Abstract
A disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13) is a multidomain metalloprotease for which until now only a single substrate has been identified. ADAMTS13 cleaves the polymeric force-sensor von Willebrand factor (VWF) that unfolds under shear stress and recruits platelets to sites of vascular injury. Shear force–dependent cleavage at a single Tyr–Met peptide bond in the unfolded VWF A2 domain serves to reduce the size of VWF polymers in circulation. In patients with immune-mediated thrombotic thrombocytopenic purpura (iTTP), a rare life-threatening disease, ADAMTS13 is targeted by autoantibodies that inhibit its activity or promote its clearance. In the absence of ADAMTS13, VWF polymers are not adequately processed, resulting in spontaneous adhesion of blood platelets, which presents as severe, life-threatening microvascular thrombosis. In healthy individuals, ADAMTS13–VWF interactions are guided by controlled conversion of ADAMTS13 from a closed, inactive to an open, active conformation through a series of interdomain contacts that are now beginning to be defined. Recently, it has been shown that ADAMTS13 adopts an open conformation in the acute phase and during subclinical disease in iTTP patients, making open ADAMTS13 a novel biomarker for iTTP. In this review, we summarize our current knowledge on ADAMTS13 conformation and speculate on potential triggers inducing conformational changes of ADAMTS13 and how these relate to the pathogenesis of iTTP.
Keywords: ADAMTS, ADAMTS13, antibody autoimmune disease, conformational change, hemostasis, thrombosis, thrombotic thrombocytopenic purpura, von Willebrand factor
Abbreviations: ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; CUB, complement C1r/C1s, Uegf, Bmp1; cys-rich, cysteine-rich; Dis, disintegrin-like; FRETS-VWF73, fluorescence resonance energy transfer substrate VWF73; iTTP, immune-mediated TTP; MP, metalloprotease; TSP, thrombospondin type 1-like; TTP, thrombotic thrombocytopenic purpura; ULVWF, ultralarge von Willebrand factor; VWF, von Willebrand factor
The increasing prevalence of autoimmune disorders provides a major challenge for the healthcare system worldwide. Our current understanding of the pathogenesis of several autoimmune disorders is incomplete. Environmental triggers have been linked to the onset of immunity. In addition, specific human leukocyte antigen loci have been identified as a risk factor for a large number of autoimmune disorders (1). Post-translational modifications such as citrullination have also been implicated in the pathogenesis of autoimmune disorders (2). Importantly, conformational changes of proteins resulting in exposure of neoepitopes may trigger the development of pathogenic autoantibodies (3). Conformational changes in beta-2 glycoprotein 1 have been linked to the antiphospholipid syndrome (4), whereas conformational changes in platelet factor 4 upon its binding to heparin trigger the development of anti-platelet factor 4 antibodies resulting in heparin-induced thrombocytopenia (5). In the current review, we explore how conformational plasticity of the protein a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13) relates to the onset of autoimmune thrombotic thrombocytopenic purpura (TTP).
TTP is a rare and life-threatening disease with an annual incidence rate of 1.5 to 6 cases per million adults per year (6), which is characterized by a severe deficiency (<10% activity) of the plasma metalloprotease (MP) ADAMTS13. The acute phase of TTP often presents itself with purpura, fever, neurological manifestations, renal dysfunctions, hemolytic anemia with schistocytes, and thrombocytopenia (7). Although TTP can be of congenital origin due to mutations in the ADAMTS13 gene, the vast majority of patients develop immune-mediated TTP (iTTP), an autoimmune disorder in which autoantibodies against ADAMTS13 develop (8). Symptoms of TTP arise because of a patient’s inability to cleave ultralarge von Willebrand factor (ULVWF) multimers on the surface of endothelial cells (9, 10) (Fig. 1). Multimeric von Willebrand factor (VWF) is produced by vascular endothelium and megakaryocytes (11, 12). VWF is stored in endothelial cell granules called Weibel–Palade bodies (13), which fuse with the cell membrane to release ULVWF multimers (14). The ability of VWF to bind circulating platelets is critically dependent on its multimeric size, with the largest multimers being most potent in capturing platelets (15) during primary hemostasis. In the absence of ADAMTS13, ULVWF multimers are not processed and platelet-rich clots spontaneously form in the microcirculation, resulting in thrombotic microangiopathy.
Figure 1.
Schematic representation of ADAMTS13 activity.A, normal hemostasis: under physiological conditions, multimeric size of the VWF is controlled by ADAMTS13, which circulates in closed conformation and only adopts a transient open conformation upon interacting with its substrate VWF. B, reduced ADAMTS13 activity leads to accumulation of ULVWF multimers, which promote formation of platelet-rich clots in the microvasculature, giving rise to thrombotic thrombocytopenic purpura. ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; VWF, von Willebrand factor; ULVWF, ultra large von Willebrand factor.
VWF multimer digestion by ADAMTS13 is dependent on shear stress, which causes the VWF protein overall shape to change from mostly globular to an elongated, stretched form (16). Upon binding to its substrate VWF, ADAMTS13 adopts a short-lived, transient “open” conformation that allows for multiple exosites within the protease, disintegrin, cysteine-rich (Cys-rich), and spacer domains to interact with complementary sites within the unfolded VWF A2 domain, a process that has been named a “molecular zipper” (17, 18). Importantly, the “open conformation” refers to the ADAMTS13 conformation that evolves upon release of autoinhibitory interactions between the ADAMTS13 C-terminal complement C1r/C1s, Uegf, Bmp1 (CUB) domains and the spacer domain (Fig. 2A). Hence, when the CUB–spacer interactions are intact, ADAMTS13 is in a “closed” conformation (Fig. 2B). The differences in these conformations can be monitored by a specifically developed mAb that can only bind the spacer domain when the protein is in an “open” conformation (19, 20). Previous literature directly correlates “open” conformation ADAMTS13 to increased proteolytic activity (21). However, subsequent findings indicate that several conformation-controlled binding events between ADAMTS13 and its substrate VWF are required for the generation of a fully catalytically active conformation of the MP domain (18). This sophisticated sequence of conformational events serves to regulate activation of ADAMTS13 as well as prevent excessive or off-target proteolysis (22).
Figure 2.
ADAMTS13 structure.A, three-dimensional model of ADAMTS13 in the “open conformation”. In this conformation, the spacer domain (blue) releases its interactions with the CUB domains (cyan and magenta). This image was created based on the full-length ADAMTS13 3D model in panel B. B, three-dimensional model of ADAMTS13 in the “closed conformation”. The MDTCS structure is determined by X-ray crystallography (18); three-dimensional structure of the CUB domains was taken from the study by Kim et al. (32); the structure of TSP2-8 was determined by homology modeling as described (49). C, the ADAMTS13 domains are depicted schematically as a signal peptide (S; black), propeptide (P; yellow), metalloprotease (MP; red) domain with active site followed by disintegrin-like (Dis; green), central thrombospondin type 1-like (TSP; gray) repeat, cysteine-rich (Cys-rich; orange), spacer (purple) domains, TSP2-8 (gray), CUB1 (cyan), and CUB2 (magenta). ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; CUB, complement C1r/C1s, Uegf, Bmp1; MDTCS, metalloprotease, disintegrin-like domain, thrombospondin type 1 repeat.
