Significance
The MYC axis is commonly disrupted in cancer, mostly by activation of the MYC oncogenes but also by genetic inactivation of MAX, the obligate partner of MYC, and of the MAX partner MGA, both of which are members of the polycomb repressive complex ncPRC1.6. While the oncogenic properties of the MYC family have been extensively studied, those of the MAX-deficient cells and the role of MGA in MAX mutant cells remain unclear. In this study, we demonstrate that MAX-deficient SCLCs cells have either ASCL1 or NEUROD1 or combined characteristics. Furthermore, our data reveal that the lack of available MAX restricts MGA occupancy in gene promoters and, although the ncPRC1.6 can still be formed, there is a deficient ncPRC1.6-mediated gene repression.
Keywords: MAX, MGA, ncPRC1.6, SCLC, tumor suppressor
Abstract
The MYC axis is disrupted in cancer, predominantly through activation of the MYC family oncogenes but also through inactivation of the MYC partner MAX or of the MAX partner MGA. MGA and MAX are also members of the polycomb repressive complex, ncPRC1.6. Here, we use genetically modified MAX-deficient small-cell lung cancer (SCLC) cells and carry out genome-wide and proteomics analyses to study the tumor suppressor function of MAX. We find that MAX mutant SCLCs have ASCL1 or NEUROD1 or combined ASCL1/NEUROD1 characteristics and lack MYC transcriptional activity. MAX restitution triggers prodifferentiation expression profiles that shift when MAX and oncogenic MYC are coexpressed. Although ncPRC1.6 can be formed, the lack of MAX restricts global MGA occupancy, selectively driving its recruitment toward E2F6-binding motifs. Conversely, MAX restitution enhances MGA occupancy to repress genes involved in different functions, including stem cell and DNA repair/replication. Collectively, these findings reveal that MAX mutant SCLCs have either ASCL1 or NEUROD1 or combined characteristics and are MYC independent and exhibit deficient ncPRC1.6-mediated gene repression.
Small-cell lung cancer (SCLC), one of the most aggressive types of lung cancer, is commonly located centrally in the lung and is thought to originate from the neuroendocrine cells of the lung epithelium (1–3). Mirroring the pattern of gene expression found in these cells, SCLCs have a high level of expression of neural-related transcripts (1, 4, 5). Recently, it has been shown that SCLCs comprise four subtypes, distinguished by the predominant expression of lineage-specific transcription factors. The most common of these are ASCL1 and NEUROD1, which target different gene sets for neuroendocrinal or neural functions (6). The genetic profile of SCLCs includes the almost universal presence of inactivating alterations at TP53. Inactivating mutations at RB1 and at PTEN, or activation of oncogenes such as MYC and PIK3CA, are also characteristic of this type of lung cancer (7). With the advent of the novel sequencing technologies, novel genes, for example, the NOTCH family, and genes coding for histone modifiers, for example, CREBBP, EP300, and MLL, have been found to be altered in SCLCs (8, 9).
The MYC axis is commonly disrupted in cancer, mostly by genetic activation of the MYC family of oncogenes. We reported that a subset of SCLCs features somatic and biallelic inactivation of MAX, a gene encoding for the obligate heterodimerization partner of the MYC family of proteins (5). Recently, it has been shown that Max deletion increases growth and transformation in cells and dramatically accelerates SCLC progression in an Rb1/Trp53-deleted mouse model (10). Previously, germline mutations of MAX had been found in patients with hereditary pheochromocytomas, another neoplasia of neuroendocrine origin (11). The presence of gene alterations at MYC or MAX and of alterations of components of the SWI/SNF complex were found to be mutually exclusive, implying a functional connection between these pathways (5).
Although the genetic and molecular data strongly suggest that the loss of function of MAX contributes to the development of SCLC, it constitutes a conundrum in the understanding of MYC biological and molecular function, which depends on its dimerization with MAX (12). Heterodimerization with MAX through the bHLH (basic helix–loop–helix) regions of both proteins allows the recognition of the DNA sequences known as E-boxes. While MAX is the only partner of the MYC proteins and lacks a transactivation domain, it has a wide variety of other putative partners with which it could heterodimerize (i.e., MXD1, MXD2, MXD3, MXD4, MNT, and MGA) (12). It is now well established that the expression of the MYC target genes is controlled by the shift between activating MYC-MAX and repressive MAX-MXDs/MNT/MGA heterodimers that bind to the same canonical E-box consensus sequences in gene promoters (12). In this regard, the role of the MAX dimerization partner MGA may be especially significant because it is known to be genetically inactivated in a subset of cancers (5, 13, 14). MGA contains a bHLH domain, through which it binds MAX, and a T-box domain (15) whose function is not understood. More recently, MAX and MGA have been shown to act as part of the Polycomb repressive complex 1 (PRC1), specifically the noncanonical PRC1, otherwise known as ncPRC1.6 (16, 17).
The current study aimed to shed light on the tumor suppressor function of MAX and its relevance to SCLC development. Given that the MAX partner MGA is also inactivated in lung cancer and that both proteins are members of the Polycomb repressive complex, ncPRC1.6, we wanted to define the possible role of MGA and of the ncPRC1.6 in cancer cells lacking MAX. Our findings demonstrate that MYC does not have any transcriptional function in MAX-deficient cells and that, in SCLC cells carrying MAX-inactivating mutations, there is a deficient ncPRC1.6-mediated gene repression which may contribute to cancer development.
Results
SCLC Cells with MAX Inactivation Have ASCL1 or NEUROD1 or Combined ASCL1/NEUROD1 Characteristics.
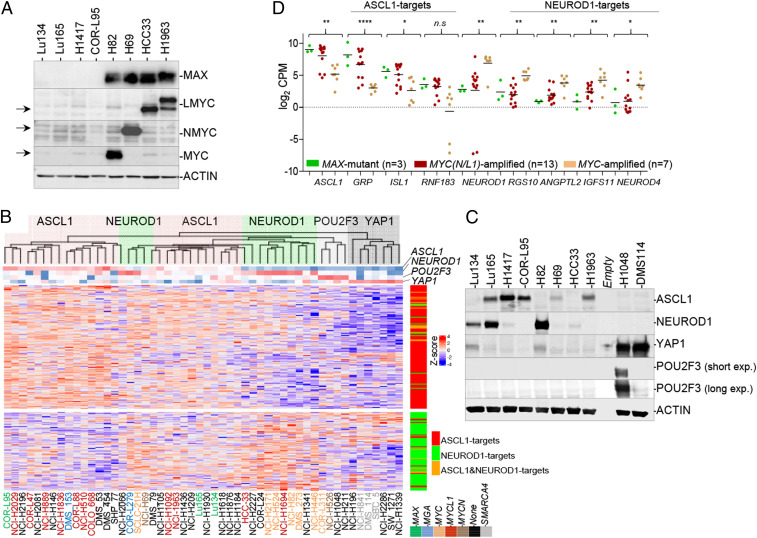
Previously, we identified four SCLC cell lines (COR-L95, H1417, Lu134, and Lu165) that lack MAX protein because of gene alterations (5). These cells have very low protein expression from the MYC family of genes (Fig. 1A). As mentioned above, SCLCs have been classified into four subtypes based on the predominant expression of lineage-specific transcription factors (6, 18). To molecularly characterize these MAX-deficient cells further, we analyzed the gene expression profiles of a panel of SCLC cell lines (n = 11), including MYC-, MYCL1-, and MYCN-amplified cells, using microarray gene expression analysis (SI Appendix, Fig. S1A) and RNA sequencing performed on the Lu134 and Lu165 cells lines and combined with RNA sequencing data from the Cancer Cell Line Encyclopedia (n = 50) (Fig. 1B and SI Appendix, Fig. S1B). The genetic status of MGA and of the SWI/SNF component SMARCA4 was also annotated. As previously reported (6), most MYC-amplified cells clustered together and showed high levels of NEUROD1 and of NEUROD1 targets, whereas most MAX-deficient and MYCL1- and MYCN-amplified cells exhibit predominantly high expression levels of ASCL1 and of ASCL1 targets. Some exceptions were the MAX mutant Lu134 cells and the MYCL1-amplified HCC33 cells, with no detectable ASCL1 levels by Western blot while expressing NEUROD1 (Fig. 1C). On the other hand, the Lu165 express both ASCL1 and NEUROD1 (Fig. 1 B and C and SI Appendix, Fig. S1B). Notably, the three SMARCA4 mutant cell lines in the study were low ASCL1/low NEUROD1 but high YAP1 expressers (Fig. 1 B and C). The high ASCL1-expressing group is enriched in various potassium channel transcripts (e.g., KCNMB2) and in other genes such as GRP, ISL1, and RNF183, among others, some of which are known targets of ASCL1 (Fig. 1B) (6). Likewise, the high NEUROD1 group features higher levels of NEUROD1 targets (e.g., ANGPTL2, NEFM, and RGS10). The comparative of the messenger RNA (mRNA) levels of selected ASCL1 and NEUROD1 targets among the MAX mutant, MYCN/MYCL1-amplififed, or MYC-amplified cell lines further evidenced this association (Fig.1D). Taken together, the results support the concept that most MAX mutant SCLC cells show ASCL1 characteristics, although some can express only NEUROD1 or both NEUROD1 and ASCL1 factors.
