Significance
Some of the most spectacular adaptive radiations of plants and animals occur on remote oceanic islands, yet such radiations are preceded by founding events that severely limit genetic variation. How genetically depauperate founder populations give rise to the spectacular phenotypic and ecological diversity characteristic of island adaptive radiations is not known. We generated genomic resources for Hawaiian Metrosideros––a hyper-variable adaptive radiation of woody taxa—for insights into the paradox of remote island radiations. We posit that divergent selection and differential sorting of an unexpectedly rich pool of ancestral variation drove the diversification of lineages. Recurring use of ancient variants from a richer-than-expected gene pool may explain how lineages can diversify to fill countless niches on remote islands.
Keywords: Metrosideros, adaptive radiation, ecological speciation, incipient speciation, speciation genomics
Abstract
Some of the most spectacular adaptive radiations begin with founder populations on remote islands. How genetically limited founder populations give rise to the striking phenotypic and ecological diversity characteristic of adaptive radiations is a paradox of evolutionary biology. We conducted an evolutionary genomics analysis of genus Metrosideros, a landscape-dominant, incipient adaptive radiation of woody plants that spans a striking range of phenotypes and environments across the Hawaiian Islands. Using nanopore-sequencing, we created a chromosome-level genome assembly for Metrosideros polymorpha var. incana and analyzed whole-genome sequences of 131 individuals from 11 taxa sampled across the islands. Demographic modeling and population genomics analyses suggested that Hawaiian Metrosideros originated from a single colonization event and subsequently spread across the archipelago following the formation of new islands. The evolutionary history of Hawaiian Metrosideros shows evidence of extensive reticulation associated with significant sharing of ancestral variation between taxa and secondarily with admixture. Taking advantage of the highly contiguous genome assembly, we investigated the genomic architecture underlying the adaptive radiation and discovered that divergent selection drove the formation of differentiation outliers in paired taxa representing early stages of speciation/divergence. Analysis of the evolutionary origins of the outlier single nucleotide polymorphisms (SNPs) showed enrichment for ancestral variations under divergent selection. Our findings suggest that Hawaiian Metrosideros possesses an unexpectedly rich pool of ancestral genetic variation, and the reassortment of these variations has fueled the island adaptive radiation.
Adaptive radiations exhibit extraordinary levels of morphological and ecological diversity (1). Although definitions of adaptive radiation vary (2–7), all center on ecological opportunity as a driver of adaptation and, ultimately, diversification (2, 8–10). Divergent selection, the primary mechanism underlying adaptive radiations, favors extreme phenotypes (11) and selects alleles that confer adaptation to unoccupied or under-utilized ecological niches. Differential adaptation results in divergence and, ultimately, reproductive isolation between populations (12). Adaptive radiations demonstrate the remarkable power of natural selection as a driver of biological diversity and provide excellent systems for studying evolutionary processes involved in diversification and speciation (13).
Adaptive radiations on remote oceanic islands are especially interesting, as colonization of remote islands is expected to involve population bottlenecks that restrict genetic variation (14). Adaptive radiations in such settings are especially impressive and even paradoxical, given the generation of high species richness from an initially limited gene pool (15). Several classic examples of adaptive radiation occur on oceanic islands, such as Darwin’s finches from the Galapagos islands (16), anole lizards from the Caribbean islands (9), Hawaiian Drosophilids (17), and Hawaiian silverswords (18), to name a few.
Recent advances in genome sequencing and analyses have greatly improved our ability to examine the genetics of speciation and adaptive radiation. By examining sequences of multiple individuals from their natural environment, it has become possible to “catch in the act” the speciation processes between incipient lineages (19). Genomic studies of early stage speciation show that differentiation accumulates in genomic regions that restrict the homogenizing effects of gene flow between incipient species (20). The number, size, and distribution of these genomic regions can shed light on evolutionary factors involved in speciation (19). Regions of high genomic differentiation can also form from evolutionary factors unrelated to speciation, such as linkage associated with recurrent background selection or selective sweeps on shared genomic features (21, 22).
Genomic studies of lineages undergoing rapid ecological diversification have begun to reveal the evolutionary mechanisms underlying adaptive radiations. Importantly, these studies highlight the pivotal role of hybridization between populations and the consequent exchange of adaptive alleles that facilitates rapid speciation and the colonization of diverse niches (23–25). Most genomic studies of adaptive radiation involve animal systems, however, in particular, birds and fishes. In plants, genomic studies of adaptive radiation are sparse (26–28), and all examine continent-wide radiations. There are no genomics studies of plant adaptive radiations in geographically restricted systems such as remote islands. Because the eco-evolutionary scenarios associated with adaptive radiations are diverse (5, 29), whether commonalities identified in adaptive radiations in animals (23, 30) are applicable to plants is an open question. For example, the genetic architecture of animal adaptive radiations typically involves differentiation at a small number of genomic regions (31–33). In contrast, the limited insights available for plants suggest a more complex genetic architecture (26).
We investigated the evolutionary genomics of adaptive radiation in Metrosideros Banks ex Gaertn. (Myrtaceae) across the Hawaiian Islands. Hawaiian Metrosideros is a landscape-dominant, hypervariable, and highly dispersible group of long-lived (possibly >650 y) (34) woody taxa that are nonrandomly distributed across Hawaii’s heterogeneous landscape, including cooled lava flows, wet forests and bogs, subalpine zones, and riparian zones (35, 36). About 25 taxa or morphotypes are distinguished by vegetative characters ranging from prostate plants that flower a few centimeters above ground to 30-m-tall trees, and leaves range dramatically in size, shape, pubescence, color, and rugosity (35, 37, 38); a majority of these forms are intraspecific varieties or races (provisional varieties) of the abundant species, Metrosideros polymorpha (35, 36, 38). Variation in leaf mass per area within the four Metrosideros taxa on Hawaii Island alone matches that observed for woody species globally (39). Common garden experiments (38, 40–44) and parent–offspring analysis (45) demonstrate heritability of taxon-diagnostic vegetative traits, indicating that taxa are distinct genetic groups and not the result of phenotypic plasticity. Metrosideros taxa display evidence of local adaptation to contrasting environments (46, 47), suggesting ecological divergent selection is responsible for diversification within the group (48). This diversification, which spans the past ∼3.1 to 3.9 million years (49, 50), has occurred despite the group’s high capacity for gene flow by way of showy bird-pollinated flowers and tiny wind-dispersed seeds (36, 51). Lastly, the presence of partial reproductive isolating barriers between taxa is consistent with the early stages of speciation (52). Here, we generated several genomic resources for Hawaiian Metrosideros and used these in population genomics analyses to gain deeper insights into the genomic architecture and evolutionary processes underlying this island adaptive radiation.
Results
An Improved Genome Assembly for Hawaiian Metrosideros.
