Significance
Arthritogenic alphaviruses, such as Chikungunya virus (CHIKV) and Mayaro virus (MAYV), cause febrile illness, rash, and a debilitating chronic polyarthritis in humans. Currently, there are no approved vaccines or antiviral therapies for the prevention or treatment of alphavirus infection. Here, we identified and characterized 33 monoclonal antibodies (mAbs) from a CHIKV-convalescent donor that cross-react with other arthritogenic alphaviruses. We demonstrate that five broadly neutralizing mAbs can inhibit multiple arthritogenic alphaviruses and map their epitopes through binding and viral escape mutant analysis. Finally, we show that two mAbs, DC2.M16 and DC2.M357, protect against alphavirus disease in mice. These studies inform our understanding of how the human immune system combats alphavirus infection and may guide the development of antiviral treatments and vaccines.
Keywords: alphaviruses, monoclonal antibodies, Mayaro virus, broadly neutralizing antibodies, heterologous immunity
Abstract
Arthritogenic alphaviruses are globally distributed, mosquito-transmitted viruses that cause rheumatological disease in humans and include Chikungunya virus (CHIKV), Mayaro virus (MAYV), and others. Although serological evidence suggests that some antibody-mediated heterologous immunity may be afforded by alphavirus infection, the extent to which broadly neutralizing antibodies that protect against multiple arthritogenic alphaviruses are elicited during natural infection remains unknown. Here, we describe the isolation and characterization of MAYV-reactive alphavirus monoclonal antibodies (mAbs) from a CHIKV-convalescent donor. We characterized 33 human mAbs that cross-reacted with CHIKV and MAYV and engaged multiple epitopes on the E1 and E2 glycoproteins. We identified five mAbs that target distinct regions of the B domain of E2 and potently neutralize multiple alphaviruses with differential breadth of inhibition. These broadly neutralizing mAbs (bNAbs) contain few somatic mutations and inferred germline–revertants retained neutralizing capacity. Two bNAbs, DC2.M16 and DC2.M357, protected against both CHIKV- and MAYV-induced musculoskeletal disease in mice. These findings enhance our understanding of the cross-reactive and cross-protective antibody response to human alphavirus infections.
Alphaviruses are enveloped, positive sense single-stranded RNA viruses that can cause significant human diseases ranging from arthritis to encephalitis (1–3). Alphaviruses that are associated with musculoskeletal disease (arthritogenic alphaviruses) include Chikungunya virus (CHIKV), Mayaro virus (MAYV), Ross River virus (RRV), O’nyong-nyong virus (ONNV), and others; these viruses are globally distributed and transmitted by mosquitos. Symptomatic infection by arthritogenic alphaviruses is characterized by fever, rash, myalgia, as well as both acute and chronic peripheral polyarthralgia (4, 5). The arthropathy can be debilitating and persist for months to years after infection. More severe manifestations of alphavirus disease—including encephalopathy and mortality—have been reported (6, 7). These viruses cause endemic disease as well as large, sporadic global outbreaks (8–10). Currently, there are no approved vaccines or antiviral therapies for the prevention or treatment of alphavirus infection.
MAYV is an arthritogenic alphavirus that was first isolated in 1954 in Trinidad, and recent outbreaks have been reported in numerous areas of Central and South America (11, 12). The primary vectors for MAYV are Haemagogus spp. mosquitoes, which transmit the virus to primates in a sylvatic cycle. However, MAYV vector competence studies have demonstrated transmission potential in multiple Aedes and Anopheles mosquitoes (13–17). The wide range and distribution of MAYV-competent vectors underscores the risk of potential urban transmission (18) and global spread (19).
The alphavirus glycoprotein is composed of heterodimers of two transmembrane subunits, E2 and E1, which mediate viral attachment and membrane fusion, respectively (20–22). The prefusion E2/E1 heterodimer forms a trimeric spike that is arranged in an icosahedral lattice on the viral particle. E2 is initially expressed as a precursor polypeptide known as p62. During virus biogenesis, p62 is processed by cellular furin to generate E2 and the peripheral E3 polypeptide. E3 remains bound to the E2/E1 heterodimer during exocytic transport and prevents premature conformational changes and membrane fusion (23, 24). The release of E3 is the final step of virus maturation and primes the glycoprotein for membrane fusion.
Both E2 and E1 proteins are targets of the neutralizing antibody response. Antibody-mediated protection by neutralizing monoclonal antibodies (mAbs) has been shown against several alphaviruses (25–31). We and others have reported the isolation of potent and protective neutralizing CHIKV mAbs targeting regions of E2, such as the β-connector region and the A domain (25–27). These mAbs neutralize viral particles via multiple mechanisms, including the prevention of attachment and membrane fusion. The alphavirus receptor Mxra8 binds to regions spanning the A and B domains of E2 protein (32), and neutralizing mAbs targeting these regions can effectively disrupt virus interaction with the host receptor (33).
Many of the identified, neutralizing human mAbs against alphaviruses are virus-specific and do not inhibit heterologous alphaviruses. Notably, most of these mAbs target CHIKV, and there are few examples of MAYV-reactive human mAbs. Recent work has demonstrated the cross-reactivity and cross-neutralization of human polyclonal sera to heterologous alphaviruses (34–36), suggesting that broadly reactive and/or broadly neutralizing monoclonal antibodies (bNAbs) may be elicited by alphavirus infection in humans. While a number of murine bNAbs have been characterized (30, 37, 38), few human bNAbs that engage multiple alphaviruses have been described (33). For example, the murine mAb CHK-265 can protect against CHIKV, MAYV, and RRV challenge in mice (38). More recently, a human mAb, RRV-12, was shown to protect mice against RRV and MAYV infection (33). Both CHK-265 and RRV-12 broadly neutralize infection by engaging the B domain of E2, but whether such protective alphavirus bNAbs are elicited commonly during the course of human CHIKV infection is unknown.
Here, we describe the isolation and characterization of cross-reactive alphavirus mAbs from a CHIKV-convalescent donor. We employed a single B cell sorting strategy using a heterologous MAYV antigen to isolate 33 cross-reactive mAbs and found that they target multiple epitopes on the E1 and E2 proteins. We identified five human bNAbs that neutralize CHIKV, MAYV, and other alphaviruses with differing potencies. Epitope binning and viral escape studies suggest that human bNAbs target related but distinct regions of the B domain of E2. Remarkably, the sequence analysis of human bNAbs showed few somatic mutations, and inferred germline variants largely retained neutralizing function. Two bNAbs demonstrated protection against both CHIKV- and MAYV-induced musculoskeletal disease in mice. Together, these studies further define heterologous humoral immunity among related alphaviruses in humans as well as the determinants of antibody-mediated cross-protection.
Results
Generation and Characterization of Recombinant MAYV p62-E1 Hybrid Protein.
