Abstract
The promoters of Drosophila genes encoding DNA replication-related proteins contain transcription regulatory elements consisting of an 8-bp palindromic DNA replication-related element (DRE) sequence (5′-TATCGATA). The specific DRE-binding factor (DREF), a homodimer of the polypeptide with 709 amino acid residues, is a positive trans-acting factor for transcription of DRE-containing genes. Both DRE binding and dimer formation are associated with residues 16 to 115 of the N-terminal region. We have established transgenic flies expressing the full-length DREF polypeptide or its N-terminal fragment (amino acid residues 1 to 125) under the control of the heat shock promoter, the salivary gland-specific promoter, or the eye imaginal disc-specific promoter. Heat shock induction of the N-terminal fragment during embryonic, larval, or pupal stages caused greater than 50% lethality. This lethality was overcome by coexpression of the full-length DREF. In salivary glands of the transgenic larvae expressing the N-terminal fragment, this fragment formed a homodimer and a heterodimer with the endogenous DREF. Ectopic expression of the N-terminal fragment in salivary gland cells reduced the contents of mRNAs for the 180-kDa subunit of DNA polymerase α and for dE2F and the extent of DNA endoreplication. Ectopic expression of the N-terminal fragment in the eye imaginal discs significantly reduced DNA replication in cells at the second mitotic wave. The lines of evidence suggest that the N-terminal fragment can impede the endogenous DREF function in a dominant negative manner and that DREF is required for normal DNA replication in both mitotic cell cycle and endo cycle.
The promoters of Drosophila genes involved in DNA replication, such as those for the 180-kDa catalytic subunit and the 73-kDa subunit of DNA polymerase α and for proliferating cell nuclear antigen (PCNA), contain DNA replication-related elements (DREs) characterized by a common 8-bp palindromic sequence (5′-TATCGATA) (13, 14, 30) in addition to E2F recognition sites (4, 20, 30, 36). The requirement of DREs for promoter activation has been confirmed with both cultured cells and transgenic flies carrying a PCNA-lacZ reporter (14, 37, 38). Introduction of mutations in the DRE sequence resulted in almost complete loss of the PCNA promoter activity in larval tissues, including the salivary gland and imaginal discs. Detailed analysis of the PCNA gene promoter with transgenic flies revealed that DRE–DRE-binding factor (DRE-DREF) is required for expression of the PCNA gene throughout development, except in the ovary of adult females (38).
We have purified a specific DREF and found it to consist of an 80-kDa polypeptide homodimer (15). Recently, we compared cDNAs and genes for DREFs from Drosophila melanogaster and Drosophila virilis (31). Elucidation of their amino acid sequences revealed three domains to be evolutionally conserved. One of the highly conserved domains corresponds to the N-terminal basic amino acid-containing region (amino acid residues 16 to 115) which is responsible for both DRE binding and homodimer formation (15). Although we have not identified the transactivation domain(s) of DREF, the C-terminal region between amino acid residues 240 and 607 is presumably involved, because a monoclonal antibody (MAb) whose epitope is located in this region inhibited in vitro transcription of the DNA polymerase α gene in Kc cell nuclear extracts (15). However, we recently found that at least two additional factors, CFDD (common regulatory factor for DNA replication and DREF genes) and BEAF-32 (boundary element-associated factor of 32 kDa) also bind to the DRE sequence in vitro (9, 11, 39). Thus, a requirement of DRE for expression of DNA replication-related genes does not necessarily indicate that DREF is the most important factor acting as a positive regulator in vivo. Therefore, we have concentrated on clarifying the contribution of DREF to regulation of DRE-containing genes in living flies.
The most direct way to address the biological roles of DREF in living flies is to analyze the phenotypes of flies with mutations in the DREF gene. However, fly lines having deletions in the 30F region, where the DREF gene is located, are not available, and we have obtained results suggesting that the region surrounding the DREF gene might be a “cold spot” for P-element insertion (unpublished results). In the present study, therefore, we tried to make transgenic fly lines expressing the N-terminal fragment of the DREF polypeptide. We expected that overexpression of the fragment in vivo might compete with the endogenous DREF for DRE binding and impede DREF function in a dominant negative manner. By expressing the N-terminal fragment of DREF by using the GAL4-UAS-targeted system, we found that DREF is required for normal DNA replication in both mitotic cell cycle and endo cycle.
MATERIALS AND METHODS
Establishment of transgenic flies and fly stocks.
Fly stocks were maintained at 25°C on standard food. The Canton S fly was used as the wild-type strain. P-element-mediated germ line transformation was carried out as described previously (29), and F1 transformants were selected on the basis of white eye color rescue (23). Multiple independent lines were obtained for each of the various transgene constructs.
Lines with UAS-DREF1–709 and UAS-DREF1–125 transgenes were obtained with pUAST constructs (1) according to standard procedures. The line expressing GAL4 under the control of the hsp70 gene promoter or the salivary gland-specific promoter has been described by Brand and Perrimon (1). Establishment of lines carrying GMR-GAL4 was described earlier (23, 31).
Ectopic expression of DREF polypeptide. (i) Heat shock induction.
The line carrying homozygous hs-GAL4 in the third chromosome, provided by Brand and Perrimon (1), was crossed with both lines carrying the homozygous P[UAS-DREF] in the second chromosome. The eggs were counted and transferred to plastic tubes. Staged embryos, larvae, and pupae were heat shocked at 37°C for 45 min and then returned to 25°C and allowed to develop into adults.
(ii) Expression in the larval salivary gland.
The GAL4 enhancer trap line has an insertion in the X chromosome and expresses GAL4 in salivary gland cells from embryonic through larval stages (1, 7). P[Sg-GAL4](l)/Binsinscy females were crossed with lines carrying homozygous P[UAS-DREF] in the second chromosome. The larvae with and without P[Sg-GAL4] were distinguished with reference to the y+ marker.
(iii) Expression in the eye imaginal disc.