Fig. 1.
SCLC cell lines with MAX inactivation have ASCL1 or NEUROD1 or combined ASCL1/NEUROD1 characteristics. (A) Western blot showing basal levels of the MYC family of proteins and MAX proteins in the different MAX-deficient SCLC cell lines as indicated. ACTIN, protein-loading control. SCLC cell lines with amplification at MYC (H82), MYCN (H69), and MYCL1 (HCC33 and H1963) are also included for comparison. The MYCL1 gene is fused with the RFL gene in the H1963 cells, rendering a larger protein. (B) Heatmap using ASCL1 and NEUROD1 gene signatures (from ref. 31) in the indicated SCLC cell lines. The gene expression has been gathered from RNA-seq (Lu134 and Lu165 are from current work, and the rest are from the Cancer Cell Line Encyclopedia [CCLE]). Dendrogram Top reflects clustering of the SCLC cell lines. ASCL1-high (red shading), NEUROD1-high (green shading), POU2F3-high (gray shading), and YAP1-high (dark gray shading) groups are indicated. The expression levels of NEUROD1, ASCL1, POU2F3, and YAP1 is indicated below the dendrogram. The genetic status (MAX, MYCN, MYC, MYCL1, MGA, and SMARCA4) of each cell line is also indicated with a color code. (Right) Position of the NEUROD1 (green), ASCL1 (red), or both (orange) transcription factors targets in the heatmap. (C) Western blot showing basal levels of the ASCL1, NEUROD1, YAP1, and POU2F3 factors in the indicated SCLC cell lines. ACTIN, protein-loading control. (D) mRNA levels from the RNA-seq analysis (CCLE) of ASCL1, NEUROD1, and of selected targets, grouped by three categories: MAX-deficient and MYCN/MYCL1- and MYC-amplified cells. Lines show mean; values from each cell line are represented. *P < 0.05, **P < 0.01, ****P < 0.001, n.s. not significant; two-sided unpaired Student’s t test (MAX mutant versus MYC-amplified groups).
Finally, we observed that, compared with the MYC-amplified cells, the MAX mutant and the MYCL1- and MYCN-amplified cells express significant low mRNA levels of the MAX-binding partner, MXD2, while showing a tendency toward higher levels of MXD3 and MXD4 (SI Appendix, Fig. S1C).
No Occupancy of MYC at Any DNA Region in MAX Mutant Cancer Cells.
As mentioned in the introduction, our current understanding of MYC biology posits that MYC's transcriptional activity depends on its dimerization with MAX (12). However, genome-wide evaluation of MYC recruitment to the DNA in cancer cells that lack MAX has never been attempted. In this context, natural MAX knockout cells are an invaluable tool for determining whether the MYC family of proteins can bind DNA in the absence of MAX. Here, we used these cells to perform chromatin immunoprecipitation sequencing (ChIP-seq) of MYC.
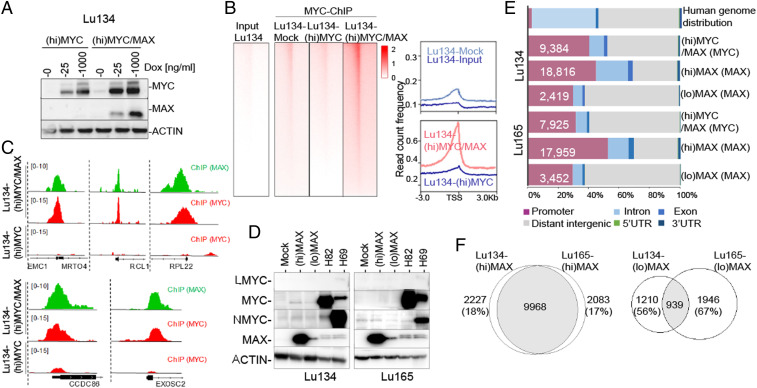
First, we used a doxycycline-inducible system to overexpress MYC (hereafter referred to as (hi)MYC cells) and the MYC and MAX proteins (hereafter (hi)MYC/MAX cells) in Lu134 cells (Fig. 2A). Next, we performed ChIP-seq of MYC and of MAX from these cells. We observed no occupancy of overexpressed MYC at any DNA region. In contrast, recruitment of MYC to the DNA could be readily detected in the (hi)MYC/MAX cells (Fig. 2 B and C). These observations support the canonical view that dimerization with MAX is required to ensure the DNA-binding activities of MYC (19–22).
Fig. 2.
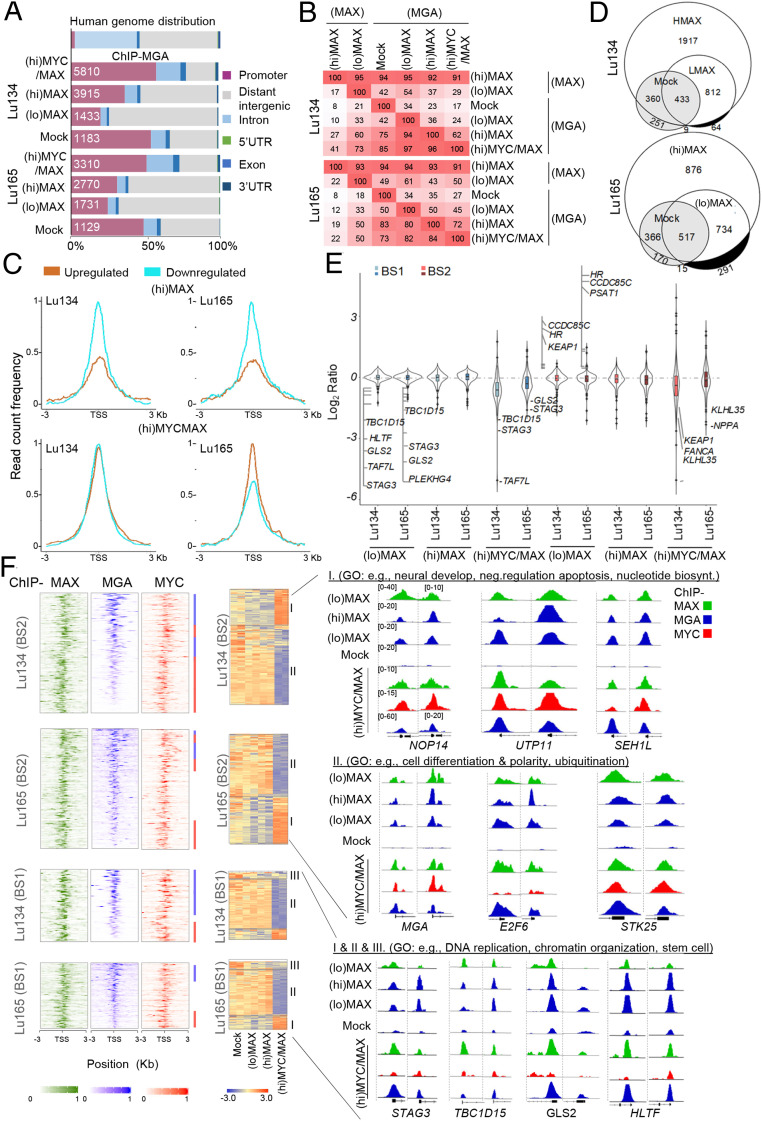
ChIP-seq of MYC and MAX in the distinct MAX-deficient cells and genetic backgrounds. (A) Western blot of total lysates to show the levels of MYC and MAX proteins in the indicated cells carrying ectopic overexpression of MYC ((hi)MYC) and of MYC and MAX simultaneously ((hi)MYC/MAX) at different concentrations of doxycycline (Dox). (B, Left) Heatmaps representing the normalized ChIP-seq intensities of ectopic MYC in Lu134-(hi)MYC cells (1,000 ng/mL Dox) and in the (hi)MYC/MAX cells (1,000 ng/mL Dox). (Right) Read count frequency of the heat maps at ±3 Kb regions centered over the TSS of MYC occupancy in the indicated cell models. (C) Representative snapshots from IGV, of ChIP-seq profiles at selected target loci, performed in the indicated cell models. (D) Western blot showing the ectopic expression of MAX ((lo)MAX cells (10 and 5 ng/mL Dox, Lu134 and Lu165, respectively), (hi)MAX cells (1,000 ng/mL Dox), and the levels of the MYC family of proteins in the indicated cells. ACTIN, protein-loading control. The H82 and the H69 cell lines are included as a control of a MYC- and MYCN-overexpressing cells, respectively. (E) Genome-wide functional annotations for peaks generated by the ChIP-seq analyses. Promoters are defined as the regions ±3 Kb around the annotated TSS. (F) Venn diagrams representing the overlap of MAX peaks in the Lu134 and Lu165 cells following expression of high ((hi)MAX) or endogenous-like ((lo)MAX) levels of MAX.