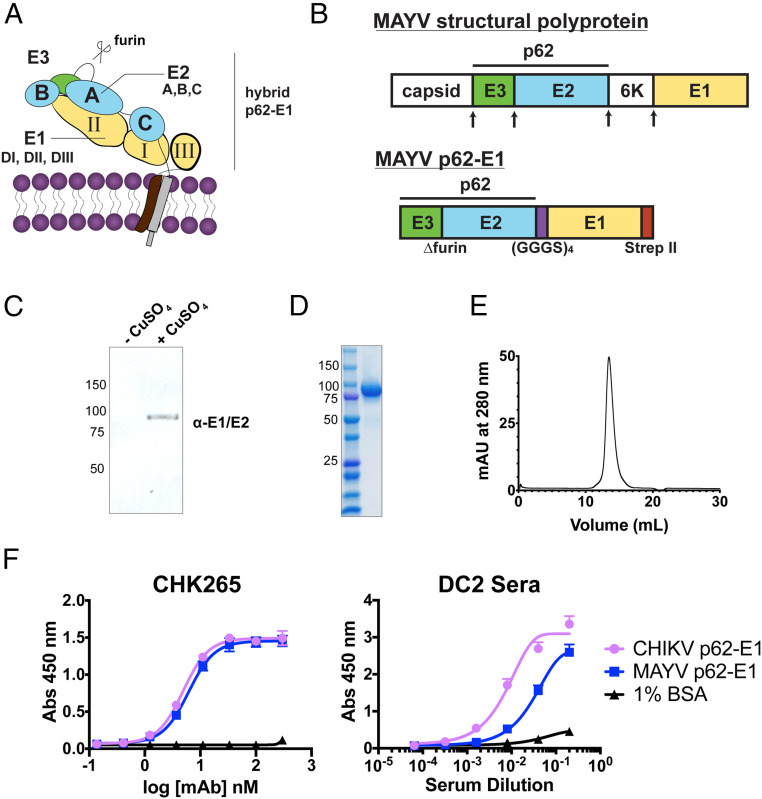
To assess the cross-reactivity of antibodies to the related alphavirus MAYV from patients who experienced heterologous CHIKV infections, we designed a recombinant protein construct containing the MAYV E2 and E1 domains tethered by a flexible glycine-serine linker for heterologous expression in S2 Drosophila cells (Fig. 1A), a strategy previously employed for the CHIKV glycoprotein (23). The MAYV ectodomain, or MAYV “p62-E1,” also contains E3 because of the introduction of a mutation at the furin cleavage site (Fig. 1B), which prevents maturation. Inducible expression of MAYV p62-E1 was confirmed by immunoblotting (Fig. 1C), and Strep-Tactin affinity purification yielded 1 to 2 mg/L of the glycoprotein at >90% purity, as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and size-exclusion chromatography (Fig. 1 D and E). We tested the reactivity of CHK-265, a previously described murine cross-reactive alphavirus mAb (38), against both MAYV and CHIKV p62-E1 glycoproteins by enzyme-linked immunosorbent assay (ELISA) (Fig. 1F). CHK-265 showed similar binding to both MAYV and CHIKV antigens, consistent with previous work (38) and confirming the MAYV p62-E1 adopts an antigenically relevant conformation. Serum from a CHIKV-convalescent donor “DC2” was tested by ELISA for reactivity against MAYV p62-E1 (Fig. 1F). Donor DC2 was exposed to CHIKV in the Dominican Republic and exhibited symptoms of fever, myalgia, and persistent arthralgia (25). Notably, MAYV has not been reported to circulate in the Dominican Republic. Serum from donor DC2 reacted with both CHIKV and MAYV glycoproteins, suggesting the presence of anti-MAYV antibodies. Together, the efficient production and antigenic analysis of recombinant MAYV p62-E1 provided a rationale for its use as a probe to isolate and characterize cross-reactive alphavirus mAbs.
Fig. 1.
Design, expression, and purification of MAYV p62-E1 heterodimer for single–B cell sorting. (A) Structural organization of the alphavirus glycoprotein. (B) Schematic of MAYV structural protein and p62-E1 construct. Arrows indicate the sites of proteolytic cleavage. MAYV p62-E1 contains a mutated furin cleavage site, a glycine-serine linker connecting p62 to E1, and a C-terminal strep II tag for purification. Immunoblot (C) and SDS-PAGE (D) of MAYV p62-E1 generated in S2 cells. MAYV p62-E1 was detected by an anti-E1/E2 antibody. (E) Size exclusion chromatogram of MAYV p62-E1. Sample was run on Superdex S200 10/300 AKTA column. A retention time of 13.5 mL was observed, consistent with molecular weight of ∼100 kDa. (F) ELISA reactivity of CHK-265 toward CHIKV/MAYV p62-E1 (Left) and serological ELISA (Right) of patient DC2 serum toward CHIKV/MAYV p62-E1. ELISAs were performed twice independently in triplicate wells (mean ± SD).
Isolation of Cross-Reactive MAYV mAbs by Single–B Cell Cloning.
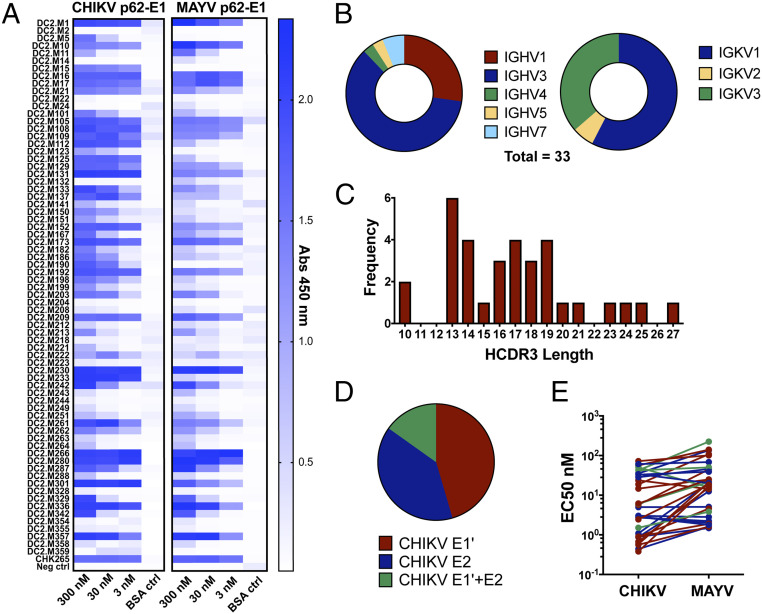
To isolate cross-reactive alphavirus mAbs from donor DC2, we employed a single B cell cloning strategy using MAYV p62-E1 as a heterologous sorting antigen (SI Appendix, Fig. S1). We sorted individual CD20+ CD27+ IgG+ MAYV p62-E1+ memory B cells and then cloned and recombinantly expressed a panel of 71 mAbs from the corresponding complementary DNA (cDNA). We focused exclusively on IGKV light chains since they generally have favorable stability and expression profiles (39). These mAbs were assessed for reactivity to CHIKV and MAYV p62-E1 by ELISA using three different concentrations (3, 30, and 300 nM) of mAb (Fig. 2A). The majority of the mAbs tested (58 of 71) bound to both the CHIKV and MAYV p62-E1 proteins. A total of 33 mAbs (termed DC2.Mx) showed strong cross-reactivity at 30 nM to both CHIKV and MAYV glycoproteins and were characterized further. Sequence analysis of the cross-reactive mAbs showed a diverse distribution of V-gene usage for both IGHV and IGKV (Fig. 2B), as well as a range of heavy chain complementarity-determining region 3 (HCDR3) lengths (Fig. 2C). To determine whether cross-reactive mAbs targeted the E1 or E2 subunit, ELISA reactivity to the ectodomains of the two independent CHIKV glycoprotein subunits (E1' or E2) was assessed (Fig. 2D). There were approximately equal numbers of cross-reactive mAbs targeting E1' and E2. The half maximal effective concentration (EC50) values for binding to CHIKV and MAYV p62-E1 ranged from 0.5 to 72 nM and 1.6 to 220 nM, respectively (Fig. 2E and SI Appendix, Table S1). Thus, a substantial subset of mAbs isolated from a CHIKV-convalescent donor cross-reacted with MAYV, and these mAbs targeted epitopes on both the E1 and E2 subunits.
Fig. 2.
Sequence and reactivity profiles of mAbs isolated from MAYV-reactive B cells. (A) ELISA three-point screen of 71 human mAbs for reactivity against CHIKV and MAYV glycoproteins. All mAbs were tested at 300, 30, and 3 nM concentrations against CHIKV p62-E1, MAYV p62-E1, and 1% BSA control. Experiment was performed twice independently in triplicate wells. (B) IGHV and IGKV gene usage of 33 cross-reactive mAbs. (C) Distribution of heavy-chain complementarity-determining region 3 (HCDR3) lengths of 33 cross-reactive mAbs. (D) Summary of CHIKV E1'/ E2 ELISA reactivity of cross-reactive mAbs. mAbs that showed signal (Abs 450 nm) greater than fivefold, relative to BSA control at 30 (E1') or 16.6 nM (E2), were considered reactive. Experiment was performed three times independently in triplicate wells. (E) Range of half maximal effective concentration values of cross-reactive mAbs for CHIKV and MAYV p62-E1. Values were determined by eight-point serial dilution series for each mAb. MAbs are colored according to the reactivity to E1' (red), E2 (blue), or both (green). The experiment was performed twice independently in duplicate wells.
