Significance
Most animals record only labile memories of single events, whereas the formation of persistent long-term memories (LTMs) usually requires recurrent experiences. Our study distinguishes these different memory types through a deconvolution of molecular/biochemical processes within specific neurons of an identified memory circuit. A training-responsive gene activator, CREBA, engages paired DAL neurons in this circuit by promoting protein synthesis–dependent LTMs, which can otherwise be antagonized by CREBB repressor proteins. Increased CREBA expression or elevated membrane excitability enhances LTMs even after only one training cycle. These findings exemplify a circuit gating mechanism via cellular changes in specific single neurons to distinguish one-time experiences from multiple sessions of learning for storage as persistent memory.
Keywords: memory consolidation, gene regulation, single neurons, CREBA, CREBB
Abstract
Episodic events are frequently consolidated into labile memory but are not necessarily transferred to persistent long-term memory (LTM). Regulatory mechanisms leading to LTM formation are poorly understood, however, especially at the resolution of identified neurons. Here, we demonstrate enhanced LTM following aversive olfactory conditioning in Drosophila when the transcription factor cyclic AMP response element binding protein A (CREBA) is induced in just two dorsal-anterior-lateral (DAL) neurons. Our experiments show that this process is regulated by protein–gene interactions in DAL neurons: (1) crebA transcription is induced by training and repressed by crebB overexpression, (2) CREBA bidirectionally modulates LTM formation, (3) crebA overexpression enhances training-induced gene transcription, and (4) increasing membrane excitability enhances LTM formation and gene expression. These findings suggest that activity-dependent gene expression in DAL neurons during LTM formation is regulated by CREB proteins.
De novo protein synthesis promoted by recurrent learning often is a distinguishing feature of long-term memory (LTM) formation (1). A predominant hypothesis is that these new proteins “consolidate” synapse-specific changes in form and function, which represent elemental features of “engrams”—the systems-wide neuronal connections that facilitate network reactivation and memory retrieval (2). Learning-induced gene expression is typically controlled through the integrated action of many cis-regulatory elements and their associated transcription factors, of which cyclic AMP response element binding (CREB) proteins are well studied (3). LTM formation involves both activator and repressor transcription factors operating in linked positive and negative feedback loops (4). Whether these molecular events operate together within identified neurons or in separate foci of a memory circuit, however, has been more difficult to establish.
The Drosophila model system has been useful for revealing the molecules and circuits that underlie memory formation. Study of aversive olfactory learning continues to identify genes, proteins, and cell signaling cascades in an increasingly more complex memory circuit (5). Flies learn to associate odor (conditioned stimulus [CS]) with electric foot shock (unconditioned stimulus [US]) mediated by projection neurons (PNs) and dopaminergic neurons (DANs), respectively, which converge on the mushroom bodies (MBs), paired neuropils in the central brain. A single training session initiates sequences of neural activity with different durations in specific subpopulations of intrinsic MB neurons (5) that support genetically distinct phases of memory (short-term memory [STM]; middle-term memory [MTM]; anesthesia-resistant memory [ARM]), all of which eventually decay away (i.e., are decremental) (6). In addition to intrinsic MB neurons, these memory phases also depend on several types of extrinsic neurons (dorsal paired medial [DPM]; anterior paired lateral [APL]; protocerebral anterior medial [PAM]; protocerebral posterior lateral [PPL] and others) that form an extended neural network involved in olfactory memory formation (7).
Long-lasting memory of the aversive olfactory task is uniquely produced after repetitive spaced training. Genetic and pharmacological dissections have established that this LTM is dependent on CREB-activated gene expression and the synthesis of new proteins (6, 8). Neural output from the MB is required for LTM formation (and retrieval), and here too several classes of extrinsic (mushroom body output [MBON]) neurons with projections to various other neuropils have been described (7). Although these efferent neurons contribute to an extended network involved in LTM formation, de novo synthesis of proteins is required in only a few of them. More recently, an integrated molecular and circuit-based regulation of memory consolidation is being realized in the fly. Our group has identified several neurons in which protein synthesis is required for LTM consolidation. We found that MB output to two DAL neurons in the larger olfactory memory circuit induces protein synthesis– and CREB-dependent LTM. Subsequent feedback from DAL to MB neurons then appears necessary for LTM retrieval (8, 9).
In Drosophila, crebA and crebB genes encode CREB family proteins, CREBA and CREBB (the latter also known as dCREB2) (10, 11). crebB generates no fewer than nine distinct isoforms that function as transcriptional repressors and one reported activator (12). Activator and repressor crebB transgenes encoding these isoforms have been shown to enhance or impair LTM formation, respectively (13–16). In contrast, crebA encodes only one protein isoform, which is homologous to mammalian CREB3L1 and has been shown to function as a leucine-zipper transcription factor essential for embryonic development (10, 17). A role for CREBA in adult memory formation has not yet been shown.
Here, we demonstrate a biochemical gating mechanism at the single neuron level within a memory circuit in which antagonistic regulatory events and changes in neural excitability lead to LTM formation. Our results show that LTM formation depends on transcriptional activation by CREBA in DAL neurons. Endogenous “reporter” genes, calcium/calmodulin-dependent protein kinase II (CaMKII) and period (per), are up-regulated specifically during recurrent learning (spaced training), by increased neuronal excitability during LTM consolidation, or by ectopic overexpression of crebA. Moreover, we show that CREBB can antagonize transcriptional activation by CREBA in DAL neurons, suggesting that interactions between CREBA and CREBB in DAL neurons together modulate LTM formation in flies.