Recently, it was shown that ADAMTS13 circulates in an “open” conformation during the acute phase and during subclinical disease in remission of iTTP, suggesting that a sustained “open” conformation of ADAMTS13 is linked to the onset and recurrence of autoimmunity in these patients (19, 20). Furthermore, in iTTP, conformational changes of ADAMTS13 are observed earlier than detectable titers of anti-ADAMTS13 autoantibodies during an acute episode of iTTP (20). This observation raises the question how conversion of ADAMTS13 from a closed to an open conformation is regulated and how it relates to the pathogenesis of iTTP. The pathological triggers inducing a switch from the closed to open conformation of ADAMTS13 are now beginning to be defined. For example, it was recently demonstrated that patient-derived anti-ADAMTS13 autoantibodies can promote conversion of ADAMTS13 from a closed to an open conformation (20, 23). Apart from autoantibodies, other pathological triggers that have not yet been identified may induce conformational changes of ADAMTS13 in patients with iTTP. In this review, we summarize the impact of key conformational changes that contribute to development of autoimmunity in patients with iTTP.
Structure and conformational flexibility of ADAMTS13
ADAMTS family members are multidomain proteins. They consist of a signal sequence, a propeptide, a metalloproteinase, a disintegrin-like (Dis), a central thrombospondin type 1-like (TSP), a Cys-rich, and a spacer domain and a set of additional TSP repeats (24) (Fig. 2C). ADAMTS13 uniquely contains two CUB domains at its carboxy terminus (24). Within the propeptide, a furin-cleavage site is localized which, when cleaved, facilitates removal of the propeptide. ADAMTS MP domains contain a zinc-binding motif with the reprolysin signature (HEXXH + HD). Furthermore, ADAMTS proteins have a conserved cysteine network inside their Cys-rich domain (24), which is involved in substrate binding in the case of ADAMTS13 (25). The primary role of the ADAMTS13 central TSP repeat seems to be spatial positioning of the VWF-binding exosites present in the surrounding domains (26). Since the isolation of ADAMTS13 from plasma (27, 28, 29) and publication of its amino acid sequence in 2001 (28, 30), researchers have intensively focused on understanding the mechanism by which ADAMTS13 processes VWF multimers. The experimental determination of the three-dimensional structure of the disintegrin-like domain, thrombospondin type 1 repeat, cysteine-rich and spacer domains of ADAMTS13 provided a framework for these studies (31). The crystal structure of the MP domain of ADAMTS13 as well as the crystal structure of the CUB domains were published more recently (18, 32).
C-terminal domains of ADAMTS family members have been distinguished as important regulators of enzymatic activity (33, 34, 35, 36, 37). Importantly, the C-terminal CUB domains are a unique feature of ADAMTS13 and have evolved as critical regulators of ADAMTS13 conformation and activity. Incubation of ADAMTS13 with the anti-CUB2 mAb 20E9 resulted in an increased activity of ADAMTS13 (21). A similar effect was observed after the addition of the D4-CK fragment derived from VWF (residues 1874–2813), which comprises the natural binding site of the VWF substrate for the ADAMTS13 CUB domains (21). In addition, mAbs directed toward the T2-T8 repeats and the CUB1/2 domains were also capable of increasing the activity of ADAMTS13 for the fluorescence resonance energy transfer substrate VWF73 (FRETS-VWF73) derived from the A2 domain of the VWF (38, 39). These findings suggested that binding of mAbs to the carboxy-terminal TSP2-8 and CUB1-2 domains induces a conformational change in ADAMTS13 that allows for more efficient processing of unfolded peptide substrates such as FRETS-VWF73. This hypothesis is strengthened further by the finding that targeting of the spacer domain by antibodies yields similar results (39).
Transmission electron microscopy of negatively stained ADAMTS13 provided evidence for being primarily present in a closed conformation, whereas a so-called gain-of-function variant in which residues Arg568, Phe592, Arg660, Tyr661, and Tyr665 were replaced for corresponding conservative residues was primarily present in an open conformation (21, 40). Small-angle X-ray scattering combined with molecular modeling provided additional evidence for intradomain interactions between the TSP8-CUB1/2 and the spacer domain (41). Follow-up studies revealed that both the CUB1 and CUB2 domains are capable of interacting with the spacer domain (Fig. 2B) (42). A crucial role for TSP8 was suggested in maintaining ADAMTS13 in a closed conformation, most likely by promoting CUB1 spacer domain interactions (42). In a recent study, Muia et al. showed that most carboxy/terminal domains with the exception of TSP3/TSP6 are needed for allosteric regulation of ADAMTS13 activity (43, 44). Deletion of TSP7 and TSP8 abrogated allosteric regulation of ADAMTS13 (44). Small-angle X-ray scattering combined with molecular modeling suggested that ADAMTS13 is present in a hairpin-like conformation with a so-called “hinge-like” region between TSP4 and TSP5 (Fig. 2B) (43). This finding is consistent with the presence of a linker region between TSP4 and TSP5 that may allow for bending of this part of ADAMTS13 while securing the more rigid disulfide-bonded configuration of the neighboring TSP4 and TSP5 repeats. In agreement with this, an important role for three linker regions after the TSP2, TSP4, and TSP8 domains was proposed to control ADAMTS13 conformation (38). Somewhat unexpected, deletion of TSP3/6 repeats in human ADAMTS13 appeared to preserve its ability to convert from a closed to an open conformation (44). This configuration was derived from a phylogenetic screen, which showed that pigeon ADAMTS13, which lacks the TSP3/TSP6 repeats, is still subject to allosteric regulation (44). To summarize, ADAMTS13 normally circulates in an autoinhibitory conformation governed by an interaction between its C-terminal domains and the spacer domain which is designated as “closed” conformation. To allow for enzymatic activity, conformational changes in ADAMTS13 are required, which will be addressed below.
