than clinical model (0.849), IHC4 model (0.682) and similar as radiomic model (0.941). Also, the AUC of the combined model in the internal (0.921) and external validation cohort (0.955) were higher than clinical model and IHC4 model. The sensitivity of combined model was higher than radiomic alone, and got the best thresholding of the AUC.
Conclusion
This study developed and validated a pretreatment multiparametric MRI-based radiomic-clinical combined model and showed good performance in predicting endocrine resistance.
Keywords: Radiomics, Endocrine resistance, Hormone receptor-positive, Breast cancer
Highlights
-
•
This study first established a model to predict endocrine resistance in non-metastatic breast cancer based on radiomic.
-
•
This model was a combined model that contain multiparametric MRI radiomics features and clinical features.
-
•
The AUC of the combined model to predict endocrine resistance was 0.975 , with great potential in clinical applications.
Abbreviations
- AI
Artificial intelligence
- AUC
Area under the ROC curve
- DCE-MRI
Dynamic contrast-enhanced magnetic resonance imaging
- DWI-ADC
Diffusion-weighted imaging quantitatively measured apparent diffusion coefficient
- DCA
Decision curve analysis
- ER
Estrogen receptor
- ET
Endocrine therapy
- HR
Hormone receptor
- HER2
Human epidermal growth factor receptor 2
- IHC
Immunohistochemistry
- MRI
Magnetic resonance imaging
- MBC
Metastatic breast cancer
- OFS
Ovarian function suppression
- PR
Progesterone receptor
- RFS
Recurrence-free survival
- RS
Recurrence score
- ROR
Risk of recurrence
- RRSs
Radiomics risk scores
- ROC
Receiver operating characteristic
- T1+C
T1-weighted imaging
- T2WI
T2-weighted imaging
1. Introduction
Breast cancer has the highest incidence rate and the second highest mortality rate among women with malignant tumor around the world [1]. Molecular subtypes based on the expression of hormone receptor (HR), human epidermal growth factor receptor 2 (HER2), and Ki67 directly determine basic treatment strategies and indicate prognosis in patients with breast cancer [2,3]. About 70–80% breast cancer cases show positive HR expression and tend to be sensitive to endocrine therapy (ET). However, one-third of HR-positive breast cancer patients experience endocrine therapy failures [4], and some patients even progress within two years of adjuvant ET toward primary endocrine resistance. Once postoperative recurrence or even metastasis occurs, these patients’ 5-year survival will be less than 30% [1]. Therefore, methods to predict the efficacy of ET need to be explored in addition to the positive expression of HR.
Models that have been used to predict the risk of endocrine therapy include Oncotype DX21-gene and the PAM50 risk score in clinical practice [5,6]. Immune infiltration was assessed with hematoxylin & eosin (H&E)-stained histology sections, and immune scores were proven to be related to recurrence-free survival (RFS) [7]. With the development of artificial intelligence (AI), radiomics models to predict tumor-related prognosis or treatment responses have become noninvasive and efficient prediction technique. Radiomics risk scores (RRSs) based on CT have been constructed to predict the efficacy of CDK 4/6 inhibitors in HR-positive metastatic breast cancer (MBC) with a receiver operating characteristic (ROC) curve of 0.675 (N = 46) [8]. However, there is still no consensus on the radiomics method to be applied for assessing ET response.
In this study, we built a model incorporating the radiomics signature of pretreatment magnetic resonance imaging (MRI) with clinical information and assessed the predictive ability.
2. Method
2.1. Study population
Clinical data and preoperative DCE-MRI from a total of 744 non-MBC patients, diagnosed between May 1, 2015 and December 31, 2018 were retrospectively collected from three hospitals in China. The study was approved by Sun Yat-sen Memorial Hospital Ethics Committee and registered with ClinicalTrials.gov (NCT04003558), Chinese Clinical Trail Registry (ChiCTR1900024020). Informed consent from the study participants was exempted as it was a retrospective study. Clinical and histopathologic data were obtained from the medical records. Clinicopathological characteristics of the patients with breast cancer included age, tumor type, estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 expression, Ki-67 proliferation index, histological grade, T stage, N stage, and ET.
The inclusion criteria were as follows: (a) non-metastatic female breast cancer patients; (b) patients underwent standardized breast cancer treatment; (c) determined by IHC as ER-positive, or/and PR-positive for breast cancer. ER or PR positive status was defined as ≥1% of the tumor cells were immunoreactive; (d) patients with complete clinicopathological characteristics, follow-up, and pre-treatment MRI data.
In this study, the definition of primary endocrine therapy resistance was that breast cancer patients progressed within two years of adjuvant ET [9]. Follow-up data were censored on January 31, 2021.