Females carrying pGMR-GAL4 (10, 31) on the X chromosome were crossed with males carrying homozygous P[UAS-DREF] in the second chromosome.
BrdU labeling.
Detection of cells in S phase was performed by a bromodeoxyuridine (BrdU)-labeling method as described previously (35), with minor modifications. For salivary gland analysis, larvae (36 h after hatching) were dissected in Grace’s medium and then incubated in the presence of 20 μg of BrdU (Boehringer) per ml for 30 min. The samples were fixed in Carnoy’s fixative (ethanol-acetic acid-chloroform [6:3:1]) for 15 min at 25°C and further fixed in 80% ethanol–50 mM glycine buffer, pH 2.0, at −20°C for 2 h. Incorporated BrdU was visualized with an anti-BrdU antibody and an alkaline phosphatase detection kit (Boehringer). The period of color development for alkaline phosphatase was precisely the same for all samples. For labeling eye imaginal discs, late-third-instar larvae were dissected in Grace’s medium and incubated in the presence of 20 μg of BrdU (Boehringer) per ml for 30 min.
Immunoprecipitation.
Third-instar larvae were dissected in phosphate-buffered saline (PBS), and salivary glands were removed. Extracts were made by sonicating salivary glands for 10 s at 4°C in solution E, containing 20 mM HEPES (pH 7.6), 150 mM NaCl, 10% glycerol, 0.3% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg each of aprotinin and leupeptin per ml, and 1 μg each of pepstatin, chymostatin, and phosphoramidon per ml. After centrifugation at 10,000 × g for 20 min, the supernatants were incubated with 10 μl of protein G-Sepharose beads (Pharmacia Biotech Inc.) for 1 h at 4°C and separated into pairs of aliquots. Each aliquot was then incubated with protein G-Sepharose beads saturated with control immunoglobulin G (IgG) or anti-DREF MAb 1. The mixtures were further incubated for 2 h at 4°C and then washed three times with solution E without proteinase inhibitors. The immunoprecipitates were boiled for 5 min in 30 μl of sample buffer for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and the eluates were subjected to SDS-polyacrylamide gel electrophoresis followed by Western immunoblotting.
Western immunoblot analysis.
Embryos of the wild type and lines carrying hs-GAL4 and UAS-DREF1–125 transgenes were dechorionated and homogenized in a solution containing 50 mM Tris-HCl (pH 7.6), 400 mM KCl, 0.1% Triton X-100, 1 mM dithiothreitol, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg each of aprotinin and leupeptin per ml, and 1 μg each of pepstatin, chymostatin, and phosphoramidon per ml at various times after heat shock. Homogenates were centrifuged at 100,000 × g at 4°C for 30 min, and polypeptides (20 μg of protein) in the supernatants were electrophoretically separated on SDS–12% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore) in a solution containing 50 mM borate-NaOH (pH 9.0) and 20% methanol at 4°C for 4 h. Blotted membranes were blocked with Tris-buffered saline (TBS) solution (50 mM Tris-HCl, pH 8.3, and 150 mM NaCl) containing 20% fetal calf serum for 30 min at room temperature and then incubated with culture supernatant of a hybridoma producing anti-DREF MAb 1 at a 1:200 dilution. The epitope for MAb 1 is located within the DNA binding domain between amino acid residues 32 and 115 of the DREF polypeptide (15). Thus, this antibody can detect the NH2-terminal region containing the DRE-binding domain in addition to detecting full-length DREF polypeptides. After extensive washing with TBS, the blots were incubated with an alkaline phosphatase-conjugated goat anti-mouse IgG (Promega) at a 1:2,000 dilution for 2 h at room temperature. After extensive washing with TBS, color was developed in a solution containing 100 mM Tris-HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.34 mg of nitroblue tetrazolium salt per ml, and 0.175 mg of 5-bromo-4-chloro-3-indolylphosphate toluidinium salt (BCIP) per ml.
Gel mobility shift assay.
Gel mobility shift assays were performed as described previously (13). Oligonucleotides used for the probe and competitor were described previously (37).
Whole-mount in situ hybridization.
pBluescript II SK(−) plasmids containing cDNA fragments for the DNA polymerase α 180-kDa subunit (12), dE2F (5, 20), and ribosomal protein 49 (rp49) (22) were used as templates for in vitro transcription with a digoxigenin (DIG) RNA-labeling kit (Boehringer). The probe length was reduced to 100 to 300 bases by alkaline hydrolysis according to the method of Cox et al. (3). Second-instar larvae of wild-type and transgenic strains were dissected in PBS. Tissues containing salivary glands and imaginal discs were fixed by treatment with 4% paraformaldehyde in PBS for 20 min on ice and with 4% paraformaldehyde–0.6% Triton X-100 in PBS for 20 min at room temperature. After being washed with PBS–0.1% Tween 20 (PBT), tissues were washed with PBT-hybridization solution (1:1) for 10 min at room temperature. The hybridization solution contained 50% deionized formamide, 5× SSC (1× SSC is 0.15 M NaCL plus 0.015 M sodium citrate), 200 μg of tRNA per ml, 100 μg of heat-denatured salmon sperm DNA per ml, and 0.1% Tween 20. After prehybridization in hybridization solution at 48°C for 1 h, the probe was added to a final concentration of 400 ng/ml. After 24 h of hybridization at 48°C, the samples were washed for 12 h at 48°C, with a change of PBT every 2 h, and then incubated for 1 h at room temperature in a 1:2,000 dilution of anti-DIG antibody conjugated to alkaline phosphatase (Boehringer) which had been preabsorbed for 1 h with fixed larval heads. Alkaline phosphatase activity was detected by incubating the tissues in a solution containing 100 mM Tris-HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.34 mg of nitroblue tetrazolium salt per ml, and 0.175 mg of BCIP per ml. The tissues were washed with PBT and mounted in 90% glycerol–PBS for microscopic observation.
RESULTS
Expression of the N-terminal fragment (amino acid residues 1 to 125) of DREF in transgenic flies.