Cross-Reactive Human mAbs Exhibit a Range of Neutralizing Potencies and Breadth.
Neutralizing activity of all 71 mAbs was tested by focus reduction neutralization test (FRNT) against CHIKV 181/25 and MAYV at two mAb concentrations (30 and 300 nM) (Fig. 3A). While many mAbs neutralized CHIKV 181/25 infection at both concentrations, only six (DC2.M16, DC2.M108, DC2.M131, DC2.M230, DC2.M336, and DC2.M357) inhibited CHIKV 181/25 and MAYV at both 300 and 30 nM concentrations. These neutralizing mAbs were all E2 reactive, and none of the E1-reactive mAbs exhibited neutralizing activity. Full neutralization curves revealed half-maximal inhibitory concentration (IC50) values ranging from 0.5 to 14 nM and 0.7 to 16 nM for CHIKV 181/25 and MAYV, respectively (Fig. 3 B and C). Next, the breadth of neutralization toward related arthritogenic alphaviruses ONNV, RRV, and Semliki Forest virus (SFV) was determined by FRNT (Fig. 3 B and C). Cross-neutralizing mAbs showed a range of inhibition against the different alphaviruses tested. DC2.M16, DC2.M131, and DC2.M336 exhibited little neutralizing activity against ONNV, RRV, or SFV, whereas DC2.M108, DC2.M230, and DC2.M357 all neutralized ONNV, the closest genetic relative to CHIKV. Furthermore, DC2.M357 also neutralized RRV and SFV, with IC50 values comparable to murine CHK-265. None of the mAbs tested neutralized the more distantly related Sindbis virus (SINV) (SI Appendix, Fig. S2). Additionally, IC50 values for DC2.M16 and DC2.M357 against the pathogenic East/Central/South African CHIKV-2006 LR-OPY1 strain were comparable to those derived against the attenuated CHIKV 181/25 Asian strain (Fig. 3C).
Fig. 3.
Identification of cross-neutralizing mAbs targeting multiple alphaviruses. (A) Two-point neutralization screen against CHIKV (181/25) and MAYV (Guyane). FRNTs were performed at 300 and 30 nM for all 71 human mAbs. Highlighted are mAbs that demonstrated >80% inhibition of both viruses at only 300 nM (black) or both 300 and 30 nM (colored as indicated). Values for each mAb represent the mean of the percent inhibition of two independent experiments performed in triplicate wells relative to untreated control. (B) Full neutralization curves of cross-neutralizing mAbs. FRNT was performed with serially diluted mAbs as indicated. Viruses tested were CHIKV 181/25, MAYV TRVL-4675, ONNV SG650, RRV T48, and SFV 88/11. Shown are experiments performed twice independently in triplicate (points represent mean ± SD) (C) IC50 values of human bNAbs. Values shown are in nanomolar and were determined by fitting nonlinear regression to FRNTs. A total of 95% confidence interval is shown for each curve.
Differential Binding Kinetics of Neutralizing and Nonneutralizing mAbs.
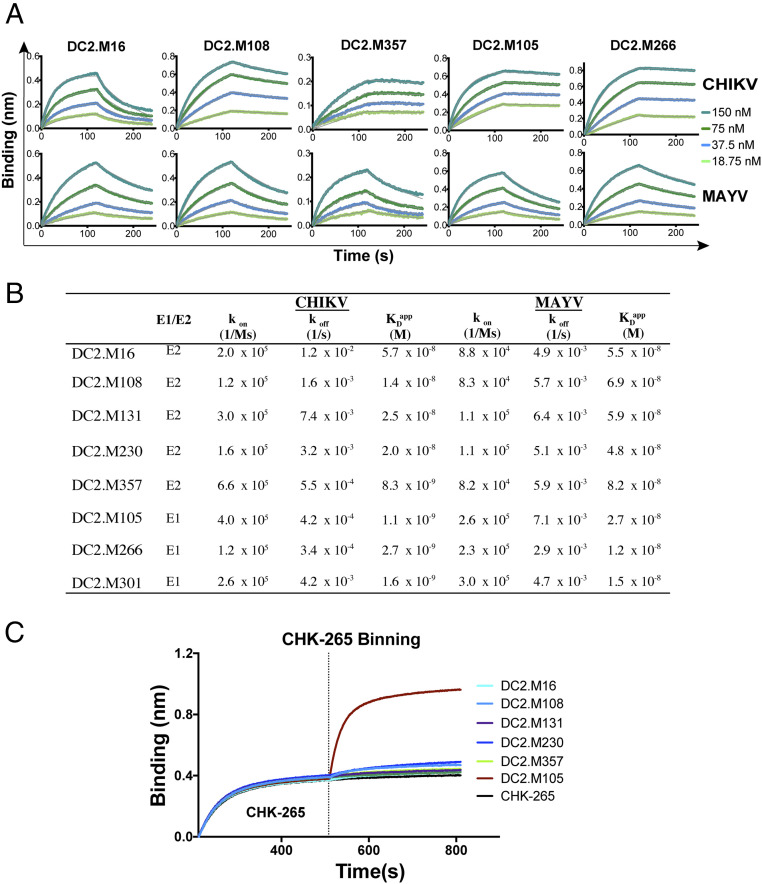
To evaluate the binding kinetics of the most potent human bNAbs for CHIKV and MAYV p62-E1 glycoproteins, we used biolayer interferometry (BLI) (Fig. 4A). The mAbs were captured on an anti-human Fc sensor and then dipped into solutions of CHIKV or MAYV p62-E1. While most sensorgrams could be fit with a 1:1 binding model, since the IgG is bivalent, we cannot rule out avidity effects and therefore have termed dissociation constants derived from on and off rates (kon and koff) as “apparent” (KDapp). The five E2-directed bNAbs DC2.M16, M108, M131, M230, and M357 had KDapp values ranging from 8 to 57 nM and 48 to 82 nM for CHIKV and MAYV p62-E1, respectively (Fig. 4B). Additionally, three E1-specific mAbs (DC2.M105, M266, and M301) were tested and all showed substantially higher affinities for both glycoproteins (1.0 to 2.7 nM for CHIKV p62-E1 and 12 to 27 nM for MAYV p62-E1) than the E2 mAbs. In all cases, the measured KDapp values are higher than the neutralizing IC50 values, possibly due to the avidity differences of mAb engagement on the monomeric recombinant glycoprotein versus the surface of the viral particle or the stoichiometry of binding required for neutralization (40). Notably, four of the eight mAbs tested (E2 mAb DC2.M357 and E1 mAbs DC2.M105, DC2.M266, and DC2.M301) had higher (∼10-fold) binding affinity for CHIKV p62-E1 than MAYV p62-E1. This enhanced binding to CHIKV p62-E1 was driven primarily by a lower off rate (koff). Thus, a range of kinetic binding profiles of E2-reactive bNAbs and nonneutralizing, cross-reactive E1-directed mAbs was observed against CHIKV and the heterologous MAYV antigen.
Fig. 4.
Binding kinetic profiles of cross-reactive mAbs and epitope binning. (A) Binding of E1 and E2 reactive mAbs to CHIKV and MAYV glycoproteins by BLI. A representative dataset from two independent experiments is shown. (B) Kinetic analysis of cross-reactive mAbs by BLI. A representative dataset from two independent experiments is shown. (C) Two-phase binding of cross-reactive mAbs to MAYV p62-E1 against CHK-265 by BLI. MAb CHK-265 was bound to a sensor loaded with MAYV p62-E1, followed by the sequential addition of the indicated second mAb. Representative data from two independent experiments are shown.
To determine the kinetics of monovalent binding, we generated antigen-binding fragments (Fabs) of bNAbs DC2.M16, DC2.M108, and DC2.M357 and tested the interaction with CHIKV and MAYV glycoproteins (SI Appendix, Fig. S3). The binding kinetics observed for the Fabs were largely consistent with the full-length mAbs, with KD values ranging from 16 to 50 nM for CHIKV p62-E1 and 56 to 64 nM for MAYV p62-E1.