2.2. Radiomic feature extraction
The radiomic workflow and MRI protocol were introduced in our first study [10]. Among the various software programs used in this study, the open-access software 3D Slicer was the main one and the license was obtained for its availability for use. N4ITK Bias Correction code of 3D Slicer version 4.10.2 was applied to normalize the whole DCE-MRIs from 3 hospitals. Radiologists who engaged in breast MRI imaging for more than five years reviewed all the preoperative DCE-MRI and semi-automatically segmented and delineated the 3D regions of interest (ROIs) of the breast tumors. All radiologists were blinded to the patients’ clinical outcomes. When there were multiple lesions, the most obvious and malignant lesion was selected. After the ROIs of the ALNs and tumors were reconstructed and segmented, the volume of interest (VOI) images (according to the Digital Imaging and Communications in Medicine [DICOM] format) were transferred to the Slicer Radiomics code. Then, an in-house texture extraction platform was developed based on the Python package PyRadiomics [11]. The voxel-based features included shape, first-order, gray-level cooccurrence matrix, gray-level size zone matrix, gray-level dependence matrix, and neighboring gray tone difference matrix. More details about the radiomic feature extraction were provided in our earlier study [10]. Eventually, 2589 radiomics features were extracted from preoperative DCE-MRI, including contrast-enhanced T1-weighted imaging (T1+C), T2-weighted imaging (T2WI), and diffusion-weighted imaging quantitatively measured apparent diffusion coefficient (DWI-ADC) imaging.
2.3. Development of radiomic signature
The random forest algorithm was used to select the most predictive 30 radiomic features for each radiomic sequence (T1+C sequence, T2WI sequence, and DWI-ADC sequence,see Supplemental Fig. 1A–1C). The number of features selected by the random forest algorithm and the optimal parameters were determined empirically based on the performance in the training cohort [12]. After normalization processing, multivariable logistic regression was used to combine the T1+C, T2WI, and DWI-ADC imaging signatures of the breast tumors for endocrine resistance prediction.
2.4. Calculation of immunohistochemistry (IHC)4 score
The IHC4 score was used according to the following algorithm: IHC4 = 94.7 × {-0.100ER10–0.079PR10 + 0.586HER2 + 0.240 ln (1 + 10 × Ki-67)} [13]. ER was quantified using the H-score. The variable ER10 was obtained by dividing the H-score by 30 to obtain a variable with a range of 0–10. PR was scored as the percentage of cells with positive staining. The positive cut-off was 10%. The variable PR10 was obtained by dividing the variable PR by 10 to obtain a variable between 0 and 10. HER2 was scored when the IHC score for HER2 was 3+ or 2+ and confirmed positive by FISH. Ki-67 was scored as the percentage of malignant cells showing positive staining [13].
2.5. Statistical analysis
Data were presented as frequency and percentage for categorical variables. The distribution of clinicopathological characteristics was presented in the three cohorts: training cohort, internal validation cohort and external validation cohort. Univariable analysis (χ2 test or Fisher's exact test) was performed to determine the associations of clinicopathological characteristics with endocrine resistance in the training cohort. Significant variables were then included in multivariable logistic regression analysis for predicting endocrine resistance.
The significant radiomic signatures and clinicopathological characteristics were included in the multivariable logistic regression to establish a combined model to endocrine resistance and presented as a nomogram in the training set. Then, the internal and external data sets were used to validate the combined model and compared with the IHC4 model using ROC curve analysis and by calculating the AUC. Additionally, the decision curve analysis was used to calculate the net benefits for a range of threshold probabilities to estimate the predictive models’ clinical utility. The statistical descriptions and analyses were performed using R software version 3.6.1, the license for which was obtained under any requirement for permission for use. All statistical tests were two-sided, with statistical significance defined as having a P-value < 0.05.
3. Result
3.1. Clinical characteristics
In total, 744 newly diagnosed female non-metastatic breast cancer patients from three hospitals in China (median age: 48 years; range: 24–74 years) were included in this study: 593 patients from Sun Yat-sen Memorial Hospital of Sun Yat-sen University, 87 patients from Shunde Hospital of Southern Medical University, and 64 patients from Tungwah Hospital of Sun Yat-sen University. In Sun Yat-sen Memorial Hospital, 433 breast cancer patients were used as the training cohort, and 160 breast cancer patients who were enrolled from an ongoing clinical trial were used as the internal validation cohort. The external validation cohort comprised 151 breast cancer patients from Shunde Hospital and Tungwah Hospital. The three cohorts’ clinical characteristics are provided in Table 1. Patient characteristics included age, histological grade, pathologic types, ER group, PR group, HER2 status, stage, T stage, N stage, drug used in chemotherapy and ET, Ovarian function suppression (OFS), and endocrine resistance. The rate of endocrine resistance was no significant differences among the three cohorts (P = 0.391), which were 4.4% (19/433) in the training cohort, 6.9% (11/160) in the internal validation cohort, and 4.0% (6/151) in the external validation cohort. There were no significant differences in histological grade, ER group, and HER2 status between the three cohorts. There were some variations regarding tumor and patient characteristics (e.g., age, PR, Ki67, stage, pathologic types, surgery, chemotherapy, ET, as presented in Table 1) between the different cohorts.
Table 1.