MAX-Reconstituted Versus MYC Oncogenic–Activated Cells: Conserved Pattern of MAX Bound to DNA but with an Antagonistic Gene Expression Profile.
Traditional DNA-binding studies have shown that MAX is bound to the same DNA sequences regardless of its dimerization partner (21–23). To investigate this at a genome-wide scale and to evaluate the influence of the MYC and MAX protein levels (physiological-like versus supraphysiological) on the dynamics and distribution of MAX genomic occupancy, we rescued the expression of MAX in Lu134 and Lu165 cells by using a doxycycline-inducible system because the stable expression of wild-type MAX in these cell lines reduces cell growth (5). It has been reported that the supraphysiological levels of MAX shift the equilibrium from MAX heterodimers toward the formation of MAX/MAX homodimers (19, 20), for which reason we tested different doxycycline doses. Doxycycline concentrations of 10 and 5 ng/mL in the Lu134 and Lu165, respectively, produced physiological-like levels of MAX (hereafter (lo)MAX cells), comparable to those in SCLC cell lines bearing wild-type MAX (Fig. 2D and SI Appendix, Fig. S2A). Instead, 1,000 ng/mL doxycycline was used to produce supraphysiological levels of MAX ((hi)MAX cells from herein). The ectopic expression of MAX was verified to be homogeneously widespread in all the cells (SI Appendix, Fig. S2B).
Next, we performed ChIP-seq of MAX in different contexts: 1) low to zero MYC and physiological-like levels of MAX ((lo)MAX cells), 2) low to zero MYC and high MAX levels ((hi)MAX cells), and 3) high MYC and physiological-like MAX levels ((hi)MYC/MAX cells). The latter cells express supraphysiological levels of MYC, with a MYC/MAX ratio almost equal to that in the MYC-amplified SCLC cell line, H82 (SI Appendix, Fig. S2C), thereby constituting a model of SCLC cells that have shifted from being MAX deficient to being MYC activated. Similar to previous observations (22–25), about half of the regions that recruited MAX were within or near gene promoters (±3 Kb around the transcription start sites [TSSs]) (Fig. 2E). There were significantly fewer MAX-bound regions in the two (lo)MAX cells, although most of the annotated promoters for single genes in the (lo)MAX cells were included among those of the (hi)MAX cells (SI Appendix, Fig. S3A). There was a significant overlap (P = 0.0001, permutation test) between peaks in annotated promoters for single genes. This was particularly pronounced between the two (hi)MAX cells (>80%) but was less between the two (lo)MAX cells (30 to 40%) (Fig. 2F). A global analysis of the ChIP-seq results and inspection of multiple regions of the genome also showed a strong overlap between the promoter regions bound by MYC and MAX in the (hi)MYC/MAX cells, reflecting their colocalization (Fig. 2C and SI Appendix, Fig. S3B).
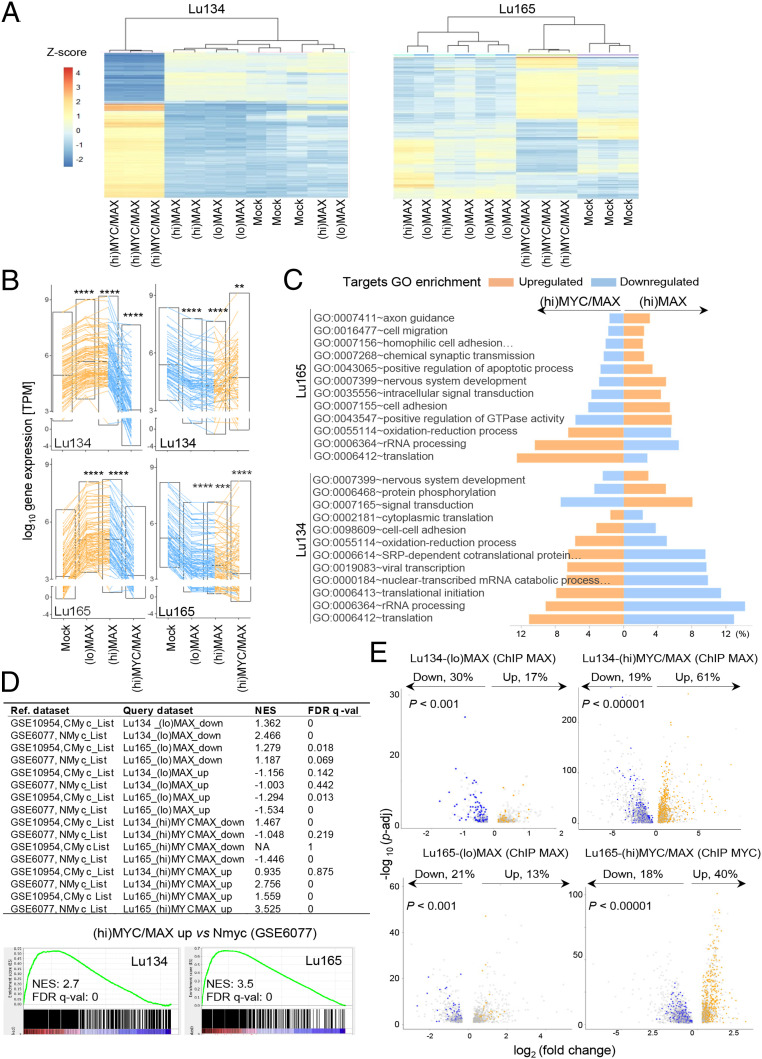
Our previous findings showed that the expression profile after restoring MAX in SCLC cells was inversely correlated with that of the lungs of mice carrying activated Myc or Nmyc (5). Supporting this, we observe here that the global changes in gene expression after rescuing MAX, for (hi)MAX and (lo)MAX cells, were opposite to those after oncogenic activation of MYC ((hi)MYC/MAX cells) (Fig. 3 A and B and Dataset S1). The overall changes were very similar among (lo)MAX- and (hi)MAX-expressing cells (SI Appendix, Fig. S4), which may be consistent with the observation that, although MAX can form homodimers, they are inhibited from binding DNA (26). Despite these similarities, it is important to mention that the up-regulation and down-regulation was more marked in the (hi)MAX than in the (lo)MAX cells. Gene Ontology (GO) enrichment analysis showed that MAX restitution activates the transcription of genes involved in differentiation while it represses genes involved in ribosome biogenesis, mitochondrial and cytoplasmic translation, and energy metabolism, which are known to be activated by MYC in MYC-transformed cells (25). These processes were inversely regulated in the (hi)MYC/MAX cells (Fig. 3C and SI Appendix, Fig. S5). Gene set enrichment analysis (GSEA) showed a direct correlation of the (hi)MYC/MAX and an inverse correlation of the (lo)MAX- and (hi)MAX-associated profiles with the previously identified targets of Myc and Nmyc (Fig. 3D).