ADAMTS13 enzymatic activation
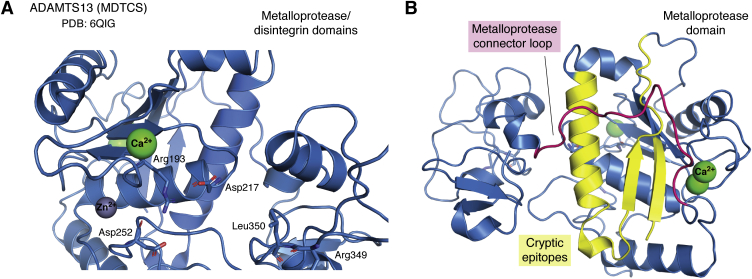
As previously introduced, multimeric VWF circulates in a globular form; however, exposure to shear forces and the binding to exposed collagen at the site of a damaged vessel promotes its unfolding (45). Single-molecule experiments using optical tweezers have provided evidence for shear force–induced unfolding of VWF that is needed for efficient cleavage of VWF polymers into smaller entities (46). ADAMTS13 can bind to globular VWF through the interaction of the C-terminal D4-CK domains of VWF with ADAMTS13 TSP7-CUB1 domains (43). This is considered to serve two functions, the first one being positioning of ADAMTS13 to the VWF through reversible association. Second, this interaction promotes exposure of the cryptic spacer domain exosite of ADAMTS13 through release of the autoinhibitory CUB–spacer domain interactions, thus inducing a transient “open” ADAMTS13 conformation. After opening, exposed exosites bind to the VWF A2 domain by an induced-fit mechanism; exosite-1 (47) (Dis domain; Arg349, Leu350) binding to the VWF1614 to 1622 and providing the juxtaposition of the active site in the MP domain and the scissile bond on VWF Tyr1605-Met1606, exosite 2 (25) (Cys-rich domain; Ala472, Ala473, Val474) binding the VWF1642 to 1651 region and exosite 3 (48, 49) (spacer domain Arg568, Phe592, Arg660, Tyr661, Tyr665) binding to VWF1660 to 1668 (18). While Akiyama et al. (31) concluded that the activity of ADAMTS13 is primarily dependent on the unfolding of the VWF molecule, a recent study by Petri et al. (18) has demonstrated that interactions between VWF and ADAMTS13 also serve to activate ADAMTS13. This activation occurs in the MP domain of ADAMTS13. A recent crystal structure of mAb 3H9-inhibited ADAMTS13 MP domain (an anti-ADAMTS13 MP domain antibody) shows that the active site of ADAMTS13 is occluded by Arg193, Asp217, and Asp252 that together comprise a so-called gatekeeper triad (18). Asp217 and Asp252 form salt bridges with Arg193 and occlude this site (Fig. 3). In normal physiology, the gatekeeper triad can transiently occlude the active site or destabilize and open up the proteolytic cleft to dock VWF Tyr1605–Met1606 scissile bond for hydrolysis. Occlusion of the proteolytic cleft by the gatekeeper triad can likely be found in circulating closed conformation ADAMTS13, as a way of avoiding off-target proteolysis while providing resistance to plasma inhibitors (18). Activation of the MP domain occurs after exosite 2 and 3 bind to the VWF, to allow Dis domain exosite 1 (R349, L350) to interact with VWF and allosterically activate the MP domain (18).
Figure 3.
Allosteric activation of the ADAMTS13 metalloprotease domain. ADAMTS13 MDTCS (metalloprotease to spacer domains) crystallized in complex with the inhibitory 3H9 Fab fragment as reported by Petri et al. (PDB ID: 6QIG) was used to create the images in this figure using PyMol. A, ADAMTS13 metalloprotease and disintegrin domains (blue), in which the gatekeeper triad is occluding the proteolytic cleft (gray sphere represents zinc, and green spheres are calcium ions). B, the position of ADAMTS13 metalloprotease connector loop (indicated in magenta), which may potentially shield cryptic epitopes (indicated in yellow) in the ADAMTS13 metalloprotease domain. ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; MDTCS, metalloprotease, disintegrin-like domain, thrombospondin type 1 repeat.
Conformational activation of the MP domain of ADAMTS13 as a result of binding of activating ADAMTS13 autoantibodies was also recently shown by Schelpe et al. (39). This exosite lies on the surface of the Dis domain, close to the MP domain active site and was shown to be crucial for ADAMTS13 activity by de Groot et al. (47). Cleavage of the scissile bond in the VWF by ADAMTS13 is preceded by docking of the VWF P1′, P1, and P3 residues in complementary subsites in the MP domain (26). Thus, activation of ADAMTS13 is a complex process that is governed by multiple interactions with the VWF substrate, following a “molecular zipper” model that ultimately leads to activation of the MP domain and cleavage of the VWF scissile bond (17). This observation suggests that MP domain conformational changes are required for development of full proteolytic activity toward its substrate VWF (18). A follow-up study revealed that mAbs directed toward the CUB1 and spacer domain promote exposure of the catalytic site (39). It is not fully understood how antibody interactions with the spacer and CUB domains yield a conformational change within the MP domain in absence of the previously described VWF-ADAMTS13 interactions. However, several conformational changes in ADAMTS13 seem to be required to optimally position the active site of ADAMTS13 for cleavage of its substrate.
Conformational consequences of the spacer–CUB domain interactions
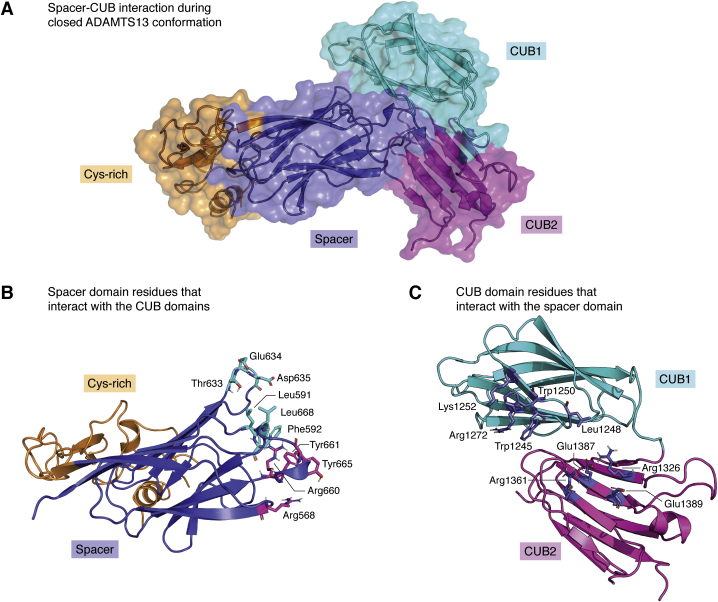
We previously discussed how in WT-ADAMTS13, the TSP2 to CUB2 domains extend like an arm, that can form an elbow-like bend between TSP4 and TSP5 and then fold back and dock CUB domains onto the spacer domain, leading ADAMTS13 to adopt a globular, closed conformation (41, 43). Gain-of-function mutations on the spacer domain (Arg568Lys/Phe592Tyr/Arg660Lys/Tyr661Phe/Tyr665Phe) disrupt the docking of CUB domains and shift the ADAMTS13 structure toward a linear, open conformation (40, 50). The 3D structure of the ADAMTS13 CUB domains were recently determined by x-ray crystallography (32). Interestingly, ADAMTS13 CUB domains do not accommodate a Ca2+-binding site in contrast to other CUB domains (32, 49). A combination of protein–protein docking between spacer domain–CUB domains and ADAMTS13 conformation assay in vitro indicated that residues Trp1245, Leu1248, Trp1250, Lys1252, and Arg1272 from the CUB1 domain and Arg1326, Arg1361, Glu1387, and Glu1389 (Fig. 4, A–C) from CUB2 domain were described as responsible for the maintenance of ADAMTS13 closed conformation (32). Based on a previous study (42), only ADAMTS13 CUB1 domains is found to be inhibiting the proteolytic activity against VWF. However, based on in silico modeling of ADAMTS13 closed conformation model (32), CUB2 was predicted to be interacting with Arg568, Arg660, Tyr661, and Tyr665 (Fig. 4A) from ADAMTS13 spacer domain, which were known to be crucial for VWF interactions. In addition, the CUB domains accommodate two N-glycosylation sites at Asn1235 and Asn1354, which both contain sialic acid–ending glycans (51). Asn to Gln mutation of these sites increased the activity of ADAMTS13 toward the FRETS-VWF73 substrate as well as shear-dependent processing of the VWF, suggesting the importance of these N-glycans in maintaining a closed conformation (52). On the other hand, the crystal structure and in silico modeling indicate the N-glycans of CUB domains are not crucial for maintaining ADAMTS13 in a closed conformation (32). It is well known that N-glycans as well as other post-translational modifications can contribute to stabilization of interdomain or intradomain interactions. We therefore speculate that in the absence of these N-glycans, CUB domains may become structurally less stable, thereby promoting conversion of N-glycan lacking ADAMTS13 from a closed to an open conformation (53). It can however not be excluded that negatively charged sialic acid residues that are present on the N-glycans linked to Asn1235 and Asn1354 might also alter the electrostatic surface potential of the CUB domains, allowing for its binding to positively charged residues in the spacer domain, thereby promoting a closed conformation of ADAMTS13 through long-range electrostatic interactions.