Clinicopathologic Characteristics and Endocrine Resistance status of patients in training, internal and external validation cohorts.
| Characteristics | Training cohort N = 433 (%) |
Internal validation cohort N = 160 (%) |
External validation cohort N = 151 (%) |
Total N = 744 (%) |
P value |
|---|---|---|---|---|---|
| age | <0.001 | ||||
| ≤35 | 27(6.2) | 29(18.1) | 8(5.3) | 64(8.6) | |
| 35–50 | 203(47.0) | 80(50.0) | 76(50.3) | 359(48.3) | |
| ≥50 | 202(46.8) | 51(31.9) | 67(44.4) | 320(43.1) | |
| ER∗ | 0.842 | ||||
| Low | 85(19.6) | 28(17.5) | 29(19.2) | 142(19.1) | |
| High | 348(80.4) | 132(82.5) | 122(80.8) | 602(80.9) | |
| PR+ | <0.001 | ||||
| Low | 188(43.4) | 95(59.4) | 51(33.8) | 334(44.9) | |
| High | 245(56.6) | 65(40.6) | 100(66.2) | 410(55.1) | |
| HER2∗∗ | 0.067 | ||||
| Negative | 311(71.8) | 99(61.9) | 104(68.9) | 514(69.1) | |
| Positive | 122(28.2) | 61(38.1) | 47(31.1) | 230(30.9) | |
| Ki-67++ | <0.001 | ||||
| Negative | 72(16.6) | 71(44.4) | 53(36.6) | 196(26.6) | |
| Positive | 361(83.4) | 89(55.6) | 92(63.4) | 542(73.4) | |
| Stage | <0.001 | ||||
| I | 84(19.4) | 5(3.1) | 37(24.5) | 126(16.9) | |
| II | 287(66.3) | 109(68.1) | 76(50.3) | 472(63.4) | |
| III | 62(14.3) | 46(28.8) | 38(25.2) | 146(19.6) | |
| T stage | <0.001 | ||||
| T1 | 95(21.9) | 5(3.1) | 59(39.1) | 159(21.4) | |
| T2 | 264(61.0) | 84(52.5) | 83(55.0) | 431(57.9) | |
| T3-4 | 74(17.1) | 71(44.4) | 9(6.0) | 154(20.7) | |
| N stage | <0.001 | ||||
| N0 | 262(60.5) | 58(36.3) | 77(51.0) | 397(53.4) | |
| N1 | 124(28.6) | 63(39.4) | 38(25.2) | 225(30.2) | |
| N2-3 | 47(10.9) | 39(24.4) | 36(23.8) | 122(16.4) | |
| Grade | 0.166 | ||||
| I-II | 249(57.5) | 101(66.0) | 93(61.6) | 443(60.1) | |
| III | 184(42.5) | 52(34.0) | 58(38.4) | 294(39.9) | |
| Pathologic type | 0.016 | ||||
| IDC | 406(93.8) | 156(97.5) | 151(100.0) | 713(95.8) | |
| ILC | 17(3.9) | 0(0.0) | 0(0.0) | 17(2.3) | |
| Other | 10(2.3) | 4(2.5) | 0(0.0) | 14(1.9) | |
| Surgery | <0.001 | ||||
| Mastectomy | 194(44.8) | 96(60.0) | 118(78.1) | 408(54.8) | |
| BCS | 239(55.2) | 64(40.0) | 33(21.9) | 336(35.2) | |
| Chemotherapy | <0.001 | ||||
| No | 82(18.9) | 0(0) | 17(11.3) | 99(13.3) | |
| Yes | 351(81.1) | 160(100) | 134(88.7) | 645(86.7) | |
| Chemotherapy treatment | <0.001 | ||||
| Anthracycline and paclitaxel-based | 246(70.1) | 160(100) | 100(74.6) | 506(78.4) | |
| Paclitaxel-based | 79(22.5) | 0(0) | 21(15.7) | 100(15.5) | |
| Anthracycline-based | 26(7.4) | 0(0) | 13(9.7) | 39(6.0) | |
| Endocrine therapy | <0.001 | ||||
| Aromatase inhibitor | 197(45.5) | 79(59.8) | 27(17.9) | 303(42.3) | |
| Tamoxifen | 236(54.5) | 53(40.2) | 124(82.1) | 413(57.7) | |
| Endocrine Resistance | 0.391 | ||||
| No | 414(95.6) | 149(93.1) | 145(96.0) | 708(95.2) | |
| Yes | 19(4.4) | 11(6.9) | 6(4.0) | 36(4.8) |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor2; IDC, Invasive ductal carcinoma; ILC, Invasive lobular carcinoma; BCS, breast conserving surgery.
∗Cases where ≥10% of tumor cells stained positive for ER with immunohistochemistry (IHC) were considered high.
+Cases where ≥20% of tumor cells stained positive for PR with IHC were considered high.
∗∗Cases that showed either 3+ IHC staining or had gene copy number>2.0 were considered HER2 positive.
++Cases where ≥14% of tumor cells stained positive for Ki-67 with IHC were considered positive.