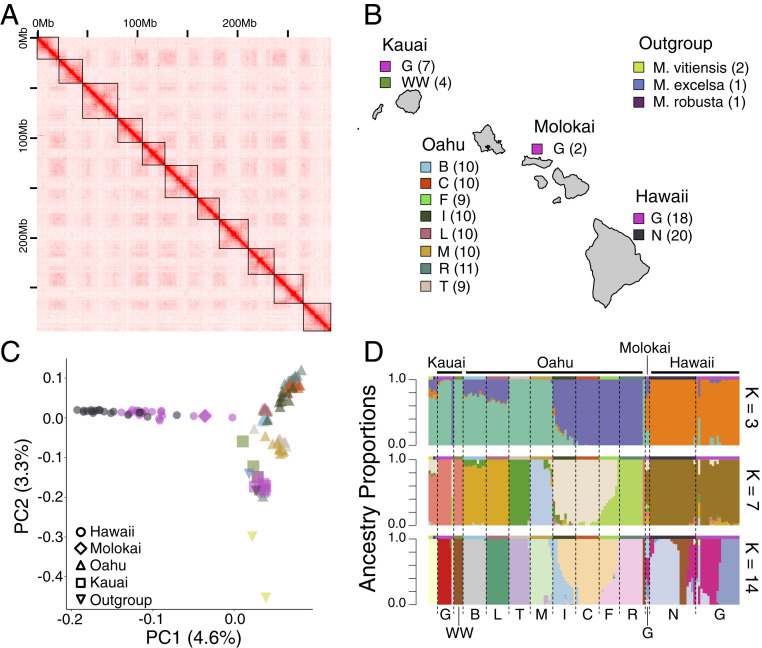
Using nanopore sequencing, an individual of Metrosideros polymorpha var. incana was sequenced to 66× coverage (refer to SI Appendix, Table S1 for genome-sequencing statistics). The reads were assembled into a draft assembly that had high contiguity with a contig N50 of 1.85 M basepair (bp) (Table 1). We implemented Pore-C sequencing (53), which combines chromosome conformation capture with long-read nanopore sequencing, to assay the Metrosideros-specific chromosome contact map and anchor contigs to their chromosomal positions (refer to SI Appendix, Table S2 for Pore-C sequencing statistics) (54). Using Pore-C contact maps, initial assembly contigs were scaffolded into 11 superscaffolds (Fig. 1A) spanning 292.8 Mbps with an N50 of 25.9 Mbp. Compared to a previous genome assembly that was based only on Illumina sequencing (55), our assembly was similar in total genome size but had significantly higher contiguity. The number of superscaffolds was consistent with the 11 chromosomes in Metrosideros (56). The assembly was evaluated with 2,326 Benchmarking Universal Single-Copy Ortholog (BUSCO) genes from eudicots, and 2,183 genes (93.8%) were present. These results suggest that our chromosome-level genome assembly is highly contiguous and complete. Gene annotation was conducted using nanopore sequencing of a complementary DNA (cDNA) library generated from leaf tissue (refer to SI Appendix, Table S3 for cDNA sequencing statistics). A total of 28,270 genes were predicted with 94.2% of the transcripts showing an annotation edit distance of less than 0.5.
Table 1.
Genome assembly statistics for M. polymorpha var. incana
| Assembly features | |
| No. of contigs | 1,035 |
| No. of scaffolded pseudomolecules | 11 |
| Total number of bases in contigs | 307,350,354 bp |
| Total number of bases scaffolded | 292,766,828 bp |
| Maximum contig length | 8,272,703 bp |
| Contig N50 length | 1.852 Mbp |
| Scaffold N50 length | 25.93 Mbp |
| BUSCO score | 93.8% |
| GC content | 39.8% |
| Repeat content | 36.3% |
| Number of annotated genes | 28,270 |
Fig. 1.
Genomics of Hawaiian Metrosideros. (A) Pore-C–based chromosome contact matrix for M. polymorpha var. incana. Black boxes indicate the 11 superscaffolds (pseudochromosomes). (B) Geographic distribution and taxon classification for the 135 samples that were analyzed in this study. Numbers in parentheses represent sample sizes. (C) PCA and individuals are color coded according to B. (D) Ancestry proportion estimates using the ADMIXTURE algorithm for K = 3, 7, and 14. Taxa/populations are separated by dotted lines and colors above admixture barplots represent taxa as in B while taxon names are labeled below.
We investigated the population genomics of Hawaiian Metrosideros by whole-genome resequencing 89 individuals from the islands of Oahu and Kauai and combining these data with previously published sequence data from Hawaii Island and Molokai (57). Our sampling from the Maui Nui complex (e.g., Molokai) included just a few samples, as past studies have shown a mixed ancestry for Metrosideros on Maui Nui involving colonization from both older and younger islands (35, 49), and our evolutionary analyses were centered on island-endemic communities. We also sequenced three Metrosideros species outside of Hawaii for use as outgroup genomes (one Metrosideros excelsa from New Zealand, one Metrosideros robusta from New Zealand, and two Metrosideros vitiensis from American Samoa and Fiji). In total, we analyzed 131 individuals belonging to 11 Hawaiian taxa, abbreviated as: M. polymorpha race B (B), M. polymorpha race C (C), M. polymorpha race F (F), M. polymorpha race L (L), M. polymorpha var. glaberrima (G), M. polymorpha var. incana (I), M. macropus (M), M. polymorpha var. newellii (N), M. rugosa (R), M. tremuloides (T), and M. polymorpha var. waialealae (WW) (Fig. 1B).
A total 10 of the Hawaiian taxa are single-island endemics (i.e., B, C, F, L, M, N, R, T, and WW) or have multi-island distributions but are sampled from just one island (i.e., I) and are thus described here as “taxa” because each of these taxa is represented in this study by a single sampled population. The 11th taxon, archipelago-wide G, is represented by three populations from three islands, which are thus described as “populations.” The median genome coverage was 14× per individual, and on average, 93% of the sequencing reads were aligned to our reference genome (SI Appendix, Table S4). The mapped reads were used to call single nucleotide polymorphisms (SNPs), and after filtering, there were 22,511,205 variable sites that were used for subsequent analysis.
Population Structure and Relationships across the Hawaiian Islands.
Using the population genomics data, we investigated the genetic structure across Hawaiian Metrosideros through principal component analysis (PCA) and ancestry analysis. PCA separated the taxa/populations by island of origin, and within islands, individuals were largely clustered by taxon (Fig. 1C). Finer scale evolutionary relationships were examined by estimating genomic ancestry proportions (K) for each individual (refer to SI Appendix, Fig. S1 for K = 3 to K = 15 results). Consistent with the PCA results, at low K, ancestries strongly reflected island of origin (Fig. 1D). With increasing K, each taxon/population showed increasingly unique ancestry, and at K = 14, with few exceptions, individuals belonging to the same taxon/population shared a single, unique ancestry. The exceptions were F and I, for which a majority of the individuals showed admixed ancestries. On Hawaii Island, G belonged to two genetic groups designated GH1 and GH2 in our previous analysis, and GH2 represented a recently admixed population with N (57). At least some individuals of F and I on Oahu and GH2 on Hawaii Island were likely to be hybrids formed from recent hybridization of genetically distinct populations (58).
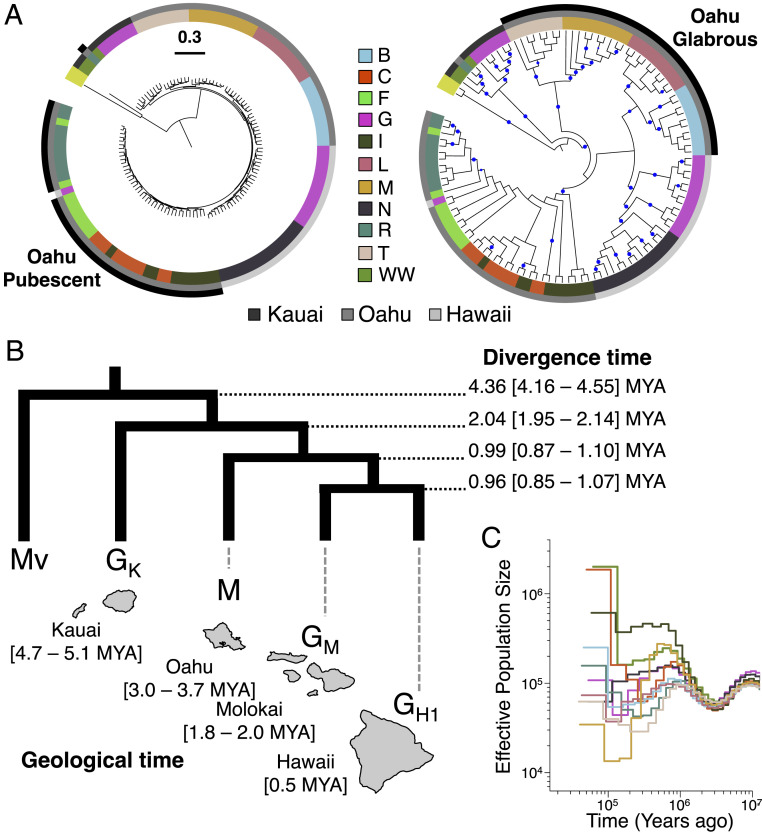
We inferred the evolutionary relationships among taxa by building a maximum-likelihood phylogenomic tree using genome-wide, fourfold-degenerate sites (Fig. 2A) and M. vitiensis as the outgroup. The internal branch lengths of all Hawaiian Metrosideros were short, consistent with a rapid radiation within the islands (Fig. 2A, Left). Differentiation among taxa was relatively low, with pairwise FST values ranging from ∼0.002 between C and I to ∼0.16 between M and GK (SI Appendix, Fig. S2). Collapsing the branch lengths to view the topological relationships revealed that individuals grouped by island with little evidence of recent migration between islands (Fig. 2A, Right). Within the phylogeny, individuals clustered according to taxon/population classification and were monophyletic with high confidence (>95% bootstrap support). Exceptions were the paraphyletic relationships among the pubescent Oahu taxa (C, F, I, and R) and between G and N on Hawaii Island. For the Oahu taxa, there was strong phylogenetic grouping for the glabrous pair B and L and for the glabrous pair M and T, but the topological relationships between these glabrous subgroups and within the pubescent group were unresolved. We used SNP and AFLP package for phylogenetic analysis (SNAPP) (59) as an independent Bayesian phylogenetic method to infer the multilocus phylogenetic trees and the uncertainty in the majority-rule topology. The SNAPP cloudogram showed a single major topology that was consistent with the maximum-likelihood tree topology (SI Appendix, Fig. S3) with the exception of the four glabrous taxa from Oahu, which showed contrasting relationships in the two trees.