Cross-Neutralizing mAbs Target the B Domain of E2.
The epitope of murine CHK-265 lies primarily within the B domain of E2. To determine whether human bNAbs also target the B domain, two-phase epitope binning was performed (Fig. 4C and SI Appendix, Fig. S4). CHK-265 was loaded onto sensors coated with MAYV p62-E1 and then added to an equimolar mixture of CHK-265 and the human mAb. All cross-neutralizing mAbs tested showed minimal binding to the MAYV glycoprotein preloaded with CHK-265, indicating that they also target the B domain. In contrast, DC2.M105 (an E1-targeting mAb) bound to the CHK-265/p62-E1 complex, since their two epitopes are spatially distant.
Viral Escape Mutants of Cross-Neutralizing mAbs Map to Distinct Regions of the E2 B Domain.
The cross-neutralizing human mAbs have similar binding affinities to recombinant MAYV and CHIKV glycoproteins and compete with B domain–specific CHK-265 but exhibited a range of potency and breadth against related alphaviruses. These observations suggest that subtle differences in their epitopes within the B domain may determine their functional activity. To test this hypothesis, we generated viral escape mutants against DC2.M108 and DC2.M357 using a replication-competent recombinant vesicular stomatitis virus bearing the CHIKV glycoproteins (rVSV-CHIKV), as previously described (25). DC2.M108 neutralizes CHIKV, ONNV, and MAYV, whereas DC2.M357 neutralizes these viruses as well as RRV and SFV.
Both DC2.M108 and DC2.M357 efficiently neutralized rVSV-CHIKV (Fig. 5A). Serial passage of rVSV-CHIKV in the presence of these mAbs resulted in a resistant population from which plaques were selected, genotyped, and tested for neutralization. For both DC2.M108 and DC2.M357, two distinct viral escape mutants each harboring a single-point mutation were isolated. All mutations mapped to the B domain of E2, consistent with binning experiments with CHK-265. For DC2.M108, escape mutants G209D or K215T resulted in complete escape from DC2.M108 as well as DC2.M16, which is clonally related (see below). However, these mutations did not confer escape for either DC2.M357 or CHK-265. For DC2.M357, the observed viral escape mutant K189N was only partially neutralized by DC2.M357 at high concentrations but was effectively neutralized by both DC2.M108 and CHK-265. Finally, the N218Y mutation conferred complete escape from both DC2.M357 and CHK-265 but not DC2.M108.
Fig. 5.
Generation of escape mutants of human bNAbs DC2.M108 and DC2.M357. (A) Neutralization curves of DC2.M16, DC2.M108, and DC2.M357 bNAbs against WT and mutant rVSV-CHIKV. Data shown are from two independent experiments performed in triplicate wells (points represent mean ± SD). IC50 values were determined by fitting a nonlinear regression, and 95% confidence interval is shown. Escape mutations for DC2.M108 and DC2.M357 mapped to the CHIKV p62-E1 heterodimer structure (B) (Protein Data Bank [PDB] ID: 3N40) and the E1/E2 heterohexamer (C) (PDB ID: 3J2W). C-α spheres indicate positions of escape mutations for DC2.M108 (cyan), DC2.M357 (magenta), as well as CHK-265 escape mutations against RRV (silver). E1 is colored gray.
Mapping of these mutations to the CHIKV p62-E1 crystal structure (Fig. 5B), and comparison with the recently described CHK-265 viral escape mutations against RRV, revealed distinct regions within the β-sheet structure of the B domain that are targeted by cross-neutralizing mAbs. All mutations we identified map to positions that are highly conserved among related alphaviruses (SI Appendix, Fig. S5). Notably, the DC2.M357 escape positions K189 and N218 are more proximal to those previously described for CHK-265 (Q183 and N219), and these mAbs share similar neutralization breadth. The DC2.M108 escape positions G209 and K215 are more distal to the CHK-265 escape mutations, which may suggest a different angle of approach for the binding of the DC2.M108 variable domains. Since some B domain mAbs can engage adjacent heterodimers on the alphavirus surface (38), we also mapped the mutations to the CHIKV-trimeric spike (Fig. 5C). The DC2.M357 (K189 and N218) escape mutations are oriented toward the crest of the crown formed by the three E2 subunits on the prefusion trimer, whereas those for DC2.M108 are more lateral. Together, these data suggest that the recognition of distinct residues within the B domain of E2 can determine the breadth of neutralization of related arthritogenic alphaviruses.
Germline Sequence and Mutagenesis Analysis of Cross-Neutralizing mAbs.
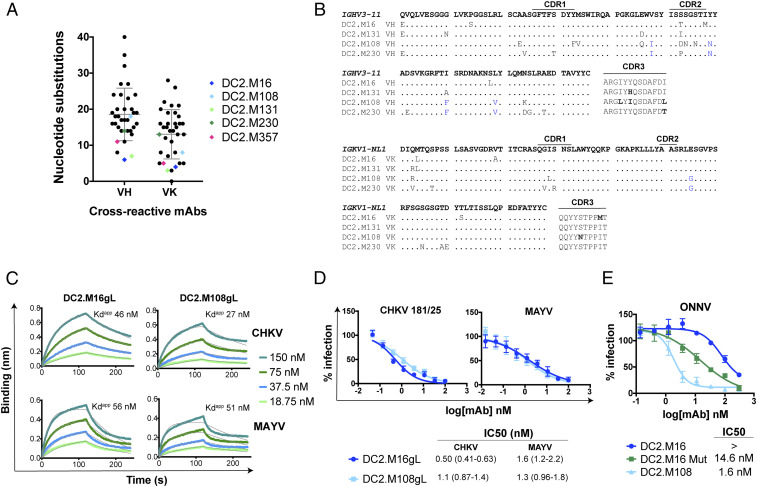
Sequence analysis of V-gene nucleotide substitutions showed that cross-reactive mAbs had a range of somatic mutations within the heavy chain variable (VH) and kappa light chain variable (VK) domains (Fig. 6A). The most potent cross-neutralizing mAbs had relatively few substitutions and had the least divergence from germline. In particular, DC2.M16, DC2.M131, and DC2.M357 all shared >96 and >98% nucleotide identity to their IGHV and IGKV germlines, respectively. MAbs DC2.M16, DC2.M131, DC2.M108, and DC2.M230 all shared the same V-gene families IGHV3-11 and IGKV1-NL1, the same CDR lengths, the same J-gene family IGHJ3 and IGKJ3, and had similar sequences (Fig. 6B). This analysis suggests that all four of these mAbs were derived from a common progenitor. DC2.M357 has an unrelated sequence derived from IGHV3-30 and IGKV3-11 (SI Appendix, Fig. S6). To determine the impact of V-gene somatic mutations on mAb function, we generated inferred germline–revertants (gL) DC2.M16gL, DC2.M108gL, and DC2.M357gL, which harbored the V-gene germline sequence while retaining the junctional diversity at the CDR3s of the heavy and light chains. DC2.M16gL and DC2.M108gL bound CHIKV and MAYV p62-E1 with similar affinities to the somatically mutated, wild-type (WT) mAbs (Fig. 6C). Furthermore, DC2.M16gL and DC2.M108gL neutralized both CHIKV 181/25 and MAYV infection with equivalent potency to the WT mAbs (Fig. 6D). In contrast, DC2.M357gL did not bind to recombinant alphavirus glycoproteins by ELISA (SI Appendix, Fig. S6) and neutralized CHIKV 181/25 and MAYV infection with ∼20-fold lower potency than the WT mAb. These results demonstrate that human mAbs from IGHV3-11 and IGKV1-NL1 lineages can possess broad alphavirus recognition and neutralizing capacity from junctional diversity alone and not V-gene somatic mutations. In the case of DC2.M357, nine total amino acid mutations in V-gene IGHV3-30 and IGKV3-11, primarily within the CDR1 and CDR2, are critical for enhancing antigen-binding and neutralizing activity (SI Appendix, Fig. S6). These data demonstrate that germline-like human mAbs harboring few somatic mutations can potently neutralize multiple alphaviruses.