P values of the comparison between 3 cohorts were generated by χ2 test for categorical variables.
3.2. Clinical model development
The association of clinicopathological characteristics and endocrine resistance in the training cohort is shown in Table 2. Patients diagnosed with higher breast cancer stages were more likely to develop endocrine resistance (P = 0.001). The rate of endocrine resistance in histological grade 3 was higher than histological grades 1 and 2 (7.1% vs 2.4%, P = 0.019). In addition, patients with PR expression <20% were more likely to develop endocrine resistance than patients with PR expression >20% (8.0% vs 1.6%, P = 0.001). The results showed that age, pathologic type, ER expression, Her-2 status, chemotherapy, endocrine drug, and OFS did not affect endocrine resistance (P > 0.05, as shown in Table 2). The clinical signature model that was incorporated with significant factors of stage, histological grade, and PR expression showed AUCs of 0.849 (95% CI: 0.785–0.914) based on logistic regression model in the training cohort (Fig. 2A).
Table 2.
The associations between Clinicopathologic Characteristics and Endocrine Resistance in training Cohort.
| Endocrine Resistance | ||||
|---|---|---|---|---|
| Characteristics | No N = 414 (%) |
Yes N = 19 (%) |
Total N = 433 (%) |
P value |
| age | 0.870 | |||
| ≤35 | 26(6.3) | 1(5.3) | 27(6.2) | |
| 35–50 | 195(47.2) | 8(42.1) | 203(47.0) | |
| ≥50 | 192(46.5) | 10(52.6) | 202(46.8) | |
| ER∗ | 0.232 | |||
| Low | 79(19.1) | 6(31.6) | 85(19.6) | |
| High | 335(80.9) | 13(68.4) | 348(80.4) | |
| PR+ | 0.001 | |||
| Low | 173(41.8) | 15(78.9) | 188(43.4) | |
| High | 241(58.2) | 4(21.1) | 245(56.6) | |
| HER2∗∗ | 0.390 | |||
| Negative | 299(72.2) | 12(63.2) | 311(71.8) | |
| Positive | 115(27.8) | 7(36.8) | 122(28.2) | |
| Ki-67++ | 0.222 | |||
| Negative | 71(17.1) | 1(5.3) | 72(16.6) | |
| Positive | 343(82.9) | 18(94.7) | 361(83.4) | |
| Stage | 0.005 | |||
| I | 81(19.6) | 3(15.8) | 84(19.4) | |
| II | 280(67.6) | 7(36.8) | 287(66.3) | |
| III | 53(12.8) | 9(47.4) | 62(14.3) | |
| T stage | 0.274 | |||
| T1 | 95(22.2) | 3(15.8) | 95(21.9) | |
| T2 | 254(61.4) | 10(52.6) | 264(61.0) | |
| T3-4 | 68(16.4) | 6(31.6) | 74(17.1) | |
| N stage | 0.001 | |||
| N0 | 254(61.4) | 8(42.1) | 262(60.5) | |
| N1 | 121(29.2) | 3(15.8) | 124(28.6) | |
| N2-3 | 39(9.4) | 8(42.1) | 47(10.9) | |
| Grade | 0.019 | |||
| I-II | 243(58.7) | 6(31.6) | 249(57.5) | |
| III | 171(41.3) | 13(68.4) | 184(42.5) | |
| Pathologic type | 0.516 | |||
| IDC | 387(93.5) | 19(100.0) | 406(93.8) | |
| ILC | 17(4.1) | 0(0.0) | 17(3.9) | |
| Other | 10(2.4) | 0(0.0) | 10(2.3) | |
| Surgery | 0.010 | |||
| Mastectomy | 180(43.5) | 14(73.7) | 194(44.8) | |
| BCS | 234(56.5) | 5(26.3) | 239(55.2) | |
| Chemotherapy | 0.064 | |||
| No | 82(19.8) | 0(0) | 82(18.9) | |
| Yes | 332(80.2) | 19(100) | 351(81.1) | |
| Chemotherapy treatment | 0.670 | |||
| Anthracycline and paclitaxel-based | 231(69.6) | 15(78.9) | 246(70.1) | |
| Paclitaxel-based | 76(22.9) | 3(15.8) | 79(22.5) | |
| Anthracycline-based | 25(7.5) | 1(5.3) | 26(7.4) | |
| Endocrine therapy | 0.267 | |||
| Aromatase inhibitor | 186(44.9) | 11(57.9) | 197(45.5) | |
| Tamoxifen | 228(55.1) | 8(42.1) | 236(54.5) | |
| Ovarian function suppression | 0.562 | |||
| No | 279(78.2) | 14(87.5) | 293(78.6) | |
| Yes | 78(21.8) | 2(12.5) | 80(21.4) | |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor2; IDC, Invasive ductal carcinoma; ILC, Invasive lobular carcinoma; BCS, breast conserving surgery.
∗Cases where ≥10% of tumor cells stained positive for ER with immunohistochemistry (IHC) were considered high.