Fig. 2.
Divergence time and demographic history of Hawaiian Metrosideros. Relative times were converted to absolute times assuming a mutation rate of 7 × 10−9 mutations per base pair per generation and a 20-y generation time. (A) Genome-wide maximum-likelihood tree built using fourfold degenerate sites. A tree with branch lengths is shown on left and a tree without branch lengths but showing phylogenetic relationships with bootstrap is on Right. Outer circle colors indicate island of origin for each sample, and inner circle colors indicate taxa as in Fig. 1B. All Hawaiian Metrosideros taxa have glabrous (hairless) leaves, except the four Oahu pubescent taxa indicated in the outer-most ring of Left tree. The four Oahu glabrous taxa are also highlighted in outer-most ring of Right tree. Nodes with greater than 95% bootstrap support are indicated with blue circles in Right. (B) G-PhoCS-estimated divergence times for representative taxa/populations GH1 (M. polymorpha var. glaberrima from Hawaii), GM (M. polymorpha var. glaberrima from Molokai), M (M. macropus from Oahu), and GK (M. polymorpha var. glaberrima from Kauai) (Above) and time of geological formation of each island based on Clague (102) (Below). Tree is rooted with outgroup M. vitiensis (Mv). (C) MSMC2-estimated effective population size for each Hawaiian taxon, color-coded as in Fig. 1B.
Reticulate Evolution Caused by Ancestral Polymorphism and Hybridization.
Results from the analysis of evolutionary relationships suggested a reticulate evolution for Hawaiian Metrosideros, which we investigated further using Patterson’s D-statistics (ABBA-BABA D test) (60, 61). Specifically, we looked for evidence of hybridization between taxa by calculating Patterson’s D-statistics for all population trios following the radiation-wide topology (Fig. 2A). M. vitiensis was used as the outgroup, specifically the sample from Fiji due to its high genome coverage. Overall, 85 of the 159 (53.5%) trio topologies had significant D-statistics (Bonferroni corrected P value < 0.05; refer to SI Appendix, Fig. S4 for results of each trio), indicating hybridization. Of the 85 trios, 53 (62.4%) topologies involved admixture between taxa on different islands, indicating that reticulate evolution was pervasive within and between islands. The relatively strongly isolated M and T were exceptions, as they had the fewest significant D-statistics and no evidence of admixture with lineages outside of Oahu (SI Appendix, Fig. S4).
Because the topological support values within the glabrous and pubescent Oahu groups were low (Fig. 2A) and indicating uncertain relationships, we examined patterns of discordance in the phylogenetic signal across the genome using TWISST (62). For a rooted four-taxon tree, there are 15 possible topologies, and we used TWISST to estimate the weight (frequency) of each topology in 10-kbp windows across the genome. Results from TWISST showed no single, dominant topology highly represented across the genome, and the difference in weight between the most common and second most common topologies was just ∼1.5% (SI Appendix, Fig. S5) for both the glabrous and pubescent groups. Simulations indicated a large ancestral effective population size (i.e., high incomplete lineage sorting) (63) for both groups, and admixture between ancestral populations could explain the topological weights observed for the Oahu taxa (SI Appendix, Fig. S6).
We next examined the topological weights (frequencies) across the Hawaiian Islands, but using just GH1, M, and GK to represent Hawaii, Oahu, and Kauai, respectively, as these taxa/populations showed no evidence of admixture with each other (SI Appendix, Fig. S4). Results showed that the topology with the highest weight was consistent with the topology generated for all taxa (Fig. 2A), but the weight of this topology was only ∼10% higher than the weights of the two alternative topologies (SI Appendix, Fig. S7). Combined, these results indicated a highly reticulated evolutionary history for Hawaiian Metrosideros, part of which was largely driven by elevated levels of shared ancestral polymorphisms that have not sorted completely among taxa.
Divergence Times of Metrosideros across the Islands.
We investigated the speciation history of Hawaiian Metrosideros through demographic modeling. The population divergence times were estimated using the generalized phylogenetic coalescent sampler (G-PhoCS) (64). Because we were interested in the colonization history of Metrosideros across the islands, we chose a single individual to represent each island. Specifically, we chose a single individual of G from each of Hawaii, Molokai and Kauai, and a single individual M from Oahu, because G samples from Oahu were not available. M showed no significant evidence of between-island admixture (SI Appendix, Fig. S4), which reduced the number of admixture models to be tested with G-PhoCS.
Initially, we ran G-PhoCS models fitting migration bands (i.e., admixture) between populations. Results showed that including admixture had no effect on estimates of divergence times (SI Appendix, Fig. S8). Given this result, we based our divergence-time analysis on the simple no-migration model. The G-PhoCS-estimated divergence time between Hawaiian Metrosideros and the outgroup, M. vitiensis, was 4.36 million years ago (MYA) (95% Highest Posterior Density 4.16 to 4.55 MYA). We also examined the effect of changing the mutation rate and generation time on absolute divergence time estimates. Using the range of mutation rates observed in plants [i.e., 7 × 10−9 in Arabidopsis (65) to 9.5 × 10−9 in Prunus (66)] and a higher generation time of 25 y, a conservative divergence time estimate for Hawaiian Metrosideros and M. vitiensis was 4 to 5.5 MYA (SI Appendix, Fig. S9). These estimates encompass the estimated timing of the subaerial appearance of Hawaii’s oldest main island, Kauai (Fig. 2B). Given that M. vitiensis is not the most closely related outgroup species to Hawaiian Metrosideros (50), however, colonization of the Hawaiian Islands likely occurred more recently than 4.36 MYA. Within the Hawaiian Islands, divergence time estimates were consistent with colonization of each island following its formation. The exception was Hawaii Island for which the divergence time of Metrosideros predated the geological formation of that island, consistent with the results of our previous study (57).
We also estimated past changes in effective population size (Ne) for each taxon/population using the program Multiple Sequentially Markovian Coalescent 2 (MSMC2) (67, 68). From each taxon/population, we chose a single individual for analysis. Results showed that all taxa had identical trajectories that included a decrease in Ne until ∼3 MYA, followed by an increase and subsequent drop in Ne in a pattern unique to each taxon (Fig. 2C). This result suggested that all Hawaiian Metrosideros taxa share the same common ancestor that experienced a population bottleneck 3 to 4 MYA, which is likely when the ancestral population initially colonized the islands. Based on G-PhoCS and MSMC2 analyses, the initial colonization of the Hawaiian Islands was estimated at ∼3 to 4.4 MYA (150,000 to 220,000 generations ago).
The Genomic Landscape of Differentiation.