Results
CREBA in DAL Neurons Positively Regulates LTM Formation.
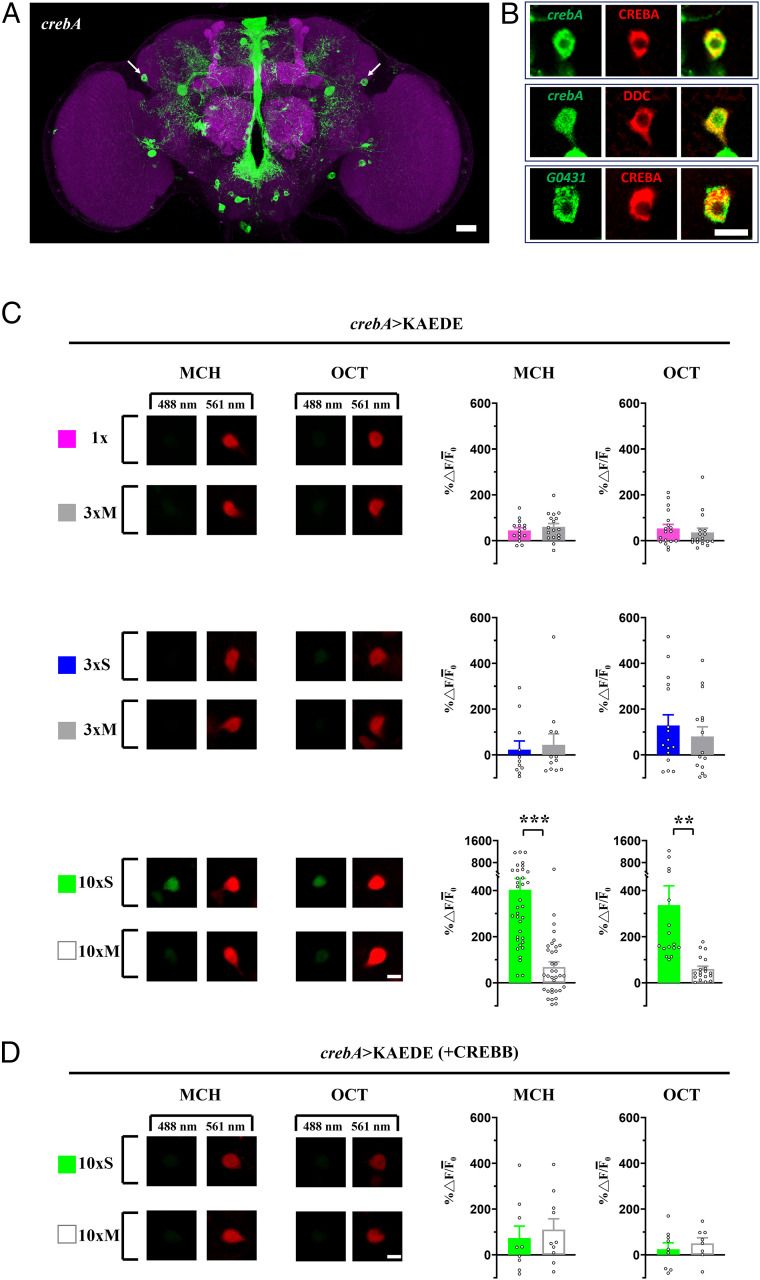
We first established a cellular function for CREBA. In a mouse F9 teratocarcinoma cell culture assay, significant PKA-dependent transcriptional activation of a CRE-luciferase transgene by CREBA was observed (SI Appendix, Fig. S1). Further, this up-regulation was antagonized by cotransfection of a CREBB repressor. To identify CREBA-positive neurons in the fly brain, we obtained and confirmed a Gal4 insertion in the crebA gene (BG00224; Berkeley Drosophila Genome Project [BDGP]) (SI Appendix, Table S1). crebA-Gal4 driving a UAS-mCD8::GFP green fluorescent protein transgene revealed a sparse neuronal pattern that included the DAL neurons (Fig. 1A). These observations were verified by (1) immunostaining with CREBA antibody (Fig. 1 B, Top and SI Appendix, Fig. S2A), (2) colocalized crebA expression with Dopa Decarboxylase (DDC) immunostaining known to label DAL neurons (Fig. 1 B, Middle), and (3) CREBA immunostaining with the DAL neuron-specific driver G0431-Gal4 (Fig. 1 B, Bottom) (8). Strong and specific expression in DAL neurons further motivated our investigation of CREBA in the context of LTM formation.
Fig. 1.
crebA expression in DAL neurons was enhanced after 10×S training and repressed by CREBB. (A) CREBA in dissected brains (green), counterstained with anti-DLG immunostaining (magenta). Arrows = DAL neurons. (Scale bar, 20 µm.) (B) Single optical slices of DAL neuron. crebA expression (green) colocalized with CREBA antibody (red) (Top) and with DDC antibody (red) (Middle) and G0431-Gal4 expression (green) colocalized with CREBA antibody (red) (Bottom). (Scale bar, 10 µm.) (C) crebA promoter activity after 1×, 3×M, and 3×S training with both MCH and OCT as reported by de novo KAEDE fluorescent protein synthesis (Left), estimated by the ratio of new (green, 488 nm) and preexisting (red, 561 nm) proteins (% ∆ F/¯F0) (Right). For each brain, single DAL neuron optical slices were imaged under identical conditions. Training effects of 1× compared with 3×M (Top) and effects of 3×S compared with 3×M (Middle) were not significant. The crebA promoter was activated after 10×S but not 10×M (Bottom). (Scale bar, 10 µm.) (D) crebA activity was repressed by crebB overexpression (+CREBB) after 10×S as compared with 10×M training. (Scale bar, 10 µm.) Bars represent mean ± SE, n ≥ 8/bar. **P < 0.01; ***P < 0.001.