Figure 4.
Three-dimensional model of ADAMTS13 closed conformation between spacer and CUB domains. The three-dimensional structures of the CUB domains (PDB ID: 7B01) and the cysteine-rich and spacer domains (PDB ID: 6QIG) as reported by Kim et al. (32) and Petri et al. (18) were used to create the images in this figure. A, three-dimensional model of the ADAMTS13 closed conformation built by protein–protein docking as previously described (49), using the residues that interact between the spacer and CUB domains determined by Kim et al. (32), which are specified in panels B and C. B, ADAMTS13 spacer residues that interact with the CUB domains in the closed conformation. The cysteine-rich (orange) and spacer (blue) domains are shown as a cartoon representation, and the CUB domain–interacting spacer residues are highlighted and shown as sticks. Highlighted spacer domain residue color corresponds to the color of the CUB domains they interact with (CUB1(cyan)/CUB2(magenta)). C, ADAMTS13 CUB domain residues which bind to the spacer domain during the closed conformation. CUB domains are shown as a cartoon representation. The residues that interact with the spacer domain are colored blue for both the CUB1 and CUB2 domains. ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; CUB, complement C1r/C1s, Uegf, Bmp1.
Shifting ADAMTS13 from a closed to an open conformation also impacts the structure and activity of the MP domain. It has been shown in several publications that the catalytic activity of ADAMTS13 is significantly enhanced upon binding of activating antibodies such as 17G2 (anti-ADAMTS13 CUB1 domain antibody) (38, 39). The underlying mechanism of this activity increase was recently linked to the conformational activation of ADAMTS13 in the MP domain, where a cryptic epitope for 6A6 (an anti-ADAMTS13 MP antibody) is exposed if ADAMTS13 is bound to the activating antibody 17G2 (39). Localization of this cryptic epitope on MP domain still remains elusive and requires more detailed mutagenesis studies to characterize this epitope or alternatively an experimentally determined 3D structure of ADAMTS13 bound to 6A6. However, we can speculate on this based on the 3D structure of the MP and disintegrin domain. The connector loop (residues 285–300) between the MP and Dis domain seems to have a high degree of flexibility. Reorientation of this connector loop as a result of 17G2 binding may expose the α-helix, the β-sheet, or the loops that reside between residues 79 and 84 or 100 and 135 (Fig. 3B). These exposed residues may contribute to the neoepitope for 6A6 (Fig. 3B).
ADAMTS13 conformational changes in immune TTP
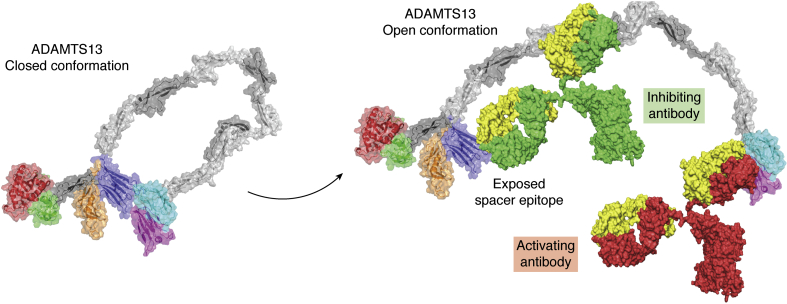
The majority of autoantibodies that develop in patients with iTTP inhibit ADAMTS13 and/or enhance its clearance from the circulation (54, 55). As mentioned above, Muia et al. and Deforche et al. showed that mAbs targeting distal ADAMTS13 regions (TSP6-CUB2) can also allosterically activate the enzyme (38, 44). Since then, activating effects for several antibodies have been reported (38, 39, 41). Physiological relevance of antibody-induced conformational changes is suggested by the ability of a plasma sample, derived IgG from an iTTP patient, to also enhance the processing activity of ADAMTS13 toward the FRETS-VWF73 small substrate (41). This observation shows that also antibodies that arise in patients with acquired TTP can enhance the proteolytic activity of ADAMTS13, most likely by promoting the conversion from a closed to an open ADAMTS13 conformation. Recently, Roose et al. (19) developed a plasma-based assay that was used to monitor the conformation of ADAMTS13 in healthy individuals as well as in patients with iTTP. This was achieved by using an mAb, designated 1C4, which binds to exosite 3 within the spacer domain that is also targeted by pathogenic antibodies that develop in patients with iTTP (40, 48). In ADAMTS13, as present in plasma of healthy individuals, this site is not accessible for 1C4 because ADAMTS13 is present in a “closed” conformation (Fig. 5) (19). Upon addition of 17G2, an mAb that binds around R1219 of the CUB1 domain of ADAMTS13 (32), the equilibrium between a closed and an open conformation is shifted toward an “open” conformation (Fig. 5). After incubation of plasma with 17G2, the binding site for 1C4 in the spacer domain becomes exposed (Fig. 5), resulting in an increase in 1C4 binding to ADAMTS13 in response to the addition of 17G2 to diluted patient plasma (19). From this, a conformation index was defined that represented the increase of 1C4 binding induced by the incubation of ADAMTS13 with 17G2 (19). Intriguingly, when using this assay to monitor ADAMTS13 conformation in plasma of acute-phase iTTP patients, it was observed that a significant part of ADAMTS13 was already able to bind 1C4 without prior incubation of the plasma sample with 17G2 (19). This observation indicates that ADAMTS13 is present in an “open” conformation in patients with iTTP. Longitudinal analysis revealed that “open” ADAMTS13 was primarily present during the acute phase of iTTP; following remission (when ADAMTS13 activity was >50%), ADAMTS13 was mainly present in a closed conformation in which the epitope for 1C4 in the spacer domain was shielded by the carboxy-terminal CUB1/2 domains (20). In a follow-up study, Roose et al. (23) showed that polyclonal antibodies isolated from plasma of patients with iTTP were capable of converting ADAMTS13 from a closed to an open conformation. This observation suggests that similar to mAb 17G2, patient-derived antibodies are capable of shifting the equilibrium from a closed to an open conformation (Fig. 5), thereby facilitating the exposure of the immunodominant spacer domain epitope (Fig. 5). How does this relate to the onset of iTTP? How does the conversion of ADAMTS13 from a closed conformation to an open conformation contribute to the onset of iTTP? These essential questions need further study to be fully answered. However, we can speculate on this. For instance, during the early autoimmune response, primarily antibodies directed toward the carboxy-terminal CUB1/2 domains may develop. Such antibodies have been shown to be present in about 20 to 50% of patients (55, 56, 57, 58, 59). Binding of these antibodies to the CUB1/2 domains of ADAMTS13 may promote exposure of the cryptic (Fig. 5), highly immunodominant spacer domain epitope. Exposure of this latent immunodominant spacer domain epitope then results in a vigorous B cell response that through the generation of high affinity spacer domain antibodies inhibits ADAMTS13 activity and subsequent processing of VWF by ADAMTS13 (Fig. 5) (40, 48). Alternatively, elevated levels of the VWF as observed, for example, during infections could shift the equilibrium between the closed and open conformations of ADAMTS13, thereby prolonging exposure of the cryptic epitope in the spacer domain. Indeed, a broad range of infections has been linked to the onset of iTTP (54, 60). Recently, two reports described the development of iTTP in patients immediately after infection with SARS-CoV2 (61, 62). Strongly elevated levels of the VWF have been observed in patients with COVID-19 (63), and these may induce conversion from a closed to an open conformation. Other factors may also contribute to conformational changes in ADAMTS13.