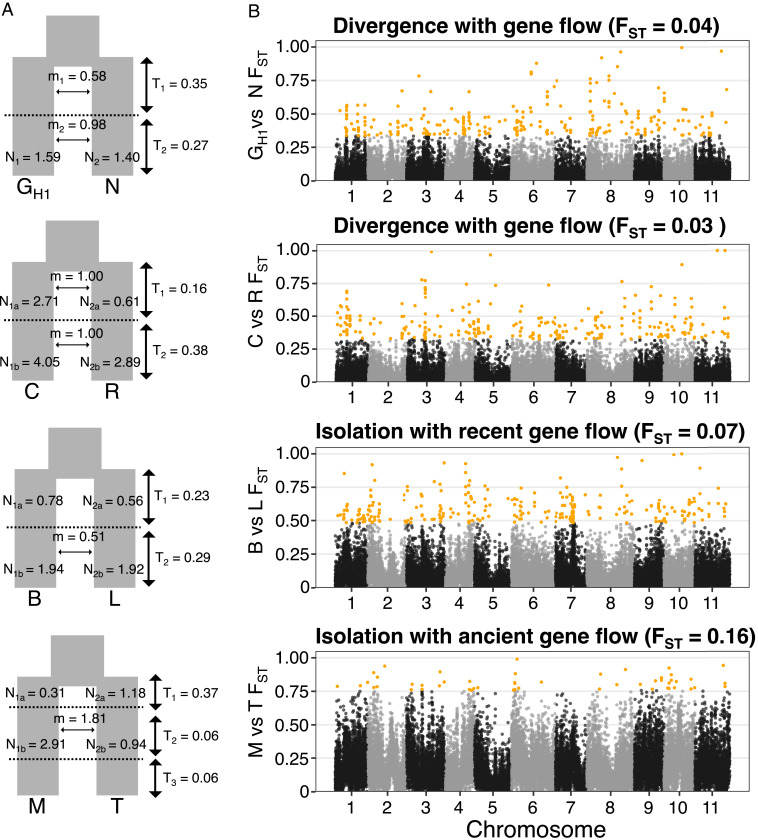
To investigate the genetic architecture underlying the Metrosideros radiation we narrowed our analysis to pairs of taxa/populations that were phylogenetic sisters (pair GH1 and N, pair C and R, pair M and T, and pair B and L), since for these sister pairs, the pattern of genome-wide differentiation reflects relatively recent divergence. We used MSMC2 to estimate the relative cross-coalescence rate (67) and population separation time for each sister pair (SI Appendix, Fig. S10). The relative cross-coalescence rates indicated the four sister pairs had comparable split times (GH1 versus N ∼635 KYA, C versus R ∼612 KYA, M versus T ∼693 KYA, and B versus L ∼888 KYA). For each sister pair, we used δaδi (69) to fit 20 different demographic models (refer to SI Appendix, Table S5 for complete δaδi results and SI Appendix, Fig. S11 for visualization of all models) to find the best-fitting model to explain its divergence history. Results showed that pair GH1 and N and pair C and R were consistent with a speciation model in which divergence occurred with continuous gene flow (i.e., primary gene flow) (70), while in pair B and L and pair M and T, the populations have been largely isolated from each other with the exception of either a recent or ancient gene flow event (Fig. 3A).
Fig. 3.
Genomic landscape of differentiation for the four phylogenetic sister pairs. (A) Best-fitting demography model based on δaδi modeling. (B) Genome-wide FST in 10-kbp windows. Yellow dots are outliers identified with z-score–transformed FST values (zFST) > 4.
We investigated the genomic architecture of the Metrosideros adaptive radiation by quantifying genome-wide patterns of differentiation and signatures of divergent selection between sister taxa. We focused on differentiation (FST) outlier regions since these regions would harbor genetic variation associated with the divergence of sister pairs (71). Results showed that, in all four sister pairs, areas of high genomic differentiation were scattered across all 11 chromosomes (Fig. 3B). Pair M and T had the fewest outlier windows (52 zFST outlier windows), while pairs GH1 and N, C and R, and B and L had over 250 zFST outlier windows each (257, 269, and 260, respectively). The median genome-wide FST between M and T (FST = 0.16) was more than twice the level of FST within the other sister pairs (GH1 and N median FST = 0.04; C and R median FST = 0.04; B and L median FST = 0.07), suggesting that increased genome-wide differentiation between M and T due to their genetic isolation may have eroded outlier windows to undetectable levels (26, 72). The genomic positions of outlier windows generally did not overlap across the four sister pairs (i.e., >84% of outlier windows were found in only one pair; SI Appendix, Fig. S12).
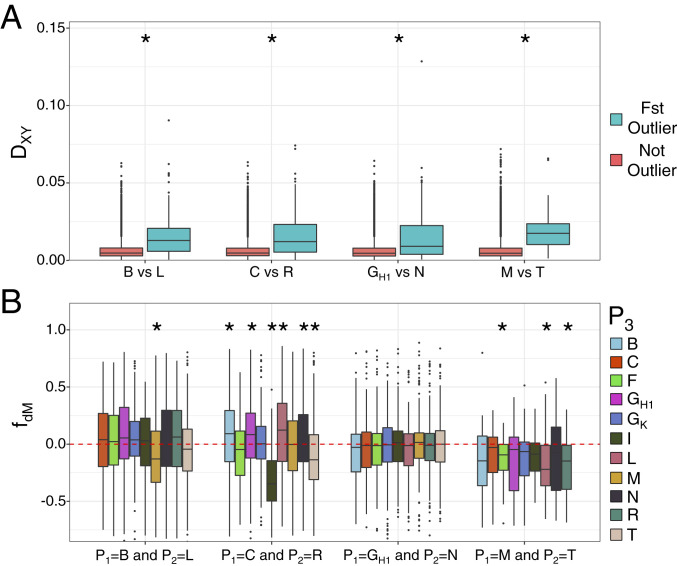
Because genomic outliers of differentiation do not always result from divergent selection (73–75) and FST may be a biased estimate of differentiation (76), we also examined levels of absolute genetic divergence (Dxy) within differentiation outlier regions. Dxy is expected to be elevated in genomic regions under divergent selection or regions acting as barriers between populations (73). Within each sister pair, Dxy levels were significantly elevated in differentiation outliers (Mann–Whitney U test P value < 0.001) relative to the genomic background (Fig. 4A). The differentiation outliers were also significantly elevated for values of relative node depth (21, 77) (SI Appendix, Fig. S13), which corrects for differences in mutation rate among loci by dividing Dxy values by the average Dxy to an outgroup (here, M. vitiensis). Differentiation outliers also had significantly lower levels of polymorphism and significantly higher values for selective sweep statistics, compared to the genomic background (SI Appendix, Fig. S14). These indicated that heterogeneity in mutation rate was not responsible for elevated Dxy in the differentiation outliers. In addition, the density of repetitive elements did not differ between the differentiation outliers and the genomic background (SI Appendix, Fig. S15), suggesting that elevated Dxy was not an artifact of misaligned and erroneous genotype calls. Instead, the differentiation outlier regions formed between Metrosideros sister taxa appear to have arisen through divergent selection.
Fig. 4.
Population genetics of the differentiation (FST) outlier regions identified in sister pairs. (A) Sequence divergence (Dxy) statistics calculated in 10-kbp windows. Red boxes are statistics from the genomic background, and green boxes are statistics from the differentiation outlier regions. (B) Localized admixture statistics (fdM) calculated in 10-kbp windows for the differentiation outlier regions identified in sister pairs (A). fdM is a rooted four-population statistic, in which P1 and P2 represent sister taxa and a positive fdM statistic indicates admixture between a third lineage, P3, and P2; while a negative fdM statistic indicates admixture between P3 and P1. For fdM, the genome-wide background is not shown to highlight the differentiation outlier region fdM values. Shown are values for median, first, and third quartiles, with whiskers representing ±1.5* interquartile range. * indicates P < 0.001 after Mann–Whitney U test comparing differentiation outlier region versus the genomic background.