Ectopic expression of the N-terminal fragment of DREF in living flies was performed by using a GAL4-mediated expression system (1, 6). The cDNA region encoding the N-terminal fragment (amino acid residues 1 to 125) in which activities for DRE binding and dimer formation are located was subcloned into the pUAST vector, and the resultant plasmid was designated UAS-DREF1–125. Four independent lines of germ line transformants carrying UAS-DREF1–125 were established and used for the analysis. Note that no phenotypic differences were observed among these lines. Transgenic flies carrying UAS-DREF1–125 were then crossed with transgenic flies carrying GAL4 cDNA put under the control of the thermoinducible hsp70 gene promoter (hs-GAL4), of the salivary gland-specific enhancer-promoter (Sg-GAL4), or of the eye imaginal disc-specific promoter (GMR-GAL4).
Ectopic expression of the N-terminal fragment in the transgenic animals was confirmed by Western immunoblotting and gel mobility shift assay with tissue extracts or immunohistochemical staining with specific antibodies. Embryos carrying single copies of hs-GAL4 and UAS-DREF1–125 before and after heat shock for 45 min at 37°C were homogenized, and amounts of DREF polypeptides in the extracts were determined with anti-DREF MAb 1. Since the epitope of MAb 1 is located in the region between amino acid residues 32 and 115, this antibody reacts to both the full-length DREF and the N-terminal fragment (15). In addition to expression of the endogenous full-length DREF, heat shock-dependent expression of the N-terminal fragment was observed (Fig. 1A). Although hardly detectable at 2 h after heat shock, it increased with time to reach a maximal level at 6 h and then gradually decreased (data not shown). The molecular number of the N-terminal fragment at 6 h after heat shock was estimated to be about 10% of that for the endogenous DREF polypeptide.
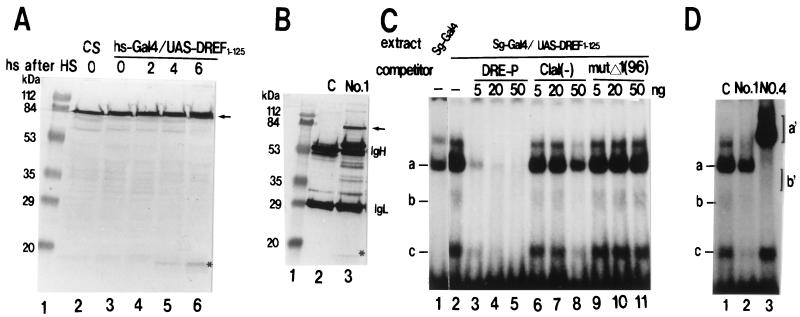
FIG. 1.
Western immunoblotting and gel mobility shift assay to detect endogenous DREF and ectopically expressed DREF1–125. (A) Extracts were prepared from embryos of Canton S (CS) flies (lane 2) and from transgenic embryos carrying hs-GAL4 and UAS-DREF1–125 without heat shock (HS) (lane 3), at 2 h after HS (lane 4), at 4 h after HS (lane 5), and at 6 h after HS (lane 6), and 20-μg aliquots of proteins were analyzed by Western immunoblotting with anti-DREF MAb 1. The arrow indicates signals for the endogenous DREF polypeptide. Signals for DREF1–125 are indicated with an asterisk. Lane 1, size markers. (B) Extracts were prepared from salivary glands from third-instar larvae carrying Sg-GAL4 and UAS-DREF1–125. Endogenous DREF and the N-terminal fragment were immunoprecipitated by using protein G-Sepharose beads with control IgG (lane 2) or anti-DREF MAb 1 (lane 3) and then analyzed by immunoblotting with anti-DREF MAb 1 (lanes 2 and 3). Samples for each lane contained 100 μg of protein. The arrow indicates signals for the endogenous DREF polypeptide. Signals for DREF1–125 are indicated with an asterisk. (C) Radiolabeled double-stranded DRE-P oligonucleotides were incubated with salivary gland extracts in the presence or absence of competitor oligonucleotide (11, 13). Lane 1, salivary gland extract of transgenic larvae carrying only Sg-GAL4; lanes 2 to 11, salivary gland extracts of transgenic larvae carrying Sg-GAL4 and UAS-DREF1–125. (D) Extracts were prepared from salivary glands from third-instar larvae carrying Sg-GAL4 and UAS-DREF1–125. Aliquots (4 μl) were preincubated with control antibody (C) (lane 1), anti-DREF MAb 1 (lane 2), or anti-DREF MAb 4 (lane 3) and then mixed with radiolabeled double-stranded DRE-P oligonucleotides.
It was difficult to quantify the amount of the N-terminal fragment of DREF by direct immunoblotting analysis with the whole extract of salivary glands of transgenic flies carrying a single copy each of Sg-GAL4 and UAS-DREF1–125. Thus, DREF polypeptides were first concentrated from salivary gland extracts (prepared from third-instar larvae) by immunoprecipitation with MAb 1 and then detected by immunoblotting with the same antibody (Fig. 1B). The amount of the N-terminal fragment of DREF was estimated to be about 20% that of the endogenous DREF.
DRE-binding activity of the N-terminal fragment in salivary glands from the transgenic flies expressing DREF1–125 was measured by a gel mobility shift assay. Three retarded bands (a, b, and c) of the DRE-P oligonucleotide probe were detected by adding salivary gland extracts from transgenic flies carrying Sg-GAL4 and UAS-DREF1–125 (Fig. 1C, lane 2). Two bands (b and c) were not detected in extracts of control salivary glands (Fig. 1C, lane 1).