+Cases where ≥20% of tumor cells stained positive for PR with IHC were considered high.
∗∗Cases that showed either 3+ IHC staining or had gene copy number>2.0 were considered HER2 positive.
++Cases where ≥14% of tumor cells stained positive for Ki-67 with IHC were considered positive.
P values of the comparison between Endocrine Resistance and non- Endocrine Resistance patients in training cohort were generated by χ2 test for categorical variables.
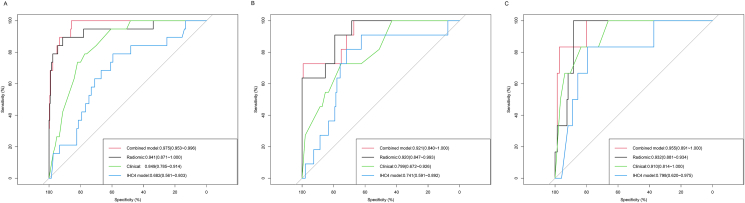
Fig. 2.
Discriminatory accuracy in predicting endocrine resistance that was assessed by ROC analysis for calculating the AUC. Training (A), internal validation (B), and external validation (C) cohorts.
3.3. Radiomics model development
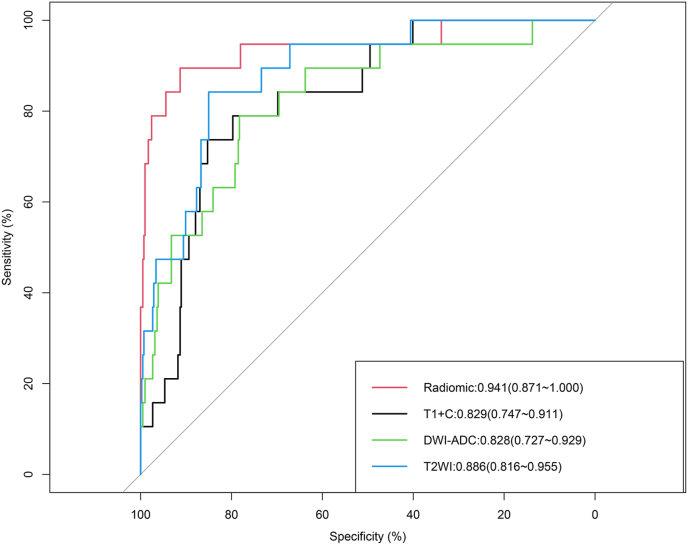
The AUC of the radiomic model with the combination of features from the T1+C, T2WI, and DWI-ADC sequences was 0.941 (95% CI: 0.871–1.000) (Fig. 1), which showed better predictive performance of endocrine resistance than the T1+C sequence alone (AUC = 0.829, 95% CI: 0.747–0.911,P = 0.031) and the DWI-ADC sequence alone (AUC = 0.828, 95% CI: 0.727–0.929, P = 0.050), and also higher than the T2WI sequence alone (AUC = 0.886, 95% CI: 0.816–0.955,P = 0.212) with no statistical significance (see Fig. 1).
Fig. 1.
Discriminatory accuracy in predicting endocrine resistance that was assessed by ROC analysis calculating the AUC using T1+C, T2WI, DWI-ADC sequences alone and combined radiomic sequences.
3.4. Radiomic-clinical model development and validation
The multivariable logistic regression model identified clinical and radiomic signatures combined to predict endocrine resistance in HR-positive patients. The radiomic-clinical combined model was presented as a nomogram in Supplemental Fig. 2 and compared with the radiomic, clinical, and IHC4 models. The AUC of the radiomic-clinical combined model to predict endocrine resistance was 0.975 (95% CI: 0.953–0.996), which was significantly higher than the clinical model (0.849, 95% CI: 0.785–0.914, P < 0.001) and the IHC4 model (0.682, 95% CI, 0.561–0.803, P < 0.001) in the training cohort, and similar as that of the radiomic model (0.941, 95% CI: 0.871–1.000,P = 0.229) (Fig. 2A). But the sensitivity and the maximal Youden index of combined model was higher than radiomic model alone (sensitivity 1.00 v.s. 0.89, Youden index 0.86 v.s. 0.81) in training cohort. Similarly, the AUC of the combined model (0.921, 95% CI: 0.840–1.000) for predicting endocrine resistance in the internal validation cohort was significantly higher than that of the clinical model (0.799, 95% CI: 0.672–0.926, P < 0.001) and IHC4 model (0.741, 95% CI: 0.591–0.892, P < 0.001), and similar as that of the radiomic model (0.920, 95% CI: 0.847–0.993,P = 0.951) (Fig. 2B). Also the sensitivity and the maximal Youden index of combined model was higher than radiomic model alone (sensitivity 0.91 v.s. 0.73, Youden index 0.72 v.s. 0.70) in the internal validation cohort. In addition, the AUC of the combined model (0.955, 95% CI: 0.891–1.000) was higher than that of the clinical model (0.910, 95% CI: 0.814–1.000, P = 0.090) and the IHC4 model (0.798, 95% CI: 0.620–0.975, P = 0.012), but similar as that of the radiomic model (0.932, 95% CI: 0.881–0.934, P = 0.440) in the external validation cohort (Fig. 2C). And the sensitivity and the maximal Youden index of combined model was higher than radiomic model alone (sensitivity 1.00 v.s. 0.83, Youden index 0.88 v.s. 0.81) in the external validation cohort.