A gene ontology (GO) enrichment analysis was done to identify any overrepresented functional categories associated with the differentiation outlier regions. Results showed that genes within the outlier windows were enriched for 19 GO terms in the “biological process” category with functions largely related to metabolic processes, cell cycle, and disease–immunity responses (SI Appendix, Fig. S16).
Evolutionary Origins of the Genomic Outliers of Differentiation.
We focused next on the evolutionary origin and history of the differentiation outlier regions as they relate directly to the genetic basis of the Hawaiian Metrosideros radiation. Due to the highly reticulated evolutionary history of Hawaiian Metrosideros, we initially examined differentiation outliers in the sister pairs for evidence of admixture from a nonsister taxon. Specifically, we asked whether the differentiation outliers that formed between the two sister taxa have evolutionary origins from a nonsister taxon. This would emphasize the importance of recent introgression from more distantly related taxa in forming the differentiation outliers observed between sister pairs (70). We calculated the fdM statistic (32), which quantifies admixture within genomic windows between both members of a sister pair and a third lineage. Results showed the Hawaii Island pair GH1 and N was the only sister pair in which the fdM statistics did not differ between the differentiation outliers and the genomic background (Fig. 4B). For each of the three other sister pairs (all on Oahu), the differentiation outliers had significant evidence of admixture with another Oahu taxon, while the pair C and R showed significant evidence of admixture with Hawaii Island taxa as well. These results suggested that hybridization from a more distantly related taxon may have contributed to the genomic regions that are significantly diverged between sister taxa in Metrosideros.
We further examined the possible role of recent introgression from nonsister taxa in the formation of differentiation outliers between sister taxa using phylogenetic analysis. Initially, the evolutionary histories of the differentiation outliers were visualized by concatenating the outlier regions and building a maximum-likelihood phylogenetic tree for each sister pair (SI Appendix, Fig. S17). In each case, the phylogenetic tree of the differentiation outliers was similar in its structure to the genome-wide phylogeny. Individuals were largely monophyletic by taxon/population in each phylogeny, and with the exception of the sister pair, the topological relationships were consistent with the genome-wide topology (Fig. 2A). For all four sister pairs, one taxon (i.e., GH1, L, R, and T) was topologically discordant (SI Appendix, Fig. S17 red star) compared to the genome-wide topology. The cause of this discordance (i.e., the evolutionary origin of the outlier regions) was uncertain due to the low phylogenetic support, and the low phylogenetic support was not due to introgression according to the fdM statistics (Fig. 4B). Moreover, with a single exception (i.e., C compared with I), these regions had significantly elevated Dxy levels in pairwise comparisons with all other Hawaiian taxa (SI Appendix, Fig. S18). Combined, these results suggested that the elevated Dxy observed in differentiation outliers between sister taxa was in fact not due to recent introgression from outside the sister pair.
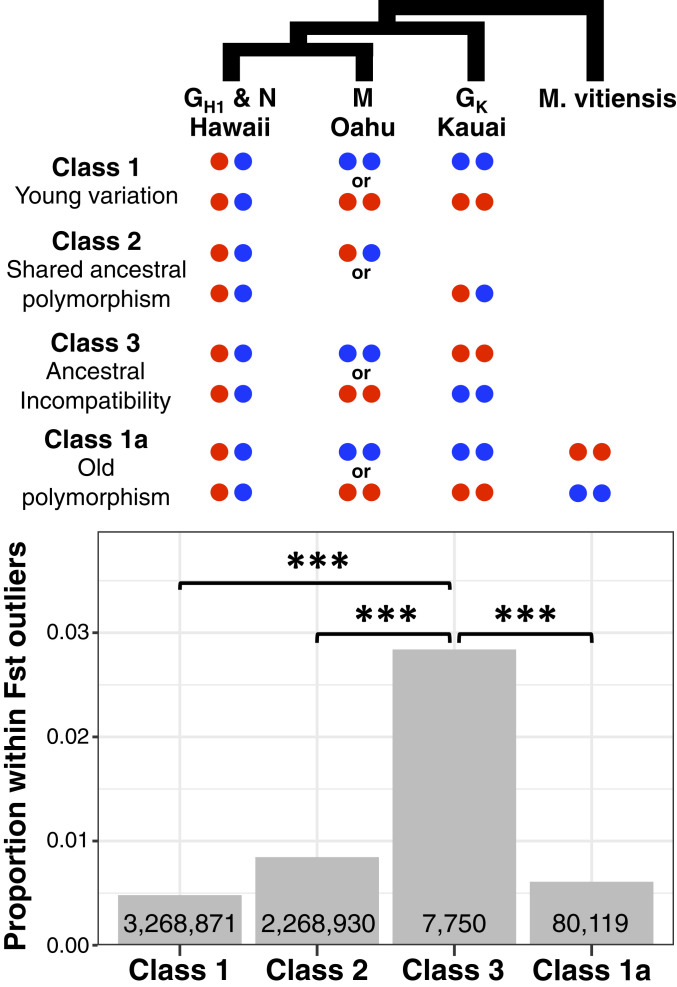
To further investigate the evolutionary origins of the differentiation outlier regions, we followed a recently developed approach that examines allele states in related populations to characterize the evolutionary history of polymorphisms in a focal group (78, 79). Specifically, for each sister pair individually, we pulled out sites that were polymorphic within the pair and determined the allele states for each within taxa or populations on other islands. This analysis divided the polymorphic sites in each sister pair into three classes that are interpreted as follows (refer to Fig. 5, Top for visual representation of the classes) 1): Sites with variants that are private to the sister pair. Such variants are likely to be young and possibly unique to the island hosting the sister pair (but see class-1a below) 2); Sites that are polymorphic within taxa/populations on two or all three of the islands examined. Such sites represent shared (unsorted) ancestral polymorphisms; and 3) Sites at which two taxa/populations from two other islands (i.e., islands not hosting the sister pair) are fixed for alternative alleles. Such sites are polymorphic in the sister pair as a result of between-island hybridization, or they represent alleles with negative epistatic interactions (i.e., Bateson-Dobzhansky-Muller incompatibilities) in the ancestral population. These three classes represent mutually exclusive categories of polymorphic sites in the sister pairs. Lastly, to allow detection of polymorphisms that may predate the initial colonization of the islands, we pulled out a subclass of class-1 sites (class-1a). Class-1a sites are sites at which the two taxa/populations from the different islands are fixed for the same allele and the outgroup M. vitiensis is fixed for the alternative allele. Given that the differentiation outliers in the sister pairs have elevated Dxy in all pairwise comparisons within the Hawaiian radiation (SI Appendix, Fig. S18), such sites are likely to represent variants that predate the Hawaiian radiation.
Fig. 5.
Evolutionary origins of the genome-wide SNPs and the differentiation (FST) outlier-region SNPs between the sister taxa, GH1 and N. The Top figure illustrates the allele state in taxa outside of Hawaii Island for a site that is polymorphic across the sister pair GH1 and N and four possible evolutionary scenarios (classes) that can result in the polymorphism. The Bottom figure shows the proportion of variants identified within the GH1-N differentiation outlier regions that are designated to each class. Numbers within the bars represent the total number of genome-wide SNPs that fall within each class. *** indicates P < 0.001 after Fisher’s exact test.
We first categorized the genome-wide polymorphisms segregating within the Hawaii Island pair GH1 and N according to allele states in M from Oahu and GK from Kauai (Fig. 5, Bottom). A majority of the GH1-N polymorphisms were categorized as class-1 (i.e., private to GH1-N) (3,268,871 SNPs). A large proportion of the polymorphic sites, however, was classified as class-2 (2,268,930 SNPs), consistent with our previous analysis that revealed extensive sharing of ancestral polymorphisms across the Hawaiian radiation. We then narrowed the analysis to just the subset of polymorphic sites that occurred within the differentiation outlier regions to test whether they were enriched for a specific class. Results showed significantly greater enrichment of class-3 sites relative to the other three classes (Fisher’s exact test P value < 0.0001). Because there was no significant evidence of between-island admixture among the four populations examined (i.e., GH1, N, M, and GK) and because the fdM statistics revealed no evidence of admixture within the differentiation outlier regions for GH1 and N, the enrichment of class-3 sites within differentiation outliers is likely due to the sorting of ancestral incompatibility alleles. Further, the GH1-N differentiation outliers were significantly more enriched for class-2 and class-1a sites compared to class-1 sites (Fisher’s exact test P value < 0.0001), providing further support for the importance of ancestral polymorphisms in the formation of differentiation outliers. We applied this analysis to the three Oahu sister pairs next and again found significant enrichment of class-3 sites in the differentiation outliers (Fisher’s exact test P value < 0.0001; SI Appendix, Fig. S19). Since recent introgression is insufficient to explain the increased Dxy in the differentiation outliers (SI Appendix, Figs. S17 and S18), these results indicate a strong role for the sorting of ancestral incompatibility alleles in the formation of differentiation outliers in all four sister pairs examined.