We used a photoconvertible fluorescent KAEDE reporter transgene in DAL neurons to monitor the effect of training on crebA expression. After converting preexisting green KAEDE into red via ultraviolet (UV) irradiation prior to training, we measured the accumulation of newly synthesized green KAEDE driven by crebA-Gal4 after training (8). New expression of crebA was not detected after one training session (1×) or three spaced training sessions (3×S) (Fig. 1 C, Top two rows) but was elevated after 10 spaced training sessions (10×S) (Fig. 1 C, Bottom row). In a manner analogous to the in vitro results above, we overexpressed crebB in DAL neurons and observed repression of 10×S training-induced crebA expression (Fig. 1D). Thus, CREBB appears to antagonize the transcriptional activity of crebA in vivo in DAL neurons.
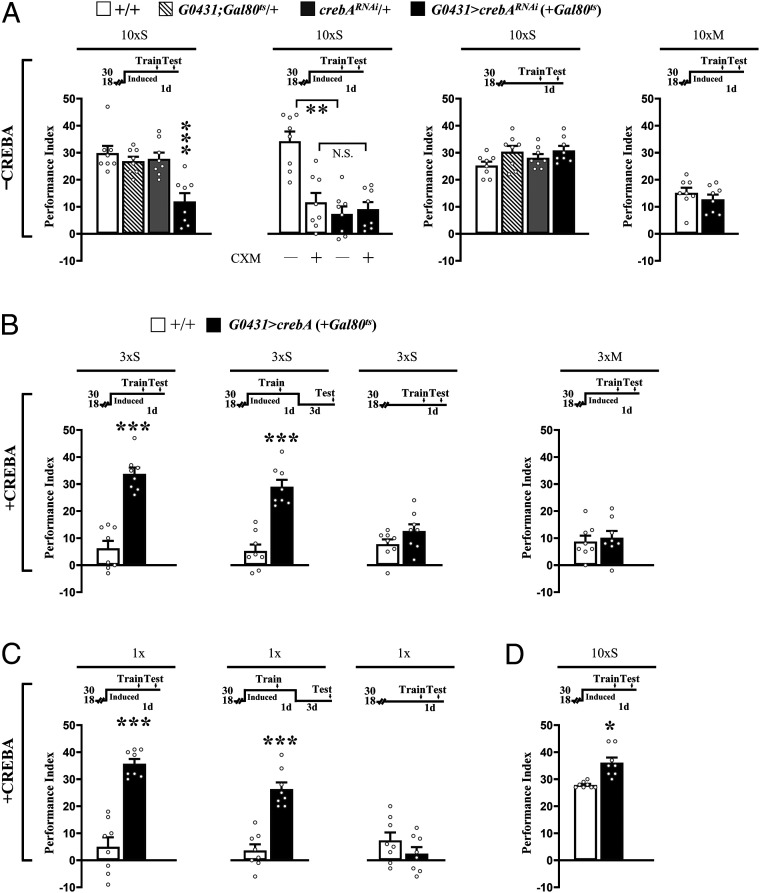
Using adult stage-specific transgenic RNA interference (RNAi) targeted by the Gal4/UAS system to knock down CREBA in DAL neurons under the temporal control of a temperature-sensitive tub-Gal80ts encoded protein (the inhibitory effect of GAL80ts protein on GAL4 expression at 18 °C is inactivated at 30 °C), we found an impairment in LTM formation after 10×S training (Fig. 2 A, Left). We observed no further reduction in 1-d memory after spaced training when flies were fed the protein synthesis inhibitor cycloheximide (CXM) (Fig. 2 A, Middle Left), indicating that the CREBA knockdown specifically blocked LTM (16). Control flies kept at 18 °C after 10×S training or at 30 °C after 10×M training were unaffected (Fig. 2 A, Middle Right and Right).
Fig. 2.
CREBA bidirectionally regulates LTM in DAL neurons. (A) Acute RNAi down-regulation of CREBA (–CREBA) in DAL impaired 1-d memory after 10×S (Left). LTM was similarly blocked by CXM feeding (Center Left). Memory was unaffected at 18 °C after 10×S (Center Right) and at 30 °C after 10×M (Right). (B) Overexpressing crebA (+CREBA) in DAL neurons at 30 °C enhanced 1-d memory after 3×S (Left) and persisted at least 4 d (Center Left). Memory was unaffected at 18 °C after 3×S (Center Right) and at 30 °C after 3×M (Right). (C) Similarly, overexpressing crebA in DAL neurons enhanced 1-d memory after 1× (Left), persisting at least 4 d (Center). Memory was unaffected at 18 °C after 1× (Right). (D) Overexpressing crebA in DAL neurons enhanced 1-d memory after 10×S. In all figures, temperature control schedules are indicated (Top). Flies were raised at 18 °C and then transferred to 30 °C to remove GAL4 inhibition by GAL80ts for 3 d before training. Bars represent mean ± SE, n = 8/bar. *P < 0.05; **P < 0.01; ***P < 0.001.