Figure 5.
Antibody-mediated ADAMTS13 structure opening. Three-dimensional model of ADAMTS13 shifting from “closed” to “open” conformation upon binding of autoantibodies. The MDTCS structure is determined by X-ray crystallography (18) (PDB ID: 6QIG); three-dimensional structure of the CUB domains was taken from the study by Kim et al. (32) (PDB ID: 7B01); the structure of TSP2-8 was determined by homology modeling as described (49). The structure of the antibodies shown in the figures is derived from PDB ID 1IGT. In this figure, the red antibody represents an activating antibody, an example of which is 17G2. Binding of an activating antibody to the CUB domains opens up the conformation of ADAMTS13, which exposes cryptic epitopes that allow the binding of inhibiting antibodies (green), which can inhibit binding to VWF. CUB, complement C1r/C1s, Uegf, Bmp1; MDTCS, metalloprotease, disintegrin-like domain, thrombospondin type 1 repeat; TSP, thrombospondin type 1-like; VWF, von Willebrand factor.
Apart from the VWF and polyclonal antibodies targeting the carboxy-terminal CUB1/2 domains, post-translational modifications may likewise impact ADAMTS13 conformation. Citrullination of arginine residues can have significant implications to protein structure, function, stability, and protein–protein interactions because of loss of positive charge of arginine residues (64). Neutrophils express peptidyl deiminase 4, an enzyme which is activated and secreted upon neutrophil extracellular trap formation (65). In a recent article, Sorvillo et al. (66) showed that peptidyl deiminase 4 can citrullinate ADAMTS13 and reduce its activity both in vitro and in vivo. Identification of citrullinated residues on ADAMTS13 with MS revealed citrullination of Arg190 and Arg636, in the MP and spacer domain, respectively (66). Citrullinated antigens play a major role in other autoimmune disorders such as rheumatoid arthritis (67). Citrullination of ADAMTS13 may therefore provide a potential link between inflammation and autoimmune TTP. Structural changes in ADAMTS13 related to citrullination may result in conformation changes that may result in exposure of cryptic epitopes in the spacer domain or other parts of the molecule. As yet, anticitrullinated protein antibodies as observed in patients with rheumatoid arthritis have not yet been observed in patients with iTTP. Uniquely for ADAMTS13, Arg190 in the MP domain forms a salt bridge with Asp182 to stabilize the Ca2+-binding loop, whereas this function is carried out by a conserved disulfide bond in other ADAMTS MP homologs (68). Citrullination of Arg190 can disrupt the salt bridge and cause loss of structural stability on the Ca2+-binding loop that may eventually lead to impaired proteolytic activity. In addition, this could disturb the conformational activation of the MP domain upon removal of the CUB–spacer interactions, yielding a “semi-open” ADAMTS13 conformation. Arg636 was not found to directly contribute to interactions between ADAMTS13 spacer and CUB domains (32); however, citrullination of this positively charged residue still might modulate electrostatic interactions with the negatively charged residues present in the CUB domains.
Apart from citrullination of ADAMTS13, other post-translational modifications such as oxidation of methionine residues by neutrophil-released hypochlorous acid may lead to conformational changes that could result in exposure of cryptic epitopes on ADAMTS13. Evidence has been provided for oxidation of Met249, Met331, and Met496 to methionine sulfoxide that resulted in a decline in ADAMTS13 activity (69). In general, oxidation of the sulfur atom of methionine residues can change the physicochemical properties of a protein (70). The sulfur atoms in methionines can form a hydrophobic interaction with the aromatic rings of tyrosines, phenylalanines, and tryptophans. This interaction is comparable with an ionic interaction and is known to stabilize protein structure but is lost upon methionine oxidation (71, 72). Whether oxidation of methionine residues plays a role in conversion of ADAMTS13 from a “closed” to an “open” conformation however remains to be established.
Concluding remarks
Current data suggest that conformational changes in ADAMTS13, in particular, its conversion from a closed to an open conformation, are involved in the pathogenesis of immune TTP. As yet, it is not fully understood how ADAMTS13 conformation is controlled in vivo. Human leukocyte antigens (73, 74, 75) provide a risk factor for immune TTP. Environmental triggers such as infections (76, 77, 78), pregnancy (79), drugs (80), and vaccines (81) may induce conformational changes that are linked to the onset of immune TTP. Future studies should reveal whether and how environmental triggers induce conformational changes in ADAMTS13 that are linked to the onset or relapses in patients with immune TTP. In addition, novel high-resolution structures corresponding to the open and closed conformations of ADAMTS13 are needed to reveal how conformational plasticity of ADAMTS13 contributes to the onset of immune TTP.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The research leading to these results has received funding from the Horizon 2020 Framework Program for Research and Innovation of the European Union under Grant Agreement No 675746, Netherlands Ministry of Health, Welfare and Sport (PPOC-18-022), and Netherlands Thrombosis Foundation (TSN2019-04).
Author contributions
B. E., T. A., J. H., and J. V. conceptualization; B. E. data curation; B. E. and J. V. investigation; B. E., T. A., J. H., and K. W. visualization; B. E., K. W., and J. V. methodology; B. E., T. A., J. H., K. W., C. P. M. R., K. V., G. A. F. N., and J. V. writing–original draft; T. A. and K. V. writing–review and editing; J. V. resources; J. V. validation.