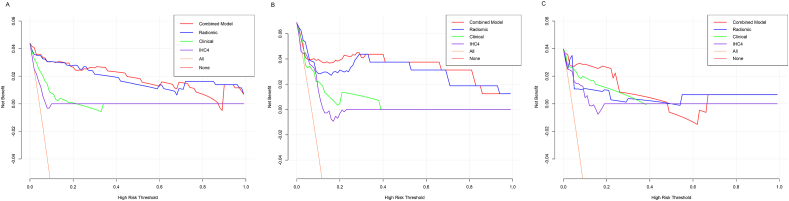
Decision curve analysis (DCA) demonstrated that the radiomic-clinical combined model and radiomic model provided a higher net benefit of threshold probabilities for predicting endocrine resistance than clinical model and IHC4 model across a wider reasonable range (7–95%) of threshold probabilities. The radiomic-clinical combined model, the radiomic model, the clinical model, and IHC4 model were shown in Fig. 3A–C for the training cohort, internal validation cohort and external validation cohort, respectively.
Fig. 3.
Decision curve analysis for the combined, radiomic, clinical, and IHC4 models. Training (A), internal validation (B), and external validation (C) cohorts.
4. Discussion
Endocrine resistance is one of the most troublesome problems of HR-positive breast cancer patients in clinical practice. Only two-thirds of patients exhibit a good response to endocrine therapy, and once postoperative recurrence or even metastasis occur, these patients would have poor prognosis [[1], [4]]. This study proposed using AI to predict endocrine resistance in HR+ non-MBC patients.
In this multicenter study, we developed and validated a pretreatment multiparametric MRI-based radiomic-clinical combined model to predict endocrine resistance in individual female patients with non-MBC. We extracted the radiomics features from the MRI images, combining the clinical features effectively (such as: stage, histological grade, PR expression), to develop a combined model with good performance in predicting endocrine resistance in the training cohort, internal validation cohort and external validation cohort, which was better than with the clinical information model alone and the IHC4 model. Although the AUC between combined model and radiomic model alone was no significance differences, the value of combined model was higher than radiomic model alone. And the sensitivity of combined model was higher than radiomic model alone, also the combined model was at the best thresholding of the AUC.
Studies have suggested that HER2 overexpression is associated with poor response to tamoxifen therapy [14]. HER2 may lead to tamoxifen resistance by activating estrogen receptor co-activator proteins [8]. Another possible cause is the loss of ER expression. It can be concluded from the results of clinical trials that Ki-67 is related to the disease-free survival and recurrence-free survival of premenopausal breast cancer patients. Dowsett et al. showed that anastrozole can reduce the Ki67 index and prolong the disease-free and recurrence-free of patient's survival [15]. Matthew. et al. studied the clinical characteristics related to the reaction of ET, which revealed that Ki-67, tumor size, nodule involvement, and ER status affect the prognosis of ET [16]. About 10–20% of initially ER-positive patients convert into negative upon relapse [8]. Some studies evaluated the role of ESR1 mutations in acquired endocrine-resistant breast cancer. ESR amplification has been shown to be significantly negatively correlated with positive HER2 status, negative ERa, T stage and number of positive lymph nodes [17]. A previous study established a predictive model for ER+/HER2- breast cancer patient response to endocrine therapy based on the 12-gene MS, with an AUC of 0.726 (95% CI: 0.60–0.85) [18]. Another study showed that after 5 years of adjuvant ET in breast cancer, tumor/lymph stage and histological grade are strongly correlated with the risk of distant recurrence [19]. In our research, the predictive ability was further improved by adding radiomic signatures using AI, and the AUC was 0.975 (95% CI: 0.953–0.996), which is higher than previous studies.
Diagnosis, prognosis, prediction, and treatment of breast cancer were improved using AI in radiomics [20]. Several studies have added clinical information, pathology information, genomics, and radiomic features to improve model performance in predicting survival prognosis of breast cancer for all molecular subtypes, and the AUC was between 0.55 and 0.90 [10,21].
This study was aimed to identify high risk endocrine resistance patients with HR-positive breast cancer in advance, provide patients with personalized therapeutic agent such as CDK4/6 inhibitors or other targeted therapies, and prolong disease-free survival [22]. Also, this study was a multi-center, retrospective study with 744 HR-positive breast cancer patients, and the radiomic-clinical combined model showed good generalizability and clinical applicability. Furthermore, we will collect multi-sequence MRI images and clinicopathological characteristics prospectively to validate the accuracy and consistency of the combined predictive model.