These results suggested that CREBA is a positive regulator of LTM formation. We tested this notion directly by overexpressing a crebA transgene and then subjecting flies to suboptimal training (3×S) (13). Along with crebA, we also tested two other putative activator transgenes and a repressor transgene encoded by crebB (13–16). Of these four candidate transgenes, adult stage-specific overexpression targeted to DAL neurons enhanced LTM only for crebA (Fig. 2 B, Left and SI Appendix, Fig. S2B). This effect of crebA persisted for at least 4 d (Fig. 2 B, Middle Left), while control flies kept at 18 °C after spaced training or at 30 °C after massed training were unaffected (Fig. 2 B, Middle Right and Right). crebA-enhanced LTM was also observed after 1× training and somewhat after 10×S training (Fig. 2 C and D).
Induced transgenic expression of crebA yielded immunopositive signals concentrated in the nuclei of DAL neurons, compared with sparse cytosolic signals in control flies (SI Appendix, Fig. S2C). Induced transgenic expression of crebARNAi, on the other hand, decreased immunopositive signals in DAL neurons (but not in another central brain structure, the ellipsoid body [EB]) (SI Appendix, Fig. S2D). Together, these data suggest that LTM formation in DAL neurons is positively regulated by CREBA.
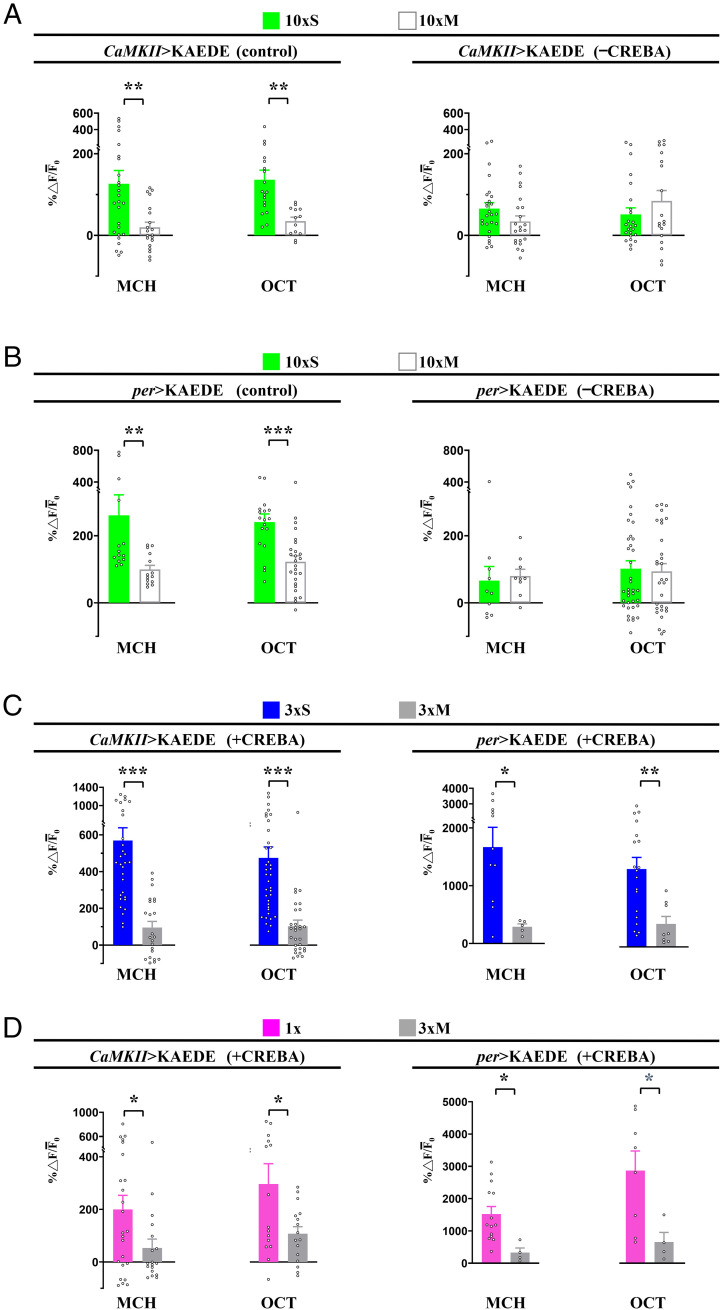
CREBA Modulates CaMKII and per Transcription.
CaMKII and per are two genes, disruption of which are known to impair LTM and which appear to be transcriptionally regulated by CREBB (8). We were interested to know whether they were regulated by CREBA in DAL neurons during LTM formation. Consistent with their established roles in LTM formation, training-induced transcription of CaMKII-KAEDE or per-KAEDE in control flies was apparent after 10×S training. When transgenic crebARNAi flies were subjected to 10×S training, however, transcription of these downstream genes was not observed (Fig. 3 A and B and SI Appendix, Fig. S3A). When transgenic crebA was overexpressed, however, CaMKII or per expression was seen after 3×S or even 1× training (Fig. 3 C and D and SI Appendix, Fig. S3B), but not without crebA overexpression after subthreshold 3×S or 1× training (SI Appendix, Fig. S4). Not surprisingly, overexpression of CaMKII or per transgenes themselves was not sufficient to enhance LTM after 3×S training, while overexpression of crebA alone was (SI Appendix, Fig. S2B). Presumably, CREBA regulates an ensemble of effector genes, the modulation of any one of which is not sufficient to enhance LTM. Nonetheless, these data show that CREBA positively regulates CREB-responsive downstream genes after space training.