All three bands were diminished by the addition of an excess amount of unlabeled DRE-P oligonucleotide as a competitor (Fig. 1C, lanes 3 to 5). Oligonucleotide ClaI(−) competed slightly with the complex formation of bands a, b, and c (Fig. 1C, lanes 6 to 8), while oligonucleotide mutΔ1(96) did not (Fig. 1C, lanes 9 to 11). Furthermore, preincubation of the extract with anti-DREF MAb 1 also reduced all three shifted bands (Fig. 1D, lane 2). The addition of anti-DREF MAb 4, on the other hand, diminished bands a and b and resulted in supershifted signals a′ and b′. However, the fastest-migrating band, band c, was not affected by the antibody (Fig. 1D, lane 4). Since the epitope of MAb 4 is located in the C-terminal half of the DREF polypeptide (15), the results indicate that bands a and b contain full-length DREF, while band c is the N-terminal fragment. These lines of evidence clearly demonstrated that bands a, b, and c correspond to DNA-protein complexes containing a homodimer of endogenous DREF (DREF1–709-DREF1–709), a heterodimer of DREF1–709-DREF1–125, and a homodimer of DREF1–125-DREF1–125, respectively. The DNA-binding activities of DREF1–125-DREF1–125 and DREF1–709-DREF1–125 in salivary gland extracts were estimated to be 35 and 15%, respectively, of that of the DREF homodimer.
Targeted expression of DREF1–125 in the eye imaginal disc was confirmed by immunostaining with MAbs 1 and 4 (Fig. 6).
FIG. 6.
Ectopic expression of DREF1–125 inhibits DNA replication of cells in the second mitotic wave. Shown are results for immunostaining of eye imaginal discs with anti-DREF MAb 1. (A) GMR-GAL4/+; +. (B) GMR-GAL4/+; UAS-DREF1–125/+. Patterns of BrdU incorporation in eye imaginal discs are apparent. (C) GMR-GAL4/+; +. (D) GMR-GAL4/+; UAS-DREF1–125/+. The eye discs from a third-instar larva were stained with an anti-BrdU antibody. Arrows indicate the position of the morphogenetic furrow (MF). The anterior (A) of the discs is on the left. P, posterior.
Expression of the N-terminal fragment of DREF causes lethality throughout developmental stages.
Biological activities of the N-terminal fragment of the DREF polypeptide during development were analyzed with transgenic flies carrying hs-GAL4 and UAS-DREF1–125. After being administered a single heat shock at 37°C for 45 min at various developmental stages, transgenic flies were incubated at 25°C so their survival to the adult stage could be monitored. Early embryos of both wild-type and transgenic flies before gastrulation (3 h after fertilization) were very sensitive to heat shock (21), while after this period more than 75% of wild-type individuals developed into adults (Fig. 2). On the other hand, less than half of the transgenic animals carrying both hs-GAL4 and UAS-DREF1–125 survived until the pupal or adult stage after heat shock at any stage. Although the surviving animals did reach adulthood, a 2-day delay in development was observed. The first-instar larvae were particularly sensitive to heat shock induction of DREF1–125.
FIG. 2.
Lethality in transgenic flies expressing DREF1–125. Eggs were counted and animals at various developmental stages were administered a single heat shock for 45 min at 37°C. The numbers of animals developing into adults were counted. The values shown were normalized for the rate of maturation into adults without heat treatment.
To assess whether overexpression of full-length DREF suppresses lethality caused by DREF1–125 expression, we established four independent transgenic lines bearing UAS-DREF1–709. Transgenic animals carrying one copy each of hs-GAL4, UAS-DREF1–125, and UAS-DREF1–709 developed as normally as wild-type Canton S, suggesting that lethality caused by the ectopic expression of DREF1–125 is rescued by overexpression of DREF1–709. The results suggest that DREF1–125 acts as a dominant negative effector in vivo and that DREF is required for normal development.
Heat shock induction of DREF1–125 caused another striking phenotype, generation of melanotic tumors (Fig. 3), which are thought to arise as a normal, heritable response to some form of abnormal development and are groups of cells that are recognized by the immune system and encapsulated in melanized cuticle (28, 34). Therefore, their formation in the ventral parts of larvae suggests that heat shock induction of DREF1–125 induced some abnormal cell proliferation or differentiation. Recently, Royzman et al. (24) reported that both E2F and DP mutant flies exhibit a dramatic delay in larval growth and the development of numerous small melanotic tumors.
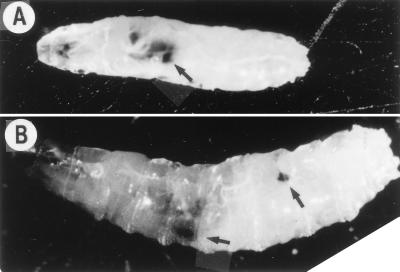
FIG. 3.
Melanotic tumors after heat shock induction of DREF1–125. (A) Melanotic tumor (arrow) observed in a second-instar larva at 24 h after heat shock. (B) Melanotic tumors (arrows) observed in a third-instar larva at 24 h after heat shock. Note that more than half of larvae had died by that time.
Expression of the N-terminal fragment reduces endoreplication in salivary gland cells.
The heat shock experiments described above suggested that DREF may be required for normal development. However, it was difficult to clarify the molecular events occurring in embryos after heat shock induction of DREF1–125 because of lethality. Therefore, we next analyzed the consequence of targeted ectopic expression of DREF1–125 in the salivary gland with an enhancer trap line in which GAL4 is expressed under the salivary gland-specific enhancer. This experiment also allowed examination of the requirement of DREF for endoreplication in this tissue.
Several transgenic lines carrying UAS-DREF1–125 were crossed with the Sg-GAL4 line (1), which exhibits GAL4 activity only in the embryonic and larval salivary glands, demonstrated by crossing with a transgenic fly line carrying the UAS-lacZ reporter (1, 2).