Fig. 6.
Germline sequence analysis and functional characteristics of inferred germline bNAbs. (A) Number of V-gene nucleotide substitutions for cross-reactive mAbs. bNAbs are highlighted as indicated. Mean and SD are shown in gray bars. (B) Alignment of mAbs DC2.M16, DC2.M131, DC2.M108, and DC2.M230 to the IGHV3-11/IGKV1-NL1 germline sequence. Mutations shared by DC2.M108 and DC2.M230 are highlighted in blue. (C) Binding of inferred germline mAbs DC2.M16gL and DC2.M108gL to CHIKV and MAYV p62-E1 proteins by BLI. (D) Neutralization of CHIKV and MAYV by inferred germline mAbs. Shown here are two independent experiments performed in triplicate wells (points represent mean ± SD). IC50 values were determined by fitting a nonlinear regression, and 95% confidence interval is shown. (E) Neutralization of ONNV by DC2.M16 after introduction of key mutations. Mutations shared by DC2.M108 and DC2.M230 were substituted into the DC2.M16 sequence (DC2.M16 Mut). Shown here are two independent experiments performed in triplicate wells (points represent mean ± SD). IC50 values were determined by fitting a nonlinear regression, and 95% CI is shown.
We observed differences in ONNV neutralization by the IGHV3-11/IGKV1-NL1 mAbs, even though they share similar sequences. Notably, DC2.M108 and DC2.M230, but not DC2.M16 and DC2.131, neutralized ONNV and shared five mutations from the germline, primarily in framework regions 2 and 3 proximal to HCDR2 (Figs. 3 B and C and 6B). Substitution of the mutations shared by DC2.M108 and DC2.M230 (Fig. 6B) into the corresponding positions in the DC2.M16 sequence (DC2.M16 Mut) substantially improved neutralization of ONNV (Fig. 6E). Thus, the functional, paratope permitting ONNV neutralization lies at least in part in these regions, and the neutralizing breadth of IGHV3-11/IGKV1-NL1 mAbs can be modulated by limited V-gene mutation.
Protective Efficacy of Cross-Neutralizing mAbs In Vivo.
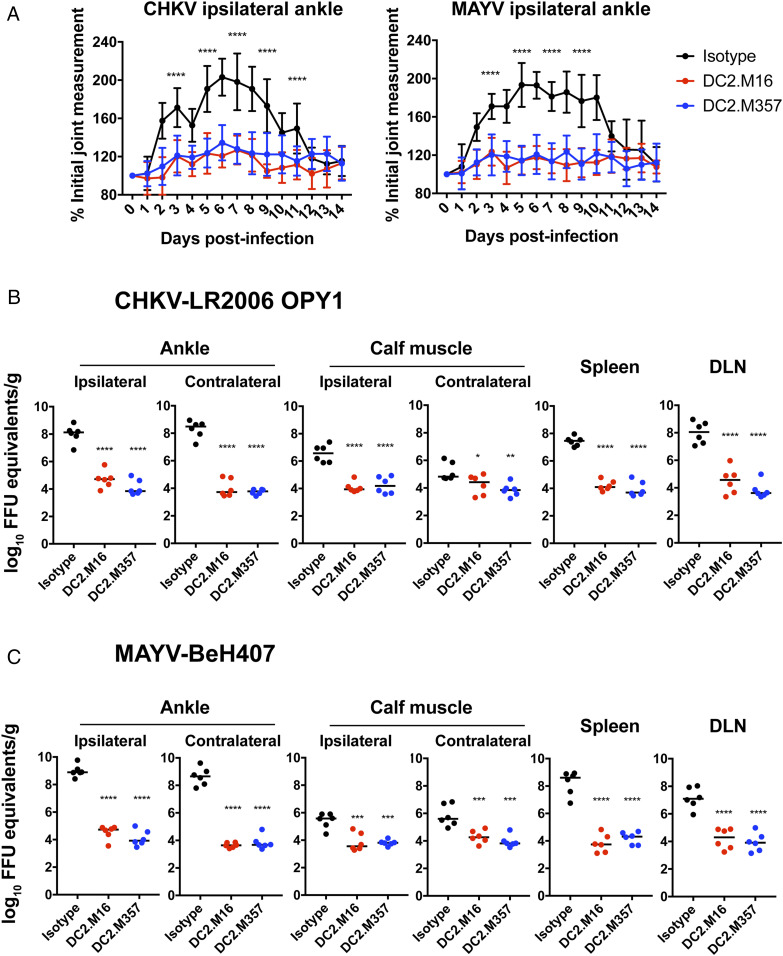
To determine whether human cross-neutralizing mAbs could confer protection against viral challenge in vivo, we tested the ability of DC2.M16 and DC2.M357 mAb treatment to mitigate joint swelling and infection induced by CHIKV and MAYV. Four-week old C57BL/6J mice were inoculated subcutaneously in the footpad with either CHIKV-LR2006 OPY1 or MAYV-BeH407 1 d following intraperitoneal mAb administration. Joint swelling was measured daily for 14 d after inoculation (Fig. 7A). Both DC2.M16 and DC2.M357 reduced CHIKV- and MAYV-induced joint swelling, compared to an isotype control mAb.
Fig. 7.
Antibody protection of CHIKV- and MAYV-induced musculoskeletal disease in vivo. (A) CHIKV- and MAYV-induced joint swelling in mAb-treated mice. The 4-wk-old C57BL/6J mice were administered 200 µg (10 mg/kg) DC2.M16, DC2.M357, or isotype control mAb by intraperitoneal route 1 d before subcutaneous inoculation of either 103 FFU CHIKV-LR2006-OPY1 or MAYV-BeH407 in the footpad. Swelling was measured in the ipsilateral ankle using digital calipers over 14 d. Two independent experiments were performed, with n = 10 mice per antibody for each group (two-way ANOVA with Tukey’s posttest). CHIKV-LR2006-OPY1 (B) and MAYV-BeH407 (C) viral titers in ankle, calf muscle, spleen, and draining lymph node (DLN) were determined 3-d postinoculation. Viral RNA levels were measured by qRT-PCR and compared to a viral RNA standard curve, generated from titered viral stock to determine FFU equivalents. Two independent experiments were performed, with n = 6 mice per antibody for each group (one-way ANOVA with Dunnett’s posttest). *P < 0.05; **P < 0.01; ***P < 0.001, and ****P < 0.0001, compared to isotype control.
To test the effect of mAb treatment on viral dissemination, mice were inoculated with CHIKV or MAYV 1 d after mAb administration, and viral RNA was measured by qRT-PCR in ankle, calf muscle, spleen, and draining lymph node tissues at 3 d after infection (Fig. 7 B and C). CHIKV and MAYV viral RNA levels in both the ipsilateral and contralateral ankle joint and calf muscle were reduced markedly in mice treated with either DC2.M16 or DC2.M357, compared to the isotype control mAb. In addition, mice treated with DC2.M16 or DC2.M357 had decreased viral RNA in the spleen and draining lymph node. These data demonstrate that DC2.M16 and DC2.M357, two near-germline human mAbs from distinct lineages, can protect against musculoskeletal infection and disease caused by CHIKV and MAYV in mice.
Discussion
Although human bNAbs have been identified and extensively characterized in the context of HIV (41), influenza (42), ebolavirus (43, 44), and other pathogens, few have been described for alphaviruses. Murine CHK-265 and RRV-12, a human mAb recently isolated from an RRV-convalescent donor, are among the only alphavirus bNAbs that have demonstrated cross-protection against multiple alphaviruses. Here, we employed a single B cell sorting strategy using a heterologous MAYV antigen to capture and profile cross-reactive human mAbs from a CHIKV-convalescent donor, with the goal of identifying cross-protective mAbs. Serological studies of CHIKV patients have recently demonstrated cross-reactivity and cross-neutralization to heterologous alphaviruses, suggesting that cross-reactive and/or broadly neutralizing mAbs may be elicited by natural CHIKV infection. We isolated 33 human mAbs that strongly reacted with both CHIKV and MAYV p62-E1. These cross-reactive mAbs had diverse sequences and targeted both the E1 and E2 subunits of the alphavirus glycoprotein. We previously characterized both neutralizing and nonneutralizing, CHIKV-specific human mAbs targeting E1 (25). Our current study identifies E1 as well as E2 as a target of human cross-reactive mAbs, although E1-specific mAbs were nonneutralizing.