Fig. 3.
CREBA bidirectionally regulates CaMKII and per transcription in DAL neurons. (A) The CaMKII promoter was activated in DAL neurons 24 h after 10×S but not after 10×M, as reported by de novo KAEDE fluorescent protein synthesis (Left) (refer to Fig. 1 for details). Elevated de novo KAEDE synthesis was suppressed following RNAi down-regulation of CREBA (–CREBA, Right). (B) The per promoter was similarly activated in DAL neurons 24 h after 10×S but not 10×M (Left). This activity was also abolished by RNAi down-regulation of CREBA (Right). (C and D) By contrast, levels of CaMKII (Left) and per (Right) gene activation were elevated by crebA overexpression (+CREBA) after 3×S and 1× but not after 3×M. Flies were raised at 18 °C and then transferred to 30 °C to remove GAL4 inhibition by GAL80ts for 3 d before training. Refer to SI Appendix, Figs. S3 and S4 for representative images. Bars represent mean ± SE, n ≥ 4/bar. *P < 0.05; **P < 0.01; ***P < 0.001.
Activity-Dependent Gene Expression in DAL Neurons Gates LTM Formation.
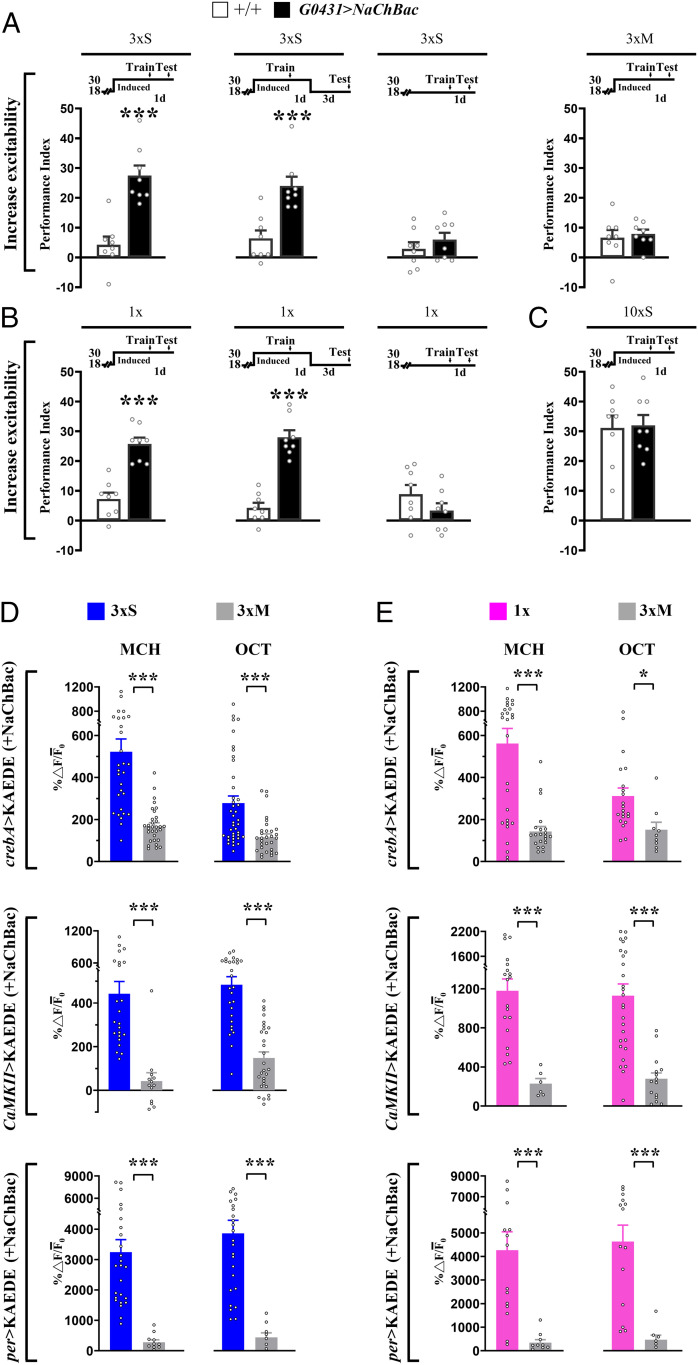
In our experiments above, training appeared to induce the transcription of crebA and its downstream effector genes, CaMKII and per. Did training, then, produce these cellular responses via an increase in neuronal activity, as is the case in other model systems (18, 19)? We explored this notion by increasing membrane excitability (and thereby the propensity of neural activity) in DAL neurons via ectopic expression of a sodium channel transgene, UAS-NaChBac, under temporal control of tub-Gal80ts. When the inhibitory effect of GAL80ts was inactivated at 30 °C before training, transgenic expression of NaChBac was sufficient to enhance 1-d memory after 3×S or 1× training (Fig. 4 A and B, Left). This manipulation had no effect on 1-d memory after 3×M training (Fig. 4 A, Right), after 10×S training (Fig. 4C) or after 3×S or 1× training in transgenic flies kept at 18 °C when active GAL80ts blocked expression of the UAS-NaChBac transgene (Fig. 4 A, Middle Right and Fig. 4 B, Right). Finally, enhanced memory after 3×S or 1× training persisted for at least 4 d, which is 3 d after flies were shifted back to 18 °C when GAL80ts again blocked further expression of UAS-NaChBac (Fig. 4 A, Middle Left and Fig. 4 B, Middle). These latter results imply that training-induced neuronal activity is required to enhance LTM during acquisition and/or memory consolidation but not during memory retrieval. Finally, we extended these observations to the cellular level by showing that crebA-KAEDE, CaMKII-KAEDE, or per-KAEDE each was induced after 3×S or 1× training when NaChBac was overexpressed (Fig. 4 D and E and SI Appendix, Fig. S5 A and B). Thus, these transcriptional responses appear to be activity-dependent during LTM formation.