There are several limitations of the study. First, it is a retrospective study, with heterogeneity in MR versions among hospitals. Therefore, several post-processing steps were conducted to normalize the images, and a standard distribution of image intensity that reduced heterogeneity was obtained. Second, genetic features such as transcriptomics, genomics, and tumor mutation burden were not included in this model due to missing data. In addition, the underlying mechanism of the interaction between tumor microenvironment and radiomic signatures warrants further investigation. Third, we only discussed primary endocrine resistance, but the number of primary endocrine resistant outcome was low with 4–9% in HR-positive patients. Secondary endocrine resistance was excluded from this study and requires further research.
In conclusion, the study established and validated a radiomic-clinical model to predict endocrine resistance in non-MBC HR positive patients. This model can assist clinicians in formulating individualized therapeutic regimens for patients.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgments
Funding: This study was supported by grant 2020ZX09201021 from the National Science and Technology Major Project, grants 82003311, 81572596 and 81972471 from the National Natural Science Foundation of China, grants 202102010272 Guangzhou Science and Technology Program, grant YXRGZN201902 from the Medical Artificial Intelligence Project of Sun Yat-Sen Memorial Hospital, grant SYS-Q-202004 and SYS-C-201801 from the Sun Yat-Sen Clinical Research Cultivating Program, grant YXQH201920 from Sun Yat-sen Memorial Hospital Yat-Sen scientific research launch project, grant A2020391 and A2020558 from the Guangdong Medical Science and Technology Program, grant 2017A030313828 from the Natural Science Foundation of Guangdong Province, grant 201704020131 from the Guangzhou Science and Technology Major Program, grant 2017B030314026 from the Guangdong Science and Technology Department, grant 2018007 from the Sun Yat-Sen University Clinical Research 5010 Program, grant SYSU-81000-20200311-0001 and SYSU-05160-20200506-0001 from Tencent Charity Foundation. And we appreciate the assistance from the Artificial Intelligence Lab and the Big Data Center of Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.breast.2021.09.005.
Contributor Information
Yaping Yang, Email: yangyap2@mail.sysu.edu.cn.
Junwei Li, Email: lijw228@mail2.sysu.edu.cn.
Yajing Liu, Email: liuyajing1030@126.com.
Ying Zhong, Email: zhongying_wing@126.com.
Wei Ren, Email: renw3@mail2.sysu.edu.cn.
Yujie Tan, Email: tanyj6@mail2.sysu.edu.cn.
Zifan He, Email: hezf5@mail2.sysu.edu.cn.
Chenchen Li, Email: lichch5@mail2.sysu.edu.cn.
Jie Ouyang, Email: kitty865@163.com.
Qiugen Hu, Email: hu6009@163.com.
Yunfang Yu, Email: yuyf9@mail.sysu.edu.cn.
Herui Yao, Email: yaoherui@mail.sysu.edu.cn.
Ethics approval and consent to participate
Ethical approval was provided by the Sun Yat-sen Memorial Hospital Ethics Committee, and the study was registered with ClinicalTrials.gov (NCT04003558) and the Chinese Clinical Trail Registry (ChiCTR1900024020). As this was a retrospective study, informed consent from the study participants was exempted.
Author contributions
Yaping Yang and Junwei Li contributed equally to this study including study design, data acquisition, data analysis and manuscript drafting, they are listed as dual first authors. Herui Yao and Yunfang Yu are listed as dual corresponding authors for the same reason. Yajing Liu,Ying Zhong, Wei Ren, Yujie Tan, Zifan He, Chenchen Li, Jie Ouyang and Qiugen Hu were responsible for the data collection and interpretation of data. Drs Yang, Yu and Herui Yao had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Identification of essential radiomic features of T1+C, T2WI, and DWI-ADC sequences with random forest algorithm (the top 30 candidates) Analysis of T1+C (A), T2WI (B), and DWI-ADC (C) sequences.