To assess whether expression of the N-terminal DREF fragment in salivary glands reduces transcription of DRE-containing genes, the levels of mRNAs for the DNA polymerase α 180-kDa subunit (12) and dE2F (5, 20) in the salivary glands were determined by in situ hybridization with or without expressing DREF1–125. As shown in Fig. 4D and F, the signals for mRNAs for the DNA polymerase α 180-kDa subunit and dE2F were obviously reduced in salivary glands with expression of DREF1–125, indicating that both genes are under the regulation of DREF in salivary glands. In a cultured cell system and by in vitro analysis, we found that the dE2F gene, as well as many DNA replication-related genes, including that for DNA polymerase α, might be regulated by DREF (27). On the other hand, the amount of mRNA for rp49, which is not related to DNA replication, was not reduced by DREF1–125 expression (Fig. 4A and B). Therefore, the reduction of mRNA seems to be specific to genes regulated by the DRE-DREF system. The N-terminal fragment might thus exert a dominant negative effect on DREF function in salivary gland cells.
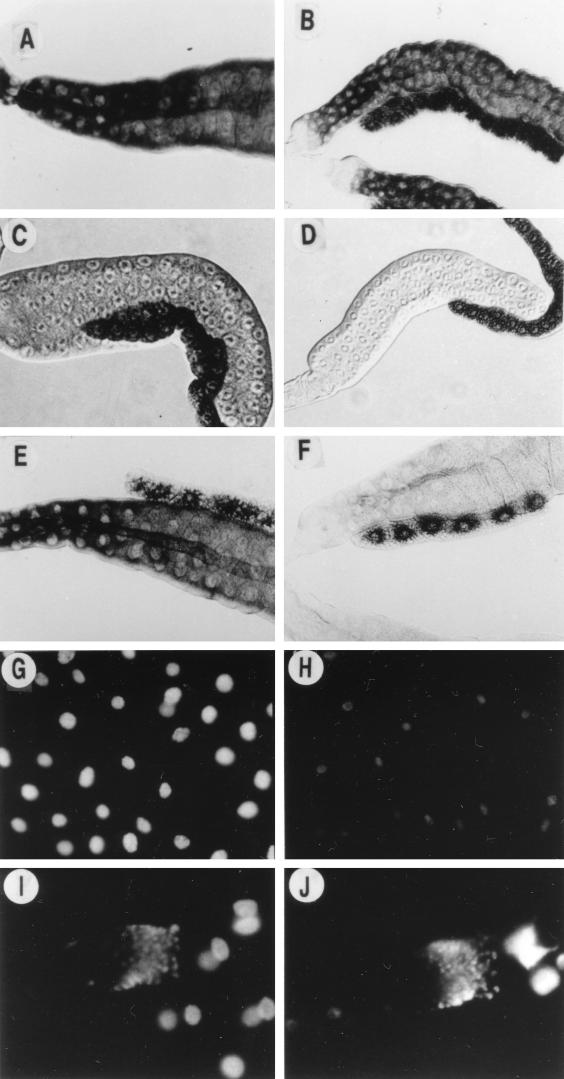
FIG. 4.
Phenotypes of salivary glands expressing DREF1–125. Transcripts of the rp49 gene (A and B), the DNA polymerase α 180-kDa subunit gene (C and D), and the dE2F gene (E and F) in salivary glands were detected by in situ hybridization. Salivary glands from third-instar larvae at 60 h after hatching were hybridized with antisense DIG-labeled RNA probes. Staining was detected with alkaline phosphatase. (G and H) DAPI staining of the salivary glands. (I and J) DAPI staining of imaginal ring cells of the same salivary glands as in panels G and H, respectively. (A, C, E, G, and I) Control fly carrying Sg-GAL4 alone; (B, D, F, H, and J) transgenic fly carrying Sg-GAL4 and UAS-DREF1–125. Magnifications: for panels A through F, ×153; for panels G and H, ×307; and for panels I and J, ×383.
Ectopic expression of DREF1–125 resulted in some reduction of the size of the salivary glands (Fig. 4B, D, and F). DAPI (4′,6-diamidino-2-phenylindole) staining of the glands from a third-instar larva revealed small nuclei with low levels of DNA in cells with DREF1–125 expression (Fig. 4H), although they were still larger than diploid cells in the imaginal ring (Fig. 4J). The results suggest that the extent of endoreplication was reduced by the expression of the N-terminal DREF fragment. This was not observed in the salivary glands expressing GAL4 only (Fig. 4G and I) or simultaneously expressing GAL4 and full-length DREF (data not shown).
To analyze the effects of the N-terminal DREF fragment on DNA replication more directly, BrdU incorporation experiments were performed. At 36 h after hatching, larvae were dissected and incubated at 25°C for 30 min in Grace’s culture medium containing BrdU, and labeled nuclei were detected by using anti-BrdU and alkaline phosphatase under identical conditions for all samples. As shown in Fig. 5, the cells in salivary glands expressing DREF1–125 incorporated BrdU to a much lesser extent than control salivary gland cells. On the other hand, non-DREF1–125-expressing diploid cells in the imaginal discs of the same animals incorporated BrdU to extents similar to those of control animals (Fig. 5A and B). It should be noted that DNA replication in the imaginal ring cells, in which the salivary gland-specific promoter used in this experiment is not active (7) and in which, therefore, DREF1–125 might not be expressed, was also indistinguishable from that of the control (Fig. 5C and D). Incorporation of BrdU appeared to be almost null in the salivary gland cells expressing DREF1–125 when the times for incubation in the BrdU-containing medium (30 min) and the color-developing reaction (10 to 15 min) were rather short. Prolonged reactions resulted in weak staining of the cells (data not shown), indicating that while expression of DREF1–125 significantly reduced endoreplication, the inhibition was not complete. Data for numbers of cells positive and negative for BrdU incorporation detected with the short-term reaction are summarized in Table 1. Although about 80% of the salivary gland cells from control larvae incorporated BrdU, less than 15% of the cells in salivary glands from flies expressing DREF1–125 were labeled.