Fig. 3.
Changes in gene expression upon MAX restitution are inversely correlated with changes in (hi)MYC/MAX-expressing cells. (A) Heatmap and dendrograms using the 500 most dynamic genes that changed expression in the RNA-seq, reflecting the gene expression profiles of the indicated cell lines. (B) Graphs showing gene expression values in transcripts per million (TPM) for the 100 most up-regulated and down-regulated genes selected from the (hi)MAX expressing cells (from Dataset S1) for each SCLC cell line and genetic context (Mock, (hi)MAX, (lo)MAX, or (hi)MYC/MAX). Bars show mean ± SEM; two-sided unpaired Student’s t test, **P < 0.01; ***P < 0.005; ****P < 0.001. (C) The common GO categories of the 20 GO most enriched categories for the up-regulated (orange) and down-regulated (blue) genes in each of the (hi)MAX (Left) and (hi)MYC/MAX cells (Right) (genes from Dataset S1 and SI Appendix, Fig. S5). (D) GSEA comparing our mRNA-seq data (query datasets) with datasets GSE6077 and GSE10954 from lungs of mice overexpressing nMyc and cMyc, respectively. (Bottom) Two selected comparatives in detail. (E) Volcano plots depicting n-fold change in gene expression for the genes bound by either MAX or MYC in each cell type. Colored dots represent the genes up-regulated (in yellow) and down-regulated (in blue) in each cell line, among the promoters that recruit MAX or MYC (from Dataset S1 and SI Appendix, Fig. S5). Changes in gene expression among genes that do not recruit MAX or MYC are indicated in gray. The percentage of bound promoters among the up-regulated (in yellow) and down-regulated (in blue) genes is also indicated. P values were determined by Pearson's χ2 test. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
The interaction of MAX with MYC activates gene expression, but its interaction with other partners has repressive effects (12, 20). Accordingly, we found stronger binding of MAX to repressed genes in (lo)MAX and (hi)MAX cells and a predominant transcriptional repression among the targets of MAX in the (lo)MAX cells. In contrast, in the (hi)MYC/MAX cells, MAX bound more strongly in the activated genes, and its recruitment to promoters was significantly associated with transactivation (Fig. 3E and SI Appendix, Fig. S6 A–C).
Taken together, these results suggest that the profile of MAX recruitment to the genomic DNA is similar when MAX is overexpressed alone and when it is concomitantly expressed with oncogenic levels of MYC. However, the global patterns of gene expression are strongly shifted under the two circumstances, possibly as a result of the different activities of MAX arising from its binding to distinct partners.
The Absence of MAX Does Not Affect the Formation of the ncPRC1.6.
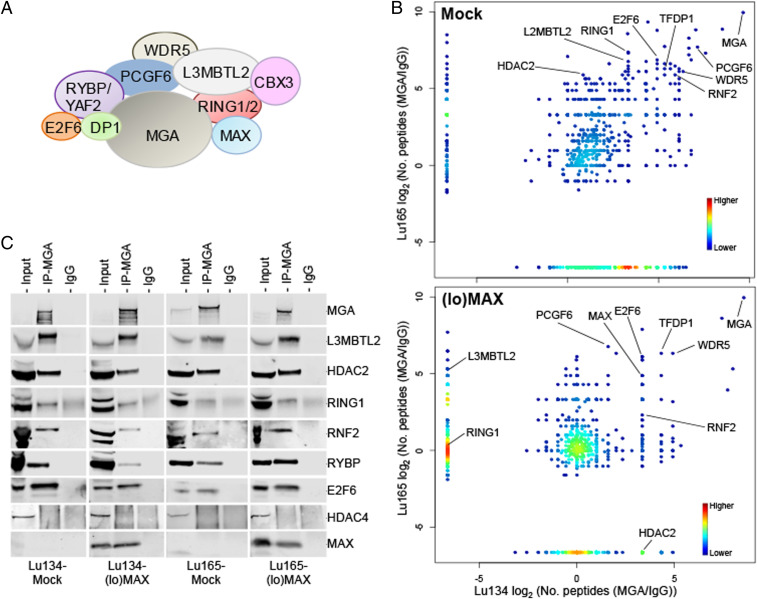
As mentioned in the introduction, the MYC-MAX and MXDs/MNT/MGA-MAX complexes have opposite or antagonistic functions in transcriptional regulation, with MAX being required for DNA binding by all the factors in the network. MAX also acts as part of the noncanonical ncPRC1, specifically ncPRC1.6, which includes the following set of proteins: E2F6, L3MBTL2, MGA, PCGF6, RING1A, RING1B, RYBP, TFDP1, YAF2, and WDR5 (Fig. 4A) (17, 27, 28). Given this, we examined whether the presence or absence of MAX affects the formation or composition of ncPRC1.6. We profiled the binding of MGA by immunoprecipitation of endogenous MGA and then carried out mass spectrometry–based proteomic analysis in the Mock, Lu134, and Lu165 cells and after restitution of MAX ((lo)MAX cells). MGA was found to associate with all components regardless of the presence of MAX (Fig. 4B). We confirmed these results by immunoprecipitating MGA followed by immunoblotting (Fig. 4C). Similar results were obtained after immunoprecipitating E2F6 followed by immunoblotting (SI Appendix, Fig. S7A). Our results demonstrate that ncPRC1.6 forms regardless of the presence or absence of MAX. In addition to the known protein constituents of ncPRC1.6, the mass spectrometry proteomics screening identified other proteins bound to MGA (Dataset S2). Additional studies are needed to assess their interaction with MGA and with the ncPRC1.6 and their functional implications.
Fig. 4.
ncPRC1.6 can be formed in the absence of MAX. (A) Schematic representation of the main components of ncPRC1.6. (B, Left) Density plots, from the immunoprecipitation–mass spectrometry (IP-MS) results, showing the proteins that form stable complexes with MGA in the Lu134 and Lu165 cells without MAX (Mock) or in cells that express ectopic and endogenous-like levels of MAX ((lo)MAX) (see also Dataset S3 for detailed information). The proteins from ncPRC1.6 are indicated. The IP-MS results were analyzed based on the enrichments of peptides precipitated by anti-MGA antibodies relative to IgG controls (transforming 0 to .1) for each indicated cell line and condition. (C) Confirmation of the MGA-containing complexes in each cell line and condition. MGA was immunoprecipitated from whole extracts followed by immunoblot of the indicated proteins. HDAC4 was included as a negative control.
As a type of PRC1, ncPRC1 catalyzes the monoubiquitination of histone H2A at lysine 119 (H2AK119ub1) through the heterodimeric E3 ligase RING1B/PCGF1–6 and thereby contributes to chromatin compaction and transcriptional silencing (28). We did not find any changes in the global levels of H2AK119ub1 upon restitution of MAX or in the (hi)MYC/MAX cells (SI Appendix, Fig. S7B).
MAX Reconstitution Enhances the Recruitment of MGA to the DNA and Represses Genes with Cell Division– and Germ Cell–Related Functions.
Since ncPRC1.6 formation is independent of MAX, we next investigated how the availability of MAX affects the DNA-binding activities of MGA. We performed ChIP-seq of MGA in the various Lu134 and Lu165 cell models (i.e., Mock, (hi)MAX, (lo)MAX, and (hi)MYC/MAX cells). Similar to previous reports (29, 30), our analysis confirmed that MGA was bound in close proximity to TSS. Furthermore, we observed that MGA was recruited to the DNA in the Mock cells, although the rescue of MAX expression leads to a global gain of MGA occupancy, including the recruitment of MGA to additional gene promoters (Fig. 5A). It was also observed that the promoters bound by MGA in any of the cell models are targets of MAX, as is evident in the (hi)MAX cells in which MAX is bound to more than 90% of the promoter targets of MGA (Fig. 5B). We found that the restitution of MAX drove moderate changes in gene expression among the MGA-associated promoters in the (hi)MAX and (lo)MAX cells, predominantly transcriptional repression (Fig. 5C and SI Appendix, Fig. S8). In contrast, in the (hi)MYC/MAX cells, the targets of MGA showed changes in gene expression consisting of both transcriptional activation and repression.