Fig. 4.
Increased DAL neuron membrane excitability activates genes and enhances LTM. (A) NaChBac overexpression (+NaChBac) increased DAL neurons excitability and elevated 1-d memory after 3×S (Left), persisting for at least 4 d (Center Left). Memory was unaffected at 18 °C after 3×S (Center Right). Memory was similarly unaffected after 3×M (Right). (B) NaChBac overexpression also elevated 1-d memory after 1× (Left), persisting for at least 4 d (Center). Memory was unaffected at 18 °C after 1× (Right). (C) NaChBac overexpression in DAL neurons had no effect on 1-d memory after 10×S. (D and E) crebA (Top), CaMKII (Middle), and per (Bottom) activity in DAL neurons as reflected by de novo KAEDE synthesis (see Fig. 1) was elevated by NaChBac overexpression after 3×S and 1×. Refer to SI Appendix, Fig. S5 for representative images. Bars represent mean ± SE, n ≥ 6/bar. *P < 0.05; ***P < 0.001.
Discussion
CREBA and CREBB both are expressed in DAL neurons (20), and transgenic manipulations of CREBB have shown an impairment of 1-d memory after 10×S training (8). These studies, however, did not investigate a functional role for CREBA in DAL neurons and, in particular, did not query whether LTM might be enhanced. Here, we have focused on a role for CREBA in LTM formation. We first established in vitro that CREBA induced expression of a CRE-luciferase reporter gene in a PKA-dependent manner and was blocked by CREBB (SI Appendix, Fig. S1). Then, we used CREBA antibody, a DAL specific Gal4 driver and a crebA-driven KAEDE reporter to confirm that CREBA not only was expressed in DAL neurons but also responded transcriptionally to 10×S (but not 3×S or 1×) training (Fig. 1 A–C). In this in vivo context, we also showed that 10×S training-induced expression of crebA in DAL neurons was antagonized by overexpression of a crebB (repressor) transgene (Fig. 1D).
These observations suggested that CREBA in DAL neurons might serve as a positive regulator of protein synthesis-dependent LTM. Indeed, inducible transgenic manipulations of crebA only in DAL neurons were sufficient to impair 1-d memory after 10×S training (similar to inhibition of protein synthesis) using crebARNAi, or to enhance 1-d memory after 1× or 3×S training by overexpressing wild-type crebA. Importantly, LTM remained enhanced 4 d after 1× or 3×S training even when induction of transgenic crebA ceased 3 d earlier (Fig. 2). Together, these results suggest that CREBA in DAL neurons is involved in learning and/or memory consolidation but not necessarily in memory retrieval.
CaMKII and per are two “downstream” genes that are CREB responsive, are expressed in DAL neurons and impair LTM when disrupted (8). Using CaMKII and per-driven KAEDE reporter transgenes, we have shown that expression of both genes is induced normally after 10×S training, is blocked after such training by induced expression of a crebARNAi and is enhanced after 1× or 3×S training when a crebA transgene is inducibly expressed (Fig. 3). Here too, these transgenic manipulations are not required for CaMKII or per expression and LTM to persist for 4 d after training and 3 d after transgenic manipulations are blocked.