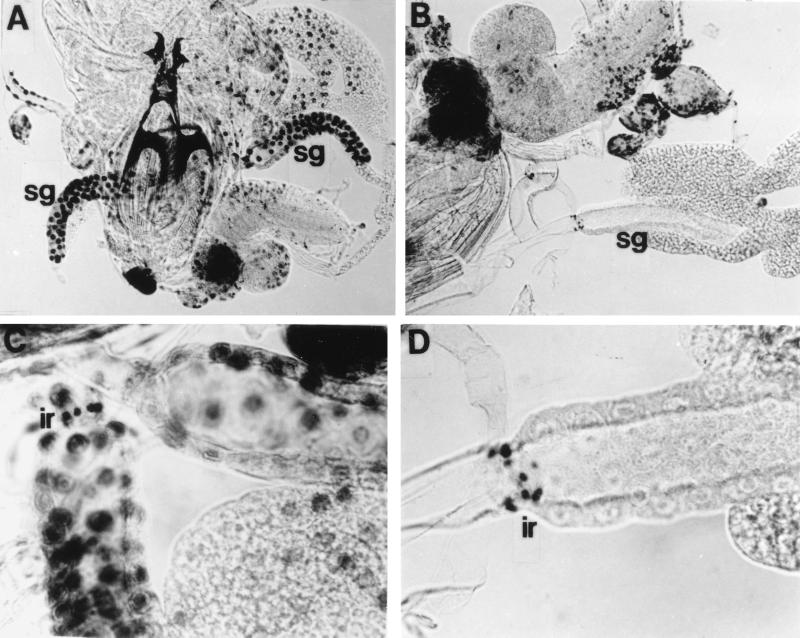
FIG. 5.
Ectopic expression of DREF1–125-reduced endoreplication. Larvae at 36 h after hatching were dissected in Drosophila Ringer’s solution, labeled with BrdU at 25°C for 30 min in Grace’s medium, and stained with anti-BrdU. (A) Control larva carrying Sg-GAL4 alone; (B) larva carrying Sg-GAL4 and UAS-DREF1–125; (C) salivary glands from a control larva carrying Sg-GAL4 alone; (D) salivary glands from a larva carrying Sg-GAL4 and UAS-DREF1–125. sg, salivary gland; ir, imaginal ring.
TABLE 1.
BrdU incorporation in salivary gland cells expressing DREF polypeptides
| Expected genotype of larvaea | No. of salivary glands (pairs) | No. of cells
|
No. of BrdU+ cells/salivary gland pair (mean ± SD) | |
|---|---|---|---|---|
| BrdU+ | BrdU− | |||
| P[Sg-Gal4]; + ; + | 12 | 1,172 | 308 | 97 ± 16 |
| w or Y + + | ||||
| P[Sg-Gal4]; P[UAS-DREF1–155]; + | 12 | 168 | 1,088 | 12 ± 4 |
| w or Y + + | ||||
| P[Sg-Gal4]; P[UAS-DREF1–155]; P[UAS-DREF1–709]TM6 or P[Sg-Gal4]; P[UAS-DREF1–155]; Pr | 12 | 618 | 816 | 52 ± 14 |
| w or Y ++w or Y ++ | ||||
P[Sg-Gal4]/Binsinscy females were mated to transgenic males carrying P[UAS-DREF]. Half of their progeny, therefore, carried the Binsinscy chromosome, while the other half did not. Larvae without Binsinscy were monitored.
In order to examine whether overexpression of the full-length DREF suppresses the inhibition of DNA replication caused by DREF1–125, we established a transgenic line carrying homozygous UAS-DREF1–125 and heterozygous UAS-DREF1–709 on the second and third chromosomes, respectively, and crossed it with the Sg-GAL4 line. Half of the progeny with the P[Sg-GAL4] chromosome would be expected to express both DREF1–709 and DREF1–125, while the other half with the P[Sg-GAL4] chromosome would be expected to express only DREF1–125 in salivary glands, depending on GAL4 expression. Of the salivary gland cells of the progeny, 43% were positive for BrdU incorporation (Table 1). Considering the relative rates of 80 and 15% for labeled nuclei in the salivary gland without and with expression of the N-terminal fragment, respectively, most, if not all, of the BrdU-labeled nuclei might have expressed full-length DREF. Thus, coexpression of DREF1–709 might have rescued DNA replication from the inhibition caused by DREF1–125 expression.
DREF1–125 expression reduces DNA replication of mitotic cell cycle.
To examine whether overexpression of DREF1–125 in cells undergoing mitotic cell cycling can inhibit DNA replication, DREF1–125 was ectopically expressed in the eye imaginal disc by using the GMR-GAL4–UAS-DREF system. In a wild-type eye disc, cells divide asynchronously anterior to the morphogenetic furrow. As they enter the furrow, they are arrested in G0/G1 phase and synchronously enter the last round of the mitotic cell cycle (second mitotic wave). Therefore, when eye discs are labeled with BrdU, the cells entering S phase appear as a clear stripe posterior to the furrow (Fig. 6C). Since the promoter carrying the glass-binding site was used for the expression of GAL4, DREF1–125 should be expressed in the region within and posterior to the morphogenetic furrow, where the cells enter the final synchronized mitotic cell cycle (Fig. 6B). In discs of larvae bearing one copy of GMR-GAL4 and one copy of UAS-DREF1–125, incorporation of BrdU in the S-phase zone corresponding to the second mitotic wave was found to be significantly reduced (Fig. 6D). Interestingly, cells ectopically labeled with BrdU were detected in the very posterior region of the eye disc. The result indicates that expression of DREF1–125 reduced or delayed S-phase entry. Therefore, it is suggested that DREF is required for normal DNA replication in the mitotic cell cycle of the eye imaginal disc.
DISCUSSION
For the analysis of DREF functions in vivo, isolation and analysis of flies with DREF gene mutations might be the most straightforward approach. However, since we have not succeeded in obtaining appropriate mutants despite extensive efforts, experiments using transgenic flies expressing dominant negative forms of the DREF polypeptide were employed in the present study. This idea arose from the finding that DREF binds to the DRE sequence as a homodimer of the 80-kDa polypeptide. p53, for example, is a transcriptional regulatory factor that binds to target sequences in the form of a homotetramer (32, 33), and expression of mutant polypeptides in vivo interferes with the wild-type p53 function in a dominant negative manner (8). This interference is thought to be dependent on hetero-oligomerization between wild-type and mutant p53 polypeptides. Such dominant negative mutations have also been reported for other transcription factors, such as Stat family members (17, 18) and the retinoic acid receptor (26).