We identified five human bNAbs that inhibited CHIKV and MAYV with IC50 values ranging from 0.5 to 4.4 nM (75 to 660 ng/mL). The epitopes engaged by these mAbs are contained in the B domain of E2, which previously was identified as the target of murine bNAb CHK-265 and more recently, the human bNAb RRV-12. While all the DC2 bNAbs we isolated competed with CHK-265, they exhibited differential neutralizing breadth and potency with respect to heterologous viruses ONNV, RRV, and SFV. Our viral escape mutant studies revealed that human bNAbs engage related but distinct regions of the B domain of E2 and that fine recognition specificity is a determinant of neutralization breadth. The bNAbs that mapped to residues G209 and K215 were more limited in breadth than DC2.M357, which targeted K189 and N218. The DC2.M357 epitope appears highly related to that of murine CHK-265, which shares similar escape mutations and neutralization profiles. RRV-12, the only reported human bNAb to protect against multiple alphaviruses, was isolated from an RRV-convalescent donor and also shares similar features with CHK-265. Although three-dimensional structures of DC2.M108 and DC2.M357 bound to E2 are not yet available, the viral escape mutations suggest different angles of mAb engagement. Given the high density of viral spikes on the alphavirus particle, this might, in part, explain their differences in potency and breadth, as mAb binding to the lateral side of the B domain by DC2.M108 would be predicted to be more sterically hindered by adjacent spikes. Thus, our work identifies distinct classes of human bNAbs that map to different regions on the E2 B domain, which may be important to consider in the design of alphavirus vaccines with broad reactivity. Recent work has demonstrated that CHIKV infection or vaccination with a vaccinia-based CHIKV vaccine confers variable and limited cross-protection to heterologous alphaviruses (45). Another study demonstrated that CHIKV and MAYV adenoviral, vector-based vaccines only partially protected against heterologous viral challenge in mice (46). Designed immunogens guided by human bNAb epitopes may aid the targeted elicitation of more robust, cross-protective antibody responses.
Sequence analysis of DC2 bNAbs revealed two distinct V-gene lineages. Four of the five human bNAbs (DC2.M16, DC2.M108, DC2.M131, and DC2.M230) were related clonally and shared IGHV3-11 and IGKV1-NL1 germline pairing, whereas DC2.M357 was distinct and utilized IGHV3-30 and IGKV3-11. Recently, IGHV3-11 usage was found to be prevalent in germline-neutralizing mAbs that target the RSV F protein (47). Although our cross-reactive mAbs showed a range of somatic mutation, bNAbs were generally close to the germline V-gene sequence with very few substitutions. Moreover, the analysis of DC2.M16- and DC2.M108-inferred germline mAbs showed equivalent binding and neutralization profiles, compared to the WT somatically mutated antibody. Germline-like, neutralizing antibodies have been recently identified for RSV (47), H7N9 Influenza virus (48), Zika virus (49, 50), Dengue virus (51), and SARS-CoV-2 (52). Such antibodies may be elicited early in the course of infection compared to those that undergo extensive affinity maturation and somatic mutation. Therefore, the induction of broadly neutralizing alphavirus antibodies may not require sustained antigen production or exposure. Furthermore, since both IGHV3-11 germline-reverted and mature alphavirus bNAbs can neutralize multiple alphaviruses, strategies to preferentially stimulate this germline could potentially result in the elicitation of higher levels of broadly neutralizing antibody responses. Recent work toward the development of germline-targeting vaccines that induce broadly protective antibody responses has been described in HIV (53–55) and influenza (56, 57). Thus, the targeted elicitation of bNAbs against alphaviruses from specific germlines may be advantageous in developing a broadly effective vaccine.
Currently, no antiviral treatments for arthritogenic alphaviruses exist, and therapeutic mAbs have shown promise in mice and nonhuman primates against homologous viruses but less so against heterologous viruses. We show that two bNAbs (DC2.M16 and DC2.M357) confer protection in mouse models of both CHIKV and MAYV infection. Both mAbs markedly reduced alphavirus-induced joint swelling, viral infection, and dissemination to a similar extent, despite having moderate (subnanomolar to low nanomolar range) neutralizing activity and having a 10-fold difference in potency. Recent work has demonstrated that, in addition to neutralizing potency, Fc effector functions are important in mediating protection against alphaviruses (30, 58, 59). CHK-265 was shown to protect against heterologous RRV challenge, despite moderate cross-neutralizing potency (37). Our study, together with these findings, demonstrates the utility of evaluating in vivo properties of moderately inhibitory mAbs, as neutralizing potency may not be the only important correlate of protection. Notably, our mAbs all contain a human IgG1 Fc that binds mouse Fc-γRI, Fc-γRIII, and Fc-γRIV, which may be important for effector functions and in vivo efficacy (60).
MAbs with fewer somatic mutations generally exhibit more favorable “drug-like” properties, such as better conformational stability and reduced hydrophobicity and polyreactivity (61). A broad antiviral antibody against arthritogenic alphaviruses may be useful because of their similar (almost indistinguishable) clinical manifestations and continued global spread. Our study identifies candidate mAbs for further development as broad immunotherapies to combat alphavirus disease and provides insight into the potential for the elicitation of similar broad responses in humans.
Materials and Methods
Ethics Statement.
The CHIKV-convalescent donor DC2 was previously identified (25), after informed written consent, blood samples were collected, and peripheral blood mononuclear cells (PBMCs) isolated by Ficoll gradient centrifugation. The study protocol was approved by the Institutional Review Board of the Albert Einstein College of Medicine (protocol IRB# 2016–6137).
The mouse challenge studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH (62). The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01) under animal use approval number 20180234.
Cells and Viruses.
Vero cells were cultured in Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum (FBS) at 5% CO2, 37 °C. ExpiCHO-S cells (Gibco) were maintained in ExpiCHO expression media as per manufacturer’s instructions. CHIKV 181/25 was obtained from Dr. Robert B. Tesh (University of Texas Medical Branch). The Mayaro Guyane virus (NR-49911) was obtained through the Biodefense and Emerging Infections Research Resources Repository, as part of the World Reference Center for Emerging Viruses and Arboviruses program. The ONNV SG650 (63), RRV T48 (64), and MAYV TRVL-4675 (65) infectious clones were obtained from Dr. Andres Merits (University of Tartu, Estonia). SFV and SINV were grown from infectious clones pSP6-SFV4 (66) and dsTE12Q (67), respectively. Viruses were propagated in baby hamster kidney fibroblast (BHK-21) WI-2 cells from Dr. Ari Helenius (ETH Zurich, Switzerland).
CHIKV and MAYV Glycoprotein Production.
CHIKV p62-E1 and E1' were expressed in Drosophila S2 cells and purified, as previously described (25, 68). CHIKV E2 residues S1-Y361 (strain LR-2006) were codon optimized, synthesized (Integrated DNA Technologies), and inserted into the pET21a vector. The plasmid construct was transformed into BL21 (DE3) competent cells (Thermo Fisher Scientific), grown to an optimal density (600 nm) of 0.8, and induced with 0.1 mM IPTG for 5 h. Cells were harvested, and inclusion bodies were isolated and refolded, as previously described (68). CHIKV E2 protein was further purified by HiLoad 16/600 Superdex 75 size-exclusion chromatography (GE Healthcare), and purity was assessed by SDS-PAGE. The MAYV p62-E1 insert was constructed in the same manner as CHIKV p62-E1 (23), based on the MAYV BeAr 20290 sequence. The sequence contained the native MAYV secretion signal, p62 ectodomain, Gly-Ser linker, E1 ectodomain, and a Strep II affinity tag. The sequence was codon optimized, synthesized (IDT), and cloned into pT353 vector for S2 expression. Protein was purified by Strep-Tactin (IBA) affinity chromatography, and purity was assessed by SDS-PAGE and size-exclusion chromatography.