Edited by Peter Cresswell
References
- 1.Dendrou C.A., Petersen J., Rossjohn J., Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 2.Curran A.M., Naik P., Giles J.T., Darrah E. PAD enzymes in rheumatoid arthritis: Pathogenic effectors and autoimmune targets. Nat. Rev. Rheumatol. 2020;16:301–315. doi: 10.1038/s41584-020-0409-1. [DOI] [PubMed] [Google Scholar]
- 3.Ludwig R.J., Vanhoorelbeke K., Leypoldt F., Kaya Z., Bieber K., McLachlan S.M., Komorowski L., Luo J., Cabral-Marques O., Hammers C.M., Lindstrom J.M., Lamprecht P., Fischer A., Riemekasten G., Tersteeg C. Mechanisms of autoantibody-induced pathology. Front. Immunol. 2017;8:603. doi: 10.3389/fimmu.2017.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Laat B., de Groot P.G. Autoantibodies directed against domain I of beta2-glycoprotein I. Curr. Rheumatol. Rep. 2011;13:70–76. doi: 10.1007/s11926-010-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreimann M., Brandt S., Krauel K., Block S., Helm C.A., Weitschies W., Greinacher A., Delcea M. Binding of anti-platelet factor 4/heparin antibodies depends on the thermodynamics of conformational changes in platelet factor 4. Blood. 2014;124:2442–2449. doi: 10.1182/blood-2014-03-559518. [DOI] [PubMed] [Google Scholar]
- 6.Sukumar S., Lämmle B., Cataland S.R. Thrombotic thrombocytopenic purpura: Pathophysiology, diagnosis, and management. J. Clin. Med. 2021;10:536. doi: 10.3390/jcm10030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kremer Hovinga J.A., Coppo P., Lämmle B., Moake J.L., Miyata T., Vanhoorelbeke K. Thrombotic thrombocytopenic purpura. Nat. Rev. Dis. Primers. 2017;3:17020. doi: 10.1038/nrdp.2017.20. [DOI] [PubMed] [Google Scholar]
- 8.Joly B.S., Coppo P., Veyradier A. Thrombotic thrombocytopenic purpura. Blood. 2017;129:2836–2846. doi: 10.1182/blood-2016-10-709857. [DOI] [PubMed] [Google Scholar]
- 9.Dong J.F., Moake J.L., Nolasco L., Bernardo A., Arceneaux W., Shrimpton C.N., Schade A.J., McIntire L.V., Fujikawa K., López J.A. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 10.Motto D.G., Chauhan A.K., Zhu G., Homeister J., Lamb C.B., Desch K.C., Zhang W., Tsai H.M., Wagner D.D., Ginsburg D. Shigatoxin triggers thrombotic thrombocytopenic purpura in genetically susceptible ADAMTS13-deficient mice. J. Clin. Invest. 2005;115:2752–2761. doi: 10.1172/JCI26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner D.D., Marder V.J. Biosynthesis of von Willebrand protein by human endothelial cells: Processing steps and their intracellular localization. J. Cell Biol. 1984;99:2123–2130. doi: 10.1083/jcb.99.6.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler J.E. von Willebrand factor. J. Biol. Chem. 1991;266:22777–22780. [PubMed] [Google Scholar]
- 13.Wagner D.D., Olmsted J.B., Marder V.J. Immunolocalization of von Willebrand protein in Weibel-Palade bodies of human endothelial cells. J. Cell Biol. 1982;95:355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karampini E., Bürgisser P.E., Olins J., Mulder A.A., Jost C.R., Geerts D., Voorberg J., Bierings R. Sec22b determines Weibel-Palade body length by controlling anterograde ER-Golgi transport. Haematologica. 2021;106:1138–1147. doi: 10.3324/haematol.2019.242727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leebeek F.W.G., Eikenboom J.C.J. Von Willebrand's disease. N. Engl. J. Med. 2017;376:701–702. doi: 10.1056/NEJMc1616060. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Halvorsen K., Zhang C.Z., Wong W.P., Springer T.A. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawley J.T., de Groot R., Xiang Y., Luken B.M., Lane D.A. Unraveling the scissile bond: How ADAMTS13 recognizes and cleaves von Willebrand factor. Blood. 2011;118:3212–3221. doi: 10.1182/blood-2011-02-306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petri A., Kim H.J., Xu Y., de Groot R., Li C., Vandenbulcke A., Vanhoorelbeke K., Emsley J., Crawley J.T.B. Crystal structure and substrate-induced activation of ADAMTS13. Nat. Commun. 2019;10:3781. doi: 10.1038/s41467-019-11474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roose E., Schelpe A.S., Joly B.S., Peetermans M., Verhamme P., Voorberg J., Greinacher A., Deckmyn H., De Meyer S.F., Coppo P., Veyradier A., Vanhoorelbeke K. An open conformation of ADAMTS-13 is a hallmark of acute acquired thrombotic thrombocytopenic purpura. J. Thromb. Haemost. 2018;16:378–388. doi: 10.1111/jth.13922. [DOI] [PubMed] [Google Scholar]
- 20.Roose E., Schelpe A.S., Tellier E., Sinkovits G., Joly B.S., Dekimpe C., Kaplanski G., Le Besnerais M., Mancini I., Falter T., Von Auer C., Feys H.B., Reti M., Rossmann H., Vandenbulcke A. Open ADAMTS13, induced by antibodies, is a biomarker for subclinical immune-mediated thrombotic thrombocytopenic purpura. Blood. 2020;136:353–361. doi: 10.1182/blood.2019004221. [DOI] [PubMed] [Google Scholar]
- 21.South K., Luken B.M., Crawley J.T., Phillips R., Thomas M., Collins R.F., Deforche L., Vanhoorelbeke K., Lane D.A. Conformational activation of ADAMTS13. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18578–18583. doi: 10.1073/pnas.1411979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.South K., Freitas M.O., Lane D.A. Conformational quiescence of ADAMTS-13 prevents proteolytic promiscuity. J. Thromb. Haemost. 2016;14:2011–2022. doi: 10.1111/jth.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roose E., Veyradier A., Vanhoorelbeke K. Insights into ADAMTS13 structure: Impact on thrombotic thrombocytopenic purpura diagnosis and management. Curr. Opin. Hematol. 2020;27:320–326. doi: 10.1097/MOH.0000000000000602. [DOI] [PubMed] [Google Scholar]
- 24.Kelwick R., Desanlis I., Wheeler G.N., Edwards D.R. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Groot R., Lane D.A., Crawley J.T. The role of the ADAMTS13 cysteine-rich domain in VWF binding and proteolysis. Blood. 2015;125:1968–1975. doi: 10.1182/blood-2014-08-594556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Y., de Groot R., Crawley J.T.B., Lane D.A. Mechanism of von Willebrand factor scissile bond cleavage by a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13) Proc. Natl. Acad. Sci. U. S. A. 2011;108:11602–11607. doi: 10.1073/pnas.1018559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furlan M., Robles R., Lämmle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87:4223–4234. [PubMed] [Google Scholar]
- 28.Fujikawa K., Suzuki H., McMullen B., Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 29.Tsai H.M. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 30.Gerritsen H.E., Robles R., Lämmle B., Furlan M. Partial amino acid sequence of purified von Willebrand factor-cleaving protease. Blood. 2001;98:1654–1661. doi: 10.1182/blood.v98.6.1654. [DOI] [PubMed] [Google Scholar]