Fig. 5.
MAX restitution enhances the recruitment of MGA to the DNA and represses cell division– and germ cell–related functions. (A) Genome-wide functional annotations for peaks generated from the ChIP-seq analyses. Promoters are defined as the regions ±3 Kb around the annotated TSS. (B) Percentage overlap of peaks at promoter regions of ChIP-seq proteins and cell line models. (C) Read count frequency of the binding of MGA among the genes up-regulated or down-regulated in each condition (from Dataset S1) ±3 Kb regions centered over the TSS, of the MGA occupancy, in each indicated cell model. (D) Venn diagrams representing the overlap of MGA peaks in the Lu134 and Lu165 cells following expression of (hi)MAX or (lo)MAX. The white and gray areas represent the BS2- and BS1-associated promoters, respectively. (E) Violin plots representing the changes in gene expression (transcripts per million) relative to the Mock cells in each cell model and group of MGA-bound promoters (top 10% each of BS1 and BS2). Some of the up-regulated or down-regulated transcripts are indicated. (F, Left) Heat maps representing the normalized ChIP-seq intensities for the MAX, MGA, and MYC proteins in the BS1 and BS2, ranked by the intensity of the MGA binding, centered ±3 Kb around the TSS. (Right) Colored bars indicate the ChIP-seq (MGA, in blue; MYC, in red) with greater intensity of binding in each of the regions. (Middle) Heatmaps of the gene expression from the BS1 and BS2 (10% greater intensity) in the indicated cell lines. Different regions have been labeled (groups I, II, and III) according to their profile of gene expression in (hi)MYC/MAX cells compared with (lo)MAX and (hi)MAX cells. (Right) Representative integrative genomics viewer screenshots for peaks generated by the ChIP-seq analyses in each cell model (screenshots Lu134 and Lu165, Left and Right, respectively). The group and the GO analyses showing selected functions for each group are also indicated.
The observations above imply that MGA can bind some promoter regions in the absence of MAX, while other promoters recruit MGA only when MAX is restored, thus constituting MAX-independent and MAX-dependent targets of MGA, respectively. Here, we classified these, respectively, as promoters with type 1– and type 2–binding sites (hereafter, BS1 and BS2). We generated lists of BS1- and BS2-associated promoters according to the criteria that BS1 were gene promoters that recruited MGA in either of the Mock cell lines and that BS2 were gene promoters that recruited MGA in the (hi)MAX cells, unless they had already been classified as BS1. The BS1 promoters were less abundant than the BS2 promoters in both cell lines (Fig. 5D). We selected approximately the top 10% BS1 and BS2 promoters with the highest intensity of binding in each group for detailed analysis (Dataset S3). GO term enrichment analysis revealed that the BS2-associated genes were enriched in cell differentiation, apoptosis, and metabolic-related features, whereas the BS1-associated genes were related to transcription and DNA replication and repair processes (SI Appendix, Fig. S9). BS1 and BS2, in (lo)MAX and (hi)MAX cells, were predominantly associated with transcriptional repression; only a few BS2-associated genes were up-regulated, whereas in (hi)MYC/MAX cells, there was up-regulation and down-regulation associated with the recruitment of MGA to both types of promoters (Fig. 5E). The presence of gene activation and repression among the targets of MGA in the (hi)MYC/MAX could be due, at least in part, to competition between MGA and MYC to bind to MAX. Consistent with this, we observe a mutually exclusive pattern of MGA and MYC intensity of binding to the DNA in (hi)MYC/MAX cells (Fig. 5F).
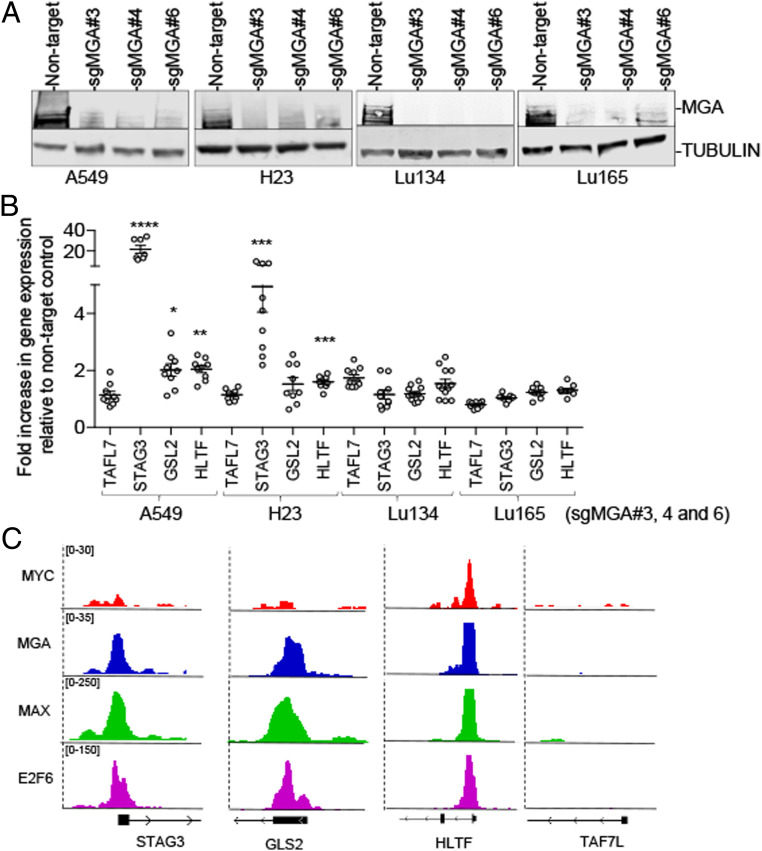
Most targets of MGA that became activated in the (hi)MYC/MAX cells were found repressed in the (lo)MAX and (hi)MAX cells (group I) (Fig. 5F). However, the opposite was not true since the genes repressed in the (hi)MYC/MAX cells barely changed in the (lo)MAX and (hi)MAX cells (group II). Group II included important tumor suppressor genes, for example, KEAP1 and FANCA, as well as components of ncPRC1.6, such as RYBP, E2F6, and MGA itself. There was a third group (III), comprising those genes that were repressed in the three cell models, such as TAF7L, GLS2, and HLTF, which were involved in germ cell–related processes (Fig. 5 E and F and Dataset S3). This is in keeping with the findings in mouse pluripotent stem cells that the heterodimeric MGA/MAX is required to repress germ cell–specific genes (29). The level of these transcripts was found to be higher in lung cancer cells carrying MGA inactivation (SI Appendix, Fig. S10). Furthermore, the generation of knockouts for MGA in the A549 and H23 lung cancer cell lines, which are wild type for MGA and for MAX (5), increased the levels of these transcripts, specially of STAG3 but not of TAF7L (Fig. 6 A and B). A ChIP-seq analysis of the A549 cells, of E2F6, MGA, and MAXs, confirmed their recruitment to the STAG3, GLS2, and HLTF, but not to TAF7L, promoters (SI Appendix, Fig. S6C). Instead, the depletion of MGA in the Lu134 and Lu165 cells rendered no significant changes in the expression of these genes. All the above demonstrate that these transcripts are repressed by MGA through the ncPRC1.6 complex.
Fig. 6.
The generation of knockouts for MGA derepressed transcripts related with division and germ cells. (A) Western blot of the immunoprecipitated MGA protein and of TUBULIN from the input as protein-loading control in the indicated lung cancer cell lines showing the knockout of MGA using three different single guide RNA. (B) Real-time quantitative PCR of the indicated transcripts, relative to IPO8 and to each corresponding nontarget control, in the indicated lung cancer cells infected with the sgMGA (sgMGA#3, sgMGA#4, sgMGA#6). Lines show mean ± SEM; values represent triplicates for each of the three different sgMGA (n = 9). *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; two-sided unpaired Student’s t test. (C) Representative integrative genomics viewer (IGV) screenshots for peaks generated from the ChIP-seq of E2F6 (Gene Expression Omnibus [GEO] accession number: GSM1010766), MAX (MAX ChIP-seq [GEO accession number: GSM935298], and MGA and MYC (GEO accession number: GSE112188) in the A549 cells.