Human mAb Isolation.
Isolation of human mAbs was performed as previously described (69). Antigen-reactive memory B cells were isolated from human PBMCs by fluorescence-activated cell sorting with the following antibodies: anti-human CD8 (PE-Cy7), CD3 (PE-Cy7), CD14 (PE-Cy7), CD20 (Pacific Blue), CD27 (APC), and IgG (FITC). MAYV p62-E1 was biotinylated using EZ-Link NHS-PEG4-Biotin (Life Technologies) and detected with streptavidin-conjugated phycoerythrin (Life Technologies). Cells were sorted into single PCR tubes, and cDNA was generated by RT-PCR. Nested PCR was performed with IgH- and IgK-specific primers and cloned into pMAZ vector (70) for sequencing and recombinant expression. Sequences were analyzed using IMGT/V-quest tool (71). The mAbs were transiently transfected in ExpiCHO cells, as per the manufacturer’s protocol (Gibco), and purified by protein A chromatography. Fab constructs containing a C-terminal His tag on the heavy chain were transiently transfected in ExpiCHO cells and purified by Ni-NTA affinity chromatography.
ELISA Binding Assays.
To assess mAb reactivity by ELISA, CHIKV and MAYV glycoproteins were coated on half-area 96-well high binding plates (Costar) at 200 ng/well. Wells were blocked with 1% bovine serum albumin (BSA) at 25 °C for 2 h and washed five times with phosphate-buffered saline (PBS)-T (PBS pH 7.4, 0.05% Tween-20). The mAbs were diluted in PBS-T (PBS pH 7.4, 0.2% BSA, 0.05% Tween) and incubated for 1 h at 37 °C. Plates were washed, and protein A conjugated to horseradish peroxidase (HRP) (Life Technologies) was added at a 1:2,000 dilution. After 1 h incubation at 37 °C, plates were washed and developed using TMB (Thermo Fisher Scientific). Absorbance at 450 nm was measured on Synergy H4 Hybrid reader (BioTek).
BLI Binding Assays.
The mAb-binding kinetics to CHIKV and MAYV p62-E1 were measured by BLI using an OctetRed96 instrument (ForteBio). The mAbs were immobilized on anti-human Fc capture sensors. Global data fitting to a 1:1 binding model was used to estimate values for the kon (association rate constant), koff (dissociation rate constant), and KD (equilibrium dissociation constant). Data were analyzed using ForteBio Data Analysis Software 9. For monovalent binding studies, Fabs were immobilized on anti-Penta-His sensors and analyzed as described above for full-length mAbs. For two-phase binning experiments, biotinylated MAYV p62-E1 was loaded first onto a streptavidin-coated sensor, and then the first mAb was bound to saturation. The sensor was then added to a well containing both the first mAb and the test mAb at equimolar concentrations (50 nM).
Focus Reduction Neutralization Assay.
Vero cells were seeded at 2.5 × 104 cells/well and incubated for 24 h at 37 °C. The mAbs were serially diluted and incubated with 100 to 150 focus-forming units (FFU) of virus for 1 h at 37 °C. Cells were inoculated with antibody–virus complexes for 1 h at 37 °C, and cells were then overlaid with 1% carboxymethylcellulose in MEM, supplemented with 2% FBS and 10 mM Hepes pH 7.4. Plates were fixed 18-h postinfection with 1% PFA diluted in PBS and permeabilized in 1× PBS with 0.1% saponin and 0.1% BSA. Cells then were treated with primary antibody followed by HRP-conjugated goat anti-mouse IgG and TrueBlue Peroxidase substrate (KPL). Developed foci where quantified on ImmunoSpot S6 Macroanalyzer (Cellular Technologies Ltd.). Infection of mAb-treated wells was determined relative to untreated control wells with virus alone. Nonlinear regression analysis was performed using Prism 7 software (GraphPad Software).
Escape Mutant Generation.
Vero cells were infected with rVSV-CHIKV in the presence of mAb at an IC90 concentration. Infection was monitored over days for cytopathic effects, and supernatants were harvested from the infected wells. After four passages under mAb selection, individual clones were plaque purified and expanded. Viral RNA isolation was performed using a Viral RNA Kit (Zymo Research), and cDNA synthesis was performed. CHIKV glycoprotein genes were amplified by PCR and sequenced to analyze for the presence of mutations.
Neutralization of rVSV-CHIKV WT and escape mutants was tested by complexing serially diluted mAb with a virus for 1 h at 37 °C, prior to the addition to cell monolayers in 96-well plates. Cells were fixed 9-h postinfection, washed with PBS, and treated with Hoescht stain. The relative infection was measured by automated counting of eGFP+ cells using a Cytation5 Imager (BioTek).
Mouse Infection Experiments.
The 4-wk-old WT C57BL/6J male mice were purchased from Jackson Laboratories (000664). A total of 200 μg (10 mg/kg) mAbs DC2.M16, DC2.M357, or an isotype control were administered to individual mice by intraperitoneal injection 1 d before inoculation in the left footpad with 103 FFU of MAYV-BeH407 or CHIKV-La Reunion OPY1 in Hank’s balanced salt solution supplemented with 1% heat-inactivated FBS. Foot swelling was monitored via measurements (width × height) using digital calipers. At 3-d postinfection, tissues were harvested after extensive perfusion with 40 mL PBS. Viral RNA was recovered and measured by qRT-PCR using a standard curve made by RNA isolated from a viral stock of known titer to determine FFU equivalents.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 AI125462 to J.R.L., R01 AI114816 and U19 AI142790 to M.S.D., contract AI201800001 to M.S.D., R01 AI075647 to M.K., and R01-AI132633 to K.C.). R.J.M. was supported by NIH Medical Scientist Training Grant T32-GM007288 and a training fellowship F30-AI150055. J.R.L. acknowledges an XSeed Award from Deerfield. We gratefully acknowledge technical assistance from the Einstein Flow Cytometry Core, which is partially supported by NIH grant P30CA013330, as well as help from Dr. Caroline Martin in the Kielian laboratory with the SFV neutralization assays.