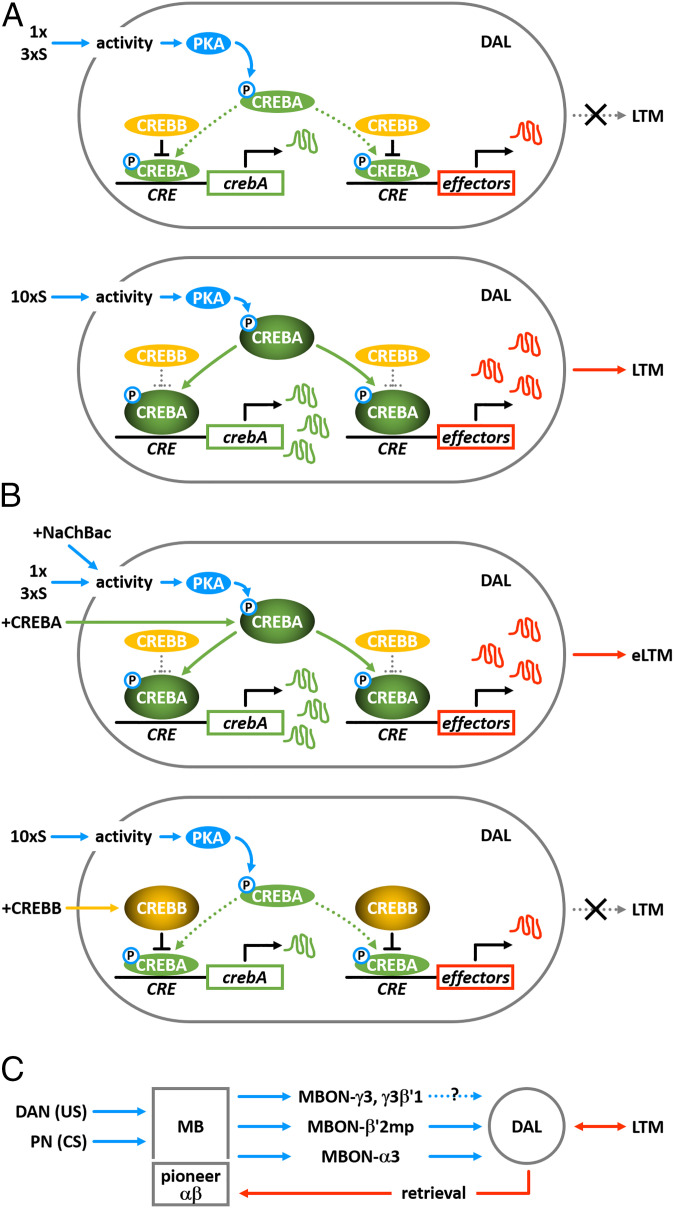
This role for CREBA in DAL neurons during learning and memory consolidation suggested that the transcriptional response might be activity dependent. We explored this possibility by expressing a NaChBac transgene in DAL neurons, which served to increase membrane excitability and presumably neural activity in response to training. We found that induced expression of NaChBac in DAL neurons was sufficient to enhance 1-d memory and to enhance expression of crebA, CaMKII, and per after 1× or 3×S training (Fig. 4). Together, these observations have suggested a model, which we describe in Fig. 5. The model illustrates how CREBA and CREBB interact to regulate transcription in DAL neurons and the activity-dependent transcriptional response to gate LTM formation.
Fig. 5.
CREBA and CREBB interactions regulate transcription in DAL neurons and gate LTM formation. (A) In normal flies, 1× or 3×S training (Top) induces neural activity in the memory circuit, which includes DAL neurons. Increased neural activity elevates cAMP signaling and activates PKA, which then directly or indirectly leads to phosphorylation and activation of CREBA. However, basal levels of CREBB repressor inhibit activated CREBA from inducing effector gene expression, thereby blocking LTM formation. With 10×S training (Bottom) activated CREBA accumulates to levels sufficient to overcome basal CREBB repression and promotes transcription of downstream effector “memory genes.” One downstream gene is crebA itself, establishing a putative positive feedback loop and contributing to further CREBA accumulation. CREBA then induces (further) transcription of downstream effector genes, which yields long-lasting changes in intrinsic neuronal excitability and/or synaptic plasticity during memory consolidation. (B) Transgenic overexpression of crebA (+CREBA) or sodium channels (+NaChBac) (Top) provides sufficient CREBA or activity (respectively) to overcome basal CREBB antagonism, establish the CREBA-positive feedback loop, and induce effector gene expression, even with subthreshold 1× or 3×S training, hence the appearance of enhanced LTM (eLTM). Conversely, transgenic overexpression of crebB (+CREBB) (Bottom) yields enough CREBB to block activation of the CREBA feedback loop and transcription of downstream effector genes even after 10×S training, thereby preventing LTM formation. (C) DAL neurons contribute to a circuit-level feedback loop among neurons intrinsic and extrinsic to MBs during olfactory memory consolidation and retrieval. The association of odor (CS) via PNs and foot shock (US) via DANs first is registered as an increase in neural activity within MBs, which then promotes a persistent neural trace among intrinsic and extrinsic MB neurons. Persistent output from MBs then drives neural activity directly or indirectly between MB and DAL neurons via several MBONs. After CREBA-induced changes in gene expression within DAL neurons is complete, output from DAL neurons projecting back to the dendrites of pioneer α/β neurons of the MB is required for memory retrieval.
CREB-dependent long-term memory formation first was shown in Drosophila using inducible transgenes, which were expressed throughout the fly (16). Acute expression of a transgenic crebB repressor blocked LTM after 10×S training, whereas similar manipulations of a synthetic crebB activator transgene enhanced LTM (13–16). An early attempt to identify specific neurons underlying LTM implicated MBs, wherein MB-specific transgenic expression of a crebB repressor was reported to impair LTM after 10×S training (21). A subsequent study revealed, however, that this behavioral impairment derived from developmental defects in MB structure due to chronic expression of the crebB transgene. In contrast, induced expression of a crebB transgene only in adult-stage MBs did not impair LTM and did not produce any developmental defects (8). In neither study was a positive (CREB) regulator identified nor was enhanced LTM evaluated.
One-trial learning is usually insufficient to produce protein synthesis–dependent LTM, except for those experiences important for survival (22, 23). Here, we have demonstrated that LTM can form after a single training session when “memory genes” in DAL neurons are genetically manipulated. Learning-related and CREB-dependent changes in membrane excitability are well known and explain aspects of neuronal plasticity underlying memory consolidation (19). Regulation of ion channel genes by CREBA and CREBB transcription factors, for example, modulate plasticity in alcohol tolerance in Drosophila (24). CREB-dependent regulation of gene expression in DAL neurons appears sufficient to promote systems memory consolidation by modulating neural excitability. Further studies may elucidate whether neural circuits involved in motivation and attention also modulate DAL neurons during LTM formation (25, 26) and whether such prolonged neural activity also produces synaptic plasticity in DAL neurons (19, 27).