MAX Restitution Shifts the DNA-Binding Profile of MGA from E2F Motifs to E-Boxes.
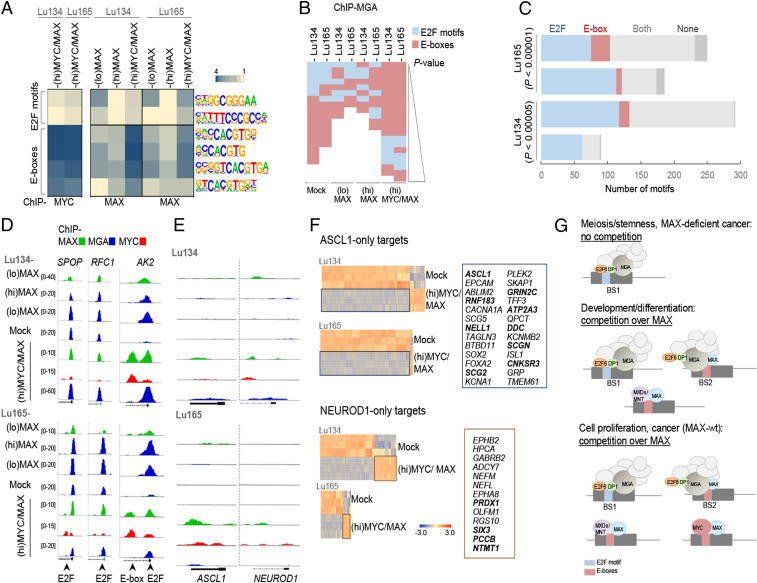
As part of ncPRC1.6, MGA also interacts with heterodimeric E2F6/DP1/2 proteins, which bind DNA in a sequence-specific manner (29, 30). Taking this into consideration, we studied the dynamics of the recruitment of MGA to E2F motifs (GCGGGA) or to E-boxes (CACGTG), depending on whether ectopic MAX is absent or present with or without oncogenic MYC. First, we determined the preferential binding of MYC, MAX, and MGA to these motifs under the different conditions. As expected, MYC was bound almost exclusively to E-boxes, whereas MAX and MGA could be recruited to E-boxes and E2F motifs (Fig. 7 A and B). A shift of MGA positioning from E2F motifs to E-boxes was observed in parallel with the restitution of MAX. This effect was stronger in the (hi)MYC/MAX cells (Fig. 7B).
Fig. 7.
MAX restitution shifts MGA DNA-binding profile from E2F sites to E-boxes and MYC-oncogenic activation decreases the levels of ASCL1. (A) Enrichment, given as the abundance relative to background, of the indicated DNA motifs (E2F sites and E-boxes) in the promoters bound by MYC or MAX, of the indicated cells and conditions (Hypergeometric Optimization of Motif EnRichment [HOMER]). (B) Enrichment, ranked by P value (P < 0.01), of E2F motifs and E-boxes found in the promoters bound by MGA, in the indicated cells and conditions (HOMER). (C) Number of E2F motifs and E-boxes in BS1 and BS2 (among the 10% selection) in the indicated cells. P values were determined by Pearson's χ2 test. (D and E) Representative integrative genomics viewer screenshots for peaks generated from the ChIP-seq analyses for each cell type and set of conditions. (F) Heat maps of the gene expression of the ASCL1-only and NEUROD1-only targets selected from ref. 30 (n = 540 for ASCL1 and n = 374 for NEUROD1) among the genes up-regulated or down-regulated in (hi)MYC/MAX cells (from Dataset S1). Selected up-regulated genes from each group are indicated on Right. Those that are common for both cell models are highlighted in bold. (G) Diagram showing scenarios in which the competition for available MAX is important in cell physiological processes and cancer development.
Studying the distribution of the E2F motifs and E-boxes among the top 10% BS1 and BS2 promoters, we found that >90% contained either one or both motifs. There was a widespread presence of the E2F motif, which was found in at least 80% of the BS1 and BS2 promoters. E-boxes were significantly overrepresented throughout the BS2, and E2F sites alone were overrepresented in BS1 (Fig. 7C). The overrepresentation of E2F motifs in BS1 indicates a preference for E2F6/DP1 binding. In this regard, it has been reported (30) that MGA is essential for recruiting ncPRC1.6 to its targets genes and that it executes its function through two different mechanisms: as a scaffold that is independent of the bHLH domain but dependent on E2F6 and by sequence-specific bHLH binding that is independent of E2F6. Many of the promoters from our BS1 lists match those regions previously found to recruit MGA in a bHLH-independent/E2F6-dependent manner (e.g., RFC1, PHF20, SPOP, and RPA2), whereas the BS2 lists include promoters that were found to recruit MGA in a bHLH-dependent/E2F6-independent manner (e.g., CDIP, ZFR) (Figs. 5F and 7D and Dataset S3). Our current findings, combined with those of previous reports, indicate that, in the cancer cells that lack MAX, MGA acts essentially as a scaffold to recruit ncPRC1.6 to E2F6/DP1-dependent binding sites. The precise transcriptional interactions between E2F6/DP1 and MGA/MAX on BS1 are not yet fully understood since both have been associated with transcriptional repression. Different affinities for the different promoters may account for the diverse and complex regulation of gene expression observed in these distinct genetic backgrounds, and competition between MGA and MYC to heterodimerize with MAX may also play a role in the case of the (hi)MYC/MAX cells. An example of these dynamics is the AK2 gene, which is repressed in (hi)MAX and (lo)MAX cells but activated in (hi)MYC/MAX cells. The AK2 promoter has an E2F motif and an E-box that are distant enough to produce independent peaks in the integrative genomics viewer. In the absence of MYC ((lo)MAX and (hi)MAX cells) the MAX/MGA heterodimer is found only in the E2F motif, whereas, after MYC overexpression ((hi)MYC/MAX cells), the MAX/MYC heterodimer is bound to the E-boxes, and the MAX/MGA is bound to the E2F motifs (Fig. 7D).
Oncogenic MYC Reduces the Level of ASCL1 and of ASCL1-Related Transcripts and Promotes NEUROD1 Characteristics.
Here, we showed that most MAX-deficient SCLCs expressed high levels of ASCL1. However, one of the two MAX-deficient cell lines studied in deep here, Lu134, predominantly express NEUROD1, while the other one, Lu165, express ASCL1 and NEUROD1 (Fig. 1 B and C). We found that when MAX was overexpressed concomitantly with oncogenic levels of MYC ((hi)MYC/MAX cells), the mRNA levels of ASCL1 were decreased by half in both cell lines, while the levels of NEUROD1 were reduced in the Lu134 but not in the Lu165 cells (Dataset S1). The effects on ASCL1 and NEUROD1 were not mediated by direct transcriptional regulation of MAX, MYC, or MGA since we were unable to detect MAX, MYC, or MGA directly occupying the promoters of ASCL1 or NEUROD1 (Fig. 6E). Concomitant with the reduction in ASCL1, there was prominent down-regulation of most ASCL1 targets and up-regulation of some NEUROD1 targets (31) in (hi)MYC/MAX cells compared with the Mock, MAX-deficient cells (Fig. 7F and SI Appendix, Fig. S11). Since NEUROD1 was not up-regulated, its relative increase over ASCL1 abundance may underlie the observed shift from ASCL1 to NEUROD1 characteristics. These findings suggest that the transformation from a MAX-deficient to a MYC-oncogenic SCLC also shifts their dependency on these two transcription factors that are associated with the development of the neural lineage.
Discussion
We have shown that most SCLC cells bearing the MAX gene inactivation have ASCL1 characteristics, as is also the case for MYCN- and MYCL1-amplified SCLC cells, suggesting that they have a degree of similarity or common origin. This is in contrast to the MYC-amplified SCLCs, which, as previously reported, have NEUROD1 characteristics (6, 31, 32). It is interesting that one MAX-deficient cell line used here express both the ASCL1 and the NEUROD1 factors, although the predominant profile was that of ASCL1. Our data also show that the expression of oncogenic MYC represses ASCL1 and triggers, to some extent, a NEUROD1 expression profile without up-regulating NEUROD1. This suggests that the two neurogenic transcription factors compete to establish a predominant genetic program. We did not observe recruitment of MYC or MAX to the ASCL1 or NEUROD1 promoters, indicating that other targets of MYC/MAX mediate the shift from ASCL1 to NEUROD1 characteristics in SCLC cells.