Footnotes
Competing interest statement: The antibodies described in this manuscript are the subject of a US Patent Application that lists R.J.M., M.K., and J.R.L. as coinventors. J.R.L. and K.C. are consultants for Celdara Medical. K.C. is a member of the Scientific Advisory Board of Integrum Scientific, LLC and Biovaxys Tecchnology Corp. M.S.D. is a consultant for Inbios, Vir Biotechnology, and Fortress Biotech and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory at Washington University School of Medicine has received sponsored unrelated research agreements from Vir Biotechnology and Emergent BioSolutions.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100104118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Holmes A. C., Basore K., Fremont D. H., Diamond M. S., A molecular understanding of alphavirus entry. PLoS Pathog. 16, e1008876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levi L. I., Vignuzzi M., Arthritogenic alphaviruses: A worldwide emerging threat? Microorganisms 7, 133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaid A., et al., Arthritogenic alphaviruses: Epidemiological and clinical perspective on emerging arboviruses. Lancet Infect. Dis. 21, e123–e133 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Chen W., et al., Arthritogenic alphaviruses: New insights into arthritis and bone pathology. Trends Microbiol. 23, 35–43 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Suhrbier A., Jaffar-Bandjee M.-C., Gasque P., Arthritogenic alphaviruses—An overview. Nat. Rev. Rheumatol. 8, 420–429 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Crosby L., et al., Severe manifestations of chikungunya virus in critically ill patients during the 2013-2014 Caribbean outbreak. Int. J. Infect. Dis. 48, 78–80 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Thiberville S.-D., et al., Chikungunya fever: Epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 99, 345–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yergolkar P. N., et al., Chikungunya outbreaks caused by African genotype, India. Emerg. Infect. Dis. 12, 1580–1583 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahid B., Ali A., Rafique S., Idrees M., Global expansion of chikungunya virus: Mapping the 64-year history. Int. J. Infect. Dis. 58, 69–76 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Yactayo S., Staples J. E., Millot V., Cibrelus L., Ramon-Pardo P., Epidemiology of Chikungunya in the Americas. J. Infect. Dis. 214 (suppl. 5), S441–S445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diagne C. T., et al., Mayaro virus pathogenesis and transmission mechanisms. Pathogens 9, 738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hozé N., et al., Reconstructing Mayaro virus circulation in French Guiana shows frequent spillovers. Nat. Commun. 11, 2842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long K. C., et al., Experimental transmission of Mayaro virus by Aedes aegypti. Am. J. Trop. Med. Hyg. 85, 750–757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira T. N., Rocha M. N., Sucupira P. H. F., Carvalho F. D., Moreira L. A., Wolbachia significantly impacts the vector competence of Aedes aegypti for Mayaro virus. Sci. Rep. 8, 6889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira T. N., Carvalho F. D., De Mendonça S. F., Rocha M. N., Moreira L. A., Vector competence of Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquitoes for Mayaro virus. PLoS Negl. Trop. Dis. 14, e0007518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brustolin M., Pujhari S., Henderson C. A., Rasgon J. L., Anopheles mosquitoes may drive invasion and transmission of Mayaro virus across geographically diverse regions. PLoS Negl. Trop. Dis. 12, e0006895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alto B. W., Civana A., Wiggins K., Eastmond B., Shin D., Effect of oral infection of Mayaro virus on fitness correlates and expression of immune related genes in Aedes aegypti. Viruses 12, 719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer M. U. G., et al., The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife 4, e08347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acosta-Ampudia Y., et al., Mayaro: An emerging viral threat? Emerg. Microbes Infect. 7, 163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jose J., Snyder J. E., Kuhn R. J., A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 4, 837–856 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-San Martín C., Liu C. Y., Kielian M., Dealing with low pH: Entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 17, 514–521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaney M.-C., Duquerroy S., Rey F. A., Alphavirus structure: Activation for entry at the target cell surface. Curr. Opin. Virol. 3, 151–158 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Voss J. E., et al., Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Uchime O., Fields W., Kielian M., The role of E3 in pH protection during alphavirus assembly and exit. J. Virol. 87, 10255–10262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiroz J. A., et al., Human monoclonal antibodies against Chikungunya virus target multiple distinct epitopes in the E1 and E2 glycoproteins. PLoS Pathog. 15, e1008061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal P., et al., Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 9, e1003312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S. A., et al., Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against Chikungunya virus. Cell Host Microbe 18, 86–95 (2015). Correction in: Cell Host Microbe 18, 382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell L. A., et al., Human monoclonal antibodies against Ross River virus target epitopes within the E2 protein and protect against disease. PLoS Pathog. 16, e1008517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selvarajah S., et al., A neutralizing monoclonal antibody targeting the acid-sensitive region in Chikungunya virus E2 protects from disease. PLoS Negl. Trop. Dis. 7, e2423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earnest J. T., et al., Neutralizing antibodies against Mayaro virus require Fc effector functions for protective activity. J. Exp. Med. 216, 2282–2301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Q. F., et al., Structural basis of Chikungunya virus inhibition by monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 117, 27637–27645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basore K., et al., Cryo-EM structure of Chikungunya virus in complex with the Mxra8 receptor. Cell 177, 1725–1737.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell L. A., et al., Human mAbs broadly protect against arthritogenic alphaviruses by recognizing conserved elements of the Mxra8 receptor-binding site. Cell Host Microbe 28, 699–711.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins K. A., et al., Neutralizing antibodies from convalescent Chikungunya virus patients can cross-neutralize Mayaro and Una viruses. Am. J. Trop. Med. Hyg. 100, 1541–1544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb E. M., et al., Effects of Chikungunya virus immunity on Mayaro virus disease and epidemic potential. Sci. Rep. 9, 20399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer C., et al., Robustness of serologic investigations for Chikungunya and Mayaro viruses following coemergence. MSphere 5, e00915-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox J. M., et al., A cross-reactive antibody protects against Ross River virus musculoskeletal disease despite rapid neutralization escape in mice. PLoS Pathog. 16, e1008743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox J. M., et al., Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell 163, 1095–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend C. L., et al., Significant differences in physicochemical properties of human immunoglobulin kappa and lambda CDR3 regions. Front. Immunol. 7, 388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierson T. C., Diamond M. S., A game of numbers: The stoichiometry of antibody-mediated neutralization of Flavivirus infection Prog. in Mol. Trans. Sci. 129, 141–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burton D. R., Hangartner L., Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 34, 635–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laursen N. S., Wilson I. A., Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 98, 476–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornholdt Z. A., et al., A two-antibody pan-ebolavirus cocktail confers broad therapeutic protection in ferrets and nonhuman primates. Cell Host Microbe 25, 49–58.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wec A. Z., et al., Development of a human antibody cocktail that deploys multiple functions to confer pan-ebolavirus protection. Cell Host Microbe 25, 39–48.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen W., et al., Arthritogenic alphavirus vaccines: Serogrouping versus cross-protection in mouse models. Vaccines (Basel) 8, 209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rafael K. C., et al., Adenoviral-vectored Mayaro and Chikungunya virus vaccine candidates afford partial cross-protection from lethal challenge in A129 mouse model. Front. Immunol. 11, 591885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodwin E., et al., Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity 48, 339–349.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu F., et al., A potent germline-like human monoclonal antibody targets a pH-sensitive epitope on H7N9 influenza hemagglutinin. Cell Host Microbe 22, 471–483.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F., et al., Development of a potent and protective germline-like antibody lineage against Zika virus in a convalescent human. Front. Immunol. 10, 2424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnani D. M., et al., A human inferred germline antibody binds to an immunodominant epitope and neutralizes Zika virus. PLoS Negl. Trop. Dis. 11, e0005655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu D., et al., A broadly neutralizing germline-like human monoclonal antibody against dengue virus envelope domain III. PLoS Pathog. 15, e1007836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreer C., et al., Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 182, 843–854.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward A. B., Wilson I. A., Innovations in structure-based antigen design and immune monitoring for next generation vaccines. Curr. Opin. Immunol. 65, 50–56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steichen J. M., et al., A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366, eaax4380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jardine J., et al., Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheung C. S.-F., et al., Identification and structure of a multidonor class of head-directed influenza-neutralizing antibodies reveal the mechanism for its recurrent elicitation. Cell Rep. 32, 108088 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Sangesland M., et al., Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity 51, 735–749.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Earnest J. T., et al., The mechanistic basis of protection by non-neutralizing anti-alphavirus antibodies. Cell Rep. 35, 108962 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fox J. M., et al., Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcγR interaction on monocytes. Sci. Immunol. 4, eaav5062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dekkers G., et al., Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs 9, 767–773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shehata L., et al., Affinity maturation enhances antibody specificity but compromises conformational stability. Cell Rep. 28, 3300–3308.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 62.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011).
- 63.Brault A. C., et al., Infection patterns of O’nyong nyong virus in the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol. Biol. 13, 625–635 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Kuhn R. J., Niesters H. G., Hong Z., Strauss J. H., Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182, 430–441 (1991). [DOI] [PubMed] [Google Scholar]
- 65.Chuong C., Bates T. A., Weger-Lucarelli J., Infectious cDNA clones of two strains of Mayaro virus for studies on viral pathogenesis and vaccine development. Virology 535, 227–231 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Liljeström P., Lusa S., Huylebroeck D., Garoff H., In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: The small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65, 4107–4113 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardwick J. M., Levine B., Sindbis virus vector system for functional analysis of apoptosis regulators. Methods Enzymol. 322, 492–508 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Sánchez-San Martín C., Nanda S., Zheng Y., Fields W., Kielian M., Cross-inhibition of Chikungunya virus fusion and infection by alphavirus E1 domain III proteins. J. Virol. 87, 7680–7687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiller T., et al., Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008). Correction in: J. Immunol. Methods 334, 142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazor Y., Barnea I., Keydar I., Benhar I., Antibody internalization studied using a novel IgG binding toxin fusion. J. Immunol. Methods 321, 41–59 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Brochet X., Lefranc M.-P., Giudicelli V., IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–W508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.