Discussion
How lineages are able to undergo rapid phenotypic and ecological diversification in isolated ecosystems is a perplexing question in evolutionary biology (15). The limited genetic variation in the founding populations that give rise to such radiations and the relatively slow rate of genetic mutation (80) are expected to restrict speciation rates in remote settings. Plant adaptive radiations on islands have been studied largely through phylogenetic approaches using DNA sequences or genomes of a single representative from each species within the radiation (81, 82). These phylogenetic analyses of island plant groups have addressed questions of monophyly and revealed significant insights into the patterns and timing of island colonization and trait evolution. We took an alternative approach to investigate the evolution of Hawaii’s landscape-dominant woody genus, Metrosideros, by constructing a chromosome-level genome assembly and sampling genome-wide variation of 131 individuals across the islands. Using a population genomics approach, we gained deeper insights into the evolutionary history of the Hawaiian Metrosideros radiation, including the demographic processes and genomic architecture underlying this island adaptive radiation. Our findings suggest that diversification of Hawaiian Metrosideros was facilitated by reassortment of an unexpectedly rich pool of ancestral polymorphisms.
Several adaptive radiations have been shown to be facilitated by the reuse of genetic variants that are older than the radiations themselves (23, 25), a recurring observation that demonstrates the evolutionary importance of standing genetic variation for adaptive radiation (83). One way that an enriched pool of standing variation can become established in an isolated setting is through colonization by an ancestor of hybrid origin (25). The “hybrid-swarm origin of adaptive radiation” model proposes that such hybrid populations are predisposed to adaptive radiation (24, 84) by 1) providing novel characters through transgressive segregation (85) and 2) breaking genetic correlations that constrain trait evolution (86). Ancient hybrid origins have been suggested for a number of adaptive radiations, in particular in fish systems such as the Alpine whitefish (87) and the East African cichlids (78, 79, 88). In plants, adaptive radiations of the allopolyploid silverswords (82) and endemic mints (89) of Hawaii are also thought to have originated from ancient hybridizations. In cases of adaptive radiation through polyploidization, however, it is not certain how much of the radiation can be attributed to the duplication of functional genetic elements (90) or to the ploidy increase itself (91).
Hawaiian Metrosideros may also have a hybrid origin. Incomplete sorting of the considerable ancestral polymorphism created through hybridization could explain the highly reticulate evolution seen within the group. A large pool of ancestral variants would have served as readily available genetic variation for adaptation, without the waiting times required for de novo adaptive mutations. Indeed, we discovered that genomic divergence (and potentially the genetic basis of reproductive isolation) between early diverging Metrosideros taxa was shaped by divergent selection targeting ancestral variations over evolutionary young de novo variations. Interestingly, the highest proportion of ancestral variations occurring within the differentiation outlier regions comprised those that were fixed for alternative alleles in other taxa from different islands and thus may have had negative epistatic interactions in the ancestral population. Differential sorting of these ancestral incompatibility alleles has been proposed as a mechanism of reproductive isolation between hybrid lineages (92), and this may have facilitated genetic divergence and the evolution of the intrinsic, postzygotic isolating barriers that have been observed within Hawaiian Metrosideros (52).
Alternatively, the patterns we observe could be explained by initial colonization of the Hawaiian Islands by a sizable population of mixed or panmictic ancestry. This scenario appears less probable, however, given that Metrosideros likely colonized the Hawaiian Islands from the Marquesas Islands south of the equator (50, 93, 94), which required traversing more than 3,000 km of open ocean against the prevailing low-altitude trade winds (94). Metrosideros in the Marquesas Islands comprises just a single extant species (Metrosideros collina) with no recognized subspecific taxa. Moreover, genetic variation within the Marquesan Metrosideros population is expected to be limited as a result of the serial founder events associated with the colonization of the more remote Pacific Islands from the south Pacific (93, 94). If in fact Hawaiian Metrosideros descends from a true founder event from the Marquesas Islands, this would suggest that the striking adaptive radiation of Metrosideros observed in Hawaii but not in other remote Pacific Island chains results simply from the greater ecological opportunity in Hawaii. The main Hawaiian Islands have the largest current and historical geographic areas, elevation, and environmental heterogeneity of any island chain in the remote Pacific (95, 96). Further genome-wide studies of this genus throughout the Pacific region will be required to uncover the sources of ancestral genetic variation in Hawaiian Metrosideros.
In both animals and plants, adaptive evolution fuels the diversification of species, but the nature of the traits under selection can lead to fundamental differences in the genomic architecture of adaptive radiation. The genome-wide distribution of differentiation outliers in this study suggests that ecological diversification within Metrosideros involves either a large number of traits of simple genetic architecture or fewer traits with a polygenic basis (97, 98), with the latter being a more likely explanation for the rich diversity of vegetative traits in the group. These patterns contrast with the genetic architecture observed for key traits in animal adaptive radiations, in which differentiation is seemingly localized to a few genomic regions with prominent, broad peaks (31–33). In animals, traits under ecological selection can often cause physical changes that ultimately become involved in mate choice and assortative mating by sexual selection (99, 100). In Metrosideros, the outlier peaks were narrow, and their distribution was heterogeneous across the genome, a pattern that was also found in the continent-wide adaptive radiation of sunflowers (26). The narrow peaks suggest that fine-scale mapping of the genes underlying divergent phenotypes may be possible.
Materials and Methods
Detailed description of materials and methods can be found in SI Appendix. Briefly, we generated a reference de novo genome assembly for Metrosideros using the Oxford Nanopore Technologies GridION sequencing platform. An M. polymorpha var. incana (NG4) was genome-sequenced, assembled using flye (101), and scaffolded using Pore-C sequencing (53). For population genomic analysis, we genome-sequenced 92 samples and combined with our previous population genomic sampling (57). Sequencing reads were aligned to the reference genome that was generated from this study, and the genome-wide variations were analyzed for determining the population relationship, demographic history, and the population genomics of the Hawaii Metrosideros adaptive radiation.
Nanopore sequencing data are available from National Center for Biotechnology Information (NCBI) bioproject ID PRJNA670777. The population genomic sequencing data are available from NCBI bioproject ID PRJNA534153, specifically with the Sequence Read Archive Run (SRR) identifiers SRR12673403 to SRR12673495. Data generated from this study, including the reference genome assembly, gene annotation, variant call file, and population genetics statistics can be found at Zenodo data repository (https://doi.org/10.5281/zenodo.4264399).
Supplementary Material
Acknowledgments
We thank the Hawaii Division of Forestry and Wildlife for permission to collect leaf samples from state forests. We also thank Jennifer Johansen, Yohan Pillon, Melissa Johnson, and Chrissen Gemmill for assistance with field collections, Tomoko Sakishima for assistance with greenhouse sample collection and DNA extractions, the College of Agriculture, Forestry, and Natural Resource Management at the University of Hawaii at Hilo for greenhouse space, and Angalee Kirby for greenhouse management. We are also grateful to the Genomics Core Facility at Princeton University for sequencing support and the New York University IT High Performance Computing for supplying the computational resources, services, and staff expertise. We thank Jean-Yves Meyer, Yohan Pillon, and the M.D.P. laboratory members, especially Jonathan Flowers, for valuable discussion of the manuscript. This research was funded by NSF Plant Genome Research Program (IOS-1546218), the Zegar Family Foundation (A16-0051), and the New York University Abu Dhabi Research Institute (G1205) to M.D.P., and the University of Nevada, Las Vegas College of Sciences, NSF Faculty Early Career Development Program (DEB0954274) (Principal Investigator) and Centers of Research Excellence in Science and Technology Program (HRD-0833211) (co-PI) to E.A.S.