The MYC family of proteins were barely expressed in the SCLC cells with genetically inactivated MAX. A recent study showed MAX deficiency to have a profound effect on MYC stability in both normal and premalignant settings (22), which supports our observations. Here, we also observed a lack of MYC recruitment to the genomic DNA, even after ectopic overexpression of MYC, implying that the transcriptional activity of the MYC proteins does not play a role in the tumorigenesis of MAX-deficient cells. The concomitant ectopic expression of MAX with oncogenic level of MYC restored the ability of MYC to bind DNA, producing a gene expression profile compatible with that of MYC-amplified cancer cells. While the requirement of MAX for the transcriptional activities of the MYC family of proteins has been known for a long time (20–23), even in a recent work using wide-genome screenings (22), our current study demonstrates this in naturally MAX-deficient cancer cells. The targets of the heterodimer MYC/MAX in these cells were associated with transactivation, whereas the targets of MAX in the MAX-restituted cells were mostly down-regulated. Competition between MYC and MXD1-4/MNT/MGA proteins for binding to MAX and to the same E-boxes is known (21–23) and is evidence that these transcriptional regulators act as functional antagonists. In this regard, we recently reported that the overexpression of MGA in lung cancer cells represses the targets of MYC, consistent with the idea that they possess competitive and antagonistic functions (33). Our results fully support this view and also suggest that, since MAX serves as a network edge, the genetic inactivation of MAX may contribute to the development of cancer by preventing the prodifferentiation transcriptional regulation exerted by its partnering with MXD/MNT/MGA.
Unlike MYC, MAX is an abundant and stable protein that is expressed in proliferating and resting normal cells (20). However, we previously showed that MAX expression can be regulated by corticoids (5). Others have shown that Max expression is transiently attenuated in germ cells undergoing meiosis in vivo and that the knockdown of Max in embryonic stem cells activates the expression of germ cell–related genes (34), indicating that the levels of MAX are regulated in some specific physiological processes. The role of MAX in regulating germ cell– and meiosis-related genes is dependent on ncPRC1.6, a PRC1 that includes MAX and MGA (27–29), the latter of which is also genetically inactivated in cancer (5, 13, 14). In this study, we found that the lack of MAX does not prevent the formation of the complex but restricts the recruitment of MGA to the DNA. We defined as BS1 those sites within promoters that can recruit MGA in a MAX-independent manner and as BS2 those that recruit MGA only after MAX restitution. We also found that BS1 are enriched in E2F motifs, compared with BS2, which have more E-boxes. Remarkably, BS1-associated promoters were coincident with those regions previously reported to recruit MGA in a bHLH-independent/E2F6-dependent manner in which MGA acts as a scaffold, whereas the BS2-associated promoters matched those that recruit MGA in a bHLH-dependent/E2F6-independent manner (30). This leads us to postulate that, in cancer cells lacking MAX, MGA has a preeminent scaffolding function. In this scenario, the regulation of gene expression from ncPRC1.6 would be directed by the E2F6/DP1 module. Interestingly, several years ago, we found that the gene coding for DP1, TFDP1, is strongly amplified in a small subset of lung tumors, leading to high levels of DP1 protein (35). Such levels of DP1 could have oncogenic potential, promoting the activities of the E2F6/DP1 module within ncPRC1.6. Our data confirms and extends previous knowledge about the various scenarios in which the competition for available MAX is important for different cell physiological processes, including cancer development (Fig. 7G).
In conclusion, we have demonstrated that most MAX-mutant SCLCs have ASCL1-like characteristics and are MYC independent and exhibit deficient ncPRC1.6-mediated gene repression.
Materials and Methods
Lung Cancer Cell Lines.
Cell lines were obtained from the American Type Culture Collection, grown under recommended conditions, and maintained at 37 °C in a humidified atmosphere. All cells tested negative for mycoplasma infection. The antibodies used are described in SI Appendix, Table S1A.
Western Blot, Immunofluorescences, and qRT-PCRs.
Antibodies and primers sequences are in SI Appendix, Table S1B. Detailed information about the methodologies are included in SI Appendix.
Construction of Expression Vectors and Infections.
The complete MAX transcript (NM_145112.2) had been previously cloned (5). Complete MYC (NM_002467.6) complementary DNA was PCR amplified from a retrotranscribed human RNA pool (Agilent Technologies) using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) following standard protocols (SI Appendix). The primers used are indicated in SI Appendix, Table S1B.
Microarray Global Gene Expression Analysis.
We followed previously described procedures (5). The analysis was undertaken at the Genomics Unit of the Center for Genomic Regulation (CRG) (SI Appendix). Expression data were analyzed using the R statistical language (R Core Team [2014], URL http://www.R-project.org/). Raw data were extracted, and the background was corrected and normalized using the quantile algorithm available in Bioconductor’s limma package (36). Normalized expression values were plotted with Bioconductor’s ggplots and Complexheatmap packages. The ASCL1 and NEUROD1 target lists were elaborated selecting common genes occupied by ASCL1 or NEUROD1 in at least two cell lines from the previous publication (6).
ChIP-Seq.
For ChIP assays, cells were fixed with 1% formaldehyde methanol free (Thermo Scientific) for 10 min at room temperature and were then quenched by 125 mmol/L glycine for 15 min at room temperature, washed with ice-cold phosphate-buffered saline twice, and centrifuged at 200 g at 4 °C for 5 min. For each ChIP reaction, 60 μL Magna ChIP Protein A+G Magnetic Beads (Merck, Millipore) was used according to the manufacturer’s protocol. Detailed information about the methodology is included in SI Appendix. At least two independent ChIP experiments were performed. Immunoprecipitated chromatin was deep sequenced at the CRG using the Illumina HiSeq 2500 system.
ChIP-Seq Data Analysis.
Reads were aligned to the human reference genome hg38 using Bowtie v1.2.2 with default parameters without allowing for multimapping (–m 1) (37). PCR duplicates were removed using PICARD (http://broadinstitute.github.io/picard/). Ambiguous mapping reads were discarded. Peaks were called using MACS2 v2.1.1 (38). To avoid false positives, peaks were discarded if they were present in the ChIP-seq of MAX in the MAX-deficient cells of the respective SCLC cells. Genomic peak annotation was performed with the ChIPpeakAnno v3.15 R package, considering the region of ±3 Kb around the TSS as the promoter (39). Unless otherwise specified, all analyses considered the peaks overlapping with promoter regions. Peak lists were then transformed to gene target lists. Permutation tests (10,000 permutations) were performed to determine associations by overlap between region sets A and B, creating random regions throughout the genome using the Bioconductor package regioneR (40).
Bedgraph files were generated using the function makeUCSCfile from HOMER with default parameters normalizing for differences in sample library size, and BigWig files were generated using the function bedGraphToBigWig from University of California, Santa Cruz. Heat maps and intensity plots were performed using the functions computeMatrix in a window of ±3 kb center in the TSS followed by plotHeatmap from deepTools (41). To homogenize the scale of all heat maps and intensity plots, signal intensity was scaled to 0 to 1 by applying the formula (X – P05)/(P98 – P05) to each matrix generated by computeMatrix.
Motif enrichment analyses were performed using HOMER motif discovery software (42). For annotated ChIP-seq peaks, a window of ±100 base pairs around the peak center was applied. Values of P < 0.01 were taken to define a motif as being significantly enriched.
RNA Sequencing.
RNA sequencing (RNA-seq) was carried out at the Spanish National Genome Analysis Center. About 2,500 ng total RNA from SCLC cell lines were used. RNA integrity values ranged from 9.0 to 10.0 when examined by a BioA RNA Nano kit (Agilent). RNA-seq paired-end reads were mapped against the human reference genome (GRCh38) using STAR version 2.5.3a with ENCODE parameters for long RNA. DEseq2 was used to normalize counts. Annotated genes (Gencode v27) were quantified using RSEM version 1.3.0 with default parameters. The RNA-seq report is provided in SI Appendix, Table S1C.