The DREF1–125 fragment lacking the transactivation domain would inhibit the normal DREF function as a transcriptional regulator through dominant negative activity for the following reasons. (i) The DREF1–125 fragment forms a homodimer by itself and a heterodimer with the endogenous DREF. Although both complexes are capable of binding to DRE sequences, they might be inactive as transcriptional activators. (ii) Expression of DREF1–125 increased lethality in flies throughout development stages and reduced the extent of DNA replication in the salivary gland and eye imaginal disc. (iii) These inhibitory effects were suppressed by simultaneous expression of the full-length DREF.
The results presented in this paper demonstrate that DREF is required for normal DNA replication in both the mitotic cell cycle and endo cycle. Effects were caused by rather low concentrations of the N-terminal fragment: the polypeptide amount and DRE-binding activity of the N-terminal fragment in transgenic flies were estimated to be only 20 and 35%, respectively, of those of endogenous DREF. Furthermore, gel mobility shift experiments with the DRE-P probe and competitor oligonucleotides carrying various mutations in the DRE sequence revealed that specific activities for DNA binding and binding specificities were almost equal for the N-terminal fragment and the full-length DREF. Therefore, it is interesting to clarify the reason why rather small amounts of the N-terminal fragment caused extensive lethality in transgenic animals and reduction of DNA replication. Several possible mechanisms can be proposed.
The first is direct down-regulation of DNA replication-related genes. As shown in Fig. 4, expression of the dominant negative DREF1–125 resulted in extensive reduction of the level of mRNA for the DNA polymerase α 180-kDa subunit. We have already demonstrated that all three DRE sequences in the regulatory region of the gene encoding this enzyme are required for high levels of promoter activity (13), and binding of dominant negative DREF to any of three DREs may result in reduction of transcription. Plural DRE copies have been detected in other replication-related genes (16).
The second possible mechanism is that decreased DREF activity causes transcription of the DNA replication-related genes to be indirectly reduced by down-regulating other transcription factors involved in their regulation. We recently analyzed the promoter region of the Drosophila E2F (dE2F) gene (27). Two mRNA species differing with respect to the first exons (exon 1-a and exon 1-b) are transcribed from this gene (5, 20). Although the transcript with exon 1-a was detected transiently only in early-stage embryos, that with exon 1-b was detected throughout all stages of development. The fluctuations of transcript b levels were similar to those for other DNA replication-related genes. Assays of transient luciferase expression with Kc cells and measurement of the promoter activity of the dE2F gene in vivo with a dE2F mutant allele in which the lacZ gene had been inserted near the translation initiation site of the dE2F gene in the same orientation (5, 20) revealed that DREF is a positive regulator of the dE2F gene (27). Eventually, the expression of DREF1–125 resulted in extensive reduction of dE2F transcription in salivary glands (Fig. 4F). Therefore, it seems probable that reduction of the endogenous DREF activity by DREF1–125 could coordinately cause decreased transcription of DNA replication-related genes through reducing dE2F activity, because many replication-related genes carry E2F-binding sites in addition to DRE.
A third possible mechanism which may bring about reduction of DNA replication can be considered. Involvement of the DRE-DREF system in regulation of a considerable variety of genes has been suggested by the results of DNA database searches (16). In about 3.5% of the Drosophila genome, 73 copies of 5′-TATCGATA sequences were found to be localized within 0.6-kb upstream regions of 61 genes, including those encoding proteins related to transcription, translation, growth signal transduction, cell cycle regulation, and transcriptional regulation, in addition to ones related to DNA replication. Recently, it was confirmed that genes for cyclin A and D-Raf are also under regulation of the DRE-DREF system (19, 25). These lines of evidence suggest that DREF is involved in transcription of a large number of genes, many of which would be directly or indirectly involved in DNA replication. Normal progression of DNA replication requires a number of factors in intact forms, and thus, inactivation of even one or a small number of genes among them by DREF1–125 might impair reactions in the complicated processes necessary for DNA replication. So far, we have not obtained clues to which of above three mechanisms contributes most to reduce DNA replication and heat-induced death during development. However, the results obtained strongly suggest that appropriate expression of DREF activity is required for normal DNA replication and development in Drosophila.
ACKNOWLEDGMENTS
We are grateful to A. Brand (University of Cambridge), Nobert Perrimon (Harvard Medical School), S. Hayashi (National Institute of Genetics), and Y. Nishida (Nagoya University) for providing strains and plasmids for the Gal4 system. We also thank Malcolm Moore for comments regarding the English language of the manuscript.
This work was supported by grants from the Ministry of Science, Education, Sports and Culture, Japan.