Footnotes
Competing interest statement: X.D., P.R., S.H., E.H., and S.J. are employees of Oxford Nanopore Technologies and are shareholders and/or share option holders.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023801118/-/DCSupplemental.
Data Availability
Genomic data have been deposited in NCBI zenodo (NCBI bioproject ID PRJNA670777, PRJNA534153; https://doi.org/10.5281/zenodo.4264399).
References
- 1.Gillespie R. G., et al., Comparing adaptive radiations across space, time, and taxa. J. Hered. 111, 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schluter D., The Ecology of Adaptive Radiation (Oxford University Press, 2000). [Google Scholar]
- 3.Futuyma D. J., Evolutionary Biology (Sinauer Associates, 1998). [Google Scholar]
- 4.Losos J. B., Adaptive radiation, ecological opportunity, and evolutionary determinism. American Society of Naturalists E. O. Wilson award address. Am. Nat. 175, 623–639 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Givnish T. J., Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: Why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207, 297–303 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Simpson G. G., Tempo and Mode in Evolution (Columbia University Press, 1944). [Google Scholar]
- 7.Glor R. E., Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 41, 251–270 (2010). [Google Scholar]
- 8.Givnish T., “Adaptive radiations and molecular systematics: Issues and approaches” in Molecular Evolution and Adaptive Radiation, Givnish T., Sytsma K., Eds. (Cambridge University Press, 1997), pp. 1–54. [Google Scholar]
- 9.Losos J., Lizards in an Evolutionary Tree: Ecology and Adaptive Radiation of Anoles (University of California Press, 2009). [Google Scholar]
- 10.Simpson G. G., The Major Features of Evolution (Columbia University Press, 1953). [Google Scholar]
- 11.Rundle H. D., Nosil P., Ecological speciation. Ecol. Lett. 8, 336–352 (2005). [Google Scholar]
- 12.Schluter D., Evidence for ecological speciation and its alternative. Science 323, 737–741 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Wilson E. O., The Diversity of Life (Harvard University Press; ) 1992). [Google Scholar]
- 14.Frankham R., Do island populations have less genetic variation than mainland populations? Heredity 78, 311–327 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Martin C. H., Richards E. J., The paradox behind the pattern of rapid adaptive radiation: How can the speciation process sustain itself through an early burst? Annu. Rev. Ecol. Evol. Syst. 50, 569–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant P. R., Grant B. R., How and Why Species Multiply: The Radiations of Darwin’s Finches (Princeton University Press, Princeton, NJ, 2008). [Google Scholar]
- 17.Carson H. L., Kaneshiro K. Y., Drosophila of Hawaii: Systematics and ecological genetics. Annu. Rev. Ecol. Syst. 7, 311–345 (1976). [Google Scholar]
- 18.Robichaux R. H., Carr G. D., Liebman M., Pearcy R. W., Adaptive radiation of the Hawaiian silversword alliance, (Compositae- Madiinae): Ecological, morphological, and physiological diversity. Ann. Mo. Bot. Gard. 77, 64–72 (1990). [Google Scholar]
- 19.Wolf J. B. W., Ellegren H., Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 18, 87–100 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Feder J. L., Egan S. P., Nosil P., The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Burri R., Interpreting differentiation landscapes in the light of long-term linked selection. Evol. Lett. 1, 118–131 (2017). [Google Scholar]
- 22.Ravinet M., et al., Interpreting the genomic landscape of speciation: A road map for finding barriers to gene flow. J. Evol. Biol. 30, 1450–1477 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Berner D., Salzburger W., The genomics of organismal diversification illuminated by adaptive radiations. Trends Genet. 31, 491–499 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Seehausen O., Conditions when hybridization might predispose populations for adaptive radiation. J. Evol. Biol. 26, 279–281 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Marques D. A., Meier J. I., Seehausen O., A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Renaut S., et al., Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat. Commun. 4, 1827 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Stankowski S., et al., Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers. PLoS Biol. 17, e3000391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pease J. B., Haak D. C., Hahn M. W., Moyle L. C., Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson M. E., Arroyo-Santos A., Thinking in continua: Beyond the “adaptive radiation” metaphor. BioEssays 31, 1337–1346 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Campbell C. R., Poelstra J. W., Yoder A. D., What is speciation genomics? The roles of ecology, gene flow, and genomic architecture in the formation of species. Biol. J. Linn. Soc. Lond. 124, 561–583 (2018). [Google Scholar]
- 31.Poelstra J. W., et al., The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344, 1410–1414 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Malinsky M., et al., Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 350, 1493–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamichhaney S., et al., Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Hart P. J., Tree growth and age in an ancient Hawaiian wet forest: Vegetation dynamics at two spatial scales. J. Trop. Ecol. 26, 1–11 (2010). [Google Scholar]
- 35.Stacy E. A., Sakishima T., Phylogeography of the highly dispersible landscape-dominant woody species complex, Metrosideros, in Hawaii. J. Biogeogr. 46, 2215–2231 (2019). [Google Scholar]
- 36.Dawson J., Stemmermann L., “Metrosideros (Gaud)” in Manual of the Flowering Plants of Hawai’i, Wagner W., Herbst D., Sohmer S., Eds. (University of Hawai’i Press, 1990), pp. 964–970. [Google Scholar]
- 37.Sur G. L., Keating R., Snow N., Stacy E. A., Leaf micromorphology aids taxonomic delineation within the hypervariable genus Metrosideros (Myrtaceae) on O‘ahu. Pac. Sci. 72, 345–361 (2018). [Google Scholar]
- 38.Stacy E. A., Sakishima T., Tharp H., Snow N., Isolation of Metrosideros ('Ohi’a) taxa on O’ahu increases with elevation and extreme environments. J. Hered. 111, 103–118 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Tsujii Y., Onoda Y., Izuno A., Isagi Y., Kitayama K., A quantitative analysis of phenotypic variations of Metrosideros polymorpha within and across populations along environmental gradients on Mauna Loa, Hawaii. Oecologia 180, 1049–1059 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Cordell S., Goldstein G., Mueller-Dombois D., Webb D., Vitousek P. M., Physiological and morphological variation in Metrosideros polymorpha, a dominant Hawaiian tree species, along an altitudinal gradient: The role of phenotypic plasticity. Oecologia 113, 188–196 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Cordell S., Goldstein G., Melcher P. J., Meinzer F. C., Photosynthesis and freezing avoidance in Ohia (Metrosideros polymorpha) at treeline in Hawaii. Arct. Antarct. Alp. Res. 32, 381–387 (2000). [Google Scholar]
- 42.Corn C. A., Hiesey W. M., Altitudinal variation in Hawaiian metrosideros. Am. J. Bot. 60, 991–1002 (1973). [Google Scholar]
- 43.Kitayama K., Pattison R., Cordell S., Webb D., Mueller-Dombois D., Ecological and genetic implications of foliar polymorphism in Metrosideros polymorpha gaud. (Myrtaceae) in a Habitat Matrix on Mauna Loa, Hawaii. Ann. Bot. 80, 491–497 (1997). [Google Scholar]
- 44.Stemmermann L., Ecological studies of Hawaiian Metrosideros in a successional context. Pac. Sci. 37, 361–373 (1983). [Google Scholar]
- 45.Stacy E. A., Johansen J. B., Sakishima T., Price D. K., Genetic analysis of an ephemeral intraspecific hybrid zone in the hypervariable tree, Metrosideros polymorpha, on Hawai’i Island. Heredity 117, 173–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ekar J. M., Price D. K., Johnson M. A., Stacy E. A., Varieties of the highly dispersible and hypervariable tree, Metrosideros polymorpha, differ in response to mechanical stress and light across a sharp ecotone. Am. J. Bot. 106, 1106–1115 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Morrison K. R., Stacy E. A., Intraspecific divergence and evolution of a life-history trade-off along a successional gradient in Hawaii’s Metrosideros polymorpha. J. Evol. Biol. 27, 1192–1204 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Stacy E. A., Johansen J. B., Sakishima T., Price D. K., Pillon Y., Incipient radiation within the dominant Hawaiian tree Metrosideros polymorpha. Heredity 113, 334–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Percy D. M., et al., Progressive island colonization and ancient origin of Hawaiian Metrosideros (Myrtaceae). Proc. Biol. Sci. 275, 1479–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupuis J. R., et al., Targeted amplicon sequencing of 40 nuclear genes supports a single introduction and rapid radiation of Hawaiian Metrosideros (Myrtaceae). Plant. Syst. Evol. 305, 961–974 (2019). [Google Scholar]