To generate the lists of up-regulated and down-regulated transcripts for each condition, we chose the following criteria: 1) transcripts induced or repressed, as indicated in each case, under each condition with respect to the Mock cell line and 2) statistical significance (see Statistical Analysis section). The genes are listed in Dataset S1.
The listed genes were subjected to several analyses, such as GO functionalities (https://david.ncifcrf.gov) or GSEA, using the indicated gene expression signatures (ranked by the n-fold values of change) as the gene set.
Immunoprecipitation and Mass Spectrometry.
For immunoprecipitation, we used previously described protocols (33). Details of the antibodies are in SI Appendix, Table S1A. A total of 10 million cells were lysed with Nonidet P-40 lysis buffer and sonicated with a tip ultrasonic homogenizer. Detailed information about the methodologies are included in SI Appendix, Methods. For mass spectrometry assays, 100 million cells were grown in suspension and harvested by centrifugation. The cell pellet was resuspended in Net2 buffer (50 mmol/L Tris HCl, 200 mmol/L NaCl, 0.1% Triton X-100, 5 mmol/L MgCl2, and 1 mmol/L ethylenediaminetetraacetic acid) and incubated while rotating at 4 °C for 1 h. Cell lysate was sonicated and centrifuged at 2,500 g at 4 °C for 3 min. Supernatant was collected and mixed with 20 μg primary MGA antibody or IgG and incubated while rotating overnight at 4 °C. Details about the mass spectrometry assays can be found in SI Appendix. Eluted and desalted peptides were resuspended in 10 µL 0.1% formic acid and loaded into the Orbitrap Velos Pro using the “STD-VL-DDA-60min-T20-CID-IT” method. Peptides were analyzed with the Proteome Discoverer v1.4. with the “STD-PWF-MASCOT-ANY-IT-DECOY” workflow. Peptides were filtered at 5% false discovery rate.
We analyzed the results based on the enrichments of peptide precipitated by anti-MGA antibodies versus IgG controls (not found in the IgG control) and that were common to the two cell models. MAX, which is known to be the canonical binding partner of MGA (15), and the MGA protein itself are among the most significantly enriched proteins in this immunoprecipitation (Dataset S2).
Generation of MGA-Depleted Cell Lines.
The A549, H23, Lu134, or Lu165 cells were infected with lentivirus using plasmids (LentiCRISPR v2 no. 52961, Addgene) expressing mammalian codon–optimized Cas9 and different single guide RNAs targeting the coding region of human MGA or a nontarget sequence as negative control. The sequences of the oligonucleotides are included in SI Appendix, Table S1B. Puromycin selection (2 μg/mL) was carried out 48 h after infection for 3 to 4 d. Cell clones were further analyzed by Western blot.
Statistical Analysis.
Data were analyzed using a two-tailed Student’s unpaired-samples t test or by Pearson's χ2 test. Group differences were presented as means and SDs. Differences were considered statistically significant for any value of P < 0.05.
Supplementary Material
Acknowledgments
We thank Isabel Bartolessis for technical assistance and the confocal facility (Carmen Casals) and the RNA-seq genomic facility (Anna Esteve) for important services. This work was supported by Spanish Grant SAF2017-82186-RAEI/FEDER (EU) (to M.S.-C.) from the Spanish Ministry of Economy and Competitiveness and a grant from the Fundación Científica Asociación Española Contra el Cancer-GCB14142170MONT (to M.S.-C.). M.T.-D. was supported by the Formacion de Personal Investigador (FPI) Fellowship BES-2012-054579, P.L. by the FPI Fellowship BES-2015-072204 and by the European Association for Cancer Research Travel Fellowship, and L.T.-D. by the FPI Fellowship (No. PRE2019-088005). B.M.J. is funded by Spanish Ministry of Science, Innovation, and Universities Project No. RTI2018-094788-A-I00 and by La Caixa Banking Foundation Junior Leader Project (LCF/BQ/PI19/11690001). The proteomics analyses were performed in the Josep Carreras Leukemia Research Institute (IJC) Proteomics Unit. The IJC Proteomics Unit is part of the Spanish Platform of Molecular and Bioinformatics Resources (ProteoRed), Instituto de Salud Carlos III (PT13/0001).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024824118/-/DCSupplemental.
Data Availability
Microarray gene expression data are available in the Gene Expression Omnibus under accession codes GSE144457 (43). The ChIP-seq and RNA-seq data obtained in this study have been uploaded to the Sequence Read Archive (National Center for Biotechnology Information) under accession number BioProject: PRJNA608275 (44). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database partner repository with the dataset identifier PXD017658. All other study data are included in the article and/or supporting information.
References
- 1.Maeda Y., Davé V., Whitsett J. A., Transcriptional control of lung morphogenesis. Physiol. Rev. 87, 219–244 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Sutherland K. D., et al., Cell of origin of small cell lung cancer: Inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19, 754–764 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Travis W. D., et al.,WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart (World Health Organization, ed. 4, 2015). [DOI] [PubMed] [Google Scholar]
- 4.Castillo S. D., et al., Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 72, 176–186 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Romero O. A., et al., MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 4, 292–303 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Rudin C. M., et al., Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco R., et al., A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 30, 1199–1206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peifer M., et al., Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44, 1104–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George J., et al., Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Augert A., et al., MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38, 97–114.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comino-Méndez I., et al., Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet. 43, 663–667 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Diolaiti D., McFerrin L., Carroll P. A., Eisenman R. N., Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta 1849, 484–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L., et al., Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 47, 1061–1066 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurlin P. J., Steingrìmsson E., Copeland N. G., Jenkins N. A., Eisenman R. N., Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 18, 7019–7028 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J., et al., The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11, 319–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauri S., et al., A high-density map for navigating the human polycomb complexome. Cell Rep. 17, 583–595 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Rudin C. M., et al., Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N., The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzner L., Blackwood E. M., Eisenman R. N., Transcriptional activities of the Myc and Max proteins in mammalian cells. Curr. Top. Microbiol. Immunol. 182, 435–443 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Littlewood T. D., Amati B., Land H., Evan G. I., Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene 7, 1783–1792 (1992). [PubMed] [Google Scholar]
- 22.Mathsyaraja H., et al., Max deletion destabilizes MYC protein and abrogates Eµ-Myc lymphomagenesis. Genes Dev. 3, 1252–1264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amati B., et al., Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359, 423–426 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Mao D. Y., et al., Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 13, 882–886 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Sabò A., et al., Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berberich S. J., Cole M. D., Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 6, 166–176 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y., A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Trojer P., et al., L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endoh M., et al., PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 6, e21064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stielow B., Finkernagel F., Stiewe T., Nist A., Suske G., MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical polycomb repressive complex PRC1.6. PLoS Genet. 14, e1007193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borromeo M. D., et al., ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 16, 1259–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollaoglu G., et al., MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 31, 270–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llabata P., et al., MGA suppresses the MYC pathway in lung adenocarcinoma. Mol. Cancer Res. 18, 574–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki A., et al., Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat. Commun. 7, 11056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castillo S. D., et al., Gene amplification of the transcription factor DP1 and CTNND1 in human lung cancer. J. Pathol. 222, 89–98 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Ritchie M. E., et al., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L. J., et al., ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gel B., et al., regioneR: An R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramírez F., et al., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S., et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Cespedes M., et al., MAX-mutant small cell lung cancers sustain impaired activities of the MGA-dependent ncPRC1.NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144457. Deposited 29 January 2020.
- 44.Sanchez-Cespedes M., et al., MAX-mutant small cell lung cancers exhibit impaired activities of MGA-dependent ncPRC1.6. NCBI Bioproject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA608275. Deposited 24 February 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray gene expression data are available in the Gene Expression Omnibus under accession codes GSE144457 (43). The ChIP-seq and RNA-seq data obtained in this study have been uploaded to the Sequence Read Archive (National Center for Biotechnology Information) under accession number BioProject: PRJNA608275 (44). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database partner repository with the dataset identifier PXD017658. All other study data are included in the article and/or supporting information.