REFERENCES
- 1.Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotype. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Brand A, Perrimon N. Raf acts downstream of the EGF receptor to determine dorso-ventral polarity during Drosophila oogenesis. Genes Dev. 1994;8:629–639. doi: 10.1101/gad.8.5.629. [DOI] [PubMed] [Google Scholar]
- 3.Cox K H, DeLeon D Y, Angerer L M, Angerer R C. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 4.Duronio R J, O’Farrell P H, Xie J E, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- 5.Dynlacht B D, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer J A, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–865. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 7.Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb M T, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 9.Hart C M, Zhao K, Laemmli U K. The scs′ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay B A, Wolff T, Rubin G M. Expression of baculovirus p35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y, Hirose F, Nishimoto Y, Shiraki M, Yamagishi M, Matsukage A, Yamaguchi M. Identification of CFDD (common regulatory factor for DNA replication and DREF genes) and role of its binding site in regulation of the proliferating cell nuclear antigen gene promoter. J Biol Chem. 1997;272:22848–22858. doi: 10.1074/jbc.272.36.22848. [DOI] [PubMed] [Google Scholar]
- 12.Hirose F, Yamaguchi M, Nishida Y, Masutani M, Miyazawa H, Hanaoka F, Matsukage A. Structure and expression during development of Drosophila melanogaster gene for DNA polymerase α. Nucleic Acids Res. 1991;19:4991–4998. doi: 10.1093/nar/19.18.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-bp sequence (DRE) and specific binding factor (DREF) involved in the expression of Drosophila genes for DNA polymerase α and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- 14.Hirose F, Yamaguchi M, Matsukage A. Repression of regulatory factor for Drosophila DNA replication-related gene promoters by zerknüllt homeodomain protein. J Biol Chem. 1994;269:2937–2942. [PubMed] [Google Scholar]
- 15.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J Biol Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 16.Matsukage A, Hirose F, Hayashi Y, Hamada K, Yamaguchi M. The DRE sequence TATCGATA, a putative promoter-activating element for Drosophila melanogaster cell proliferation-related genes. Gene. 1995;166:233–236. doi: 10.1016/0378-1119(95)00586-2. [DOI] [PubMed] [Google Scholar]
- 17.Mui A L, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima K, Yamanaka Y, Nakae K, Kojoma H, Ichiba M, Kiuchi N, Kataoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 19.Ohno K, Hirose F, Sakaguchi K, Nishida Y, Matsukage A. Transcriptional regulation of the Drosophila CycA gene by the DNA replication-related element (DRE) and DRE-binding factor (DREF) Nucleic Acids Res. 1996;24:3942–3946. doi: 10.1093/nar/24.20.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtani K, Nevins J R. Functional properties of a Drosophila homolog of the E2F1 gene. Mol Cell Biol. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson N S. Effects of heat and chemical stress on development. Adv Genet. 1990;28:275–296. doi: 10.1016/s0065-2660(08)60529-5. [DOI] [PubMed] [Google Scholar]
- 22.Ramos-Onsins S, Segarra C, Rozas J, Aguade M. Molecular and chromosomal phylogeny in the obscura group of Drosophila inferred from sequences of the rp49 gene region. Mol Phylogenet Evol. 1998;9:22–41. doi: 10.1006/mpev.1997.0438. [DOI] [PubMed] [Google Scholar]
- 23.Robertson H M, Preston C R, Philips R W, Johnson-Shlitz D M, Benz W K, Engels W R. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Royzman I, Whittaker A J, Orr-Weaver T L. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcription program. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu J-R, Choi T-Y, Kwon E-J, Lee W-H, Nishida Y, Hayashi Y, Matsukage A, Yamaguchi M, Yoo M-A. Transcriptional regulation of the Drosophila-raf proto-oncogene by the DNA replication-related element (DRE)/DRE-binding factor (DREF) system. Nucleic Acids Res. 1997;25:794–799. doi: 10.1093/nar/25.4.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou M, Sugai S, Tanaka H, Shimouchi K, Fuchs E, Narumiya S, Kakizuka A. Inhibition of skin development by targeted expression of a dominant-negative retinoic acid receptor. Nature. 1995;374:159–162. doi: 10.1038/374159a0. [DOI] [PubMed] [Google Scholar]
- 27.Sawado T, Hirose F, Takahashi Y, Sasaki T, Shinomiya T, Sakaguchi K, Matsukage A, Yamaguchi M. Roles of DNA replication related elements (DRE) in regulation of Drosophila E2F (dE2F) gene. J Biol Chem. 1998;273:26042–26051. doi: 10.1074/jbc.273.40.26042. [DOI] [PubMed] [Google Scholar]
- 28.Sparrow J C. Melanotic “tumours,”. In: Ashburner M, Wright T W, editors. The genetics and biology of Drosophila. 2b. London, United Kingdom: Academic Press; 1978. pp. 277–313. [Google Scholar]
- 29.Spradling A C. P element-mediated transformation. In: Roberts D B, editor. Drosophila: a practical approach. Oxford, United Kingdom: IRL Press; 1986. pp. 175–197. [Google Scholar]
- 30.Takahashi Y, Yamaguchi M, Hirose F, Cotterill S, Kobayashi J, Miyajima S, Matsukage A. DNA replication-related elements cooperate to enhance promoter activity of the Drosophila DNA polymerase α 73-kDa subunit gene. J Biol Chem. 1996;271:14541–14547. doi: 10.1074/jbc.271.24.14541. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi Y, Hirose F, Matsukage A, Yamaguchi M. Identification of three conserved regions in the DREF transcription factors from Drosophila melanogaster and Drosophila virilis. Nucleic Acids Res. 1999;27:510–516. doi: 10.1093/nar/27.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarunina M, Jenkins J R. Human p53 binds DNA as a protein homodimer but monomeric variants retain full transcription transactivation activity. Oncogene. 1993;8:3165–3173. [PubMed] [Google Scholar]
- 33.Waterman J L, Shenk J L, Halazonetis T D. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson K L, Johnson T K, Denell R E. Lethal (l) aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev Genet. 1991;12:173–187. doi: 10.1002/dvg.1020120302. [DOI] [PubMed] [Google Scholar]
- 35.Wilder E L, Perrimon N. Dual function of wingless in the Drosophila leg imaginal disc. Development. 1995;121:477–488. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi M, Hayashi Y, Matsukage A. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J Biol Chem. 1995;270:25159–25165. doi: 10.1074/jbc.270.42.25159. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi M, Hayashi Y, Nishimoto Y, Hirose F, Matsukage A. A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNA replication-related genes. J Biol Chem. 1995;270:15808–15814. doi: 10.1074/jbc.270.26.15808. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi M, Hirose F, Matsukage A. Roles of multiple promoter elements of the proliferating cell nuclear antigen gene during Drosophila development. Genes Cells. 1996;1:47–58. doi: 10.1046/j.1365-2443.1996.03003.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhao K, Hart C M, Laemmli U K. Visualization of chromosome domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]