- 51.Drake D. R., Seed dispersal of Metrosideros polymorpha (Myrtaceae): A pioneer tree of Hawaiian lava flows. Am. J. Bot. 79, 1224–1228 (1992). [Google Scholar]
- 52.Stacy E. A., Paritosh B., Johnson M. A., Price D. K., Incipient ecological speciation between successional varieties of a dominant tree involves intrinsic postzygotic isolating barriers. Ecol. Evol. 7, 2501–2512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulahannan N., et al., Nanopore sequencing of DNA concatemers reveals higher-order features of chromatin structure. bioRxiv [Preprint] (2019). 10.1101/833590. Accessed 7 November 2019. [DOI]
- 54.Kaplan N., Dekker J., High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat. Biotechnol. 31, 1143–1147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izuno A., et al., Genome sequencing of Metrosideros polymorpha (Myrtaceae), a dominant species in various habitats in the Hawaiian Islands with remarkable phenotypic variations. J. Plant Res. 129, 727–736 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Carr G. D., Chromosome numbers of Hawaiian flowering plants and the significance of cytology in selected taxa. Am. J. Bot. 65, 236–242 (1978). [Google Scholar]
- 57.Choi J. Y., Purugganan M., Stacy E. A., Divergent selection and primary gene flow shape incipient speciation of a riparian tree on Hawaii Island. Mol. Biol. Evol. 37, 695–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mallet J., Hybrid speciation. Nature 446, 279–283 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Bryant D., Bouckaert R., Felsenstein J., Rosenberg N. A., RoyChoudhury A., Inferring species trees directly from biallelic genetic markers: Bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 29, 1917–1932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green R. E., et al., A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin S. H., Van Belleghem S. M., Exploring evolutionary relationships across the genome using topology weighting. Genetics 206, 429–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pamilo P., Nei M., Relationships between gene trees and species trees. Mol. Biol. Evol. 5, 568–583 (1988). [DOI] [PubMed] [Google Scholar]
- 64.Gronau I., Hubisz M. J., Gulko B., Danko C. G., Siepel A., Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43, 1031–1034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ossowski S., et al., The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie Z., et al., Mutation rate analysis via parent-progeny sequencing of the perennial peach. I. A low rate in woody perennials and a higher mutagenicity in hybrids. Proc. Biol. Sci. 283, 20161016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schiffels S., Durbin R., Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46, 919–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schiffels S., Wang K., “MSMC and MSMC2: The multiple sequentially Markovian coalescent” in Statistical Population Genomics, Methods in Molecular Biology, Dutheil J. Y., Ed. (Springer US, 2020), pp. 147–166. [DOI] [PubMed] [Google Scholar]
- 69.Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richards E. J., Servedio M. R., Martin C. H., Searching for sympatric speciation in the genomic era. BioEssays 41, e1900047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Via S., Natural selection in action during speciation. Proc. Natl. Acad. Sci. U.S.A. 106 (suppl. 1), 9939–9946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han F., et al., Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin’s finches. Genome Res. 27, 1004–1015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cruickshank T. E., Hahn M. W., Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23, 3133–3157 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Nachman M. W., Payseur B. A., Recombination rate variation and speciation: Theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 409–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noor M. A., Bennett S. M., Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103, 439–444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charlesworth B., Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15, 538–543 (1998). [DOI] [PubMed] [Google Scholar]
- 77.Feder J. L., et al., Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc. Natl. Acad. Sci. U.S.A. 102 (suppl. 1), 6573–6580 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meier J. I., et al., Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Svardal H., et al., Ancestral hybridization facilitated species diversification in the Lake Malawi cichlid fish adaptive radiation. Mol. Biol. Evol. 37, 1100–1113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrett R. D. H., Schluter D., Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Nevado B., Wong E. L. Y., Osborne O. G., Filatov D. A., Adaptive evolution is common in rapid evolutionary radiations. Curr. Biol. 29, 3081–3086.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Barrier M., Baldwin B. G., Robichaux R. H., Purugganan M. D., Interspecific hybrid ancestry of a plant adaptive radiation: Allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol. Biol. Evol. 16, 1105–1113 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Hedrick P. W., Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Seehausen O., Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 85.Rieseberg L. H., Archer M. A., Wayne R. K., Transgressive segregation, adaptation and speciation. Heredity 83, 363–372 (1999). [DOI] [PubMed] [Google Scholar]
- 86.Gomulkiewicz R., Houle D., Demographic and genetic constraints on evolution. Am. Nat. 174, E218–E229 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Hudson A. G., Vonlanthen P., Seehausen O., Rapid parallel adaptive radiations from a single hybridogenic ancestral population. Proc. Biol. Sci. 278, 58–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Irisarri I., et al., Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 9, 3159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindqvist C., Albert V. A., Origin of the Hawaiian endemic mints within North American Stachys (Lamiaceae). Am. J. Bot. 89, 1709–1724 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Barrier M., Robichaux R. H., Purugganan M. D., Accelerated regulatory gene evolution in an adaptive radiation. Proc. Natl. Acad. Sci. U.S.A. 98, 10208–10213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Bodt S., Maere S., Van de Peer Y., Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597 (2005). [DOI] [PubMed] [Google Scholar]
- 92.Schumer M., Cui R., Rosenthal G. G., Andolfatto P., Reproductive isolation of hybrid populations driven by genetic incompatibilities. PLoS Genet. 11, e1005041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wright S. D., Yong C. G., Dawson J. W., Whittaker D. J., Gardner R. C., Riding the Ice Age El Nino? Pacific biogeography and evolution of Metrosideros subg. Metrosideros (Myrtaceae) inferred from nuclear ribosomal DNA. Proc. Natl. Acad. Sci. U.S.A. 97, 4118–4123 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright S. D., Yong C. G., Wichman S. R., Dawson J. W., Gardner R. C., Stepping stones to Hawaii: A trans-equatorial dispersal pathway for Metrosideros (Myrtaceae) inferred from nrDNA (ITS+ETS). J. Biogeogr. 28, 769–774 (2001). [Google Scholar]
- 95.Neall V. E., Trewick S. A., The age and origin of the Pacific islands: A geological overview. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 3293–3308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Filmer P. E., McNutt M. K., Webb H. F., Dixon D. J., Volcanism and archipelagic aprons in the Marquesas and Hawaiian Islands. Mar. Geophys. Res. 16, 385–406 (1994). [Google Scholar]
- 97.Hall M. C., Basten C. J., Willis J. H., Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics 172, 1829–1844 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pérez-Pérez J. M., Serrano-Cartagena J., Micol J. L., Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162, 893–915 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wagner C. E., Harmon L. J., Seehausen O., Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Butler M. A., Sawyer S. A., Losos J. B., Sexual dimorphism and adaptive radiation in Anolis lizards. Nature 447, 202–205 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A., Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Clague D., “The growth and subsidence of the Hawaiian-Emperor volcanic chain” in The Origin and Evolution of Pacific Island Biotas, New Guinea to Eastern Polynesia: Patterns and Processes, Keast A., Miller S. E., Eds. (SPB Academic Publishing, 1996), pp. 35–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data have been deposited in NCBI zenodo (NCBI bioproject ID PRJNA670777, PRJNA534153; https://doi.org/10.5281/zenodo.4264399).