Materials and Methods
In this report we used an automated olfactory aversive learning task (6) and assessed LTM after elevating or blocking CREBA-dependent gene activation in temporally and spatially targeted domains. Similarly, we bidirectionally modulated the expression of CREBA in DAL neurons to examine the effects on training-responsive gene transcription using a Gal4-targeted UV-sensitive KAEDE reporter system (8). Spatial and temporal regulation of Na+ channel activity with transgene overexpression in DAL neurons was used to evaluate impacts of elevated membrane excitability on LTM and on training-responsive genes using the KAEDE reporter system (8). See SI Appendix, Materials and Methods for details of fly strains, reagents, and all procedures.
Supplementary Material
Acknowledgments
We thank Dr. Allan C. Spradling, Dr. Amita Sehgal, Dr. Jay Hirsh, Dr. Jeffrey C. Hall, Dr. Ronald L. Davis, Dr. Yoshiki Takamatsu, Dr. Hugo J. Bellen, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and the Kyoto Drosophila Genomics Resource Center for fly stocks; the Developmental Studies Hybridoma Bank for antibodies; and D. Anderson and A. Fayyazuddin (Dart NeuroScience LLC) for performing the CRE-luciferase cell culture experiments. This work was financially supported by the Brain Research Center under the Higher Education Sprout Project co-funded by the Ministry of Education and the Ministry of Science and Technology in Taiwan, the Yushan Scholar Program from the Ministry of Education in Taiwan, and Dart NeuroScience LLC.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100624118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Hernandez P. J., Abel T., The role of protein synthesis in memory consolidation: Progress amid decades of debate. Neurobiol. Learn. Mem. 89, 293–311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonegawa S., Morrissey M. D., Kitamura T., The role of engram cells in the systems consolidation of memory. Nat. Rev. Neurosci. 19, 485–498 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Alberini C. M., Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 89, 121–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao L., Yang Z., Bi Y., Stochasticity and bifurcations in a reduced model with interlinked positive and negative feedback loops of CREB1 and CREB2 stimulated by 5-HT. Math. Biosci. 274, 73–82 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Aso Y., Rubin G. M., Toward nanoscale localization of memory engrams in Drosophila. J. Neurogenet. 34, 151–155 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Margulies C., Tully T., Dubnau J., Deconstructing memory in Drosophila. Curr. Biol. 15, R700–R713 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perisse E., Burke C., Huetteroth W., Waddell S., Shocking revelations and saccharin sweetness in the study of Drosophila olfactory memory. Curr. Biol. 23, R752–R763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C. C., et al., Visualizing long-term memory formation in two neurons of the Drosophila brain. Science 335, 678–685 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Wu J. K., et al., Long-term memory requires sequential protein synthesis in three subsets of mushroom body output neurons in Drosophila. Sci. Rep., 10.1038/s41598-017-07600-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolik S. M., Rose R. E., Goodman R. H., A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol. Cell. Biol. 12, 4123–4131 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usui T., Smolik S. M., Goodman R. H., Isolation of Drosophila CREB-B: A novel CRE-binding protein. DNA Cell Biol. 12, 589–595 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Yin J. C., et al., A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol. Cell. Biol. 15, 5123–5130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin J. C., Del Vecchio M., Zhou H., Tully T., CREB as a memory modulator: Induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81, 107–115 (1995). [DOI] [PubMed] [Google Scholar]
- 14.T. C.TubonJr., et al., dCREB2-mediated enhancement of memory formation. J. Neurosci. 33, 7475–7487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fropf R., et al., Time of day influences memory formation and dCREB2 proteins in Drosophila. Front. Syst. Neurosci. 8, 43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tully T., Regulation of gene expression and its role in long-term memory and synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 94, 4239–4241 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose R. E., Gallaher N. M., Andrew D. J., Goodman R. H., Smolik S. M., The CRE-binding protein dCREB-A is required for Drosophila embryonic development. Genetics 146, 595–606 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis R. L., Traces of Drosophila memory. Neuron 70, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., et al., The role of intrinsic excitability in the evolution of memory: Significance in memory allocation, consolidation, and updating. Neurobiol. Learn. Mem. 173, 107266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crocker A., Guan X. J., Murphy C. T., Murthy M., Cell-type-specific transcriptome analysis in the Drosophila mushroom body reveals memory-related changes in gene expression. Cell Rep. 15, 1580–1596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D., Akalal D. B., Davis R. L., Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron 52, 845–855 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krashes M. J., Waddell S., Rapid consolidation to a radish and protein synthesis-dependent long-term memory after single-session appetitive olfactory conditioning in Drosophila. J. Neurosci. 28, 3103–3113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano Y., et al., Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science 339, 443–446 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Ghezzi A., Yin J. C., Atkinson N. S., CREB regulation of BK channel gene expression underlies rapid drug tolerance. Genes Brain Behav. 8, 369–376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krashes M. J., et al., A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139, 416–427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Swinderen B., et al., Shared visual attention and memory systems in the Drosophila brain. PLoS One 4, e5989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baltruschat L., et al., Circuit reorganization in the Drosophila mushroom body calyx accompanies memory consolidation. Cell Rep. 34, 108871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.