- 31.Akiyama M., Takeda S., Kokame K., Takagi J., Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H.J., Xu Y., Petri A., Vanhoorelbeke K., Crawley J.T.B., Emsley J. Crystal structure of ADAMTS13 CUB domains reveals their role in global latency. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abg4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tortorella M., Pratta M., Liu R.Q., Abbaszade I., Ross H., Burn T., Arner E. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J. Biol. Chem. 2000;275:25791–25797. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 34.Kashiwagi M., Enghild J.J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 35.Gendron C., Kashiwagi M., Lim N.H., Enghild J.J., Thøgersen I.B., Hughes C., Caterson B., Nagase H. Proteolytic activities of human ADAMTS-5: Comparative studies with ADAMTS-4. J. Biol. Chem. 2007;282:18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 36.Flannery C.R., Zeng W., Corcoran C., Collins-Racie L.A., Chockalingam P.S., Hebert T., Mackie S.A., McDonagh T., Crawford T.K., Tomkinson K.N., LaVallie E.R., Morris E.A. Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J. Biol. Chem. 2002;277:42775–42780. doi: 10.1074/jbc.M205309200. [DOI] [PubMed] [Google Scholar]
- 37.Fushimi K., Troeberg L., Nakamura H., Lim N.H., Nagase H. Functional differences of the catalytic and non-catalytic domains in human ADAMTS-4 and ADAMTS-5 in aggrecanolytic activity. J. Biol. Chem. 2008;283:6706–6716. doi: 10.1074/jbc.M708647200. [DOI] [PubMed] [Google Scholar]
- 38.Deforche L., Roose E., Vandenbulcke A., Vandeputte N., Feys H.B., Springer T.A., Mi L.Z., Muia J., Sadler J.E., Soejima K., Rottensteiner H., Deckmyn H., De Meyer S.F., Vanhoorelbeke K. Linker regions and flexibility around the metalloprotease domain account for conformational activation of ADAMTS-13. J. Thromb. Haemost. 2015;13:2063–2075. doi: 10.1111/jth.13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schelpe A.-S., Petri A., Roose E., Pareyn I., Deckmyn H., De Meyer S.F., Crawley J.T.B., Vanhoorelbeke K. Antibodies that conformationally activate ADAMTS13 allosterically enhance metalloprotease domain function. Blood Adv. 2020;4:1072–1080. doi: 10.1182/bloodadvances.2019001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jian C., Xiao J., Gong L., Skipwith C.G., Jin S.Y., Kwaan H.C., Zheng X.L. Gain-of-function ADAMTS13 variants that are resistant to autoantibodies against ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Blood. 2012;119:3836–3843. doi: 10.1182/blood-2011-12-399501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muia J., Zhu J., Gupta G., Haberichter S.L., Friedman K.D., Feys H.B., Deforche L., Vanhoorelbeke K., Westfield L.A., Roth R., Tolia N.H., Heuser J.E., Sadler J.E. Allosteric activation of ADAMTS13 by von Willebrand factor. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18584–18589. doi: 10.1073/pnas.1413282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.South K., Freitas M.O., Lane D.A. A model for the conformational activation of the structurally quiescent metalloprotease ADAMTS13 by von Willebrand factor. J. Biol. Chem. 2017;292:5760–5769. doi: 10.1074/jbc.M117.776732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J., Muia J., Gupta G., Westfield L.A., Vanhoorelbeke K., Tolia N.H., Sadler J.E. Exploring the “minimal” structure of a functional ADAMTS13 by mutagenesis and small-angle X-ray scattering. Blood. 2019;133:1909–1918. doi: 10.1182/blood-2018-11-886309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muia J., Zhu J., Greco S.C., Vanhoorelbeke K., Gupta G., Westfield L.A., Sadler J.E. Phylogenetic and functional analysis of ADAMTS13 identifies highly conserved domains essential for allosteric regulation. Blood. 2019;133:1899–1908. doi: 10.1182/blood-2018-11-886275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu H., Jiang Y., Yang D., Scheiflinger F., Wong W.P., Springer T.A. Flow-induced elongation of von Willebrand factor precedes tension-dependent activation. Nat. Commun. 2017;8:324. doi: 10.1038/s41467-017-00230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q., Zhou Y.F., Zhang C.Z., Zhang X., Lu C., Springer T.A. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Groot R., Bardhan A., Ramroop N., Lane D.A., Crawley J.T.B. Essential role of the disintegrin-like domain in ADAMTS13 function. Blood. 2009;113:5609–5616. doi: 10.1182/blood-2008-11-187914. [DOI] [PubMed] [Google Scholar]
- 48.Pos W., Crawley J.T., Fijnheer R., Voorberg J., Lane D.A., Luken B.M. An autoantibody epitope comprising residues R660, Y661, and Y665 in the ADAMTS13 spacer domain identifies a binding site for the A2 domain of VWF. Blood. 2010;115:1640–1649. doi: 10.1182/blood-2009-06-229203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ercig B., Wichapong K., Reutelingsperger C.P.M., Vanhoorelbeke K., Voorberg J., Nicolaes G.A.F. Insights into 3D structure of ADAMTS13: A stepping stone towards novel therapeutic treatment of thrombotic thrombocytopenic purpura. Thromb. Haemost. 2018;118:28–41. doi: 10.1160/TH17-06-0404. [DOI] [PubMed] [Google Scholar]
- 50.Yu S., Liu W., Fang J., Shi X., Wu J., Fang Y., Lin J. AFM imaging reveals multiple conformational states of ADAMTS13. J. Biol. Eng. 2019;13:9. doi: 10.1186/s13036-018-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verbij F.C., Stokhuijzen E., Kaijen P.H., van Alphen F., Meijer A.B., Voorberg J. Identification of glycans on plasma-derived ADAMTS13. Blood. 2016;128:e51–e58. doi: 10.1182/blood-2016-06-720912. [DOI] [PubMed] [Google Scholar]
- 52.Nowak A.A., O'Brien H.E.R., Henne P., Doerr A., Vanhoorelbeke K., Laffan M.A., McKinnon T.A.J. ADAMTS-13 glycans and conformation-dependent activity. J. Thromb. Haemost. 2017;15:1155–1166. doi: 10.1111/jth.13688. [DOI] [PubMed] [Google Scholar]
- 53.Nagae M., Yamaguchi Y. Function and 3D structure of the N-glycans on glycoproteins. Int. J. Mol. Sci. 2012;13:8398–8429. doi: 10.3390/ijms13078398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrdinová J., D'Angelo S., Graça N.A.G., Ercig B., Vanhoorelbeke K., Veyradier A., Voorberg J., Coppo P. Dissecting the pathophysiology of immune thrombotic thrombocytopenic purpura: Interplay between genes and environmental triggers. Haematologica. 2018;103:1099–1109. doi: 10.3324/haematol.2016.151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas M.R., de Groot R., Scully M.A., Crawley J.T. Pathogenicity of anti-ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. EBioMedicine. 2015;2:942–952. doi: 10.1016/j.ebiom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng X.L., Wu H., Shang D., Falls E., Skipwith C., Cataland S., Bennett C., Kwaan H. Multiple domains of ADAMTS13 are targeted by autoantibodies against ADAMTS13 in patients with acquired idiopathic thrombotic thrombocytopenic purpura. Haematologica. 2010;95:1555–1562. doi: 10.3324/haematol.2009.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pos W., Sorvillo N., Fijnheer R., Feys H.B., Kaijen P.H., Vidarsson G., Voorberg J. Residues Arg568 and Phe592 contribute to an antigenic surface for anti-ADAMTS13 antibodies in the spacer domain. Haematologica. 2011;96:1670–1677. doi: 10.3324/haematol.2010.