Clinical-radiomic nomogram developed using the training cohort to predict endocrine resistance
References
- 1.Rebecca L., Siegel K.D.M., Jemal Ahmedin. Cancer statistics, 2020. Cancer. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Perou C.M., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lønning P.E., Børresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6769):747–752. doi: 10.1038/35021093. TS. [DOI] [PubMed] [Google Scholar]
- 3.Goldhirsch A., Winer E.P., Coates A.S., Gelber R.D., Piccart-Gebhart M., Thürlimann B., Senn H.J., Albain K.S., André F., Bergh J. Personalizing the treatment of women with early breast cancer: highlights of the st gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurebayashi J. Endocrine-resistant breast cancer: underlying mechanisms and strategies for overcoming resistance. Breast Cancer. 2003;10(2):112–119. doi: 10.1007/BF02967635. [DOI] [PubMed] [Google Scholar]
- 5.Soonmyung Paik 1 S.S., Gong Tang, Kim Chungyeul, Baker Joffre, Cronin Maureen, Baehner Frederick L., Walker Michael G., Watson Drew, Park Taesung, Hiller William, Fisher Edwin R., Wickerham D Lawrence, Bryant John, Norman Wolmark. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;352(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen T.O., Parker J.S., Leung S., Voduc D., Ebbert M., Vickery T., Davies S.R., Snider J., Stijleman I.J., Reed J. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Canc Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heindl A., Sestak I., Naidoo K., Cuzick J., Dowsett M., Yuan Y. Relevance of spatial heterogeneity of immune infiltration for predicting risk of recurrence after endocrine therapy of ER+ breast cancer. J Natl Cancer Inst. 2018;110(2) doi: 10.1093/jnci/djx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunte S., Braman N., Bera K., Leo P., Abraham J., Montero A.J., Madabhushi A. Radiomics risk score (RRS) on CT to predict survival and response to CDK 4/6 inhibitors in hormone receptor (HR) positive metastatic breast cancer (MBC) 2020;38(15_suppl) e13041-e13041. [Google Scholar]
- 9.Cardoso F., Costa A., Senkus E., Aapro M., André F., Barrios C.H., Bergh J., Bhattacharyya G., Biganzoli L., Cardoso M.J. 3rd ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 3) Ann Oncol : official journal of the European Society for Medical Oncology. 2017;28(1):16–33. doi: 10.1093/annonc/mdw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Y., Tan Y., Xie C., Hu Q., Ouyang J., Chen Y., Gu Y., Li A., Lu N., He Z. Development and validation of a preoperative magnetic resonance imaging radiomics–based signature to predict axillary lymph node metastasis and disease-free survival in patients with early-stage breast cancer. JAMA Network Open. 2020;3(12) doi: 10.1001/jamanetworkopen.2020.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H. Computational radiomics system to decode the radiographic phenotype. Canc Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patrick J., Heagerty T.L., Pepe Margaret S. Time-dependent roc curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 13.Cuzick J., Dowsett M., Pineda S., Wale C., Salter J., Quinn E., Zabaglo L., Mallon E., Green A.R., Ellis I.O. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol : official journal of the American Society of Clinical Oncology. 2011;29(32):4273–4278. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 14.Dowsett M. Overexpression of HER-2 as a resistance mechanism to hormonal therapy for breast cancer. Endocr Relat Canc. 2001;8(3):191–195. doi: 10.1677/erc.0.0080191. [DOI] [PubMed] [Google Scholar]
- 15.Dowsett M., Smith I.E., Ebbs S.R., Dixon J.M., Skene A., Griffith C., Boeddinghaus I., Salter J., Detre S., Hills M. Short-term changes in Ki-67 during neoadjuvant treatment of primary breast cancer with anastrozole or tamoxifen alone or combined correlate with recurrence-free survival. Clin Canc Res. 2005;11(2 Pt 2):951s–958s. [PubMed] [Google Scholar]
- 16.Ellis M.J., Tao Y., Luo J., A'Hern R., Evans D.B., Bhatnagar A.S., Chaudri Ross H.A., von Kameke A., Miller W.R., Smith I. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100(19):1380–1388. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen K.V., Ejlertsen B., Müller S., Møller S., Rasmussen B.B., Balslev E., Lænkholm A.V., Christiansen P., Mouridsen H.T. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Canc Res Treat. 2011;127(2):345–355. doi: 10.1007/s10549-010-0984-y. [DOI] [PubMed] [Google Scholar]
- 18.Singer C.F., Pfeiler G., Hubalek M., Bartsch R., Stöger H., Pichler A., Petru E., Bjelic-Radisic V., Greil R., Rudas M. Efficacy and safety of the therapeutic cancer vaccine tecemotide (L-BLP25) in early breast cancer: results from a prospective, randomised, neoadjuvant phase II study (ABCSG 34) Eur J Canc. 2020;132:43–52. doi: 10.1016/j.ejca.2020.03.018. (Oxford, England : 1990) [DOI] [PubMed] [Google Scholar]
- 19.Pan H., Gray R., Braybrooke J., Davies C., Taylor C., McGale P., Peto R., Pritchard K.I., Bergh J., Dowsett M. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med. 2017;377(19):1836–1846. doi: 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R., Even A.J.G., Jochems A. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Zhu Y., Burnside E.S., Drukker K., Hoadley K.A., Fan C., Conzen S.D., Whitman G.J., Sutton E.J., Net J.M. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston S.R.D., Harbeck N., Hegg R., Toi M., Martin M., Shao Z.M., Zhang Q.Y., Martinez Rodriguez J.L., Campone M., Hamilton E. Abemaciclib combined with endocrine therapy for the adjuvant treatment of HR+, HER2-, node-positive, high-risk, early breast cancer (monarchE) J Clin Oncol : official journal of the American Society of Clinical Oncology. 2020;38(34):3987–3998. doi: 10.1200/JCO.20.02514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of essential radiomic features of T1+C, T2WI, and DWI-ADC sequences with random forest algorithm (the top 30 candidates) Analysis of T1+C (A), T2WI (B), and DWI-ADC (C) sequences.
Clinical-radiomic nomogram developed using the training cohort to predict endocrine resistance