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klaus C., Plaimauer B., Studt J.D., Dorner F., Lämmle B., Mannucci P.M., Scheiflinger F. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103:4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 59.Kangro K., Roose E., Joly B S., Sinkovits G., Falter T., von Auer C., Rossmann H., Reti M., Voorberg J., Prohaszka Z., Lämmle B., Coppo P., Veyradier A., de Meyer S.F., Männik A. Anti-ADAMTS13 autoantibody profiling in immune-mediated thrombotic thrombocytopenic purpura patients. Blood Adv. 2021;5:3427–3435. doi: 10.1182/bloodadvances.2020004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verbij F.C., Fijnheer R., Voorberg J., Sorvillo N. Acquired TTP: ADAMTS13 meets the immune system. Blood Rev. 2014;28:227–234. doi: 10.1016/j.blre.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 61.Albiol N., Awol R., Martino R. Autoimmune thrombotic thrombocytopenic purpura (TTP) associated with COVID-19. Ann. Hematol. 2020;99:1673–1674. doi: 10.1007/s00277-020-04097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marco C., Cristina M., Chiara A., Ilaria M., Daniele P., Flora P. Dramatic presentation of acquired thombotic thrombocytopenic purpura associated with COVID-19. Haematologica. 2020;105 doi: 10.3324/haematol.2020.262345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goshua G., Pine A.B., Meizlish M.L., Chang C.H., Zhang H., Bahel P., Baluha A., Bar N., Bona R.D., Burns A.J., Dela Cruz C.S., Dumont A., Halene S., Hwa J., Koff J. Endotheliopathy in COVID-19-associated coagulopathy: Evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7:e575–e582. doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Witalison E.E., Thompson P.R., Hofseth L.J. Protein arginine deiminases and associated citrullination: Physiological functions and diseases associated with dysregulation. Curr. Drug Targets. 2015;16:700–710. doi: 10.2174/1389450116666150202160954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohrbach A.S., Slade D.J., Thompson P.R., Mowen K.A. Activation of PAD4 in NET formation. Front. Immunol. 2012;3:360. doi: 10.3389/fimmu.2012.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorvillo N., Mizurini D.M., Coxon C., Martinod K., Tilvawala R., Cherpokova D., Salinger A.J., Seward R.J., Staudinger C., Weerapana E., Shapiro N.I., Costello C.E., Thompson P.R., Wagner D.D. Plasma peptidylarginine deiminase IV promotes VWF-platelet string formation and accelerates thrombosis after vessel injury. Circ. Res. 2019;125:507–519. doi: 10.1161/CIRCRESAHA.118.314571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darrah E., Andrade F. Rheumatoid arthritis and citrullination. Curr. Opin. Rheumatol. 2018;30:72–78. doi: 10.1097/BOR.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lancellotti S., Peyvandi F., Pagliari M.T., Cairo A., Abdel-Azeim S., Chermak E., Lazzareschi I., Mastrangelo S., Cavallo L., Oliva R., De Cristofaro R. The D173G mutation in ADAMTS-13 causes a severe form of congenital thrombotic thrombocytopenic purpura. A clinical, biochemical and in silico study. Thromb. Haemost. 2016;115:51–62. doi: 10.1160/TH15-02-0119. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y., Chen J., Ling M., López J.A., Chung D.W., Fu X. Hypochlorous acid generated by neutrophils inactivates ADAMTS13: An oxidative mechanism for regulating ADAMTS13 proteolytic activity during inflammation. J. Biol. Chem. 2015;290:1422–1431. doi: 10.1074/jbc.M114.599084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aledo J.C., Cantón F.R., Veredas F.J. Sulphur atoms from methionines interacting with aromatic residues are less prone to oxidation. Sci. Rep. 2015;5:16955. doi: 10.1038/srep16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valley C.C., Cembran A., Perlmutter J.D., Lewis A.K., Labello N.P., Gao J., Sachs J.N. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim G., Weiss S.J., Levine R.L. Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta. 2014;1840:901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joly B.S., Loiseau P., Darmon M., Leblanc T., Chambost H., Fouyssac F., Guigonis V., Harambat J., Stepanian A., Coppo P., Veyradier A. HLA-DRB1∗11 is a strong risk factor for acquired thrombotic thrombocytopenic purpura in children. Haematologica. 2020;105 doi: 10.3324/haematol.2019.241968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verbij F.C., Turksma A.W., de Heij F., Kaijen P., Lardy N., Fijnheer R., Sorvillo N., ten Brinke A., Voorberg J. CD4+ T cells from patients with acquired thrombotic thrombocytopenic purpura recognize CUB2 domain-derived peptides. Blood. 2016;127:1606–1609. doi: 10.1182/blood-2015-10-668053. [DOI] [PubMed] [Google Scholar]
- 75.Sakai K., Kuwana M., Tanaka H., Hosomichi K., Hasegawa A., Uyama H., Nishio K., Omae T., Hishizawa M., Matsui M., Iwato K., Okamoto A., Okuhiro K., Yamashita Y., Itoh M. HLA loci predisposing to immune TTP in Japanese: Potential role of the shared ADAMTS13 peptide bound to different HLA-DR. Blood. 2020;135:2413–2419. doi: 10.1182/blood.2020005395. [DOI] [PubMed] [Google Scholar]
- 76.Koh Y.R., Hwang S.H., Chang C.L., Lee E.Y., Son H.C., Kim H.H. Thrombotic thrombocytopenic purpura triggered by influenza A virus subtype H1N1 infection. Transfus. Apher. Sci. 2012;46:25–28. doi: 10.1016/j.transci.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 77.Deepanjali S., Naik R.R., Mailankody S., Kalaimani S., Kadhiravan T. Dengue virus infection triggering thrombotic thrombocytopenic purpura in pregnancy. Am. J. Trop. Med. Hyg. 2015;93:1028–1030. doi: 10.4269/ajtmh.15-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saab K.R., Elhadad S., Copertino D., Laurence J. Thrombotic microangiopathy in the setting of HIV infection: A case report and review of the differential diagnosis and therapy. AIDS Patient Care STDs. 2016;30:359–364. doi: 10.1089/apc.2016.0124. [DOI] [PubMed] [Google Scholar]
- 79.von Auer C., von Krogh A.S., Kremer Hovinga J.A., Lämmle B. Current insights into thrombotic microangiopathies: Thrombotic thrombocytopenic purpura and pregnancy. Thromb. Res. 2015;135 Suppl 1:S30–S33. doi: 10.1016/S0049-3848(15)50437-4. [DOI] [PubMed] [Google Scholar]
- 80.Jacob S., Dunn B.L., Qureshi Z.P., Bandarenko N., Kwaan H.C., Pandey D.K., McKoy J.M., Barnato S.E., Winters J.L., Cursio J.F., Weiss I., Raife T.J., Carey P.M., Sarode R., Kiss J.E. Ticlopidine-, clopidogrel-, and prasugrel-associated thrombotic thrombocytopenic purpura: A 20-year review from the southern network on adverse reactions (SONAR) Semin. Thromb. Hemost. 2012;38:845–853. doi: 10.1055/s-0032-1328894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sissa C., Al-Khaffaf A., Frattini F., Gaiardoni R., Mimiola E., Montorsi P., Melara B., Amato M., Peyvandi F., Franchini M. Relapse of thrombotic thrombocytopenic purpura after COVID-19 vaccine. Transfus. Apher. Sci. 2021;60:103145. doi: 10.1016/j.transci.2021.103145. [DOI] [PMC free article] [PubMed] [Google Scholar]